We narrowed to 9,909 results for: tre promoter
-
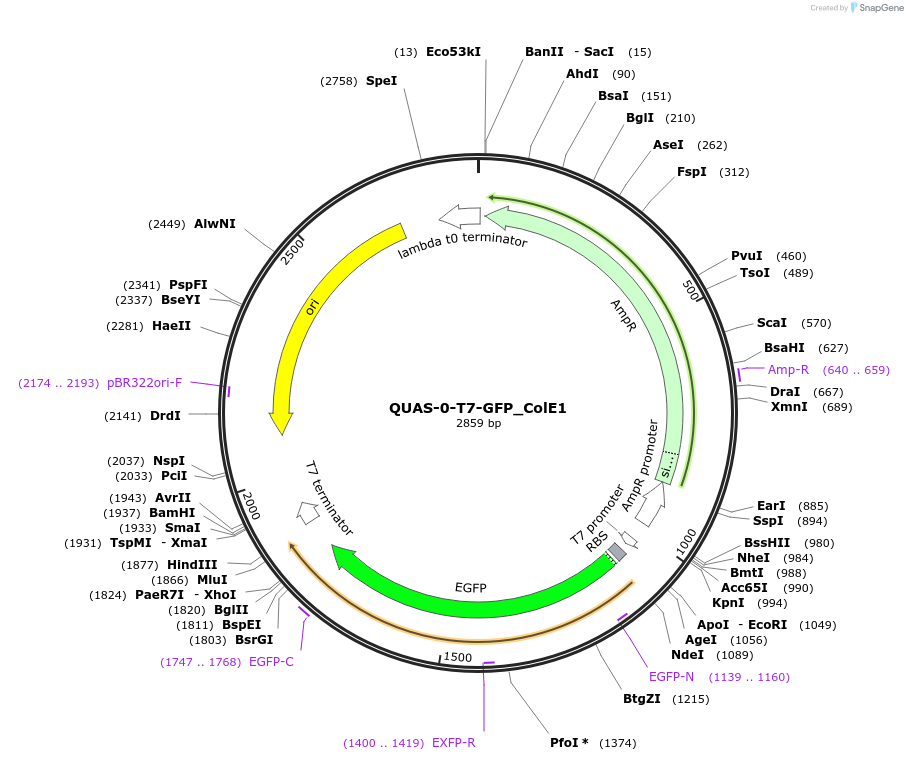
Plasmid#171649PurposeQUAS directly upstream of a T7 promoter. Expresses GFP in BL21(DE3) E. coli. Contains ColE1 origin of replication.DepositorInsertQUAS-0-T7
TagsssrA degradation tag (DAS+4)ExpressionBacterialPromoterT7Available SinceOct. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
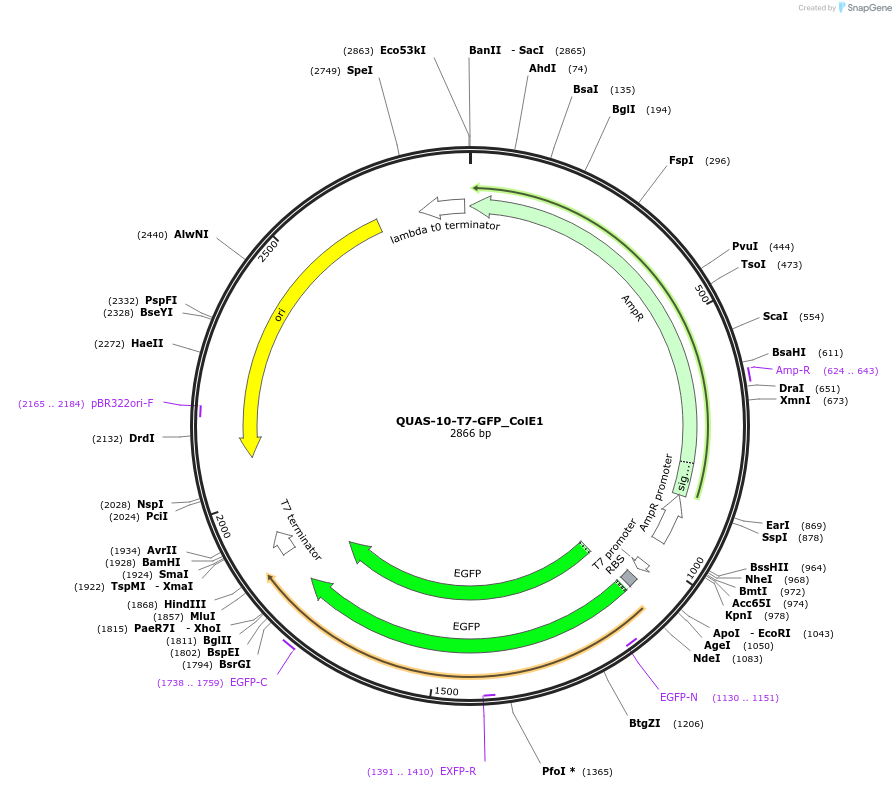
QUAS-10-T7-GFP_ColE1
Plasmid#171654PurposeQUAS ten base pairs upstream of a T7 promoter. Expresses GFP in BL21(DE3) E. coli. Contains ColE1 origin of replication.DepositorInsertQUAS-10-T7
TagsssrA degradation tag (DAS+4)ExpressionBacterialPromoterT7Available SinceOct. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
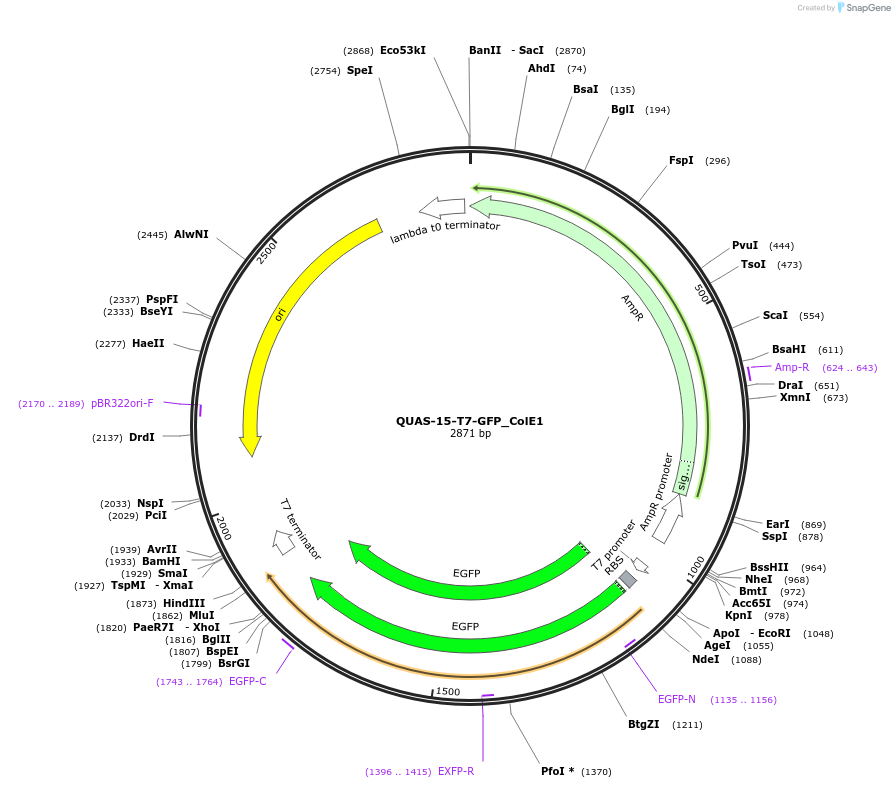
QUAS-15-T7-GFP_ColE1
Plasmid#171655PurposeQUAS fifteen base pairs upstream of a T7 promoter. Expresses GFP in BL21(DE3) E. coli. Contains ColE1 origin of replication.DepositorInsertQUAS-15-T7
TagsssrA degradation tag (DAS+4)ExpressionBacterialPromoterT7Available SinceOct. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
-
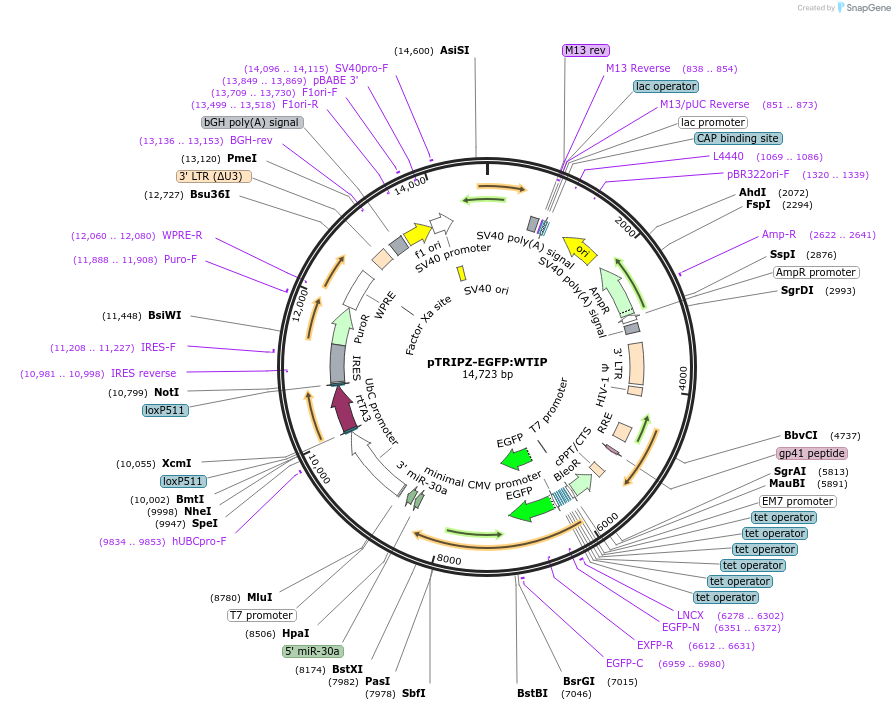
pTRIPZ-EGFP:WTIP
Plasmid#108231PurposeLentiviral vector for Tet-inducible WTIP expressionDepositorInsertWTIP (WTIP Human)
UseLentiviralTagsEGFPExpressionMammalianPromoterTet/ON Minimal CMV promoterAvailable SinceJuly 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
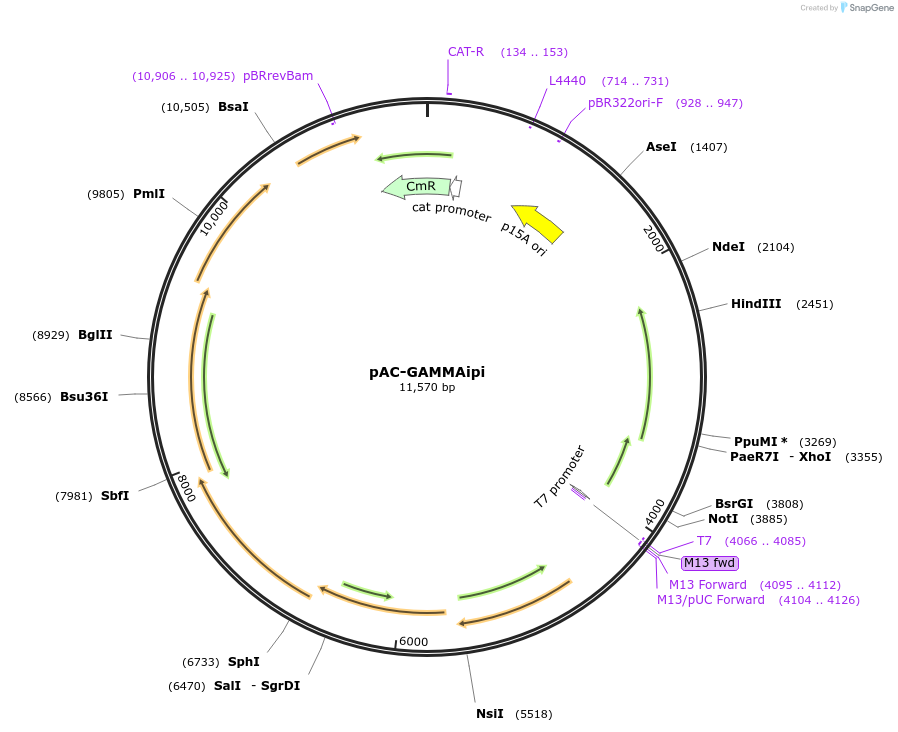
pAC-GAMMAipi
Plasmid#53278PurposeHas ctrE, crtB, crtI, crtY, and idi genes of Erwinia herbicola (Pantoea agglomerans) Eho10 and crtYm gene of marine bacterium P99-3. and produces gamma-carotene in Escherichia coliDepositorInsertcrtYm
UseLow copy number bacterial cloning vectorPromoterendogenous promoterAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
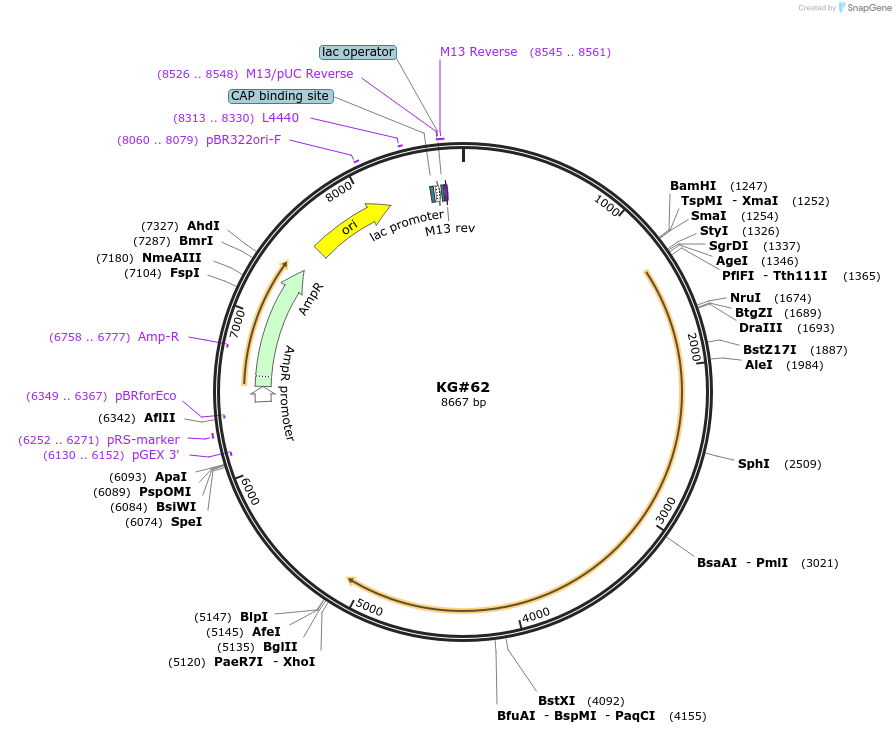
KG#62
Plasmid#110874PurposeExpresses the C. elegans acy-1 cDNA pan-neuronallyDepositorInsertsrab-3 promoter
acy-1 cDNA
unc-54 3' control region with 1 intron just upstream and 1 intron in control region
ExpressionBacterialAvailable SinceJune 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
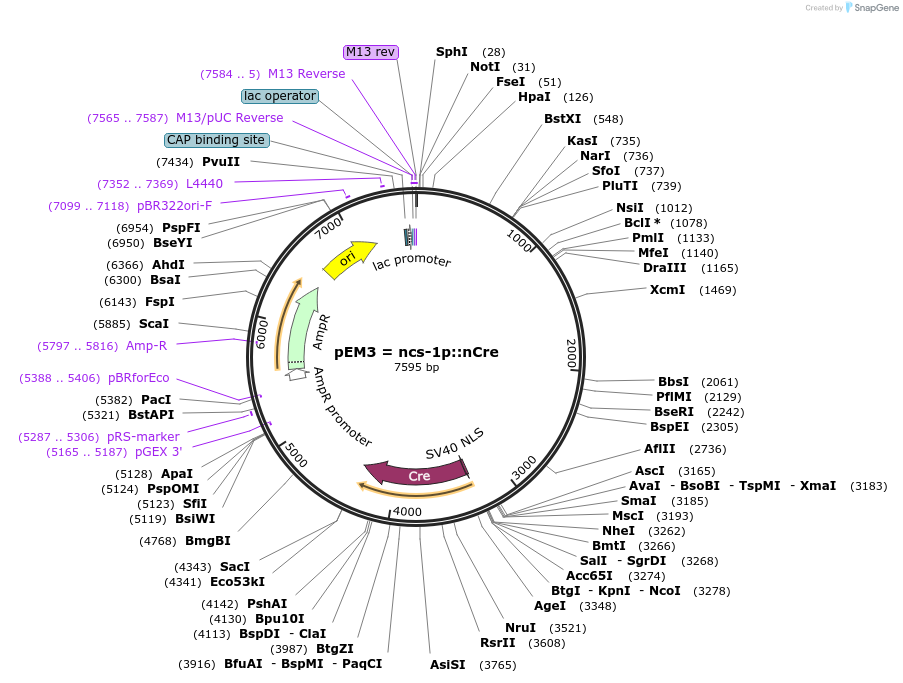
pEM3 = ncs-1p::nCre
Plasmid#24034DepositorAvailable SinceFeb. 24, 2010AvailabilityAcademic Institutions and Nonprofits only -
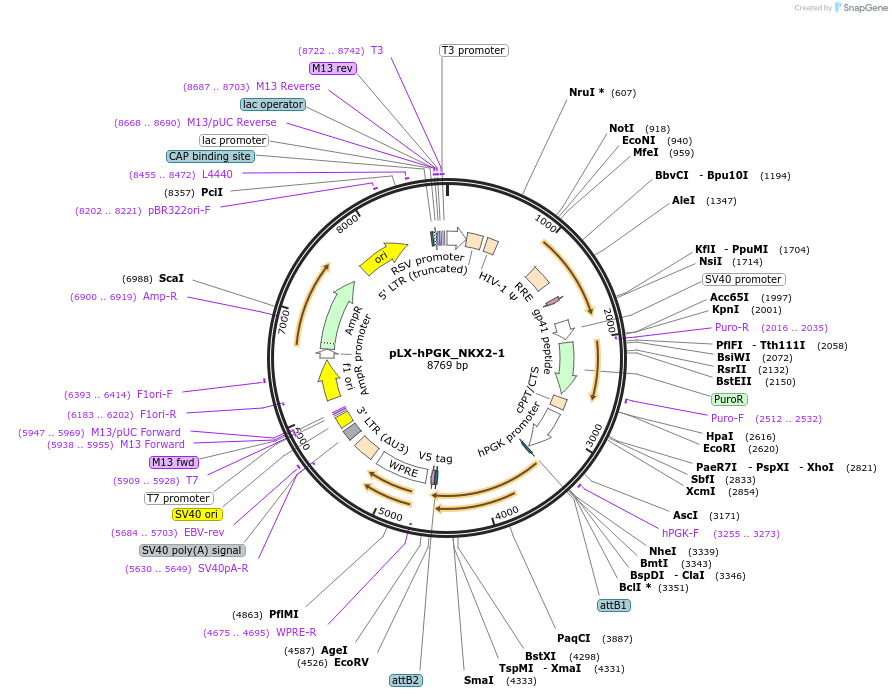
pLX-hPGK_NKX2-1
Plasmid#221494PurposeNKX2-1 short isoform lentiviral overexpression using hPGK promoterDepositorAvailable SinceJuly 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
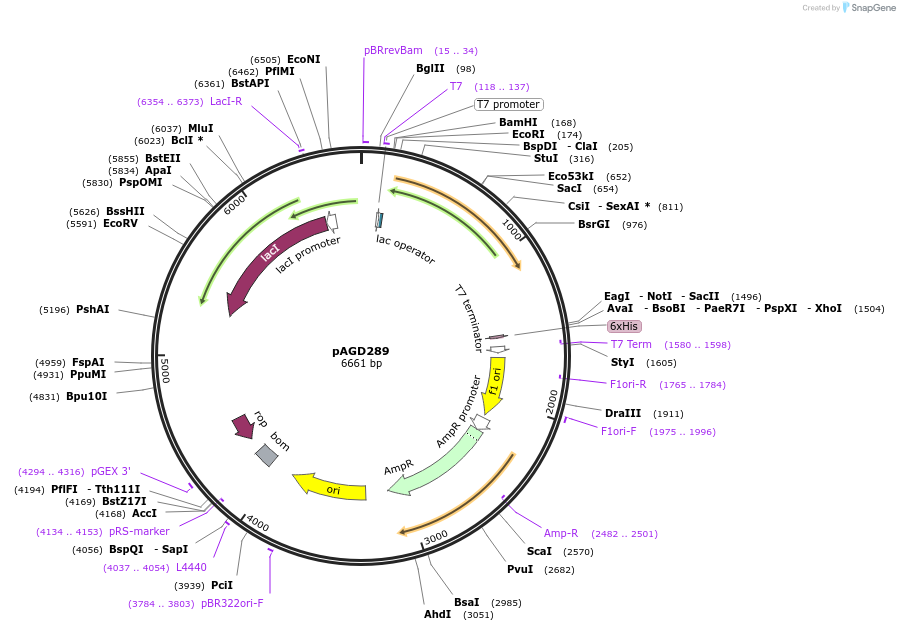
pAGD289
Plasmid#228113PurposepET-21(+) deriviative with retron Kva1 containing a donor to edit E. coli rpoB gene cloned downstream of a T7 promoterDepositorInsertKva1 retron, ncRNA with 90 bp rpoB donor
ExpressionBacterialPromoterT7Available SinceDec. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
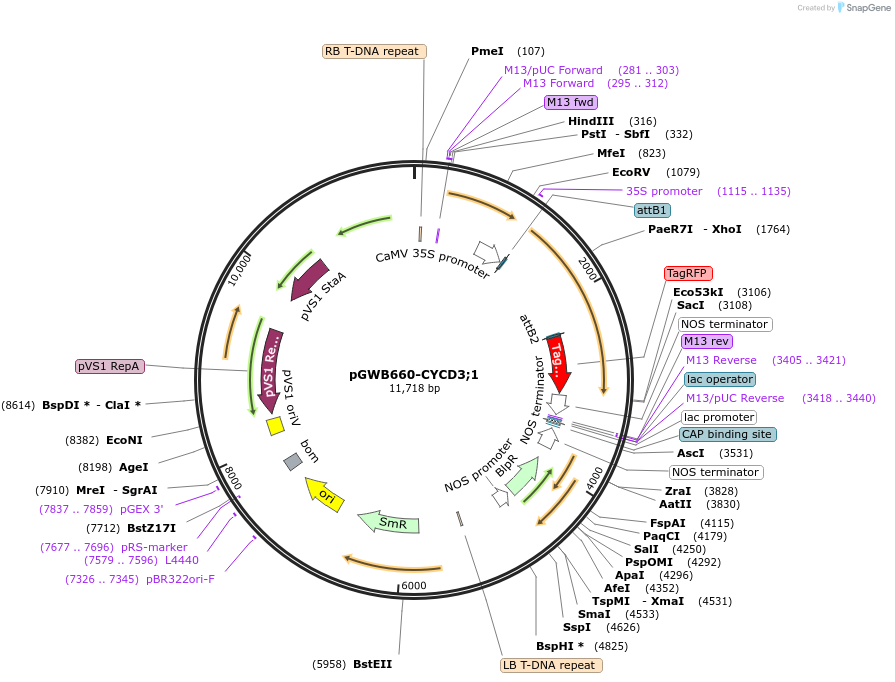
pGWB660-CYCD3;1
Plasmid#140751PurposeInducing plant cells to enter mitosisDepositorAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
pGL3-LRF
Plasmid#66936PurposeDestination vector; gateway cassette cloned upstream of basal promoter driving luciferase. Forward orientation (attR1, attR2).DepositorTypeEmpty backboneUseLuciferaseExpressionMammalianPromoterSV-40Available SinceAug. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
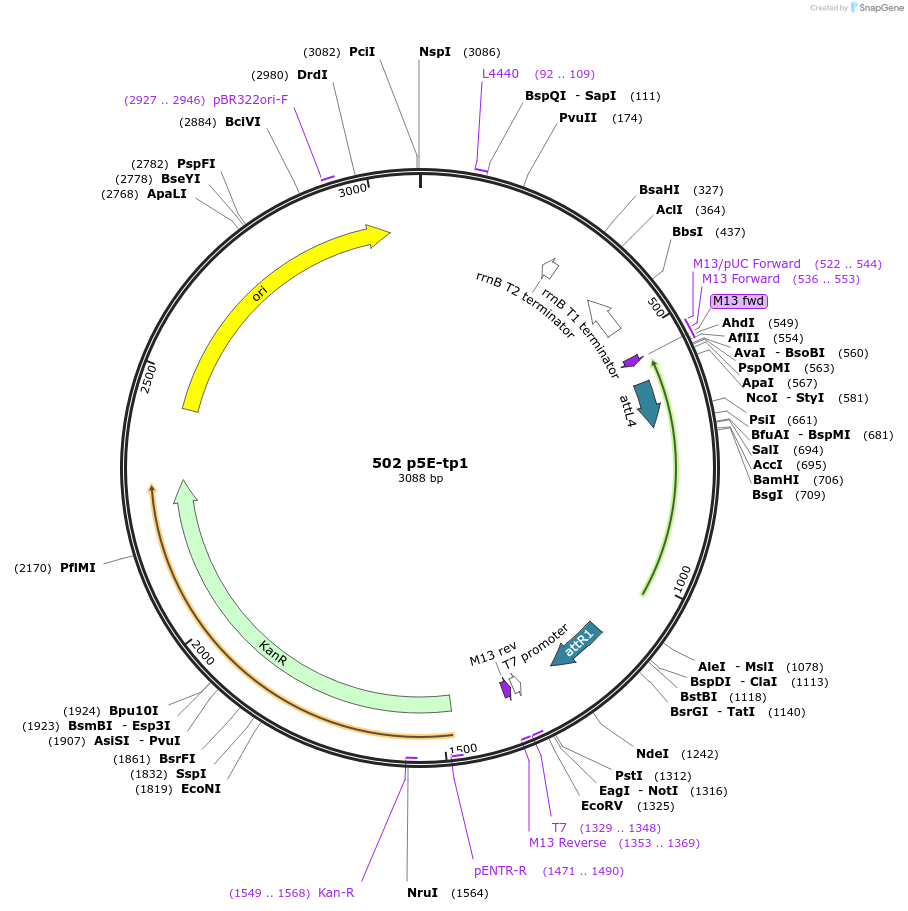
502 p5E-tp1
Plasmid#73585Purposeentry vector (L4-R1) containing the Notch responsive tp1 element upstream of a basal promoterDepositorInserttp1 element
Available SinceFeb. 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
RSV-betaGal
Plasmid#24058DepositorAvailable SinceOct. 27, 2010AvailabilityAcademic Institutions and Nonprofits only -
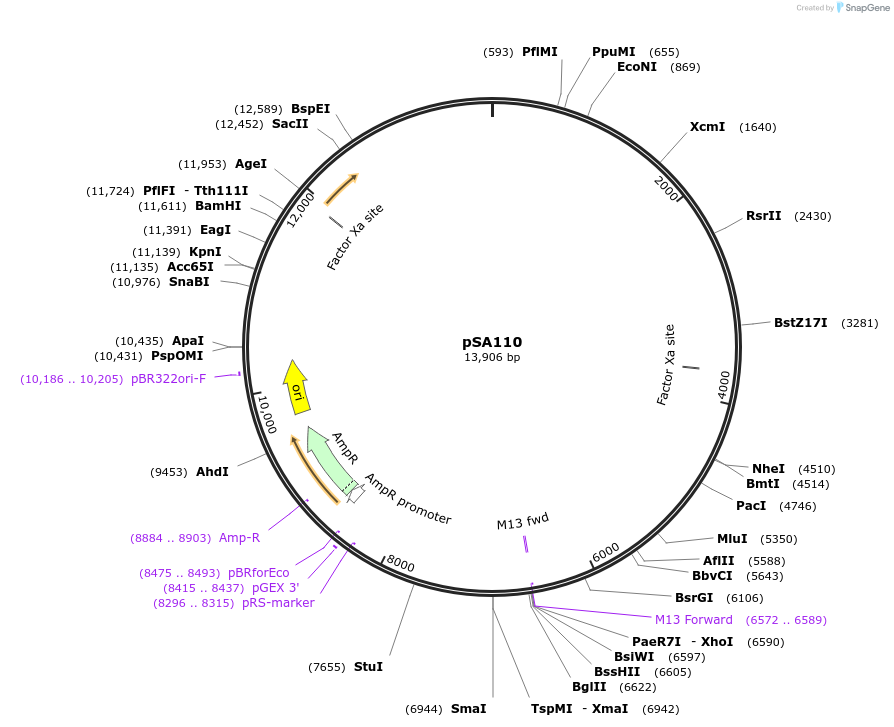
pSA110
Plasmid#59784Purposedrives mCherry in C. elegans intestinal cellsDepositorInsertsExpressionBacterialAvailable SinceNov. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
Pdx1 Cre-ER (DM#255)
Plasmid#15022DepositorInsertPdx1 promoter (Pdx1 Mouse)
UseCre/Lox; Mouse transgenicTagsCre-ER(TM)ExpressionMammalianAvailable SinceOct. 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
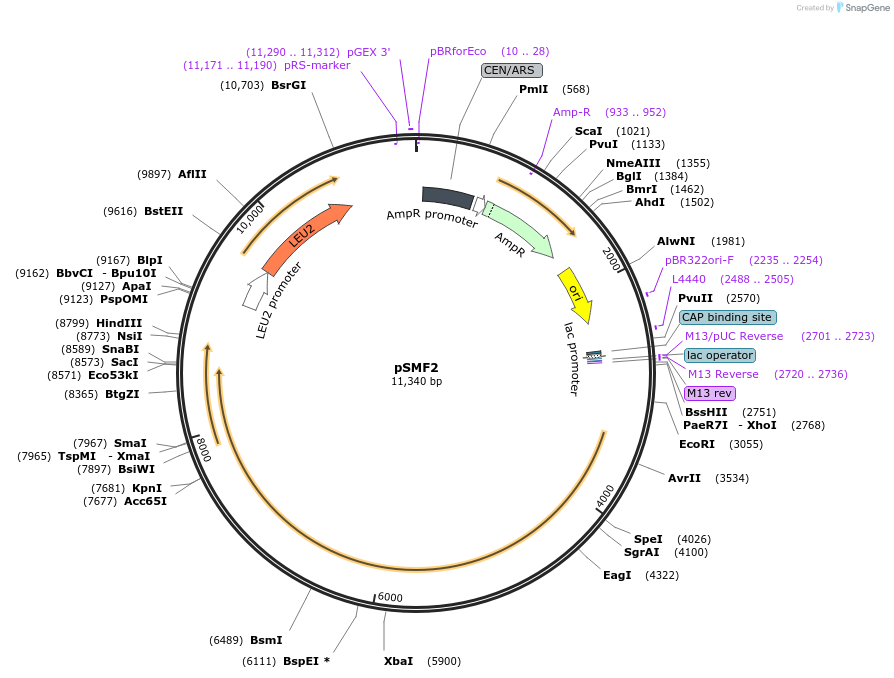
pSMF2
Plasmid#112819PurposeExpresses wild-type Rpb1 in yeast cellsDepositorAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
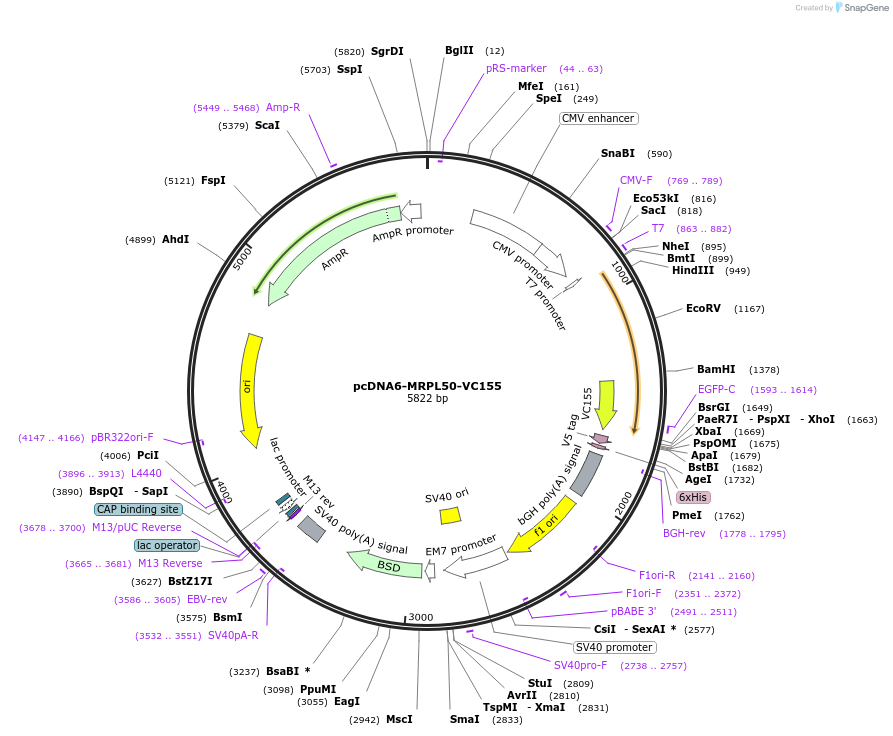
pcDNA6-MRPL50-VC155
Plasmid#182373PurposemVenus-based mito-riboBiFC vector (Negative control)DepositorAvailable SinceMarch 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
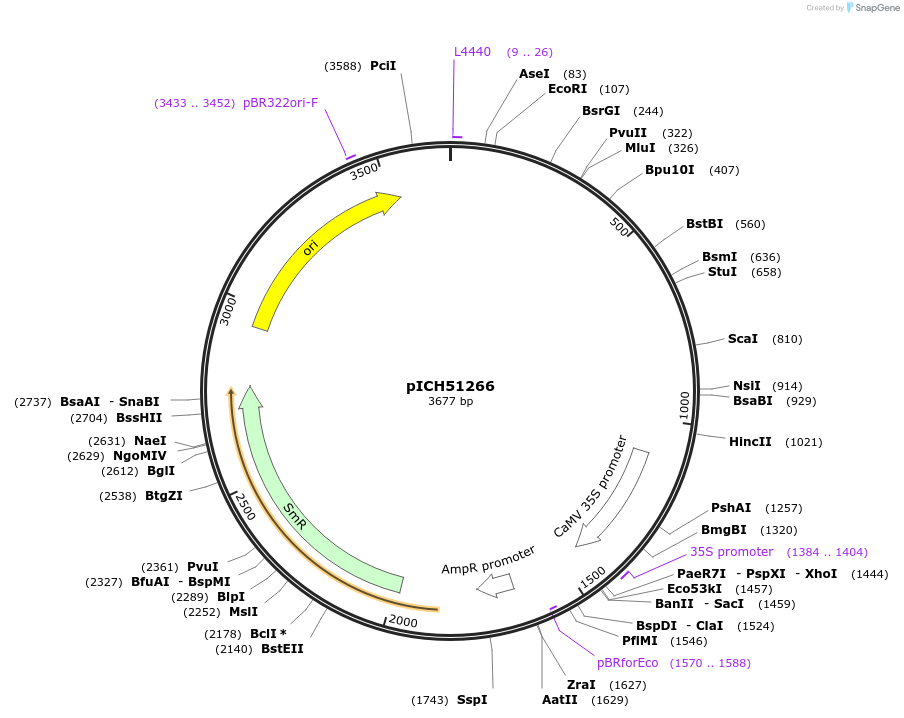
pICH51266
Plasmid#50267DepositorInsertpromoter (1.3 kb ), 35s (Cauliflower Mosaic Virus) + 5'UTR omega (Tobacco Mosaic Virus)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
Pdx1-RFP puro (DM#235)
Plasmid#15085DepositorAvailable SinceOct. 18, 2007AvailabilityAcademic Institutions and Nonprofits only