We narrowed to 5,490 results for: crispr cas9 grna plasmid
-
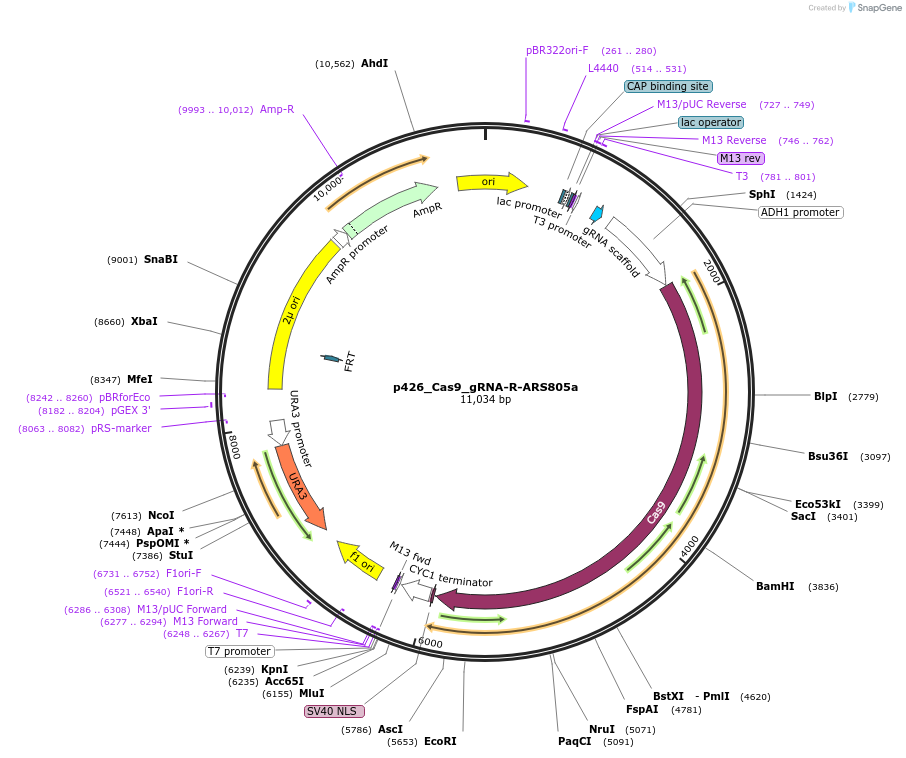
Plasmid#87408Purposep426_Cas9_gRNA-ARS805a without ribozyme All-in-one CRISPR plasmid for integrating, deleting, or replacing a gene in S. cerevisiae. High-copy, URA3 plasmid contains human codon-optimized Cas9 and a guide RNA targeting ARS805a sequence TTATTTGAATGATATTTAGT in yeast chromosome 8.DepositorInsertHuman Optimized S. pyogenes Cas9 and guide RNA targeting ARS805a
UseCRISPRPromoterADH1, pTyrosineAvailable SinceApril 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
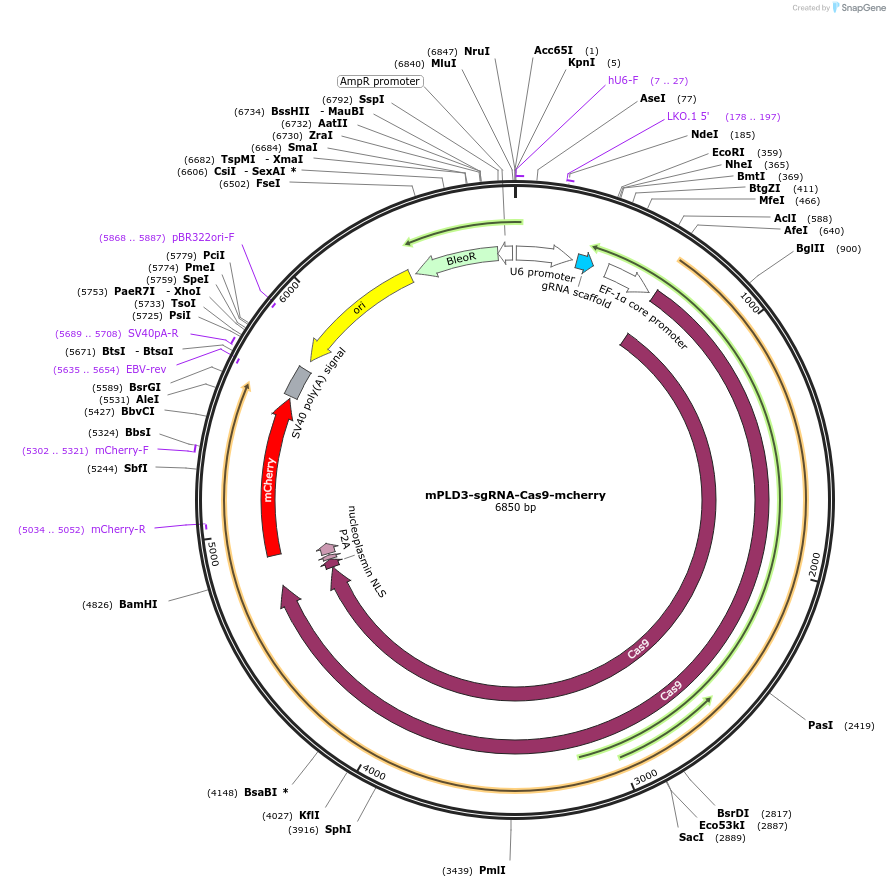
mPLD3-sgRNA-Cas9-mcherry
Plasmid#199700Purposeencodes sgRNA for mouse PLD3 KO, (target 217-223aa) plus Cas9-P2A-mCherryDepositorInserthSpCas9
UseCRISPRTagsP2A-RFPExpressionMammalianPromoterU6 promoterAvailable SinceMay 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
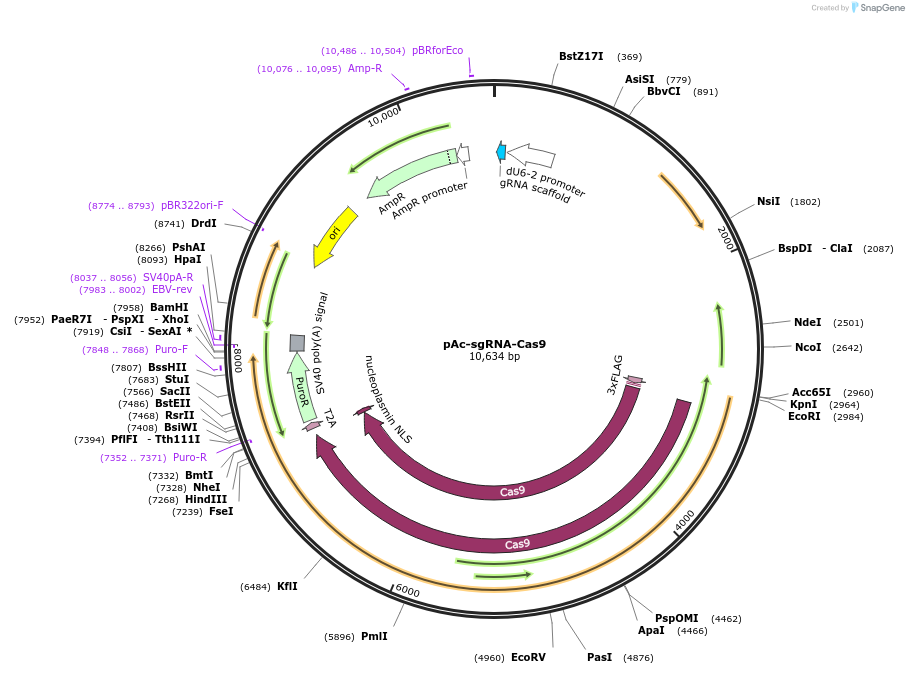
pAc-sgRNA-Cas9
Plasmid#49330PurposeExpresses sgRNA and Cas9-Puro in Drosophila S2 cellsDepositorInsertsCas9
dU6-sgRNA
UseCRISPRTags3xFLAG and NLSExpressionInsectMutationHuman codon optimisedPromoterActin-5c and Drosophila U6Available SinceDec. 19, 2013AvailabilityAcademic Institutions and Nonprofits only -
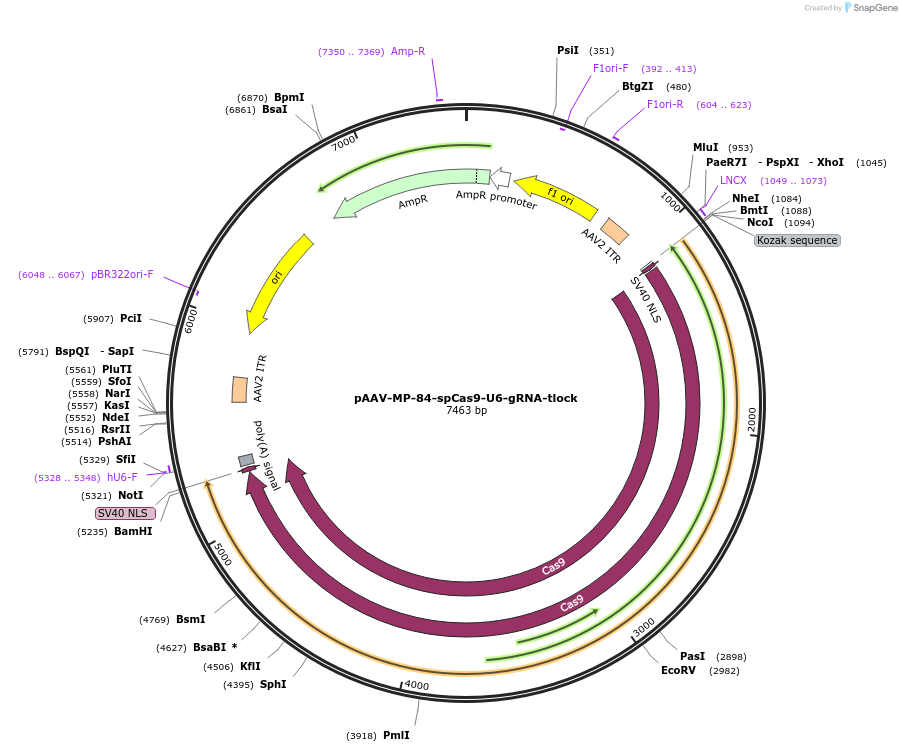
pAAV-MP-84-spCas9-U6-gRNA-tlock
Plasmid#206936PurposeExpression of spCas9 endonuclease and gRNA in an AAV vector allowing packaging of viral genome smaller than 5 kb.DepositorInsertspCas9
UseAAVTagsSV40 NLSExpressionMammalianPromoterMP-84Available SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
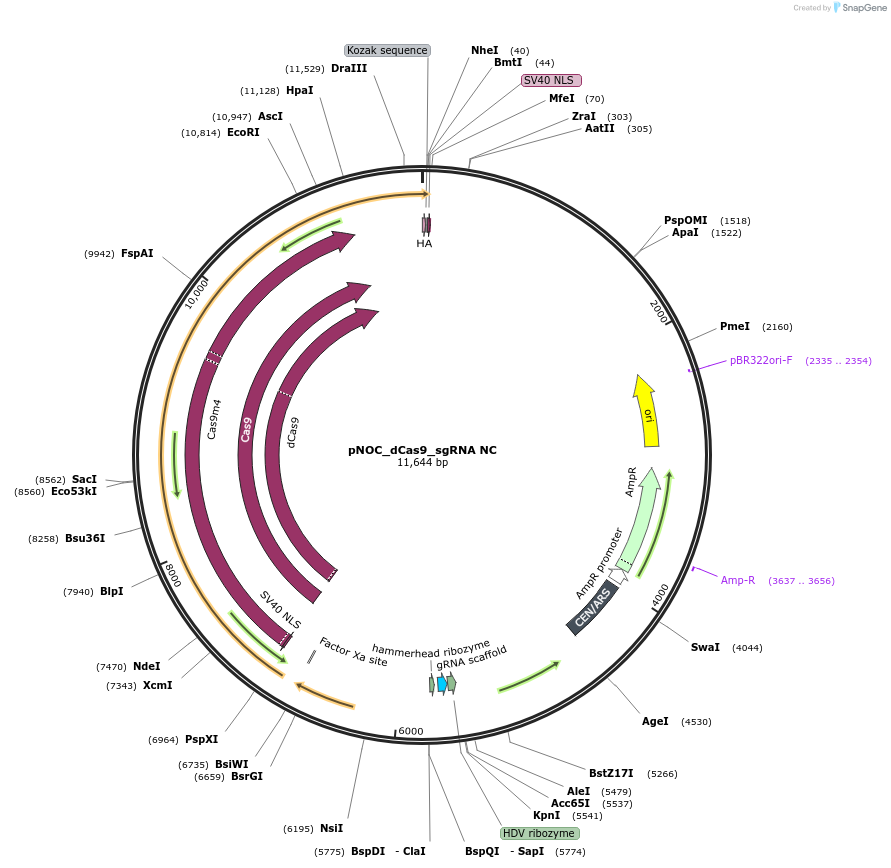
pNOC_dCas9_sgRNA NC
Plasmid#176259PurposepNOC episomal plasmid harboring the dead version of humanized spCas9 gene sequence tagged with Nlux and spacer sequence on sgRNA is replaced by the type IIS restriction site for endonuclease BaeI andDepositorInsertdead spCas9
UseCRISPR, Luciferase, and Synthetic Biology ; Expre…TagsluciferaseMutationH840A and D10A mutations on spCas9 to inactivate …Available SinceOct. 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
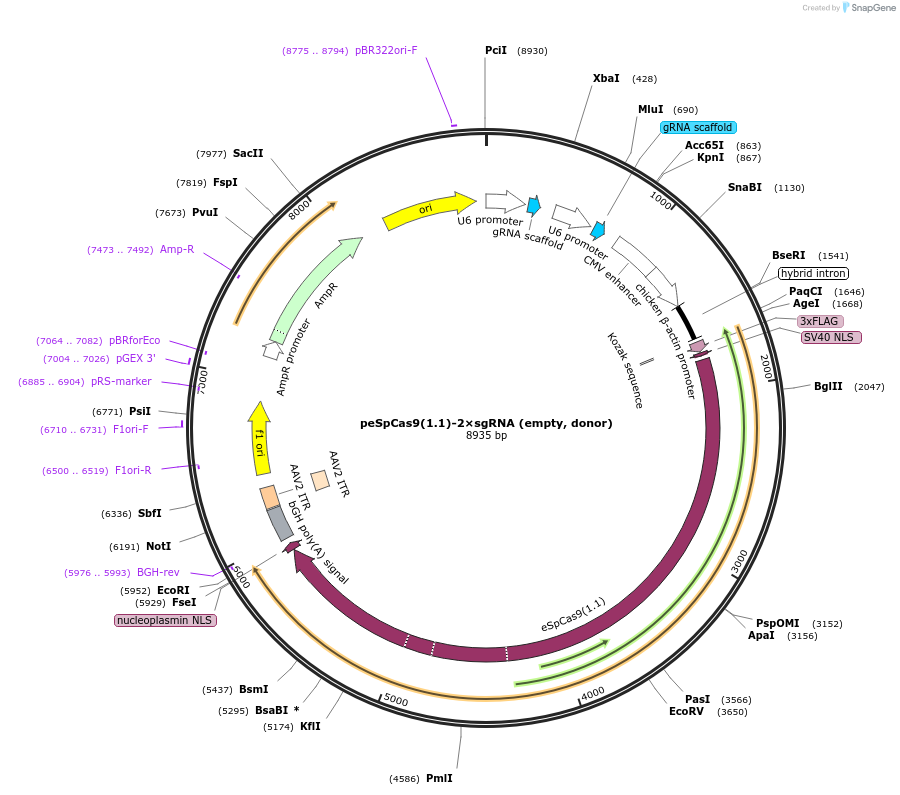
peSpCas9(1.1)-2×sgRNA (empty, donor)
Plasmid#80768PurposeAll-in-one vector for CRISPR/Cas9-mediated homology-independent knock-in system. The plasmid contains eSpCas9(1.1) and two sgRNA expression cassettes. The first gRNA cloning site is empty.DepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceMarch 15, 2017AvailabilityAcademic Institutions and Nonprofits only -
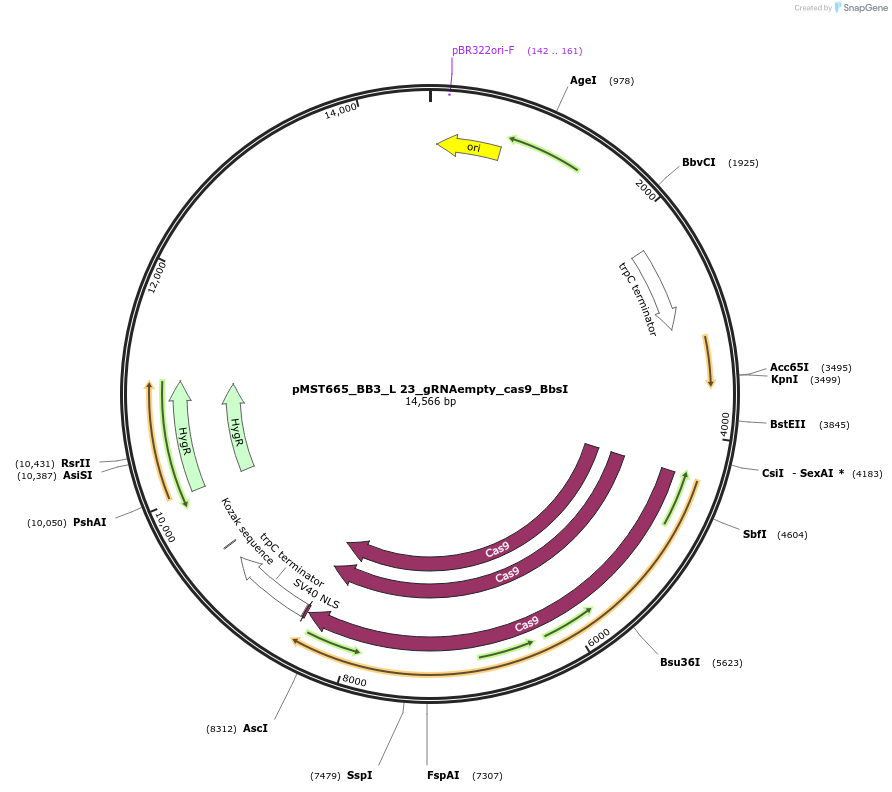
pMST665_BB3_L 23_gRNAempty_cas9_BbsI
Plasmid#90278PurposeBB3_L 23_gRNAempty_cas9_BbsI; CRISPR plasmid with empty gRNA spacer to incorporate gRNA, Cas9DepositorInsertCas9
UseA. nigerExpressionBacterialAvailable SinceJuly 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
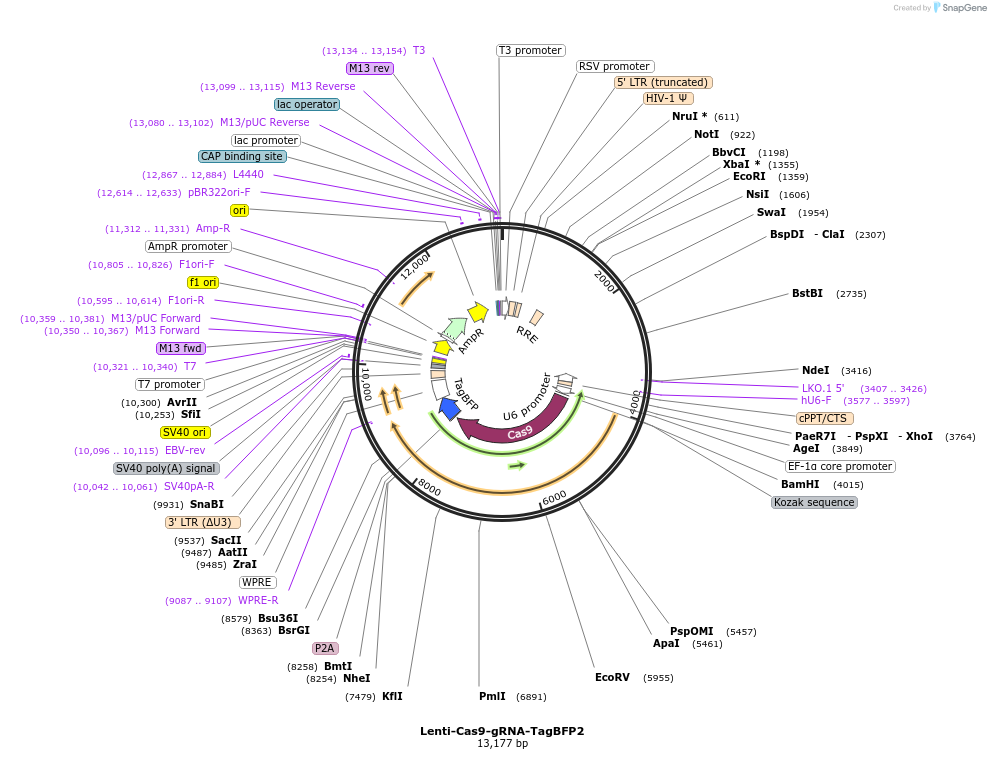
Lenti-Cas9-gRNA-TagBFP2
Plasmid#124774Purpose3rd generation lentiviral plasmid encoding Cas9, TagBFP2, and a gRNA backboneDepositorTypeEmpty backboneUseCRISPR and LentiviralExpressionMammalianAvailable SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
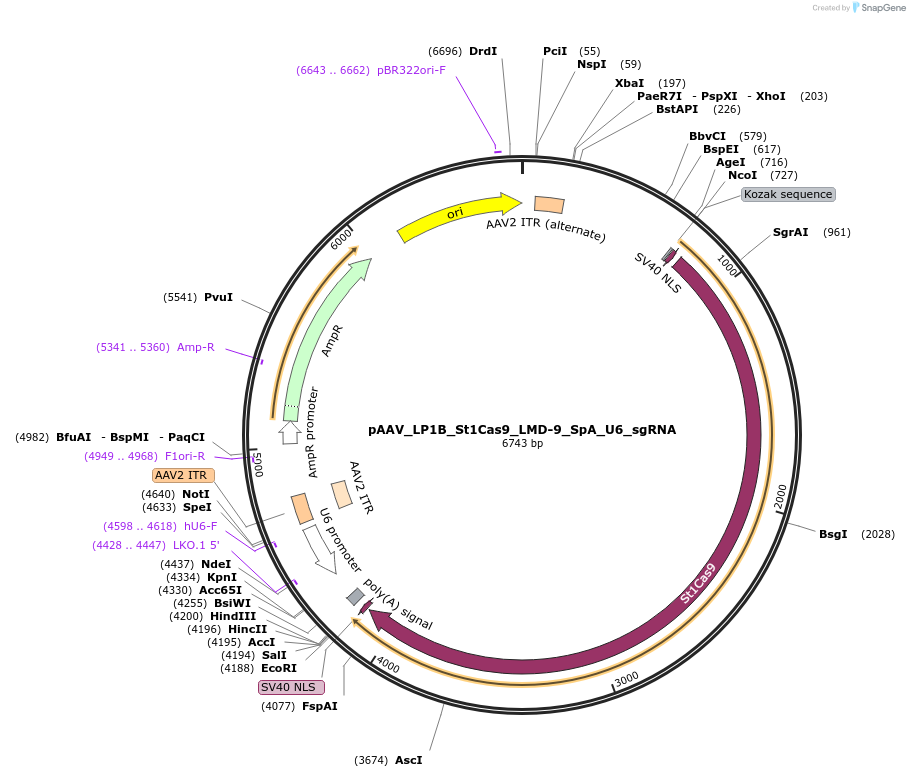
pAAV_LP1B_St1Cas9_LMD-9_SpA_U6_sgRNA
Plasmid#110624PurposeA single vector AAV-Cas9 system containing a liver-specific promoter with Cas9 from Streptococcus thermophilus CRISPR1 (St1Cas9 LMD-9) and its U6-driven sgRNADepositorInsertsSt1Cas9 LMD-9
sgRNA for St1Cas9
UseAAV, CRISPR, and Mouse TargetingTagsSV40 NLSExpressionMammalianPromoterLP1B and hU6Available SinceMay 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
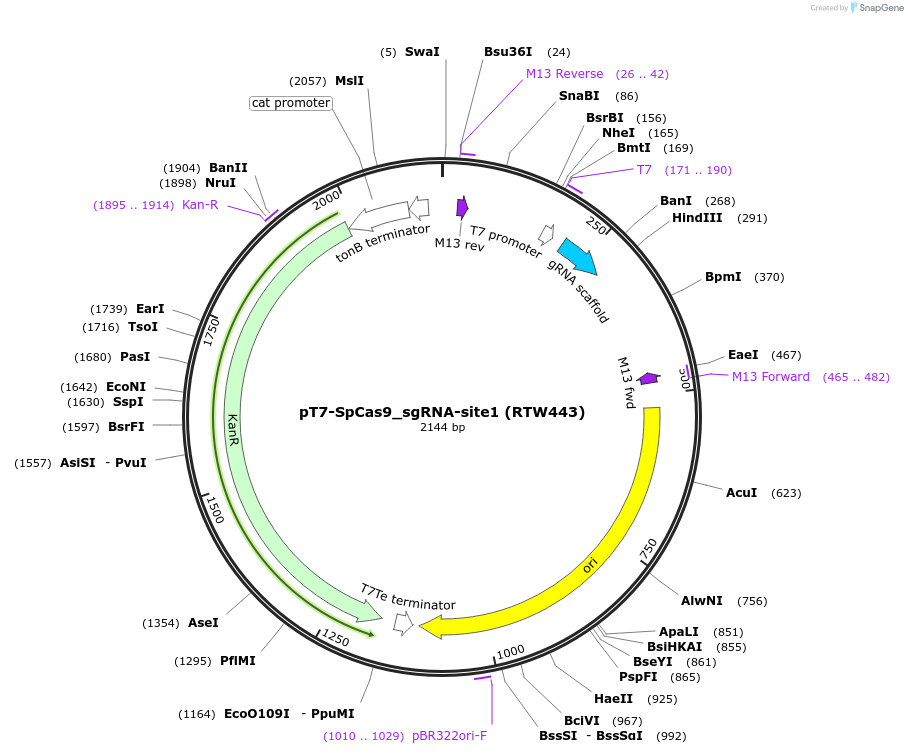
pT7-SpCas9_sgRNA-site1 (RTW443)
Plasmid#160136PurposeT7 promoter expression plasmid for in vitro transcription of SpCas9 sgRNA with spacer #1DepositorInsertSpCas9 sgRNA with spacer #1 (spacer=GGGCACGGGCAGCTTGCCGG)
UseIn vitro transcription of sgrna from t7 promoterPromoterT7Available SinceFeb. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
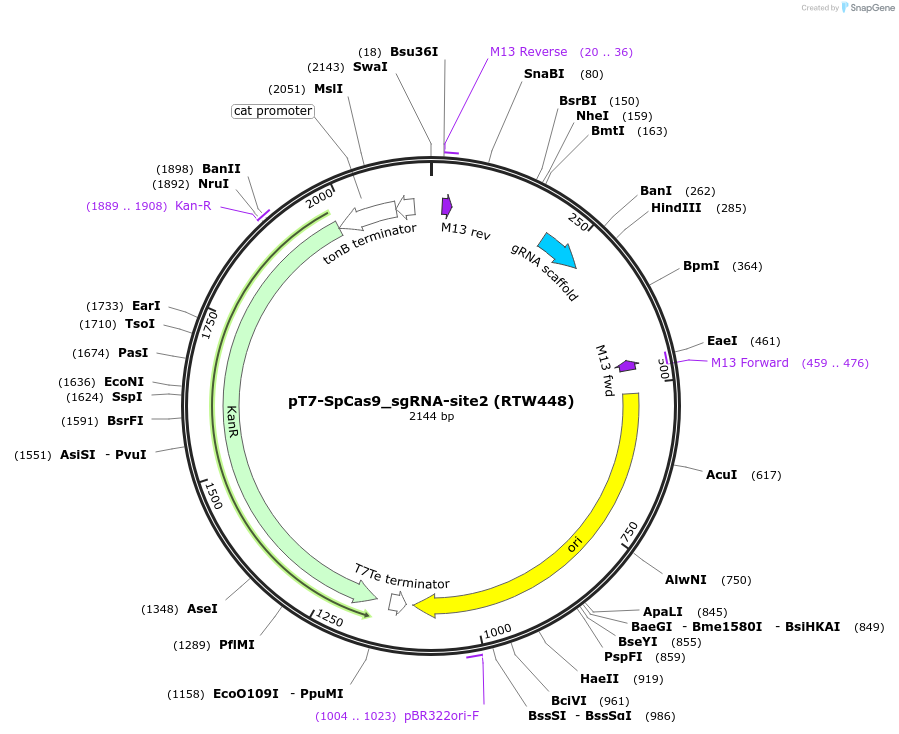
pT7-SpCas9_sgRNA-site2 (RTW448)
Plasmid#160137PurposeT7 promoter expression plasmid for in vitro transcription of SpCas9 sgRNA with spacer #2DepositorInsertSpCas9 sgRNA with spacer #2 (spacer=GTCGCCCTCGAACTTCACCT)
UseIn vitro transcription of sgrna from t7 promoterPromoterT7Available SinceFeb. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
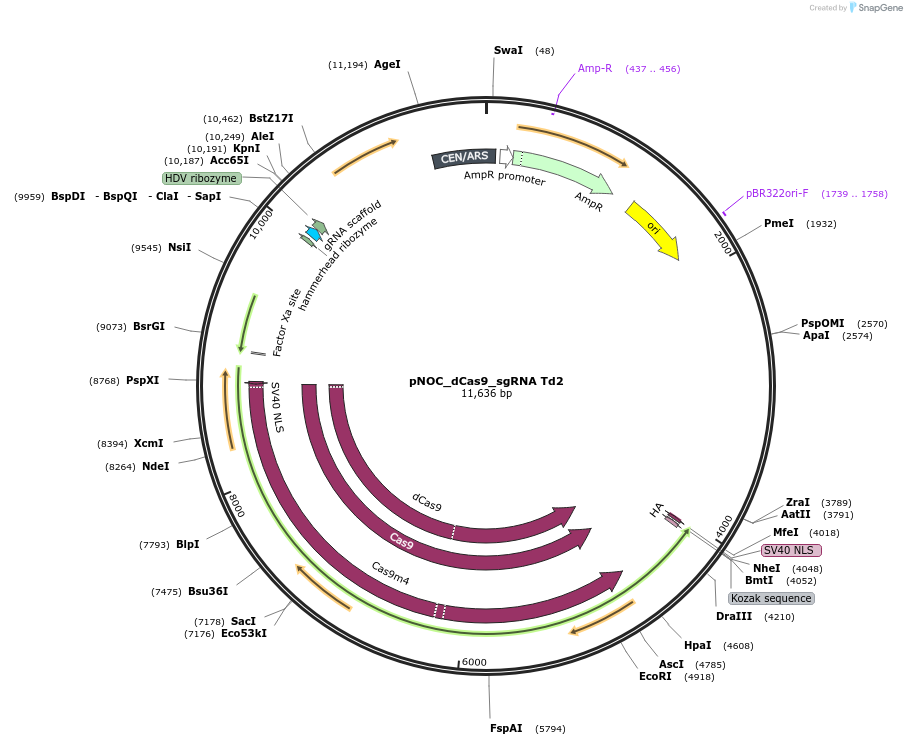
pNOC_dCas9_sgRNA Td2
Plasmid#176258PurposepNOC episomal plasmid harboring the dead version of humanized spCas9 gene sequence tagged with Nlux and sgRNA targeting the fluorescent gene tdtomato with spacer 2DepositorInsertdead spCas9
UseCRISPR, Luciferase, and Synthetic Biology ; Expre…TagsluciferaseMutationH840A and D10A mutations on spCas9 to inactivate …Available SinceNov. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
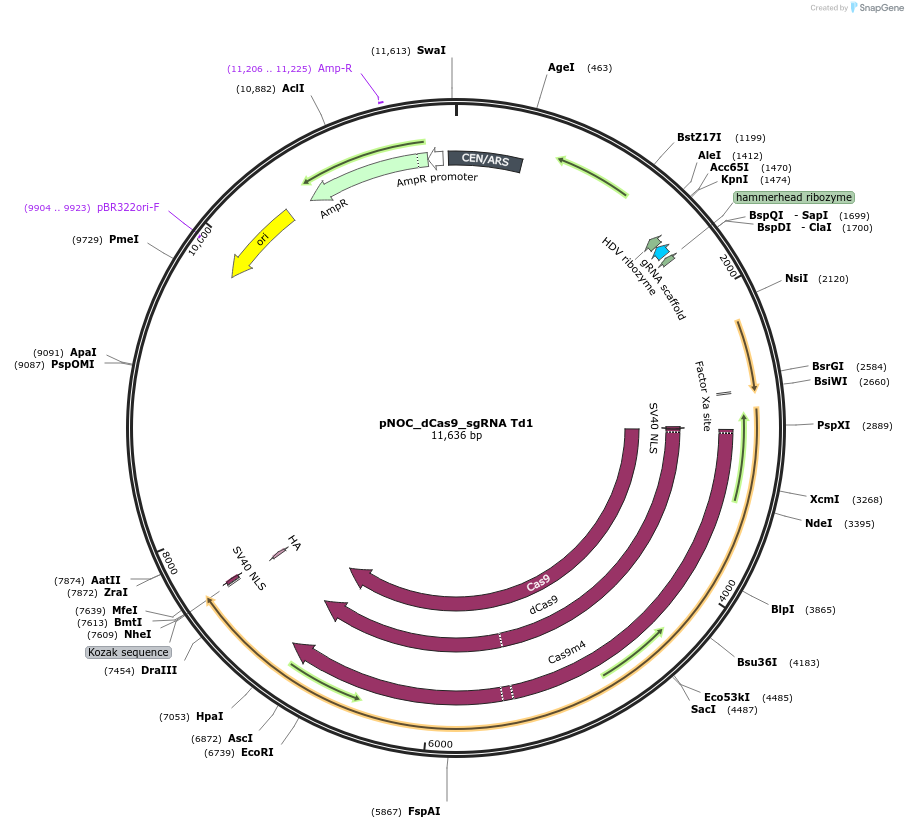
pNOC_dCas9_sgRNA Td1
Plasmid#176257PurposepNOC episomal plasmid harboring the dead version of humanized spCas9 gene sequence tagged with Nlux and sgRNA targeting the fluorescent gene tdtomato with spacer 1DepositorInsertdead spCas9
UseCRISPR, Luciferase, and Synthetic Biology ; Expre…TagsluciferaseAvailable SinceNov. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
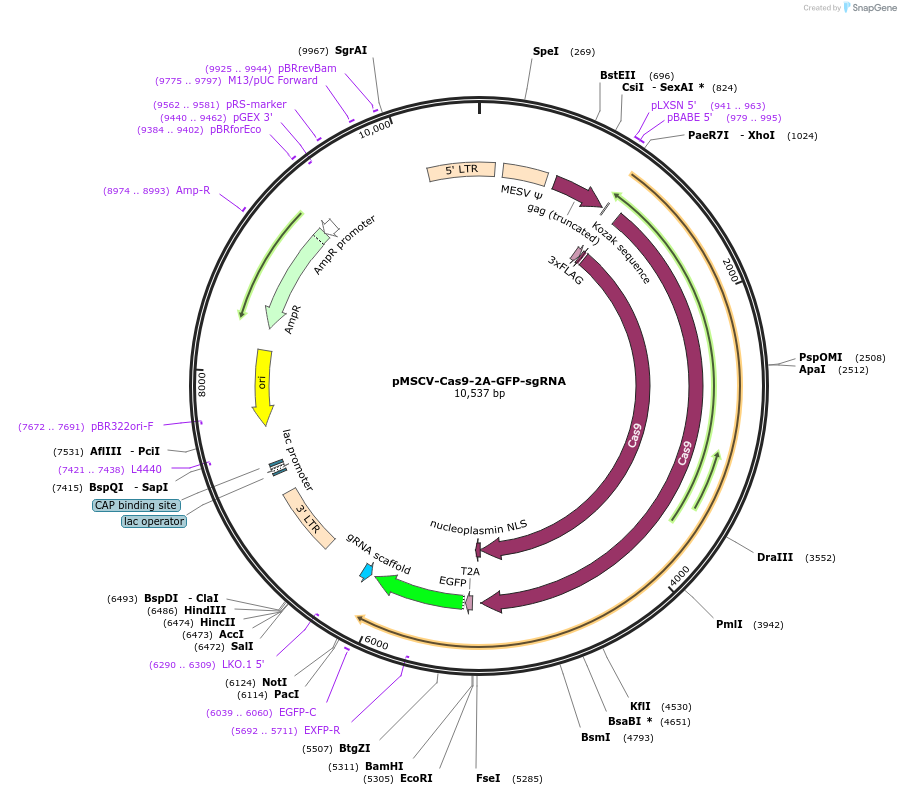
pMSCV-Cas9-2A-GFP-sgRNA
Plasmid#124889PurposeContains spCas9-2A-GFP and guide RNA scaffold in single retroviral vectorDepositorInsertCas9
UseRetroviralTags2A-GFPPromoterMSCVAvailable SinceMay 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
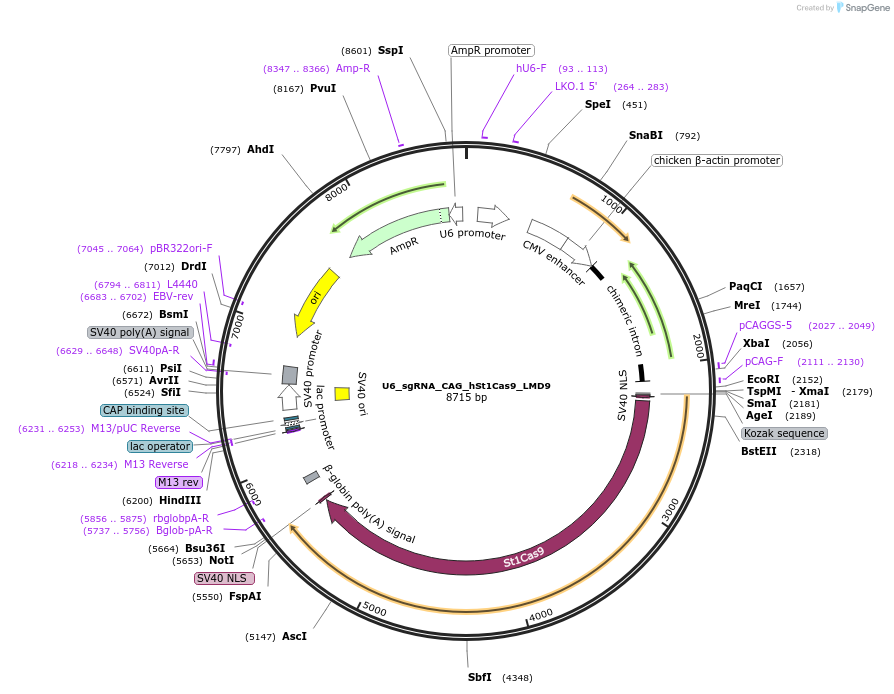
U6_sgRNA_CAG_hSt1Cas9_LMD9
Plasmid#110626PurposeA single vector mammalian expression system containing a CAG promoter with Cas9 from Streptococcus thermophilus CRISPR1 (St1Cas9 LMD-9) and its U6-driven sgRNA.DepositorInsertsSt1Cas9 LMD-9
sgRNA for St1Cas9
UseCRISPR and Synthetic BiologyTagsSV40 NLSExpressionMammalianPromoterCAG and hU6Available SinceMay 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
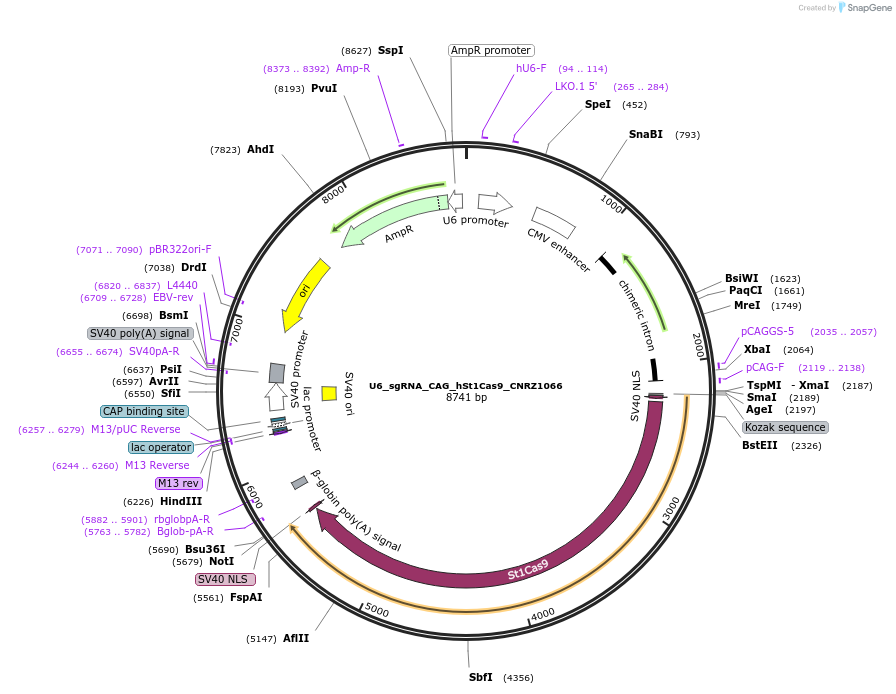
U6_sgRNA_CAG_hSt1Cas9_CNRZ1066
Plasmid#136651PurposeA single vector mammalian expression system containing a CAG promoter with Cas9 from Streptococcus thermophilus CRISPR1 (St1Cas9 LMD9:CNRZ1066 chimera) and its U6-driven sgRNA.DepositorInsertsSt1Cas9 LMD-9:CNRZ_1066 Chimera
sgRNA for St1Cas9
UseCRISPR and Synthetic BiologyTagsSV40 NLSExpressionMammalianPromoterCAG and hU6Available SinceFeb. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
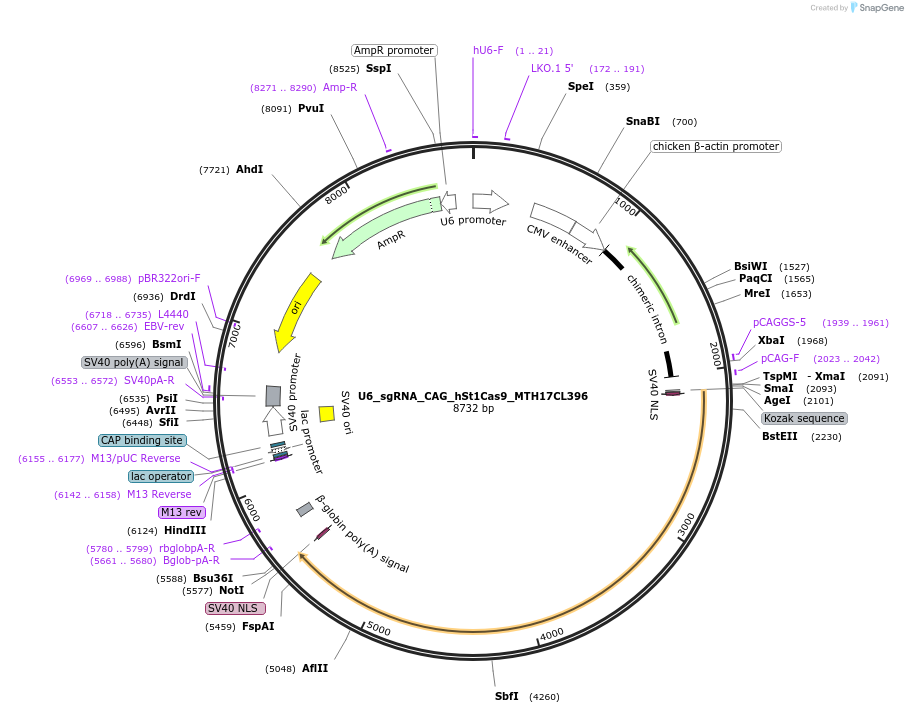
U6_sgRNA_CAG_hSt1Cas9_MTH17CL396
Plasmid#136656PurposeA single vector mammalian expression system containing a CAG promoter with Cas9 from Streptococcus thermophilus CRISPR1 (St1Cas9 LMD9:MTH17CL396 chimera) and its U6-driven sgRNA.DepositorInsertsSt1Cas9 LMD-9:MTH17CL396 Chimera
sgRNA for St1Cas9
UseCRISPR and Synthetic BiologyTagsSV40 NLSExpressionMammalianPromoterCAG and hU6Available SinceMarch 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
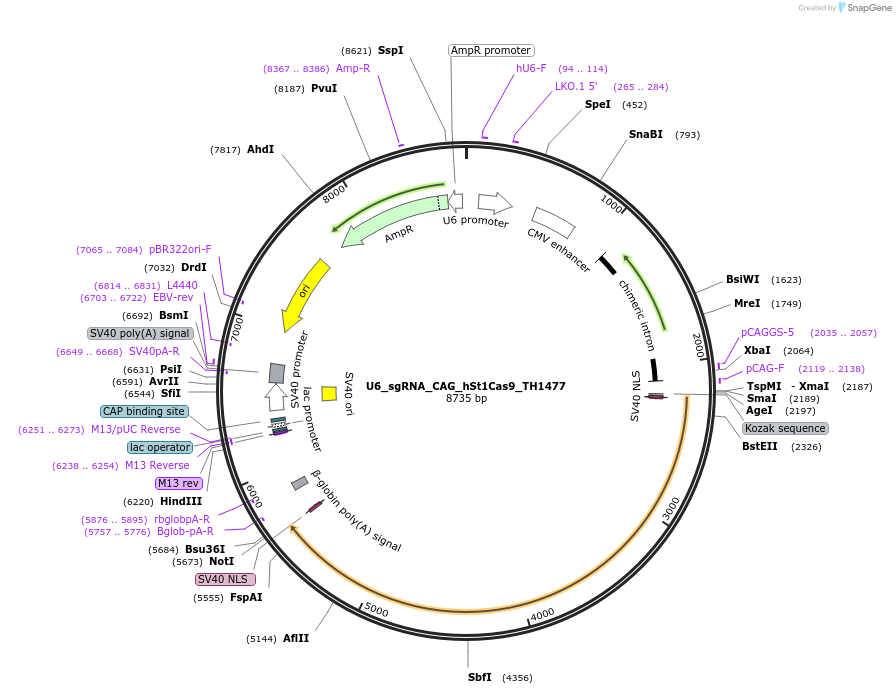
U6_sgRNA_CAG_hSt1Cas9_TH1477
Plasmid#136655PurposeA single vector mammalian expression system containing a CAG promoter with Cas9 from Streptococcus thermophilus CRISPR1 (St1Cas9 LMD9:TH1477 chimera) and its U6-driven sgRNA.DepositorInsertsSt1Cas9 LMD-9:TH1477 Chimera
sgRNA for St1Cas9
UseCRISPR and Synthetic BiologyTagsSV40 NLSExpressionMammalianPromoterCAG and hU6Available SinceFeb. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
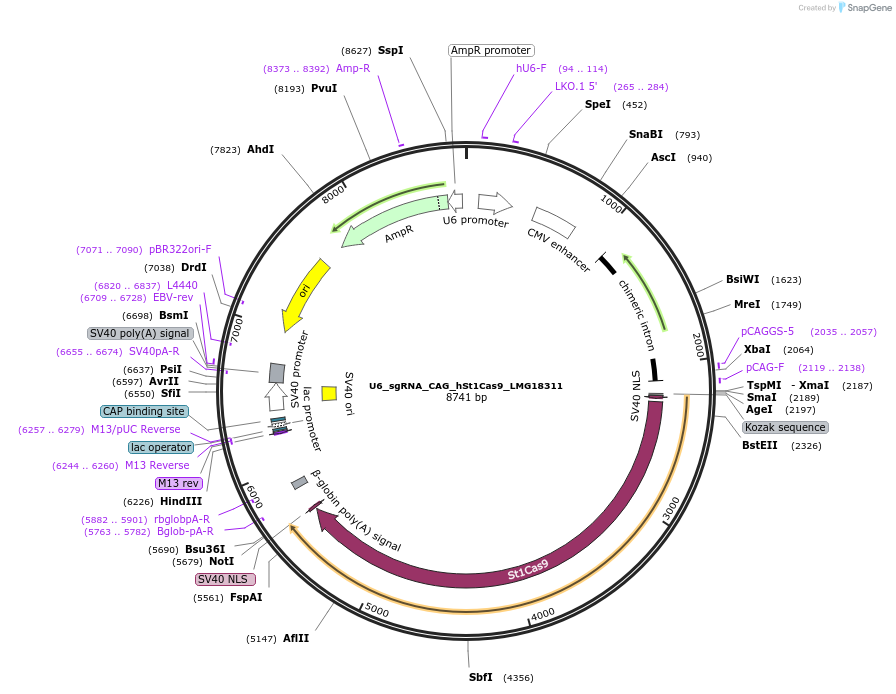
U6_sgRNA_CAG_hSt1Cas9_LMG18311
Plasmid#136653PurposeA single vector mammalian expression system containing a CAG promoter with Cas9 from Streptococcus thermophilus CRISPR1 (St1Cas9 LMD9:LMG18311 chimera) and its U6-driven sgRNA.DepositorInsertsSt1Cas9 LMD9:LMG18311 chimera
sgRNA for St1Cas9
UseCRISPR and Synthetic BiologyTagsSV40 NLSExpressionMammalianPromoterCAG and hU6Available SinceFeb. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
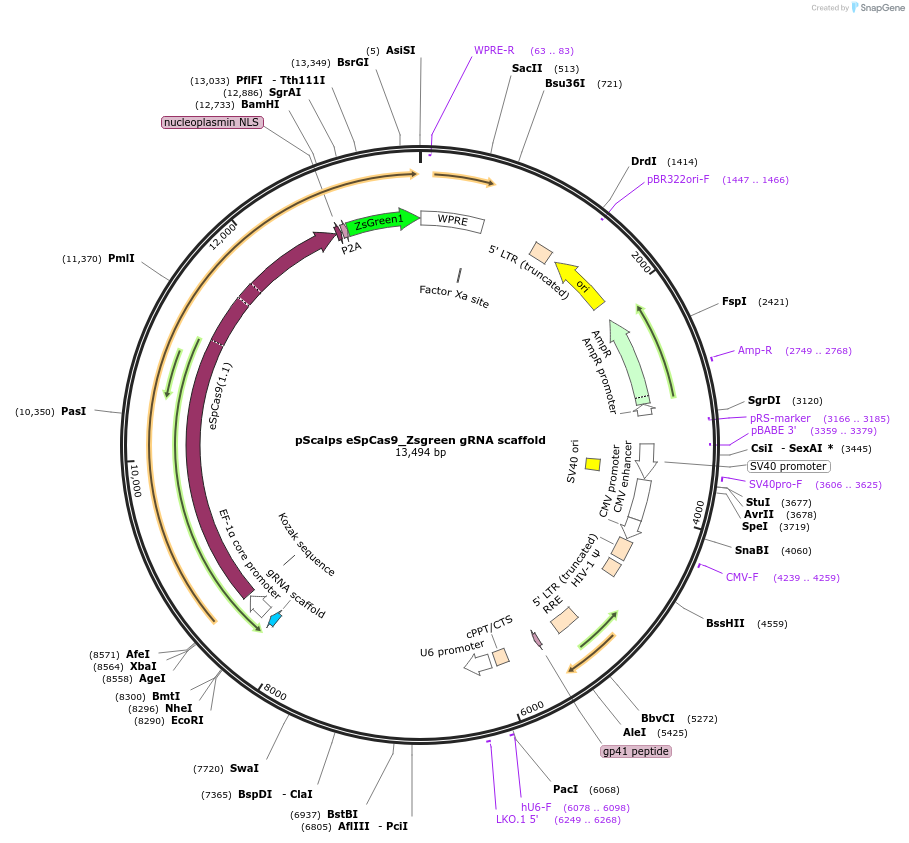
pScalps eSpCas9_Zsgreen gRNA scaffold
Plasmid#188449PurposeExpression of eSpCas9 tagged with ZsgreenDepositorTypeEmpty backboneUseCRISPR and LentiviralPromoterEF1-alphaAvailable SinceJan. 10, 2023AvailabilityAcademic Institutions and Nonprofits only