295,694 results
-
Plasmid#144114PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 8, 2023AvailabilityAcademic Institutions and Nonprofits only
-
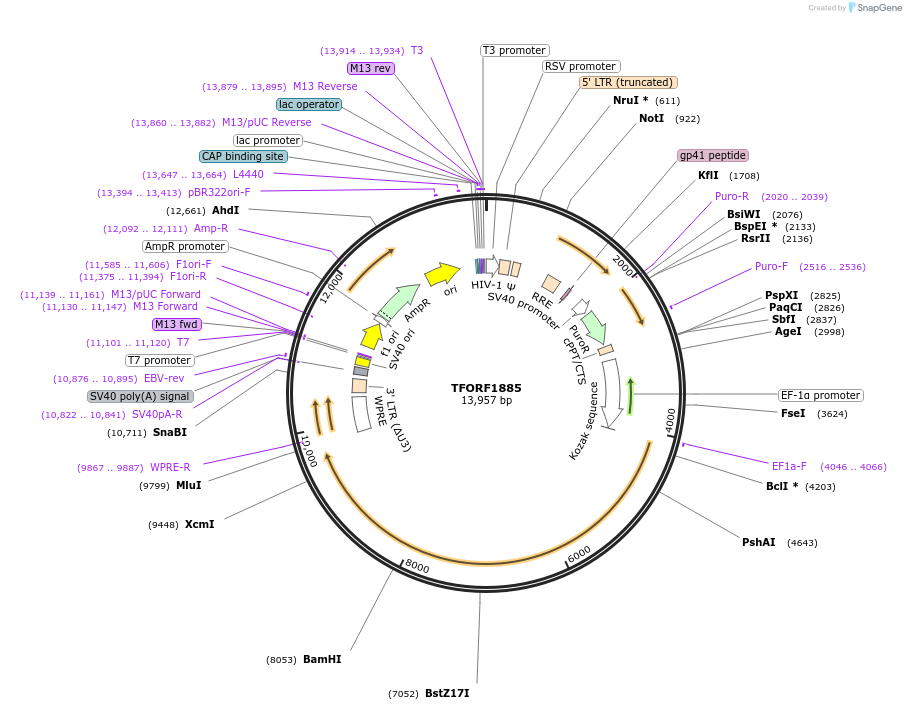
pRK5 Flag-RagC(S75N)
Plasmid#99724PurposeMammalian expression of Flag-RagC(S75N)DepositorAvailable SinceNov. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
pRK5 Flag-RagC(S75N)
Plasmid#99724PurposeMammalian expression of Flag-RagC(S75N)DepositorAvailable SinceNov. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
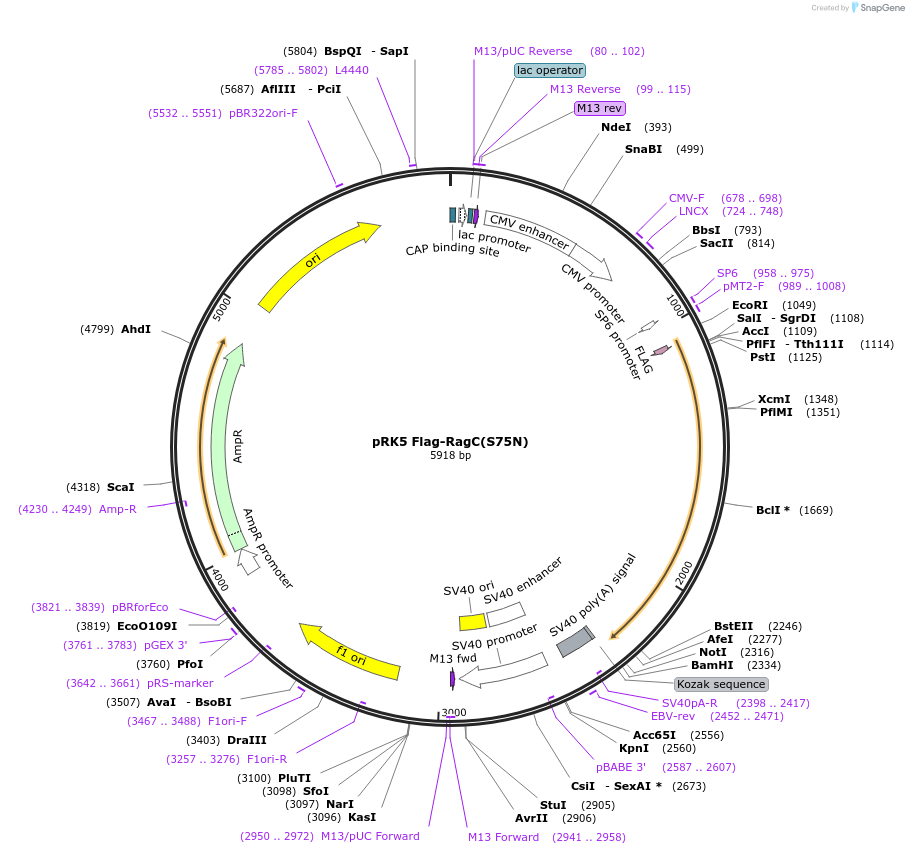
EK0395 mCherry-SG(s70587)SG-EGFP (pEAQ)
Plasmid#191170PurposeEK0285 in plant-expressable format: Sensor 1 (for EK0208); mCherry marker, EGFP output.DepositorInsertmCherry:s70587:EGFP (plant)
ExpressionPlantMutationWTAvailable SinceOct. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
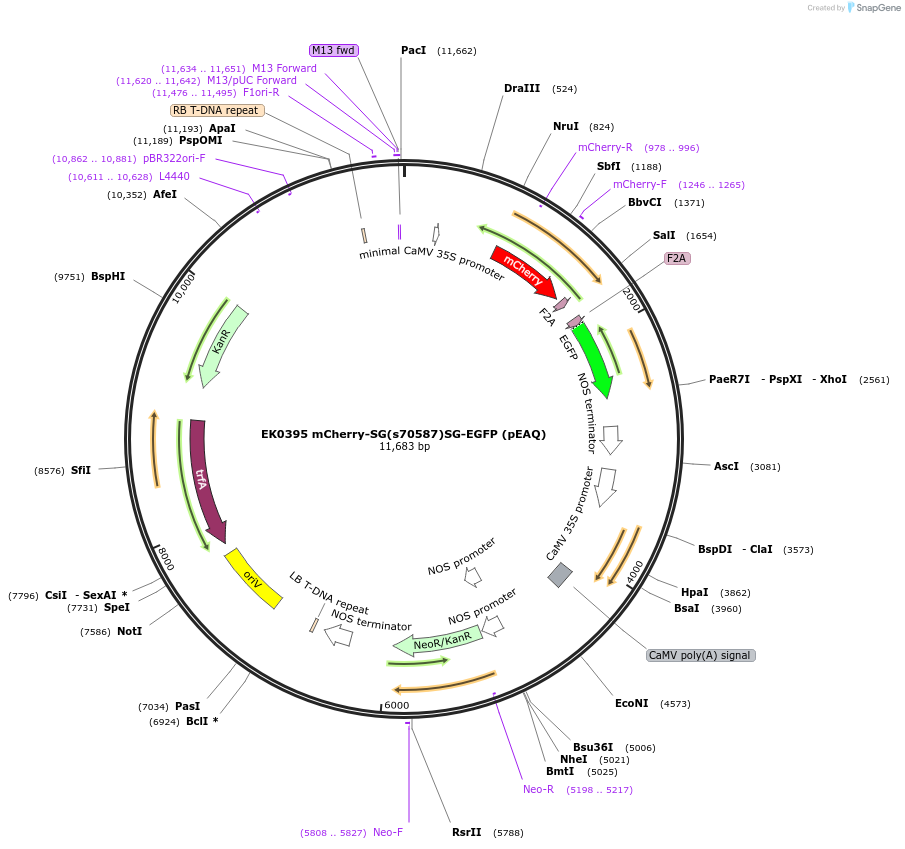
FUGW-H2B-mCherry
Plasmid#132333PurposeThird generation lentiviral vector for hUbC-driven H2B-mCherry expressionDepositorInsertH2B-mCherry
UseLentiviralExpressionMammalianPromoterhUbCAvailable SinceJuly 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
FUGW-H2B-mCherry
Plasmid#132333PurposeThird generation lentiviral vector for hUbC-driven H2B-mCherry expressionDepositorInsertH2B-mCherry
UseLentiviralExpressionMammalianPromoterhUbCAvailable SinceJuly 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
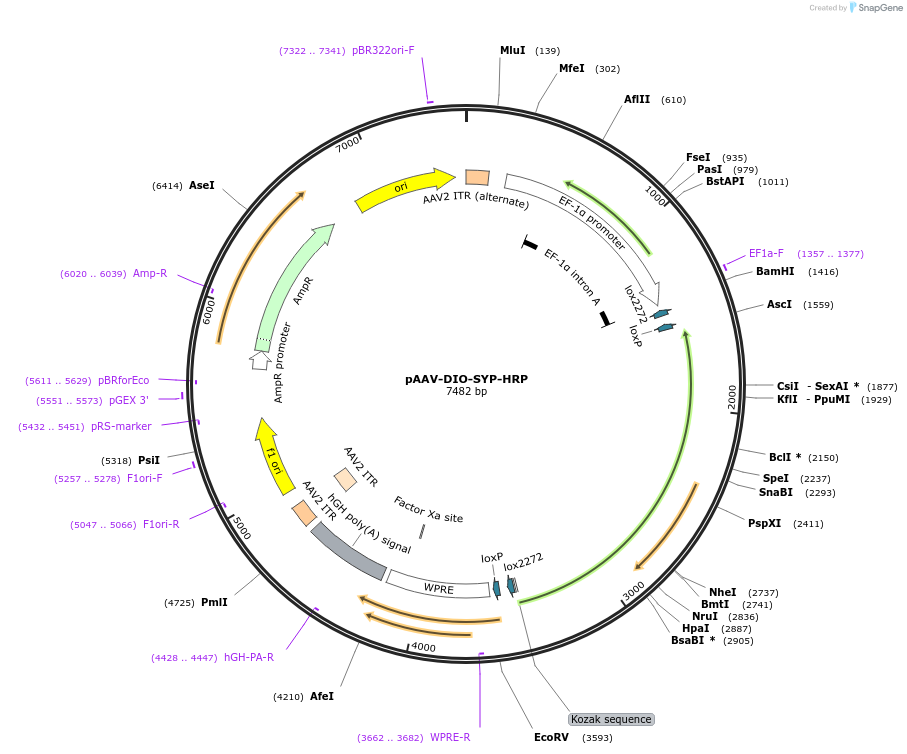
pAAV-DIO-SYP-HRP
Plasmid#117186PurposePlasmid name in publication: pAAV-DIO-SV-HRP. Peroxidase reporter for multiplexed electron microscopy labelingDepositorInsertSYP-HRP
UseAAV and Cre/LoxExpressionMammalianPromoterEF1aAvailable SinceMarch 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
pAAV-DIO-SYP-HRP
Plasmid#117186PurposePlasmid name in publication: pAAV-DIO-SV-HRP. Peroxidase reporter for multiplexed electron microscopy labelingDepositorInsertSYP-HRP
UseAAV and Cre/LoxExpressionMammalianPromoterEF1aAvailable SinceMarch 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
pLKO mouse shRNA 1 raptor
Plasmid#21339DepositorAvailable SinceJuly 7, 2009AvailabilityAcademic Institutions and Nonprofits only -
pLKO mouse shRNA 1 raptor
Plasmid#21339DepositorAvailable SinceJuly 7, 2009AvailabilityAcademic Institutions and Nonprofits only -
TFORF2875
Plasmid#143650PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
TFORF2875
Plasmid#143650PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
pEGFPC3-mSufu
Plasmid#65431PurposeExpresses mouse Sufu protein in mammalian cellsDepositorAvailable SinceJune 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
pEGFPC3-mSufu
Plasmid#65431PurposeExpresses mouse Sufu protein in mammalian cellsDepositorAvailable SinceJune 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
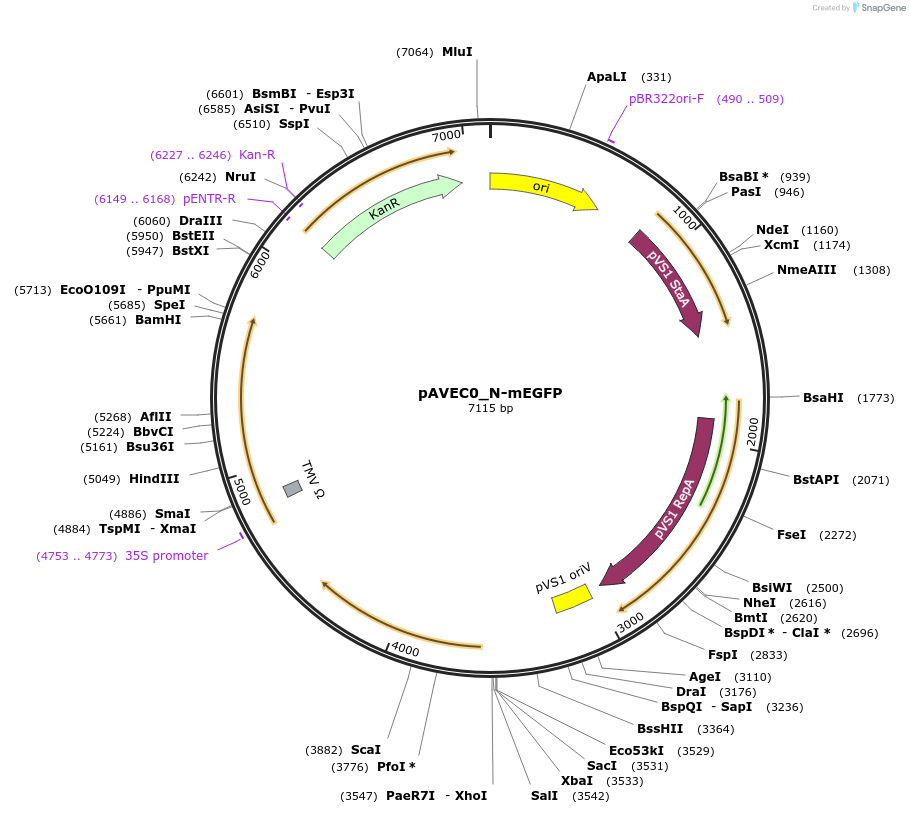
pAVEC0_N-mEGFP
Plasmid#231255PurposeFor expressing fusion proteins with N terminally tagged mEGFP in plantsDepositorTypeEmpty backboneTagsmEGFPExpressionPlantMutationN/AAvailable SinceAug. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
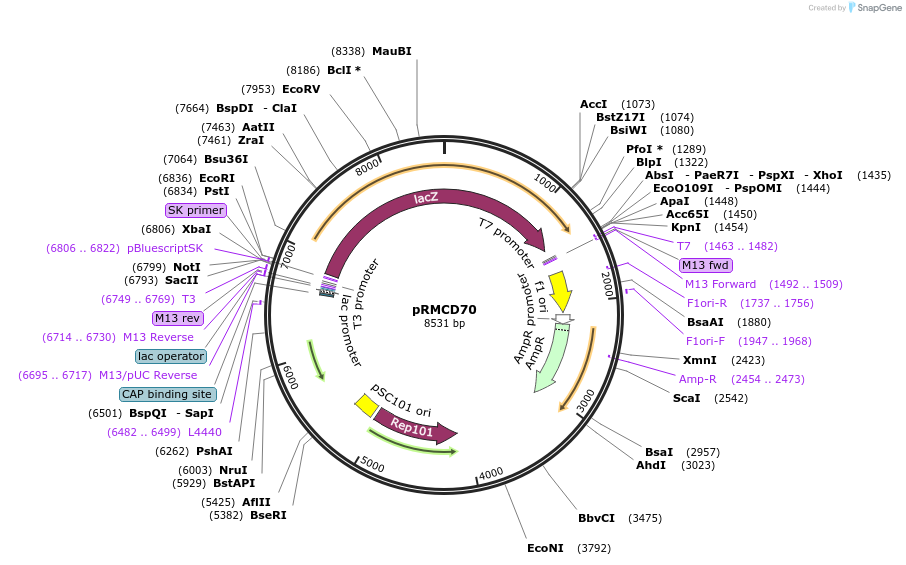
pRMCD70
Plasmid#167557PurposePlasmid pWSK29 encoding E. coli LacZ' for making reporter fusion proteins for topology mappingDepositorTypeEmpty backboneTagsLacZ'ExpressionBacterialAvailable SinceMay 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
pRMCD70
Plasmid#167557PurposePlasmid pWSK29 encoding E. coli LacZ' for making reporter fusion proteins for topology mappingDepositorTypeEmpty backboneTagsLacZ'ExpressionBacterialAvailable SinceMay 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
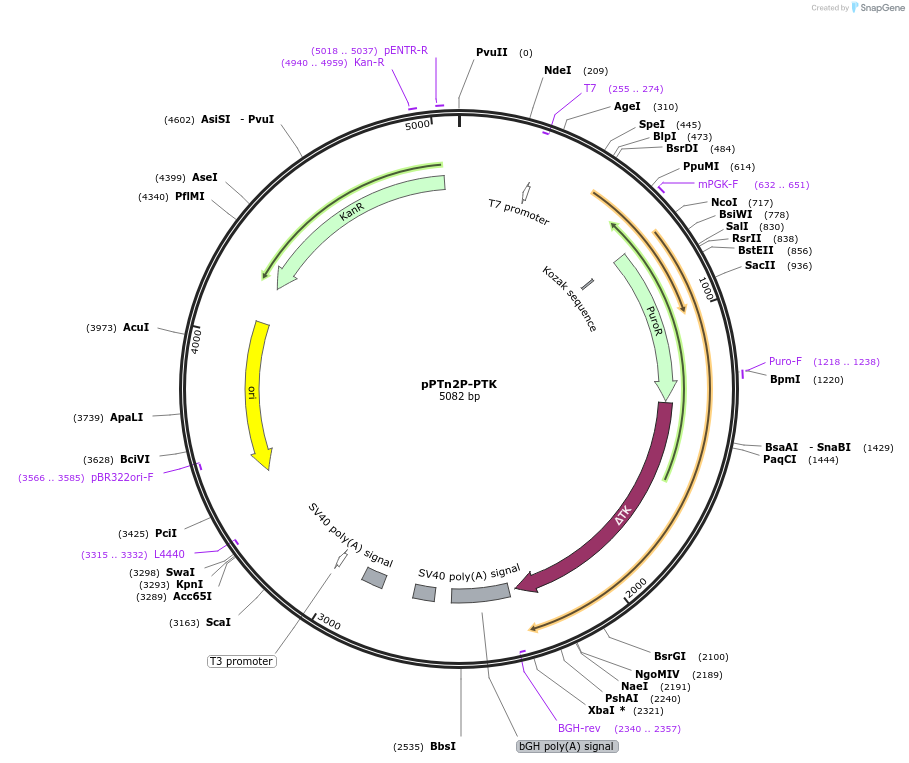
pPTn2P-PTK
Plasmid#85604PurposePassport transposon with a puromycin-thymidine kinase fusion driven by a PGK promoter.DepositorInsertPassport transposon with a puromycin-thymidine kinase fusion
ExpressionMammalianPromoterPGKAvailable SinceDec. 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
pPTn2P-PTK
Plasmid#85604PurposePassport transposon with a puromycin-thymidine kinase fusion driven by a PGK promoter.DepositorInsertPassport transposon with a puromycin-thymidine kinase fusion
ExpressionMammalianPromoterPGKAvailable SinceDec. 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
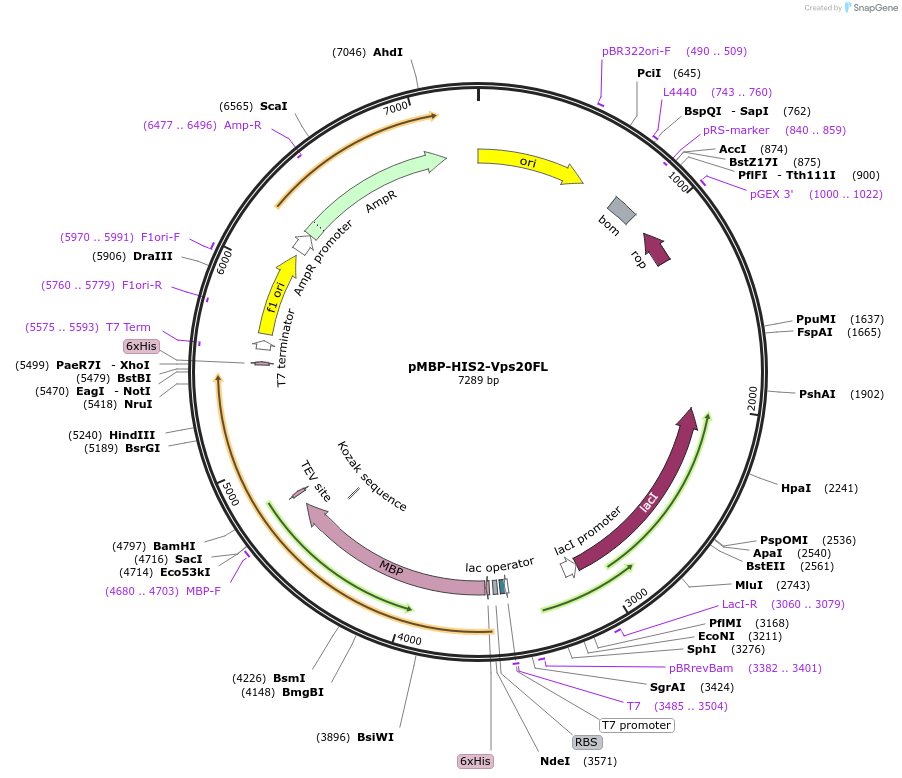
pMBP-HIS2-Vps20FL
Plasmid#21490DepositorAvailable SinceSept. 9, 2009AvailabilityAcademic Institutions and Nonprofits only -
pMBP-HIS2-Vps20FL
Plasmid#21490DepositorAvailable SinceSept. 9, 2009AvailabilityAcademic Institutions and Nonprofits only -
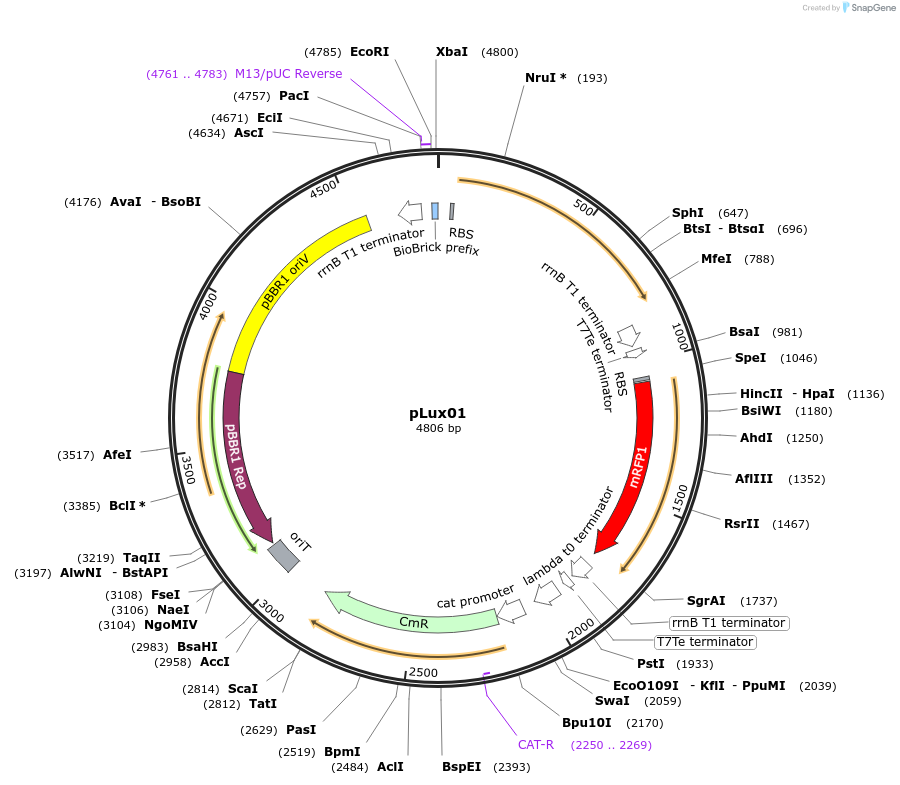
pLux01
Plasmid#78281PurposemRFP1 expressed behind an AHL-inducible constructDepositorInsertAHL inducible mRFP1
Available SinceNov. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
pLux01
Plasmid#78281PurposemRFP1 expressed behind an AHL-inducible constructDepositorInsertAHL inducible mRFP1
Available SinceNov. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
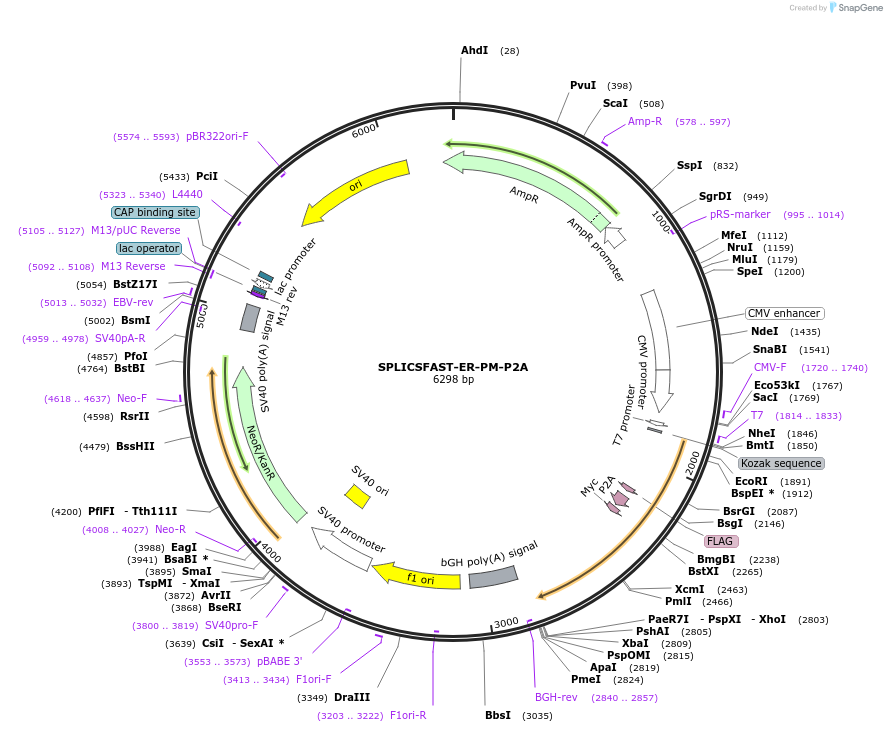
SPLICSFAST-ER-PM-P2A
Plasmid#214893PurposeExpresses equimolar Plasma Membrane- and Endoplasmic Reticulum- targeted split-FAST fragments for Short-range ER-PM Contact sitesDepositorInsertSPLICSFAST-ER-PM-P2A
TagsnoneExpressionBacterial and MammalianAvailable SinceFeb. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
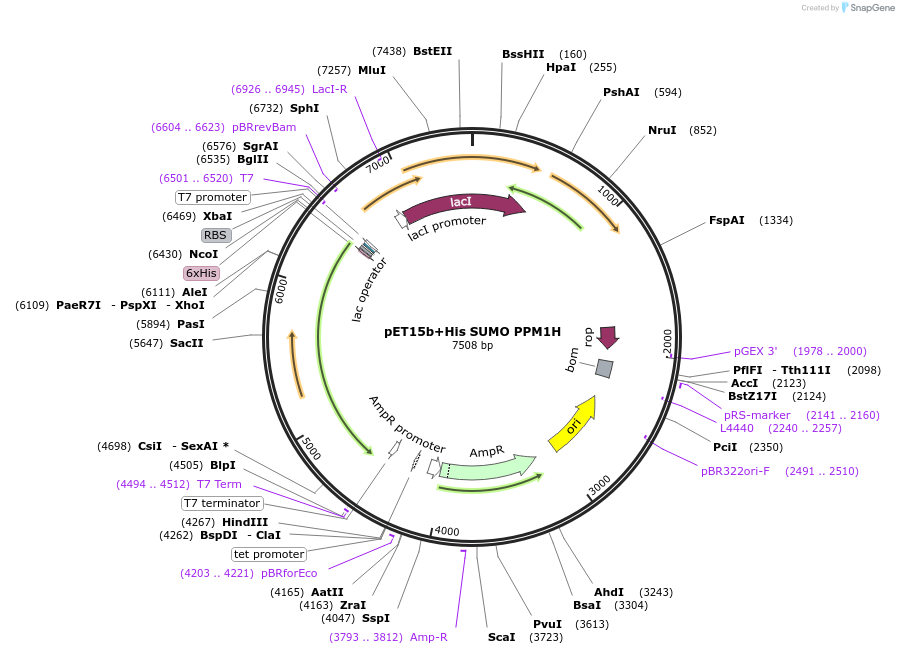
pET15b+His SUMO PPM1H
Plasmid#207333PurposeBacterial expression of His SUMO PPM1HDepositorAvailable SinceNov. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
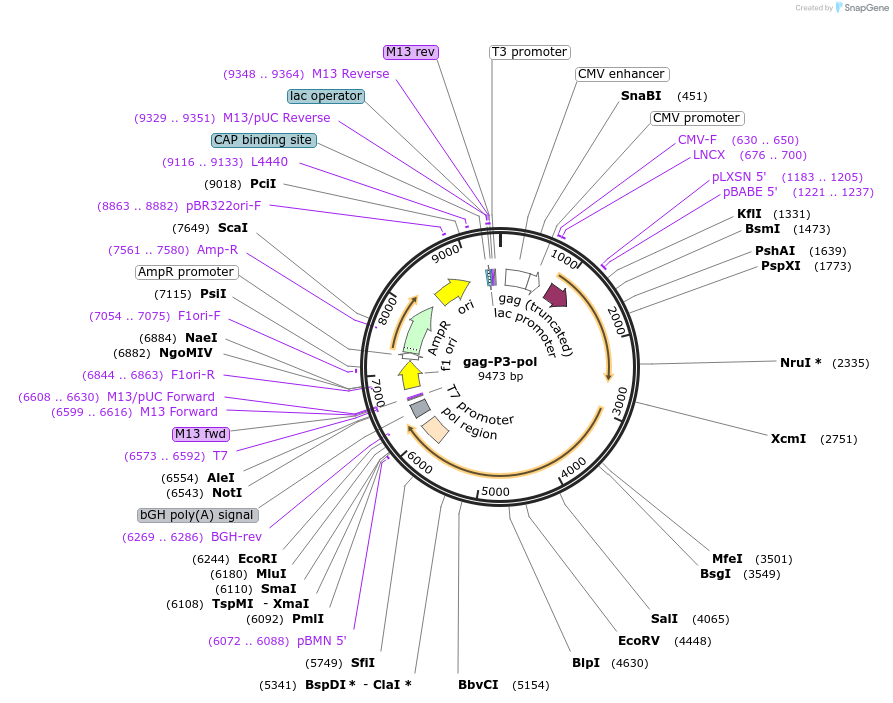
gag-P3-pol
Plasmid#211374PurposePackaging plasmid for v3b PE-eVLP productionDepositorInsertMMLVgag-P3-pol
ExpressionMammalianAvailable SinceJan. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
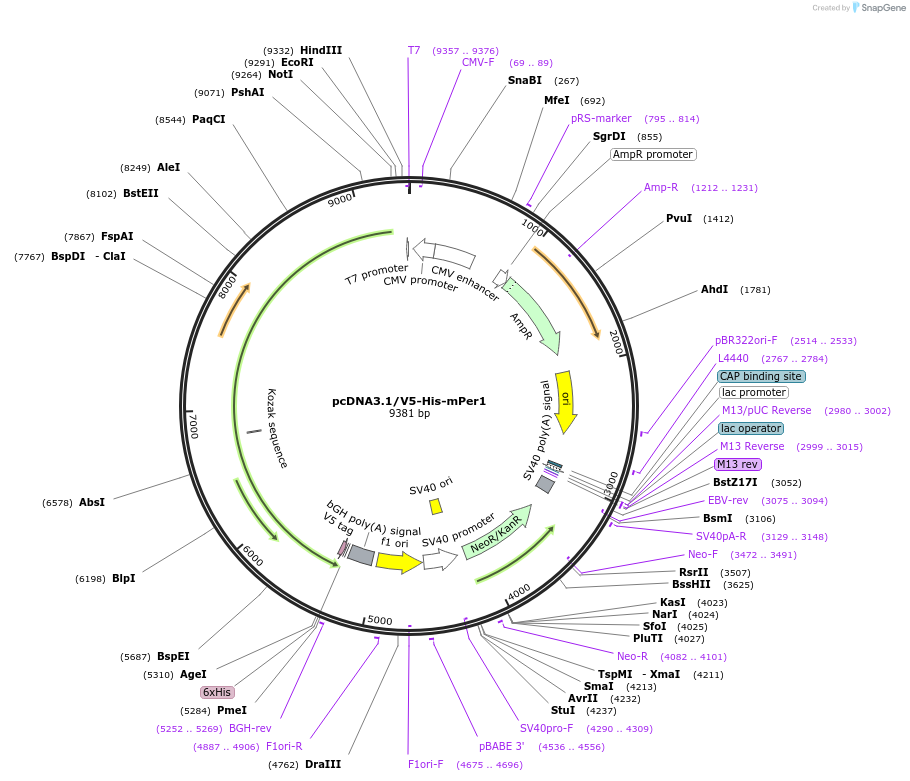
pcDNA3.1/V5-His-mPer1
Plasmid#219421PurposeExpresses 3873 bp of mPer1 coding sequence in E. coli.DepositorAvailable SinceDec. 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
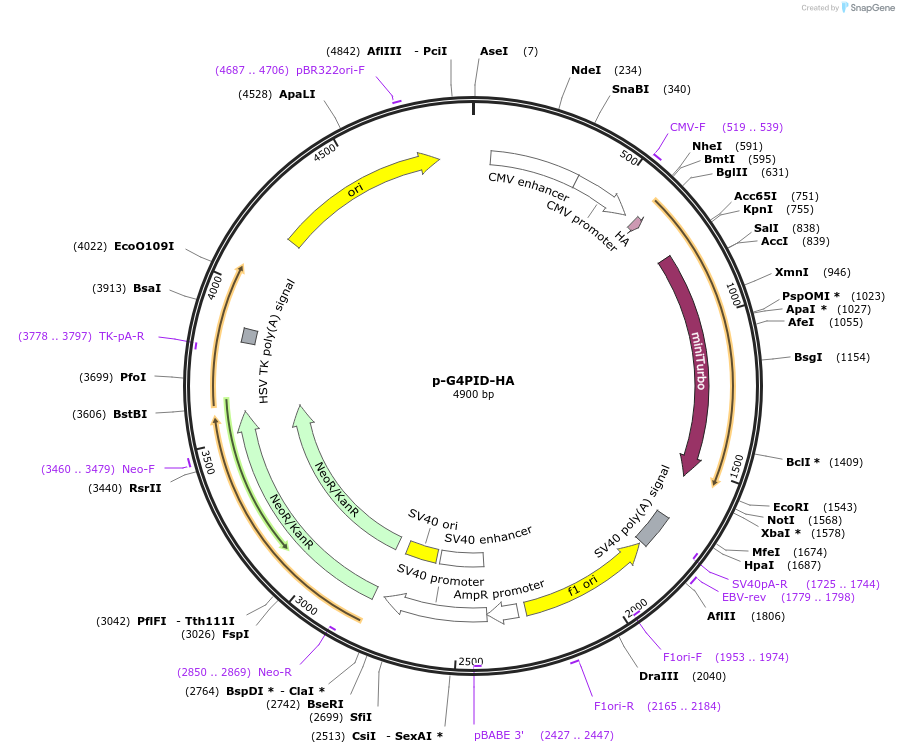
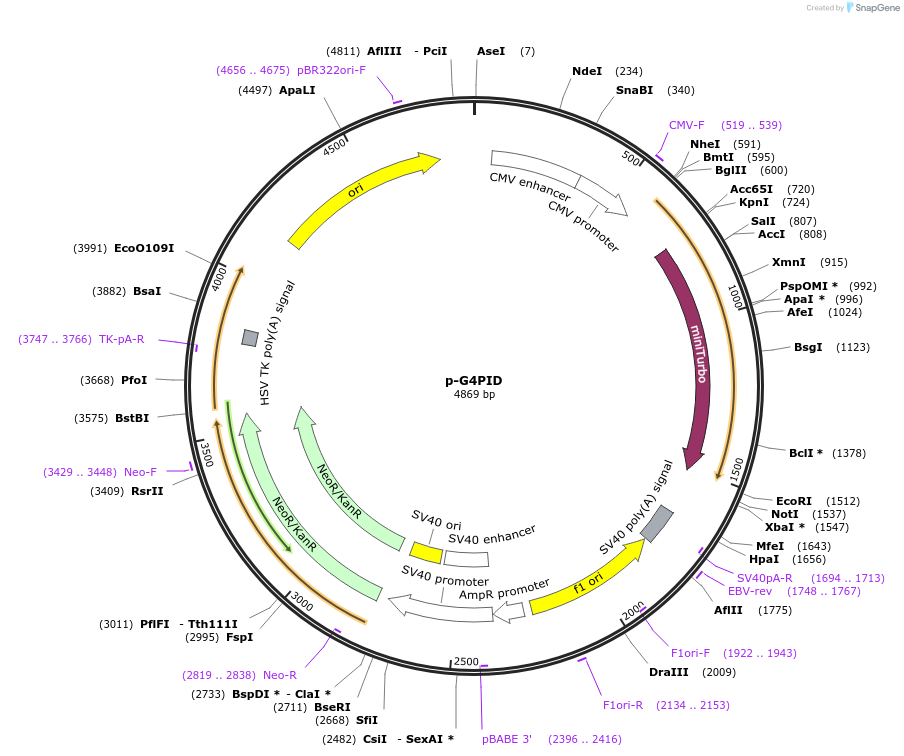
p-G4PID-HA
Plasmid#209639PurposeExpression G4PID-HA in mammalian cellsDepositorInsertG4PID (DHX36 Human)
ExpressionMammalianAvailable SinceApril 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
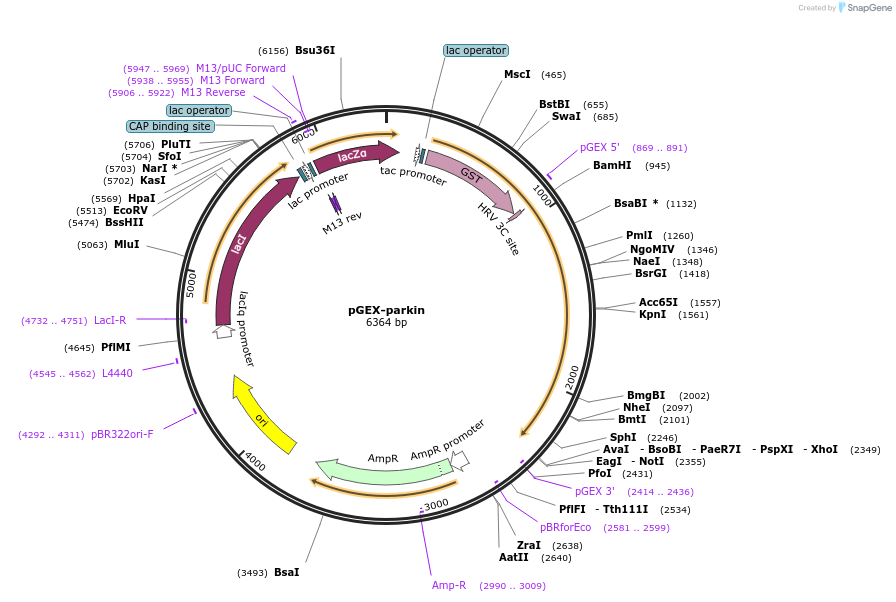
pGEX-parkin
Plasmid#227367PurposeFor bacterial expression of parkinDepositorInsertParkin (PRKN Human)
TagsGSTExpressionBacterialMutationCodon usage optimized for E.coli expression.Available SinceNov. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
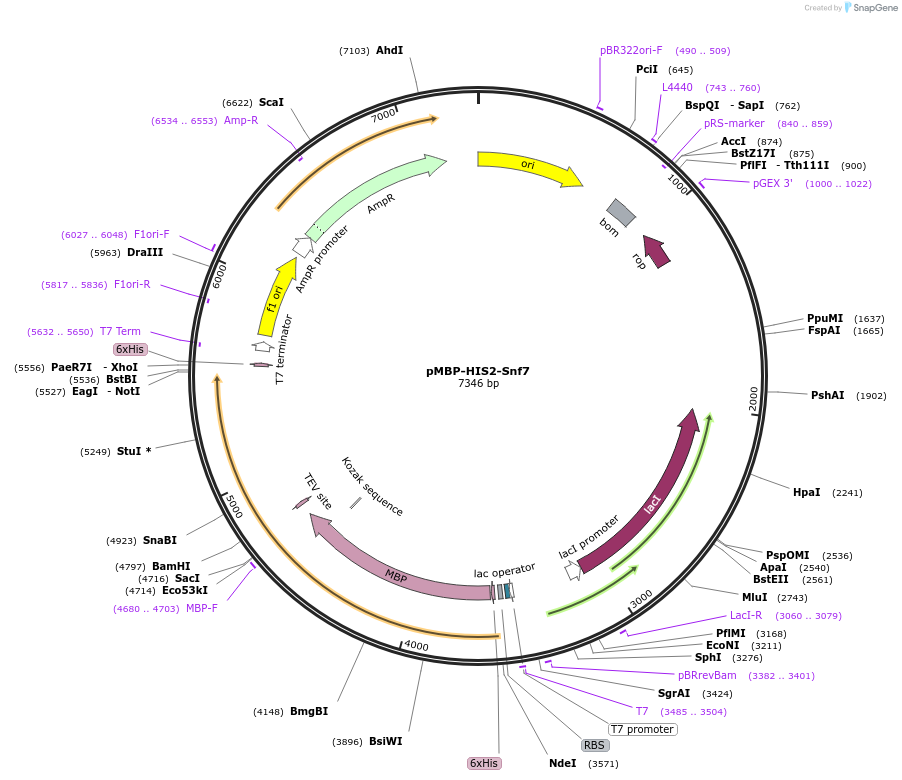
pMBP-HIS2-Snf7
Plasmid#21492DepositorAvailable SinceSept. 9, 2009AvailabilityAcademic Institutions and Nonprofits only -
pMBP-HIS2-Snf7
Plasmid#21492DepositorAvailable SinceSept. 9, 2009AvailabilityAcademic Institutions and Nonprofits only -
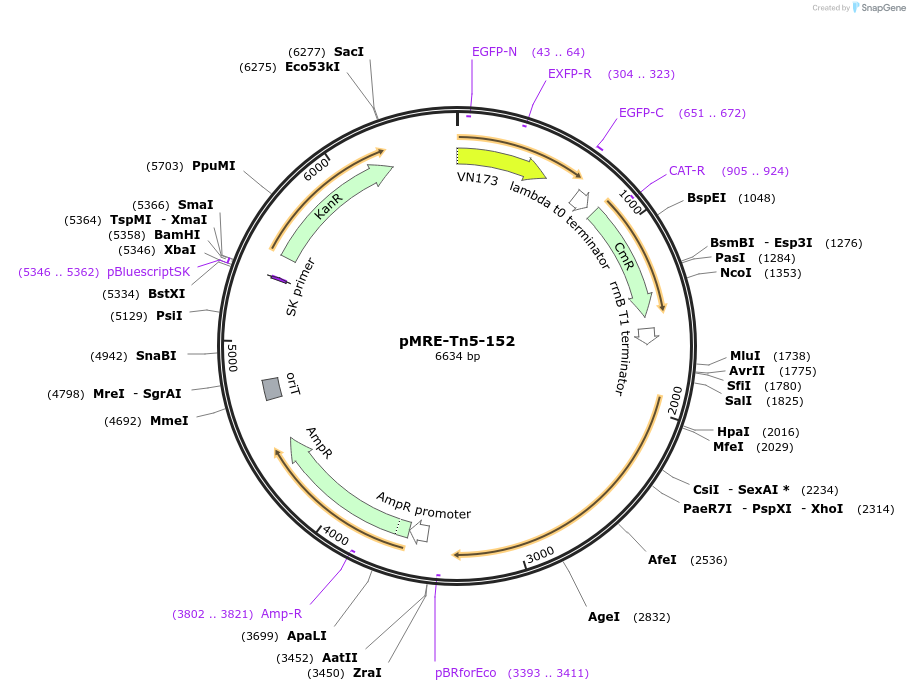
pMRE-Tn5-152
Plasmid#118534PurposeminiTn5 plasmid to deliver constitutively expressed fluorescent protein genes in bacteriaDepositorInsertsGFP2
UseMinitn5 delivery vectorExpressionBacterialAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
pMRE-Tn5-152
Plasmid#118534PurposeminiTn5 plasmid to deliver constitutively expressed fluorescent protein genes in bacteriaDepositorInsertsGFP2
UseMinitn5 delivery vectorExpressionBacterialAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
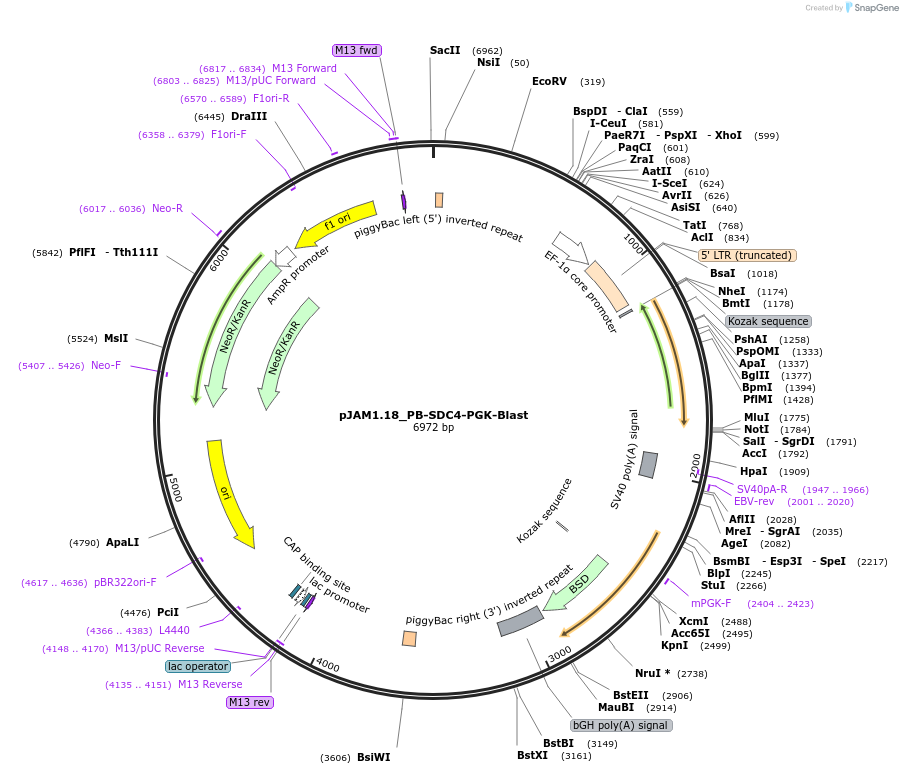
pJAM1.18_PB-SDC4-PGK-Blast
Plasmid#205563PurposeExpress human SDC4DepositorAvailable SinceSept. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
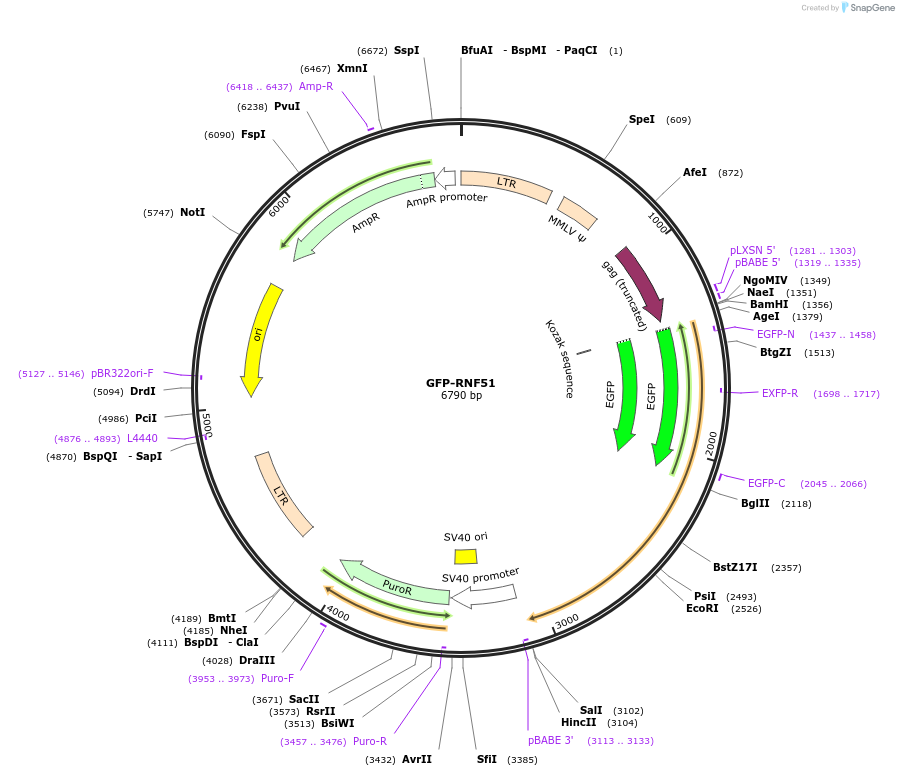
GFP-RNF51
Plasmid#126546PurposeExpresses GFP-tagged RNF51 in mammalian cellsDepositorAvailable SinceJuly 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
GFP-RNF51
Plasmid#126546PurposeExpresses GFP-tagged RNF51 in mammalian cellsDepositorAvailable SinceJuly 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
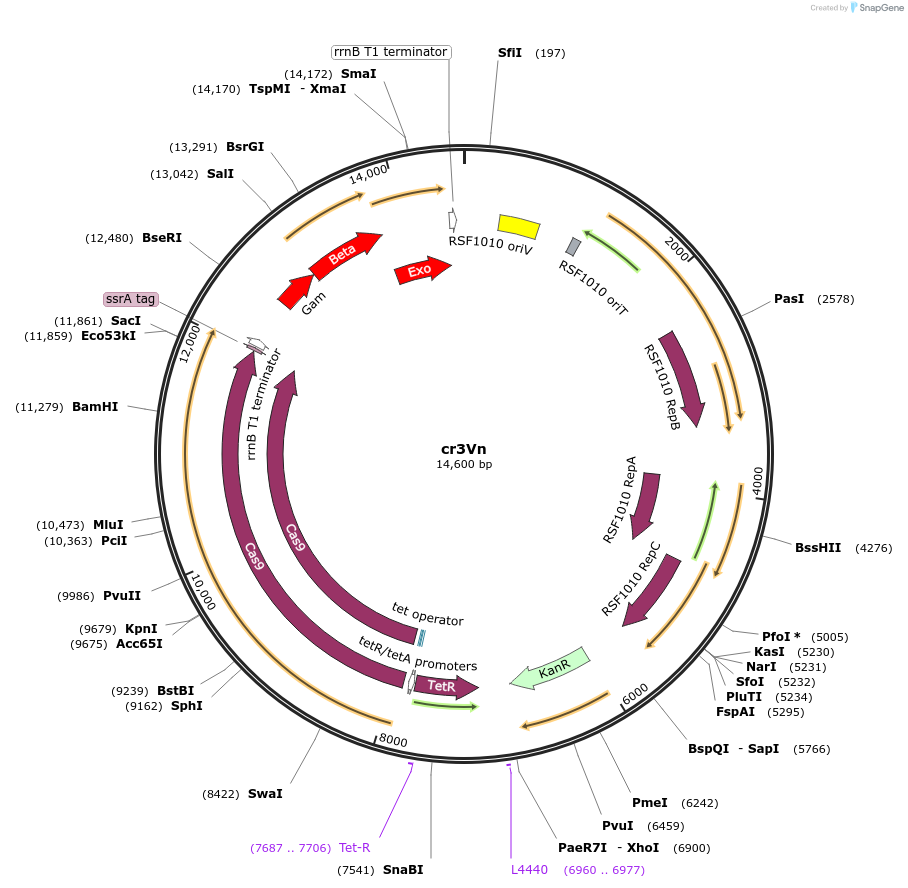
cr3Vn
Plasmid#184861Purposecr3 helper plasmid for recombination and Cas9 induction; harbors the Cas9 endonuclease and an V. natriegens-specific recombination systemDepositorInsertCas9, gam, bet, exo
TagsCas9 ssrA degradation tagExpressionBacterialAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
cr3Vn
Plasmid#184861Purposecr3 helper plasmid for recombination and Cas9 induction; harbors the Cas9 endonuclease and an V. natriegens-specific recombination systemDepositorInsertCas9, gam, bet, exo
TagsCas9 ssrA degradation tagExpressionBacterialAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
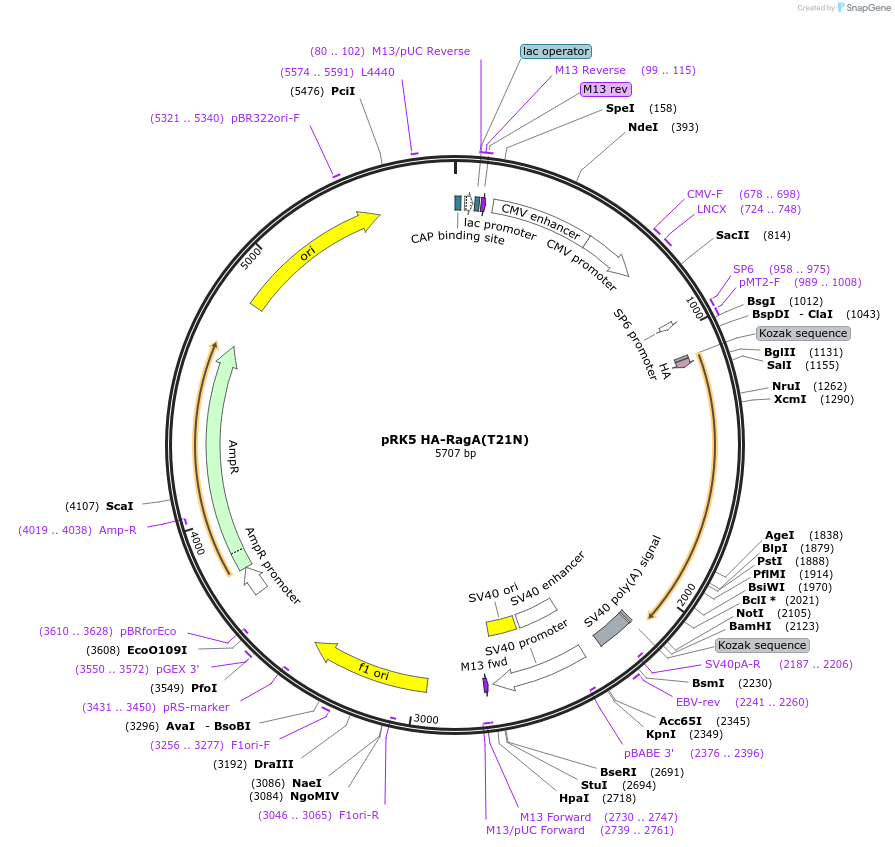
pRK5 HA-RagA(T21N)
Plasmid#99711PurposeMammalian expression of HA-RagA(T21N)DepositorAvailable SinceNov. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
pRK5 HA-RagA(T21N)
Plasmid#99711PurposeMammalian expression of HA-RagA(T21N)DepositorAvailable SinceNov. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
-
-
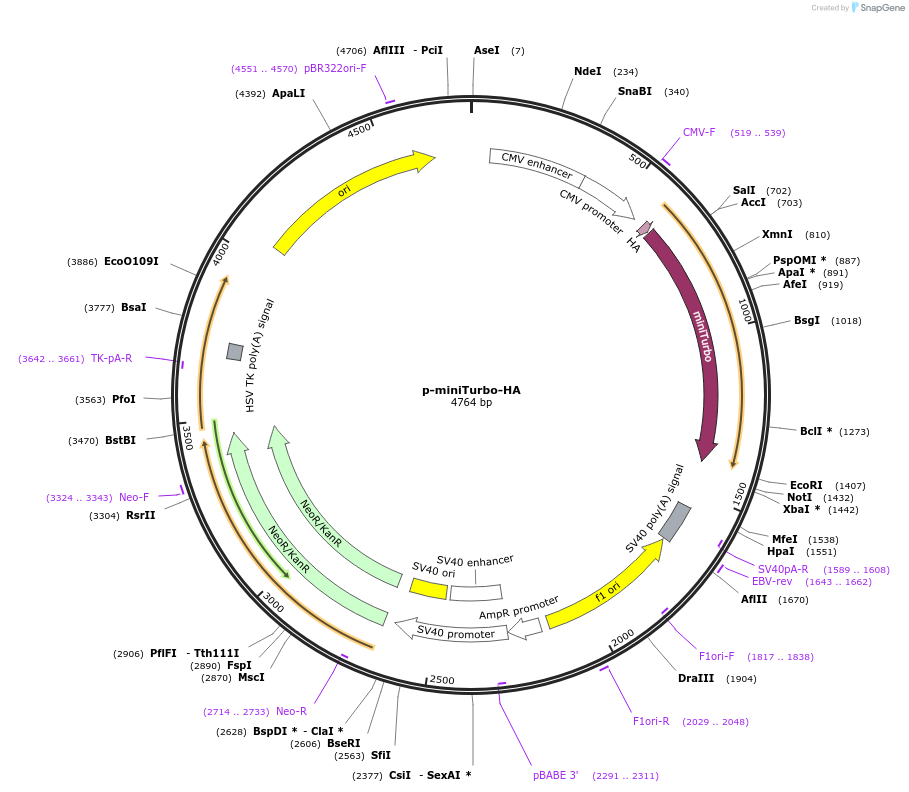
p-miniTurbo-HA
Plasmid#209637PurposeExpression miniTurbo-HA in mammalian cellsDepositorInsertminiTurbo
ExpressionMammalianAvailable SinceApril 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
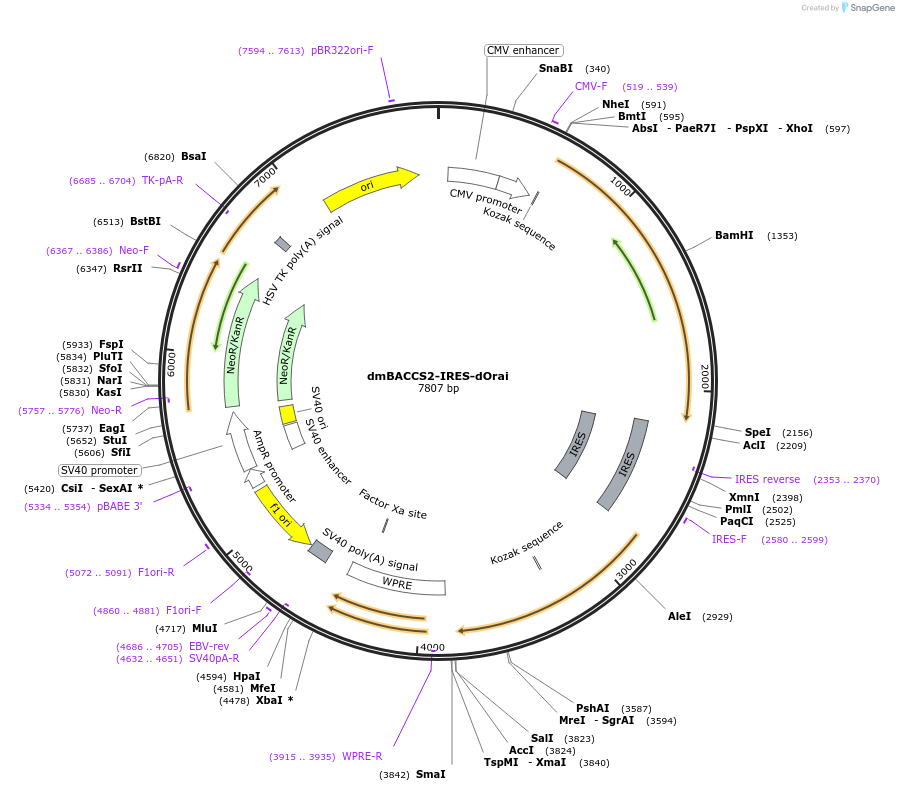
dmBACCS2-IRES-dOrai
Plasmid#67627PurposeMammalian expression of dmBACCS2, a Drosophila melanogaster blue light-activated Ca2+ channel switch, and dOraiDepositorInsertdmBACCS2-IRES-dOrai
ExpressionMammalianPromoterCMVAvailable SinceAug. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
dmBACCS2-IRES-dOrai
Plasmid#67627PurposeMammalian expression of dmBACCS2, a Drosophila melanogaster blue light-activated Ca2+ channel switch, and dOraiDepositorInsertdmBACCS2-IRES-dOrai
ExpressionMammalianPromoterCMVAvailable SinceAug. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
pLKO mouse shRNA 2 raptor
Plasmid#21340DepositorAvailable SinceJuly 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
pLKO mouse shRNA 2 raptor
Plasmid#21340DepositorAvailable SinceJuly 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
-
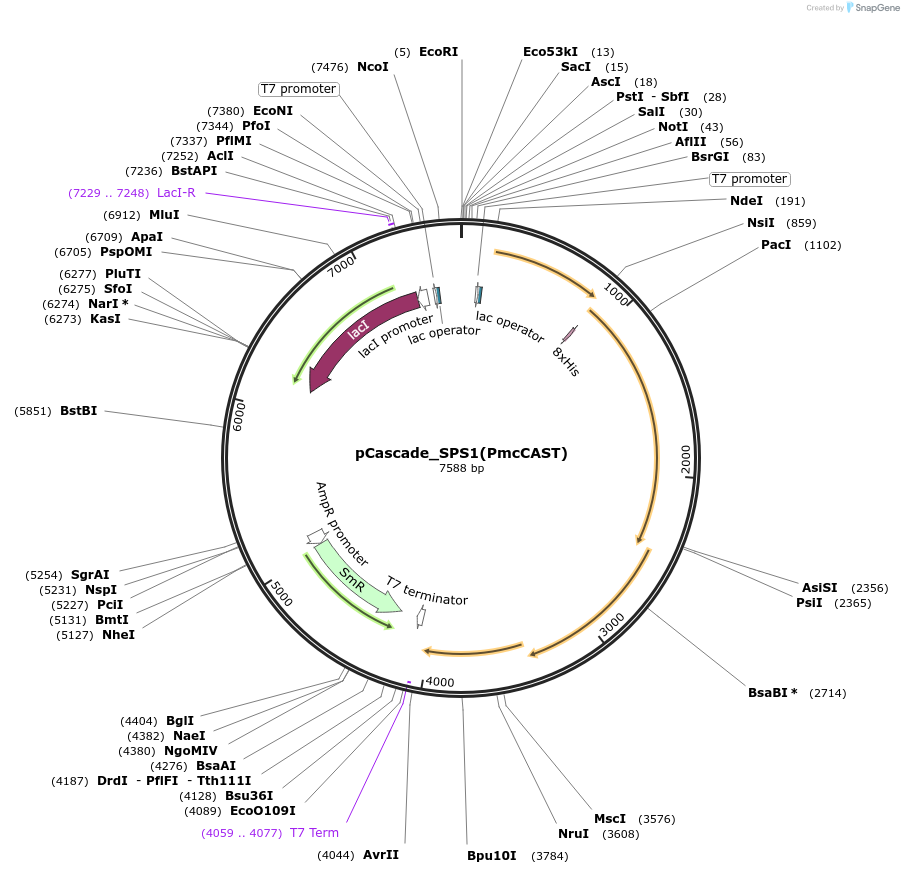
pCascade_SPS1(PmcCAST)
Plasmid#224926PurposeExpression and purification of type I-B2 Cascade in CRISPR-associated transposon of RomsettaDepositorInsertsCas5, Cas6, Cas7, Cas8, Cas11
CRISPR assay for crRNA expression
UseCRISPRExpressionBacterialAvailable SinceApril 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
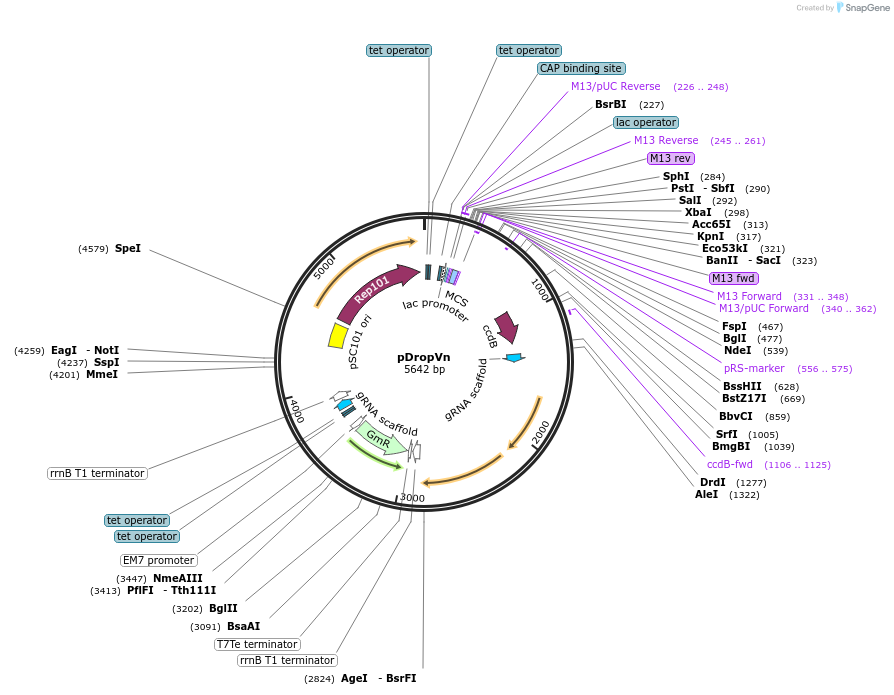
pDropVn
Plasmid#184873PurposeChromosomal transfer helper plasmid with sgRNAs (one fixed, one flexible) for Vibrio natriegens genome editing protocol.DepositorInsertlacZalpha,ccdB
ExpressionBacterialAvailable SinceJan. 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits