We narrowed to 51,543 results for: des.1
-
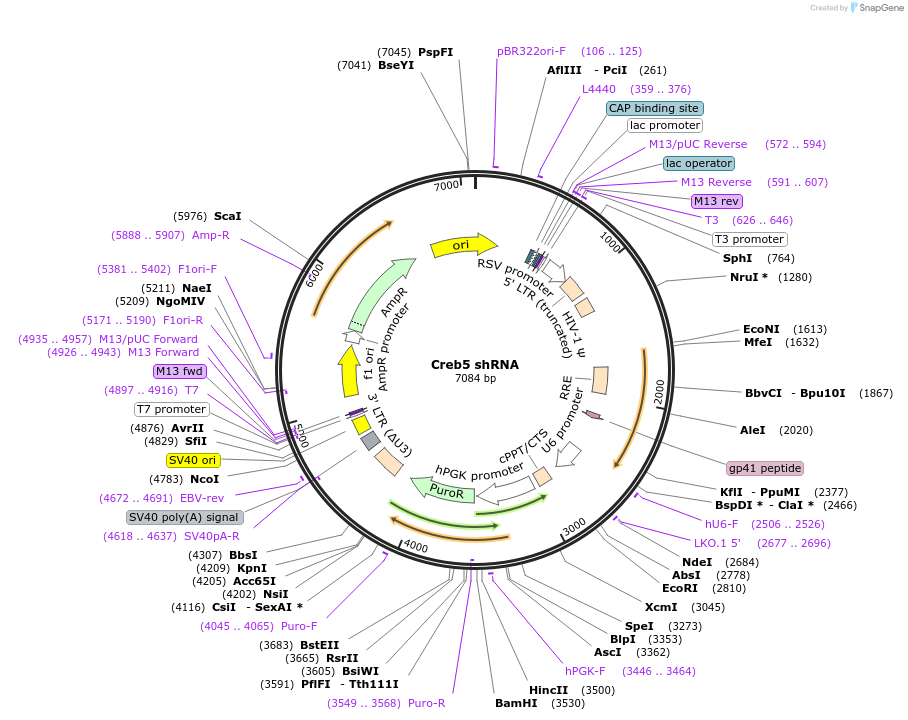
Plasmid#195020PurposeBovine Creb5 shRNA targeting the Creb5 3′UTRDepositorInsertCreb5 shRNA (CREB5 Bovine)
ExpressionMammalianAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
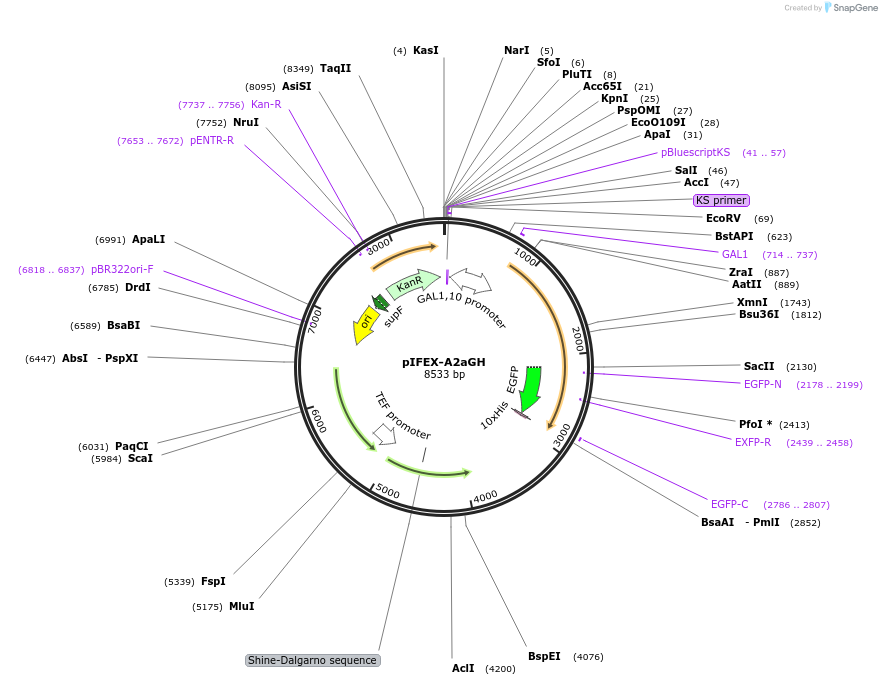
pIFEX-A2aGH
Plasmid#160542PurposepITY4 derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
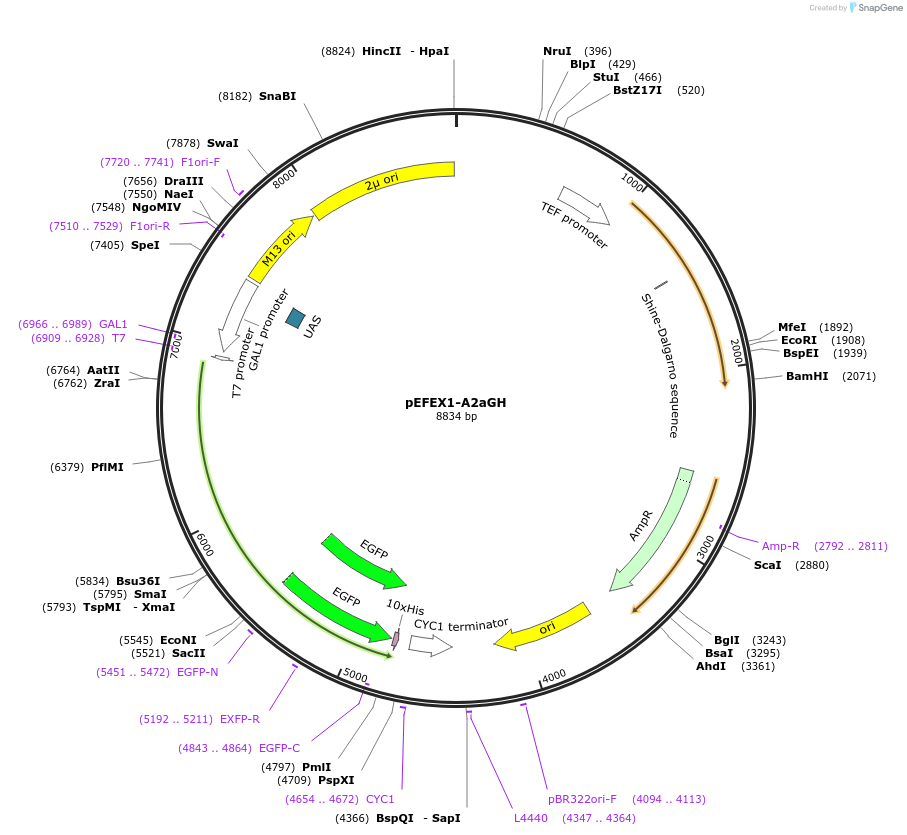
pEFEX1-A2aGH
Plasmid#160543PurposepYES2 derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
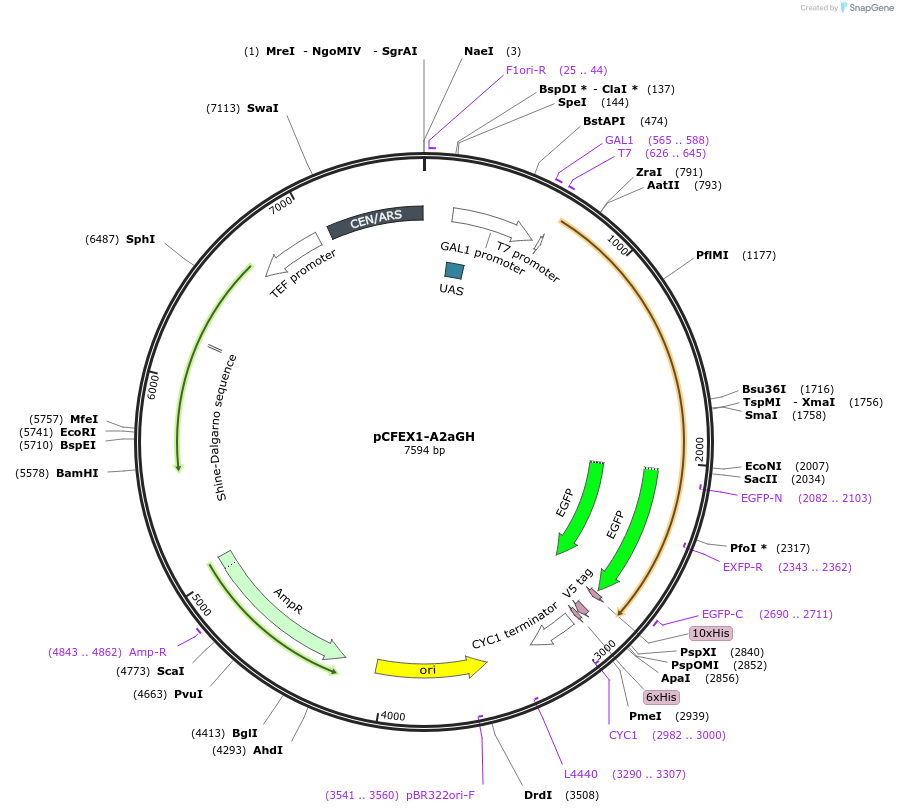
pCFEX1-A2aGH
Plasmid#160546PurposepYC2/CT derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
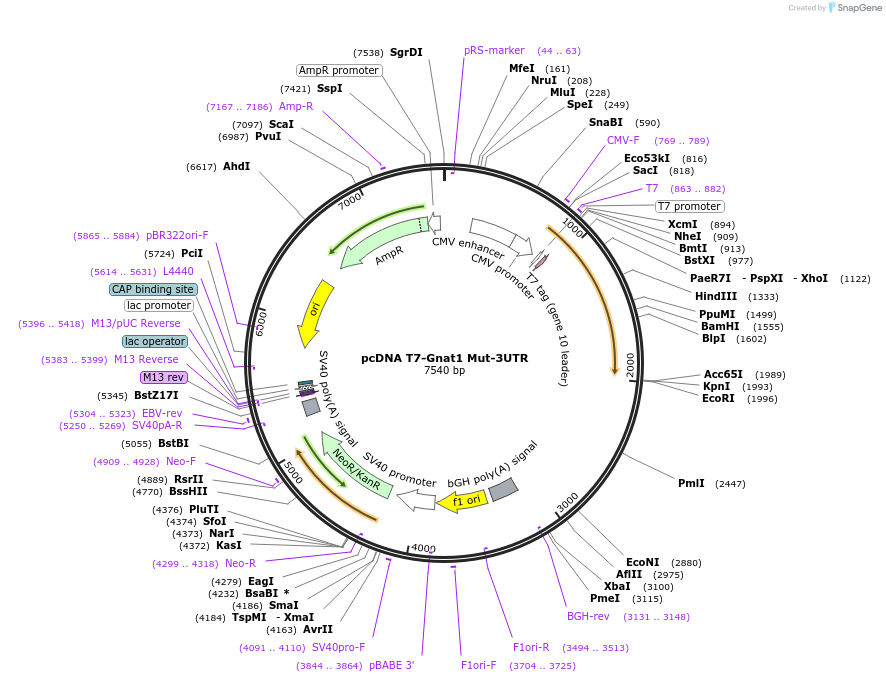
pcDNA T7-Gnat1 Mut-3UTR
Plasmid#184027PurposeExpresses mouse Gnat1 protein with N-terminal T7 tag from cDNA that contains the mutant 3'-UTR where the TAG binding sites for MSI1 are mutated to TGADepositorInsertGnat1 (Gnat1 Mouse)
TagsT7ExpressionMammalianMutationTAG sites in the 3'-UTR are mutated to TGAPromoterCMVAvailable SinceMay 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
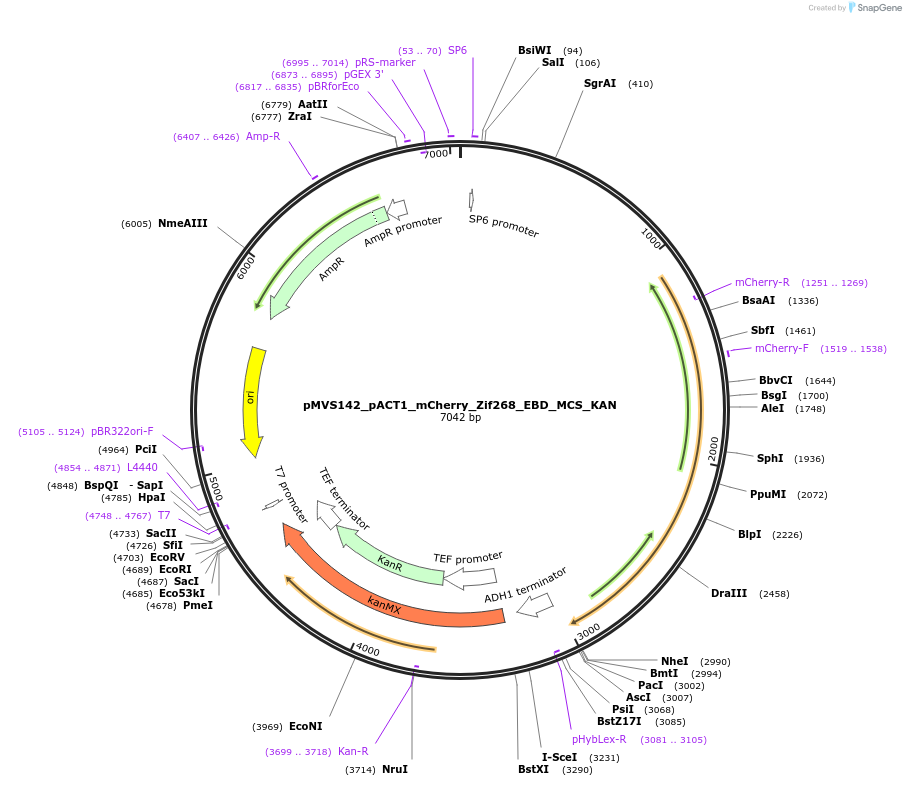
pMVS142_pACT1_mCherry_Zif268_EBD_MCS_KAN
Plasmid#99049PurposeTranscription Factor chassis for activation domains. mCherry, Zif268 DNA binding domain, estrogen response domainDepositorInsertsExpressionYeastAvailable SinceMarch 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
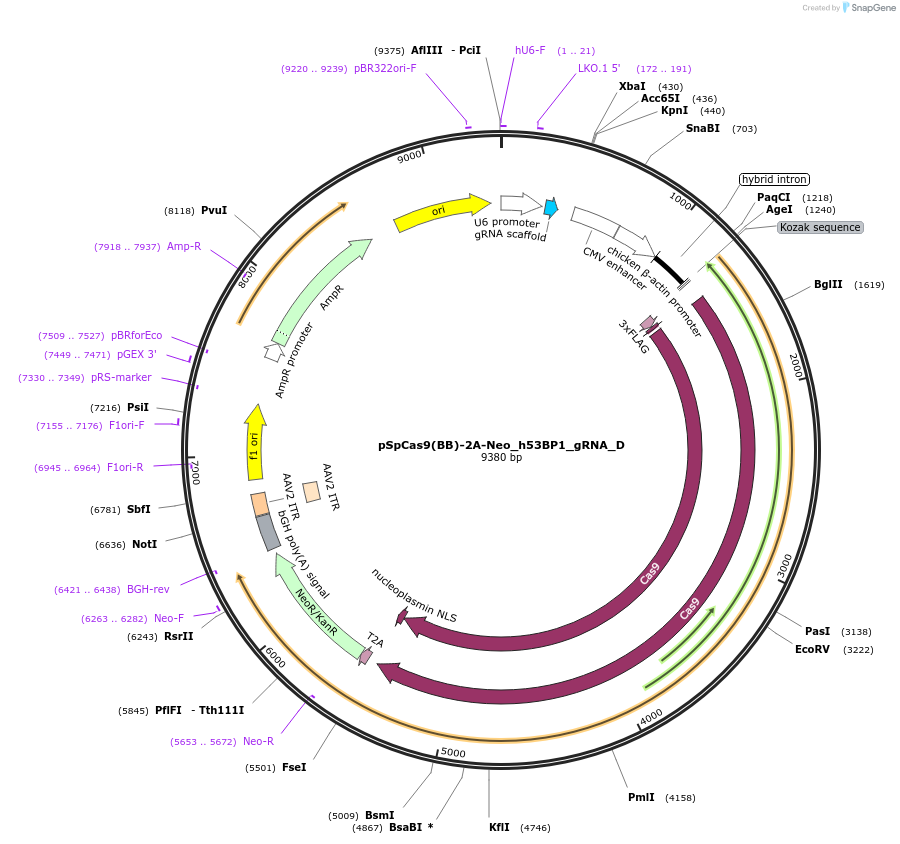
pSpCas9(BB)-2A-Neo_h53BP1_gRNA_D
Plasmid#110300PurposeExpresssion of Cas9-T2A-neomycin resistant gene and a gRNA targeting exon 10 of human 53BP1DepositorInsertp53-binding protein 1 (TP53BP1 Human)
ExpressionMammalianAvailable SinceFeb. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
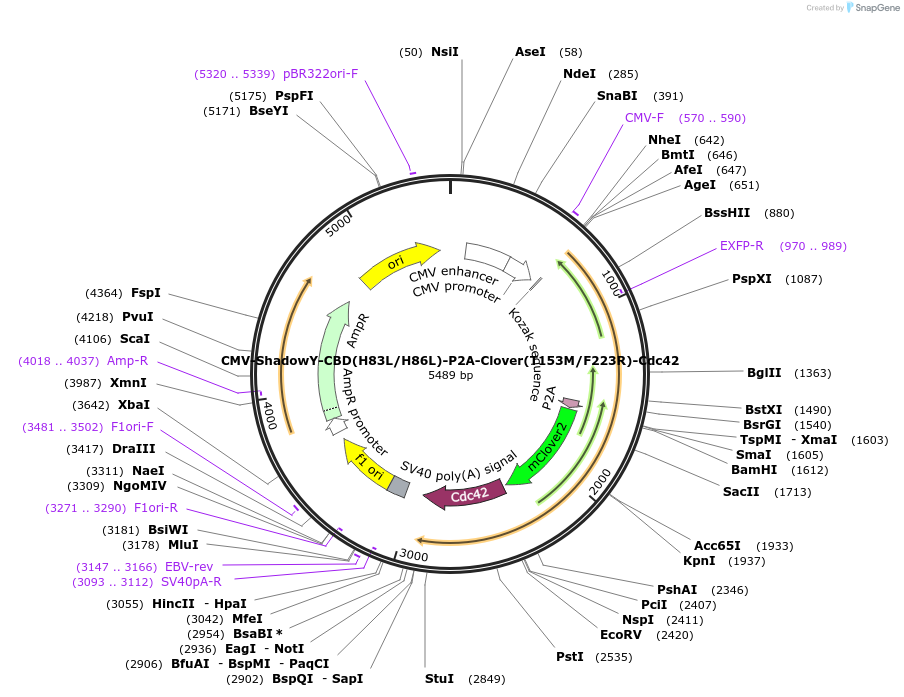
CMV-ShadowY-CBD(H83L/H86L)-P2A-Clover(T153M/F223R)-Cdc42
Plasmid#165436PurposeExpresses ShadowY-CBD(H83L/H86L) and Clover(T153M/F223R)-Cdc42 in mammalian cells as a negative control for FLIM imaging.DepositorInsertShadowY-CBD(H83L/H86L)-P2A-Clover(T153M/F223R)-Cdc42
TagsShadowY, Clover(T153M/F223R)ExpressionMammalianMutationH83L/H86L: CDB(Cdc42 Binding Domain, Pak3, residu…PromoterCMVAvailable SinceMarch 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
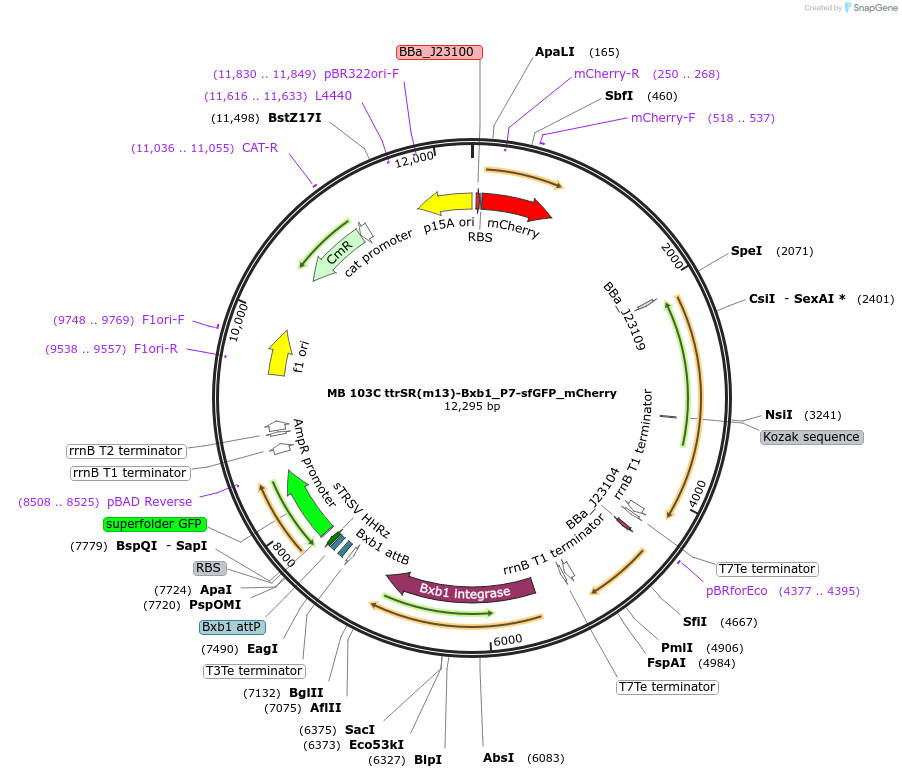
MB 103C ttrSR(m13)-Bxb1_P7-sfGFP_mCherry
Plasmid#232475PurposeOptimized tetrathionate sensor with recombinase switch and fluorescent reporter (GFP), constitutive mCherryDepositorInsertsttrS
ttrR
PttrB185-269
sfGFP
AxeTxe
mCherry
Bxb1 integrase
TagsssrA degradation tagExpressionBacterialPromoterConstitutive (TTGATAGCTAGCTCAGTCCTAGGTATTGTGCTAGC…Available SinceNov. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
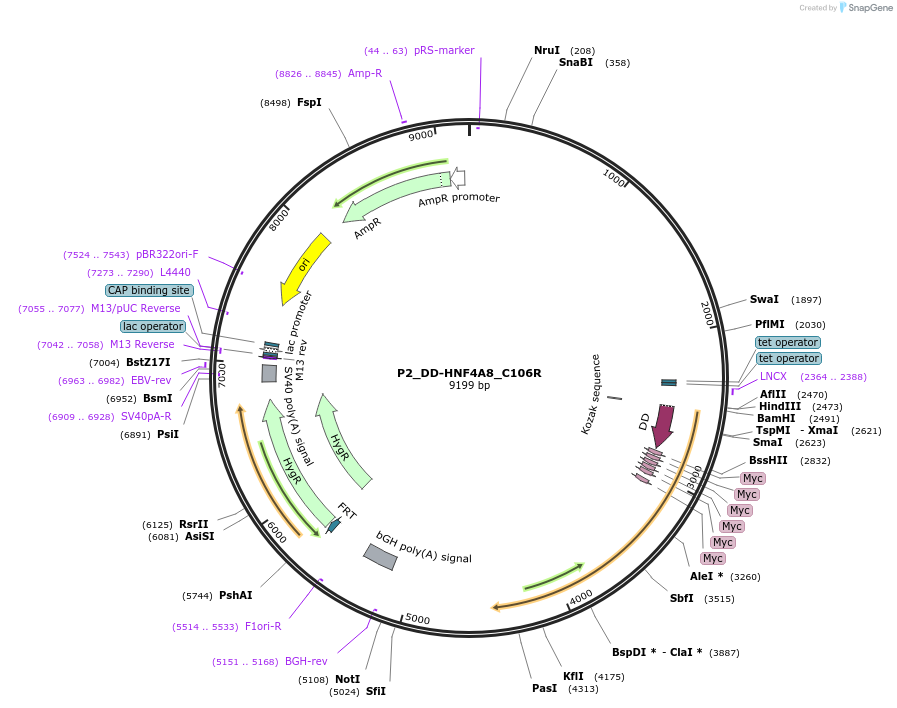
P2_DD-HNF4A8_C106R
Plasmid#31096DepositorInsertHNF4A (HNF4A Human)
UseFlp/frtTagsFKBP L106P and mycExpressionMammalianMutationsplice variant 8 derived from promoter P2 with zi…Available SinceOct. 14, 2011AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 B
Plasmid#11325DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation B from figure 4A of paper.Available SinceJuly 18, 2006AvailabilityAcademic Institutions and Nonprofits only -
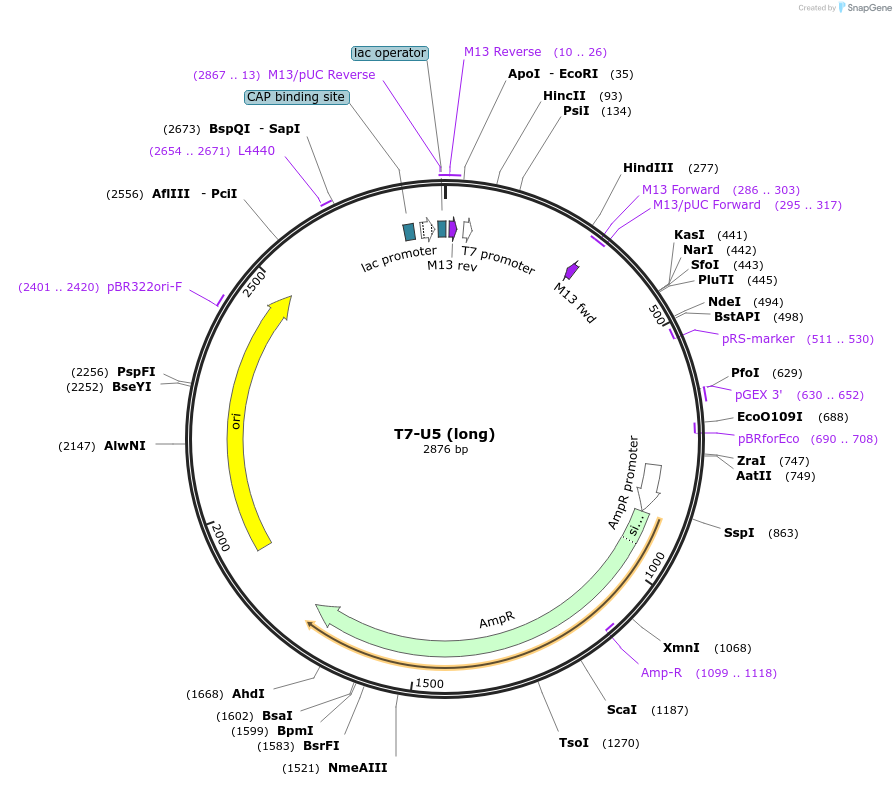
T7-U5 (long)
Plasmid#111337Purposefor in vitro snRNA transcriptionDepositorInsertsnR7-L
ExpressionBacterialAvailable SinceAug. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 D
Plasmid#11327DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation D from figure 4A of paper.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 C
Plasmid#11326DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation C from figure 4A of paper.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 I
Plasmid#11332DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation I from figure 4A of paper.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 E
Plasmid#11328DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation E from figure 4A of paper.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 F
Plasmid#11329DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation F from figure 4A of paper.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 G
Plasmid#11330DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation G from figure 4A of paper.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK let7 H
Plasmid#11331DepositorInsert2 binding sites for let-7a miRNA
UseLuciferaseTagsRr-lucMutationMutation H from figure 4A of paper.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
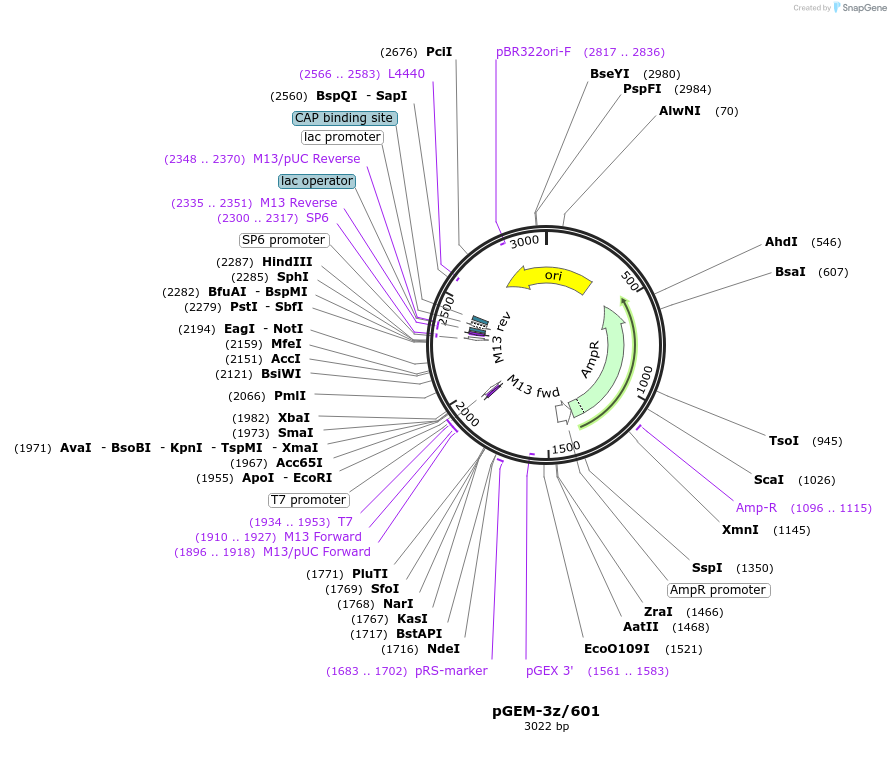
pGEM-3z/601
Plasmid#26656DepositorInsert
ExpressionBacterialAvailable SinceNov. 2, 2010AvailabilityAcademic Institutions and Nonprofits only