We narrowed to 4,223 results for: phage
-
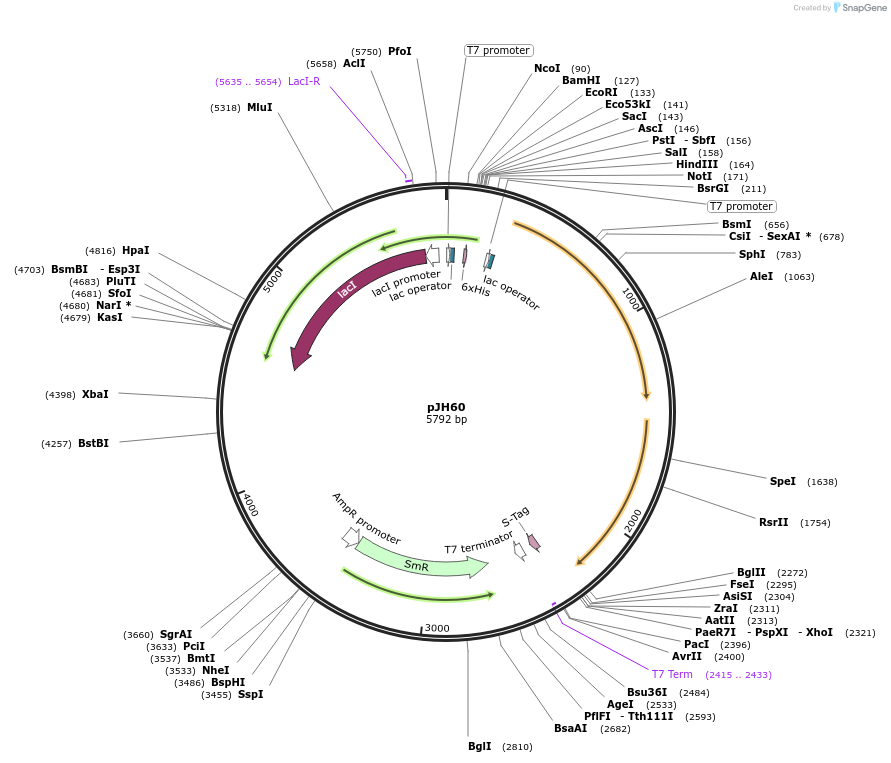
Plasmid#172142PurposeMicroscopy inclusion body assayDepositorInsertpCDF.CD
ExpressionBacterialMutationPylCD partial operon (PylB deleted to assess impa…Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
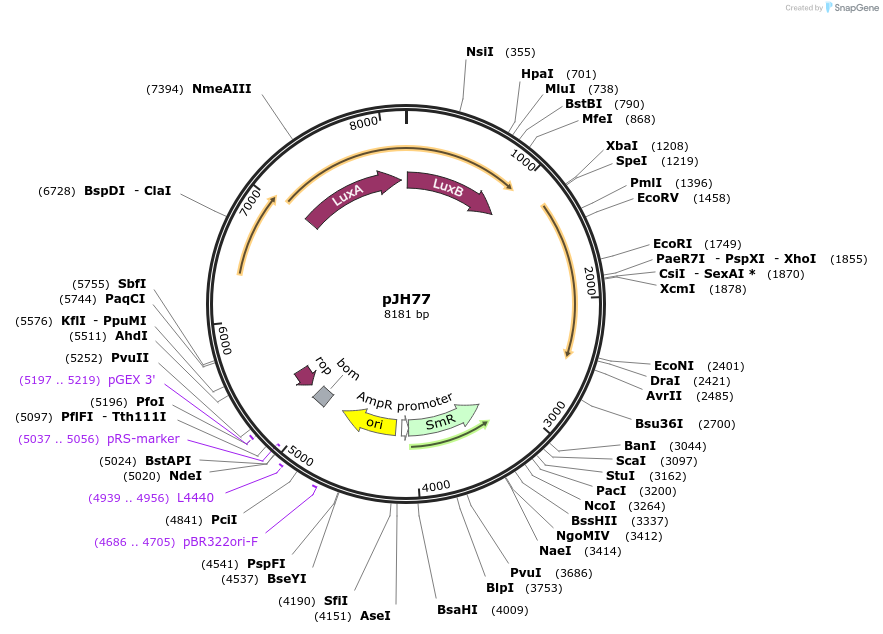
pJH77
Plasmid#172145PurposeAccessory Plasmid for PACE of pylBCD variantsDepositorInsertPpsp gIII.3TAG, luxAB; Plpp pylS; PproK pylT
ExpressionBacterialMutationgIII(P29*, P83*, Y184*)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
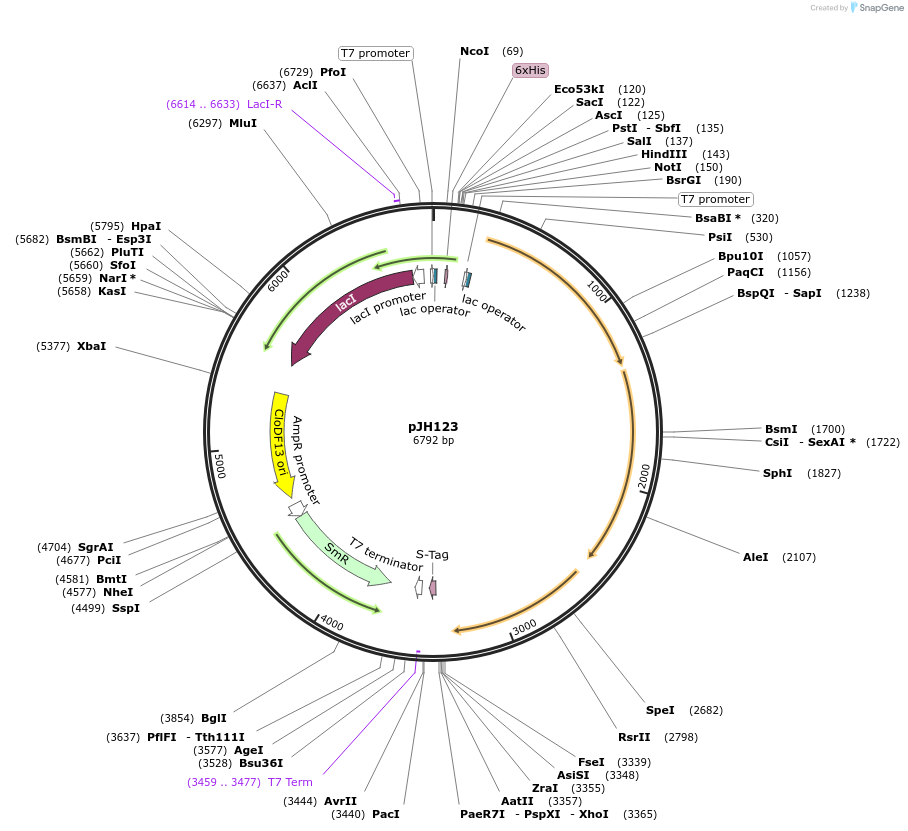
pJH123
Plasmid#172146PurposeMicroscopy inclusion body assayDepositorInsertpCDF.B.(SD4)CD
ExpressionBacterialMutationPylBCD operon (RBS inserted for PylC)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
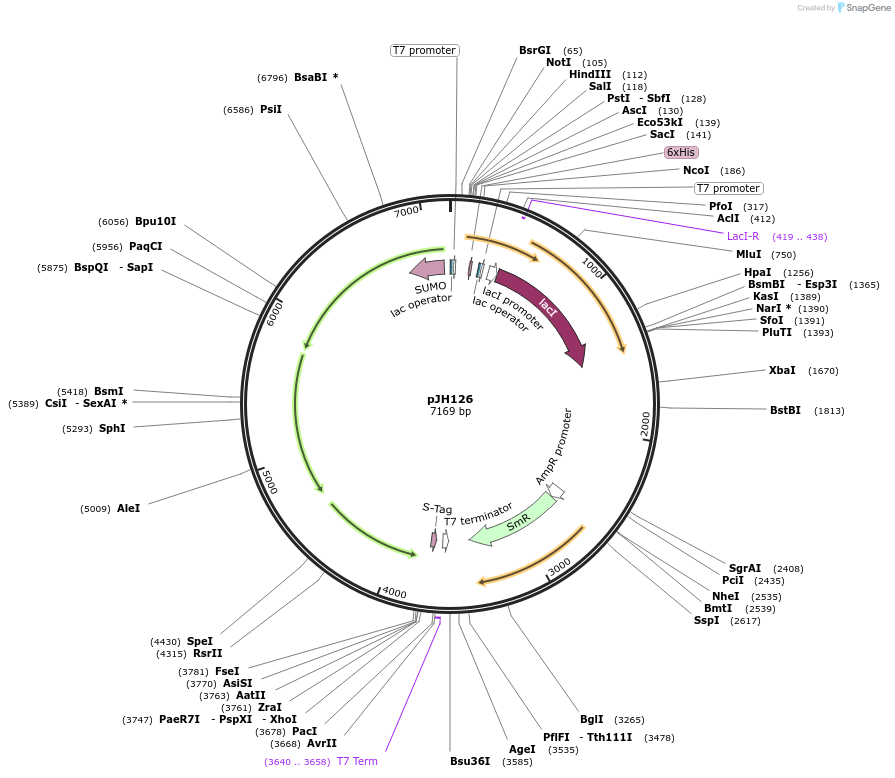
pJH126
Plasmid#172147PurposeMicroscopy inclusion body assayDepositorInsertpCDF.SUMO-B.(SD4).CD
ExpressionBacterialMutationPylBCD operon (SUMO tag inserted for PylB; RBS in…Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
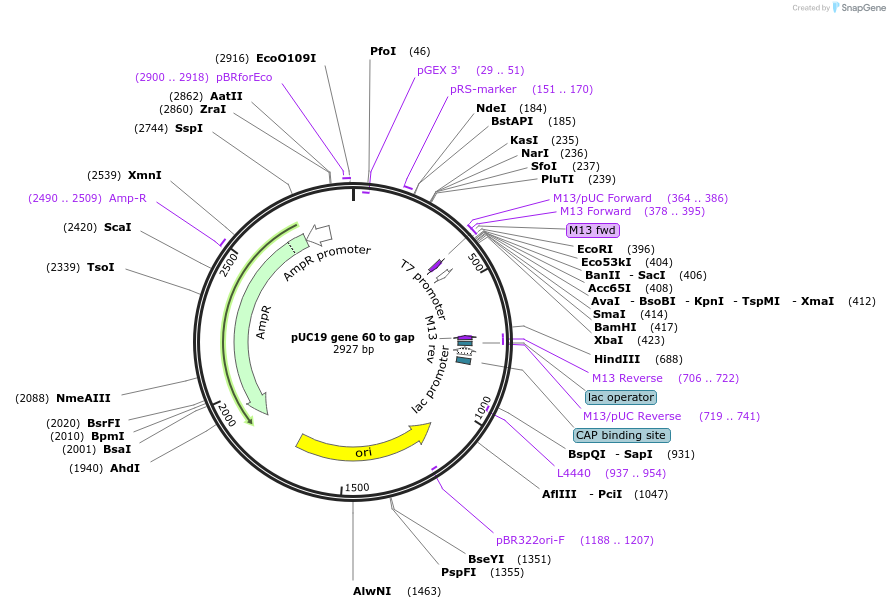
pUC19 gene 60 to gap
Plasmid#49158PurposeTemplate for generating gene 60 mRNA, sequence 5' of coding gap onlyDepositorInsertBacteriophage T4 gene 60
MutationCoding gap and downstream sequence have been del…PromoterT7Available SinceJan. 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
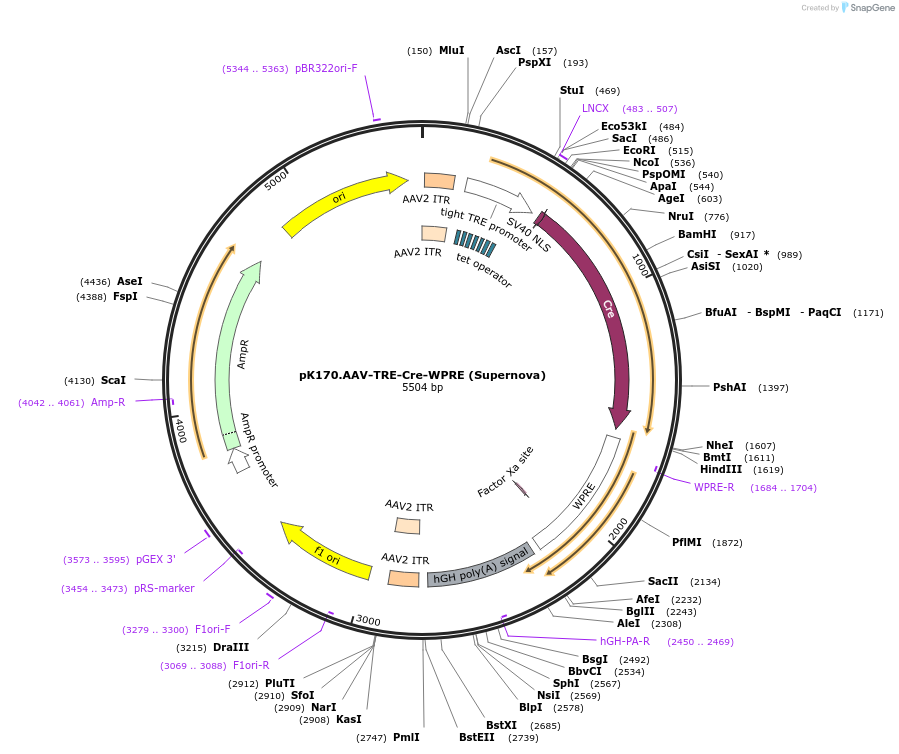
pK170.AAV-TRE-Cre-WPRE (Supernova)
Plasmid#85040PurposeFor AAV-TRE-Cre based Supernova, pK170 should be used with pK168 etc.DepositorInsertnlsCre-WPRE
UseAAVExpressionMammalianAvailable SinceNov. 30, 2016AvailabilityAcademic Institutions and Nonprofits only -
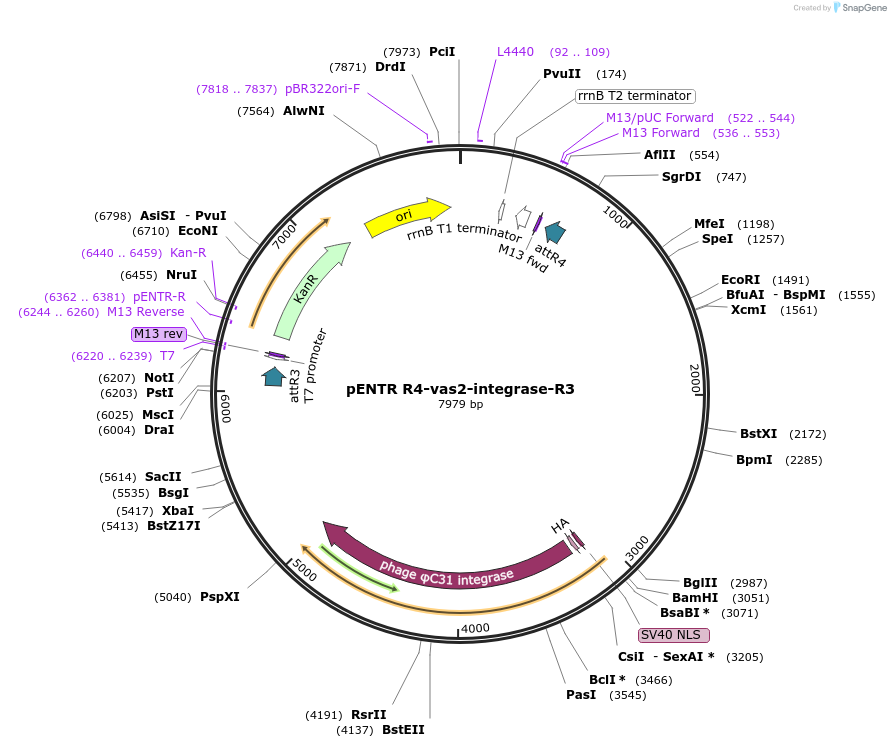
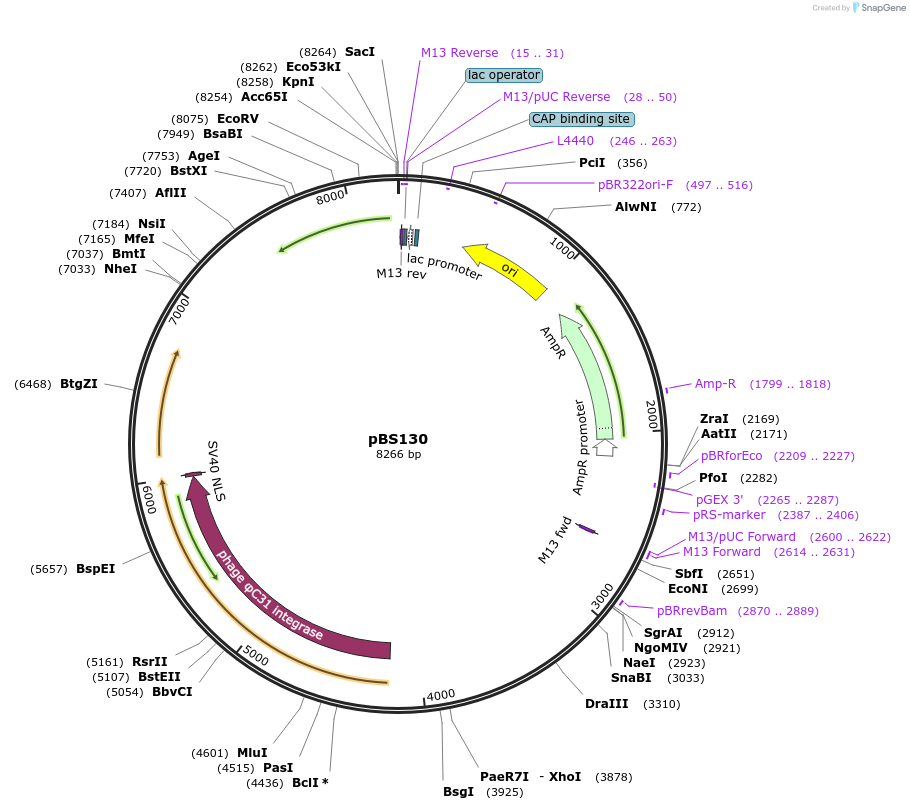
pENTR R4-vas2-integrase-R3
Plasmid#62299PurposephageC31 integrase-expressing helper plasmid for Anopheles transgenesis, vasa promoterDepositorInsertphageC31 integrase under control of vasa promoter and terminator
TagsHA and SV40 NLSExpressionInsectPromotervasaAvailable SinceMay 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
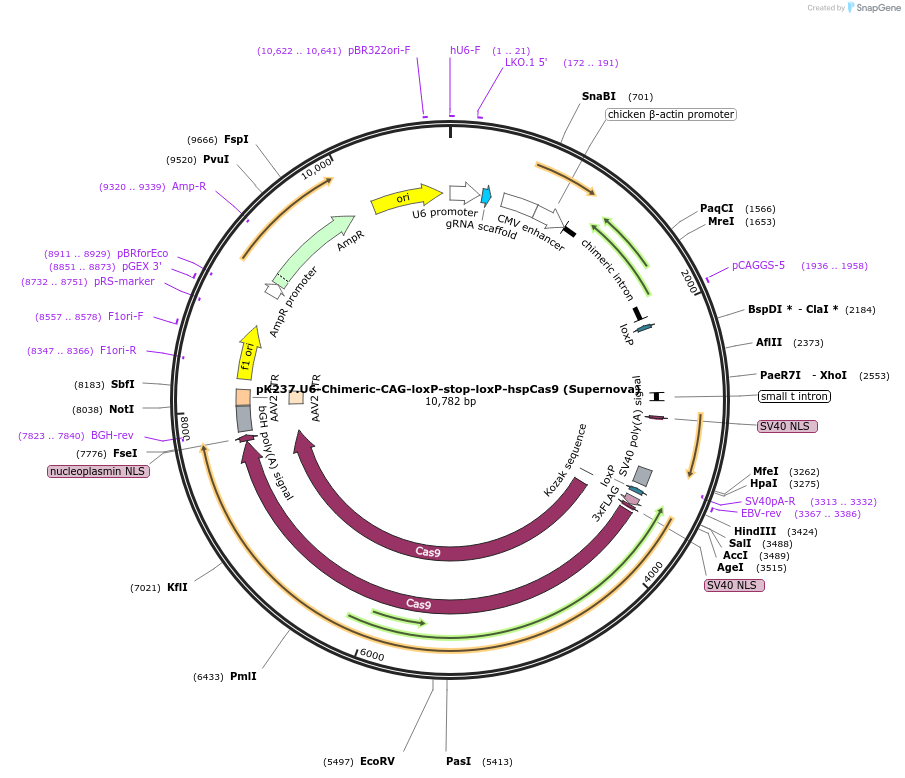
pK237.U6-Chimeric-CAG-loxP-stop-loxP-hspCas9 (Supernova)
Plasmid#85041PurposeThis plasmid should be used for Cre/loxP-based Supernova-CRISPR/Cas9.DepositorInsertloxP-stop-loxP
UseCRISPRExpressionMammalianAvailable SinceNov. 30, 2016AvailabilityAcademic Institutions and Nonprofits only -
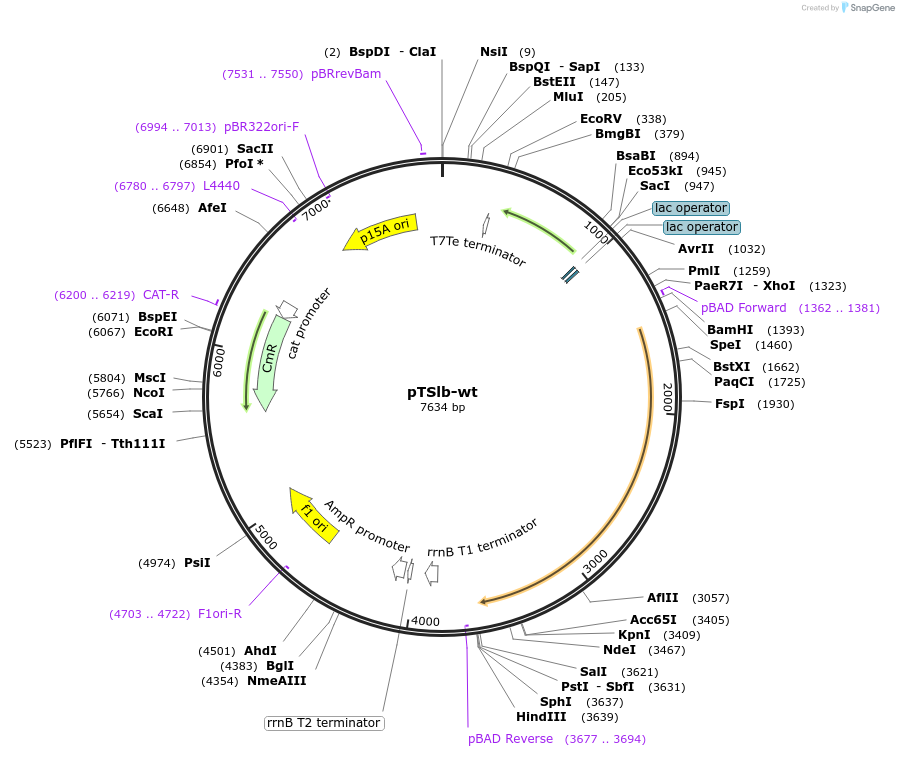
pTSlb-wt
Plasmid#60728PurposeExpresses both fragments of T7 RNAP split at position 179. The N-terminal fragment is driven by PLac while the C-terminal fragment is driven by PBAD.DepositorInsertsResidues 1-179 of split T7 RNAP
Residues 180-880 of split T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationResidues 1-180 of split T7 RNAP and Residues 180-…Available SinceNov. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
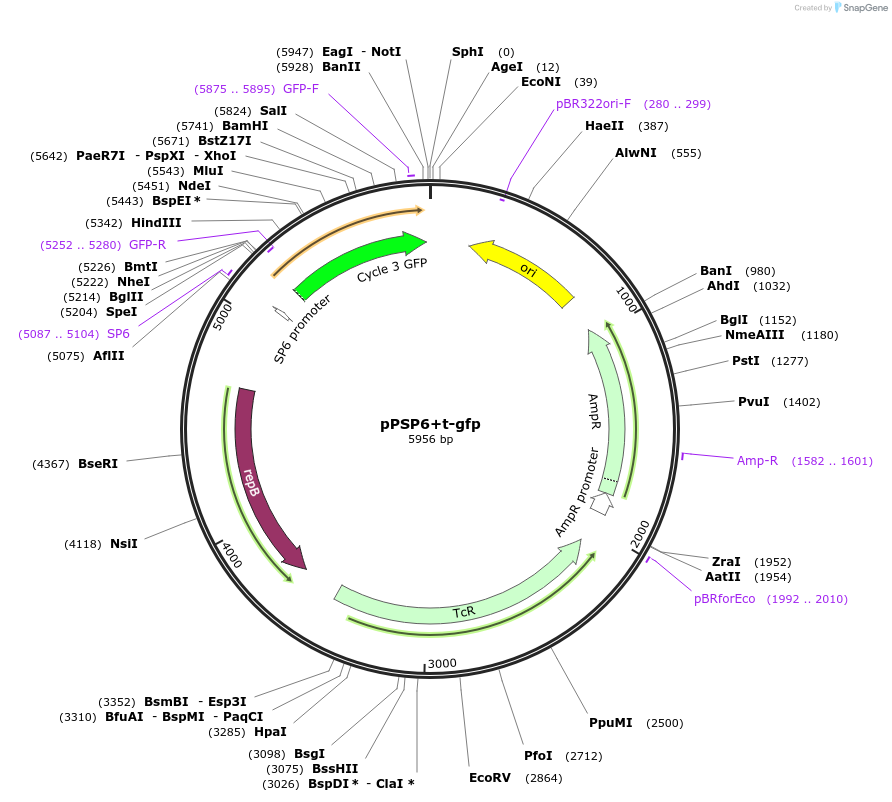
pPSP6+t-gfp
Plasmid#48130PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-SP6-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
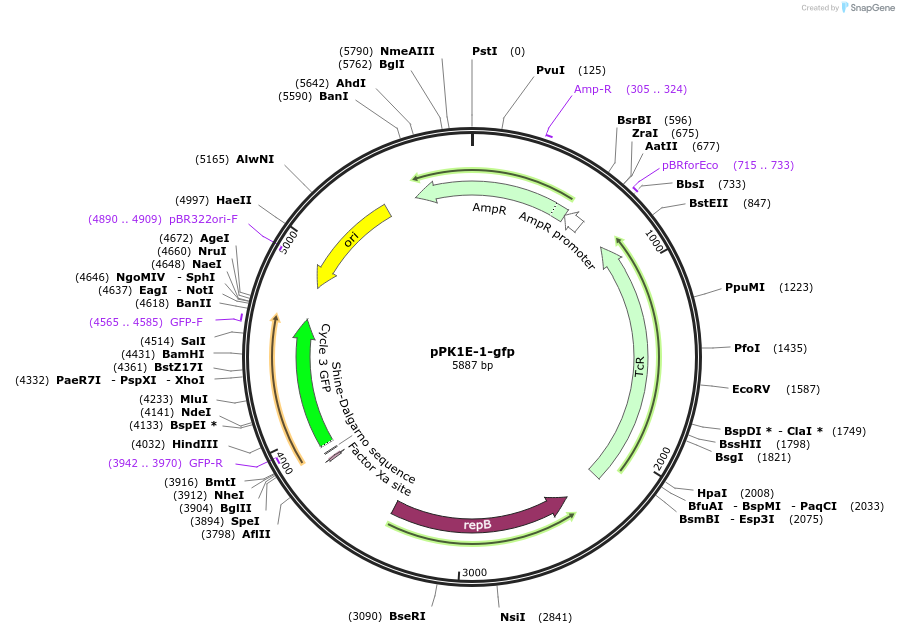
pPK1E-1-gfp
Plasmid#48133PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-K1E-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
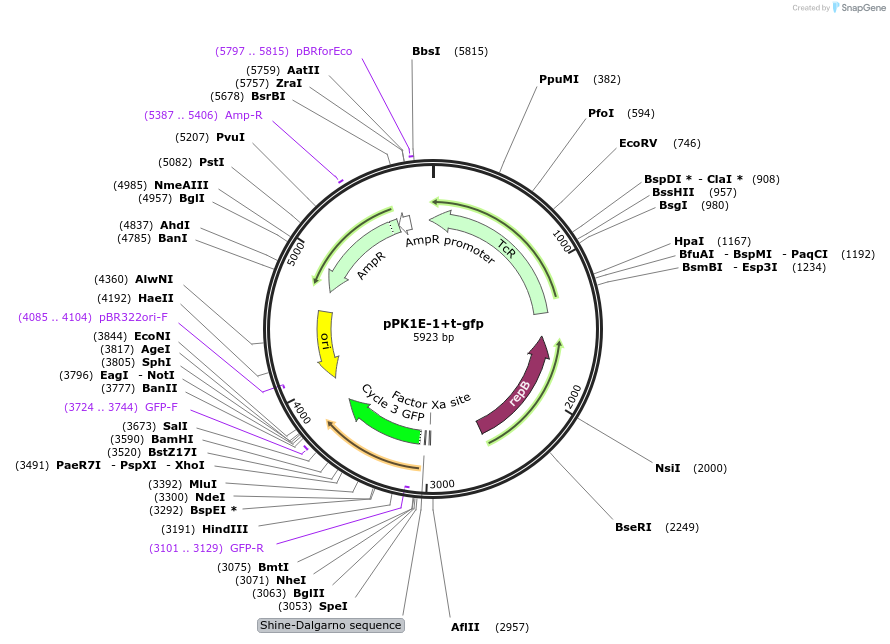
pPK1E-1+t-gfp
Plasmid#48131PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-K1E-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 11, 2013AvailabilityAcademic Institutions and Nonprofits only -
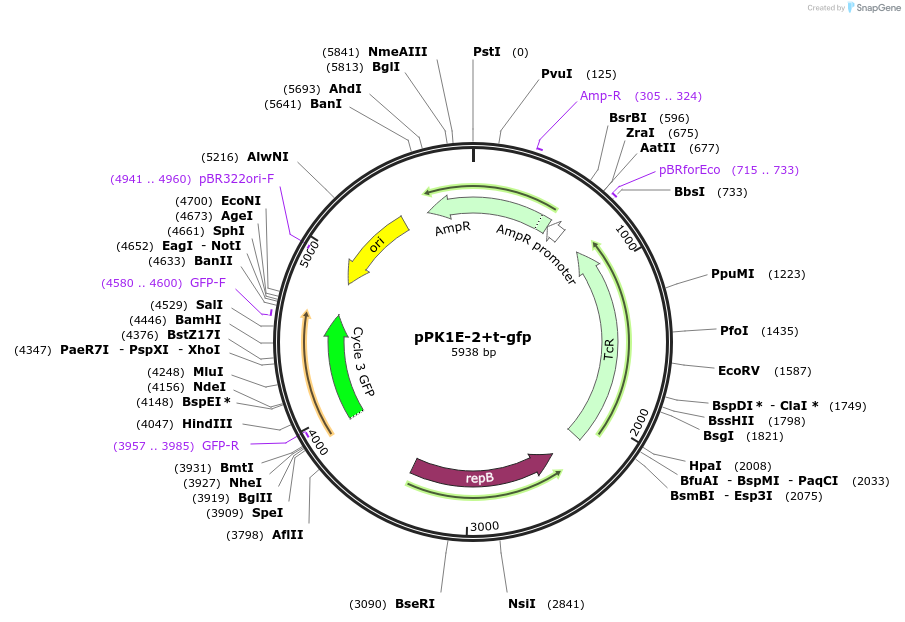
pPK1E-2+t-gfp
Plasmid#48132PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-K1E-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
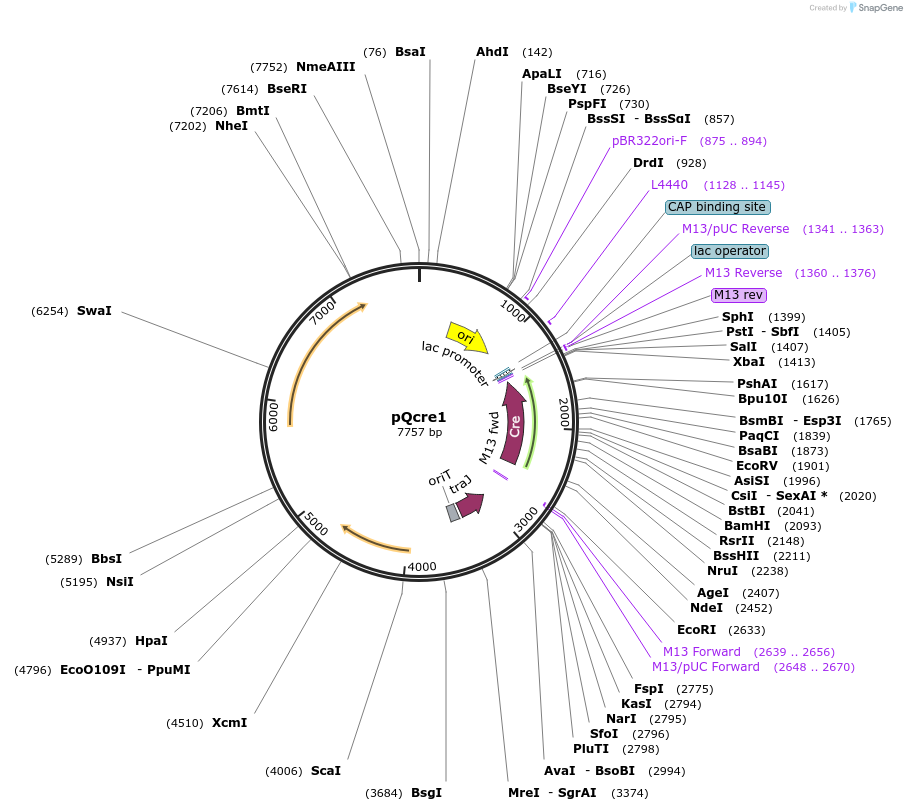
pQcre1
Plasmid#135659PurposeClostridial vector, encoding the Cre protein, which mediates recombination between lox sites.DepositorInsertCre recombinase
UseCre/LoxExpressionBacterialPromoterppag promoterAvailable SinceMarch 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
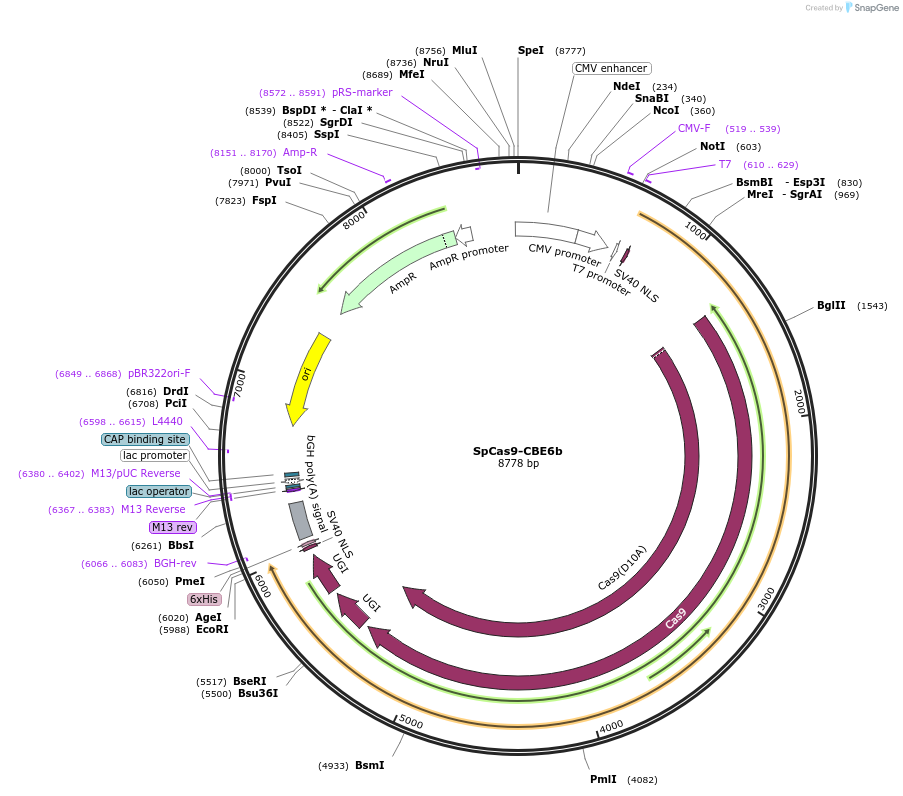
SpCas9-CBE6b
Plasmid#215820PurposeExpress CBE6b (with SpCas9) in mammalian cellsDepositorInsertCBE6b-SpCas9-2xUGI (cas9 )
ExpressionMammalianAvailable SinceMarch 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
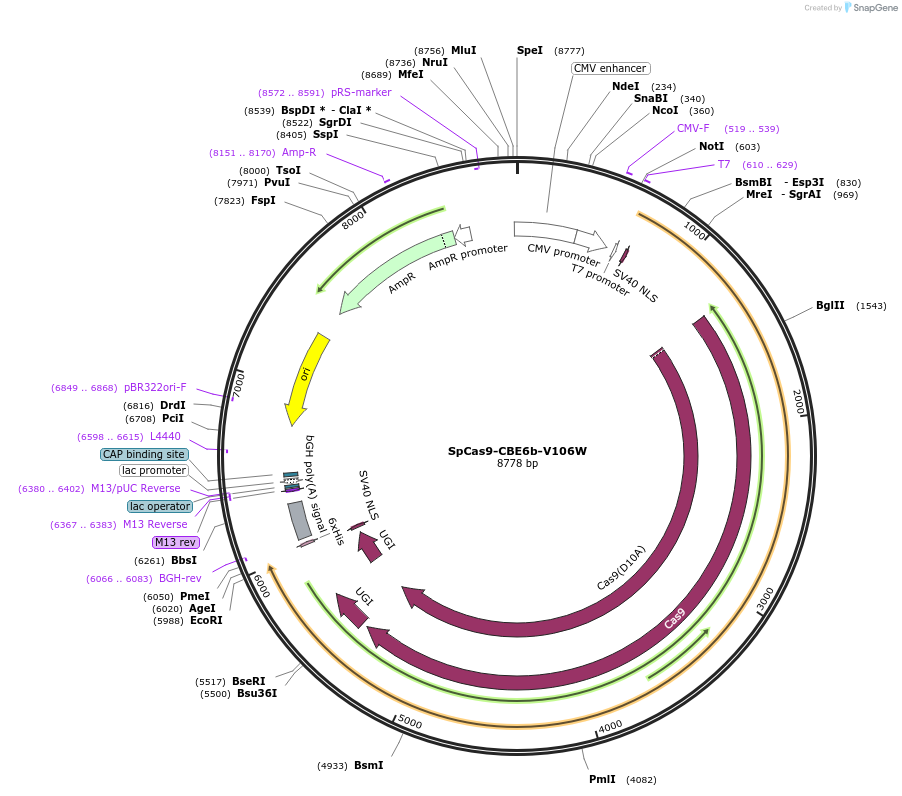
SpCas9-CBE6b-V106W
Plasmid#215825PurposeExpress CBE6b V106W (with SpCas9) in mammalian cellsDepositorInsertCBE6b V106W-SpCas9-2xUGI (cas9 )
UseIn vitro transcription templateAvailable SinceMarch 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
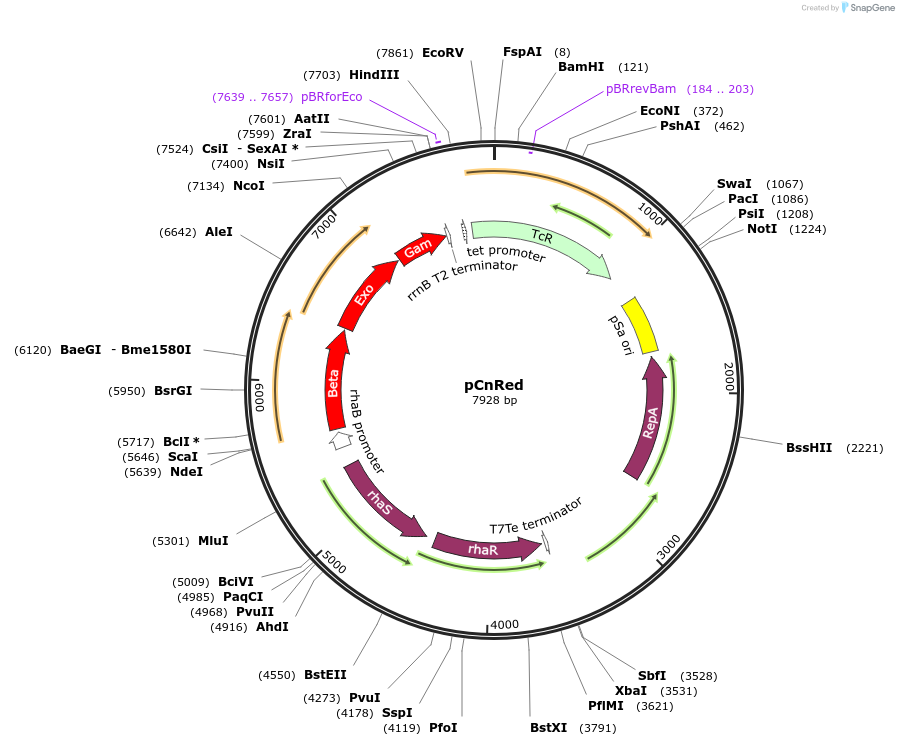
pCnRed
Plasmid#232908PurposeElectroporation vector for expression of the lambda Red recombineering system in Cupriavidus necator.DepositorAvailable SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
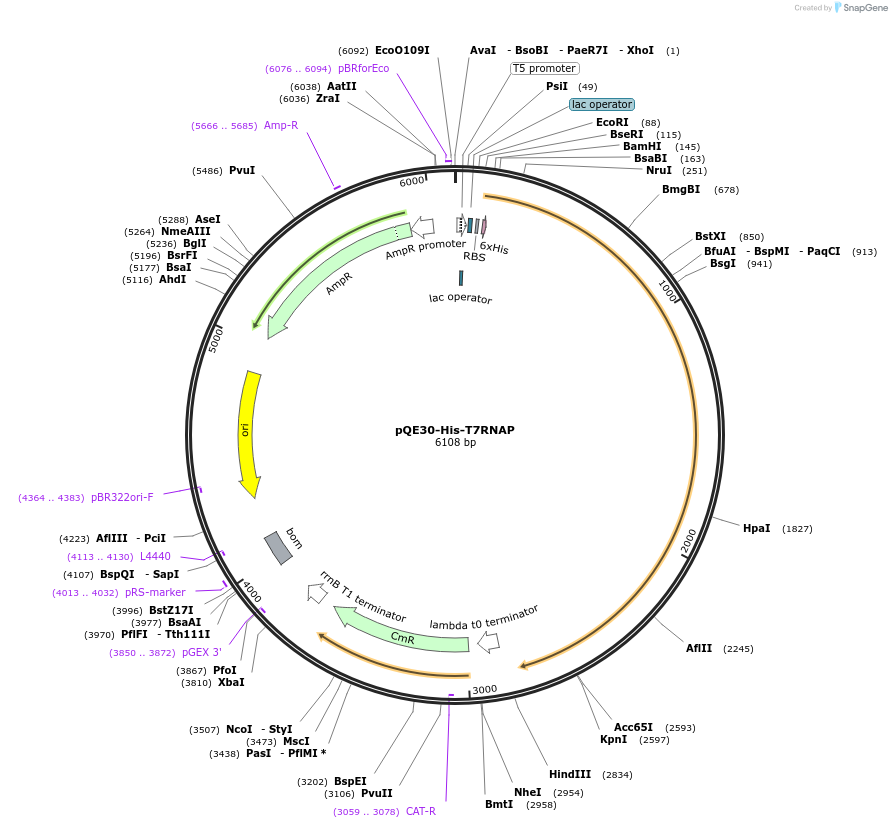
pQE30-His-T7RNAP
Plasmid#124138PurposeBacterial expression plasmid for PURE system componentsDepositorAvailable SinceApril 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
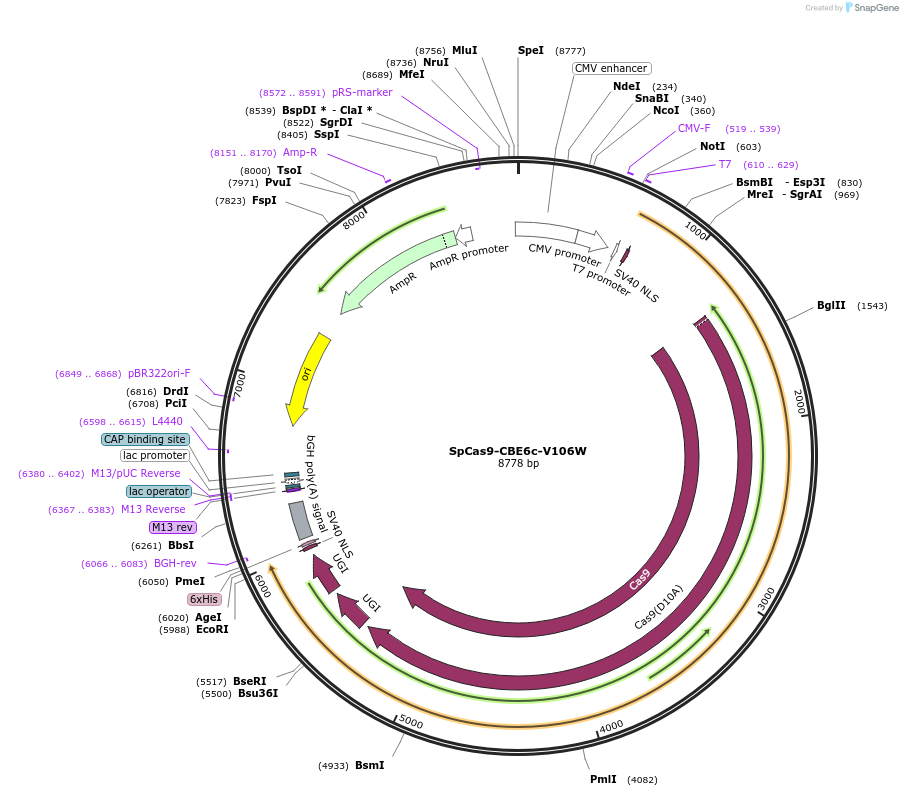
SpCas9-CBE6c-V106W
Plasmid#215826PurposeExpress CBE6c V106W (with SpCas9) in mammalian cellsDepositorInsertCBE6c V106W-SpCas9-2xUGI (cas9 )
UseIn vitro transcription templateAvailable SinceMarch 8, 2024AvailabilityAcademic Institutions and Nonprofits only