We narrowed to 9,586 results for: Coli
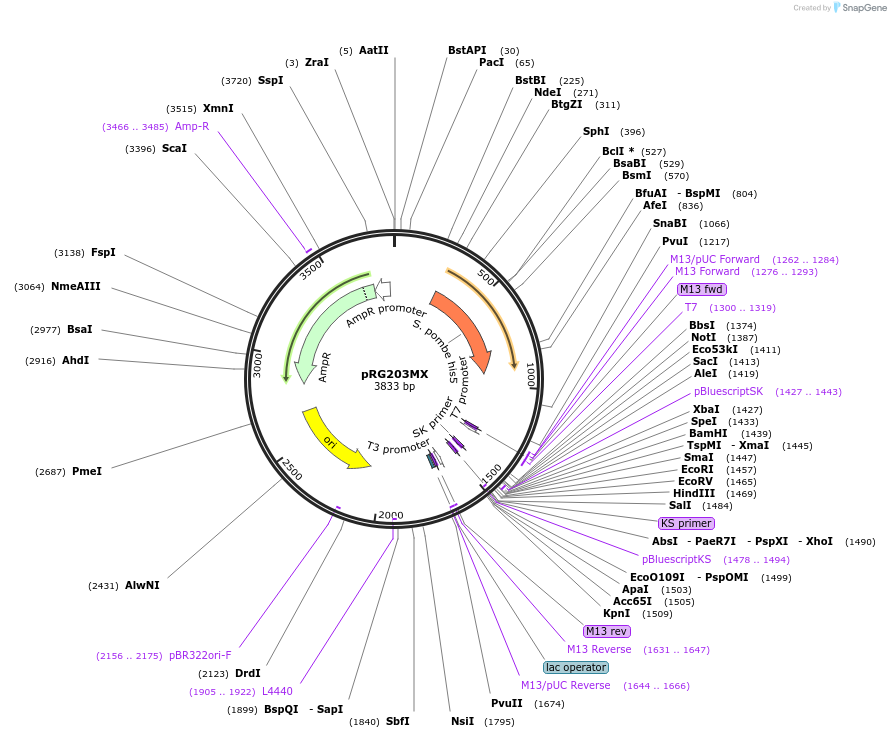
-
Plasmid#64521PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only
-
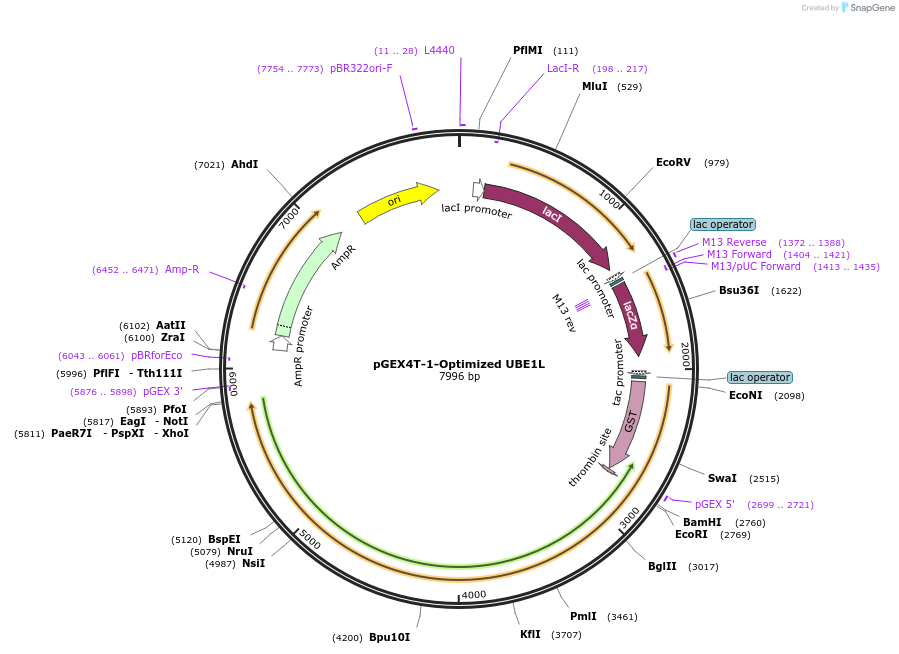
pGEX4T-1-Optimized UBE1L
Plasmid#165099Purposeecoli optimized expression of GST-fusion of human Ube1LDepositorInsertUbeE1L (UBA7 Human)
TagsGST from plasmid.ExpressionBacterialMutationcodon optimized for E. coli expressionAvailable SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
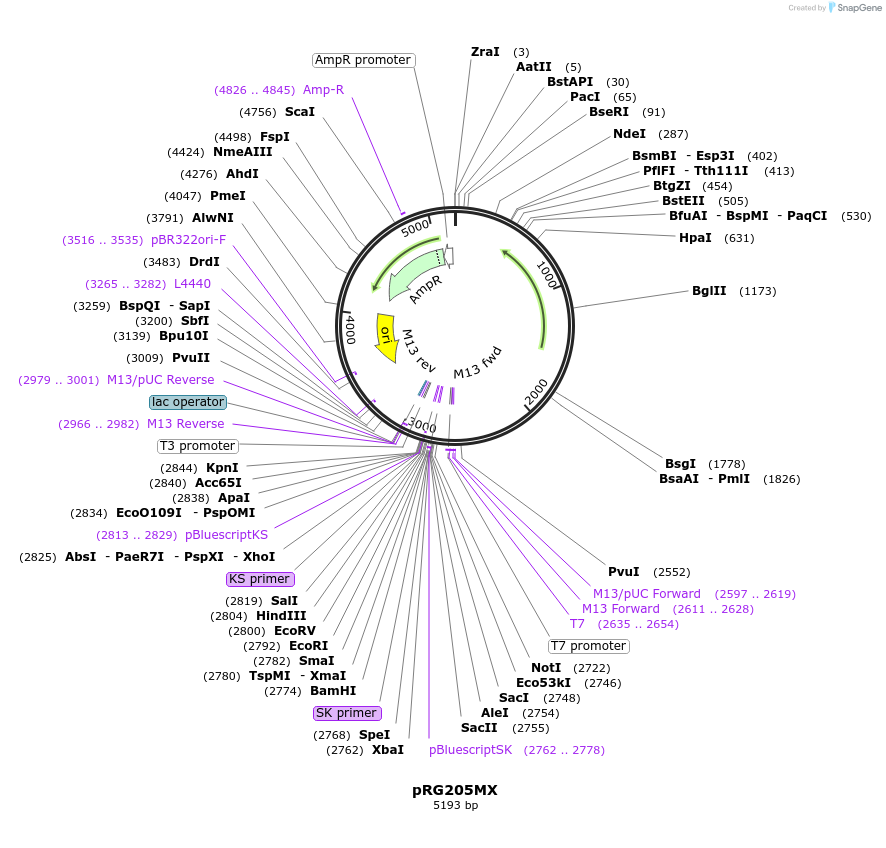
pRG205MX
Plasmid#64535PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
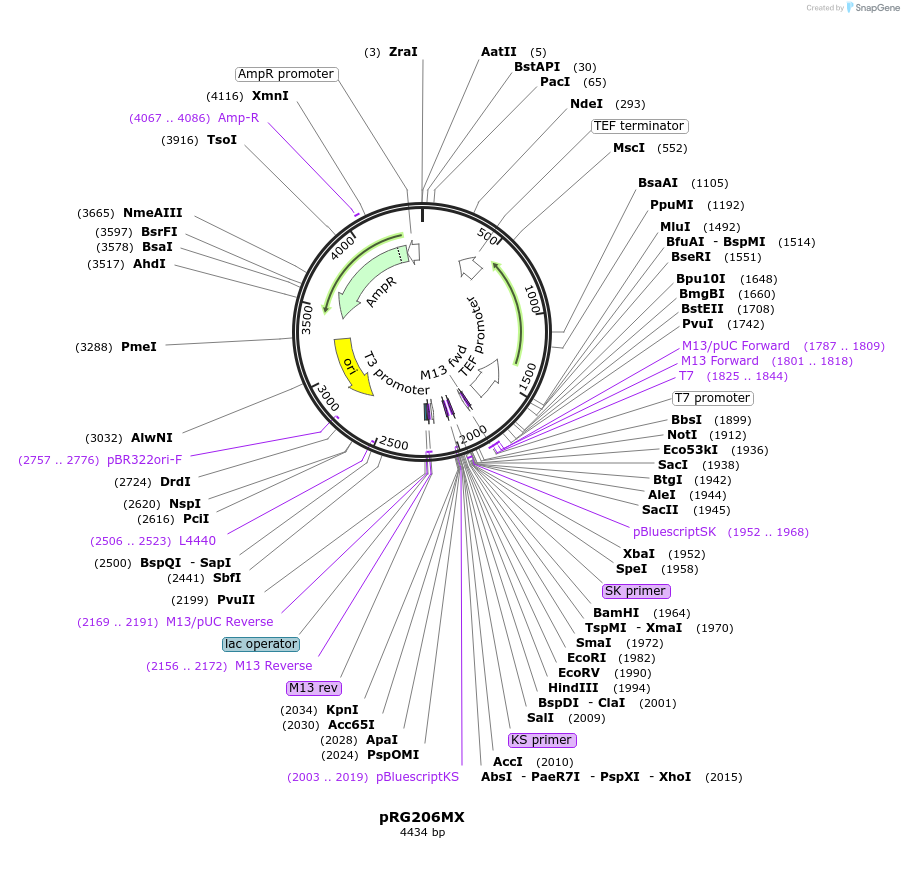
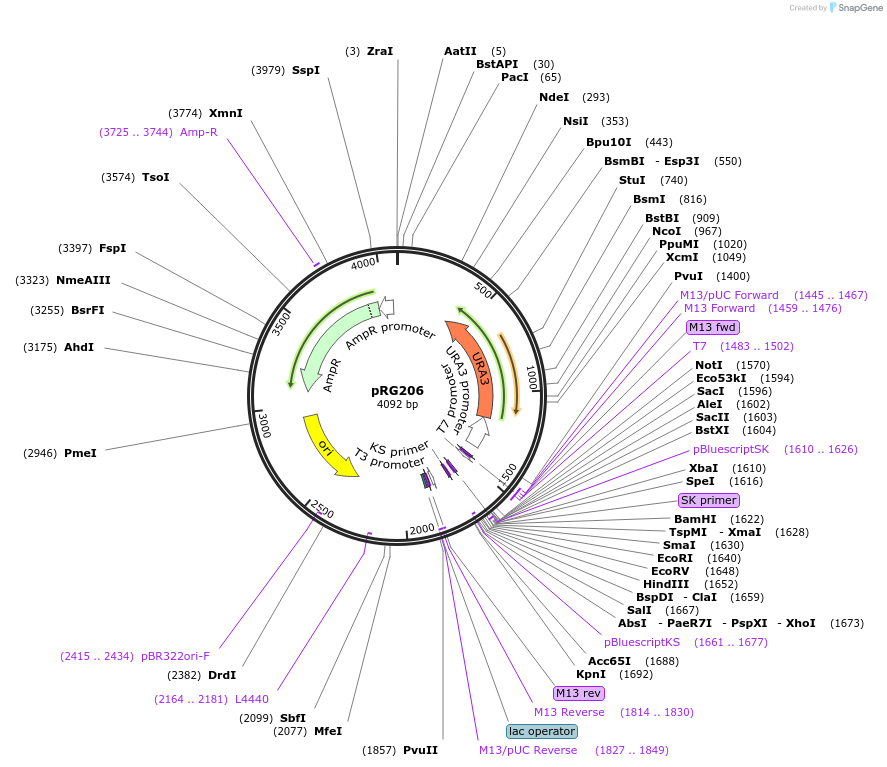
pRG206MX
Plasmid#64536PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
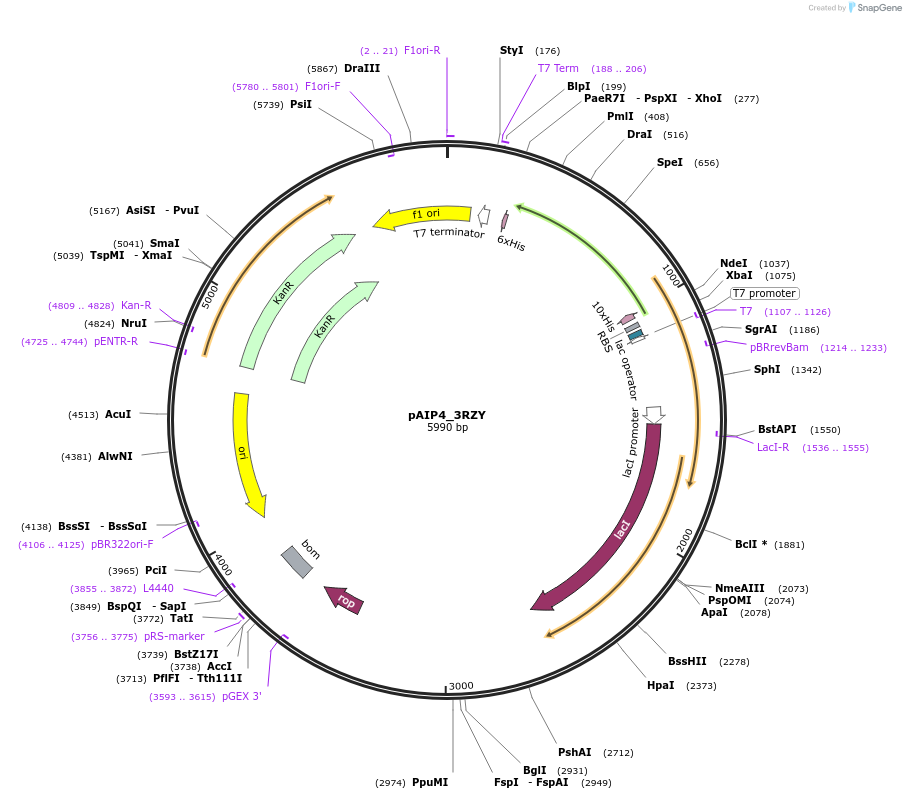
pAIP4_3RZY
Plasmid#234156PurposeHeterologous protein expression of fatty acid-binding protein, adipocyte (ALBP) in Escherichia coliDepositorAvailable SinceMarch 6, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
pRG235
Plasmid#64533PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
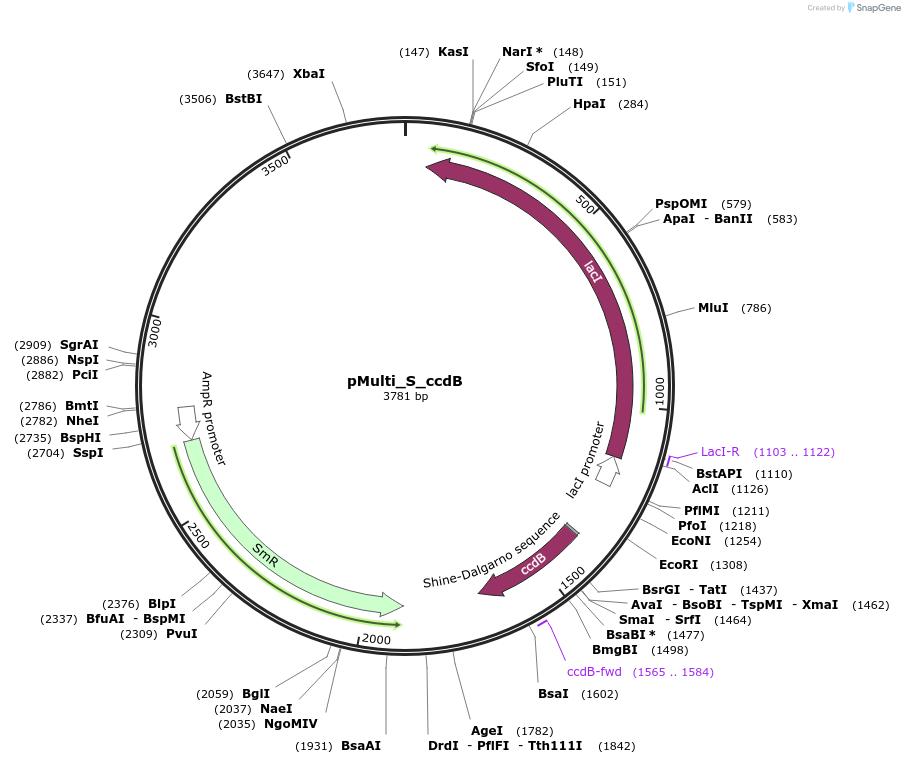
pMulti_S_ccdB
Plasmid#223853PurposeGEC-acceptor for 4G-cloning, designed for expression in E. coli (Streptomycin resistance)DepositorTypeEmpty backboneUseGolden-gate acceptorExpressionBacterialAvailable SinceDec. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
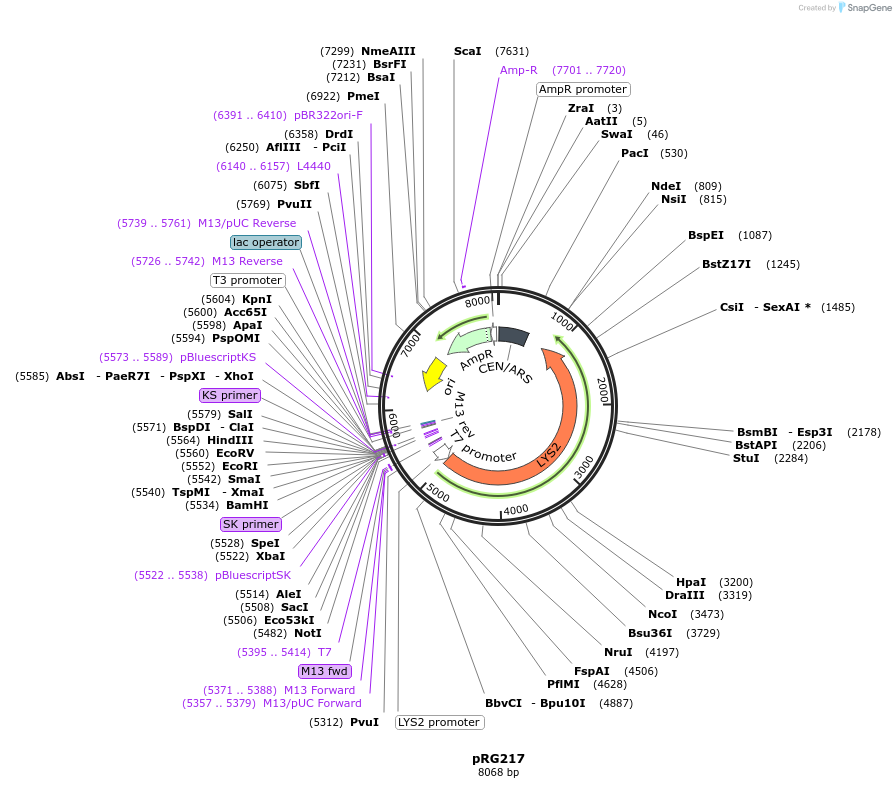
pRG217
Plasmid#64531PurposeCentromeric E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
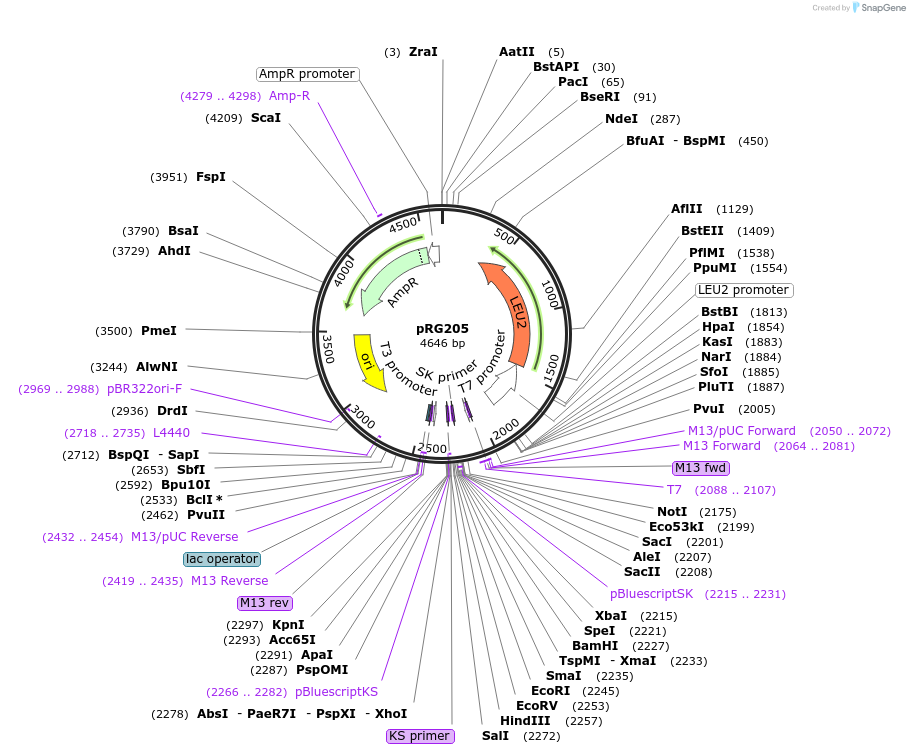
pRG205
Plasmid#64524PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRG206
Plasmid#64527PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
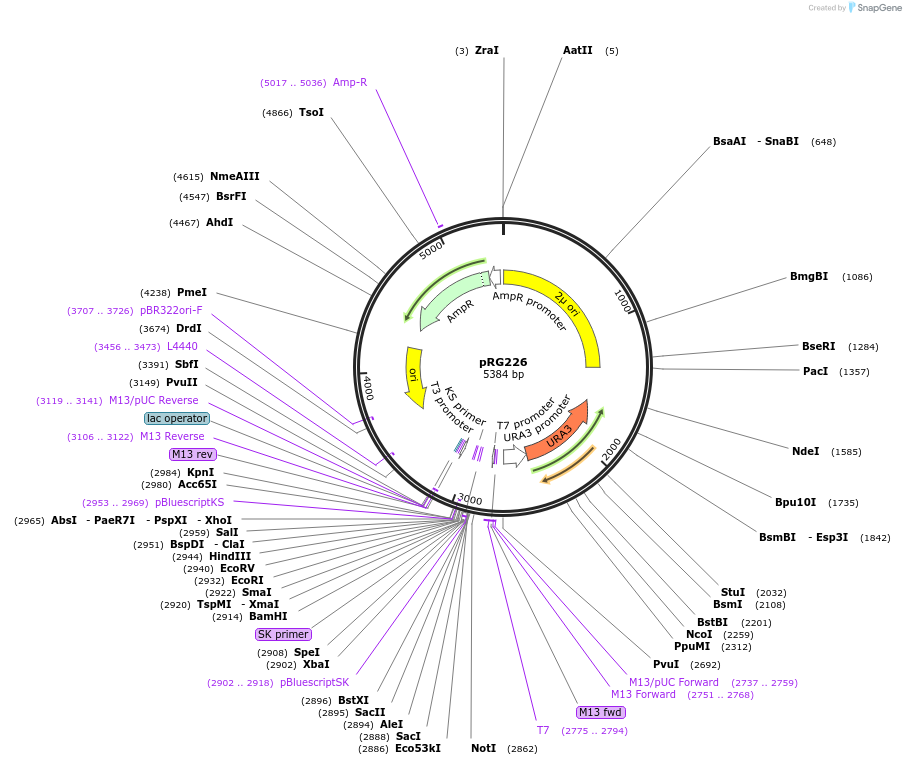
pRG226
Plasmid#64529PurposeEpisomal E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
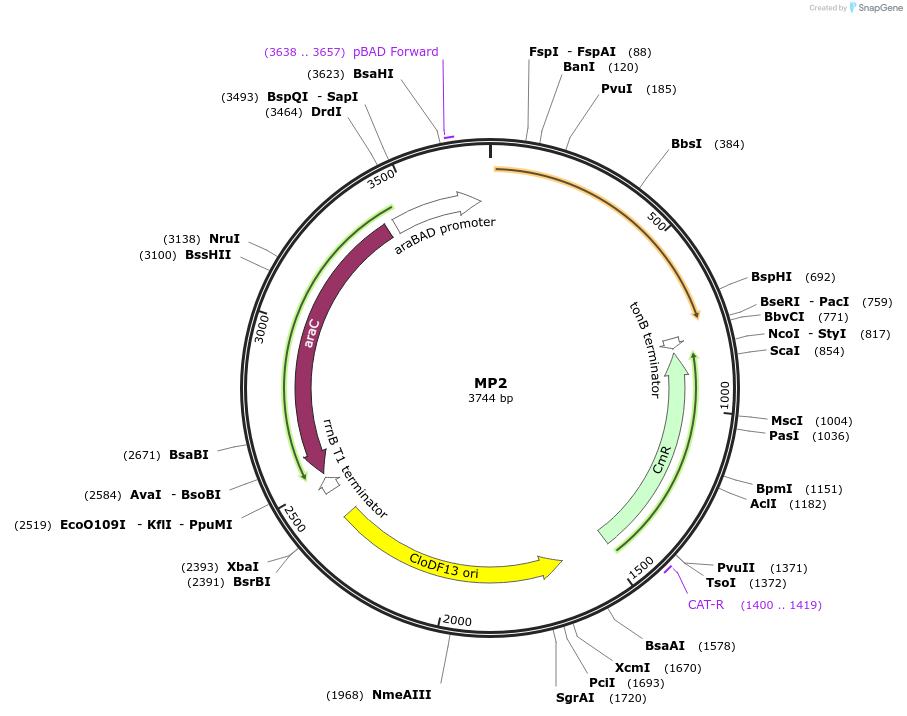
MP2
Plasmid#69628PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
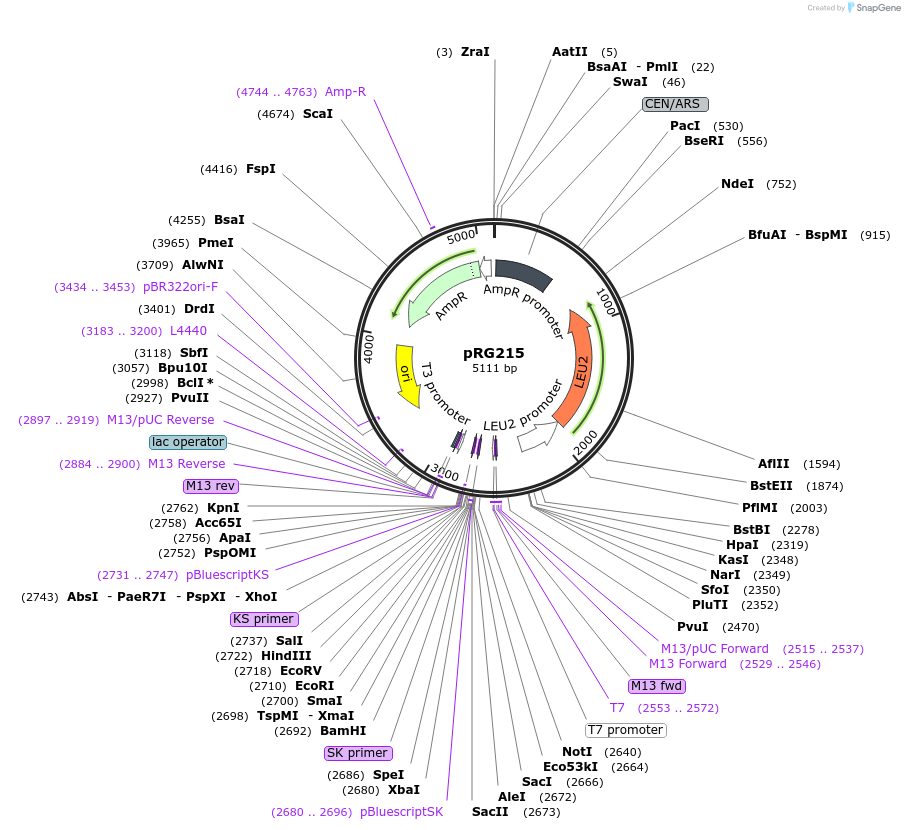
pRG215
Plasmid#64525PurposeCentromeric E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
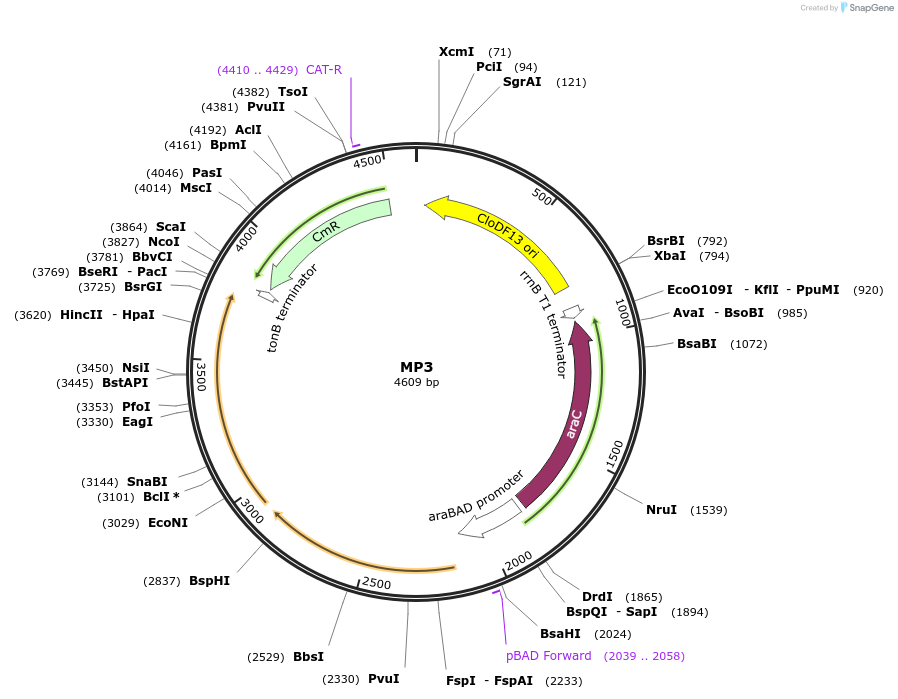
MP3
Plasmid#69650PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
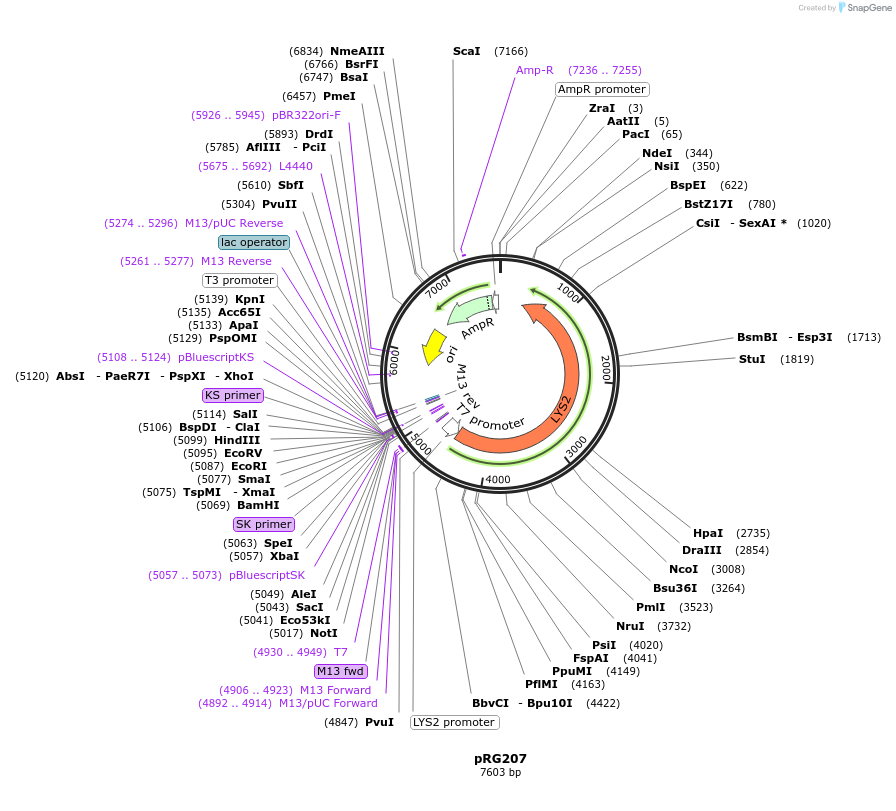
pRG207
Plasmid#64530PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
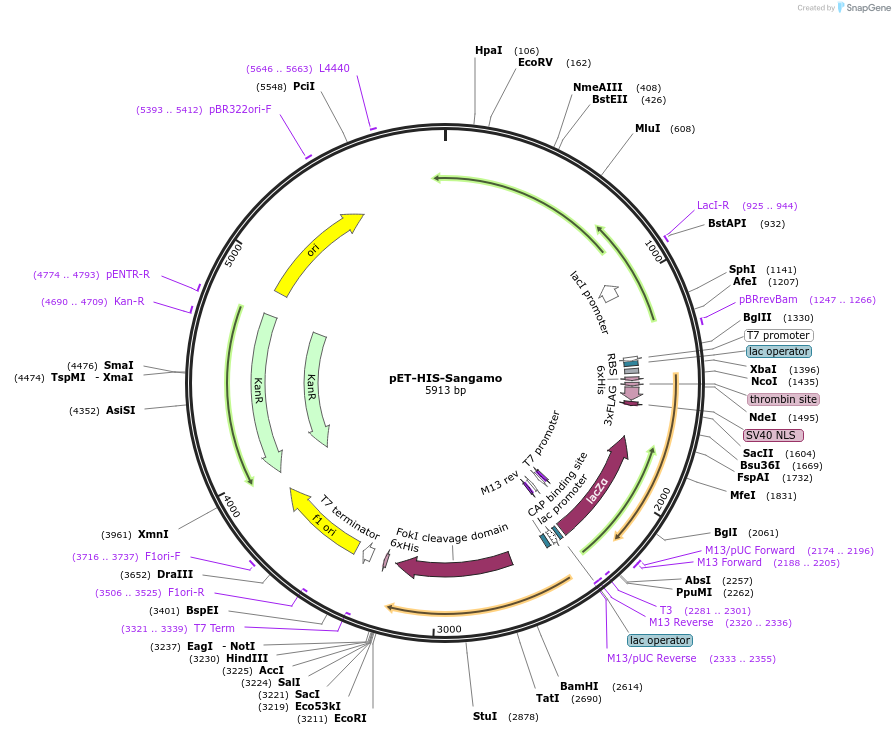
pET-HIS-Sangamo
Plasmid#40786Purposefor e. coli expression, Golden Gate Compatible TALEN incorporated with N Terminal 6xHIS Tag, Codon Optimized FokIDepositorInsertTranscription Activator Like Effector Nuclease
UseTALENTagsN Terminal 6xHISExpressionBacterialMutation+63 C Terminus, Codon Optimized FokI EndonucleasePromoterT7Available SinceDec. 12, 2013AvailabilityIndustry, Academic Institutions, and Nonprofits -
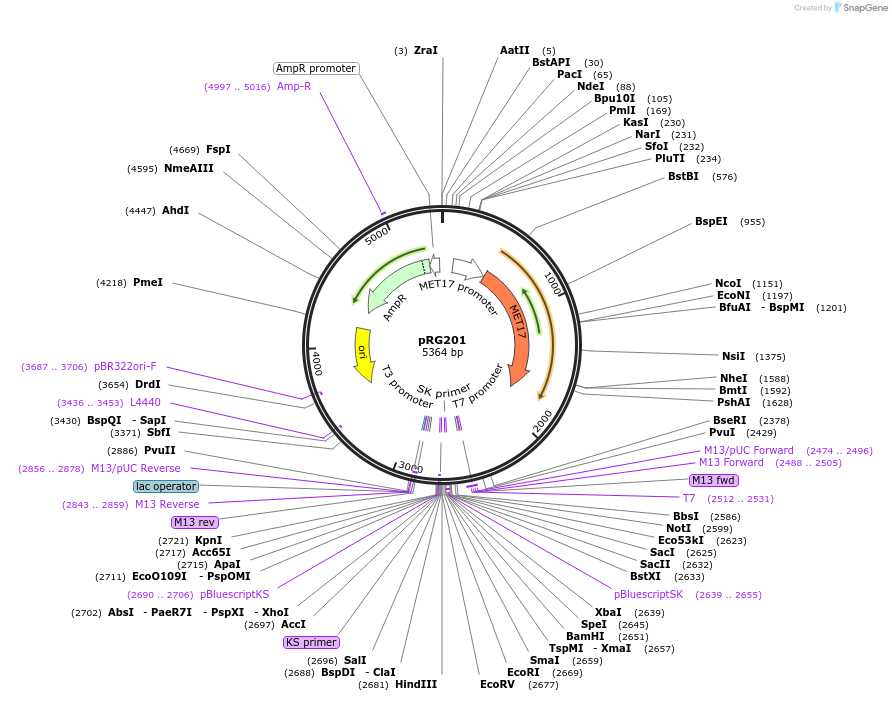
pRG201
Plasmid#64394PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
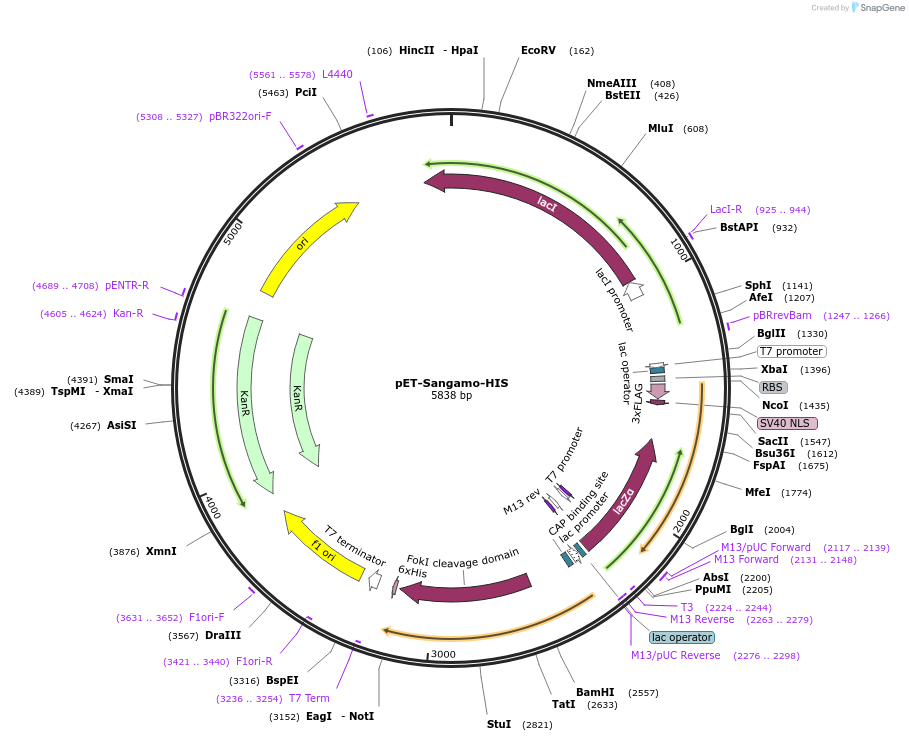
pET-Sangamo-HIS
Plasmid#40787Purposefor e. coli expression, Golden Gate Compatible TALEN incorporated with C Terminal 6xHIS Tag, Codon Optimized FokIDepositorInsertTranscription Activator Like Effector Nuclease
UseTALENTagsC Terminal 6xHISExpressionBacterialMutation+63 C Terminus,Codon Optimized FokI EndonucleasePromoterT7Available SinceDec. 12, 2013AvailabilityIndustry, Academic Institutions, and Nonprofits -
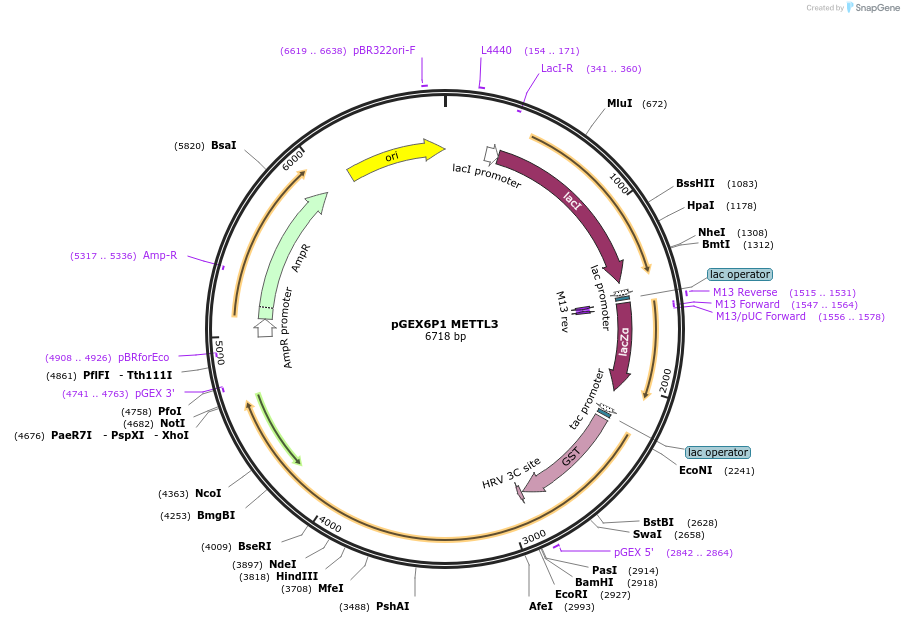
pGEX6P1 METTL3
Plasmid#200731PurposeBacterial expression of GST-Tagged codon optimized human METTL3DepositorInsertMETTL3 (METTL3 Human)
TagsGST from backbone plasmidExpressionBacterialMutationCodon optimized for expression in E. coliAvailable SinceJuly 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
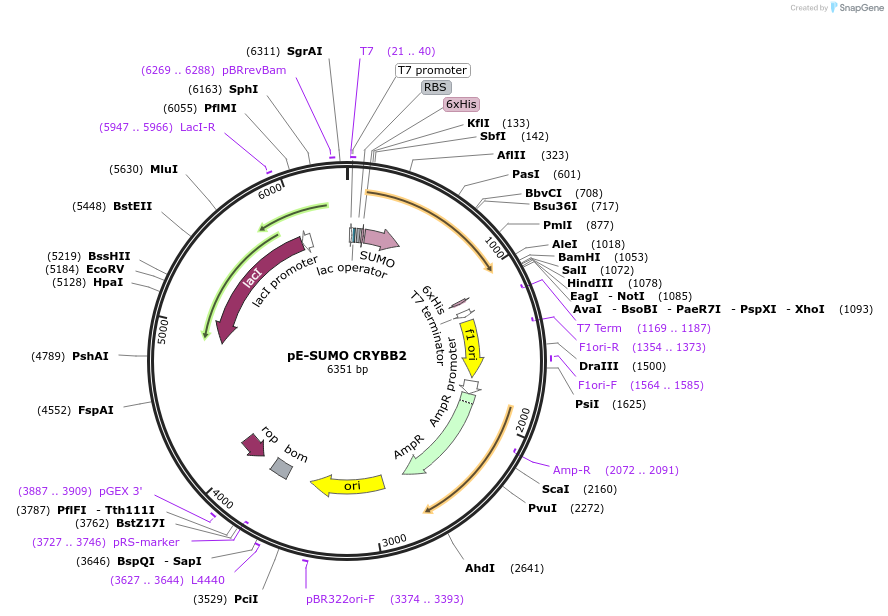
pE-SUMO CRYBB2
Plasmid#80749PurposeExpression of human beta B2-crystallin in E. coliDepositorAvailable SinceMarch 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
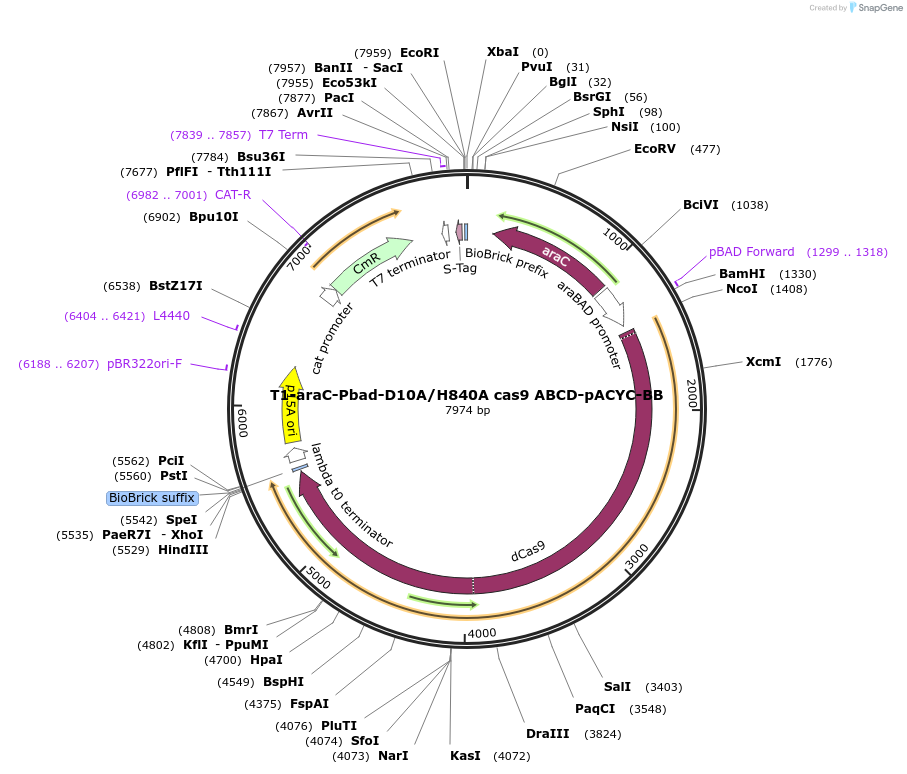
T1-araC-Pbad-D10A/H840A cas9 ABCD-pACYC-BB
Plasmid#110547PurposeCRISPR interference; works in conjunction with an sgRNA expression vector to silence genes in the E. coli chromosome.DepositorInsertdCas9
ExpressionBacterialMutationD10A, H840APromoterpBADAvailable SinceJuly 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
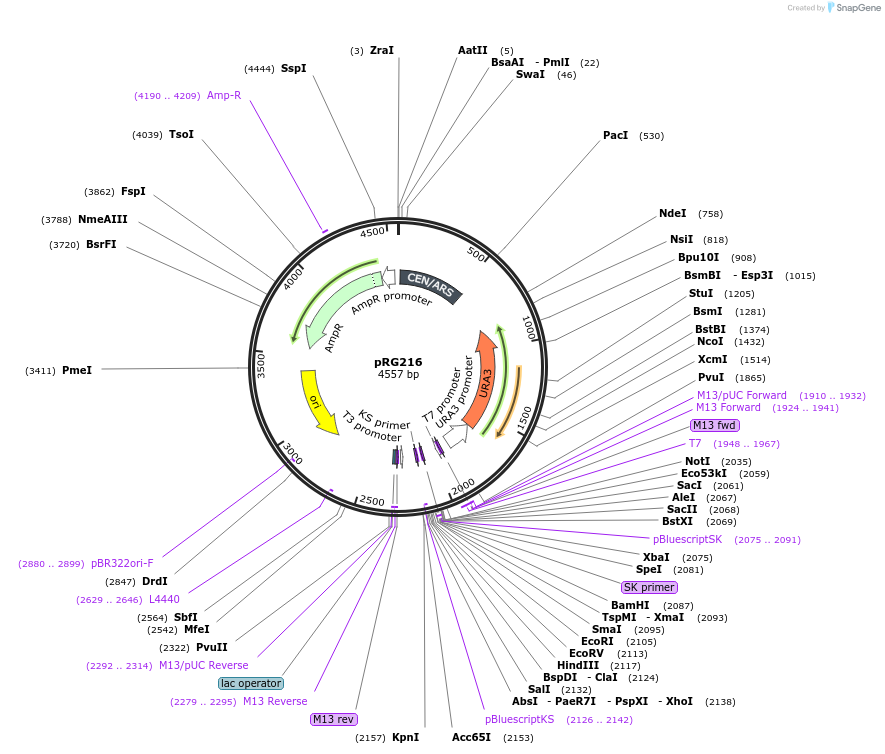
pRG216
Plasmid#64528PurposeCentromeric E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
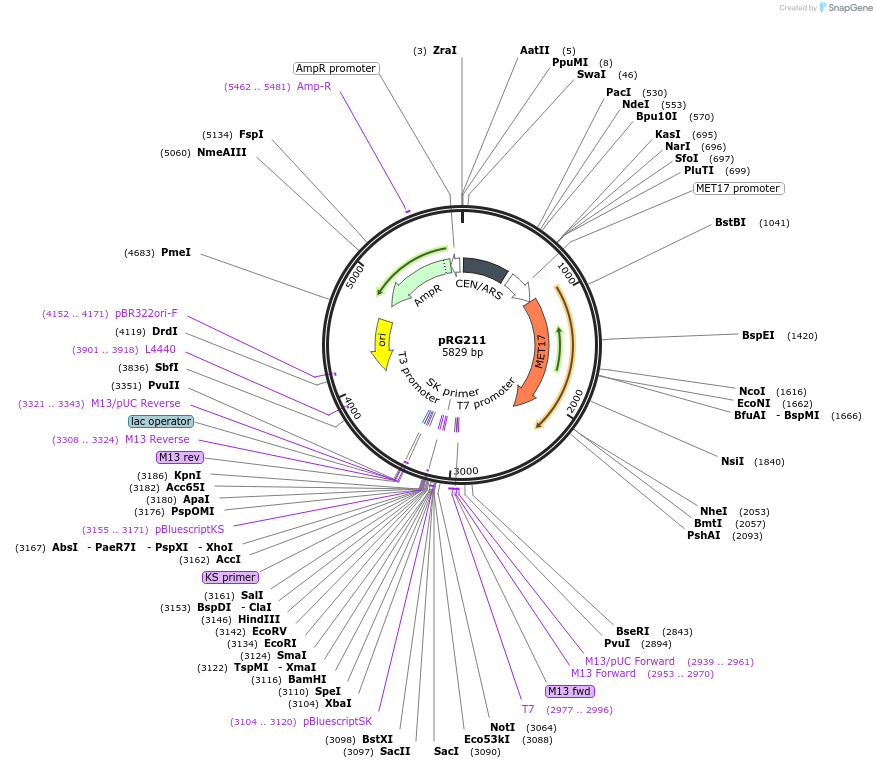
pRG211
Plasmid#64519PurposeCentromeric E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
5407 pClpB
Plasmid#1235DepositorInsertClpB
ExpressionBacterialMutationnoneAvailable SinceJan. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
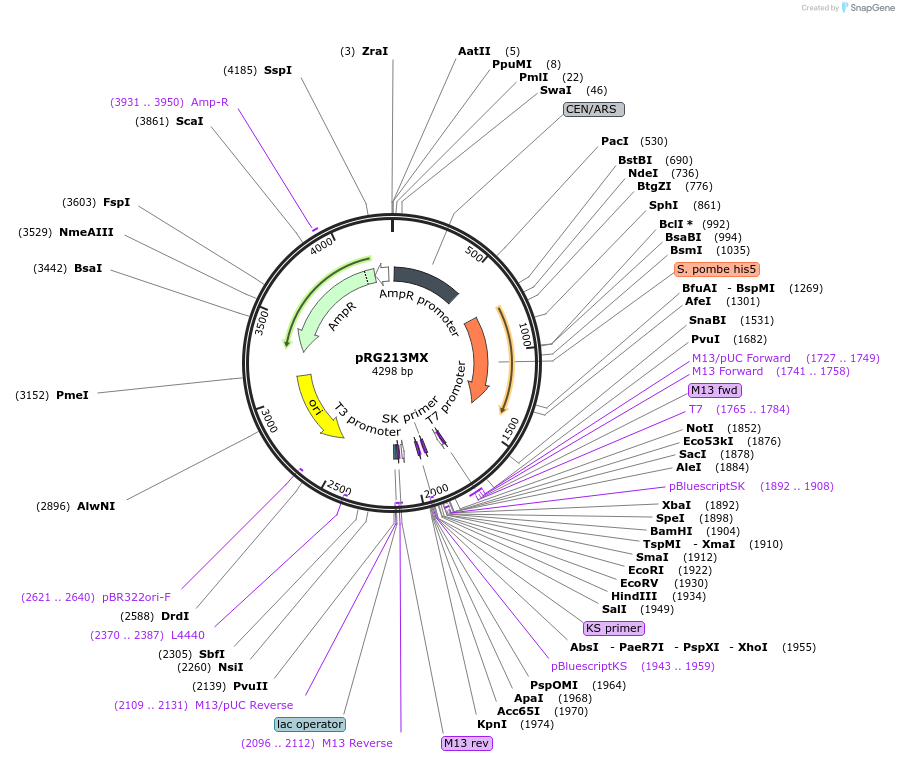
pRG213MX
Plasmid#64522PurposeCentromeric E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
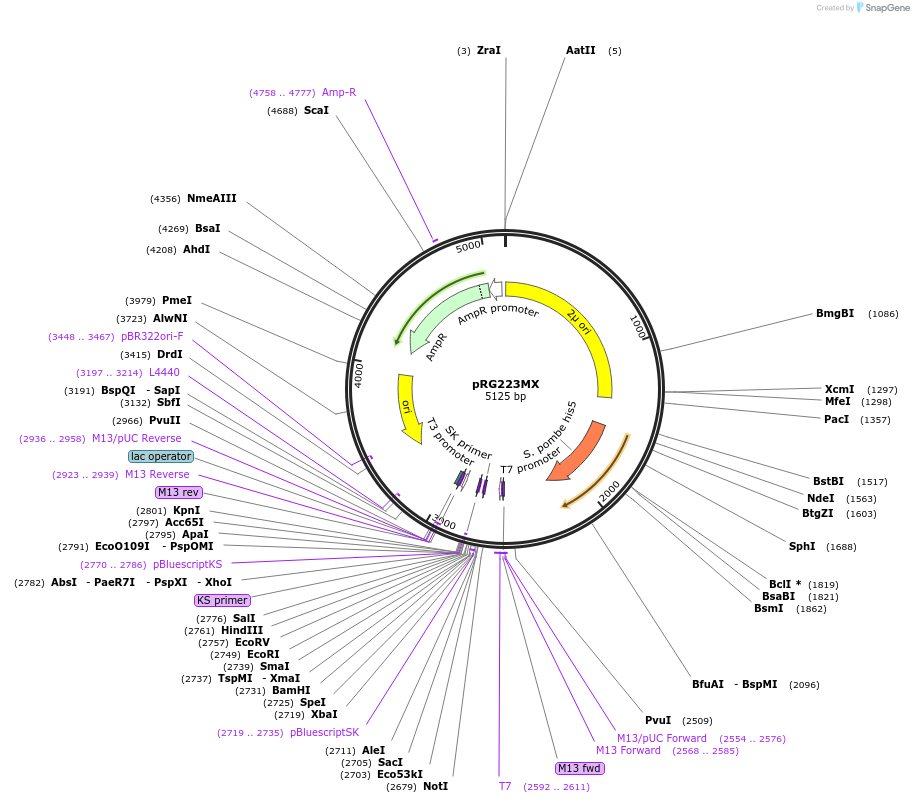
pRG223MX
Plasmid#64523PurposeEpisomal E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
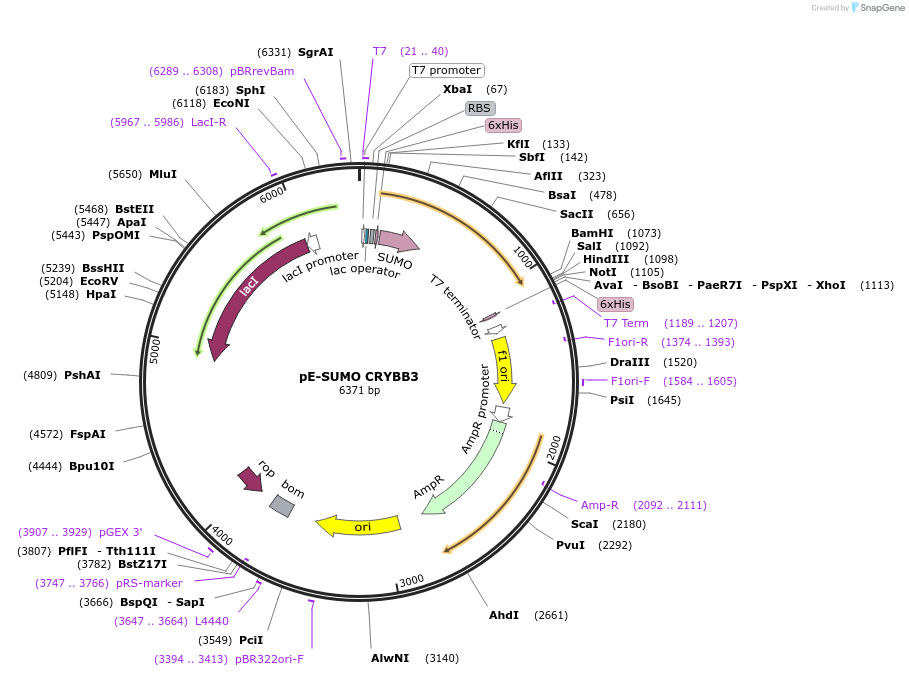
pE-SUMO CRYBB3
Plasmid#80750PurposeExpression of human beta B3-crystallin in E. coliDepositorAvailable SinceMarch 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
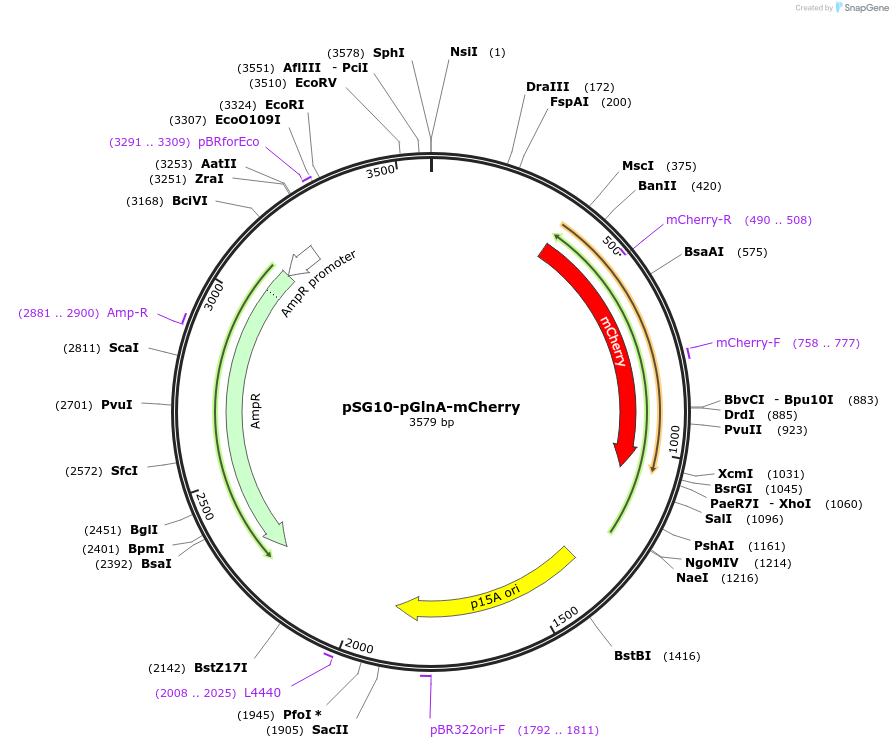
pSG10-pGlnA-mCherry
Plasmid#102467PurposeExpresses mCherry fluorescent protein in E. coli. The expression is controlled by a pGlnA promoter.DepositorArticleInsertmCherry
ExpressionBacterialPromoterpGlnAAvailable SinceOct. 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
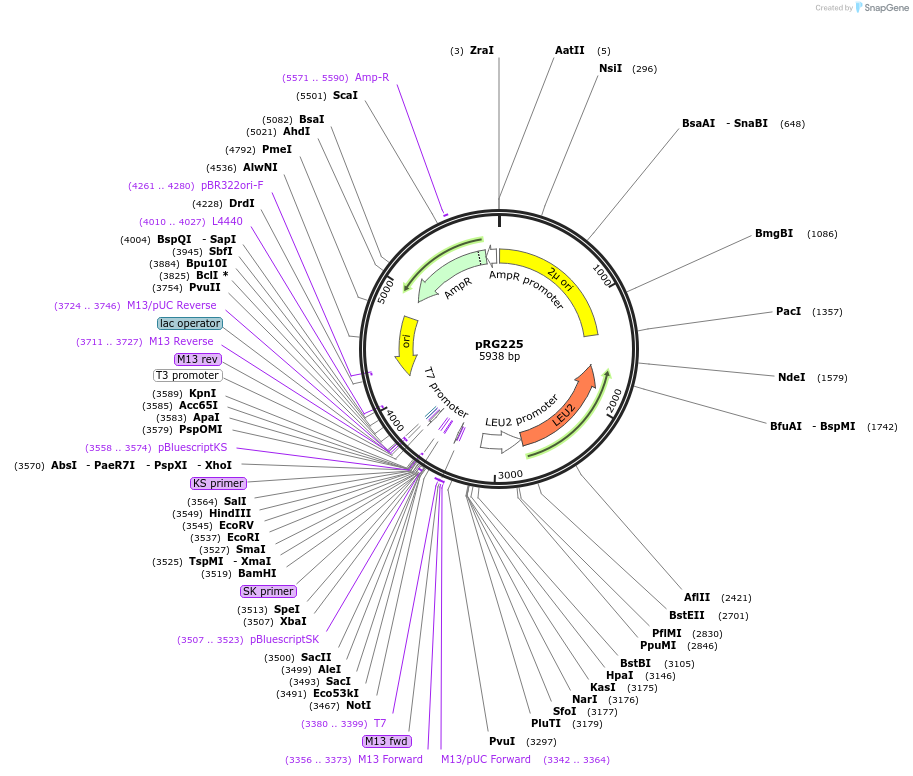
pRG225
Plasmid#64526PurposeEpisomal E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
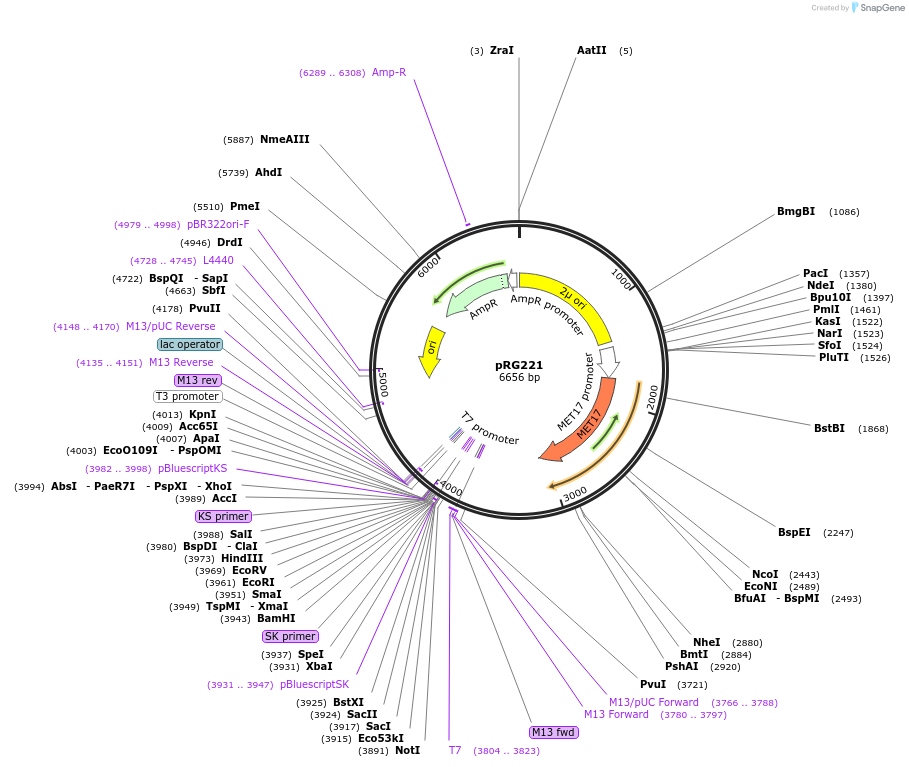
pRG221
Plasmid#64520PurposeEpisomal E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
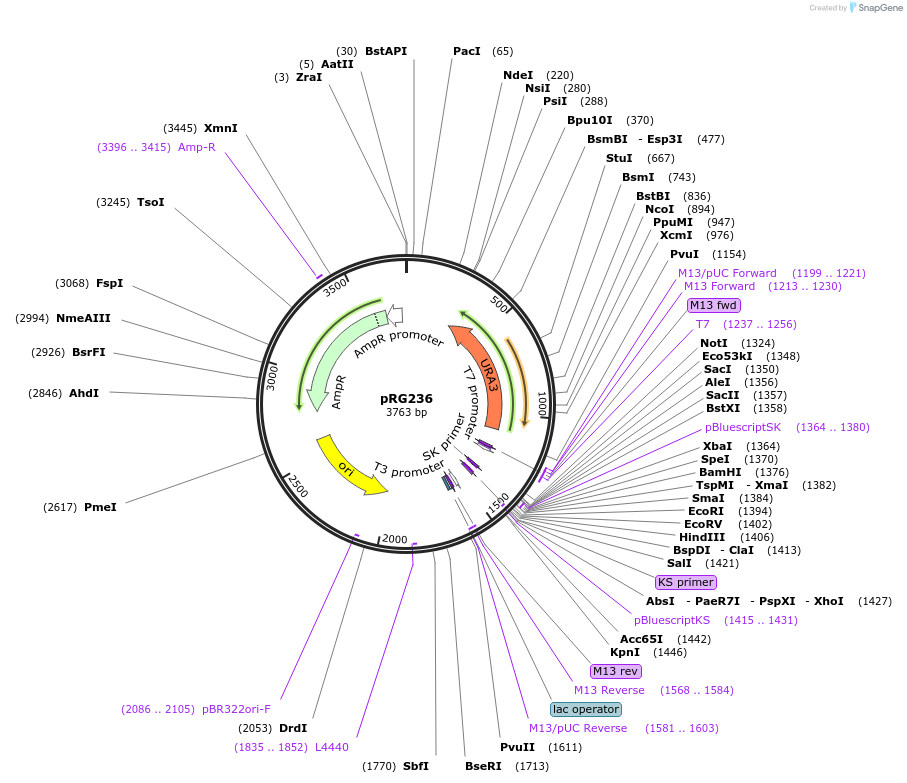
pRG236
Plasmid#64534PurposeIntegrative E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
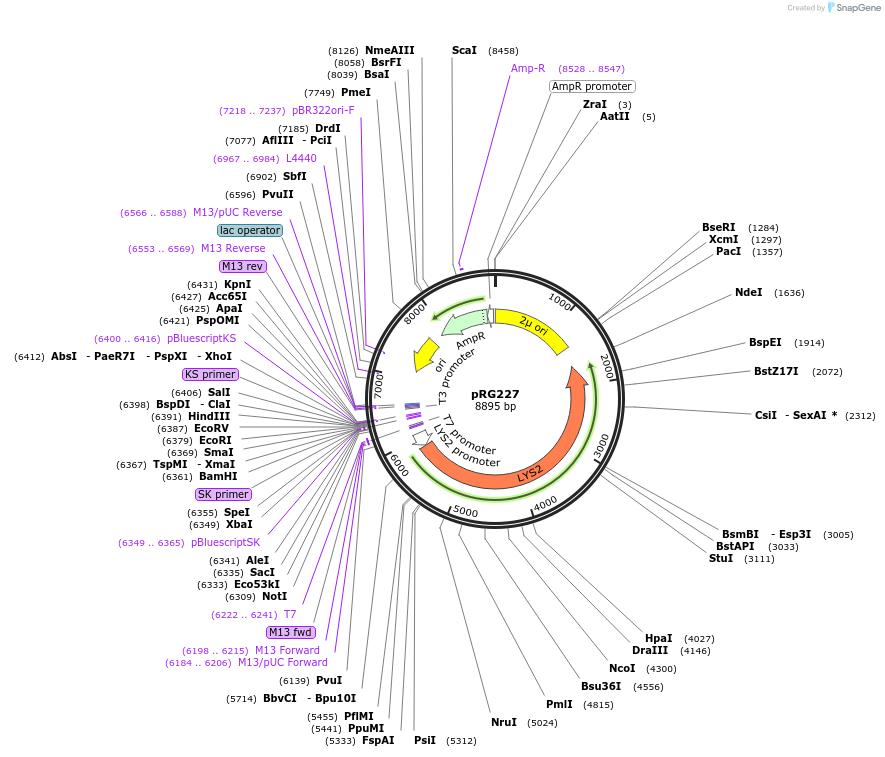
pRG227
Plasmid#64532PurposeEpisomal E. coli/S. cerevisiae shuttle vectorDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
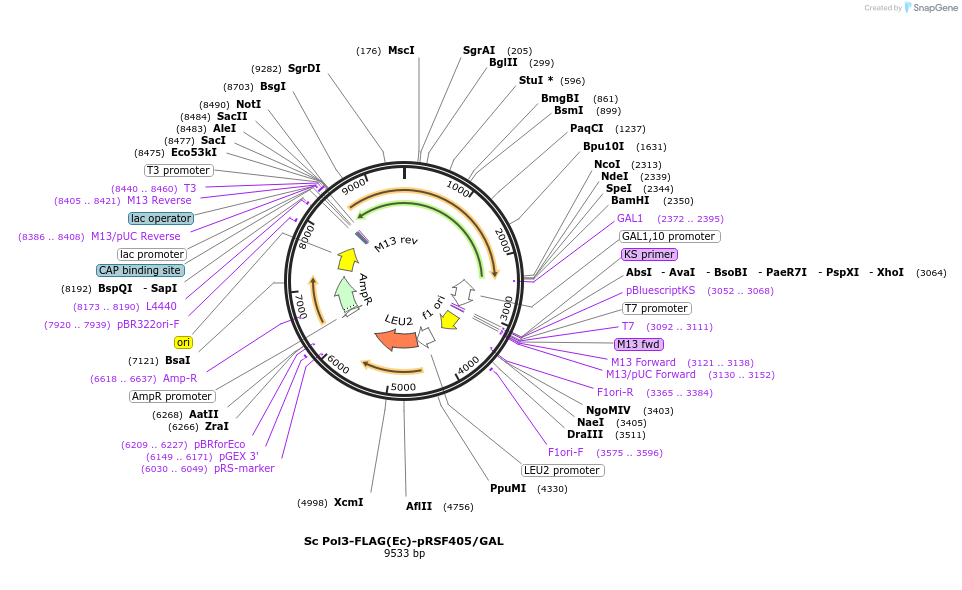
Sc Pol3-FLAG(Ec)-pRSF405/GAL
Plasmid#239198PurposeOverexpress Sc Pol3(C-terminal 3X FLAG tag) when integrated into yeast (S. cer)DepositorInsertPol3 (POL3 Budding Yeast)
Tags3X FLAGExpressionYeastMutationE. coli codon bias (synthetic gene), but same ami…Available SinceJune 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
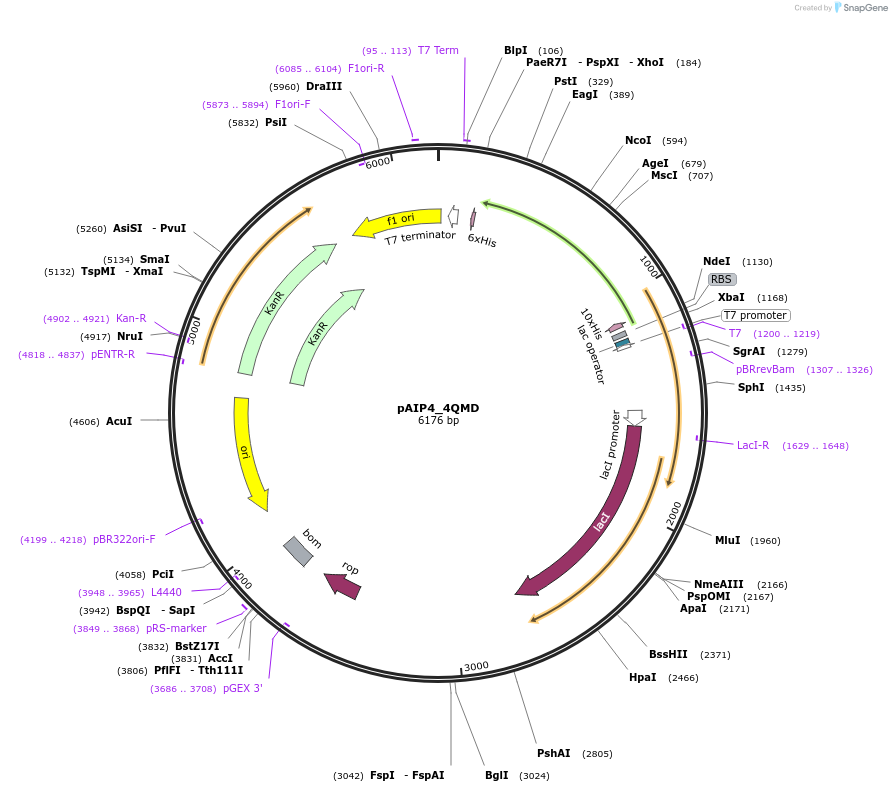
pAIP4_4QMD
Plasmid#234174PurposeHeterologous protein expression of envoplakin in Escherichia coliDepositorAvailable SinceMarch 6, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
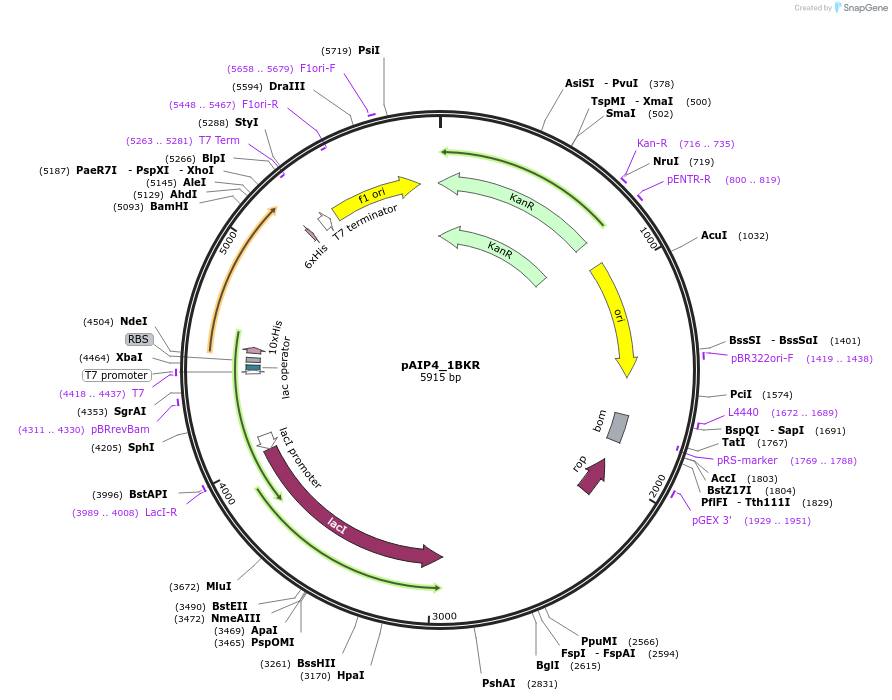
pAIP4_1BKR
Plasmid#234168PurposeHeterologous protein expression of spectrin beta chain, non-erythrocytic 1 (Beta-II spectrin) in Escherichia coliDepositorAvailable SinceMarch 6, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
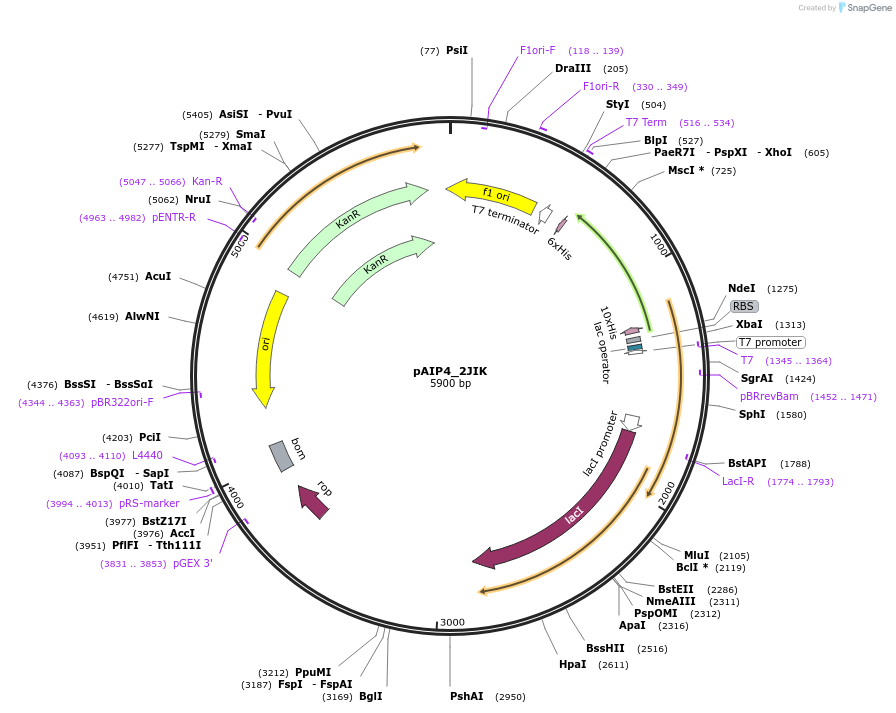
pAIP4_2JIK
Plasmid#234170PurposeHeterologous protein expression of synaptojanin-2-binding protein in Escherichia coliDepositorAvailable SinceMarch 6, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
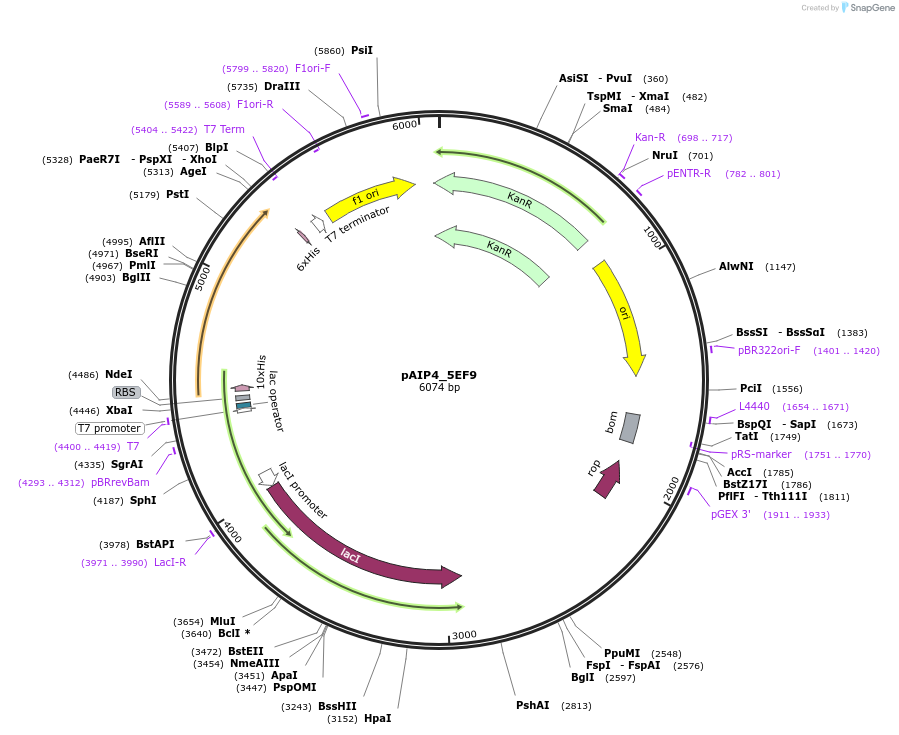
pAIP4_5EF9
Plasmid#234154PurposeHeterologous protein expression of polymerase basic protein 2 (RNA-directed RNA polymerase subunit P3) in Escherichia coliDepositorInsert5EF9 (PB2 Influenza B virus (B/Lee/1940))
Tags10x HisTag-Smt3ExpressionBacterialPromoterT7Available SinceMarch 6, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
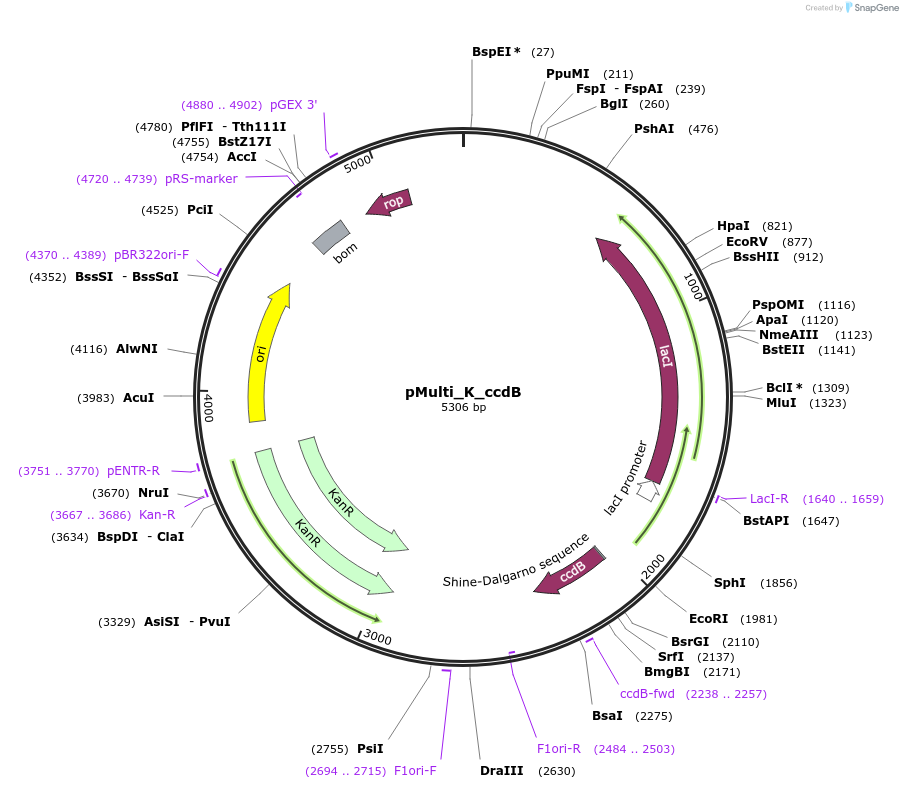
pMulti_K_ccdB
Plasmid#223852PurposeGEC-acceptor for 4G-cloning, designed for expression in E. coli (Kanamyicin resistance)DepositorTypeEmpty backboneUseGolden-gate acceptorExpressionBacterialAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
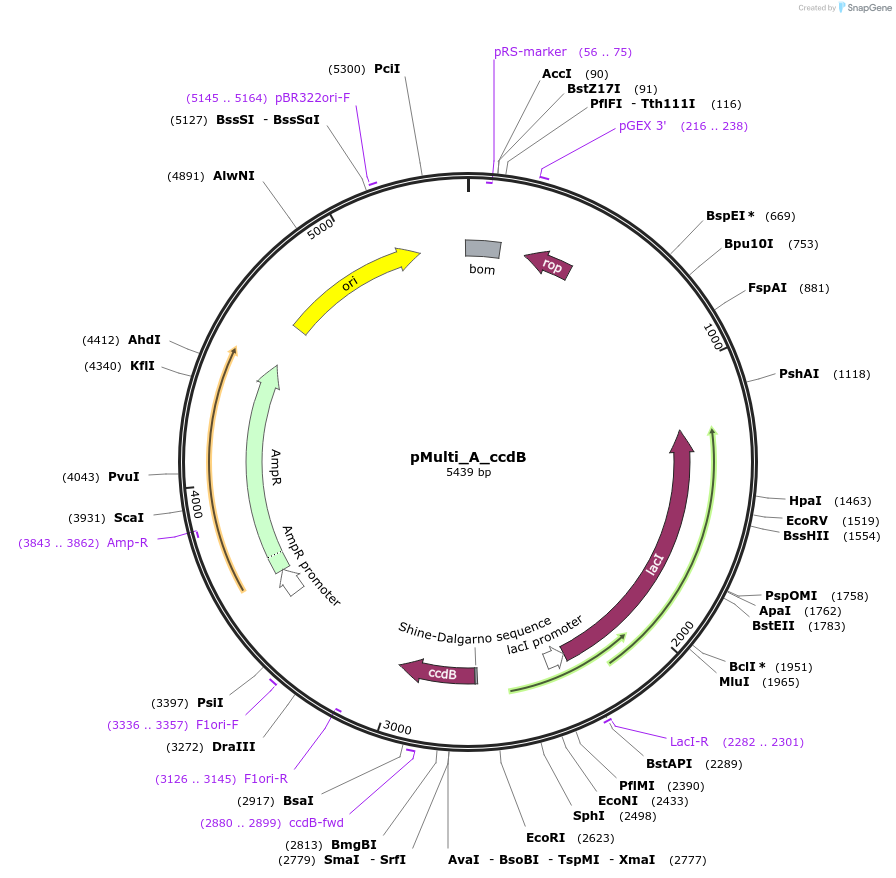
pMulti_A_ccdB
Plasmid#223851PurposeGEC-acceptor for 4G-cloning, designed for expression in E. coli (Ampicillin resistance)DepositorTypeEmpty backboneUseGolden-gate acceptorExpressionBacterialAvailable SinceOct. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
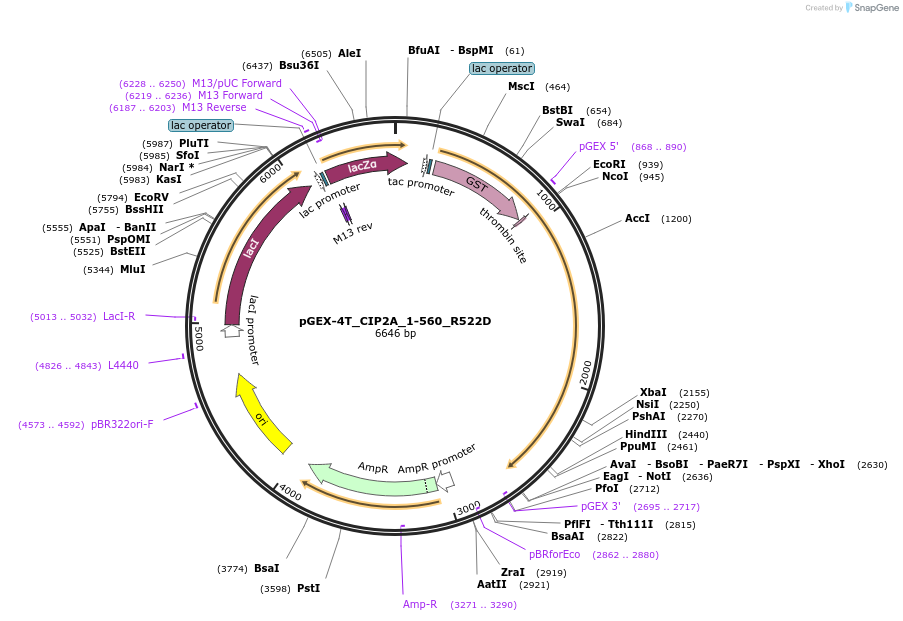
pGEX-4T_CIP2A_1-560_R522D
Plasmid#217923PurposeE. coli expression of GST-tagged CIP2A_1-560 with R522D mutationDepositorAvailable SinceMay 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
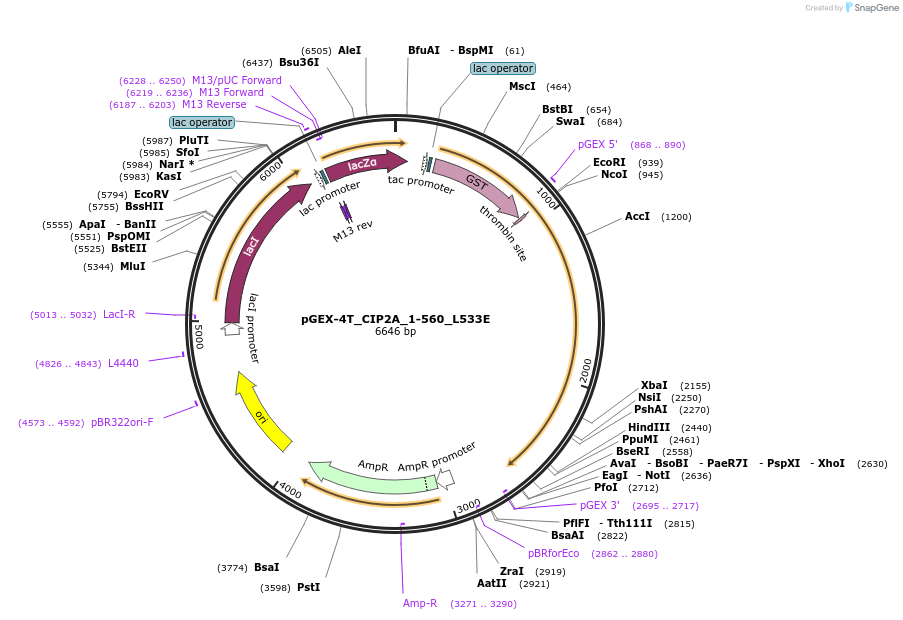
pGEX-4T_CIP2A_1-560_L533E
Plasmid#217924PurposeE. coli expression of GST-tagged CIP2A_1-560 with L533E mutationDepositorAvailable SinceMay 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
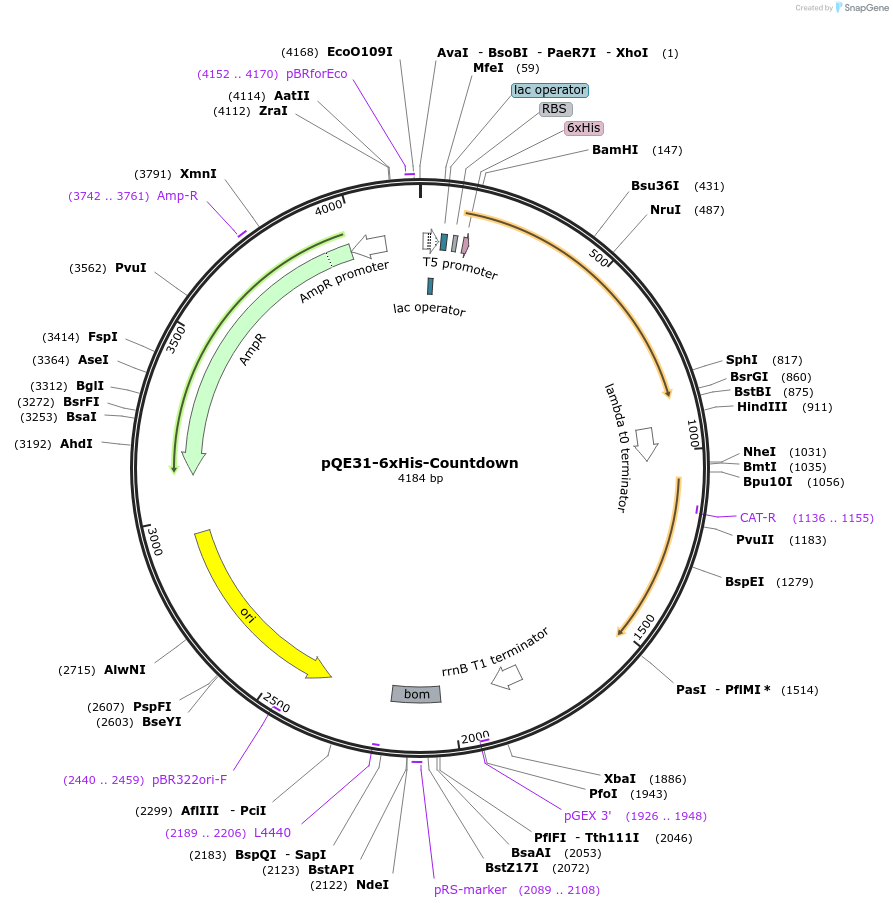
pQE31-6xHis-Countdown
Plasmid#182299PurposeE. coli plasmid encoding Countdown with a hexahistidine tagDepositorInsertCountdown
Tagshistidine tagExpressionBacterialPromoterT5Available SinceApril 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
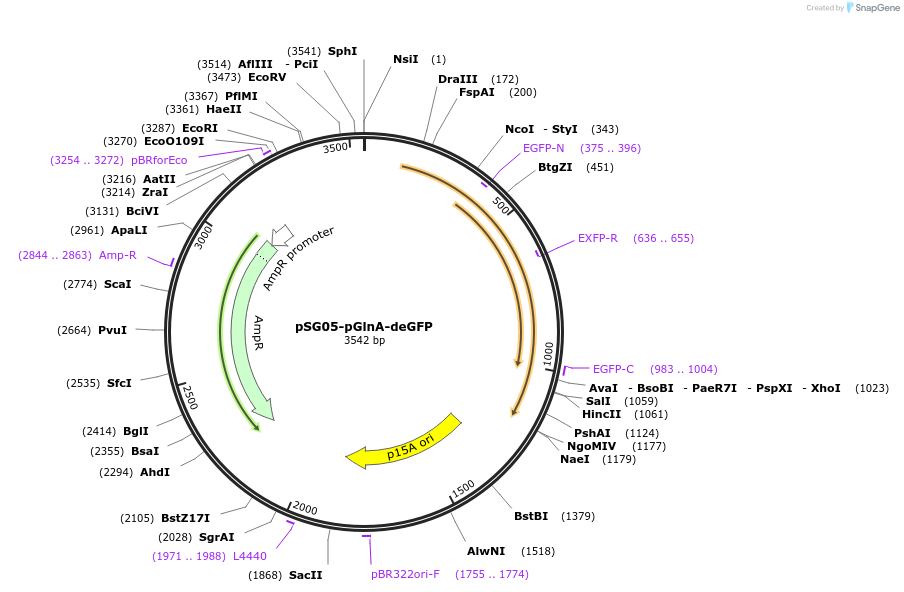
pSG05-pGlnA-deGFP
Plasmid#102462PurposeExpresses deGFP fluorescent protein in E. coli. The expression is controlled by a pGlnA promoter.DepositorArticleInsertdeGFP
ExpressionBacterialPromoterpGlnAAvailable SinceOct. 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
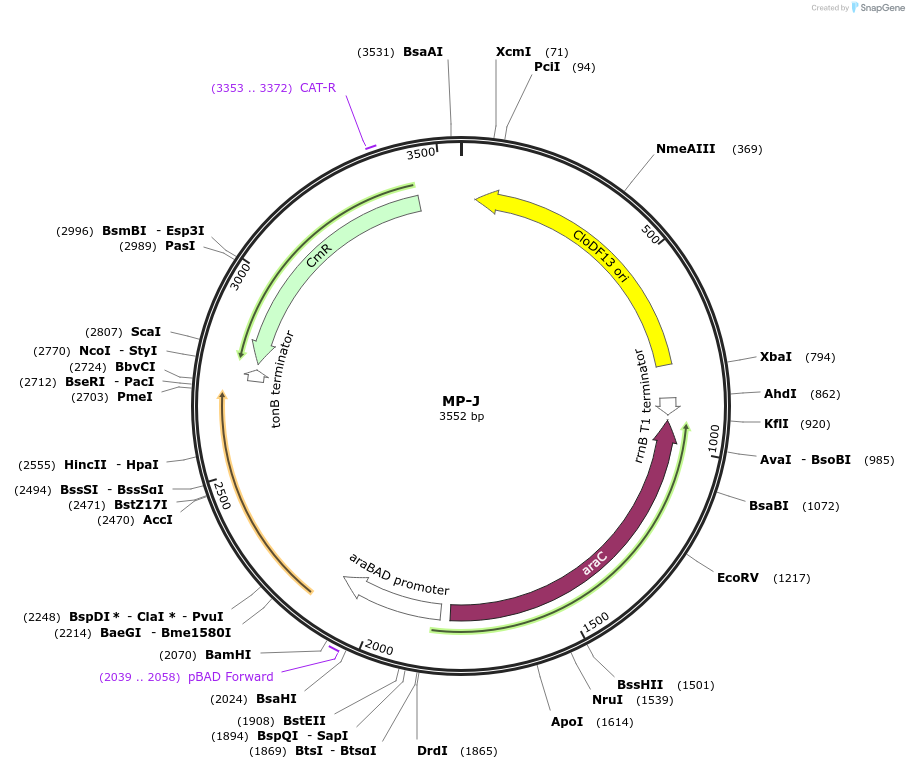
MP-J
Plasmid#69647PurposeUntargetted Bacterial MutagenesisDepositorInsertrpsD12
ExpressionBacterialPromoterE. coli PBADAvailable SinceNov. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
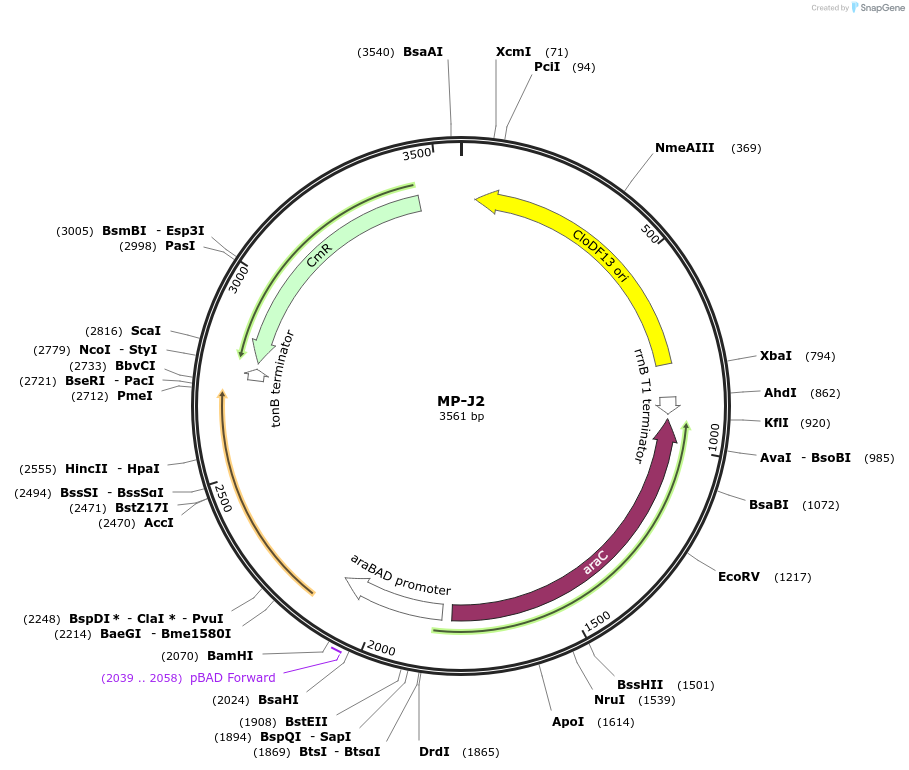
MP-J2
Plasmid#69648PurposeUntargetted Bacterial MutagenesisDepositorInsertrpsD14
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 22, 2015AvailabilityAcademic Institutions and Nonprofits only -
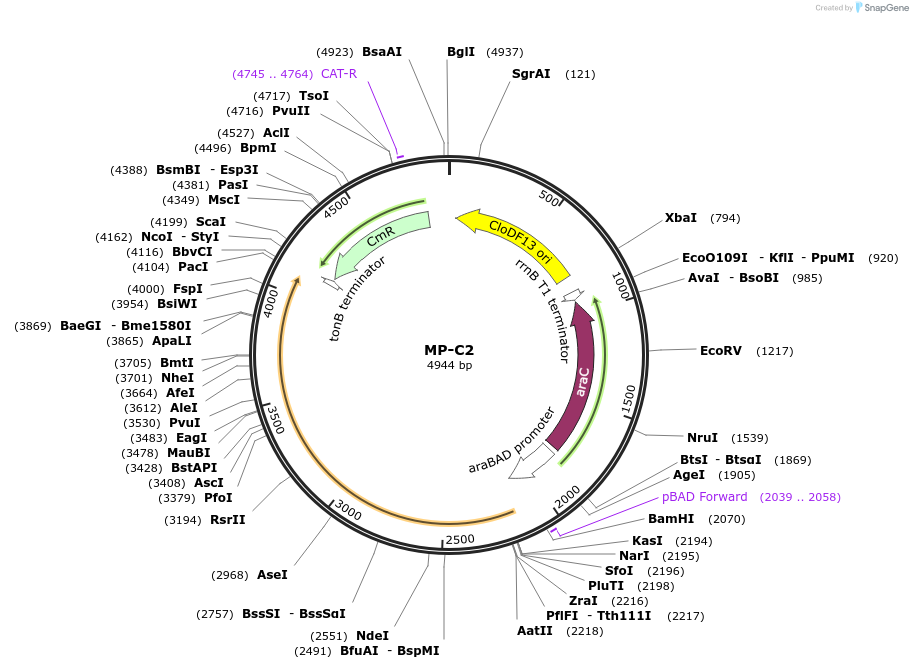
MP-C2
Plasmid#69632PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaX36
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
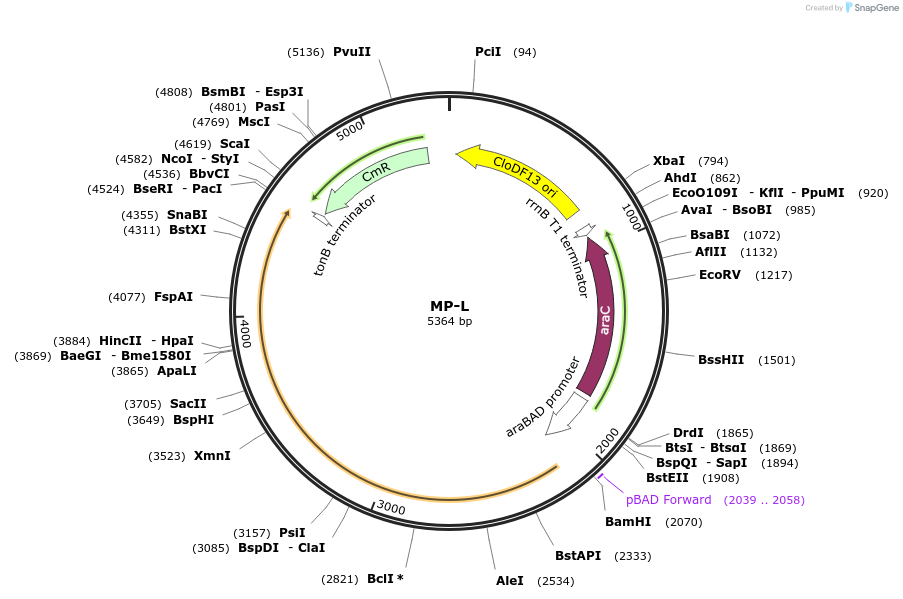
MP-L
Plasmid#69659PurposeUntargetted Bacterial MutagenesisDepositorInsertpolB
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
MP-J3
Plasmid#69649PurposeUntargetted Bacterial MutagenesisDepositorInsertrpsD16
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
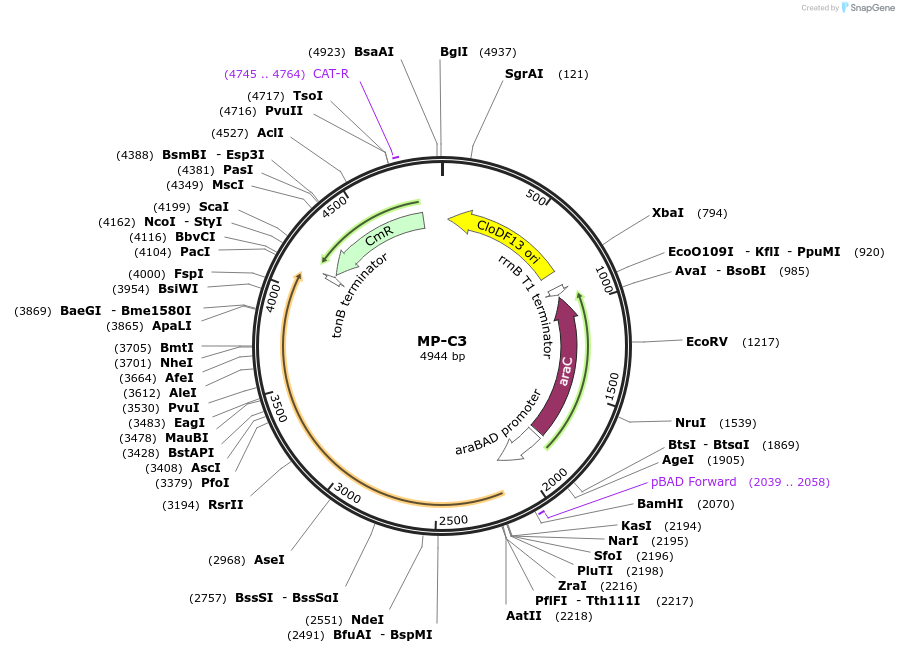
MP-C3
Plasmid#69633PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaX2016
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
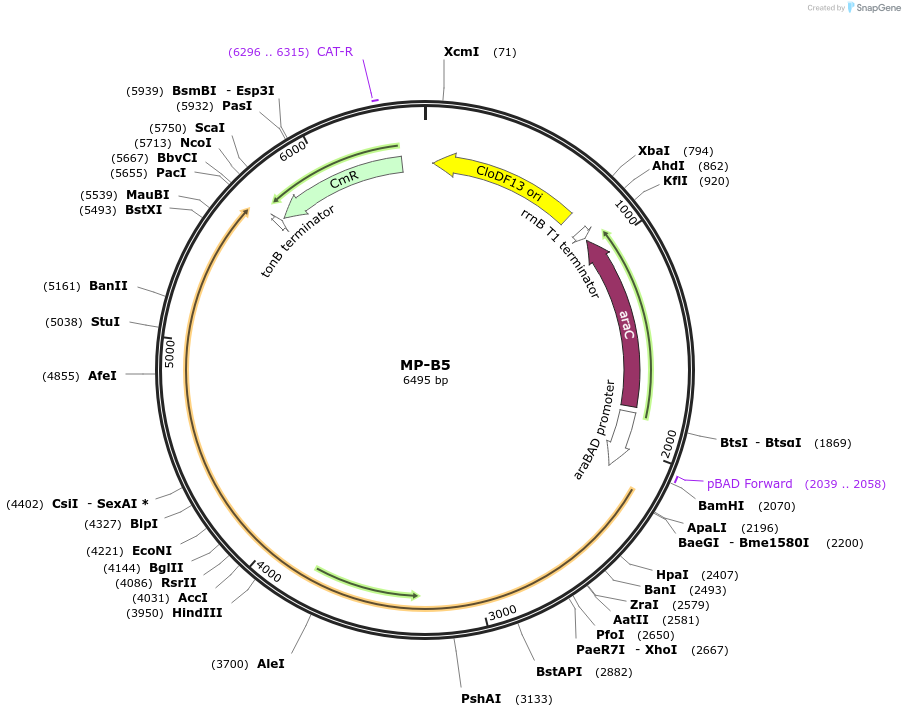
MP-B5
Plasmid#69631PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaE1026
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only