We narrowed to 7,680 results for: CCH
-
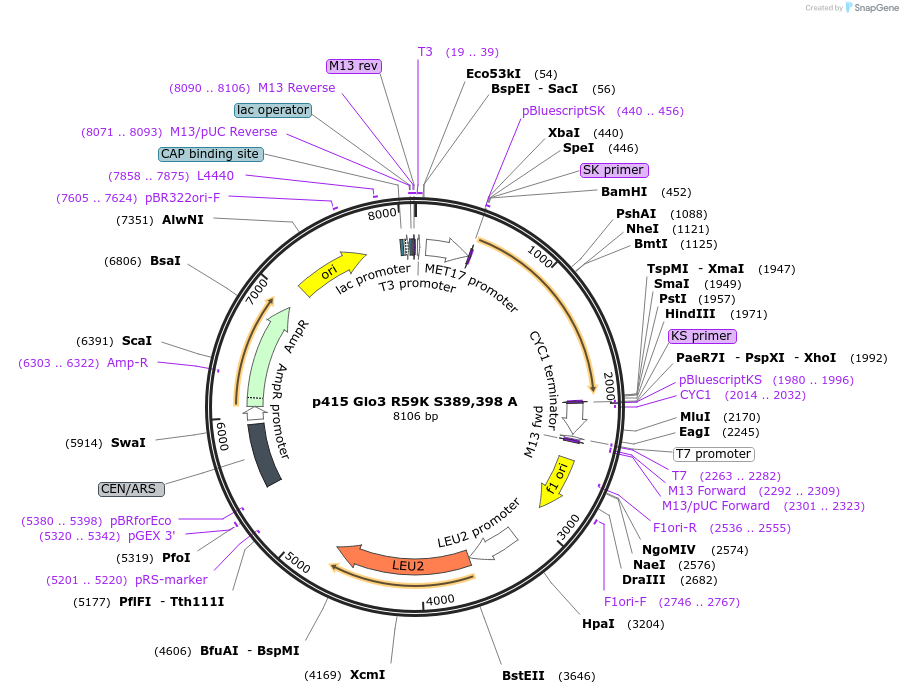
Plasmid#112656PurposeExpression S.cerevisiaeDepositorInsertGlo3 R59K S389,398A
ExpressionYeastMutationR59K, mutations (S389A, S398A) in Glo3 motif, non…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
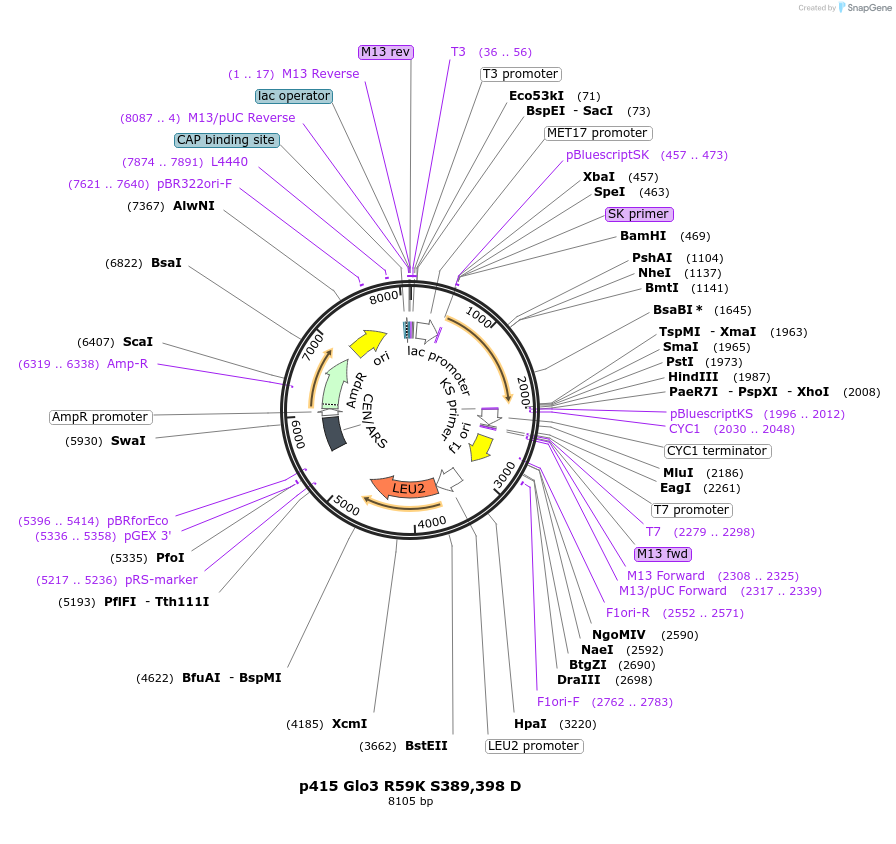
p415 Glo3 R59K S389,398 D
Plasmid#112657PurposeExpression S.cerevisiaeDepositorInsertGlo3 R59K S389,398D
ExpressionYeastMutationR59K, mutations (S389D, S398D) in Glo3 motif, pho…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
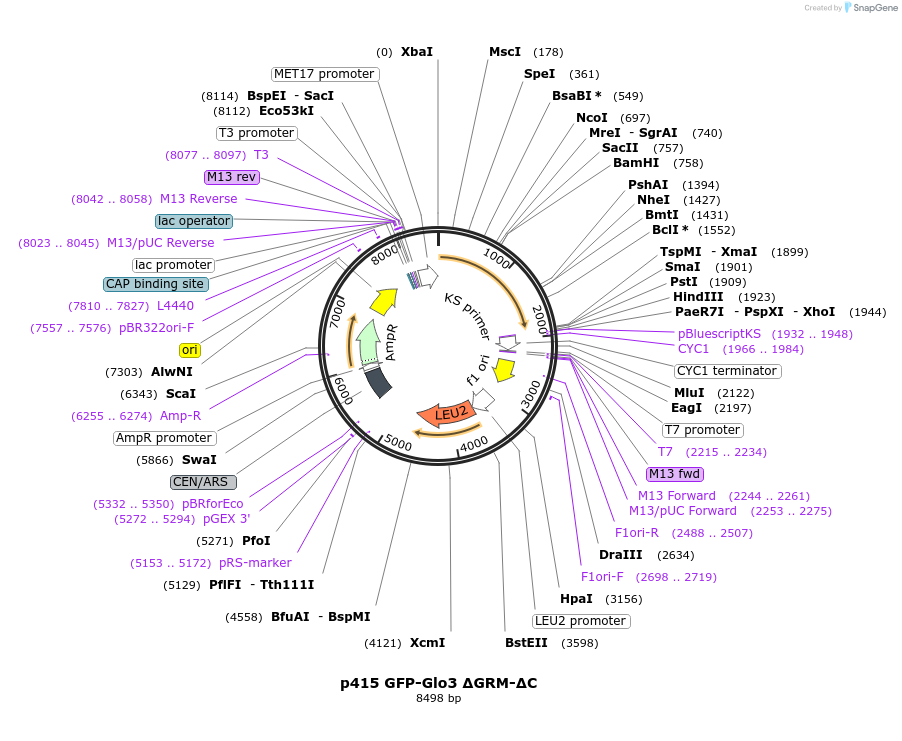
p415 GFP-Glo3 ΔGRM-ΔC
Plasmid#112659PurposeExpression S.cerevisiaeDepositorInsertGFP-Glo3 ΔGRM ΔC
TagssfGFPExpressionYeastMutationΔ376-493 (deletion of Glo3 motif & C-terminal…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
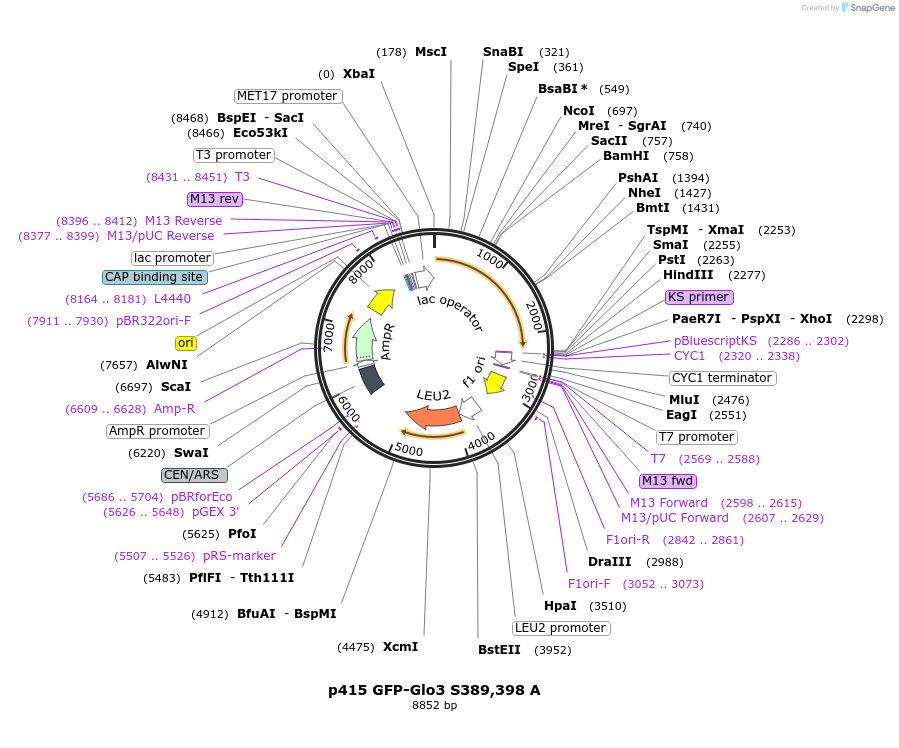
p415 GFP-Glo3 S389,398 A
Plasmid#112660PurposeExpression S.cerevisiaeDepositorInsertGFP- Glo3 S389,398A
TagssfGFPExpressionYeastMutationmutations (S389A, S398A) in Glo3 motif, non-phosp…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
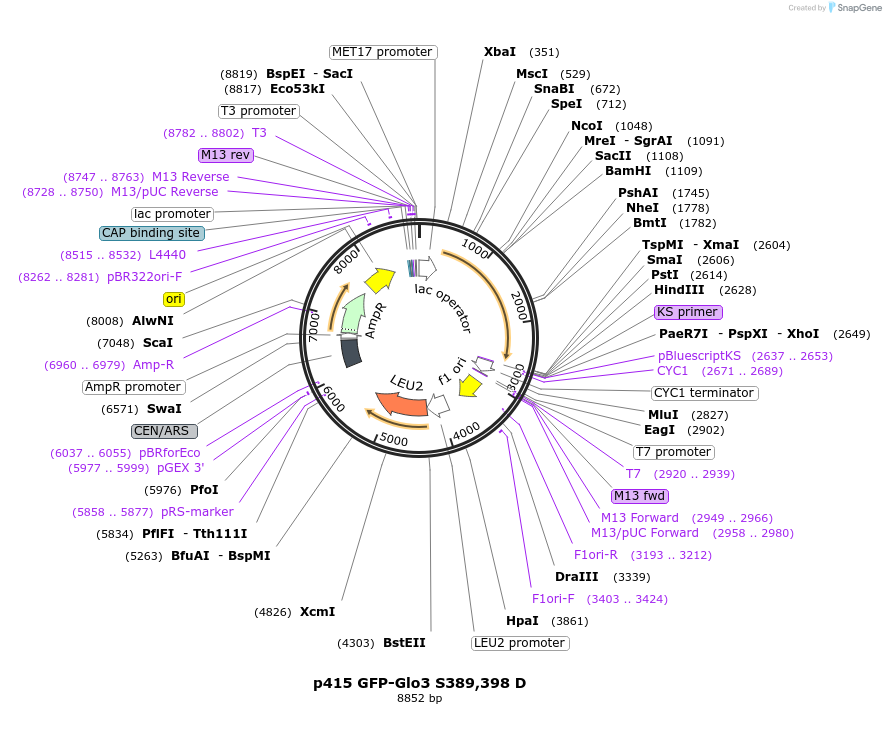
p415 GFP-Glo3 S389,398 D
Plasmid#112661PurposeExpression S.cerevisiaeDepositorInsertGFP-Glo3 S389,398D
TagssfGFPExpressionYeastMutationmutations (S389D, S398D) in Glo3 motif, phosphomi…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
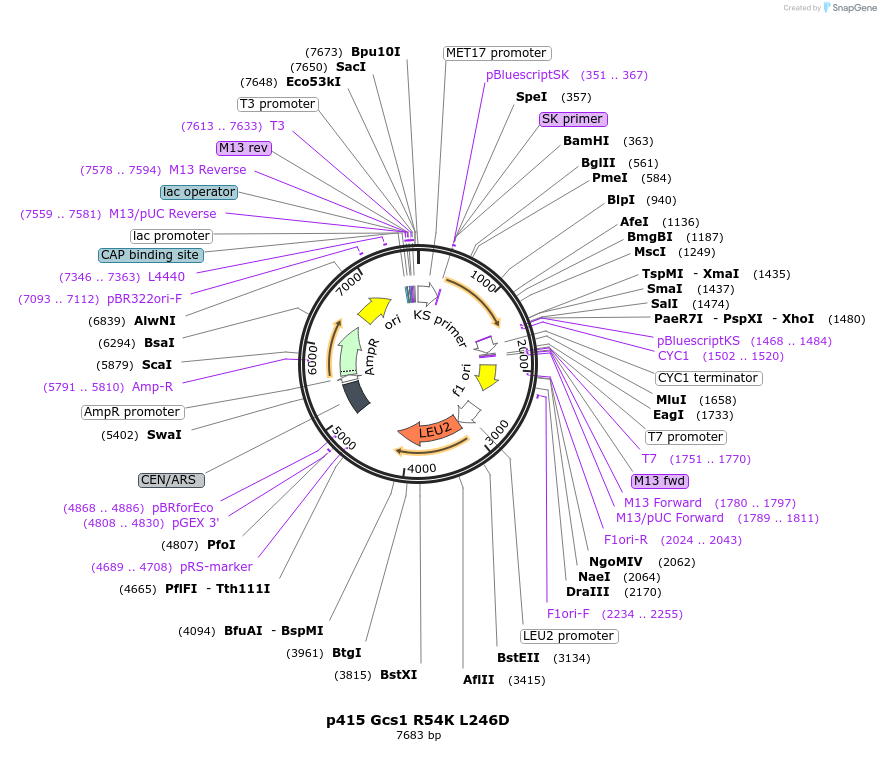
p415 Gcs1 R54K L246D
Plasmid#112648PurposeExpression S.cerevisiaeDepositorInsertGcs1 R54K L246D
ExpressionYeastMutationR54K & L246D (in ALPS motif)PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
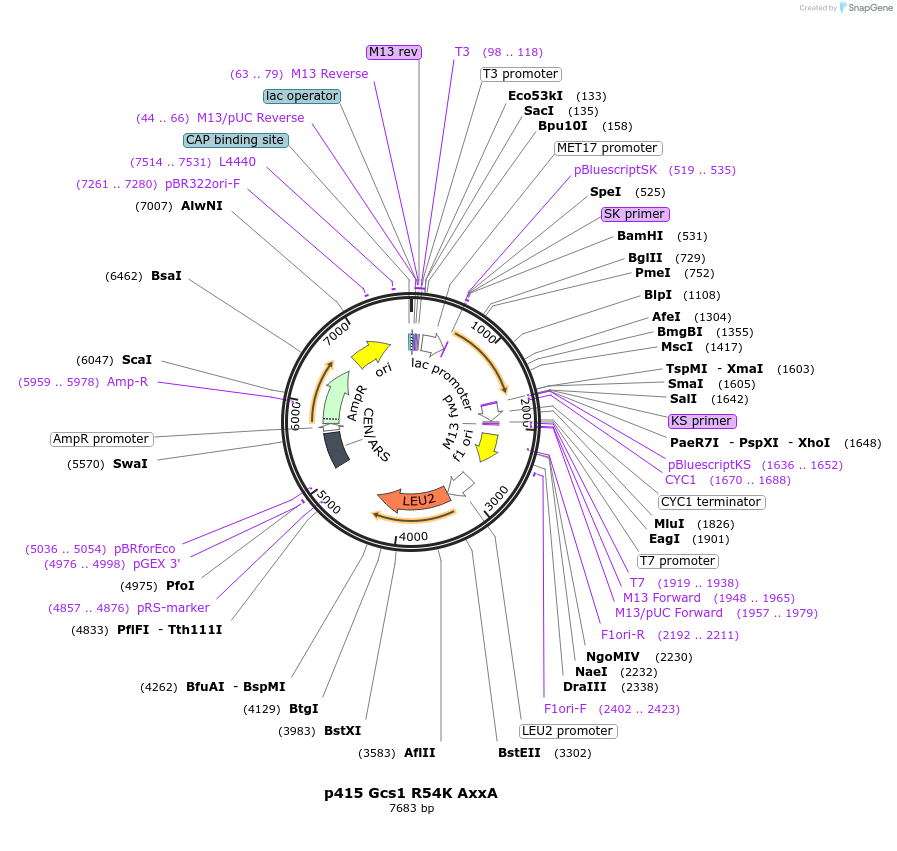
p415 Gcs1 R54K AxxA
Plasmid#112649PurposeExpression S.cerevisiaeDepositorInsertGcs1 R54K AxxA
ExpressionYeastMutationR54K & Alanine substitution of C-terminal δL …PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
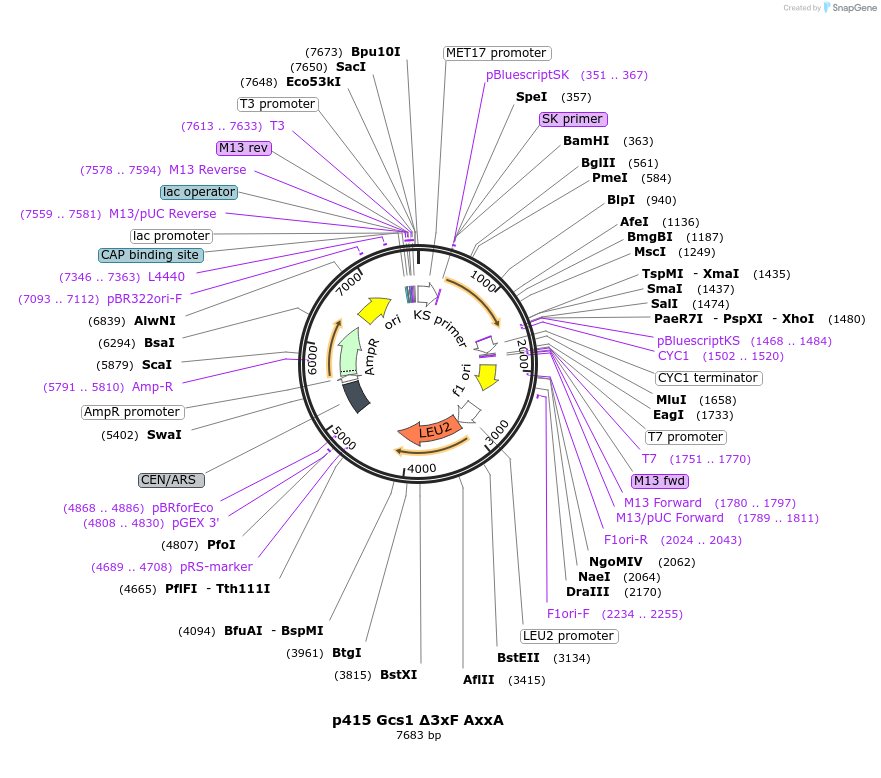
p415 Gcs1 Δ3xF AxxA
Plasmid#112650PurposeExpression S.cerevisiaeDepositorInsertGcs1 Δ3xF AxxA
ExpressionYeastMutationR54K, W304A, W307A, W310A, W349A, F353APromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
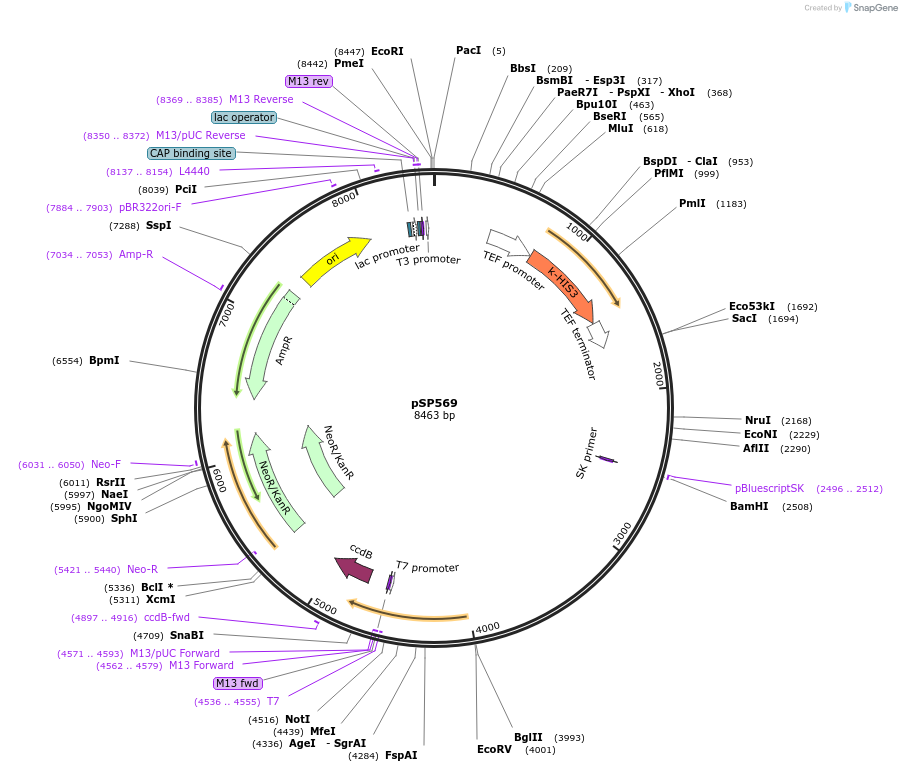
pSP569
Plasmid#139499PurposeRepair DNA plasmid to insert PP7 stem loops in the endogenous STL1 locus after double strand break in S. cerevisiaeDepositorInsertpSTL1 24xPP7sl STL1 100 - 600
ExpressionYeastMutationWTPromoterpSTL1Available SinceJune 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
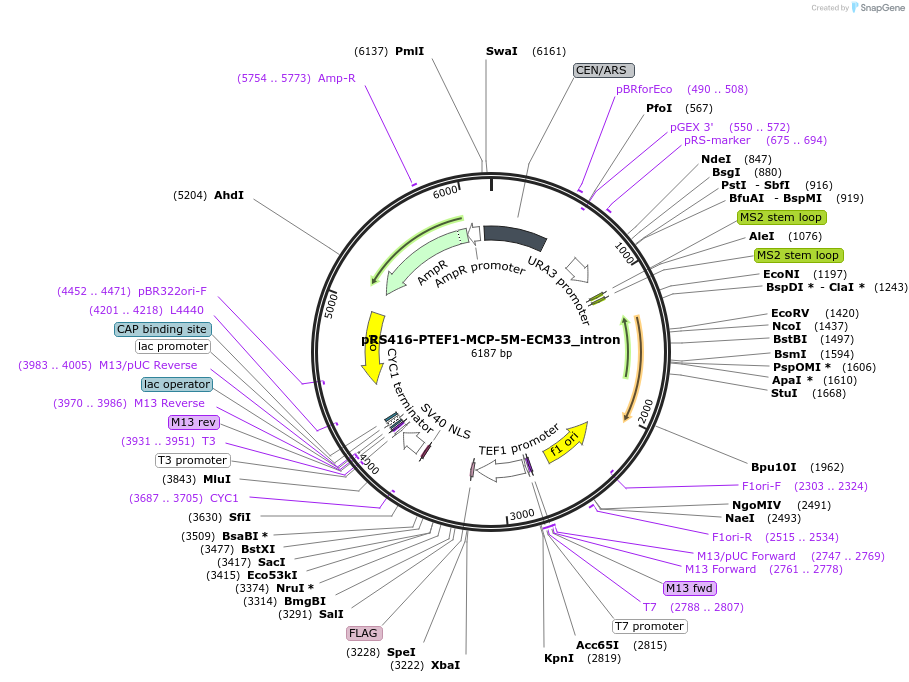
pRS416-PTEF1-MCP-5M-ECM33_intron
Plasmid#127600Purposelow-copy URA-selectable plasmid, 1xFLAG-MCP under TEF1 promoter, 5M-ECM33 intron inserted into URA3DepositorInsertECM33 intron with MS2 hairpins replacing sequence near 5′ end of intron
ExpressionYeastAvailable SinceApril 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
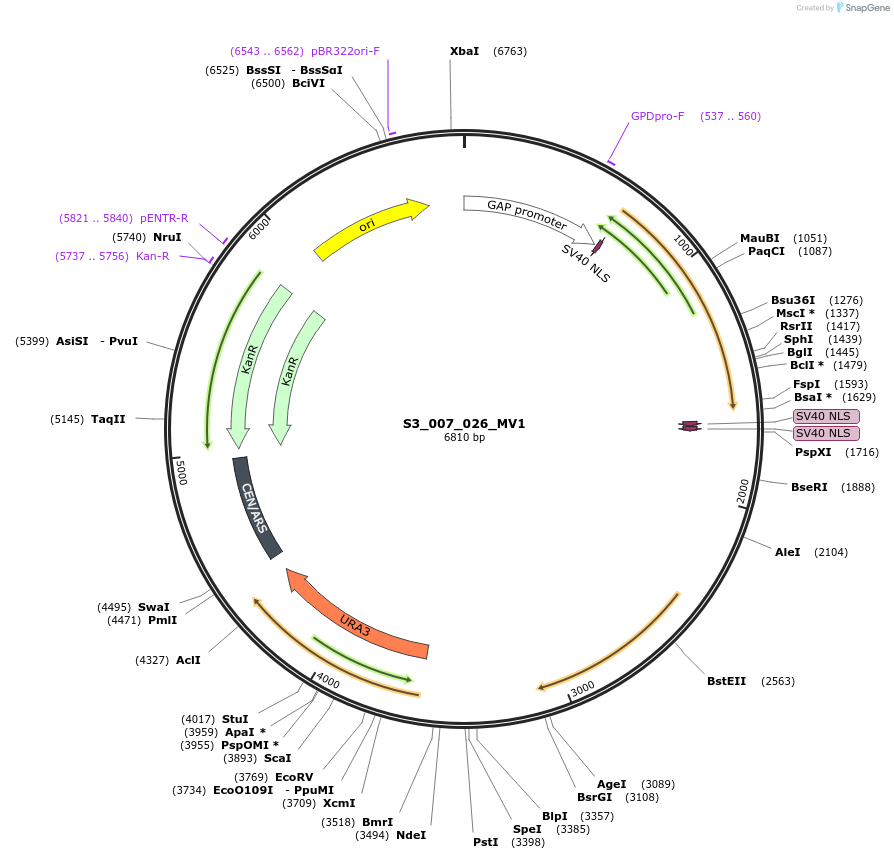
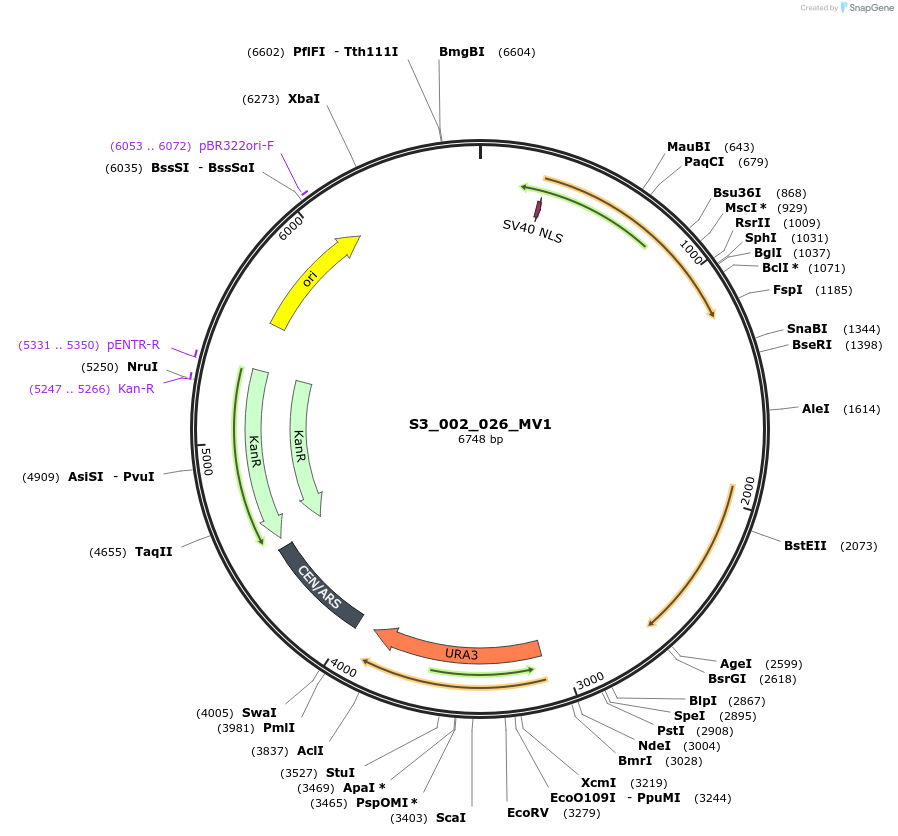
S3_007_026_MV1
Plasmid#134419PurposepYTKMV1-pTDH3_ NLS_FdeR-N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pTDH3Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
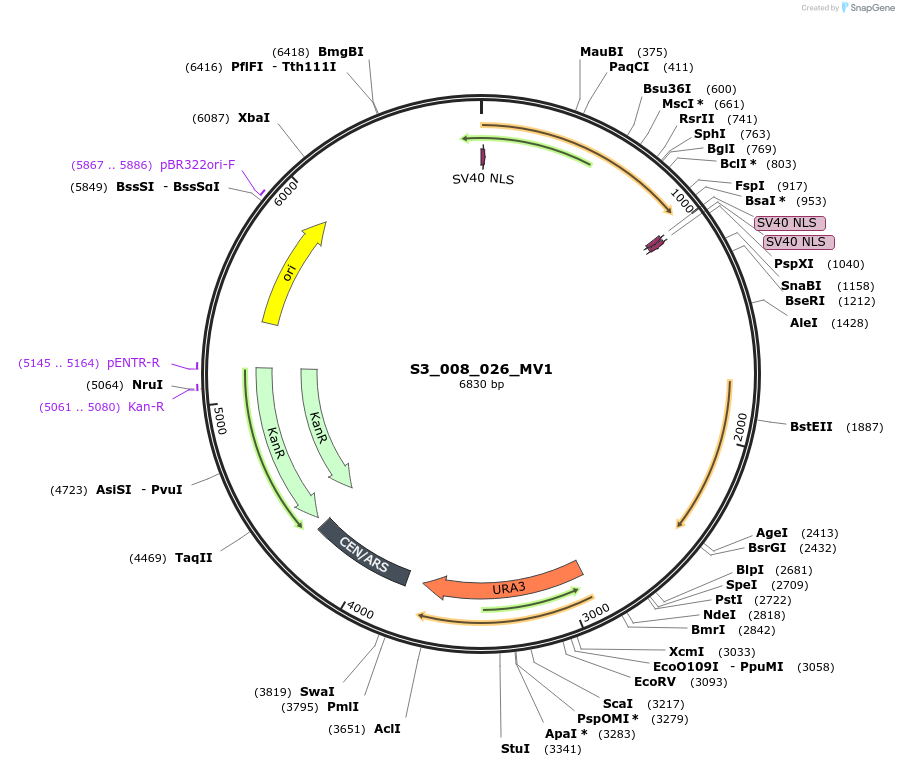
S3_008_026_MV1
Plasmid#134420PurposepYTKMV1-pRPL18B_ NLS_FdeR_N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pRPL18BAvailable SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
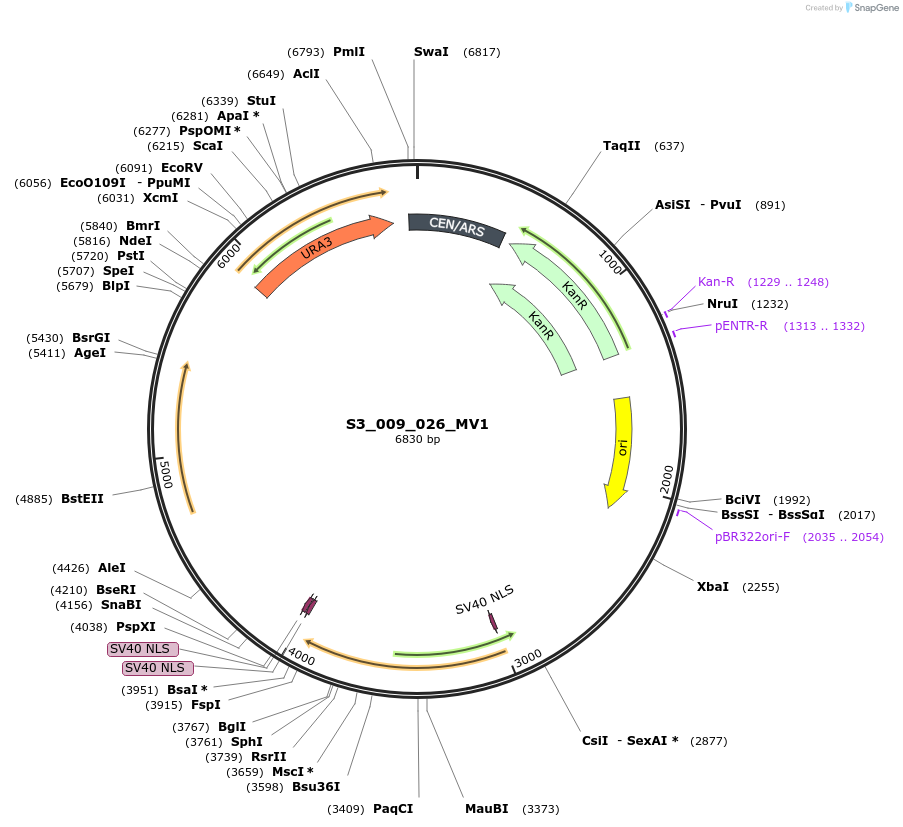
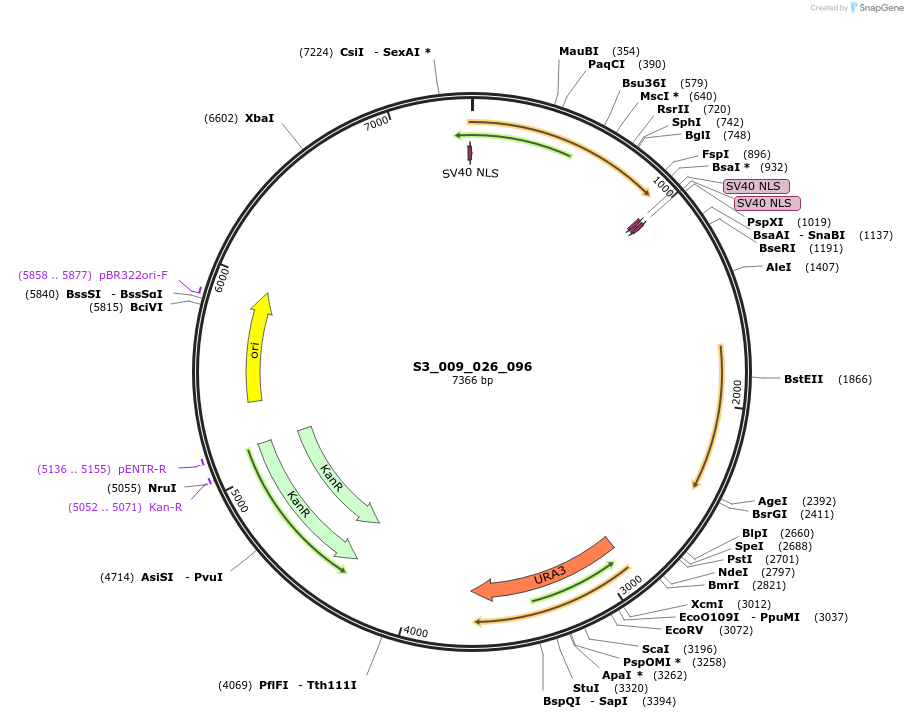
S3_009_026_MV1
Plasmid#134421PurposepYTKMV1-pREV1_ NLS_FdeR-N_2NLS-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
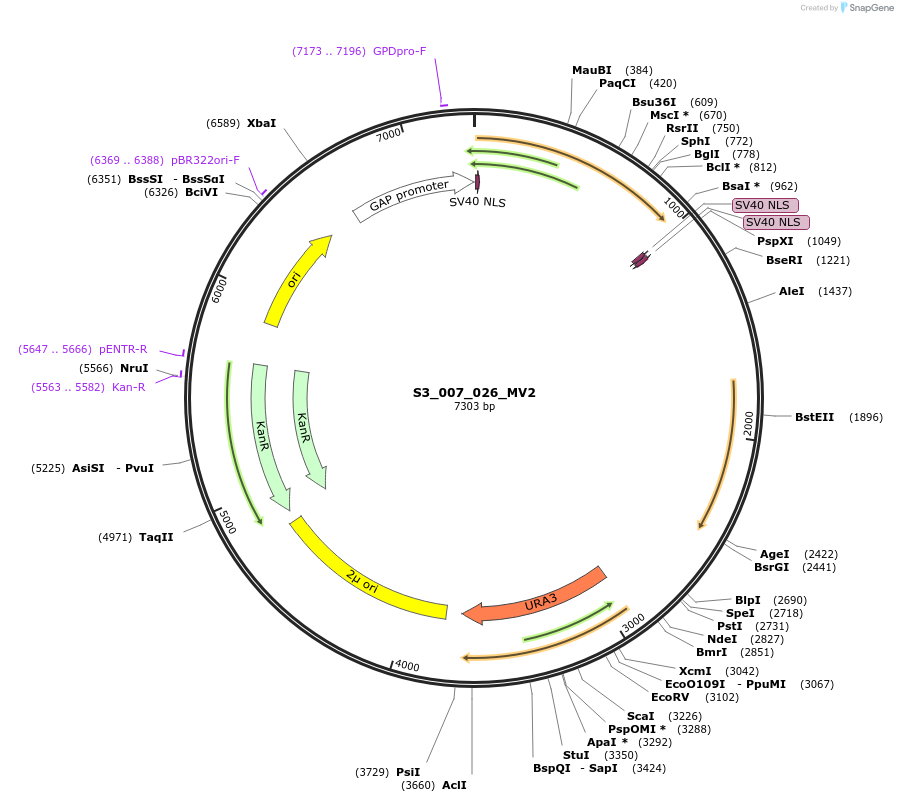
S3_007_026_MV2
Plasmid#134424PurposepYTKMV2-pTDH3_ NLS_FdeR-N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pTDH3Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
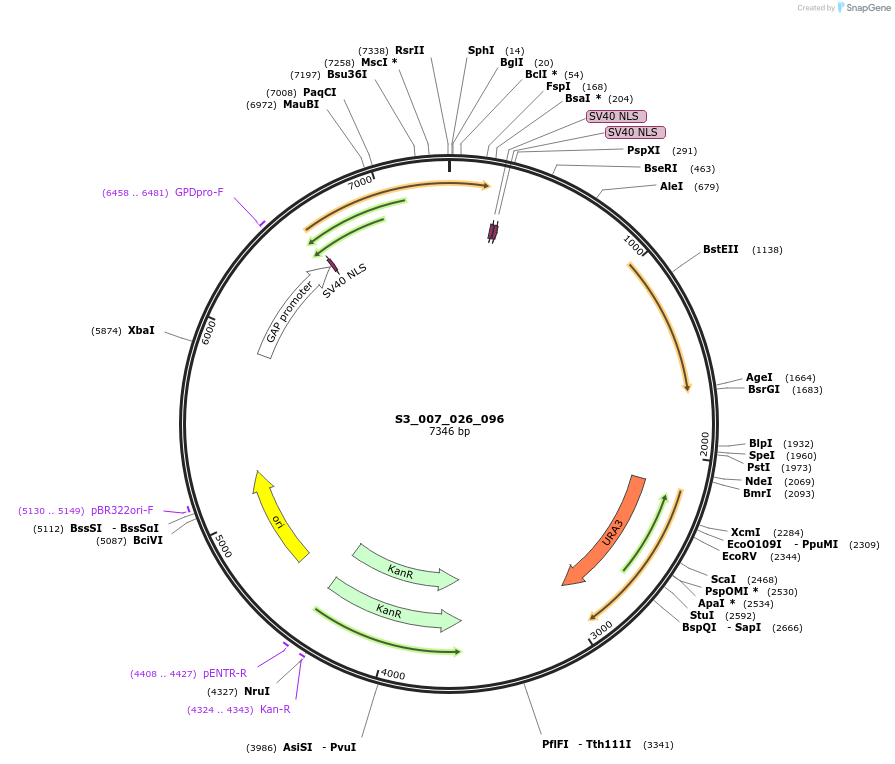
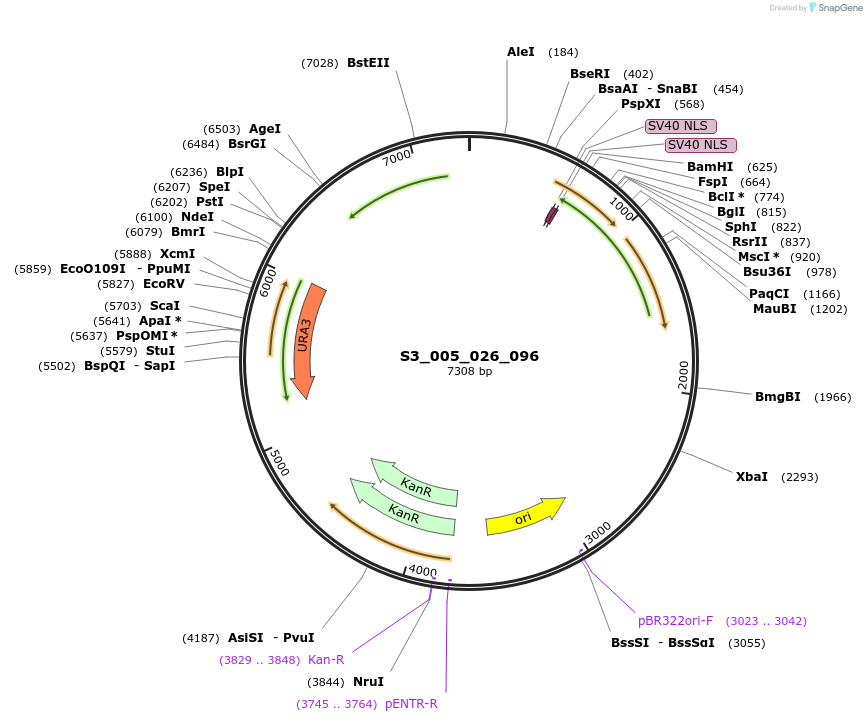
S3_007_026_096
Plasmid#134410PurposepYTK096-pTDH3_ NLS_FdeR-N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
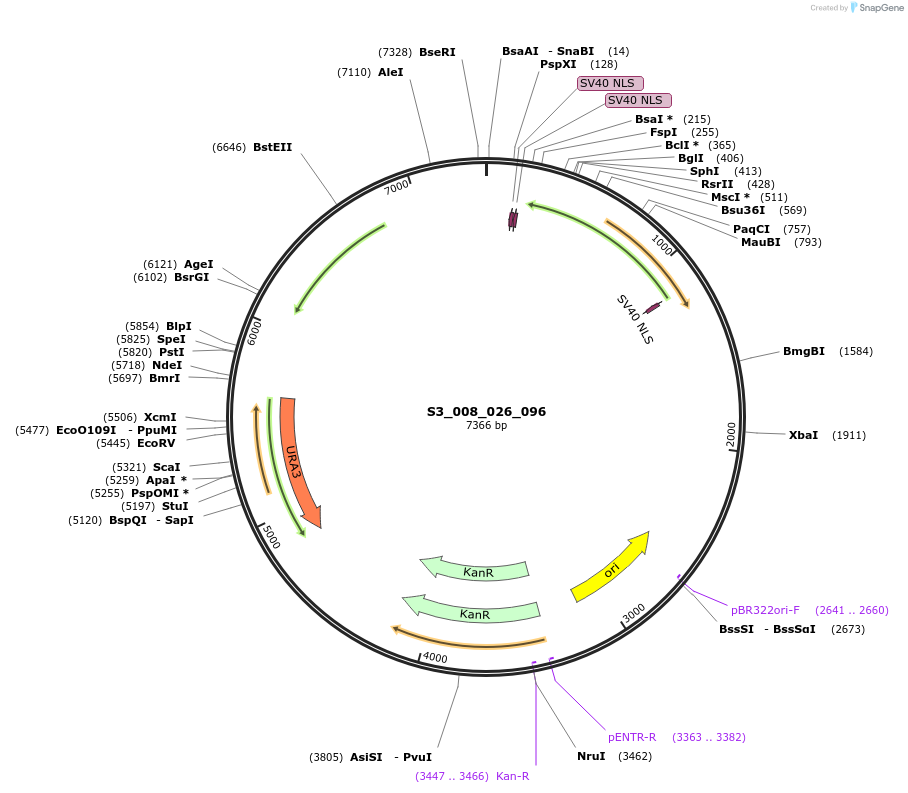
S3_008_026_096
Plasmid#134411PurposepYTK096-pRPL18B_ NLS_FdeR_N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pRPL18BAvailable SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
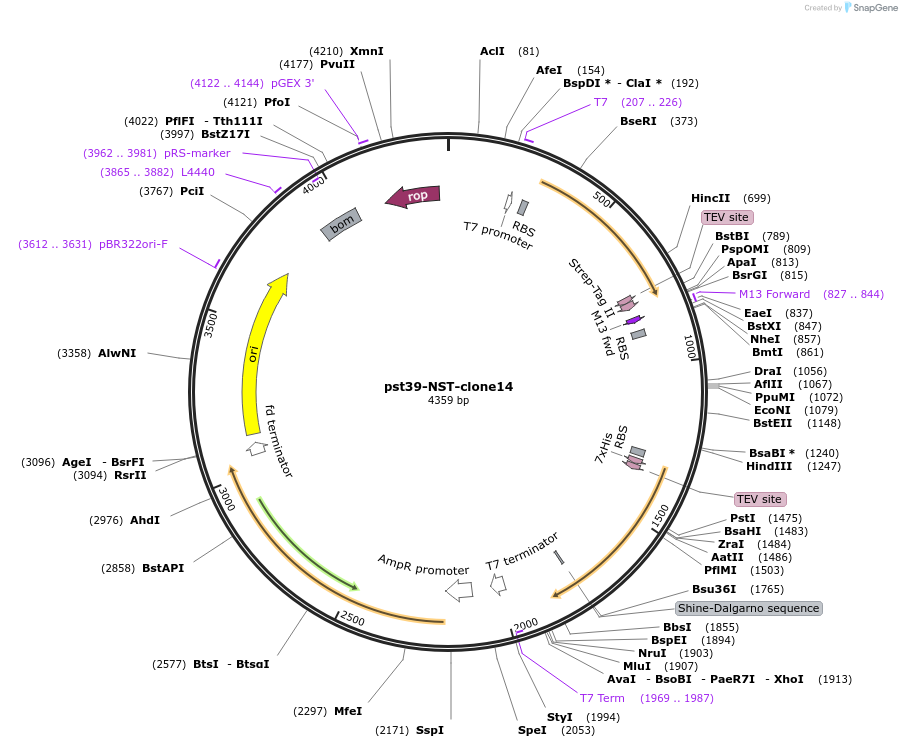
pst39-NST-clone14
Plasmid#133421PurposeS. cerevisiae Nip1 (with C-terminal TEV cleavage site and strep-II tag), Sui1 and Tif5B6 (with N-terminal 7xHis tag and TEV cleavage set) coding sequences with intervening ribosome binding sites and other cis-elements for expression as a polycistron, T7 theta 10 promoter (and corresponding terminator after polycistron).DepositorExpressionBacterialMutationORF for Nip1 encodes aa 4-156 (not complete ORF),…Available SinceJan. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
S3_009_026_096
Plasmid#134412PurposepYTK096-pREV1_ NLS_FdeR-N_2NLS-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
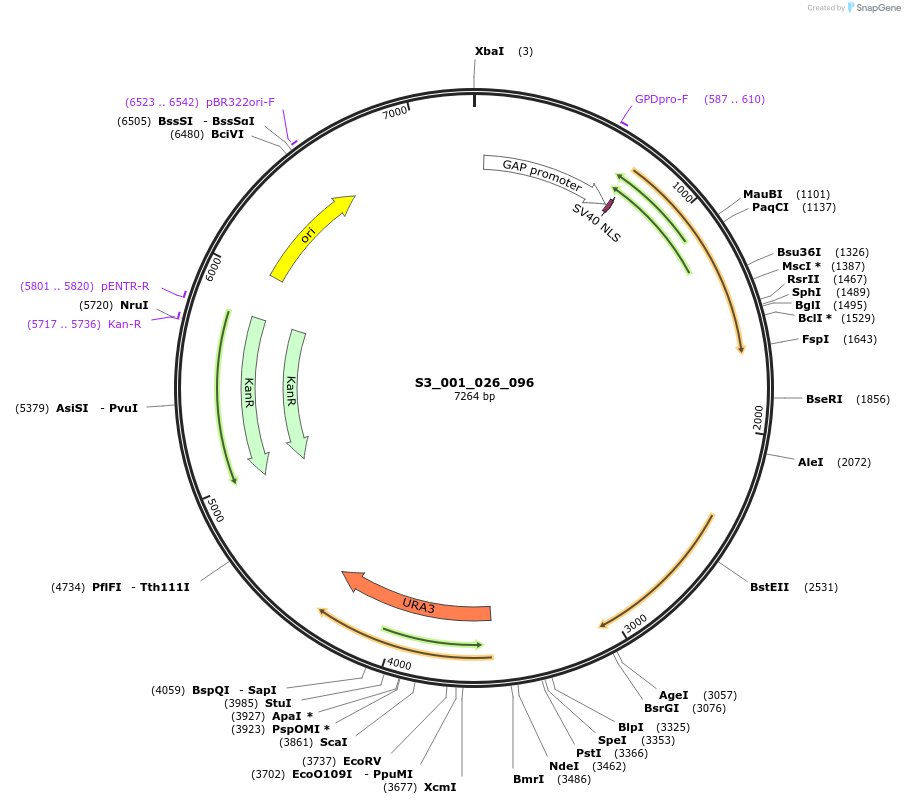
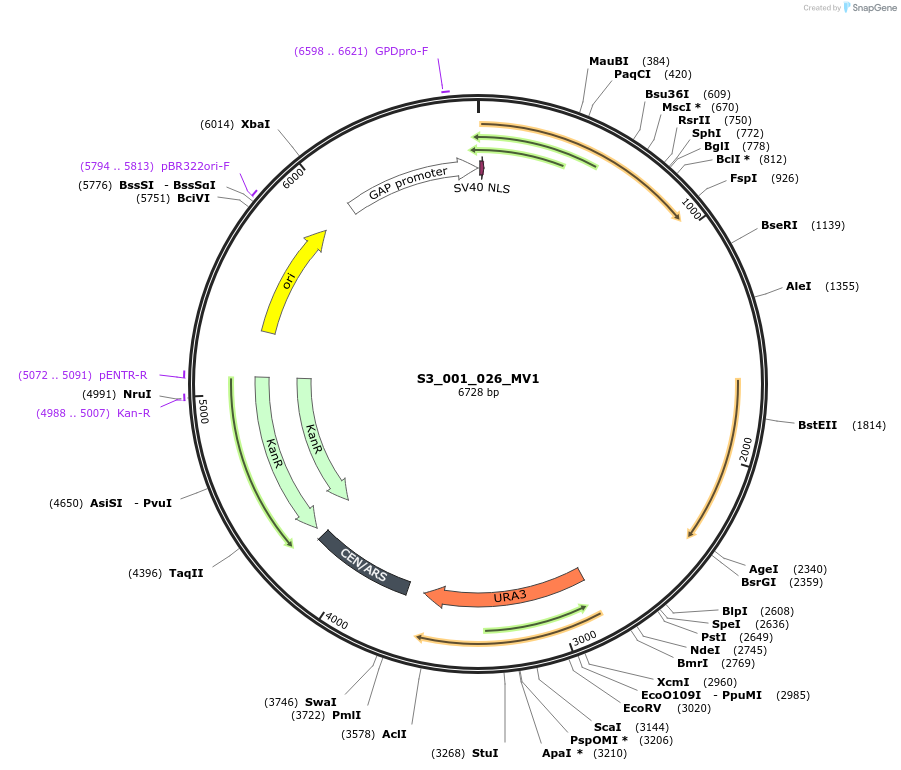
S3_001_026_096
Plasmid#134402PurposepYTK096-pTDH3_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
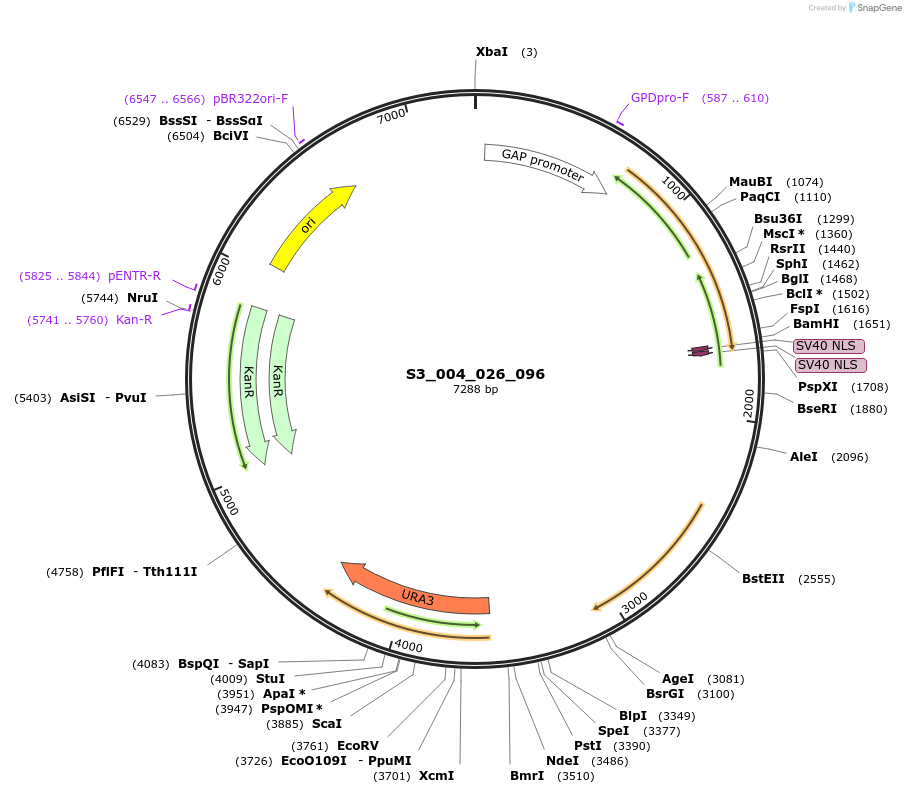
S3_004_026_096
Plasmid#134407PurposepYTK096-pTDH3_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
S3_002_026_MV1
Plasmid#134414PurposepYTKMV1-pRPL18B_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pRPL18BAvailable SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
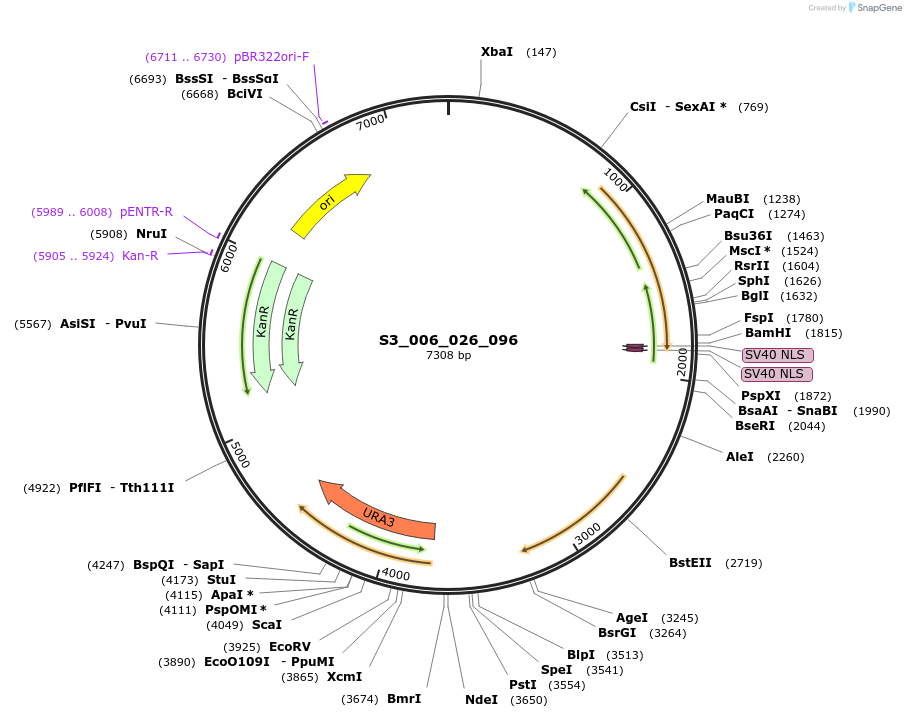
S3_006_026_096
Plasmid#134409PurposepYTK096-pREV1_FdeR-N_2NLS_tPGK1- pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pREV1Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
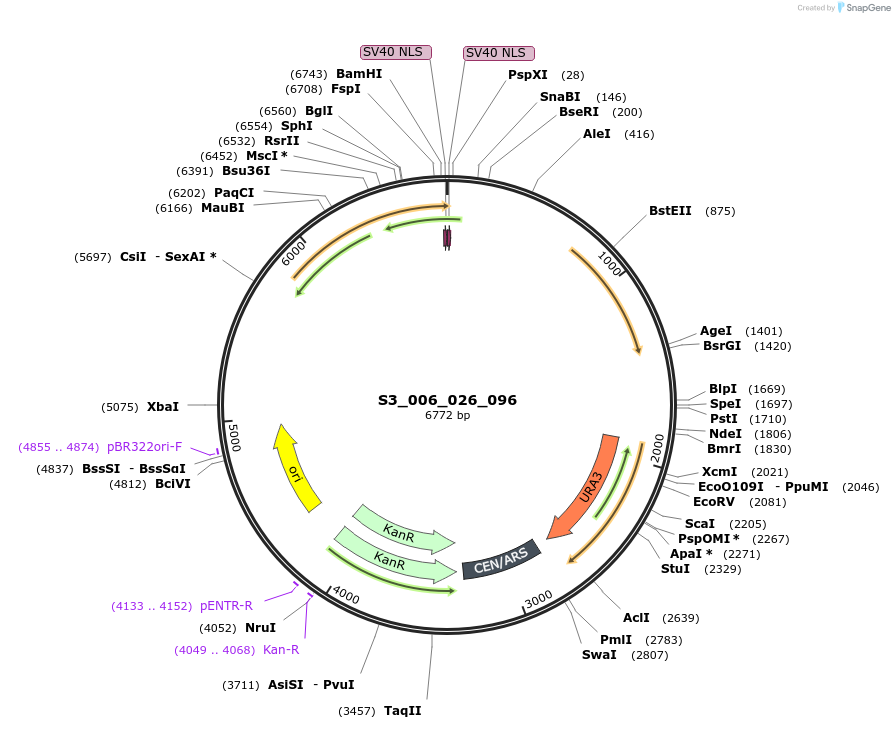
S3_006_026_096
Plasmid#134418PurposepYTK096-pREV1_FdeR-N_2NLS_tPGK1- pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
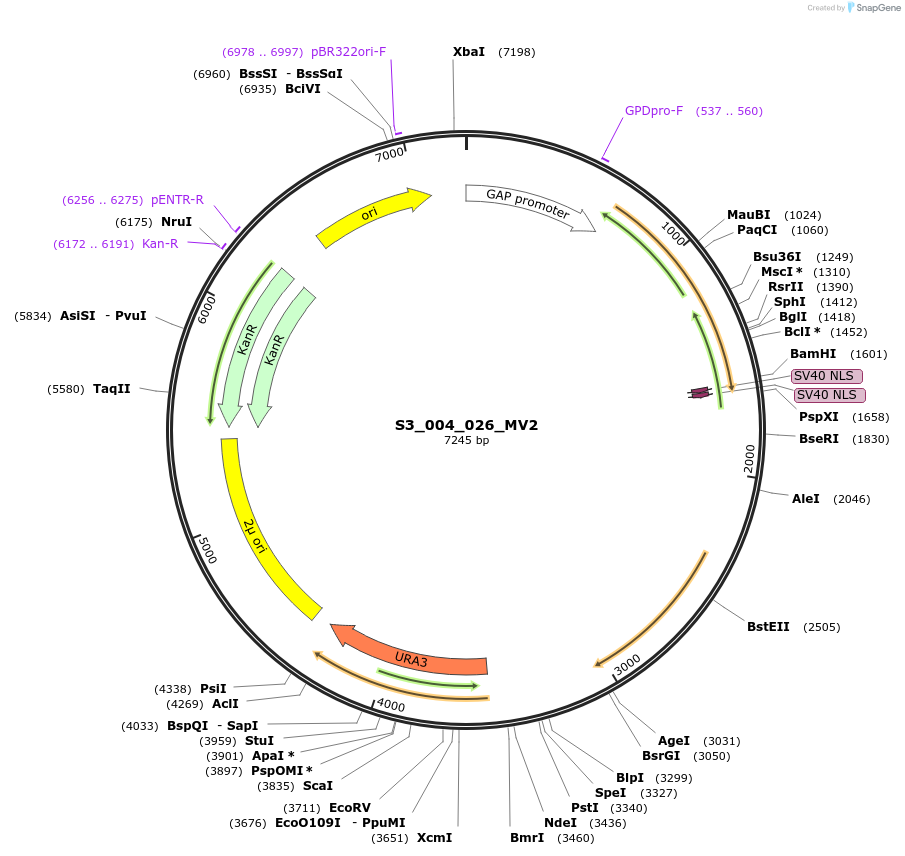
S3_004_026_MV2
Plasmid#134423PurposepYTKMV2-pTDH3_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pTDH3Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
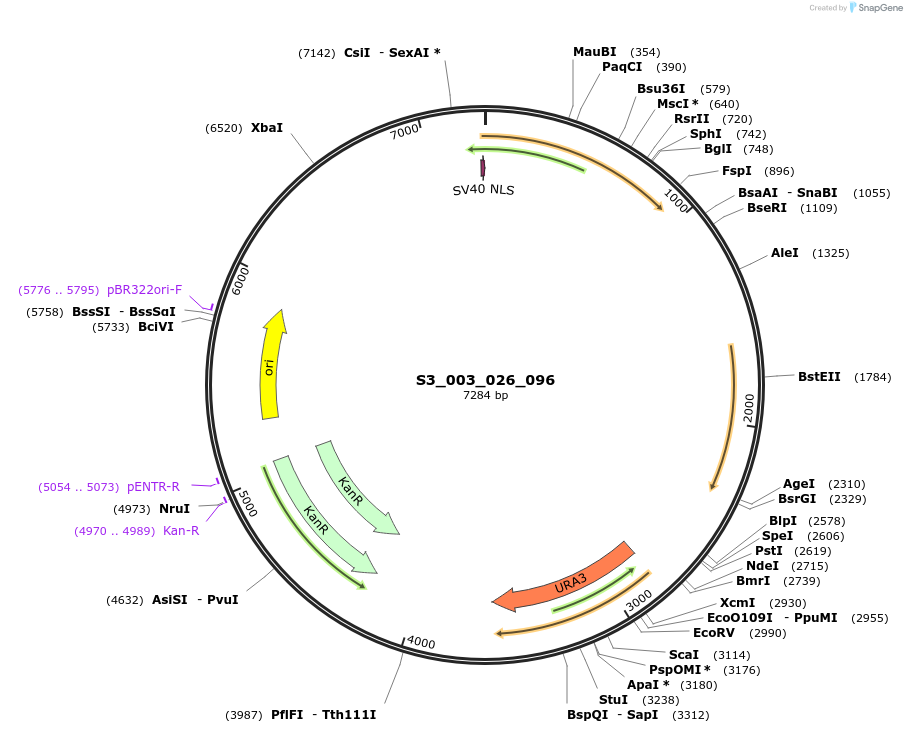
S3_003_026_096
Plasmid#134406PurposepYTK096-pREV1_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pREV1Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
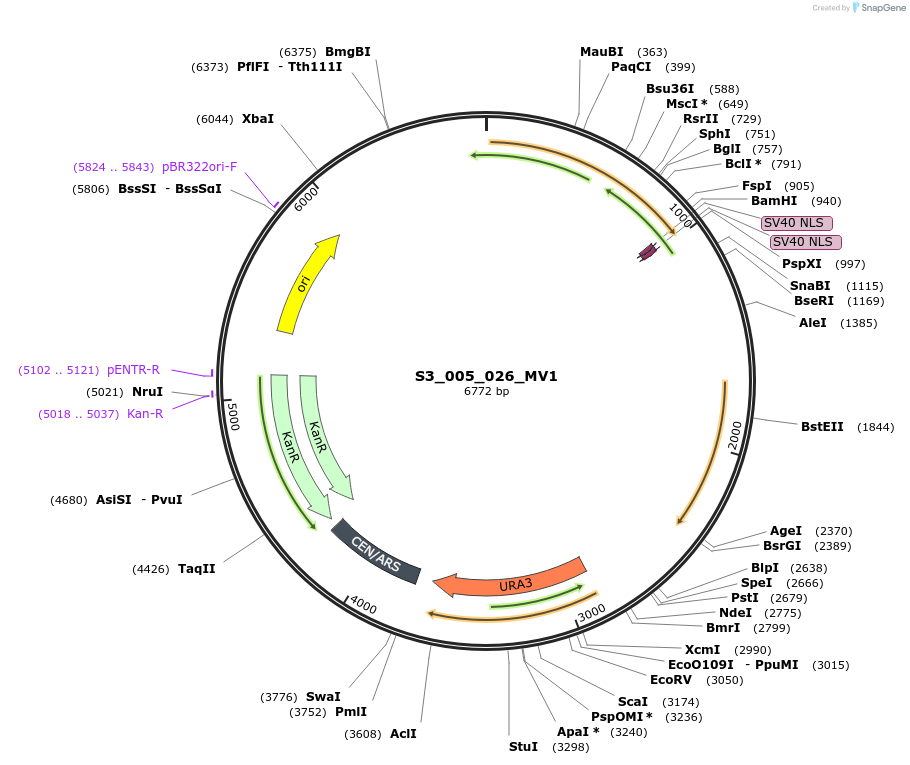
S3_005_026_MV1
Plasmid#134417PurposepYTKMV1-pRPL18B_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pRPL18BAvailable SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
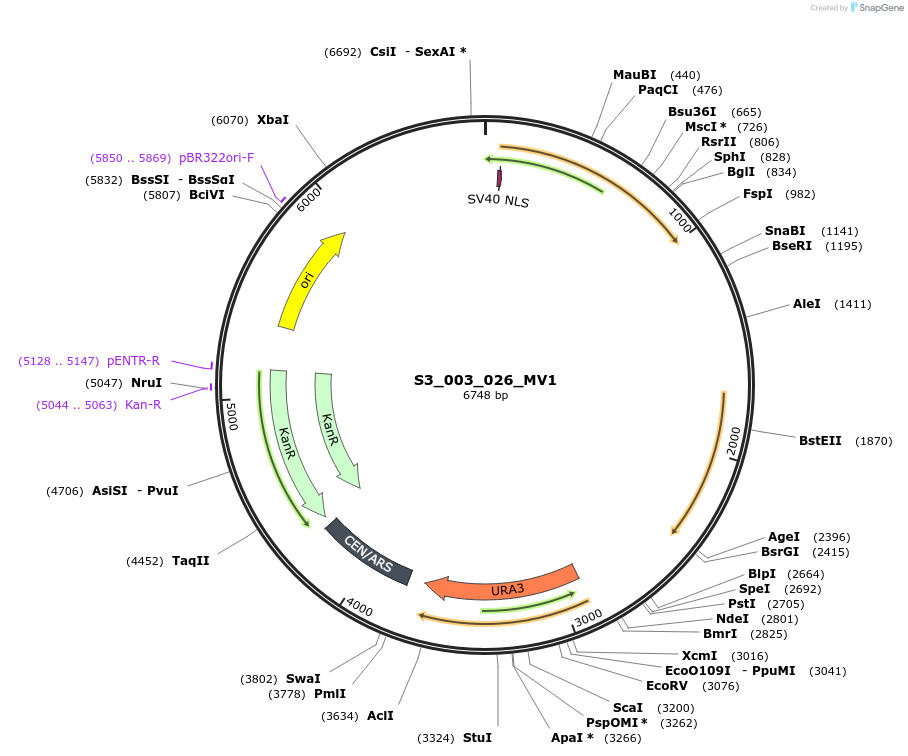
S3_003_026_MV1
Plasmid#134415PurposepYTKMV1-pREV1_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
S3_001_026_MV1
Plasmid#134413PurposepYTKMV1-pTDH3_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
S3_005_026_096
Plasmid#134408PurposepYTK096-pRPL18B_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pRPL18BAvailable SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
pGS-BacA-21222-FLP-3xFLAG
Plasmid#109165PurposeEncodes yeast full-length Flp with C-terminal 3xFLAG tag to be expressed in a baculovirus/insect cell expression systemDepositorInsertFlp
Tags3xFLAGExpressionInsectPromoterPolyhedrinAvailable SinceApril 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
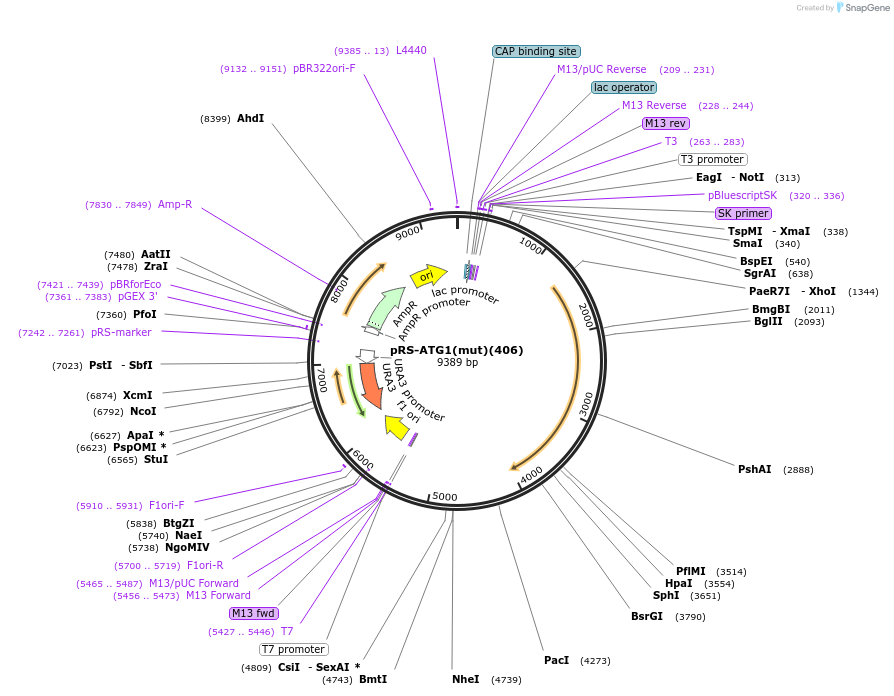
pRS-ATG1(mut)(406)
Plasmid#122069PurposeExpressing a mutant ATG1DepositorInsertATG1 (ATG1 Budding Yeast)
ExpressionYeastMutationCodon changes without changing the amino acid seq…Available SinceApril 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
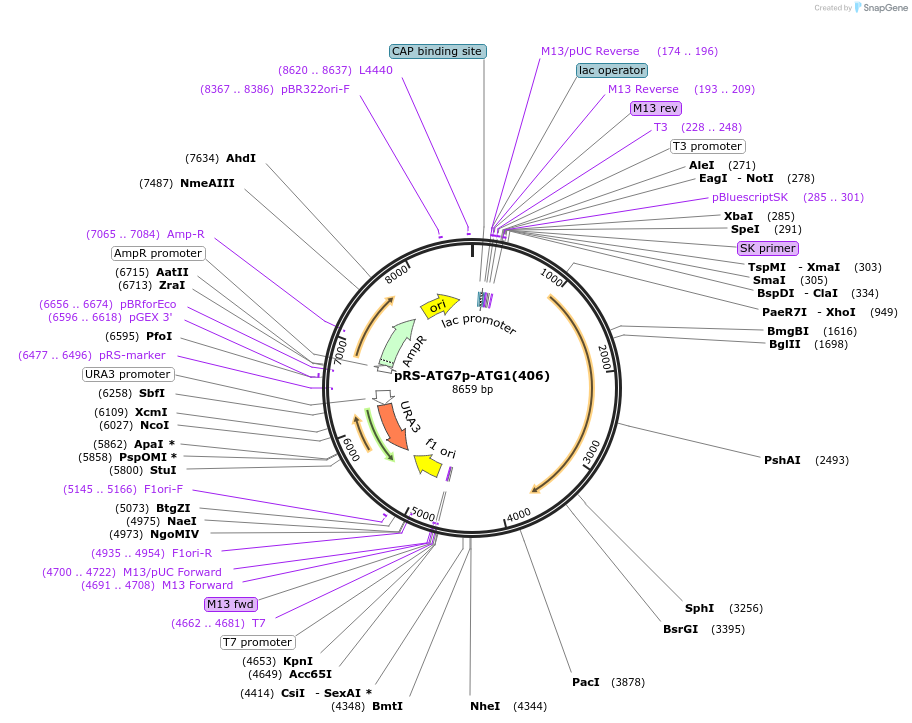
pRS-ATG7p-ATG1(406)
Plasmid#122071PurposeExpressing ATG1DepositorInsertATG1 (ATG1 Budding Yeast)
ExpressionYeastMutationThe native promoter was replaced by ATG7 promoter…Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
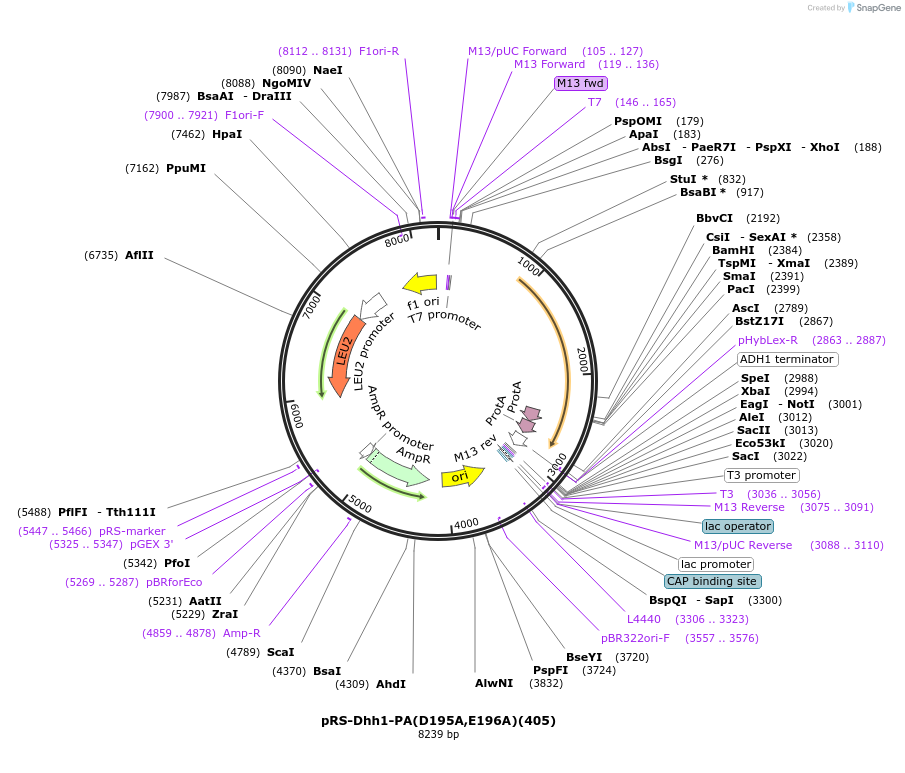
pRS-Dhh1-PA(D195A,E196A)(405)
Plasmid#122065PurposeExpressing Dhh1-PADepositorInsertDhh1-PA (DHH1 Budding Yeast)
TagsPAExpressionYeastMutationAspartic acid 195 to alanine; Glutamic acid 196 t…Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
pRS-Dhh1-PA(S226A,T228A)(405)
Plasmid#122066PurposeExpressing Dhh1-PADepositorInsertDhh1-PA (DHH1 Budding Yeast)
TagsPAExpressionYeastMutationSerine 226 to alanine; Threonine 228 to alanineAvailable SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
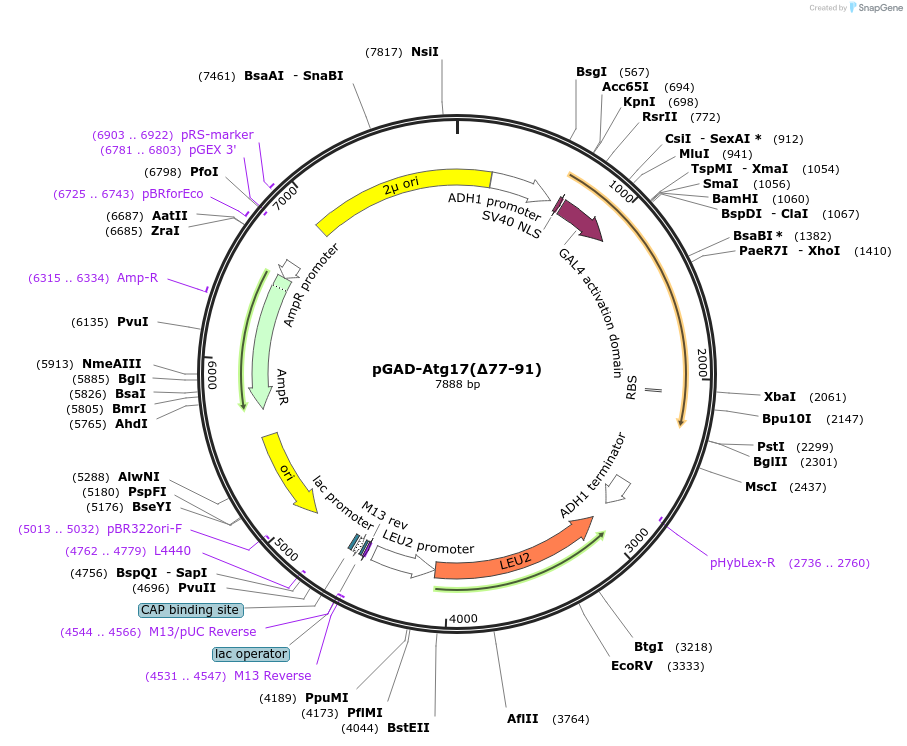
pGAD-Atg17(∆77-91)
Plasmid#121516PurposeFor Yeast-two-hybrid analysisDepositorInsertATG17
ExpressionYeastMutationdeleted amino acids 77-91Available SinceMarch 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
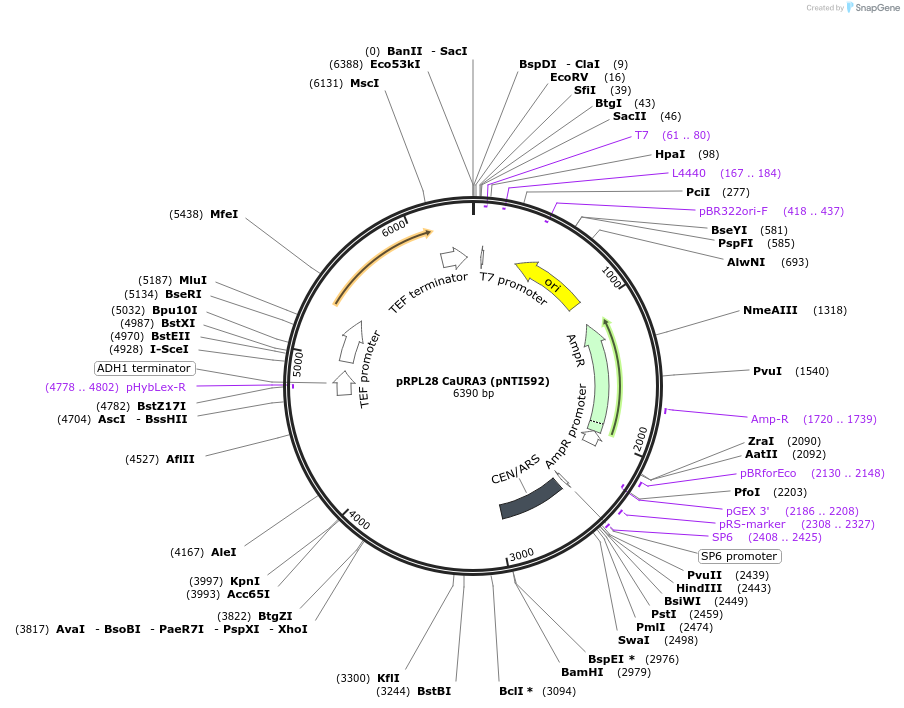
pRPL28 CaURA3 (pNTI592)
Plasmid#115431PurposeURA3-marked RPS28DepositorInsertsExpressionYeastMutationKpnI site in intronPromoterAshbya gossypii and endogenousAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
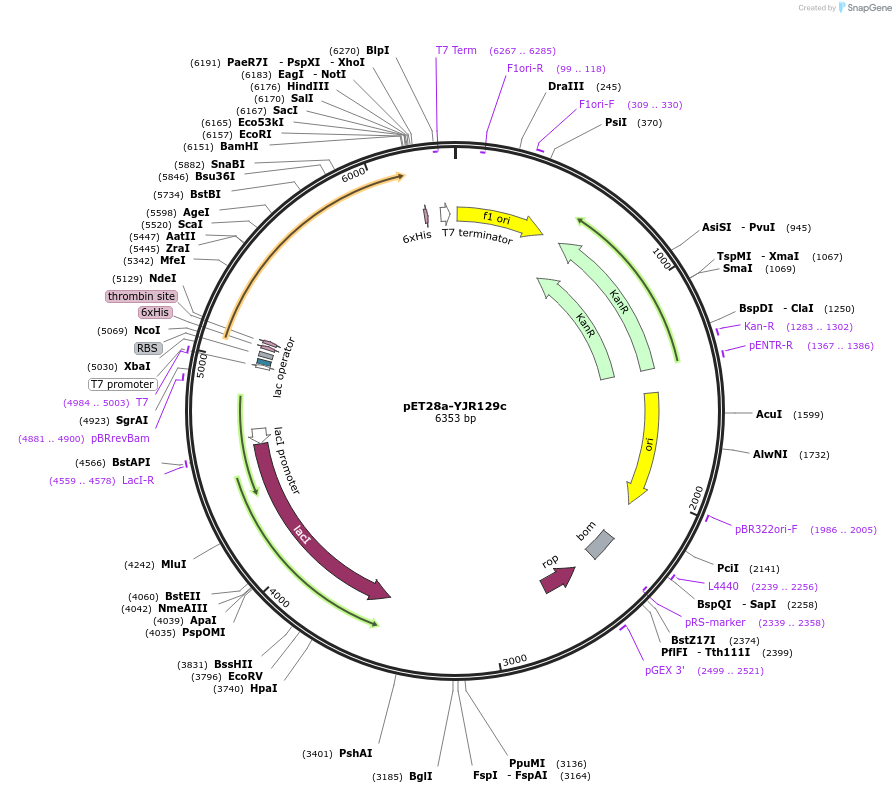
pET28a-YJR129c
Plasmid#85118Purposebacterial expression of YJR129cDepositorAvailable SinceNov. 12, 2018AvailabilityAcademic Institutions and Nonprofits only -
Spt2-K166R-GFP
Plasmid#115573PurposeExpresses yeast Spt2-GFP fusion protein mutated from lysine to arginine at site 166DepositorInsertSPT2
TagsGFPExpressionBacterial and YeastMutationLysine 166 mutated to ArginineAvailable SinceOct. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
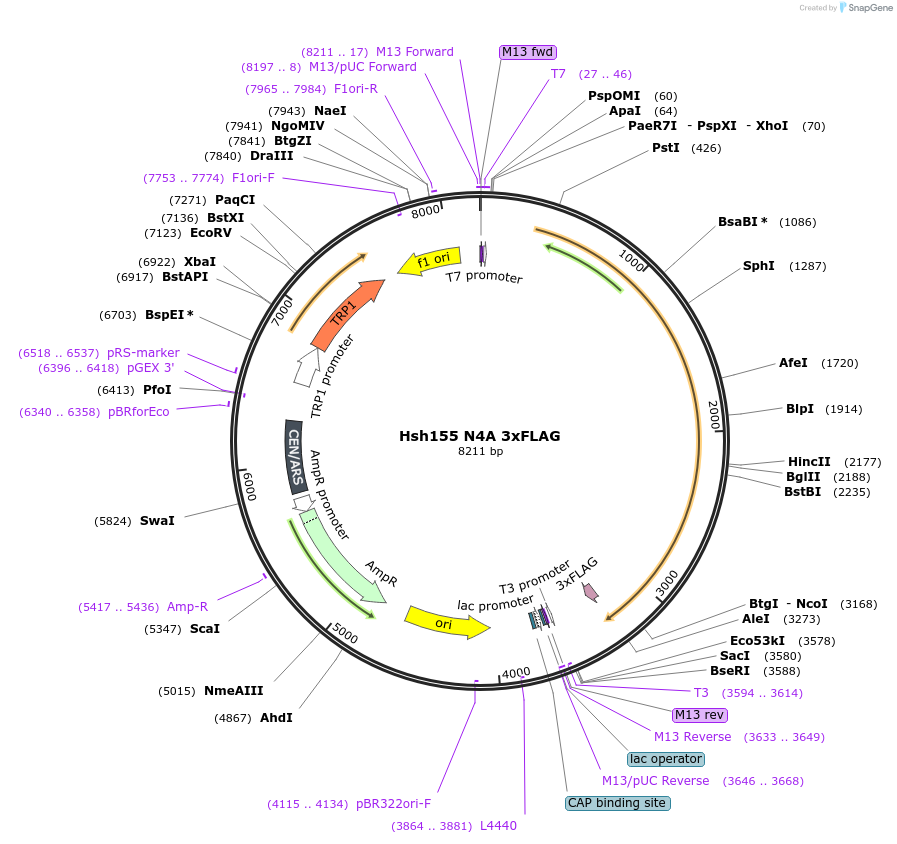
Hsh155 N4A 3xFLAG
Plasmid#111975PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker. It has 3xFLAG at the C-terminus. Contains the N4A mutation.DepositorInsertHsh155 N4A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationT178A R181A R182A R186AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
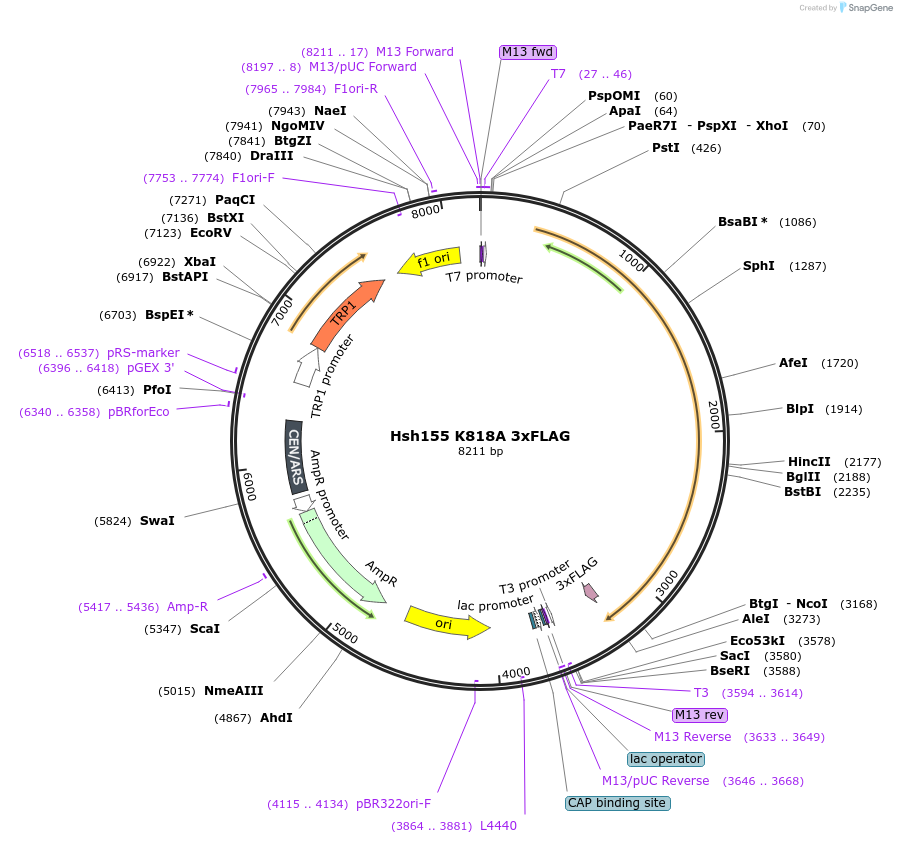
Hsh155 K818A 3xFLAG
Plasmid#111982PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker for shuffle into a HSH155 deletion strain. It has 3xFLAG at the C-terminus. Contains K818A mutation. LethalDepositorInsertHsh155 K818A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationK818AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
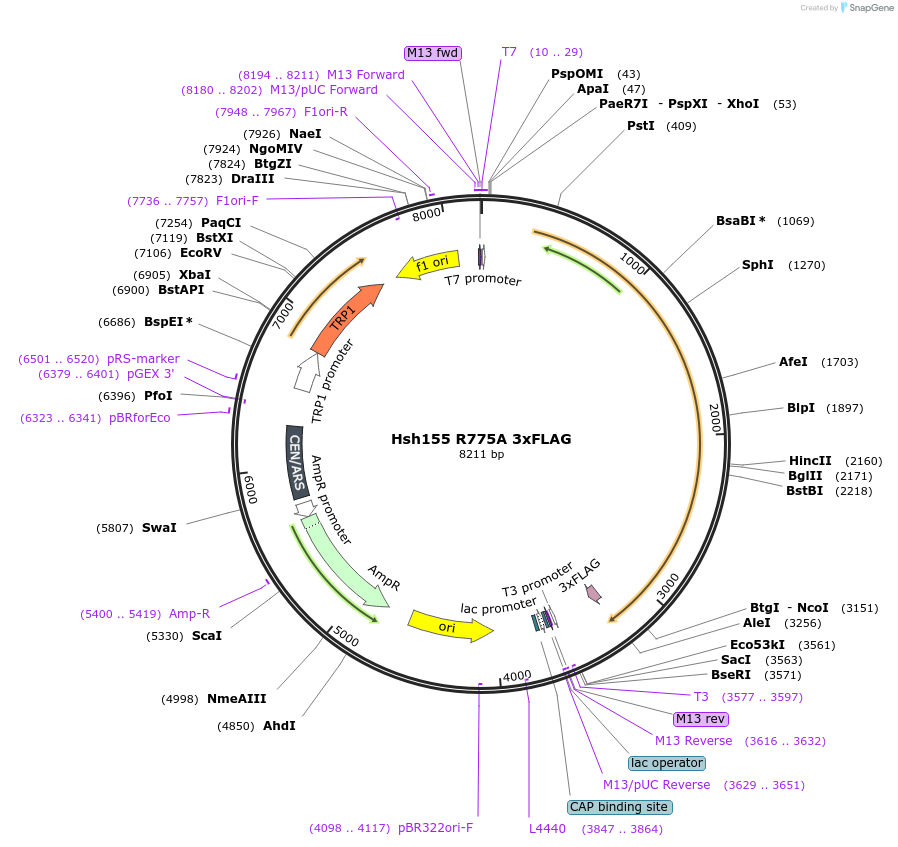
Hsh155 R775A 3xFLAG
Plasmid#111977PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker for shuffle into a HSH155 deletion strain. It has 3xFLAG at the C-terminus. Contains R775A mutation. LethalDepositorInsertHsh155 R775A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationR775AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
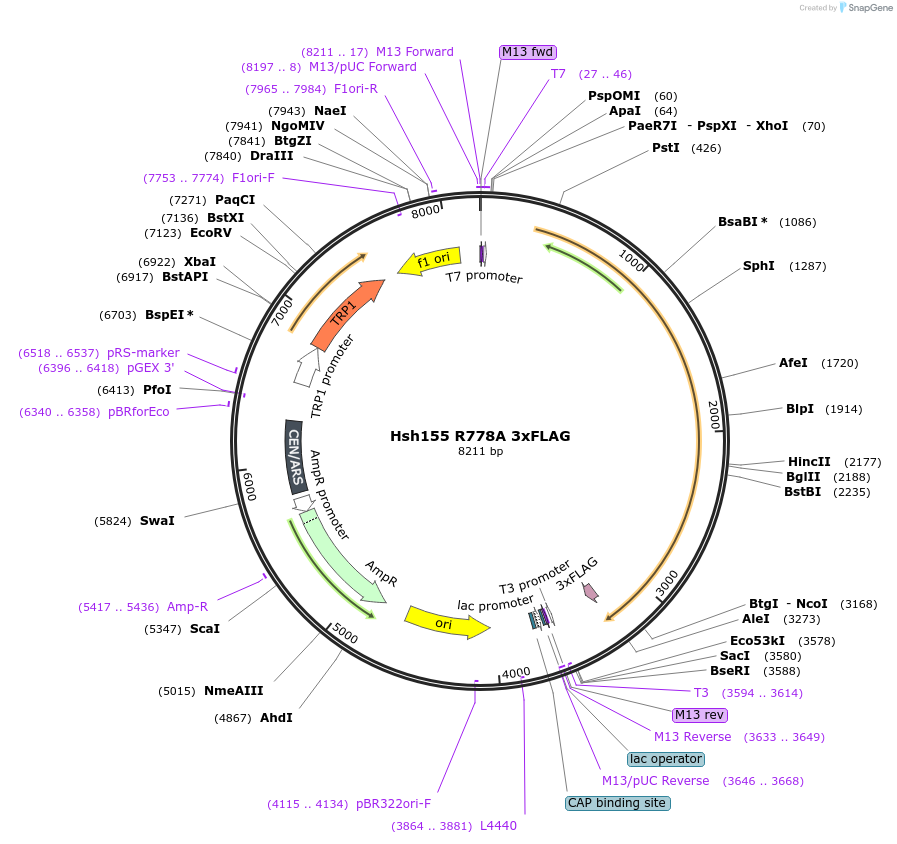
Hsh155 R778A 3xFLAG
Plasmid#111979PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker for shuffle into a HSH155 deletion strain. It has 3xFLAG at the C-terminus. Contains R778A mutation. LethalDepositorInsertHsh155 R778A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationR778AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
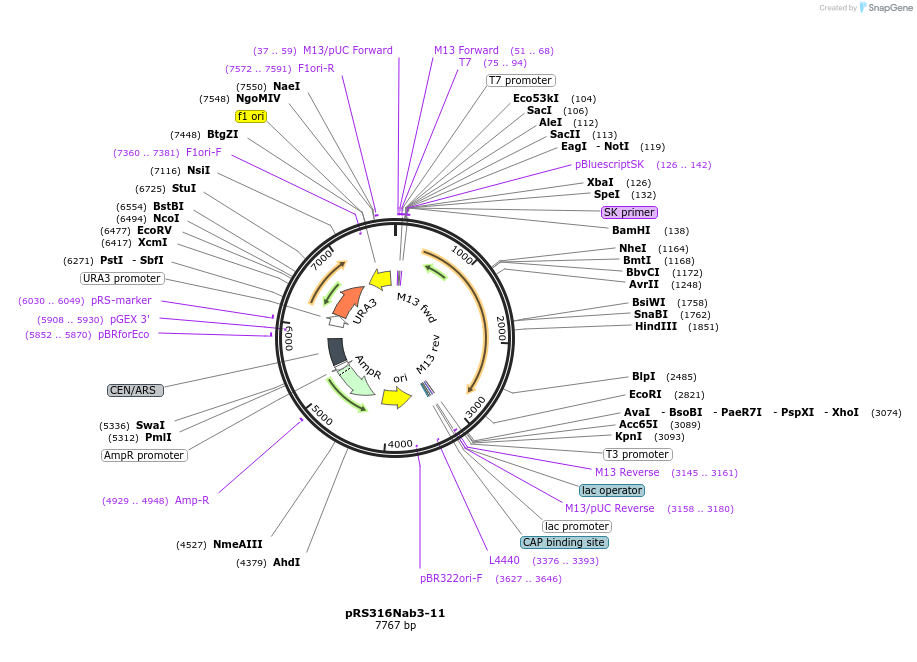
pRS316Nab3-11
Plasmid#110535PurposeExpresses yeast NAB3 using the endogenous promoterDepositorAvailable SinceJune 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
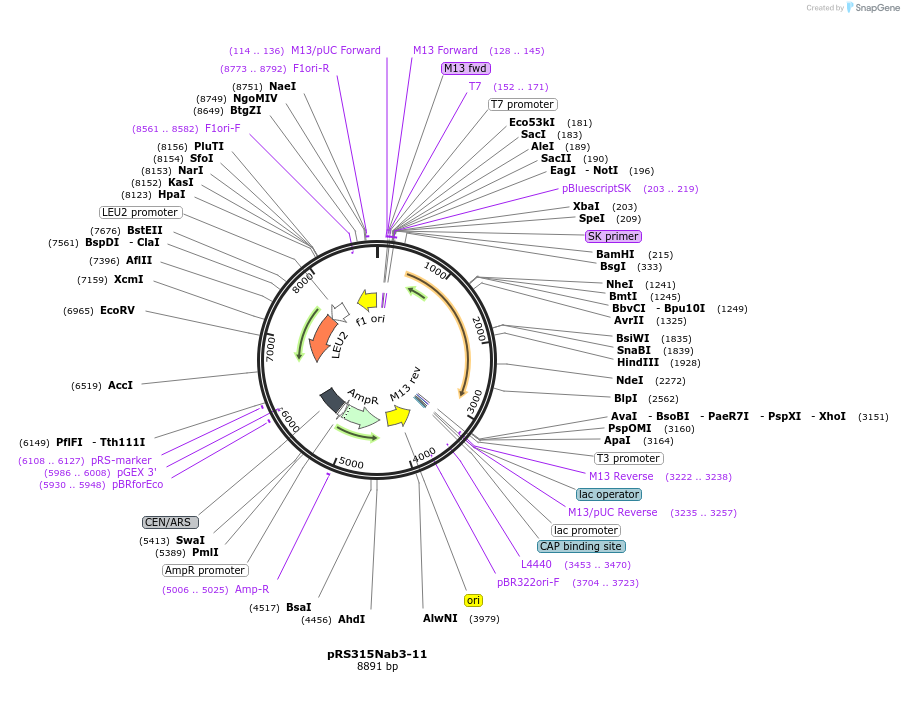
pRS315Nab3-11
Plasmid#110534PurposeExpresses yeast NAB3 using the endogenous promoterDepositorAvailable SinceMay 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
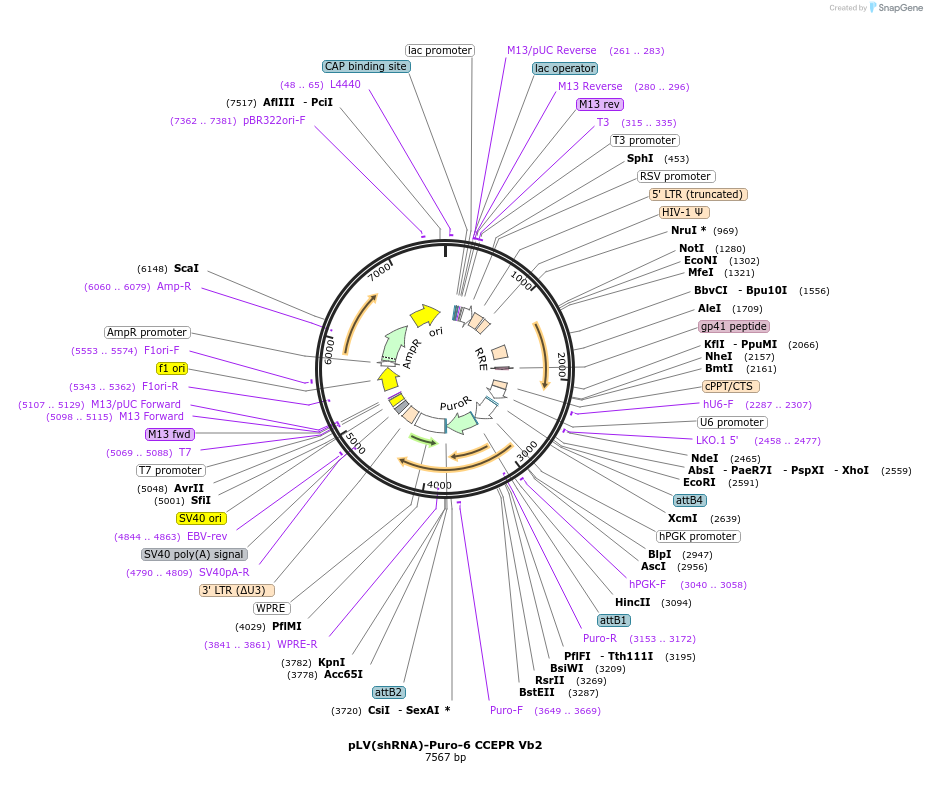
pLV(shRNA)-Puro-6 CCEPR Vb2
Plasmid#108278PurposeshRNA for CCEPR lncRNADepositorAvailable SinceApril 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
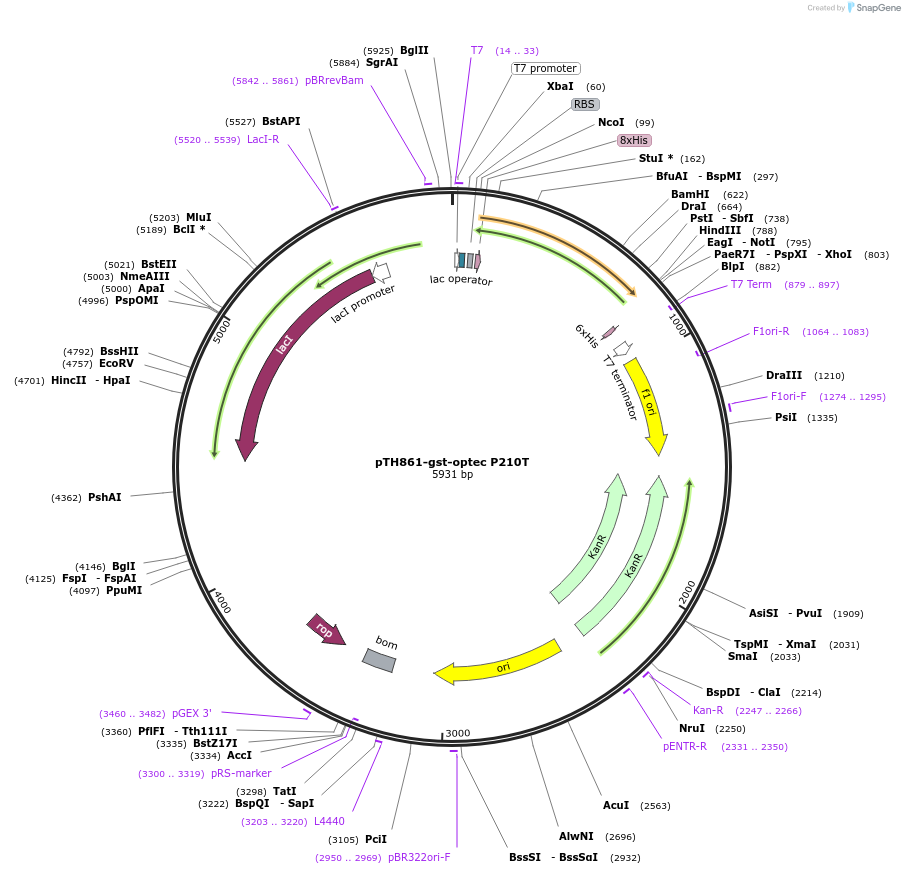
pTH861-gst-optec P210T
Plasmid#89595PurposePlasmid for bacterial expression of a His-tagged P210T mutant of GST encoded by preferred codonsDepositorArticleInsertglutathione-S transferase P210T mutant, codon-optimised for bacterial expression
Tags8xhistidineExpressionBacterialMutationchanged proline 210 to threoninePromoterT7Available SinceMarch 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
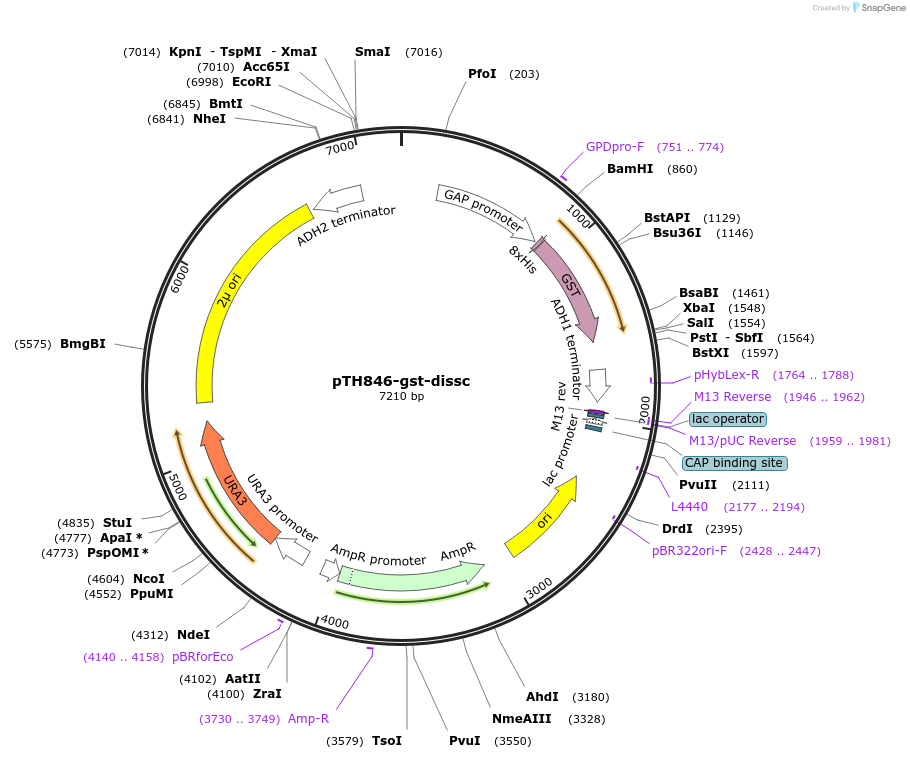
pTH846-gst-dissc
Plasmid#96907PurposePlasmid for yeast expression of GST encoded by non-preferred codonsDepositorArticleInsertyeast codon dis-optimised glutathione-S transferase
Tags8xhistidineExpressionYeastPromoterTDH3Available SinceMarch 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
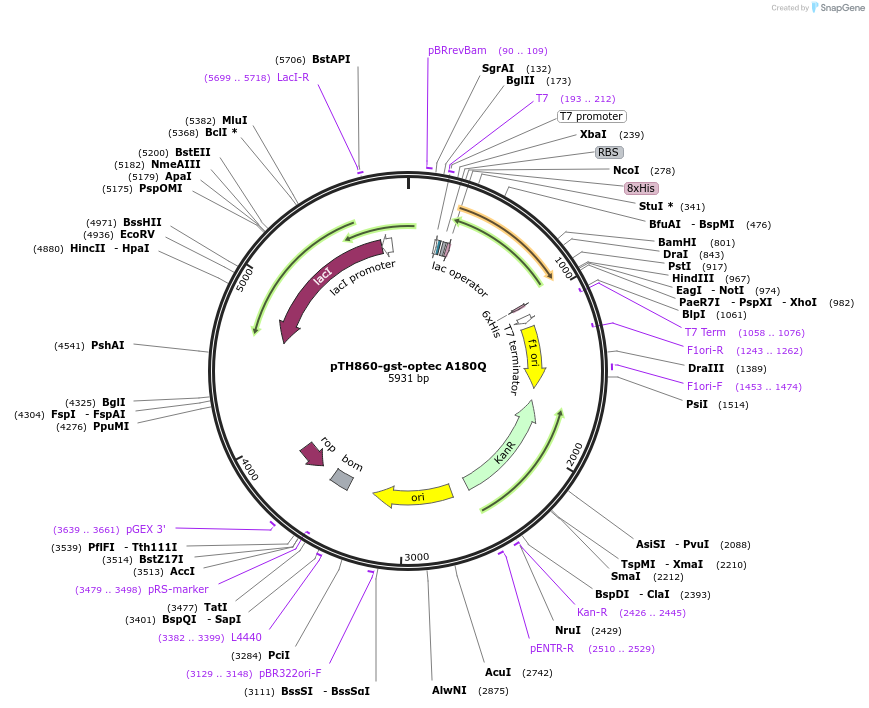
pTH860-gst-optec A180Q
Plasmid#89594PurposePlasmid for bacterial expression of a His-tagged A180Q mutant of GST encoded by preferred codonsDepositorArticleInsertglutathione-S transferase A180Q mutant, codon-optimised for bacterial expression
Tags8xhistidineExpressionBacterialMutationchanged alanine 180 to glutaminePromoterT7Available SinceMarch 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
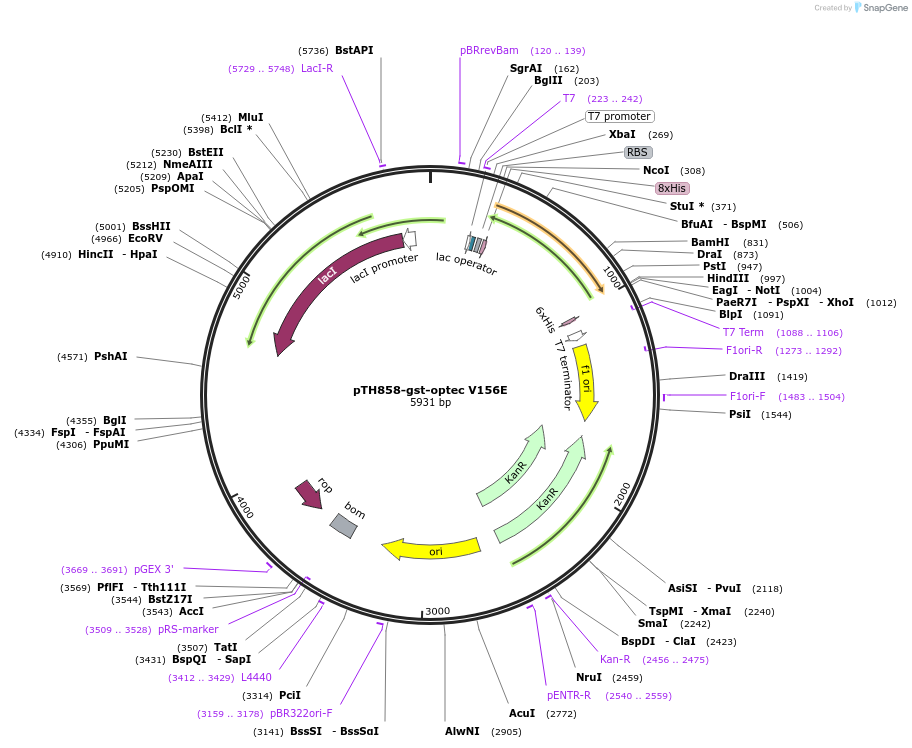
pTH858-gst-optec V156E
Plasmid#89592PurposePlasmid for bacterial expression of a His-tagged V156E mutant of GST encoded by preferred codonsDepositorArticleInsertglutathione-S transferase V156E mutant, codon-optimised for bacterial expression
Tags8xhistidineExpressionBacterialMutationchanged valine 156 to glutamic acidPromoterT7Available SinceMarch 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
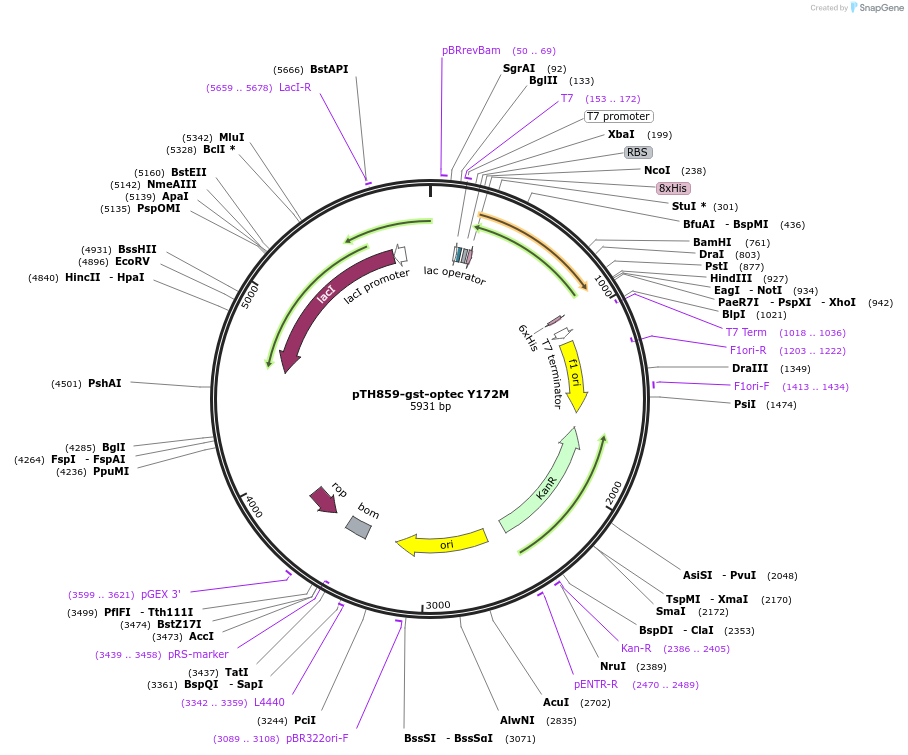
pTH859-gst-optec Y172M
Plasmid#89593PurposePlasmid for bacterial expression of a His-tagged Y172M mutant of GST encoded by preferred codonsDepositorArticleInsertglutathione-S transferase Y172M mutant, codon-optimised for bacterial expression
Tags8xhistidineExpressionBacterialMutationchanged tyrosine 172 to methioninePromoterT7Available SinceMarch 20, 2018AvailabilityAcademic Institutions and Nonprofits only