We narrowed to 561 results for: prime editing
-
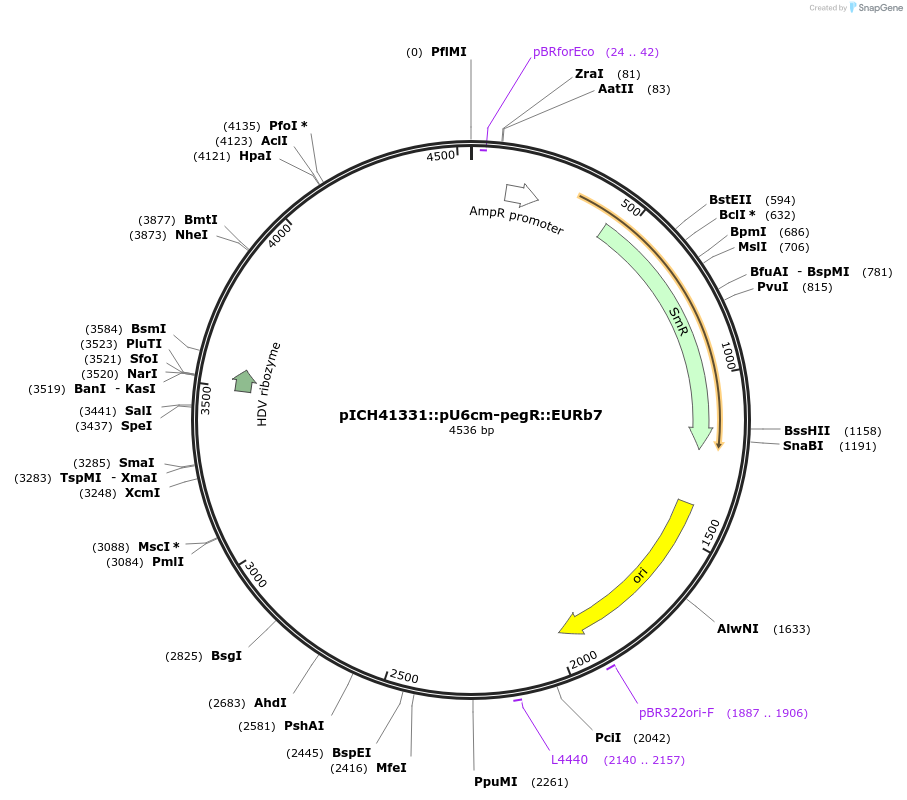
Plasmid#227695PurposeGolden Gate level 0 acceptor vector for cloning altered epegRNA: altered epegRNA being driven by U6 composite promoter (pU6cm) and terminated by a dual terminator (EURb7).DepositorInsertAltered epegRNA backbone with cloning sites for gRNA and RTT.
UseCRISPRExpressionPlantPromoterU6 compositeAvailable SinceNov. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
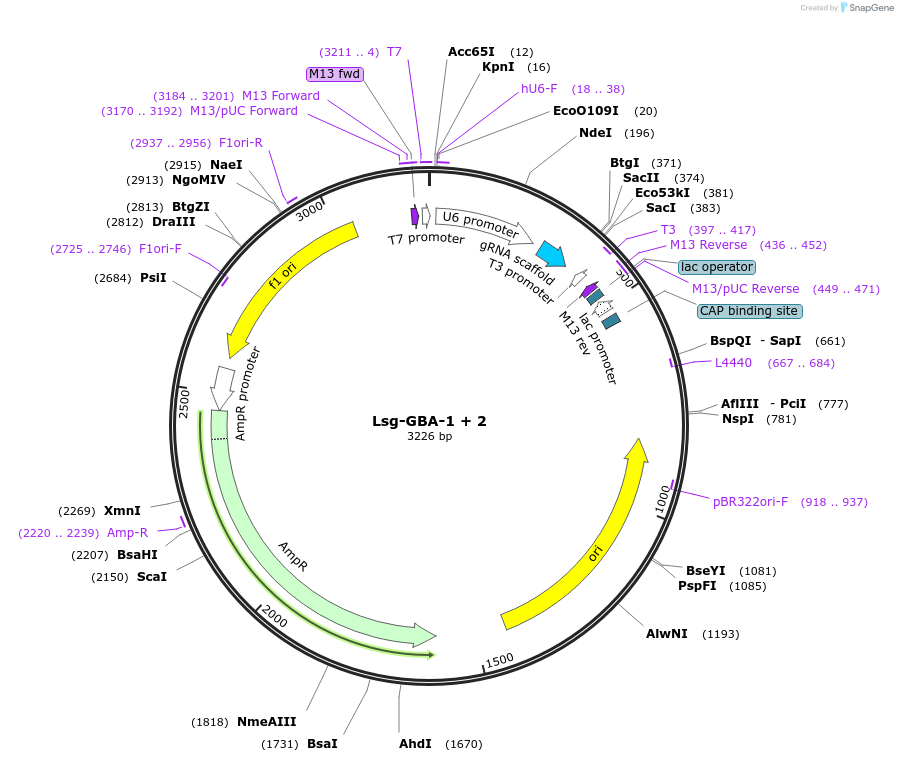
Lsg-GBA-1 + 2
Plasmid#199276Purposenicking sgRNA to induce or correct GBA (c. 1226 A > G) mutation using PE3DepositorInsertGBA nick
ExpressionMammalianAvailable SinceApril 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
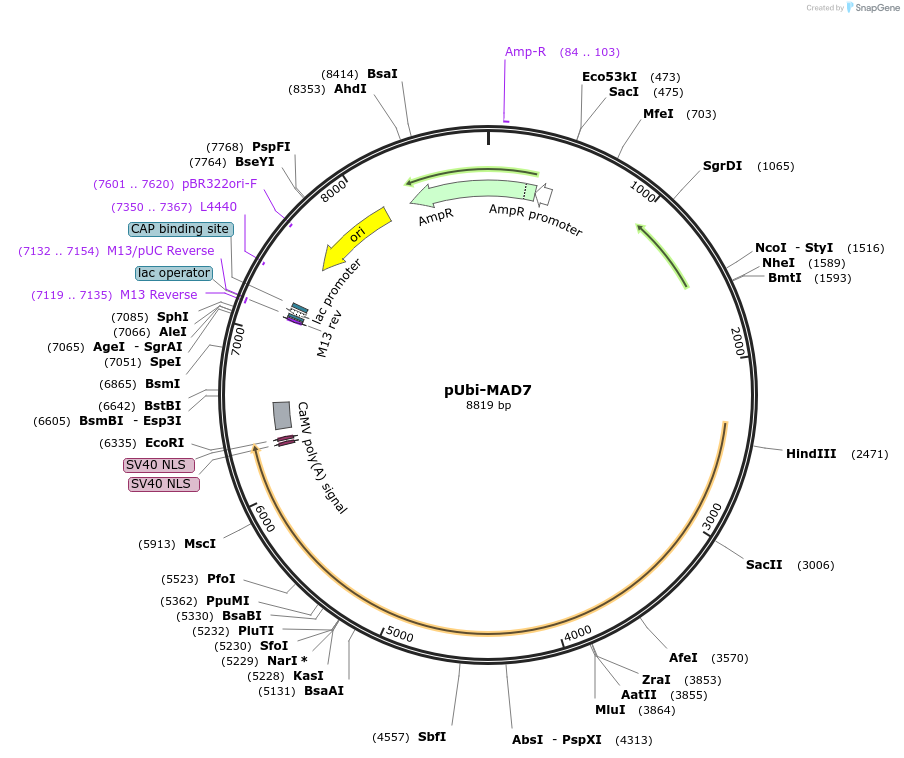
pUbi-MAD7
Plasmid#170133PurposeFor plant prime editing in wheat plants or monocotyledons protoplastsDepositorInsertMAD7(ErCas12a)
UseCRISPRExpressionPlantPromotermaize Ubiquitin-1Available SinceSept. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
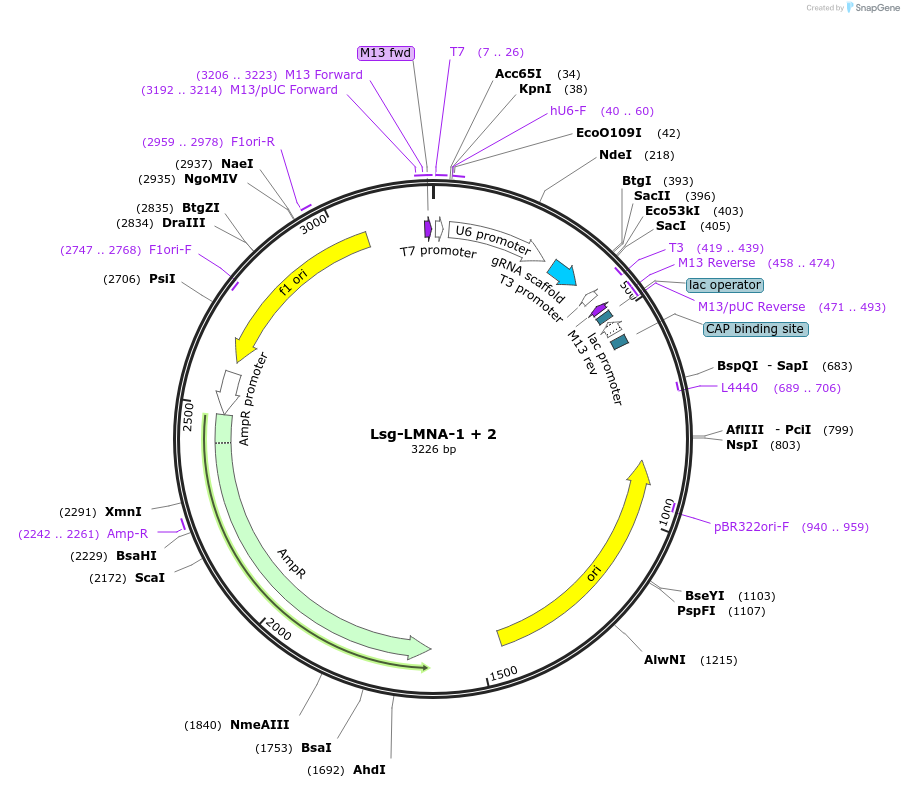
Lsg-LMNA-1 + 2
Plasmid#199278Purposenicking sgRNA to induce LMNA (c.1824C > T) mutation using PE3DepositorInsertLMNA nick
ExpressionMammalianAvailable SinceApril 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
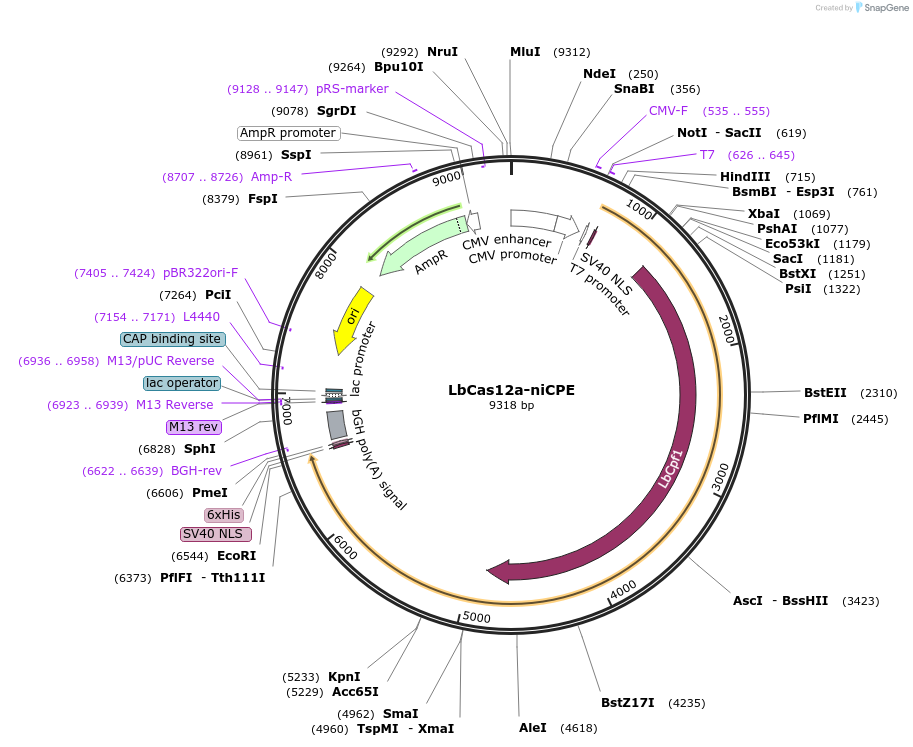
LbCas12a-niCPE
Plasmid#213747PurposeFor circular RNA-mediated prime editor using LbCas12a-niCPE in HEK293T cellsDepositorInsertMCP-nLbCas12a-M-MLV RT∆RNase H
UseCRISPRTagsBPNLSExpressionMammalianMutationD156R, R1138A in LbCas12aPromoterCMVAvailable SinceFeb. 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
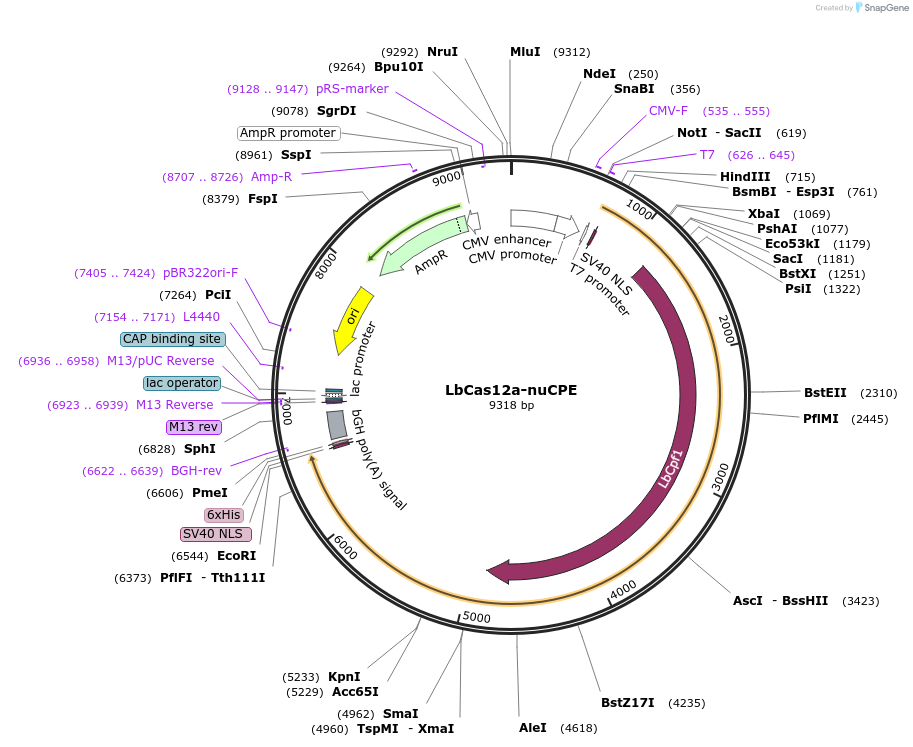
LbCas12a-nuCPE
Plasmid#213748PurposeFor circular RNA-mediated prime editor using LbCas12a-nuCPE in HEK293T cellsDepositorInsertMCP-LbCas12a-M-MLV RT∆RNase H
UseCRISPRTagsBPNLSExpressionMammalianMutationD156R in LbCas12aPromoterCMVAvailable SinceFeb. 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
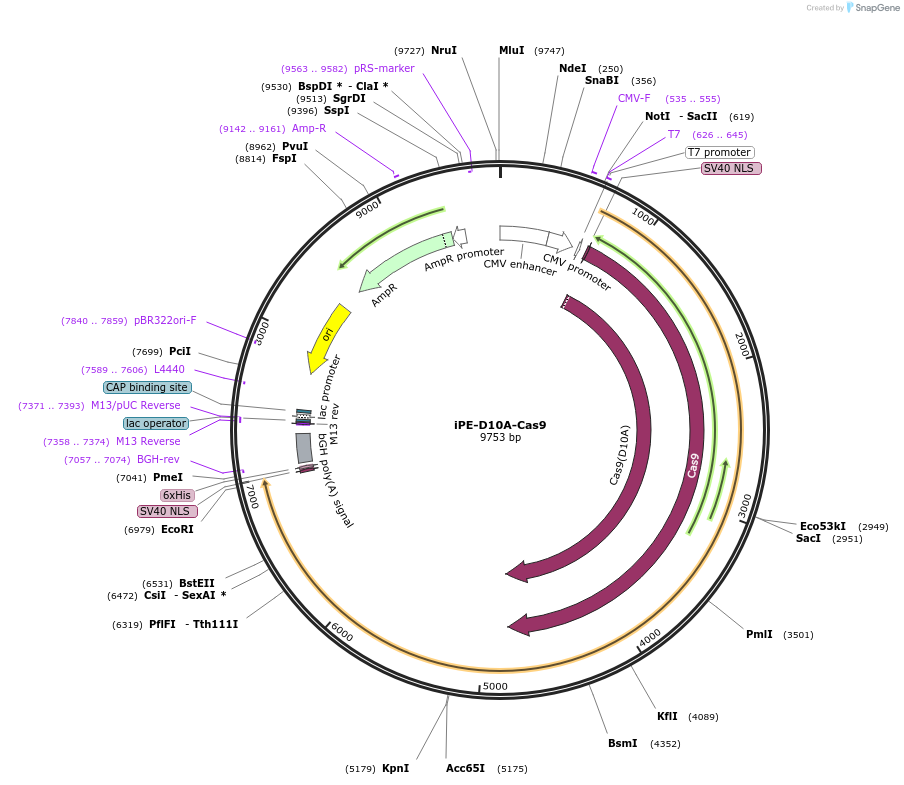
iPE-D10A-Cas9
Plasmid#239380PurposeFor inverse prime editor using nCas9-D10A in HEK293T cellsDepositorInsertnCas9-D10A, M-MLV RT
UseCRISPRTagsBPNLSExpressionMammalianMutationD10A in Cas9PromoterCMVAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
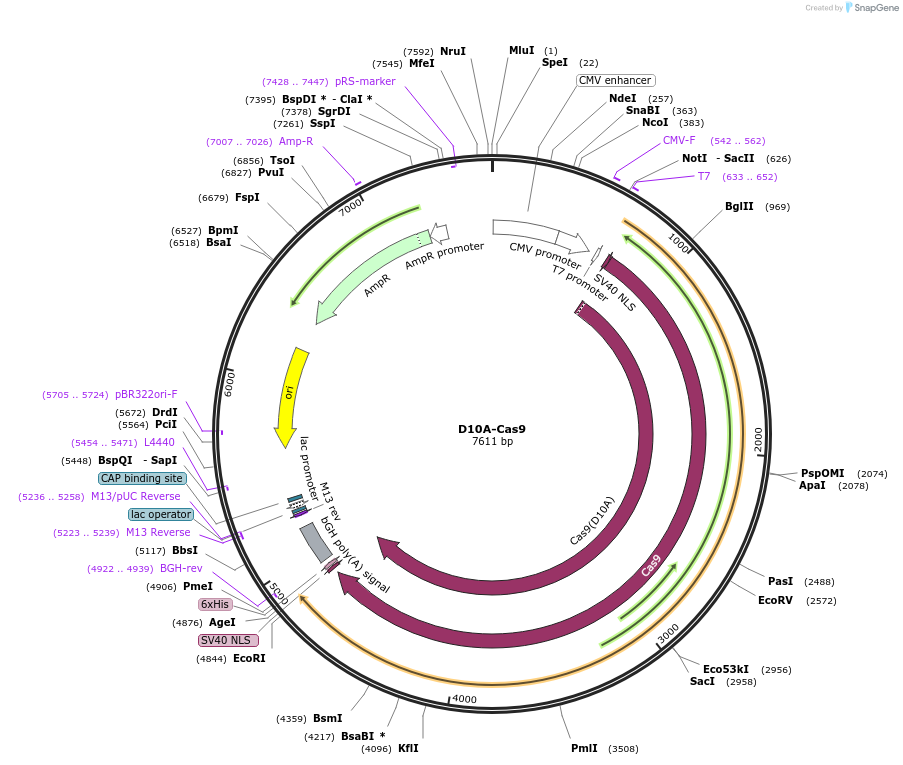
D10A-Cas9
Plasmid#239383PurposeFor circular RNA-mediated inverse prime editor using nCas9-D10A in HEK294T cellsDepositorInsertnCas9-D10A
UseCRISPRTagsBPNLS and SV40 NLSExpressionMammalianMutationD10A in Cas9PromoterCMVAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
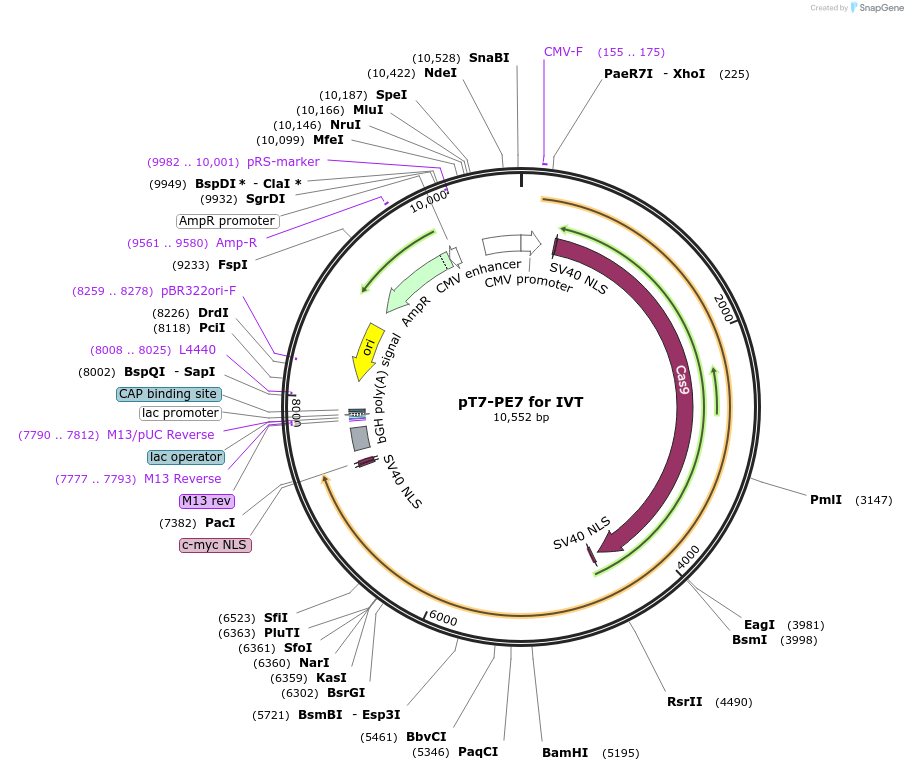
pT7-PE7 for IVT
Plasmid#214813PurposeTemplate for in vitro transcription of PE7DepositorInsertPE7
UseTemplate for in vitro transcriptionTagsSV40 bpNLS and c-Myc NLSPromoterT7 (inactivated)Available SinceMarch 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
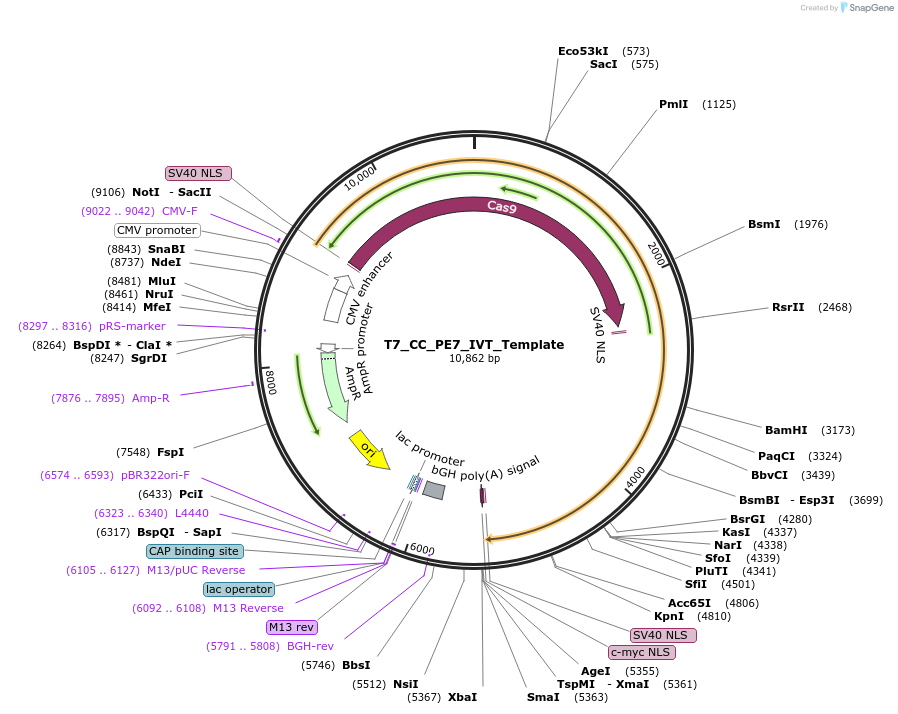
T7_CC_PE7_IVT_Template
Plasmid#223022PurposeTemplate for in vitro transcription of PE7. For HSPC-related experiments.DepositorInsertPE7
UseTemplate for in vitro transcriptionTagsSV40 bpNLS and c-Myc NLSPromoterT7Available SinceAug. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
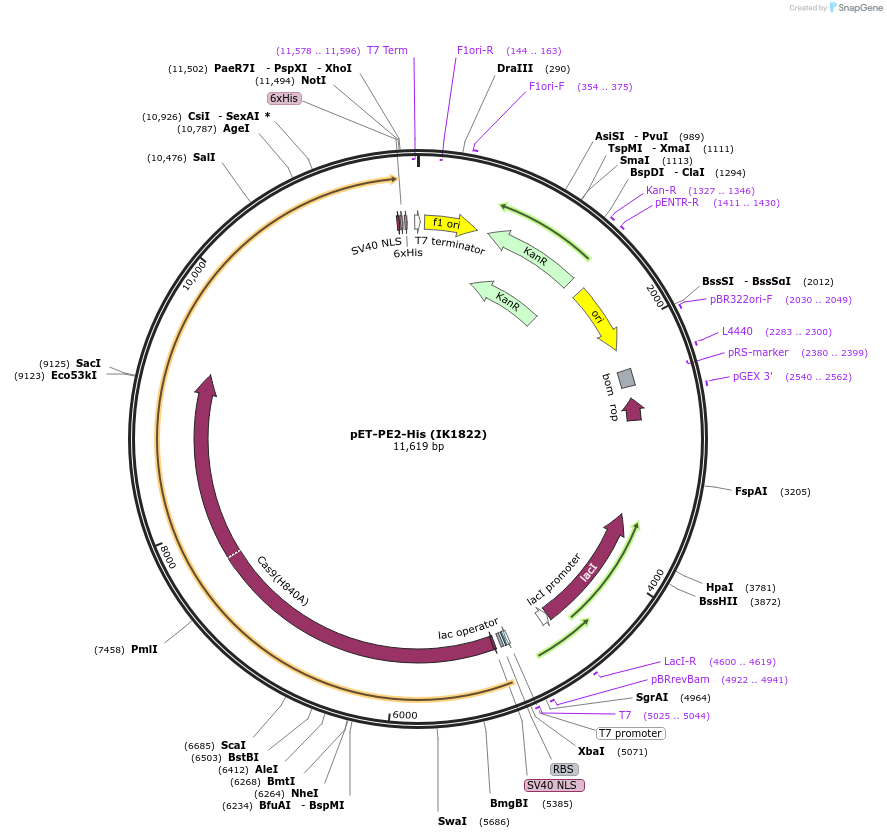
pET-PE2-His (IK1822)
Plasmid#170103PurposeT7 promoter bacterial expression plasmid for PE2 (nSpCas9(H840A)-MMLV-RT) with C-terminal 6xHis-tag and bipartite NLSDepositorInsertbpNLS-nSpCas9(H840A)-MMLVRT-bpNLS-6xHis
UseCRISPRTags6xHis tagExpressionBacterialMutationBacteria codon-optimized nSpCas9 (H840A) & M-…PromoterT7Available SinceMay 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
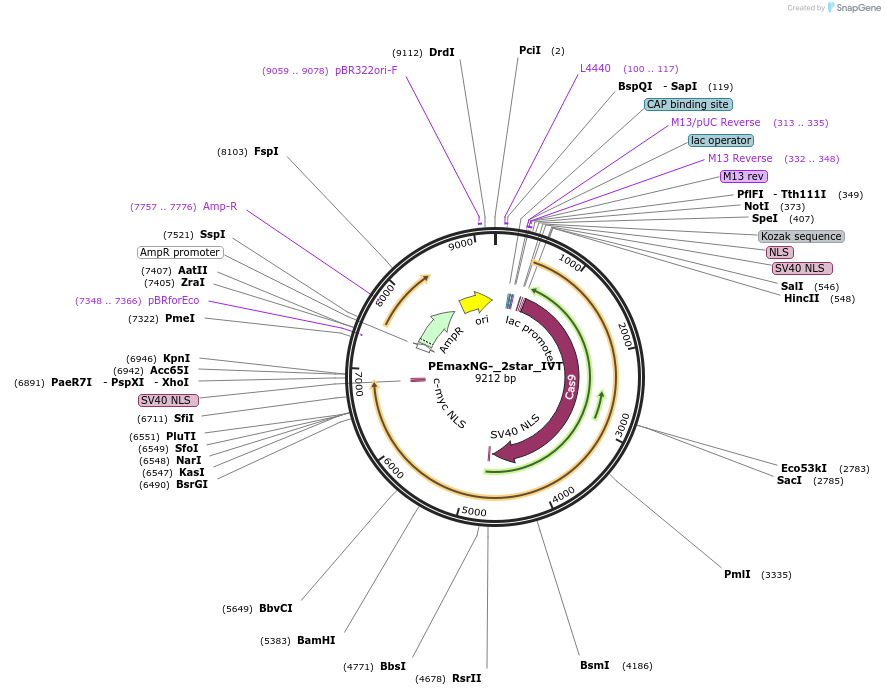
PEmaxNG-_2star_IVT
Plasmid#227678Purposeplasmid for SpCas9-NG-PEmax** mRNA IVTDepositorInsertNG-PEmax_2_star
UseCRISPRTagsnoExpressionMammalianPromoterT7Available SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
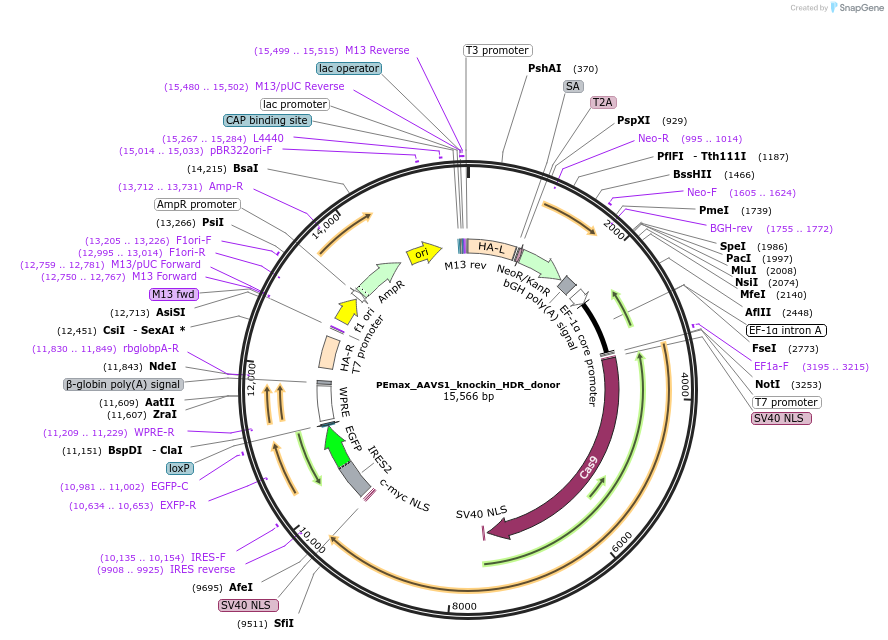
PEmax_AAVS1_knockin_HDR_donor
Plasmid#223000PurposeHDR donor plasmid for PEmax knockin at AAVS1 locus. Left_homology_arm-splicing_acceptor-T2A-NeoR-bGHpA-pEF1α-PEmax-IRES2-EGFP-WPRE-βglobinpA-right_homology_armDepositorInsertPEmax
UseHdr donorTagsSV40 bpNLS and c-Myc NLSExpressionMammalianPromoterEF1αAvailable SinceAug. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
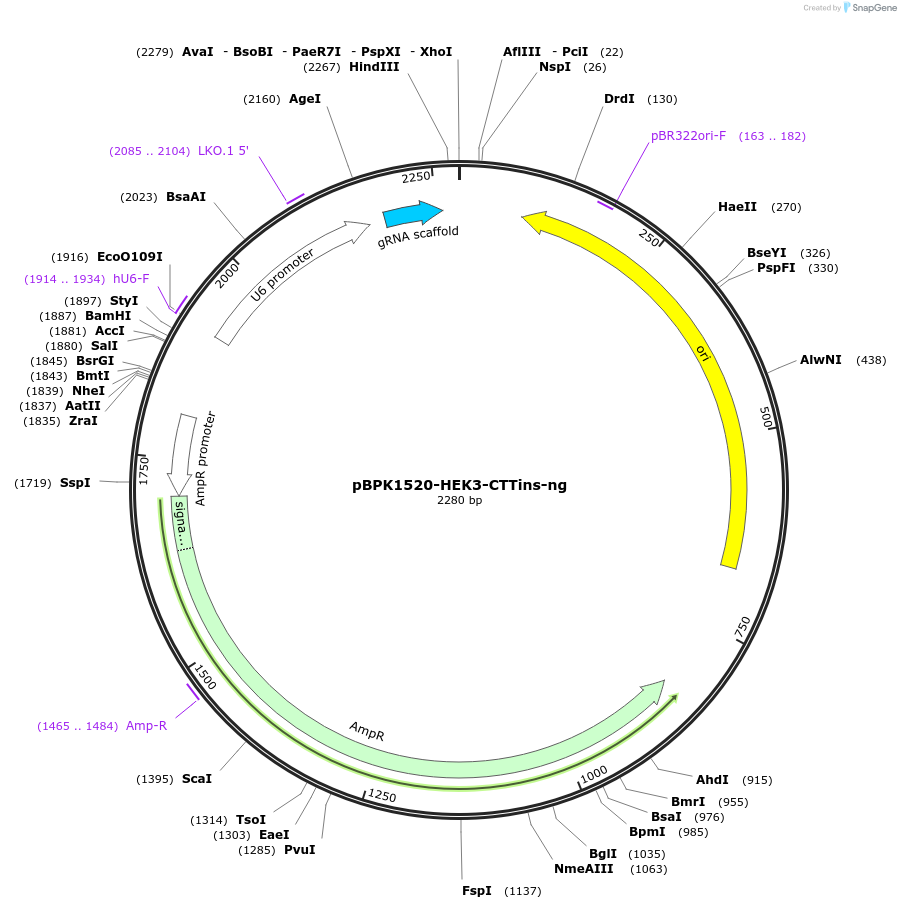
pBPK1520-HEK3-CTTins-ng
Plasmid#180019PurposeTransiently expressing a nicking sgRNA to facilitate the introduction of HEK3 CTT insertion in PE3 approachDepositorInsertPrime editing ngRNA for HEK3-CTTins (EPHA8 Human)
UseCRISPRExpressionMammalianPromoterHuman U7Available SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
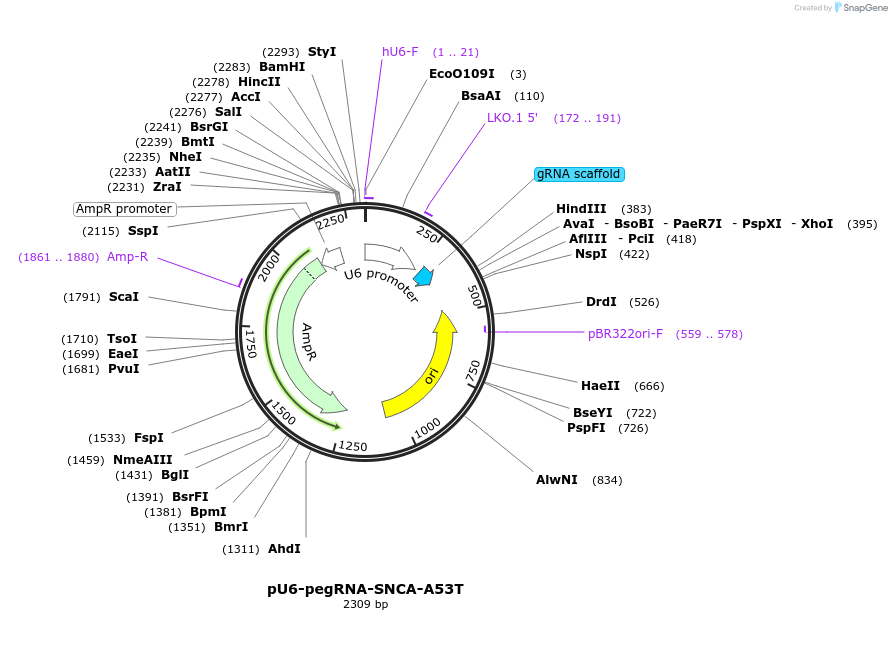
pU6-pegRNA-SNCA-A53T
Plasmid#181738PurposeTransiently expressing a pegRNA to introduce SNCA-A53T mutation in human cellsDepositorInsertPrime editing pegRNA for SNCA-A53T (SNCA Synthetic)
UseCRISPRExpressionMammalianPromoterHuman U6Available SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
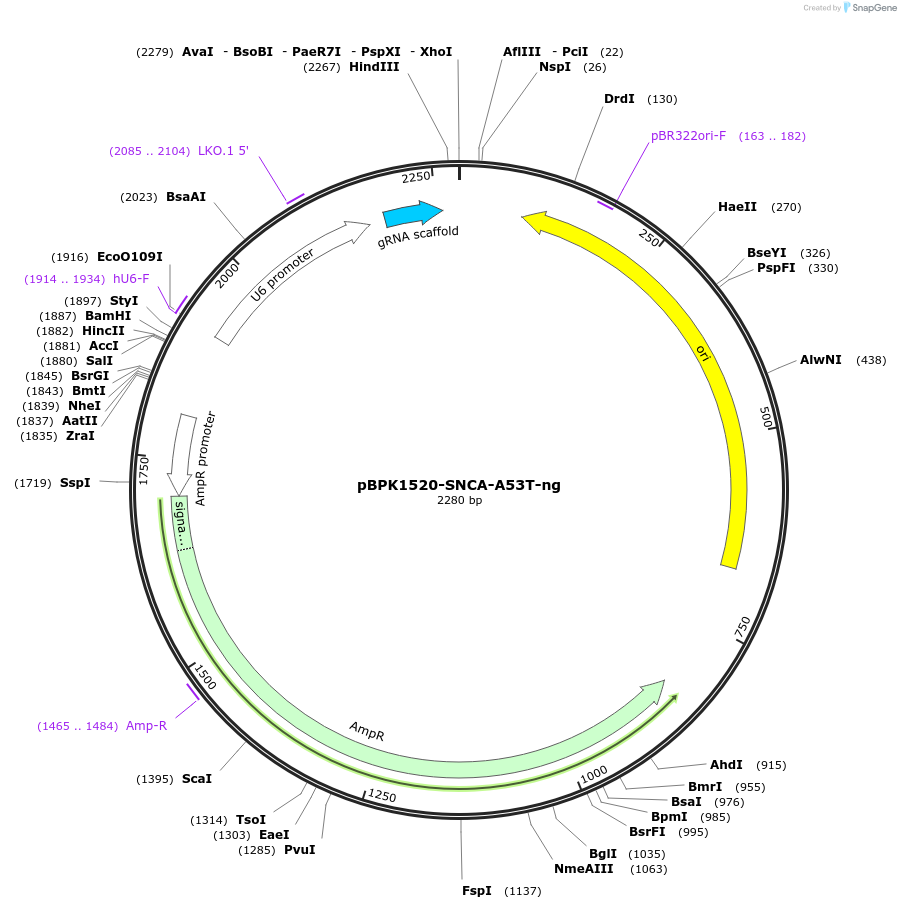
pBPK1520-SNCA-A53T-ng
Plasmid#181739PurposeTransiently expressing a nicking sgRNA to facilitate the introduction of SNCA-A53T mutation in PE3 approachDepositorInsertPrime editing ngRNA for SNCA-A53T (SNCA Human)
UseCRISPRExpressionMammalianPromoterHuman U6Available SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
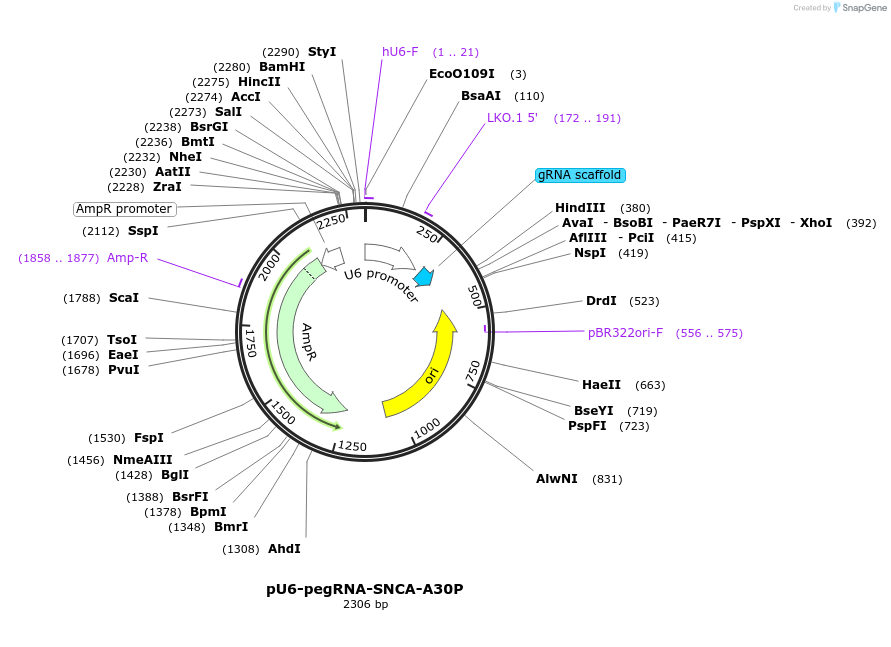
pU6-pegRNA-SNCA-A30P
Plasmid#180016PurposeTransiently expressing a pegRNA to introduce SNCA-A30P mutation in human cellsDepositorInsertPrime editing pegRNA for SNCA-A30P (SNCA Synthetic)
UseCRISPRExpressionMammalianPromoterHuman U6Available SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
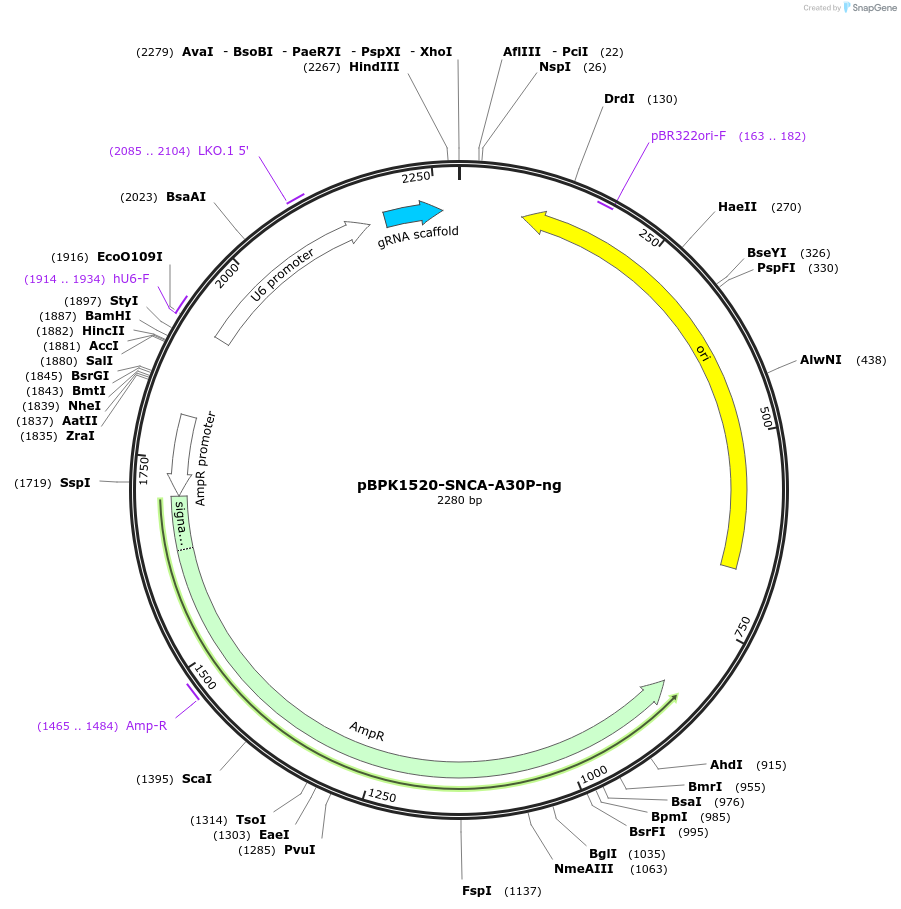
pBPK1520-SNCA-A30P-ng
Plasmid#180018PurposeTransiently expressing a nicking sgRNA to facilitate the introduction of SNCA-A30P mutation in PE3 approachDepositorInsertPrime editing ngRNA for SNCA-A30P (SNCA Human)
UseCRISPRExpressionMammalianPromoterHuman U6Available SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
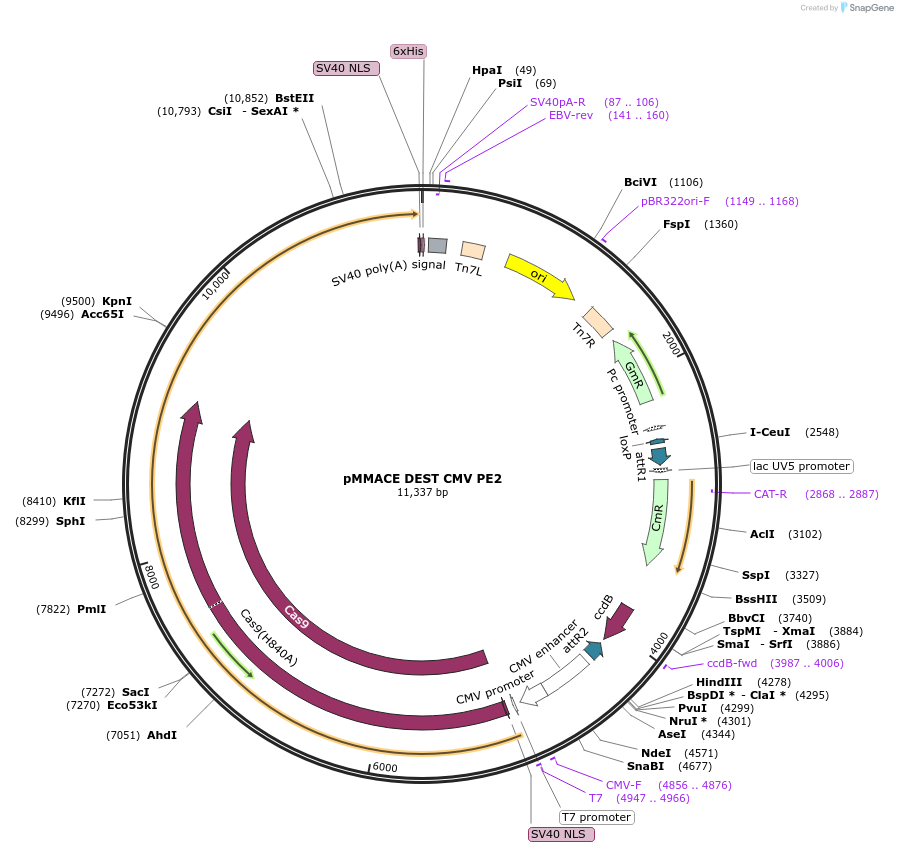
pMMACE DEST CMV PE2
Plasmid#206276PurposeDEST vector for MultiSite Gateway assembly and production of recombinant baculovirus using MultiMate assembly. Encodes PE2 under the control of CMV promoter. CRE-Acceptor in MultiBac system.DepositorInsertPE2
UseCRISPR; Recombinant baculovirus production, multi…ExpressionMammalianAvailable SinceSept. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
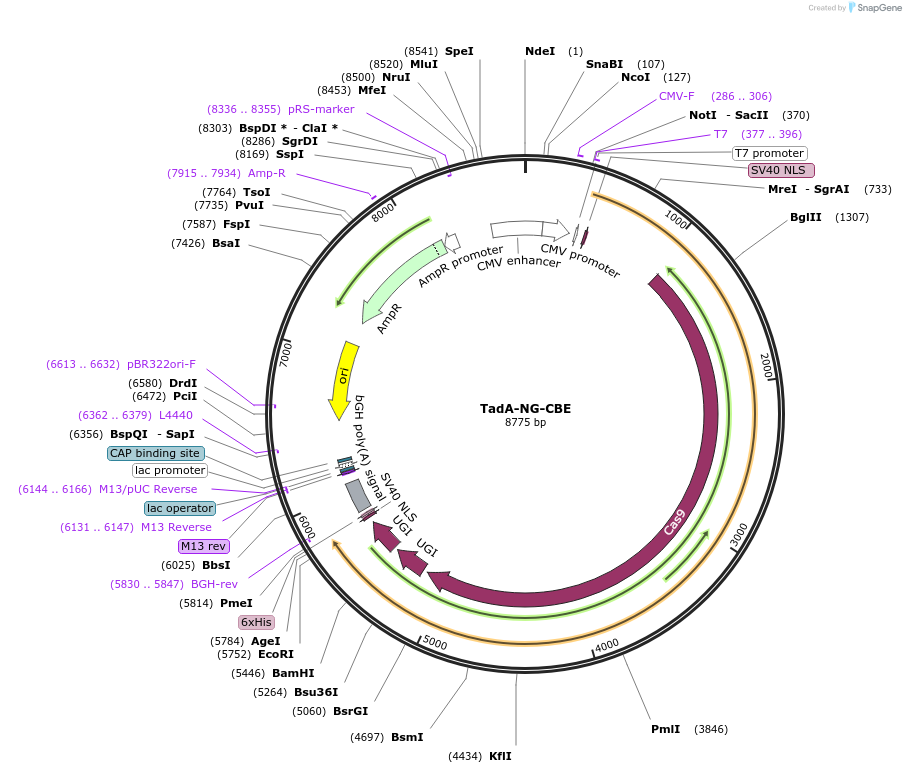
TadA-NG-CBE
Plasmid#227420PurposeC-to-T base editing in mammalian cellsDepositorAvailable SinceOct. 25, 2024AvailabilityAcademic Institutions and Nonprofits only