We narrowed to 18,548 results for: MUT
-
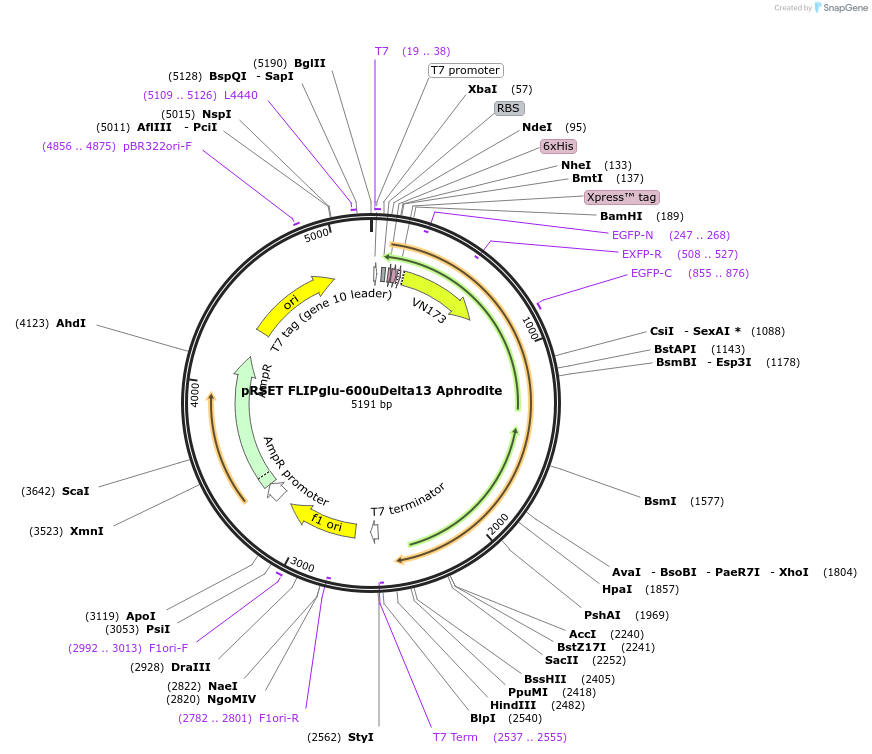
Plasmid#13570DepositorInsertFLIPglu F16A Delta13 Aphrodite
TagsAphrodite and ECFPExpressionBacterialMutationFluorophore-derived linker shortened. F16A mutati…Available SinceMarch 2, 2007AvailabilityAcademic Institutions and Nonprofits only -
pSP64R1 SHP2 D61A
Plasmid#8326DepositorAvailable SinceJune 17, 2005AvailabilityAcademic Institutions and Nonprofits only -
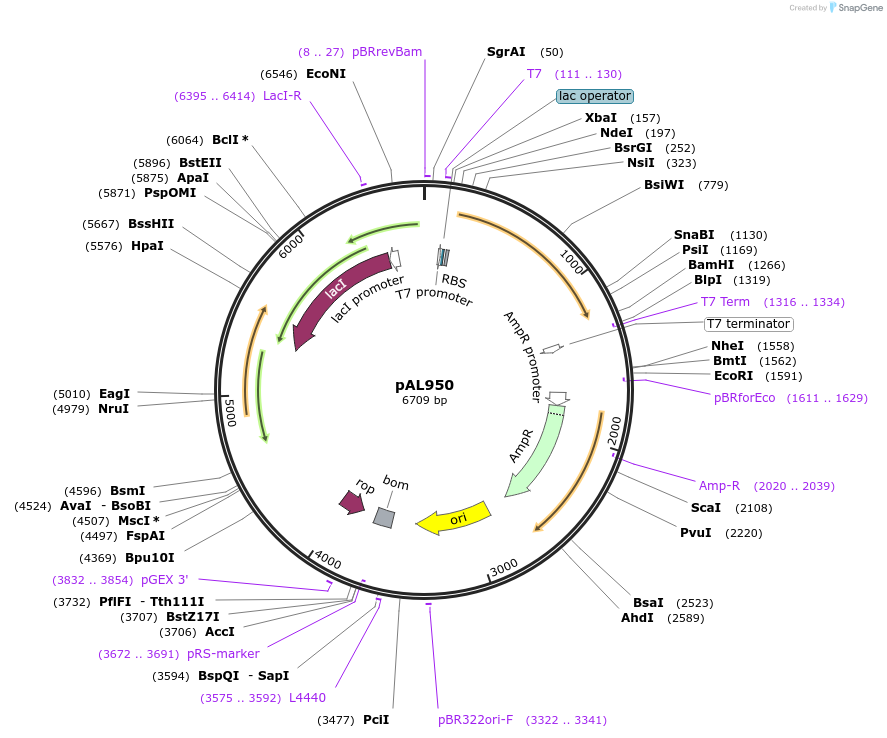
pAL950
Plasmid#119748Purposeprecursor outer membrane pore protein E, phoE with mutations Alanine 249 to Cysteine and Glutamine 307 to CysteineDepositorInsertprecusor Outer membrane pore protein E, phoE, with mutations Alanine 249 to Cysteine and Glutamine 307 to Cysteine
TagsnoneExpressionBacterialMutationInternal NdeI site removed and mutations Alanine …PromoterT7Available SinceJan. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
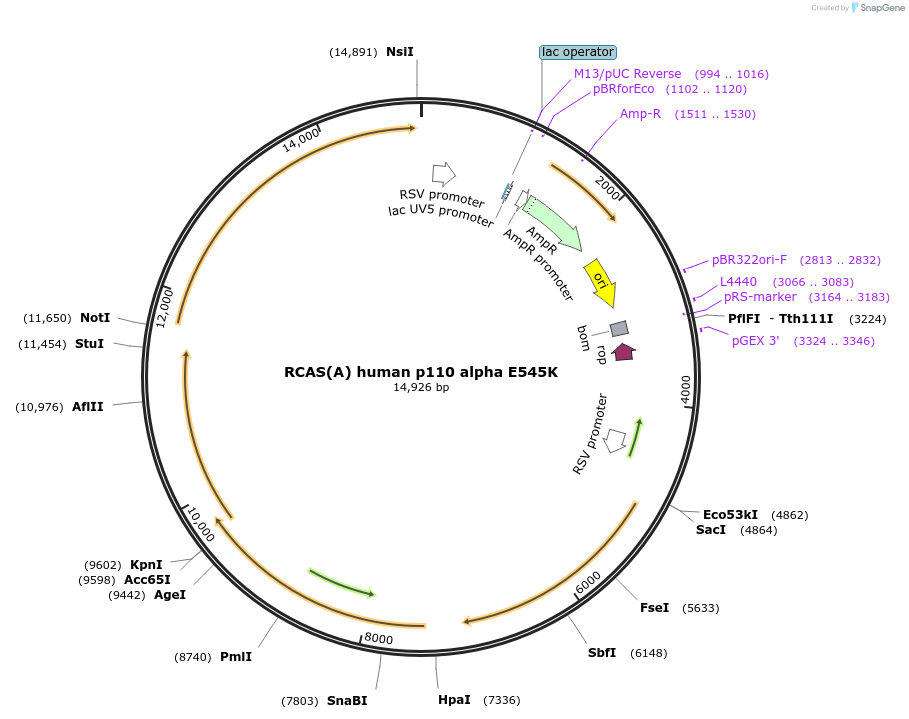
RCAS(A) human p110 alpha E545K
Plasmid#52211Purposeexpresses human p110 alpha with E545K mutation in avian cellsDepositorAvailable SinceJuly 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
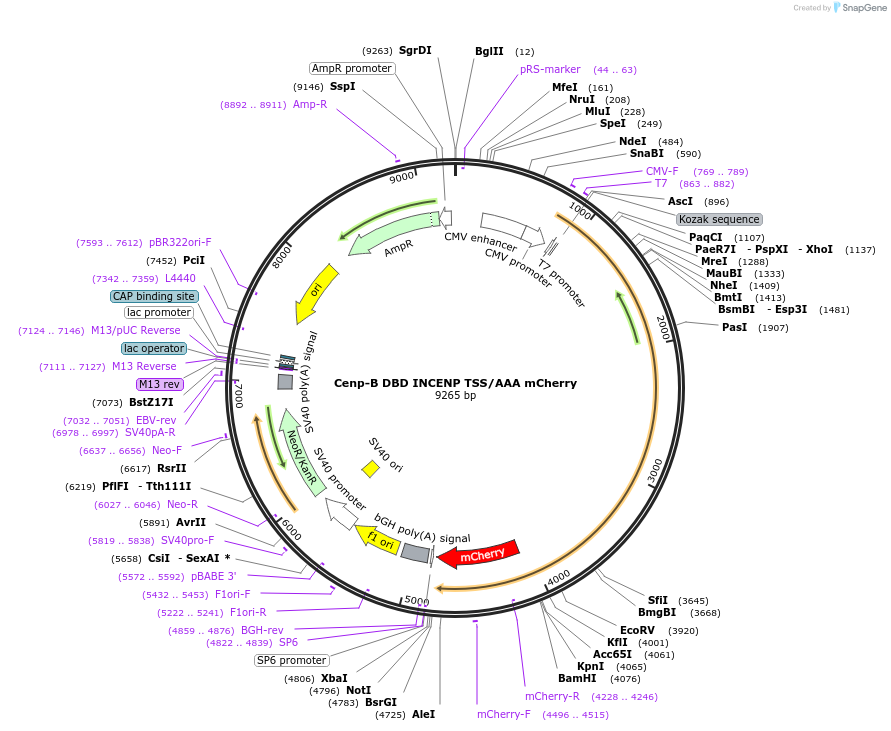
Cenp-B DBD INCENP TSS/AAA mCherry
Plasmid#45234PurposeTarget INCENP (with TSS mutated to AAA) to centromeres via CENP-B DNA binding domain (DBD)DepositorInsertCenp-B DBD, INCENP-ΔCEN with AAA mutation, mCherry
TagsCENP-B DBD, DNA binding domain of CENP-B and mChe…ExpressionMammalianMutationINCENP C-terminal TSS mutated to AAAPromotert7Available SinceSept. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
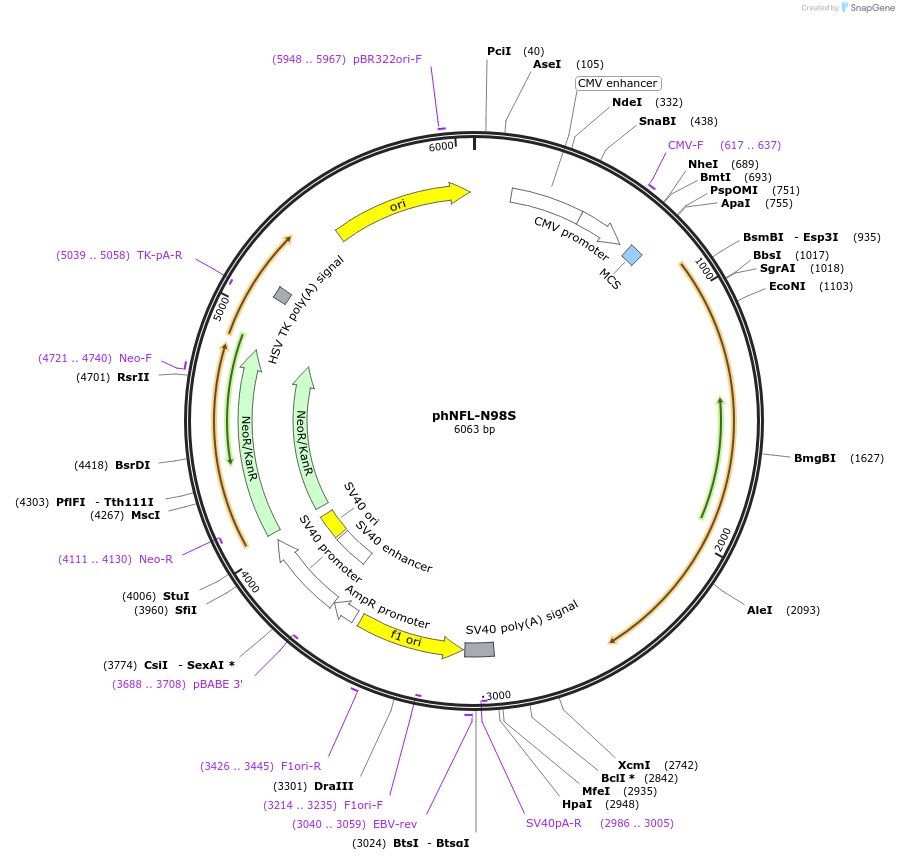
phNFL-N98S
Plasmid#132592PurposeExpresses human NFL containing the Charcot-Marie-Tooth disease type 2E N98S mutationDepositorAvailable SinceNov. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pBabe hygro NFS1 CD
Plasmid#102978PurposeExpression of NFS1 C381A mutantDepositorInsertNFS1 C381A (NFS1 Human)
UseRetroviralExpressionMammalianMutationC381A catalytic site mutant, Silent mutations to …Available SinceDec. 18, 2017AvailabilityAcademic Institutions and Nonprofits only -
pSP64R1 SHP2 E76A
Plasmid#8327DepositorAvailable SinceJune 17, 2005AvailabilityAcademic Institutions and Nonprofits only -
PGL3-Ogt (-178 to +233 bp)
Plasmid#79266PurposeTo identify the Cis and Trans acting elements that control Ogt gene transcription.DepositorInsertOgt promoter element (Ogt Mouse)
ExpressionMammalianAvailable SinceSept. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
FLAG-mPar6a WT
Plasmid#86148PurposeFLAG tagged mouse Par6 wild type - Binds to aPKC, Par3, Lgl, Cdc42DepositorAvailable SinceJune 6, 2017AvailabilityAcademic Institutions and Nonprofits only -
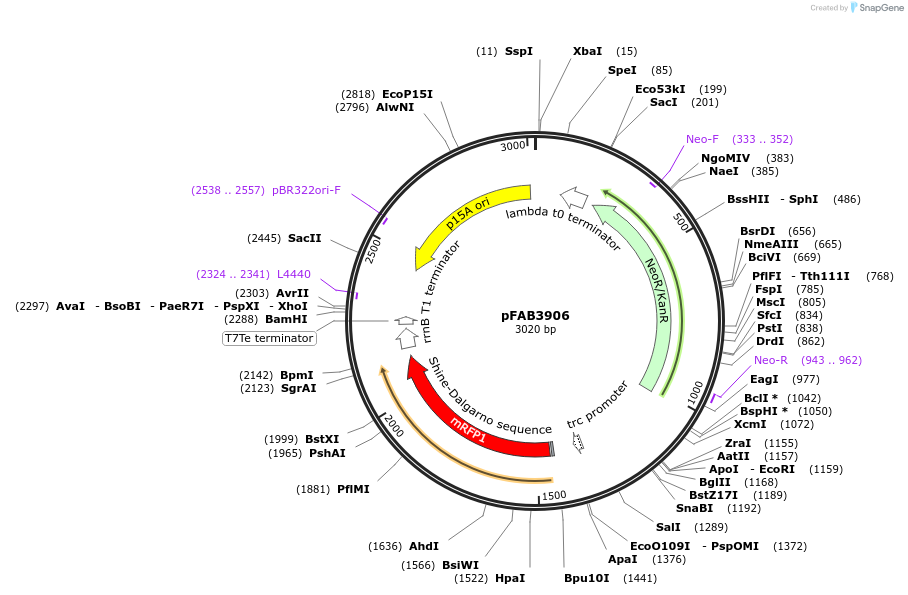
pFAB3906
Plasmid#47809PurposeBIOFAB RFP reporter plasmid for measuring promoter 14 + BCD2 efficiency.DepositorInsertpromoter 14 and BCD2
UseSynthetic BiologyExpressionBacterialPromotersee insertAvailable SinceAug. 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
-
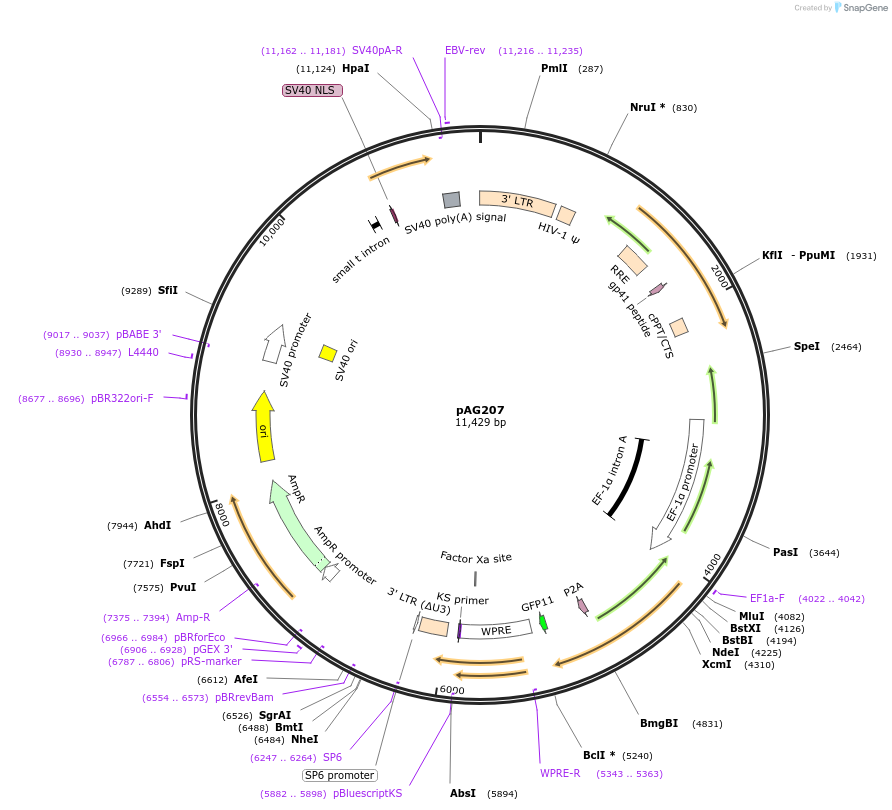
pAG207
Plasmid#226738PurposeExpresses a reporter for an alpha-helical OMM proteinDepositorAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
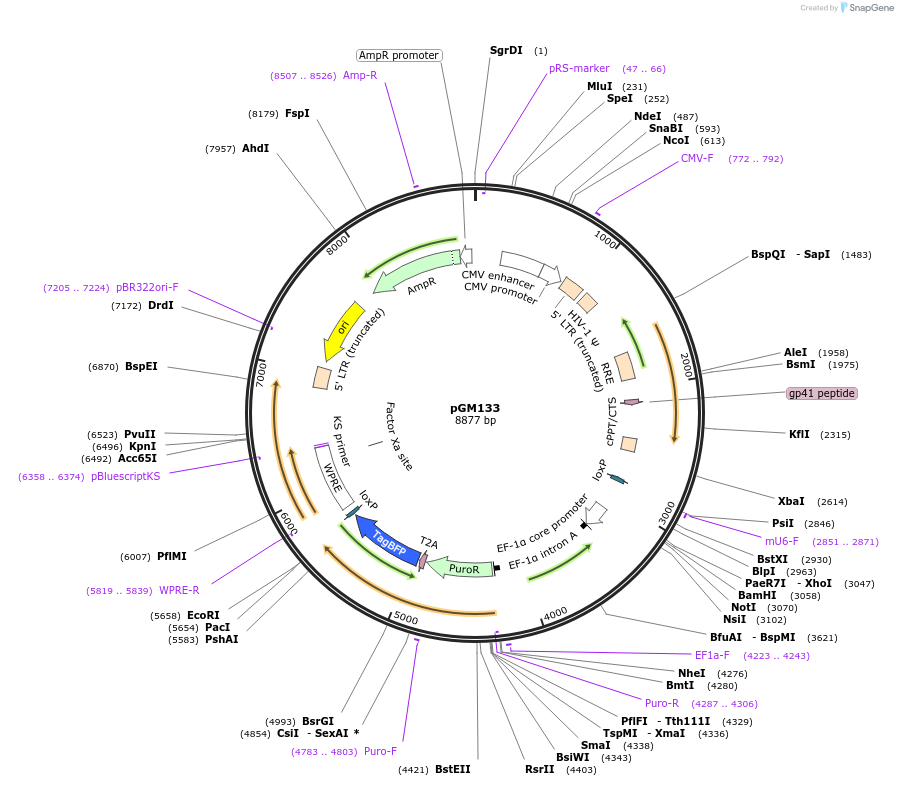
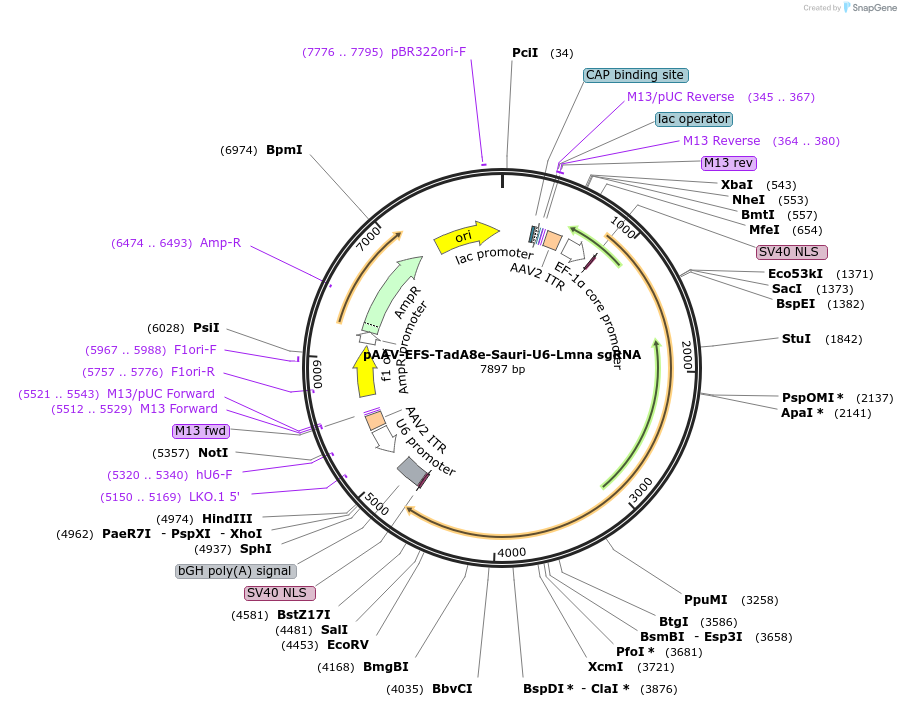
pAAV-EFS-TadA8e-Sauri-U6-Lmna sgRNA
Plasmid#206979PurposeExpresses SauriABE8e by EFS promoter and sgRNA targeting murine Lmna c.1621T mutation by U6 promoterDepositorInsertLmna sgRNA
UseAAV; Adenine base editorPromoterU6Available SinceFeb. 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
-
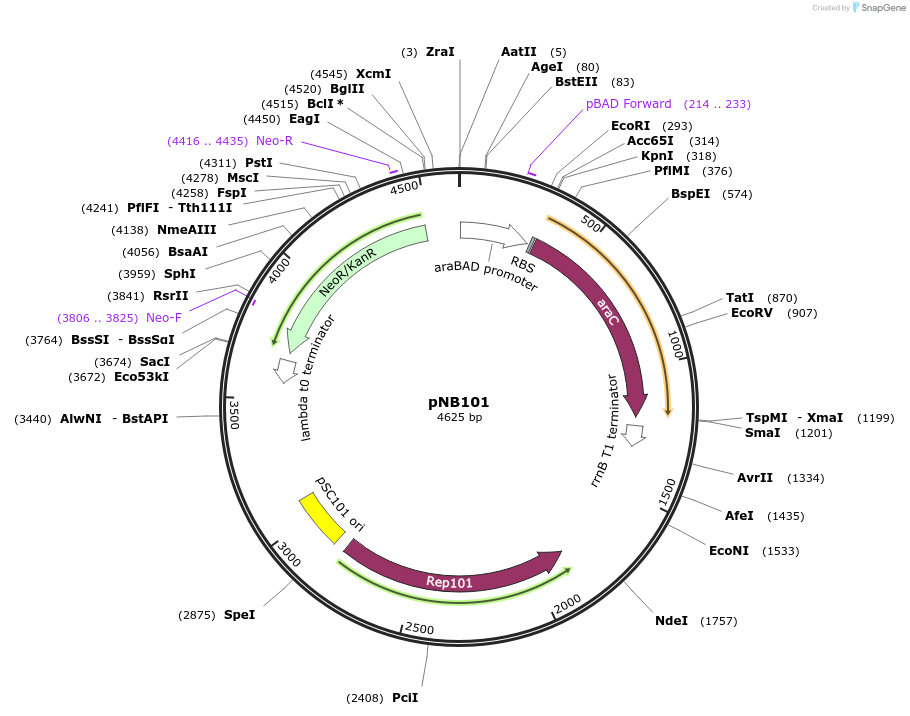
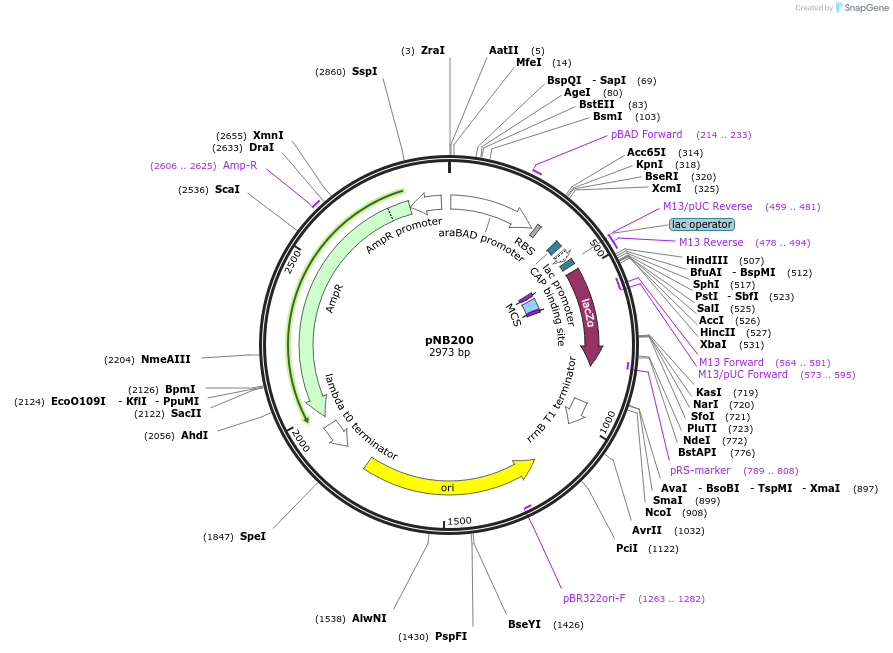
pNB201
Plasmid#169112PurposeTESEC expression vector for Alr from M. tuberculosisDepositorInsertalr (alr Mycobacterium tuberculosis H37Rv)
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E. coliPromoterpBADAvailable SinceJuly 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
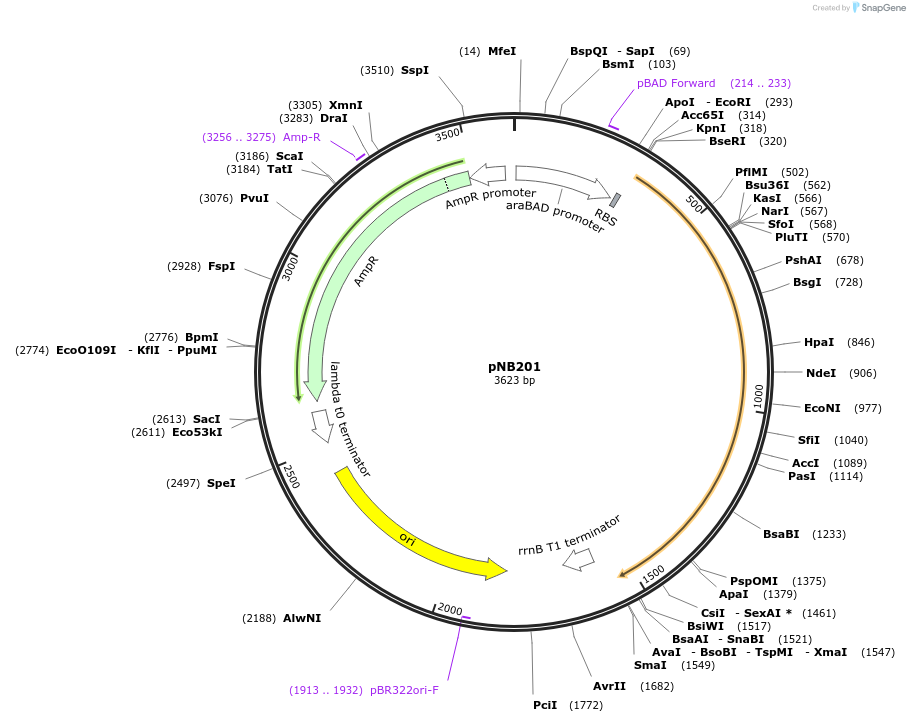
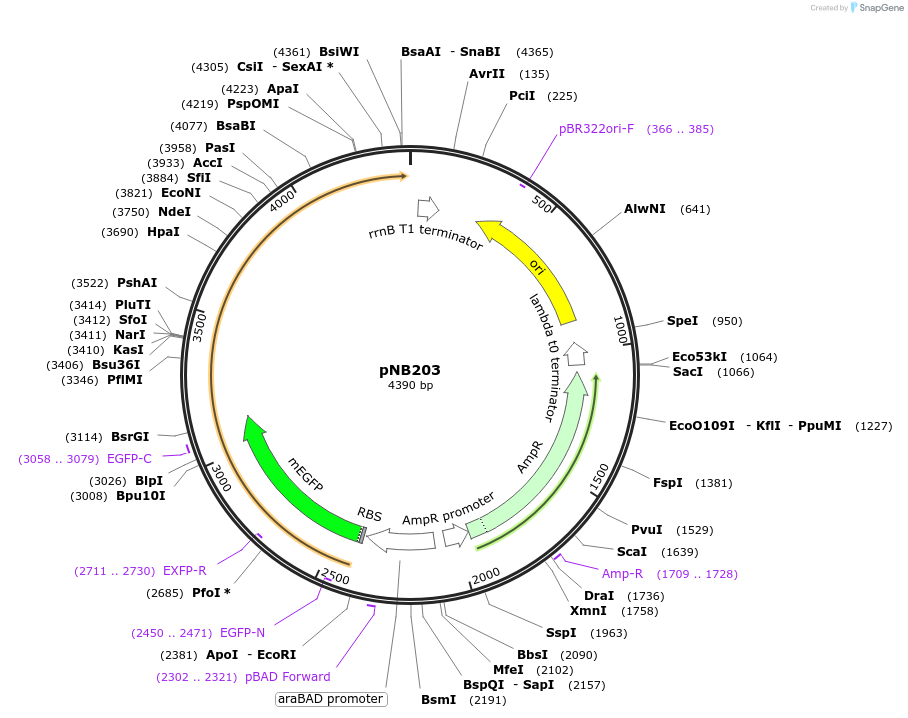
pNB203
Plasmid#169114PurposeTESEC expression vector for mEGFP-Alr fusion constructDepositorInsertmEGFP-alr (alr Mycobacterium tuberculosis H37Rv, Synthetic)
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E. coliPromoterpBADAvailable SinceJuly 8, 2022AvailabilityAcademic Institutions and Nonprofits only