We narrowed to 44,705 results for: cha;
-
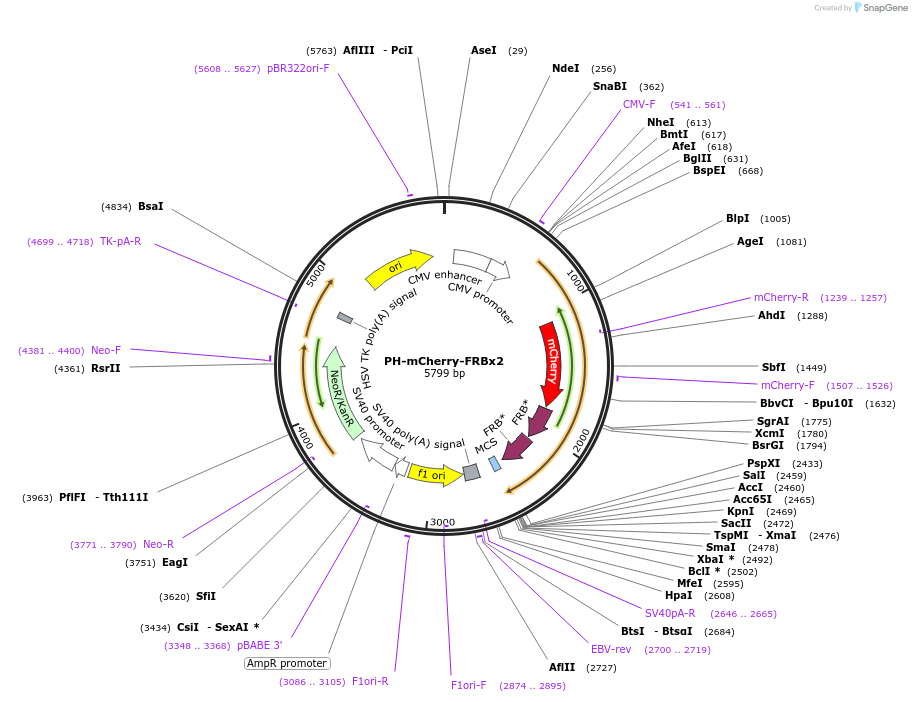
Plasmid#193570PurposeExpresses PH domain from PLC delta1 tagged with mCherry and two FRB mutants (T2098L) in mammalian cells.DepositorInsertPH domain from PLC delta1 (Plcd1 Rat)
TagsFRB (T2098L) and mCherryExpressionMammalianMutationPH domainPromoterCMVAvailable SinceFeb. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
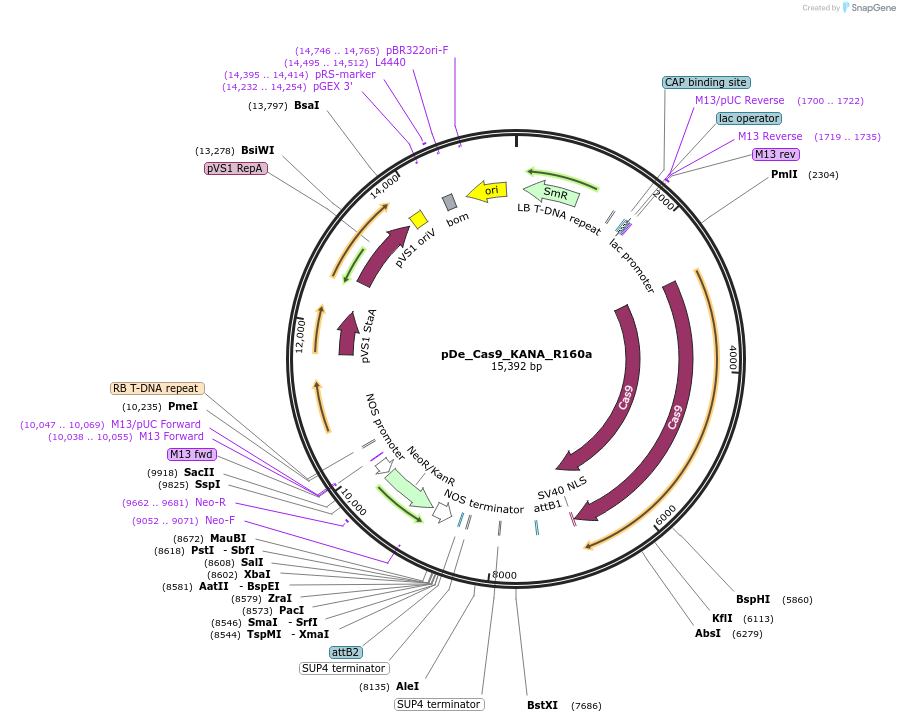
pDe_Cas9_KANA_R160a
Plasmid#173756PurposeBinary expression vector with A.th. codon-optimized Cas9 and sgRNA for miRNA160aDepositorInsertCas9_sgRNA1/R160a_sgRNA2/R160a
UseCRISPRPromoterAtU6 &StU6Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
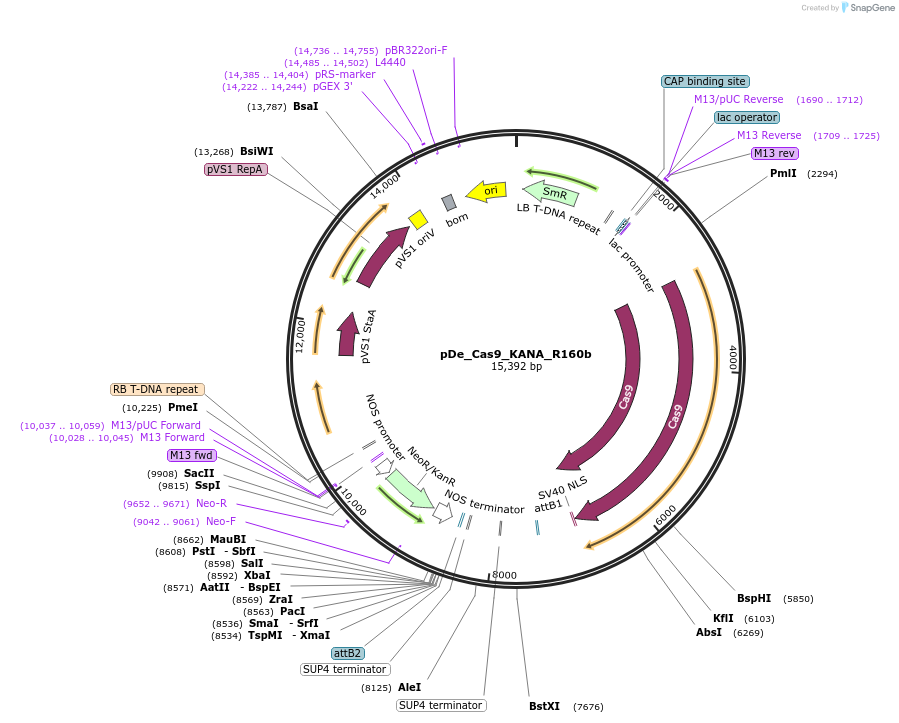
pDe_Cas9_KANA_R160b
Plasmid#173757PurposeBinary expression vector with A.th. codon-optimized Cas9 and sgRNA for miRNA160bDepositorInsertCas9_sgRNA1/R160b_sgRNA2/160b
UseCRISPRPromoterAtU6 &StU6Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
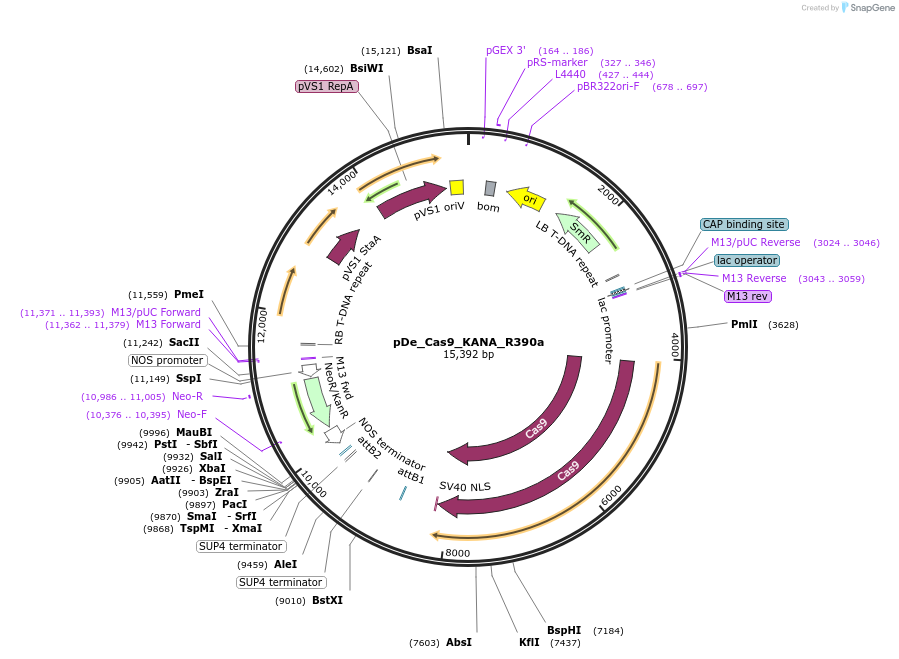
pDe_Cas9_KANA_R390a
Plasmid#173758PurposeBinary expression vector with A.th. codon-optimized Cas9 and sgRNA for miRNA390aDepositorInsertCas9_sgRNA1/390a_sgRNA2/390a
UseCRISPRPromoterAtU6 &StU6Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-MUC2 D1
Plasmid#181812PurposeExpresses human MUC2 D1, residues 21-389DepositorAvailable SinceOct. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
pILGFP4D41
Plasmid#185841PurposeTesting the CYC1 core promoter inserted with 4 Z268 elements using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PCYC1+[Z268]>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
pILGFP10CF72 (B11)
Plasmid#185859PurposeTesting the SkGAL2 promoter inserted with 2 PZ4 element using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1- PSkGAL2M3(2*PZ4)>(BamHI)yEGFP>TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
pILGFP8EFA
Plasmid#185854PurposeTesting a heat inducible protein degradation tag -DHFR P66L using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PTEF1>UBI4>RHGSGTMV>DHFRP66L>yEGFP>TPGK1-TURA3
ExpressionYeastMutationDHFR P66LAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
pCfB9339 (pgRNA_VII-1_NatMX)
Plasmid#161591PurposeEasyClone-MarkerFree guiding RNA vector to direct Cas9 to cut at site VII-1DepositorInsertguiding RNA
UseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
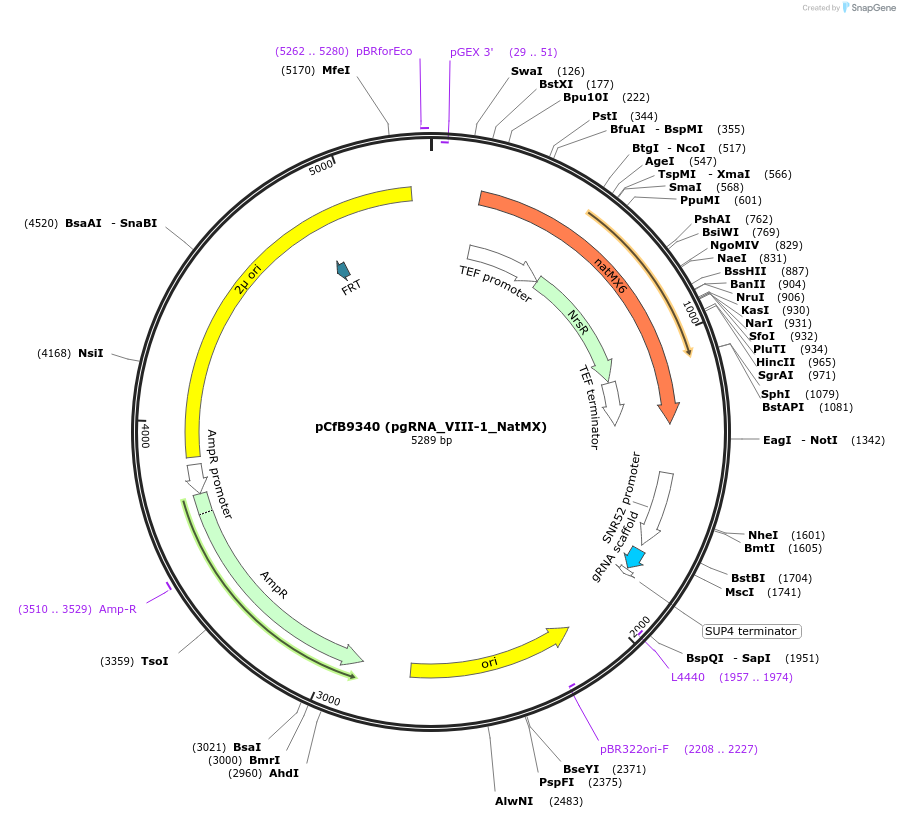
pCfB9340 (pgRNA_VIII-1_NatMX)
Plasmid#161592PurposeEasyClone-MarkerFree guiding RNA vector to direct Cas9 to cut at site VIII-1DepositorInsertguiding RNA
UseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
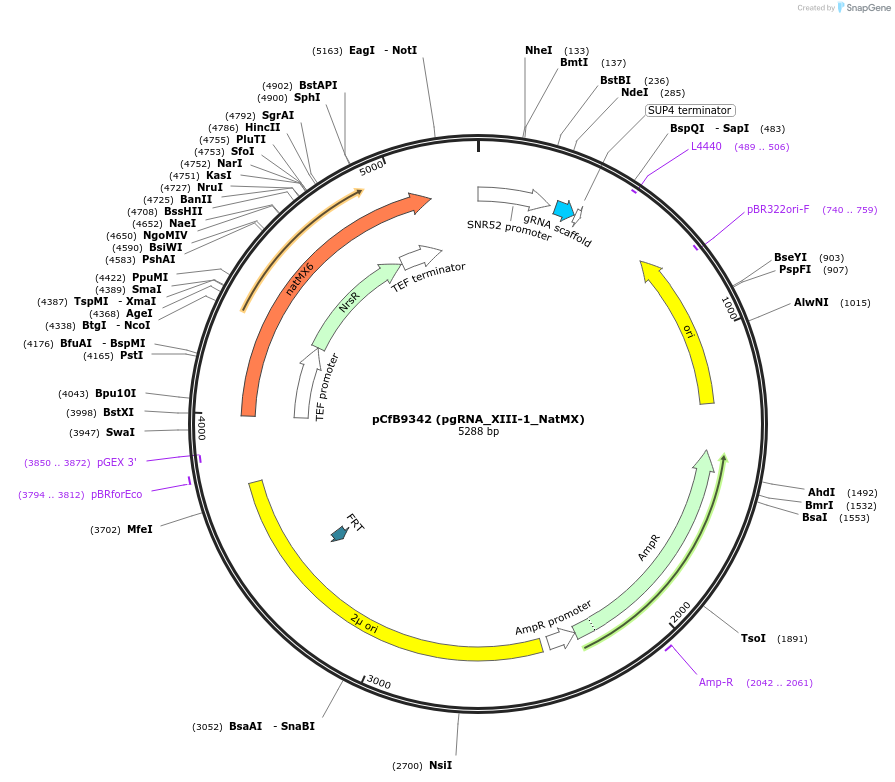
pCfB9342 (pgRNA_XIII-1_NatMX)
Plasmid#161594PurposeEasyClone-MarkerFree guiding RNA vector to direct Cas9 to cut at site XIII-1DepositorInsertguiding RNA
UseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
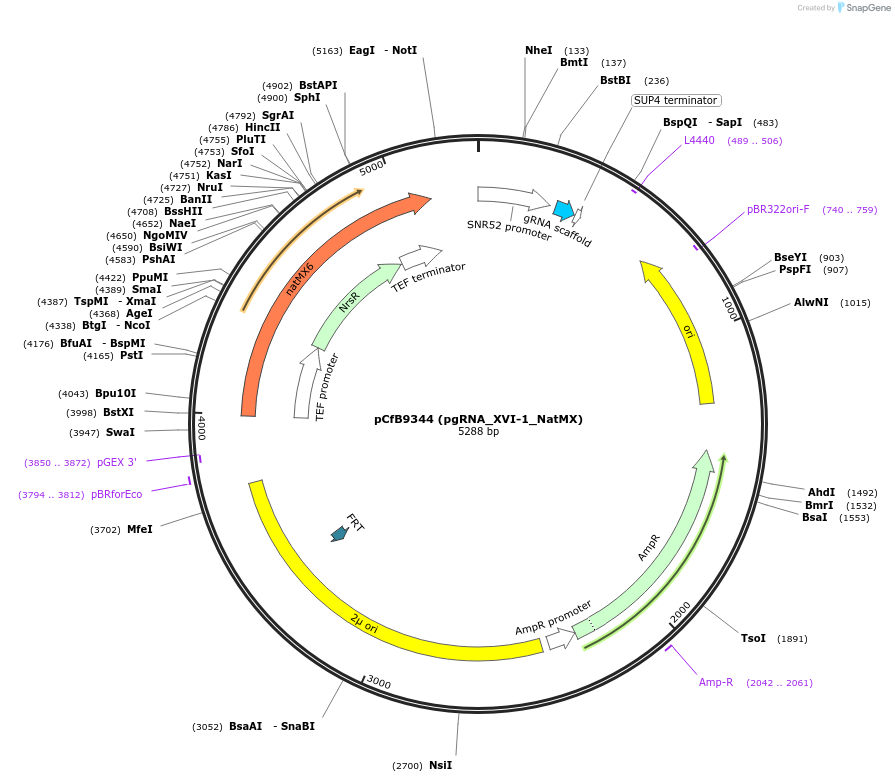
pCfB9344 (pgRNA_XVI-1_NatMX)
Plasmid#161596PurposeEasyClone-MarkerFree guiding RNA vector to direct Cas9 to cut at site XVI-1DepositorInsertguiding RNA
UseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
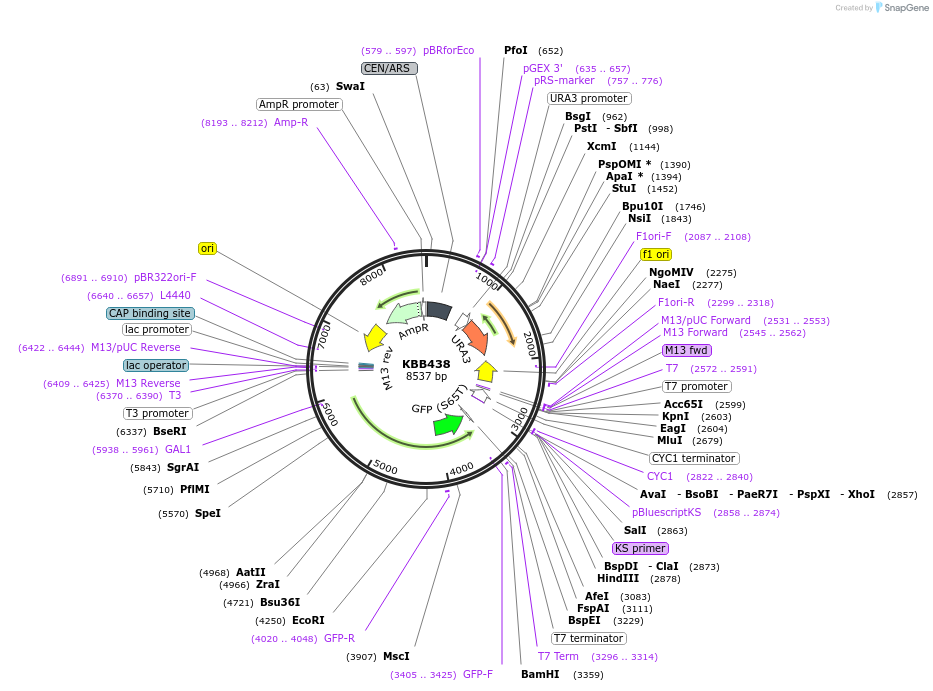
KBB438
Plasmid#185092PurposeGAL1::Rfa1[1-337, 344-621]-GFP with NLS removedDepositorInsertRFA1
ExpressionYeastMutationRfa1 with amino acids 338-343 deletedPromoterGAL1Available SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
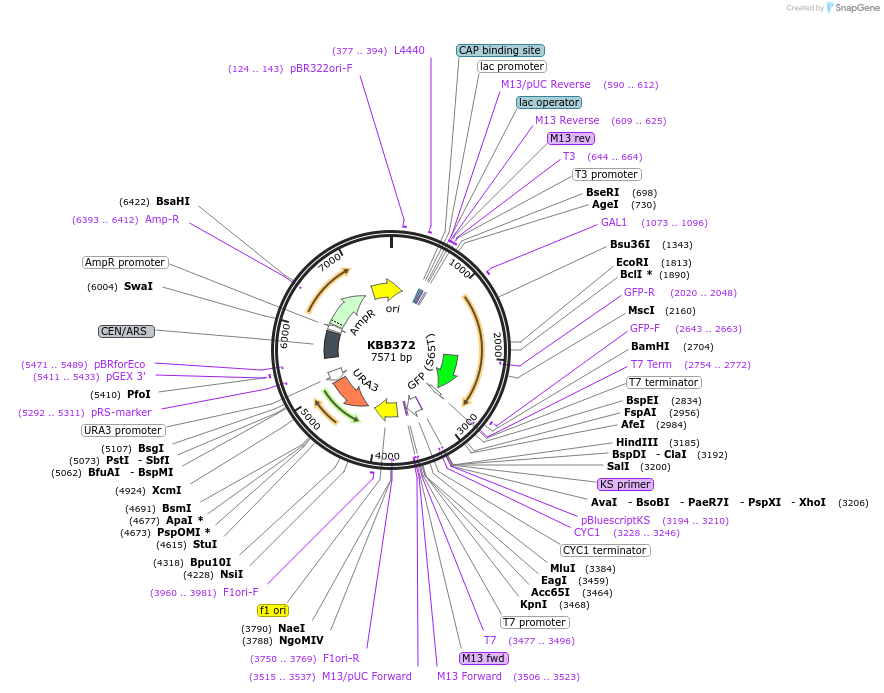
KBB372
Plasmid#185084PurposeGAL::Rfa1[330-621]-GFP under control of GAL1 promoterDepositorInsertRFA1
TagsGFPExpressionYeastMutationRfa1 amino acids 330-621PromoterGAL1Available SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
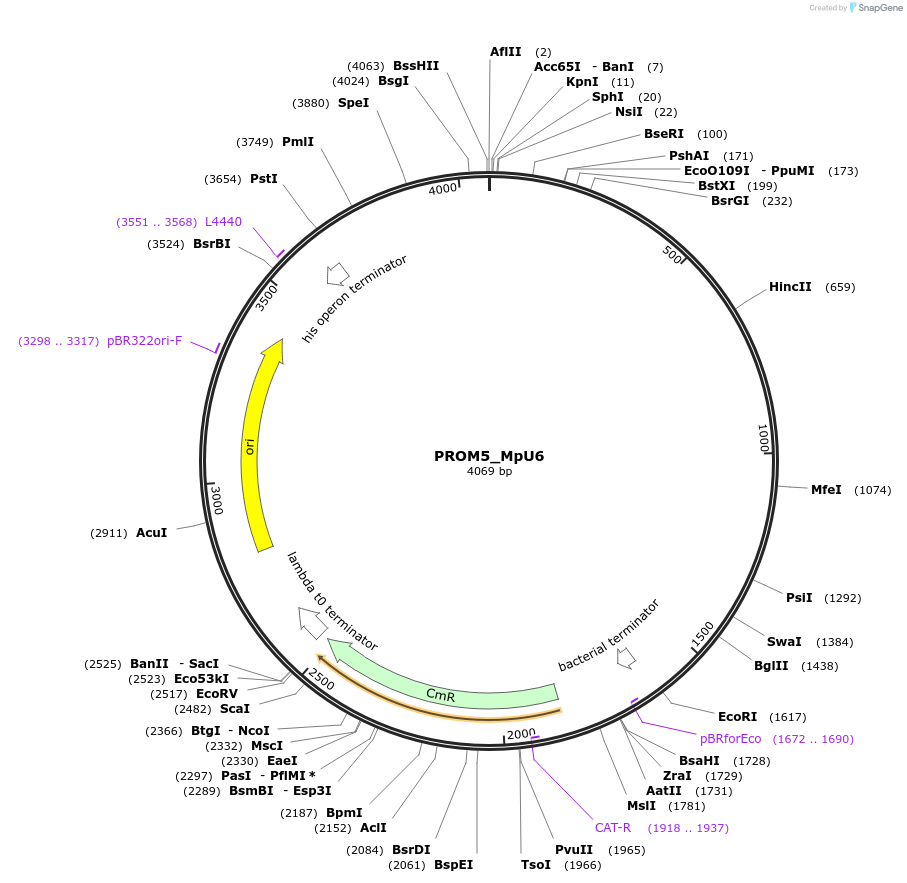
PROM5_MpU6
Plasmid#136120PurposeMp U6 promoter (type III RNA polymerase promoter) to drive expression of gRNA for CRISPR/Cas9DepositorInsertPROM5_MpU6
UseSynthetic BiologyExpressionPlantMutationBsaI/ SapI domesticatedAvailable SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
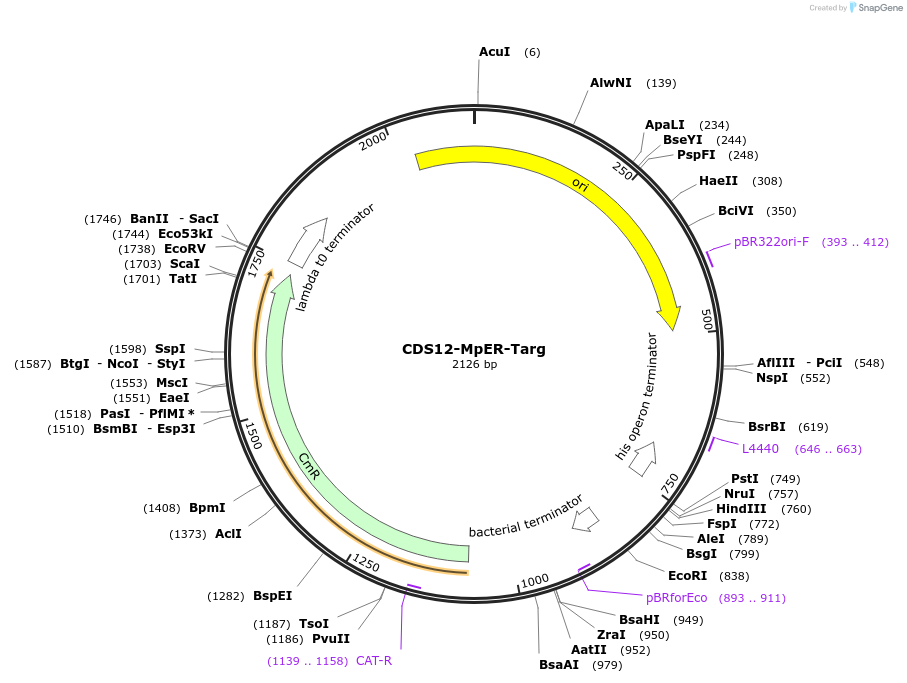
CDS12-MpER-Targ
Plasmid#136094PurposeCDS12 with ER targeting peptide to fuse with CTAG containing HDEL ER retention signal at the 3'endDepositorInsertER targeting peptide from predicted chitinase Mapoly0069s0092
UseSynthetic BiologyExpressionPlantMutationBsaI/ SapI domesticatedAvailable SinceNov. 30, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
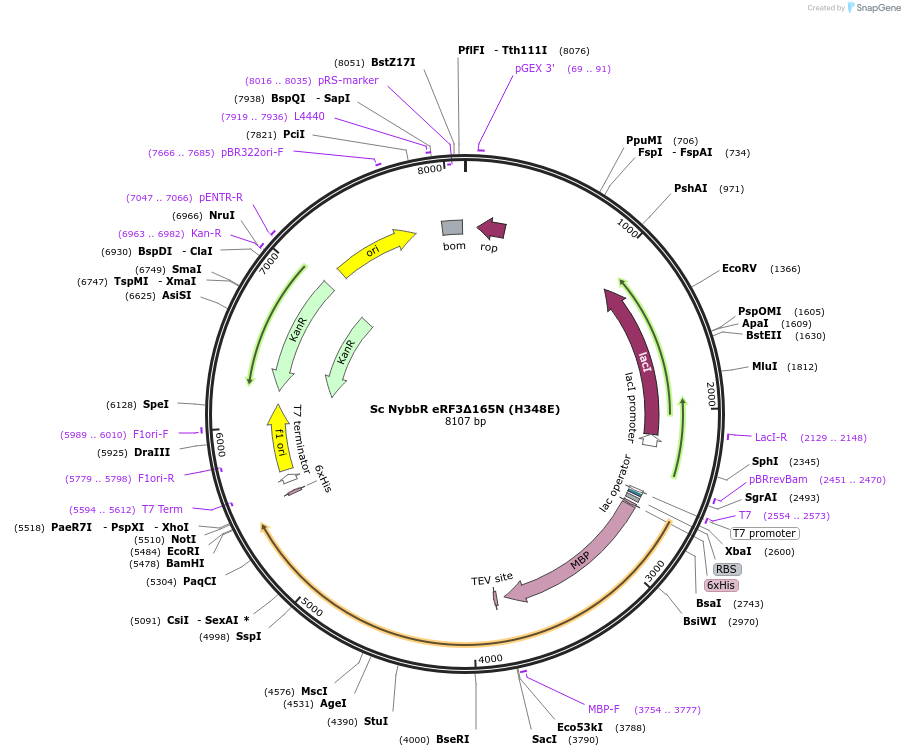
Sc NybbR eRF3∆165N (H348E)
Plasmid#175420PurposeEncodes His-MBP-TEV-ybbR-H348E eRF3∆165NDepositorInserteRF3 (SUP35 Budding Yeast)
TagsHis-MBP-TEV and ybbRExpressionBacterialMutation∆165N, H348EPromoterT7Available SinceOct. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
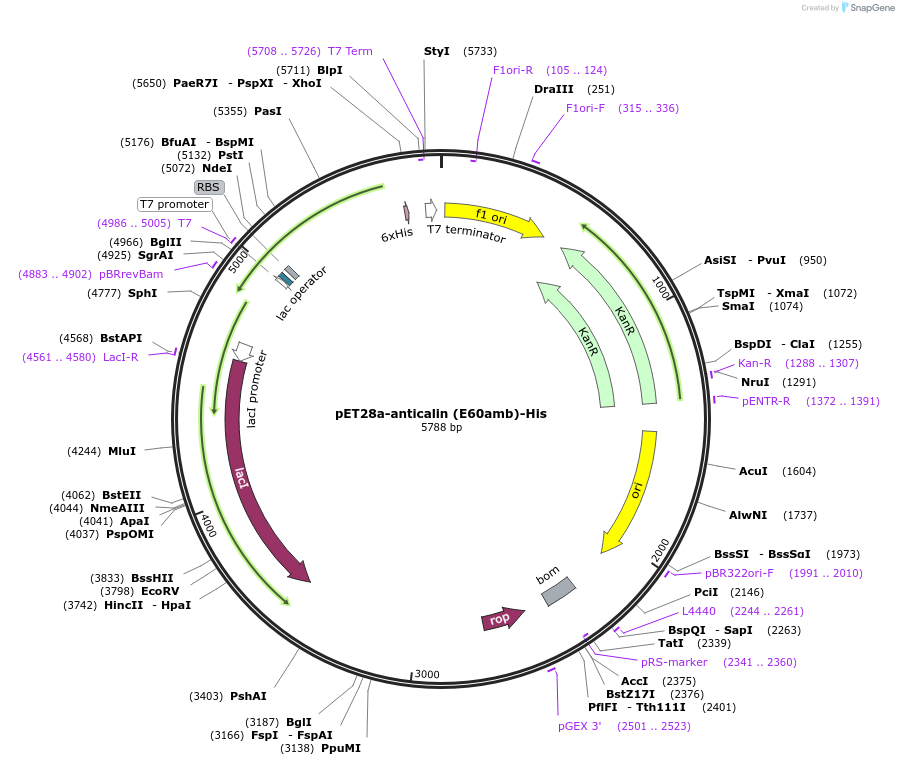
pET28a-anticalin (E60amb)-His
Plasmid#168043PurposeExpresses anticalin targeting CTLA-4 with the codon encoding E60 changed to amber codon (TAG).DepositorInsertAnticalin
Tags6x HistagExpressionBacterialMutationGlu 60 was mutated to amber codon (TAG)PromoterT7 promoterAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
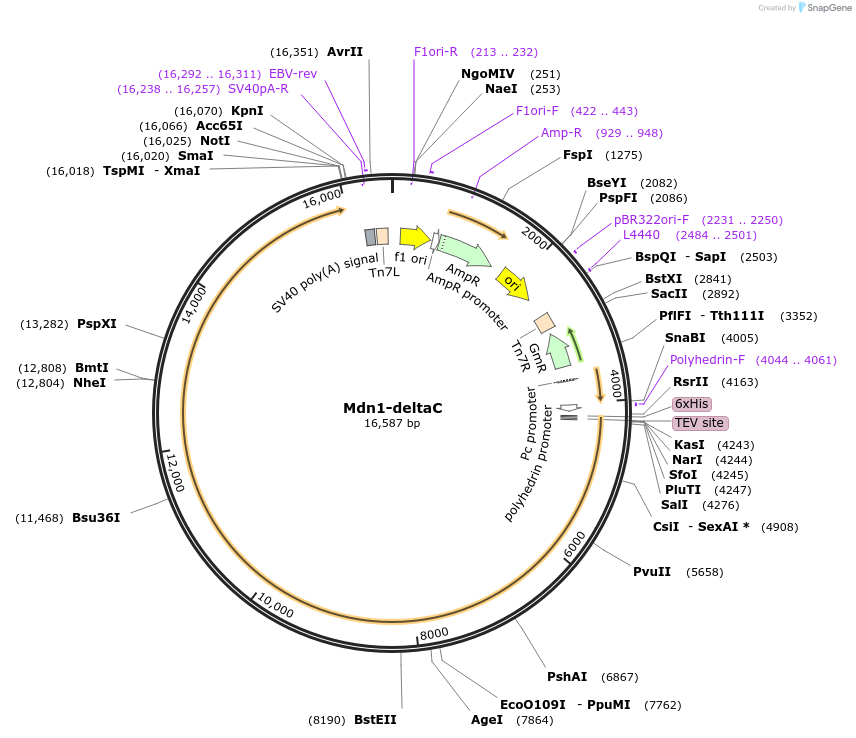
Mdn1-deltaC
Plasmid#171603PurposeExpresses S. pombe Mdn1 a.a. 1-3911 in insect cells.DepositorInsertMdn1
TagsHis6 tagExpressionInsectMutationC-terminal deletion; a.a. 1-3911 onlyAvailable SinceJuly 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
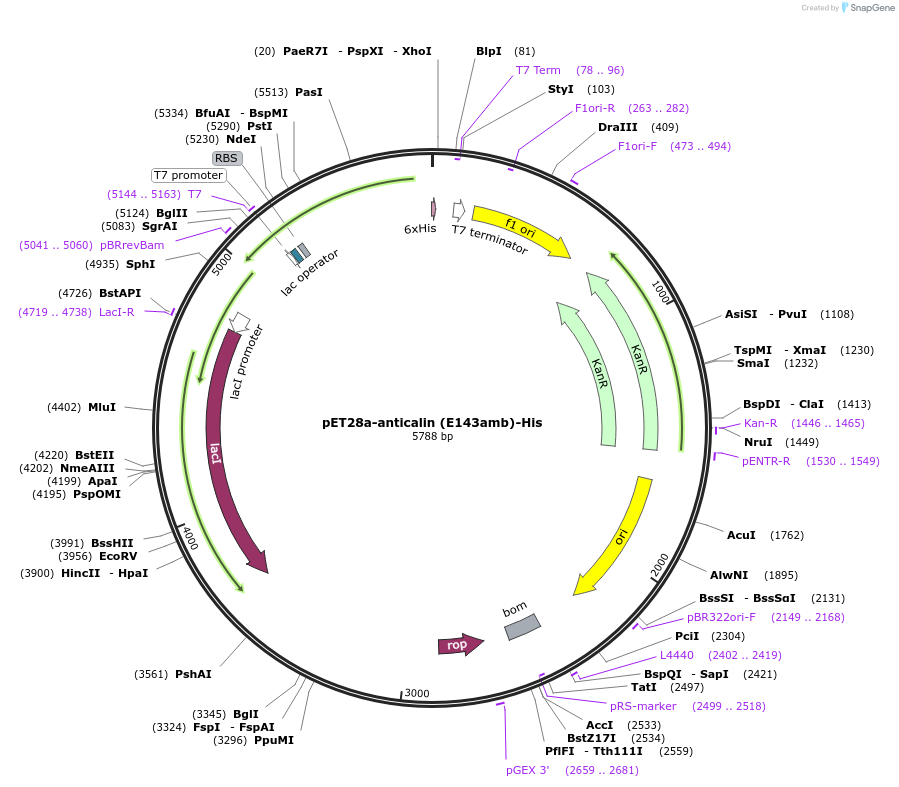
pET28a-anticalin (E143amb)-His
Plasmid#168046PurposeExpresses anticalin targeting CTLA-4 with the codon encoding E143 changed to amber codon (TAG).DepositorInsertAnticalin
Tags6x HistagExpressionBacterialMutationGlu 143 was mutated to amber codon (TAG)PromoterT7 promoterAvailable SinceJuly 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
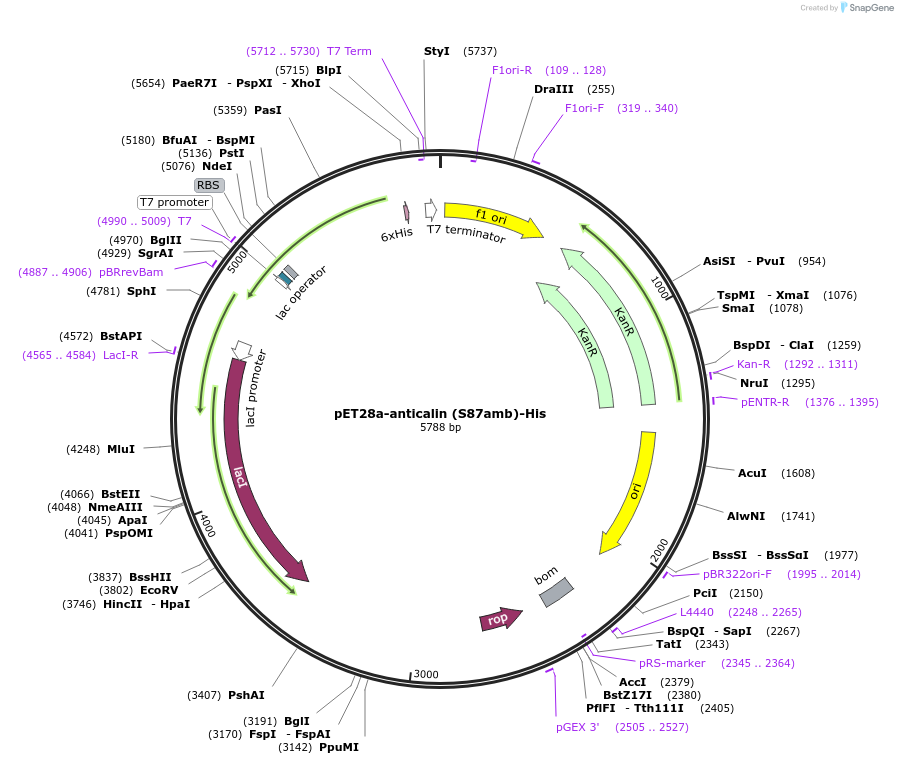
pET28a-anticalin (S87amb)-His
Plasmid#168044PurposeExpresses anticalin targeting CTLA-4 with the codon encoding S87 changed to amber codon (TAG).DepositorInsertAnticalin
Tags6x HistagExpressionBacterialMutationSer 87 was mutated to amber codon (TAG)PromoterT7 promoterAvailable SinceJuly 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
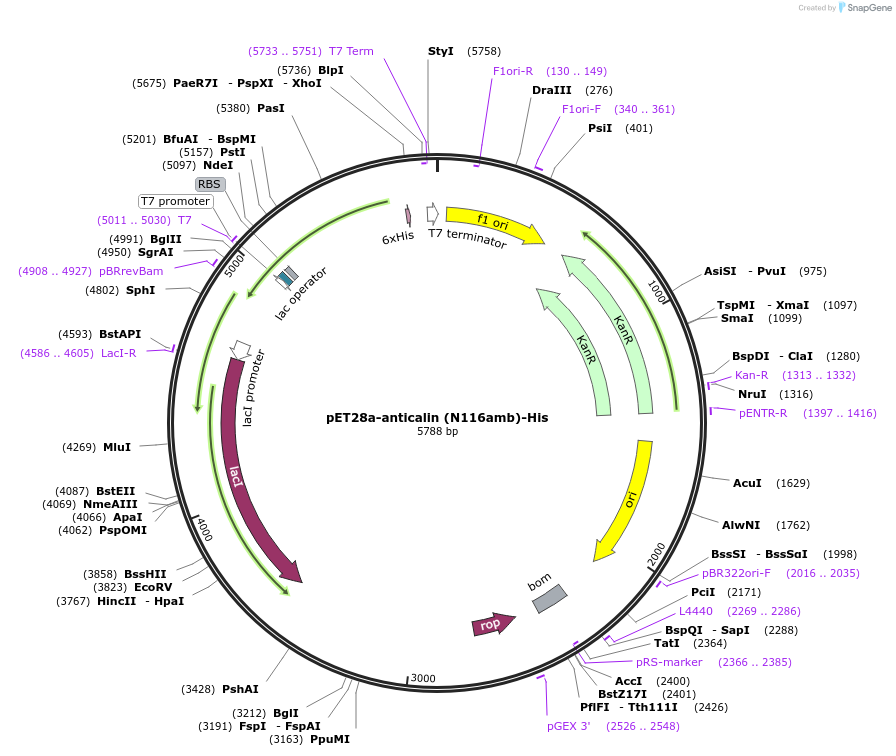
pET28a-anticalin (N116amb)-His
Plasmid#168045PurposeExpresses anticalin targeting CTLA-4 with the codon encoding N116 changed to amber codon (TAG).DepositorInsertAnticalin
Tags6x HistagExpressionBacterialMutationAsn 116 was mutated to amber codon (TAG)PromoterT7 promoterAvailable SinceJuly 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
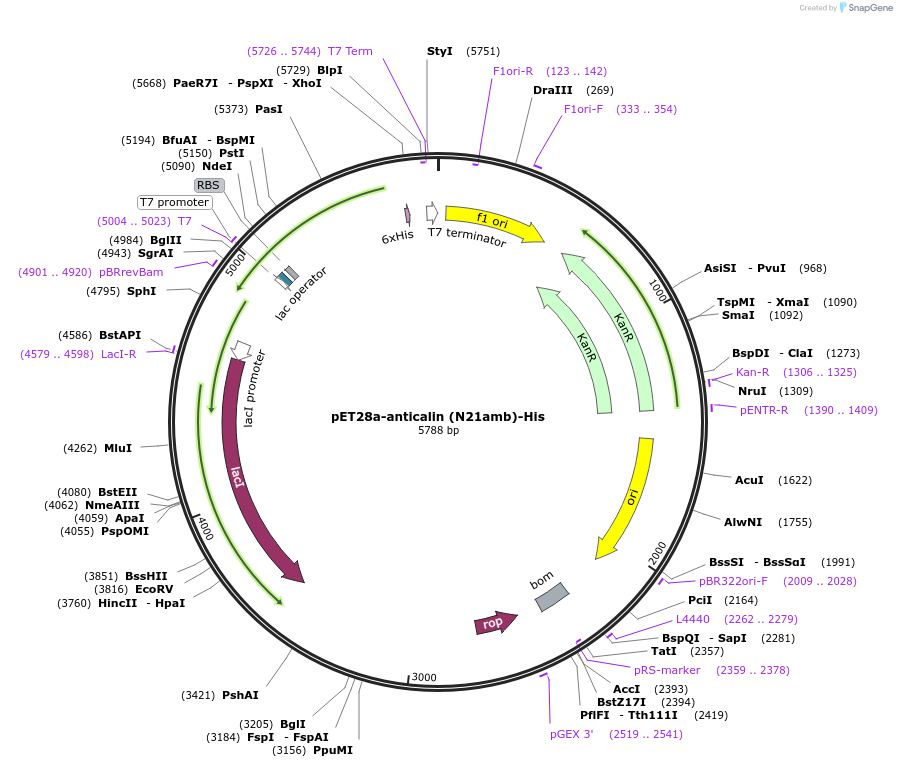
pET28a-anticalin (N21amb)-His
Plasmid#168041PurposeExpresses anticalin targeting CTLA-4 with the codon encoding N21 changed to amber codon (TAG).DepositorInsertAnticalin
Tags6x HistagExpressionBacterialMutationAsn 21 was mutated to amber codon (TAG)PromoterT7 promoterAvailable SinceJuly 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
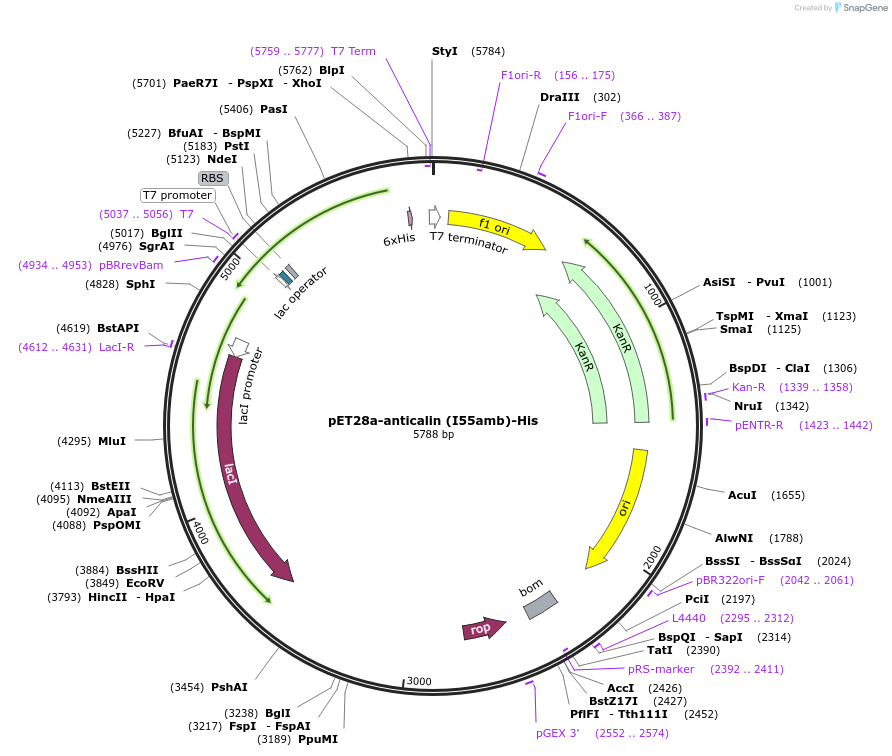
pET28a-anticalin (I55amb)-His
Plasmid#168042PurposeExpresses anticalin targeting CTLA-4 with the codon encoding I55 changed to amber codon (TAG).DepositorInsertAnticalin
Tags6x HistagExpressionBacterialMutationIle 55 was mutated to amber codon (TAG)PromoterT7 promoterAvailable SinceJune 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
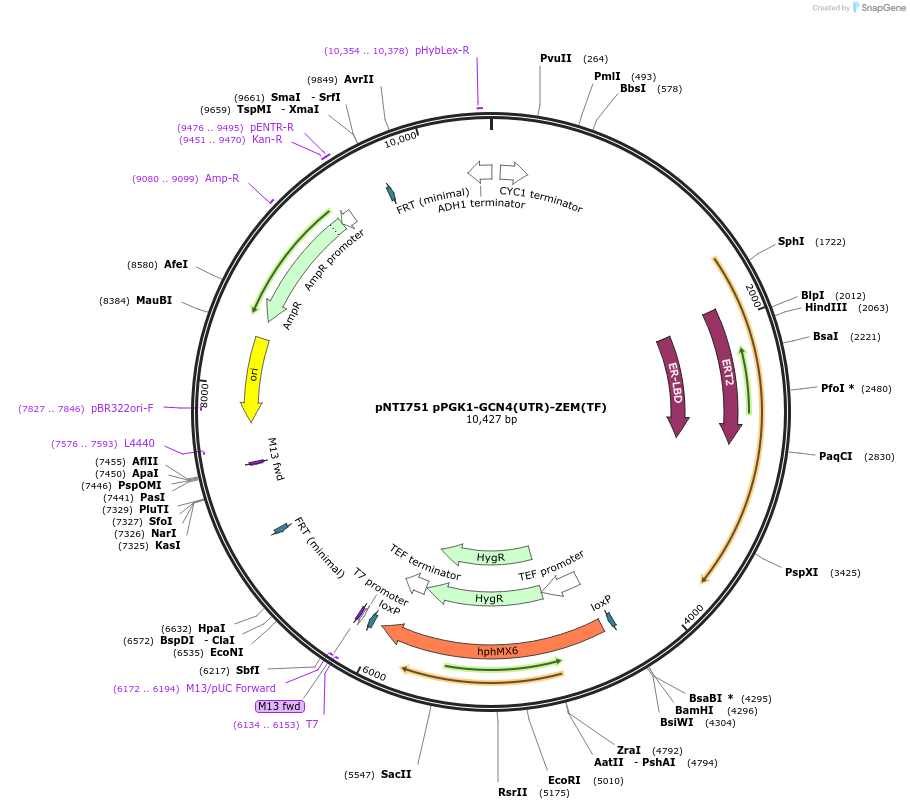
pNTI751 pPGK1-GCN4(UTR)-ZEM(TF)
Plasmid#164916PurposeExpresses the ZEM transcription factor under the translational control of the yeast GCN4 5'UTRDepositorInsertGCN4(UTR)-ZEM(TF)
UseSynthetic BiologyExpressionYeastPromoterPGK1 (yeast)Available SinceFeb. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
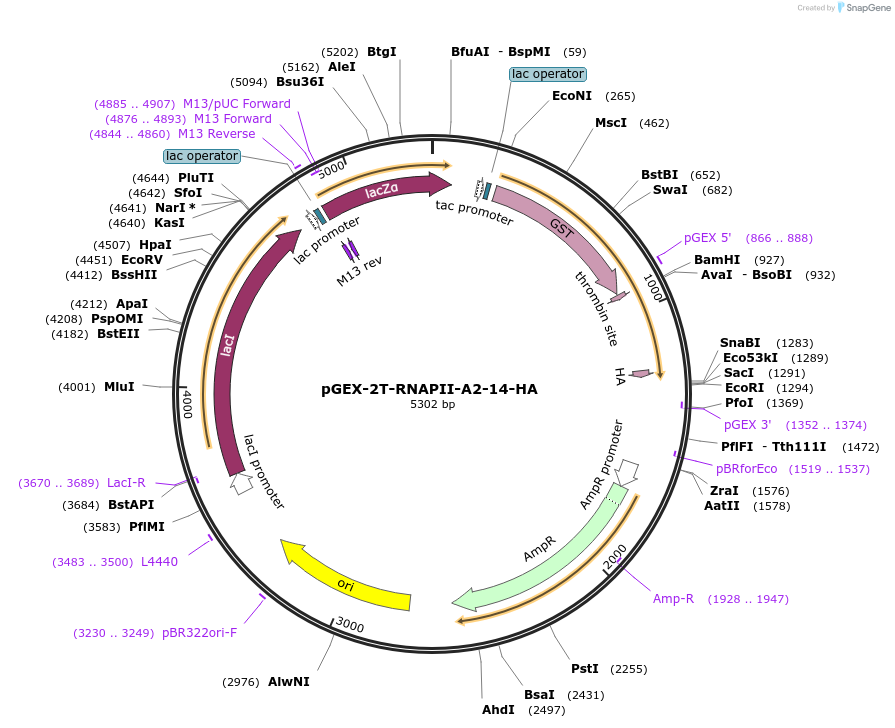
pGEX-2T-RNAPII-A2-14-HA
Plasmid#160689PurposeExpression of GST-RNA Polymerase II C-terminal domain fusion protein. Contains an S-to-A mutation in position 2 of 14 of the consensus heptapeptide repeats shown to be phosphorylated by Cdc2 kinase.DepositorInsertRNA polymerase II CTD
ExpressionBacterialMutationContains an S-to-A mutation in position 2 of 14 o…Available SinceNov. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
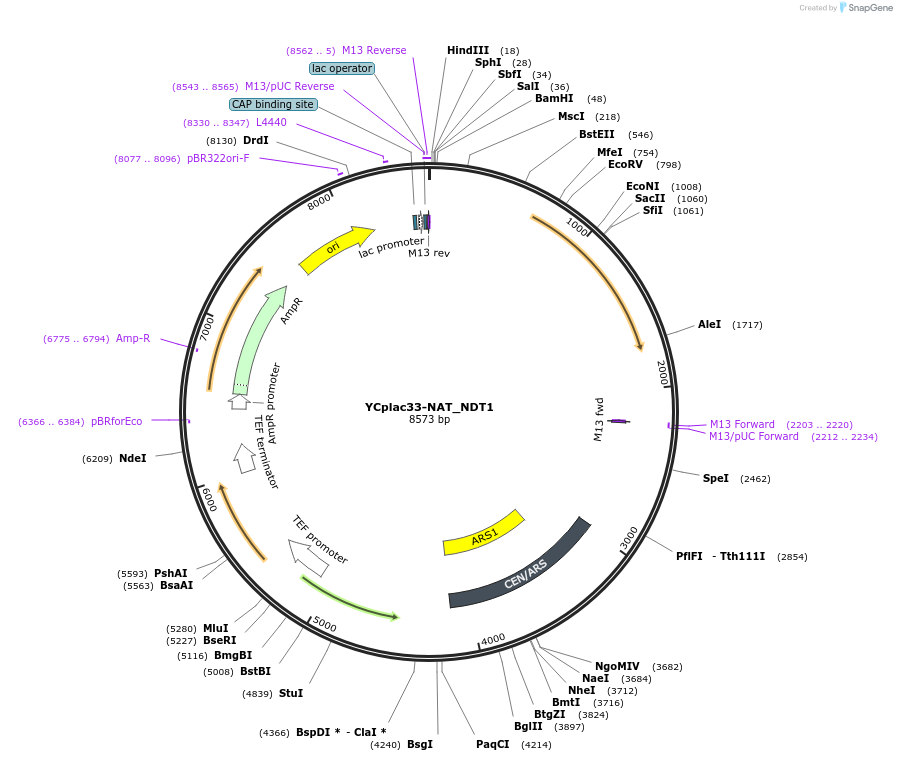
YCplac33-NAT_NDT1
Plasmid#140593PurposeExpress NDT1 in S. cerevisiaeDepositorInsertNDT1 with 5' and 3' UTRs (YIA6 Budding Yeast)
ExpressionYeastPromoterNDT1 promoter and 3'UTRAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
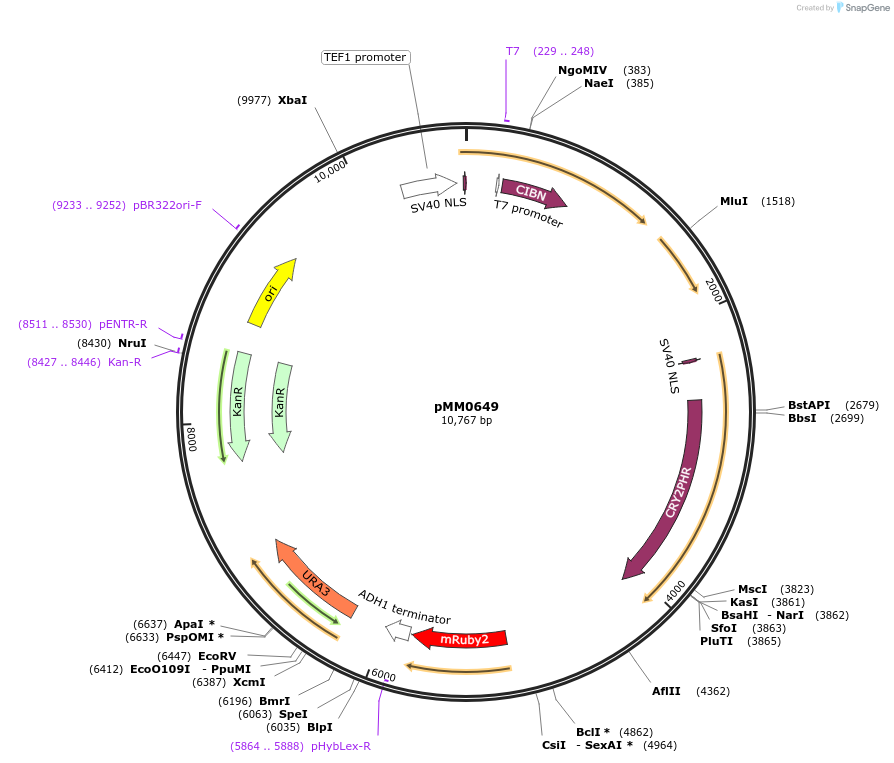
pMM0649
Plasmid#129010PurposeControl for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) integration with pWEAK-mRUBY2 reporterDepositorInsert5' URA3-pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZiF268DBD-CRY2PHR-tSSA1-pREV1-mRUBY2-tADH1-URA3 URA3 3'-KanR-ColE1
ExpressionYeastAvailable SinceFeb. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
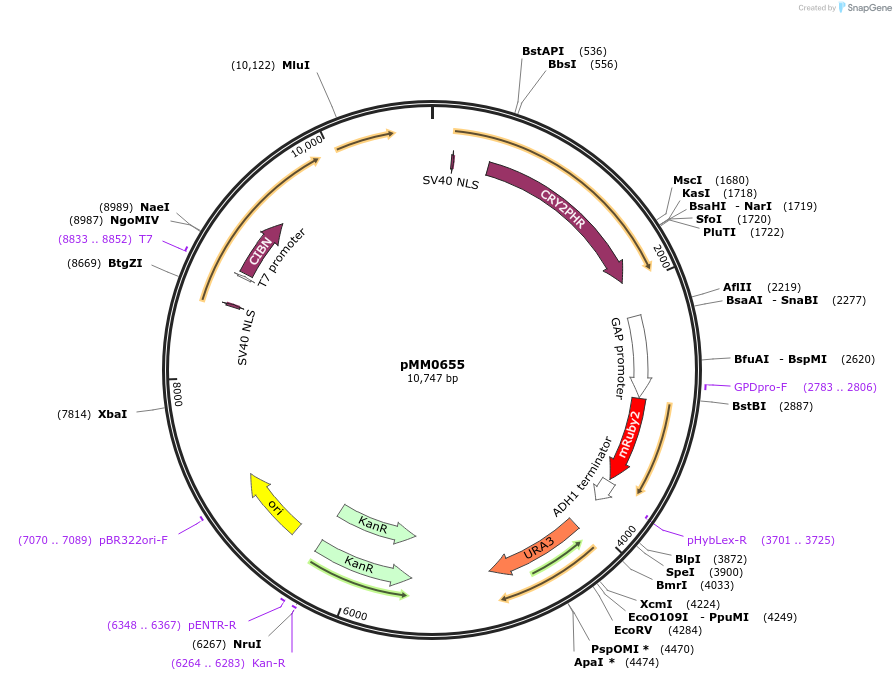
pMM0655
Plasmid#129016PurposeControl for optogenetic machinery (pMEDIUM-VP16-CIB1/pMEDIUM-ZCRY2PHR) integration with pSTRONG-mRUBY2 reporterDepositorInsert5' URA3-pRPL18B-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pTDH3-mRUBY2-tADH1-URA3 URA3 3'-KanR-ColE1
ExpressionYeastAvailable SinceOct. 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMM0654
Plasmid#129015PurposeControl for optogenetic machinery (pMEDIUM-VP16-CIB1/pMEDIUM-ZCRY2PHR) integration with pMEDIUM-mRUBY2 reporterDepositorInsert5' URA3-pRPL18B-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pRPL18B-mRUBY2-tADH1-URA3 URA3 3'-KanR-ColE1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
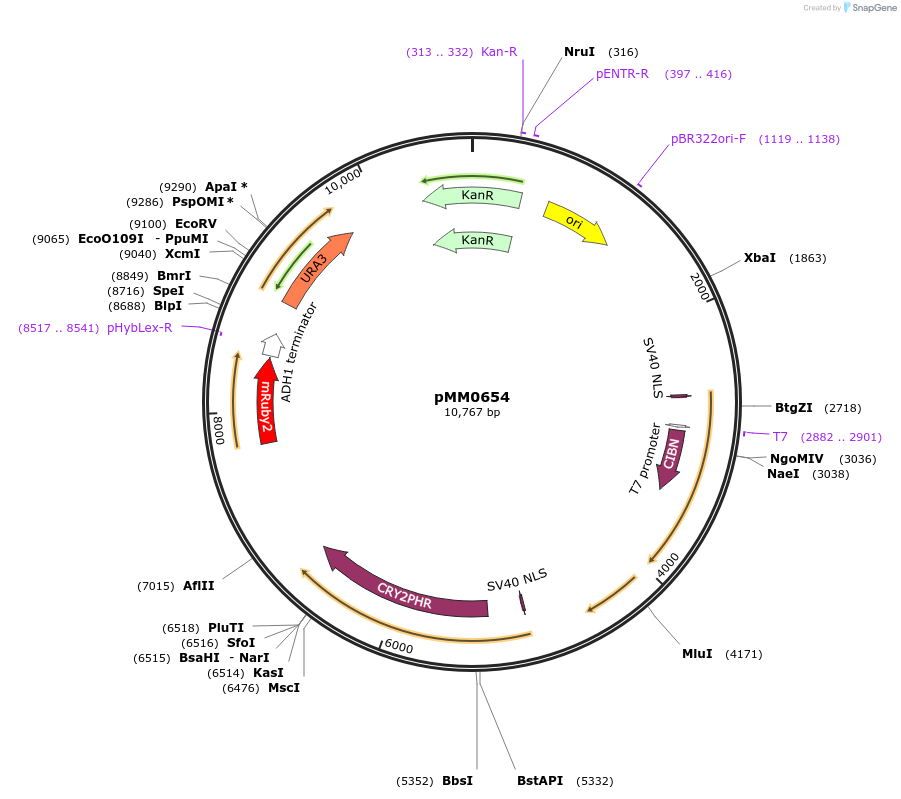
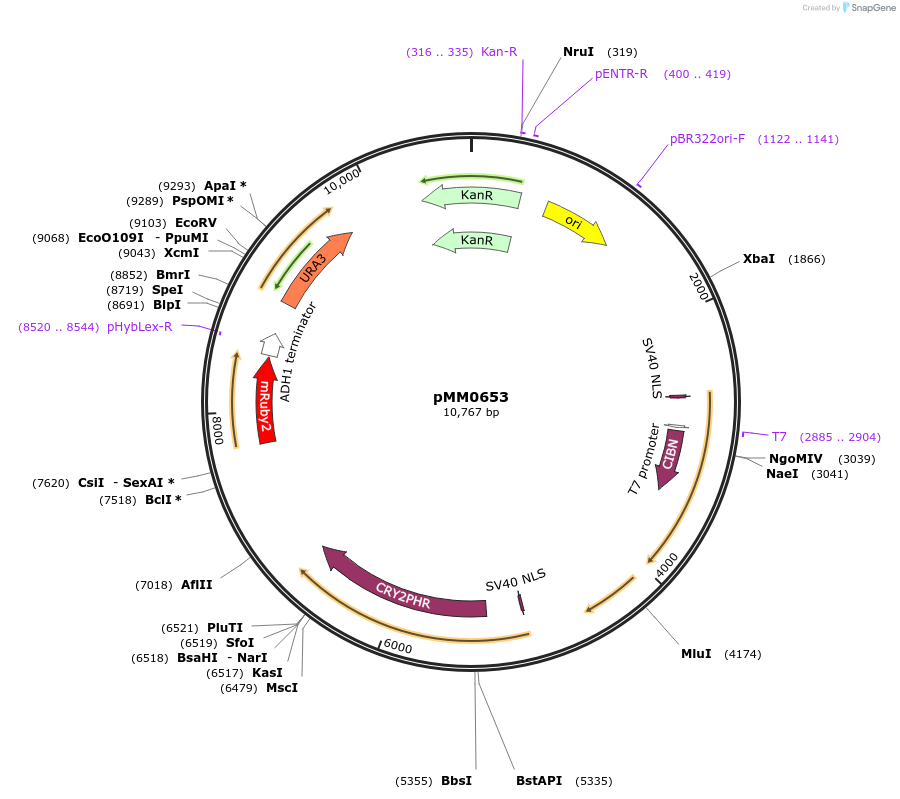
pMM0653
Plasmid#129014PurposeControl for optogenetic machinery (pMEDIUM-VP16-CIB1/pMEDIUM-ZCRY2PHR) integration with pWEAK-mRUBY2 reporterDepositorInsert5' URA3-pRPL18B-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pREV1-mRUBY2-tADH1-URA3 URA3 3'-KanR-ColE1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
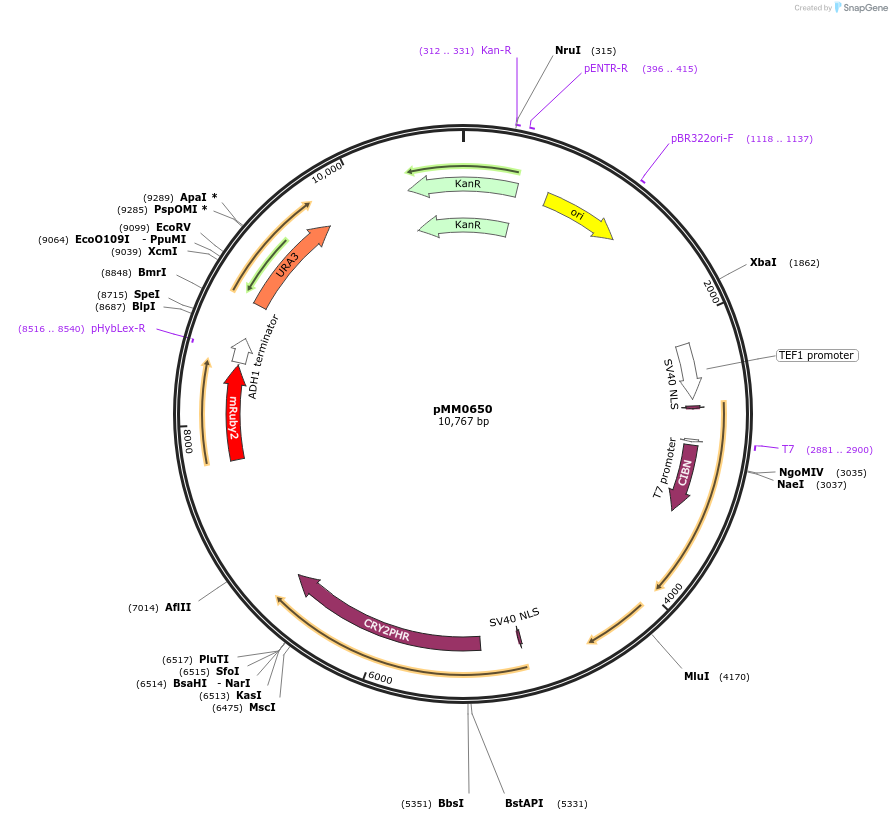
pMM0650
Plasmid#129011PurposeControl for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) integration with pMEDIUM-mRUBY2 reporterDepositorInsert5' URA3-pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pRPL18B-mRUBY2-tADH1-URA3 URA3 3'-KanR-ColE1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
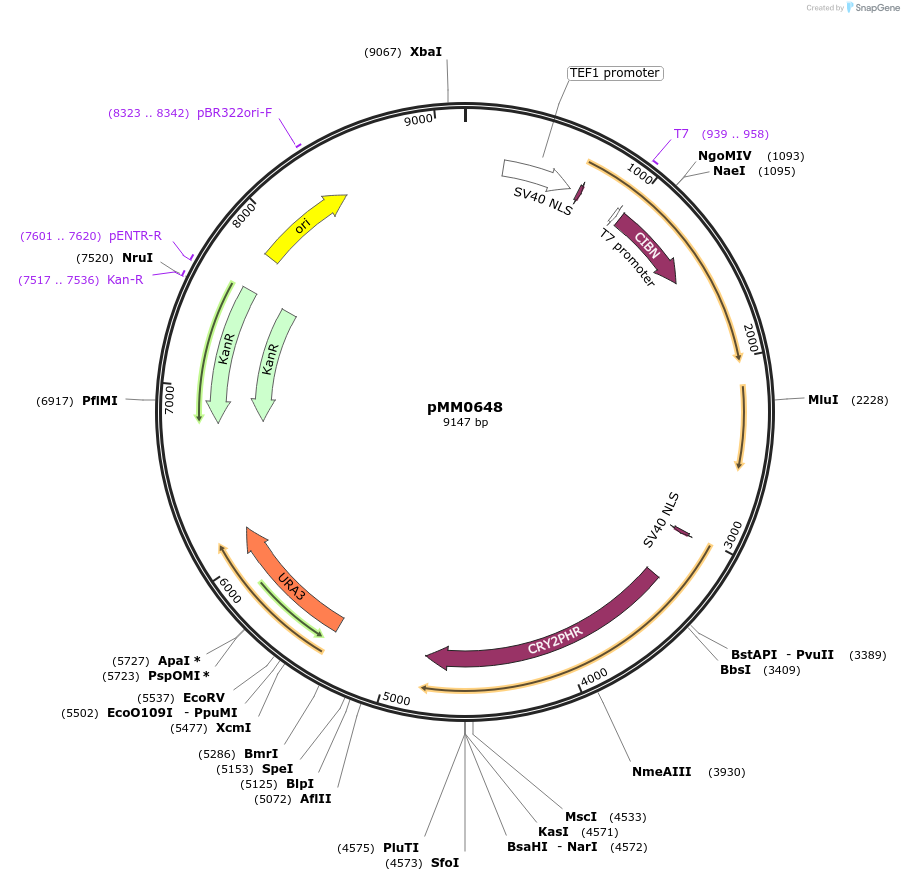
pMM0648
Plasmid#129009PurposeControl for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) integration effectsDepositorInsert5' URA3 homology-pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-Spacer-URA3 URA3 3'-homology KanR-ColE1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
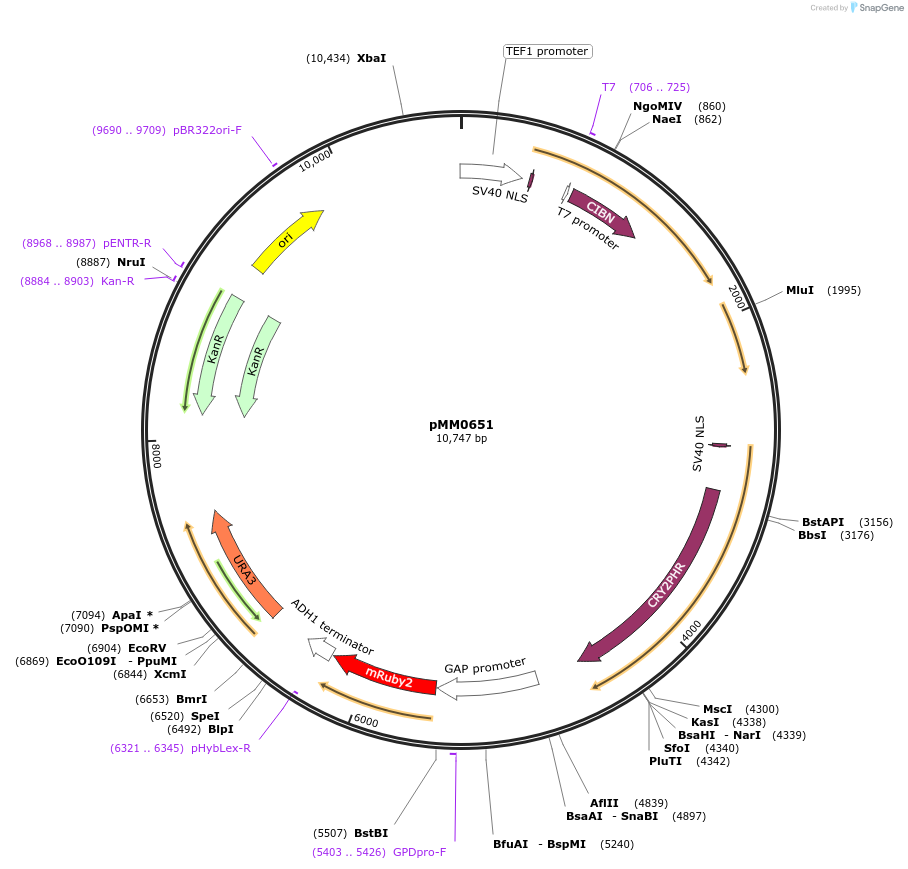
pMM0651
Plasmid#129012PurposeControl for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) integration with pSTRONG-mRUBY2 reporterDepositorInsert5' URA3-pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pTDH3-mRUBY2-tADH1-URA3 URA3 3'-KanR-ColE1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
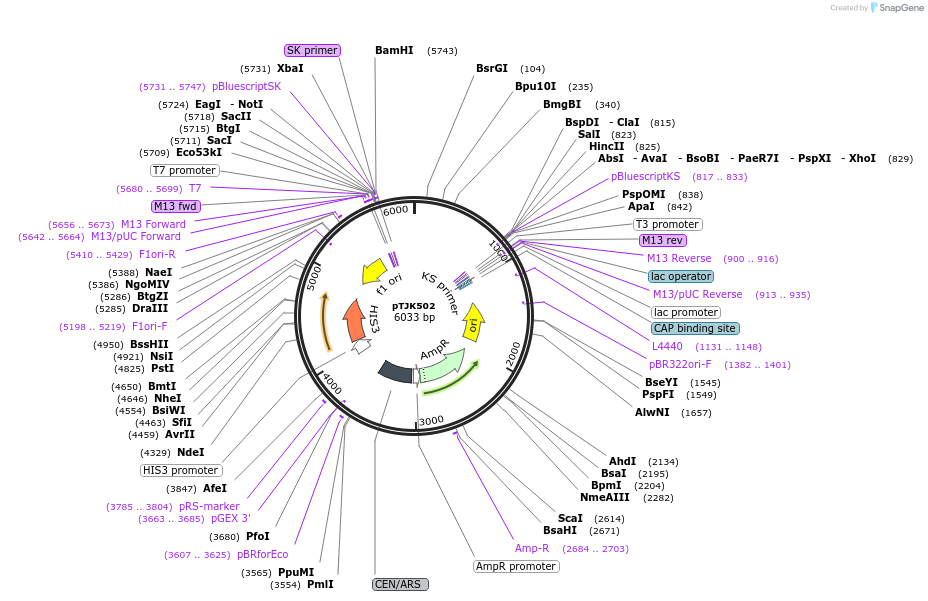
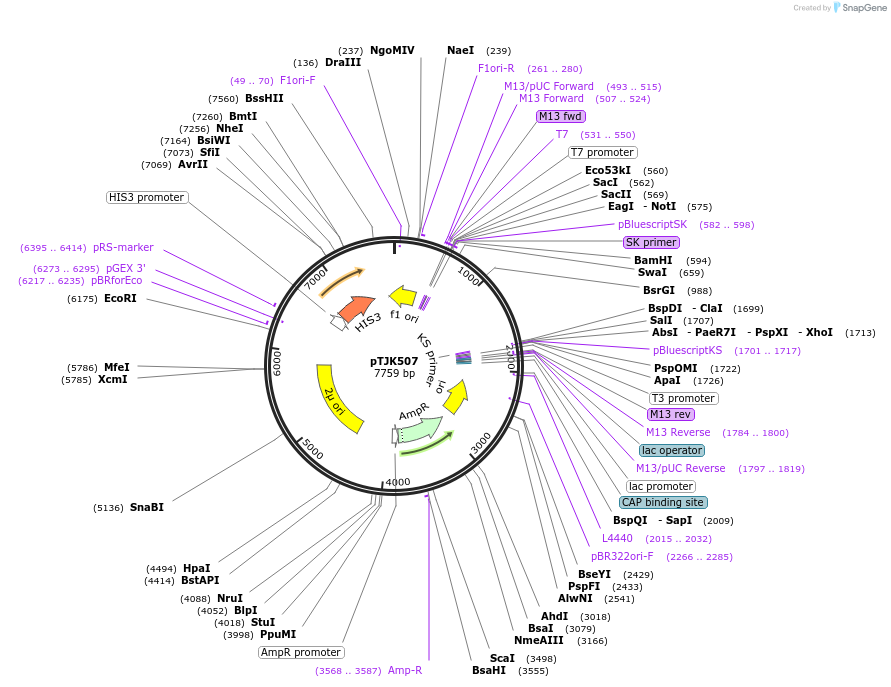
pTJK502
Plasmid#121457PurposeCEN HIS3 CNB1DepositorAvailable SinceMay 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
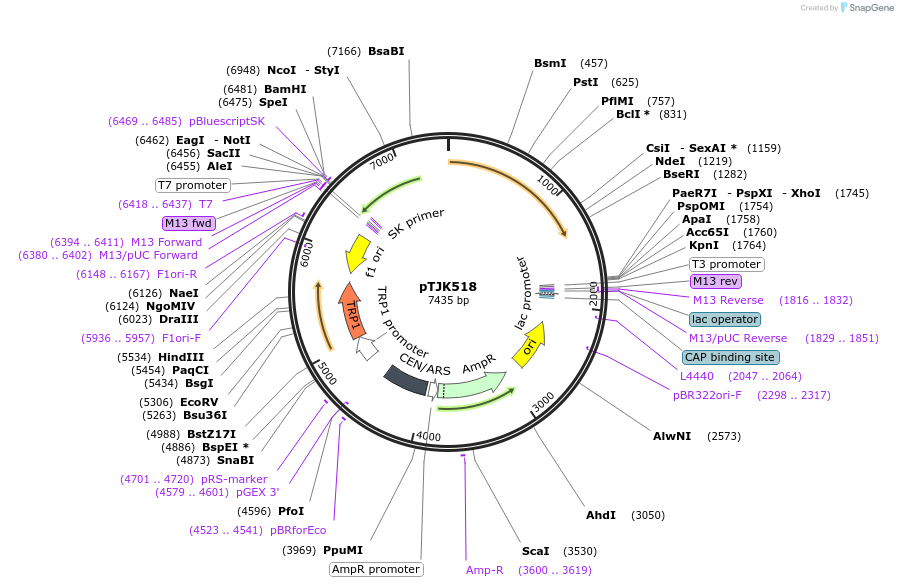
pTJK518
Plasmid#121475PurposeCEN TRP1 CNA1ΔAIDDepositorInsertCNA1deltaAID
UseYeast cen vectorMutationTruncated after amino acid 508PromoterEndogenous genomic promoter regionAvailable SinceApril 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
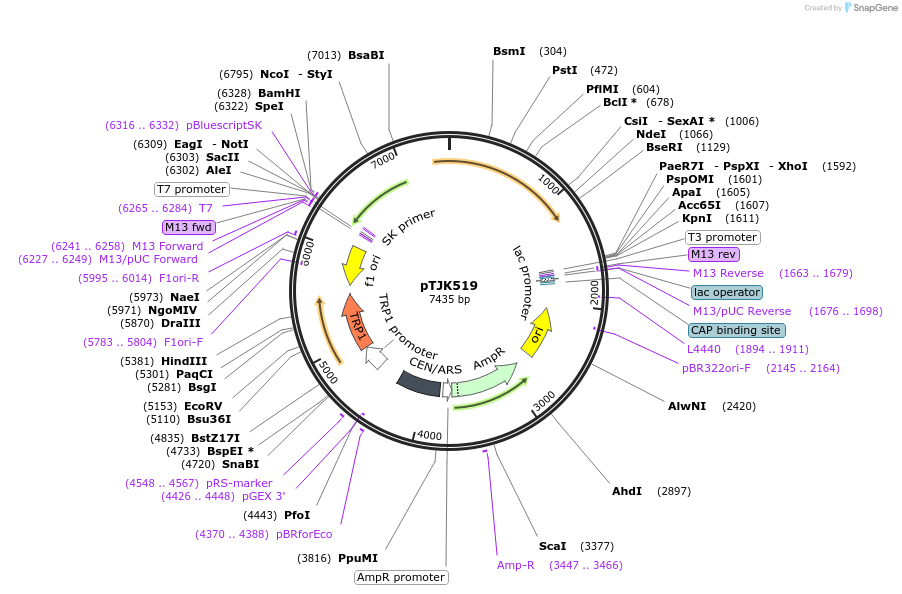
pTJK519
Plasmid#121476PurposeCEN TRP1 CNA1ΔCBDDepositorInsertCNA1deltaCBD
UseYeast cen vectorMutationTruncated after amino acid 453PromoterEndogenous genomic promoter regionAvailable SinceMarch 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
-
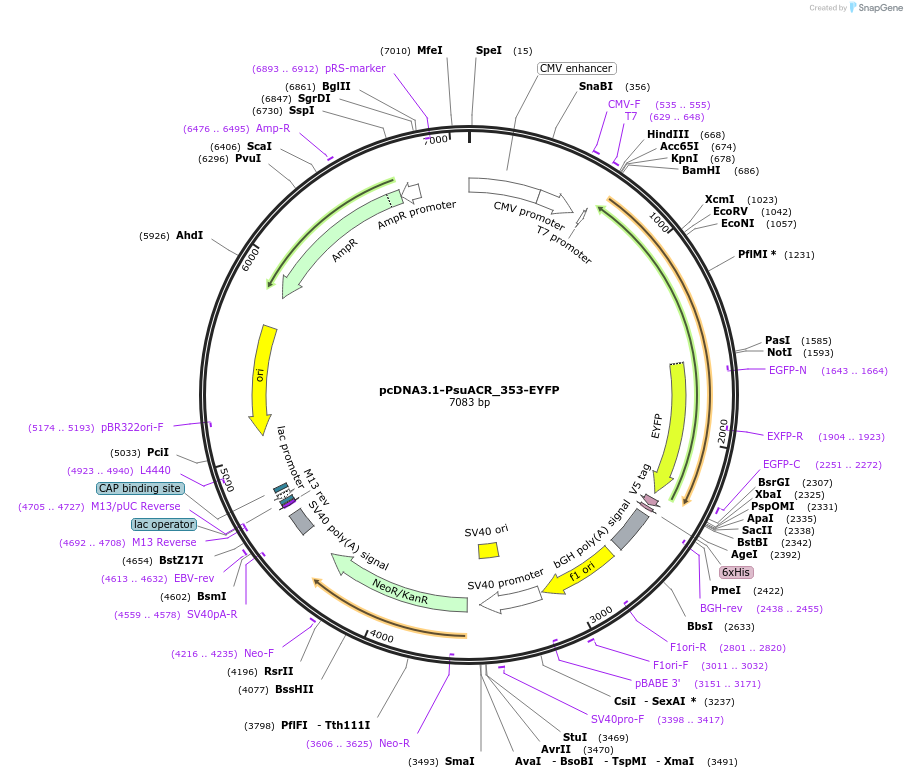
pcDNA3.1-PsuACR_353-EYFP
Plasmid#111076PurposeAnion channelrhodopsin from Proteomonas sulcata (CCMP704) for expression in mammalian cellsDepositorInsertAnion channelrhodopsin PsuACR_353
TagsEYFPExpressionMammalianAvailable SinceJune 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
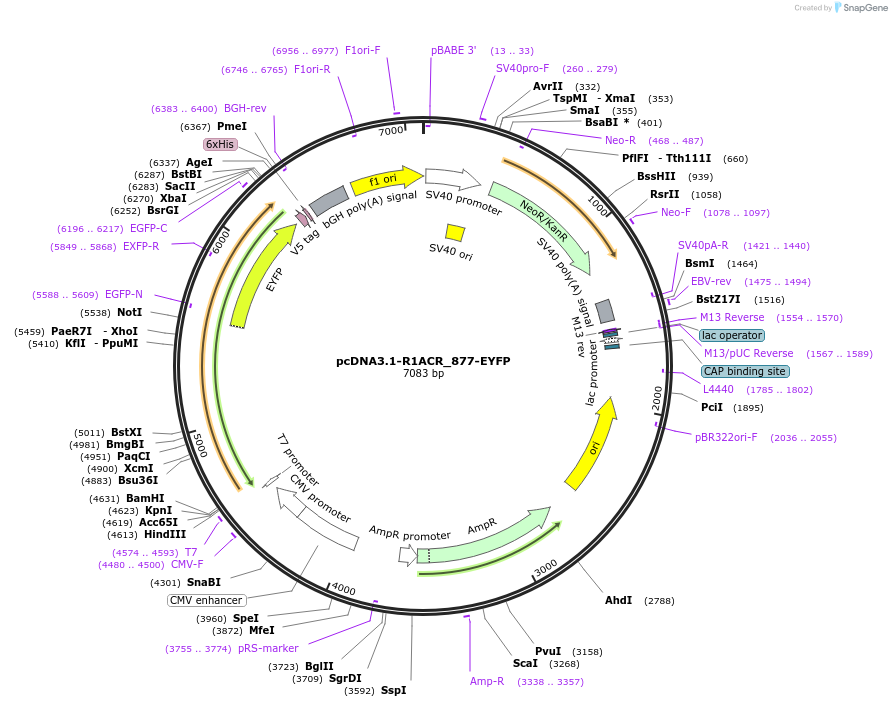
pcDNA3.1-R1ACR_877-EYFP
Plasmid#103883PurposeAnion channelrhodopsin from Rhodomonas sp. (CCMP768) for expression in mammalian cellsDepositorInsertAnion channelrhodopsin R1ACR_877
TagsEYFPExpressionMammalianMutationhuman codon optimizedPromoterCMVAvailable SinceNov. 30, 2017AvailabilityAcademic Institutions and Nonprofits only