We narrowed to 14,204 results for: RING;
-
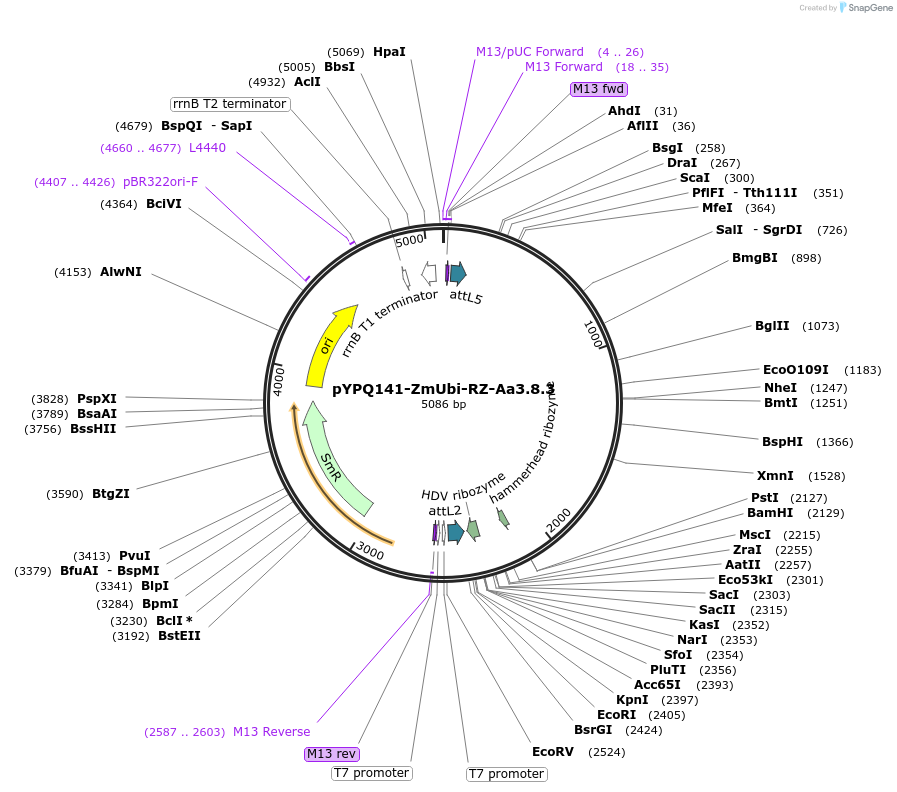
Plasmid#136374PurposeGateway entry clone for AaCas12b sgRNA expression under ZmUbi promoter with ribozyme processing; sgRNA scaffold 3.8 with MS2 at the third stem loop is usedDepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceFeb. 18, 2020AvailabilityAcademic Institutions and Nonprofits only
-
pYPQ141-ZmUbi-RZ-Aac.3
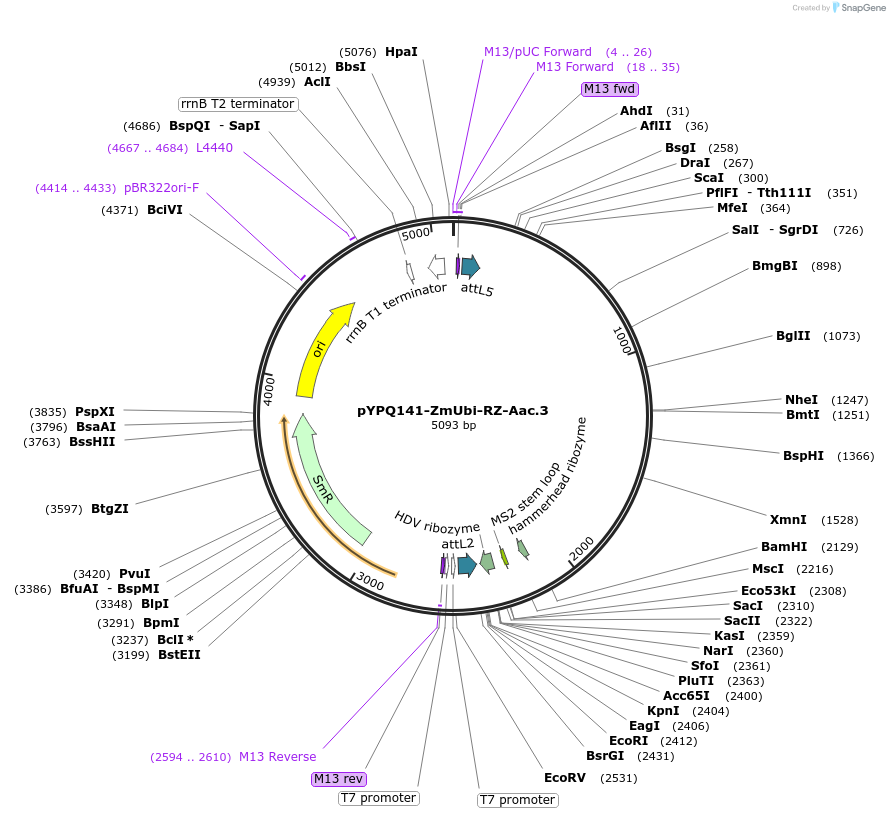
Plasmid#136377PurposeGateway entry clone for AacCas12b sgRNA expression under ZmUbi promoter with ribozyme processing; sgRNA scaffold with MS2 at the third stem loop is usedDepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceFeb. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
pYPQ141-ZmUbi-RZ-Aa1.2.3
Plasmid#136372PurposeGateway entry clone for AaCas12b sgRNA expression under ZmUbi promoter with ribozyme processing; sgRNA scaffold 1.2 with MS2 at the third stem loop is usedDepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceFeb. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
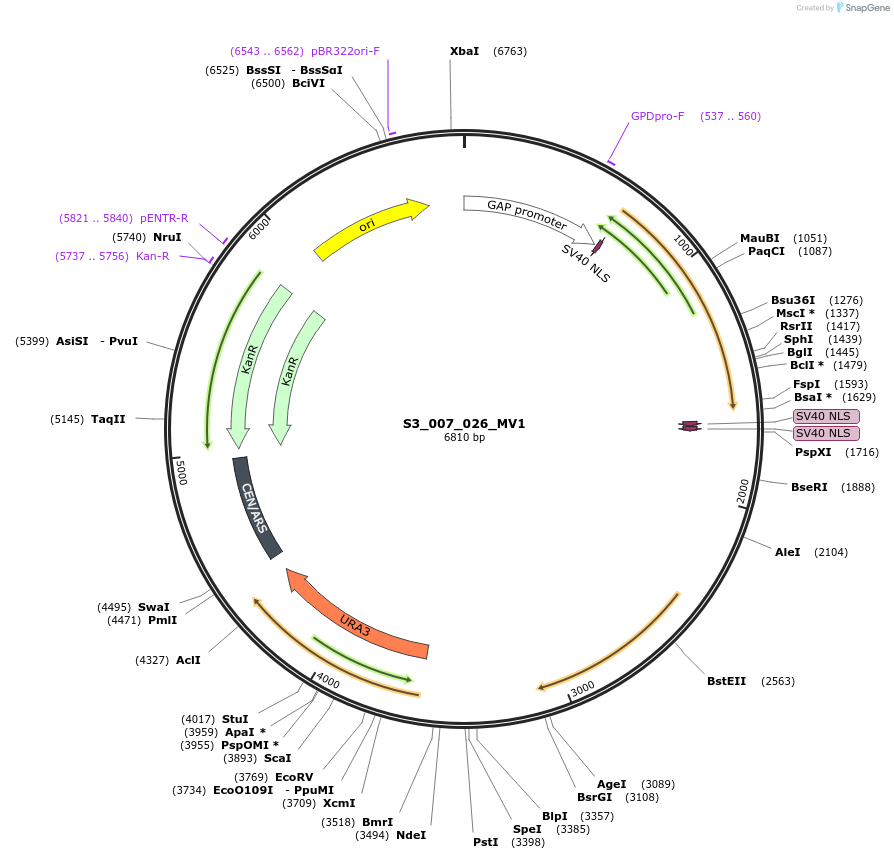
S3_007_026_MV1
Plasmid#134419PurposepYTKMV1-pTDH3_ NLS_FdeR-N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pTDH3Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
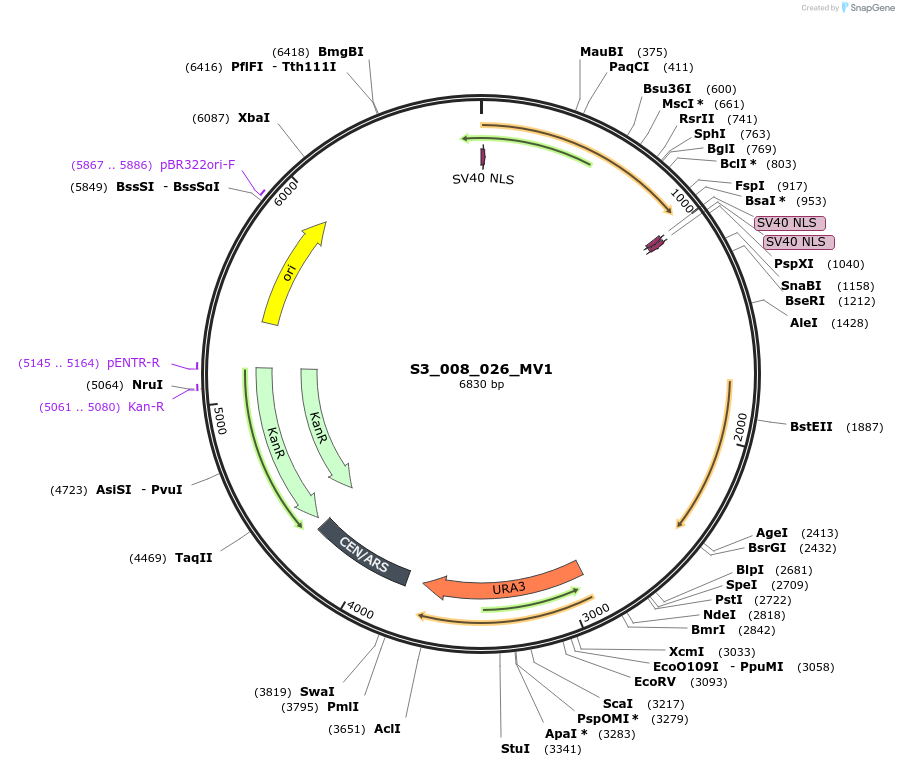
S3_008_026_MV1
Plasmid#134420PurposepYTKMV1-pRPL18B_ NLS_FdeR_N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pRPL18BAvailable SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
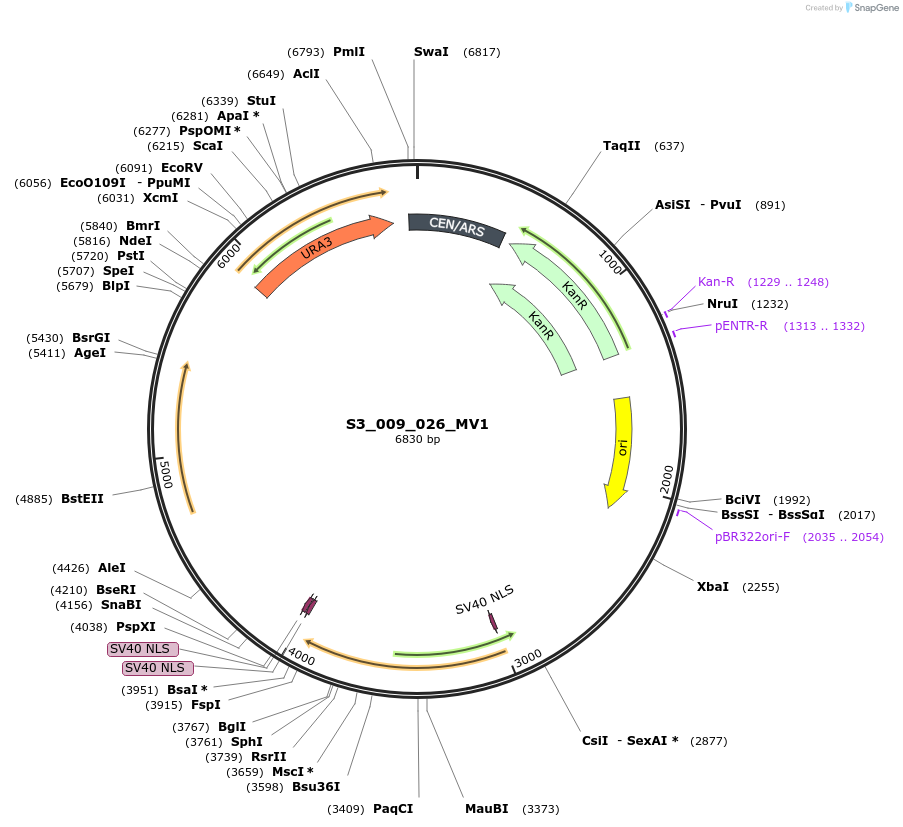
S3_009_026_MV1
Plasmid#134421PurposepYTKMV1-pREV1_ NLS_FdeR-N_2NLS-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
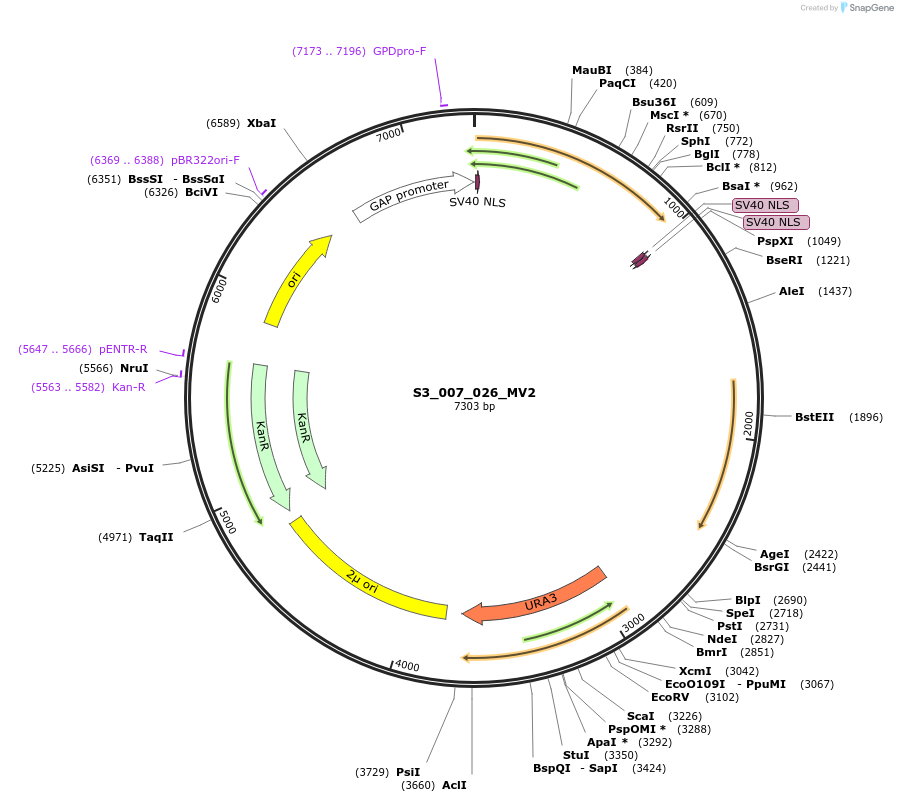
S3_007_026_MV2
Plasmid#134424PurposepYTKMV2-pTDH3_ NLS_FdeR-N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pTDH3Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
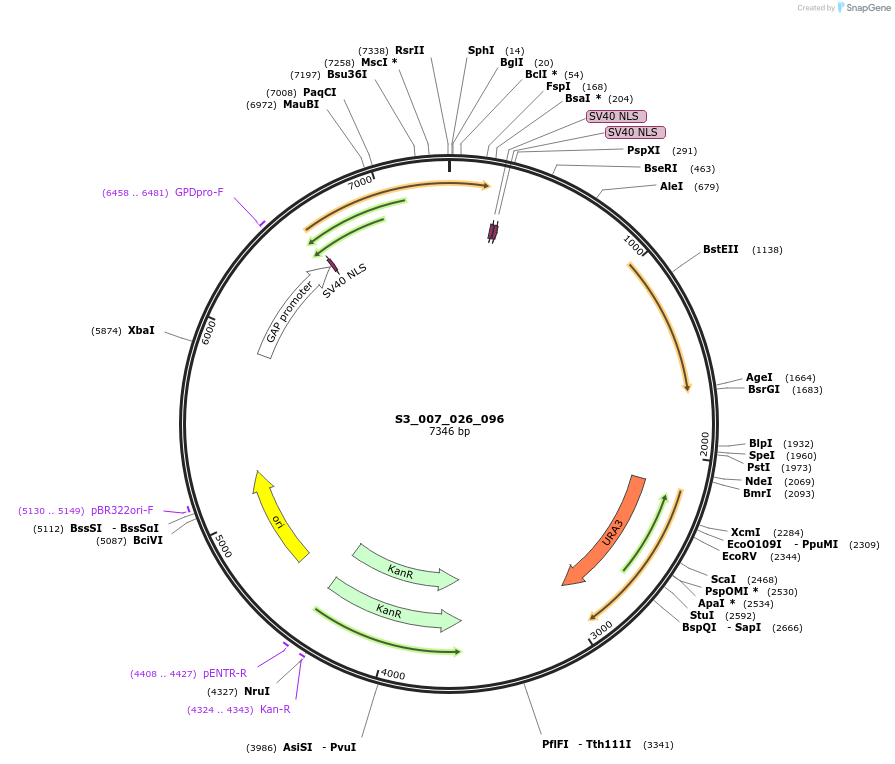
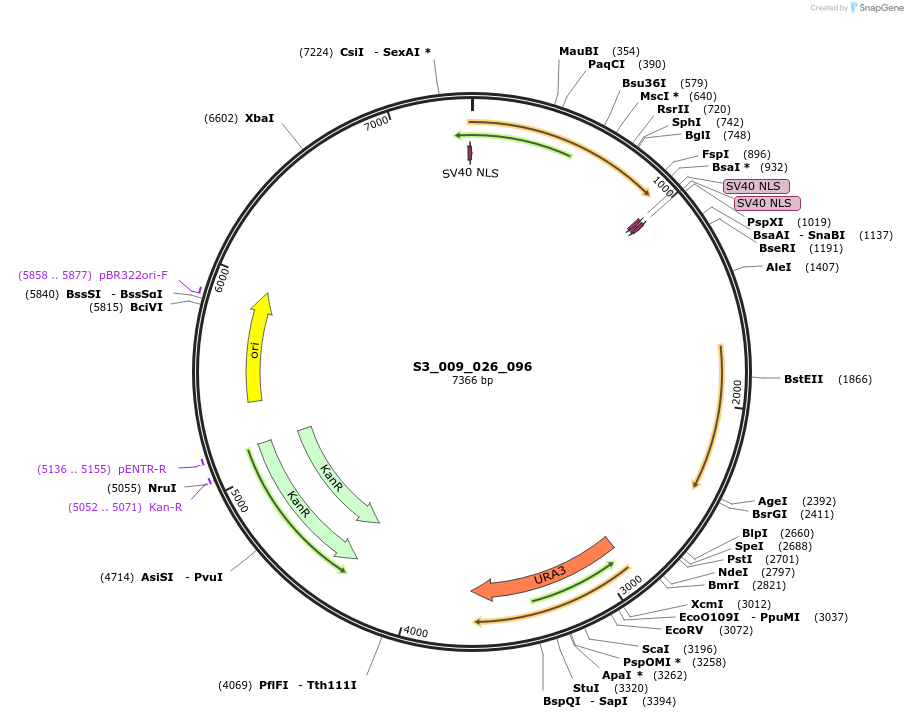
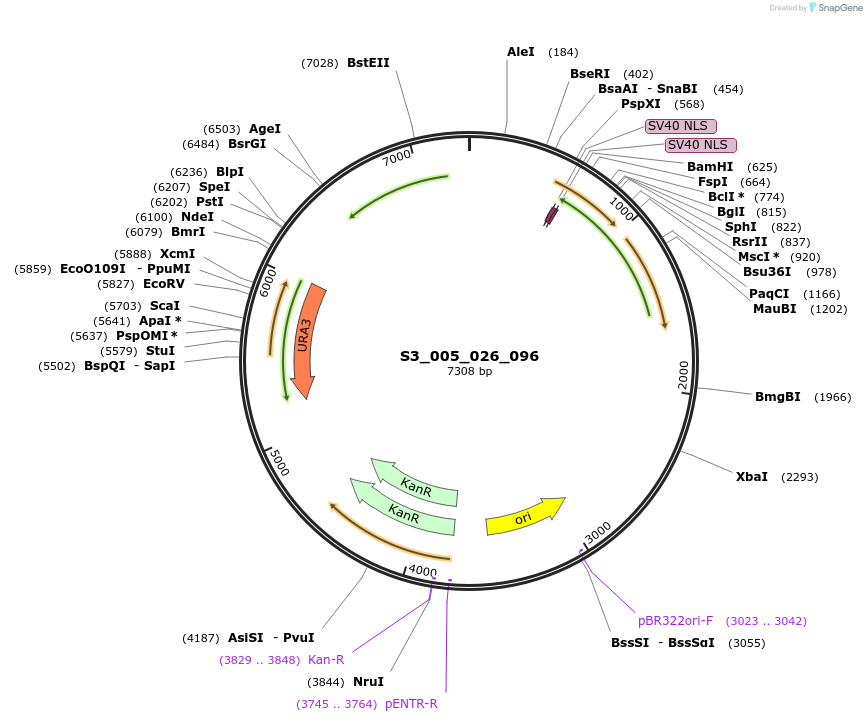
S3_007_026_096
Plasmid#134410PurposepYTK096-pTDH3_ NLS_FdeR-N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
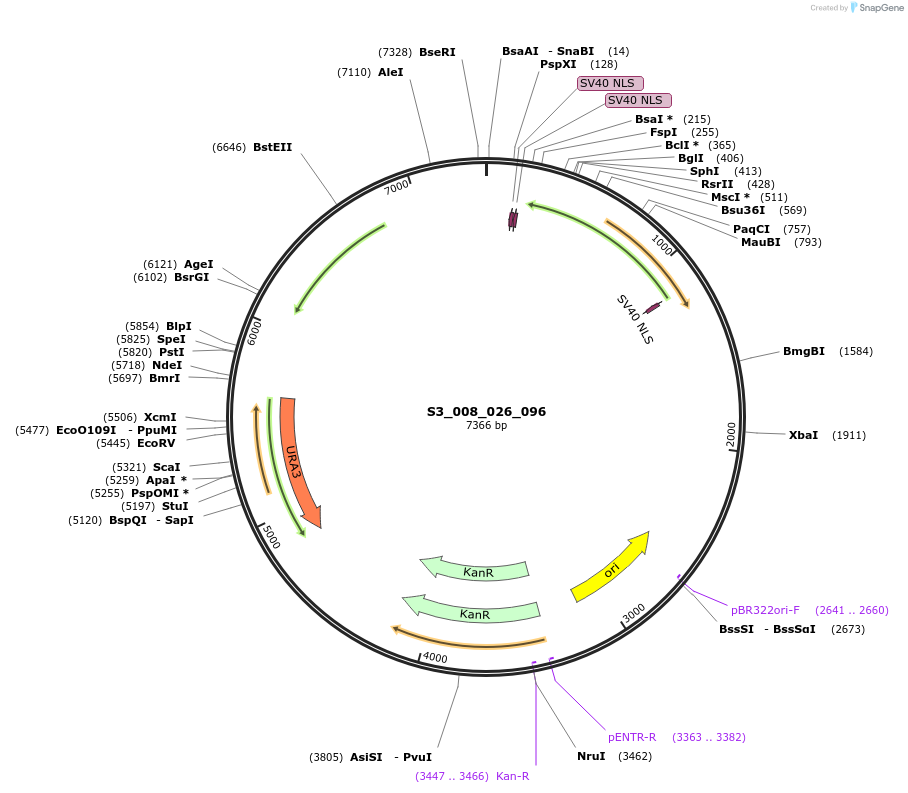
S3_008_026_096
Plasmid#134411PurposepYTK096-pRPL18B_ NLS_FdeR_N_2NLS _tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pRPL18BAvailable SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
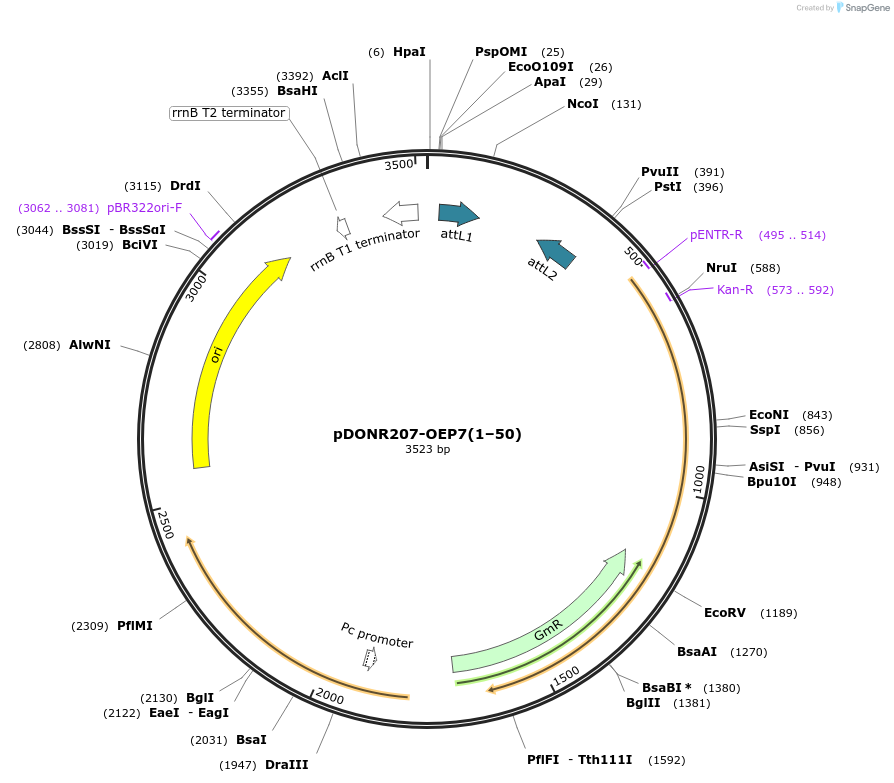
pDONR207-OEP7(1–50)
Plasmid#100601PurposeGateway entry vector for AtOEP7(1–50)DepositorInsertOEP7(1–50) (OEP7 Mustard Weed)
UseGateway entry vectorAvailable SinceFeb. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
S3_009_026_096
Plasmid#134412PurposepYTK096-pREV1_ NLS_FdeR-N_2NLS-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
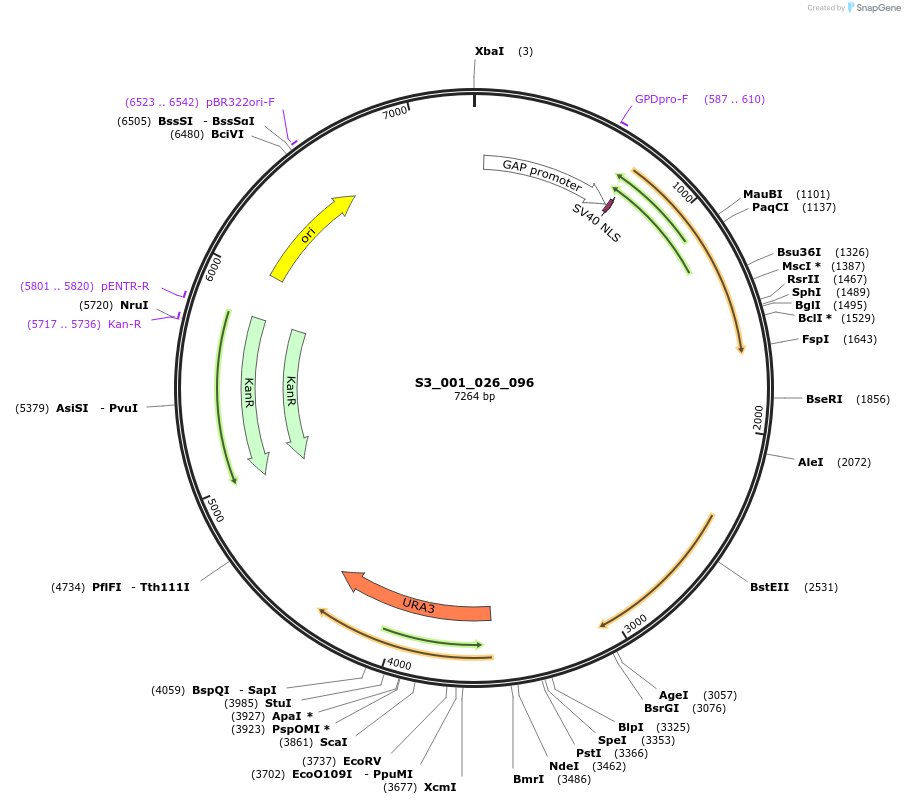
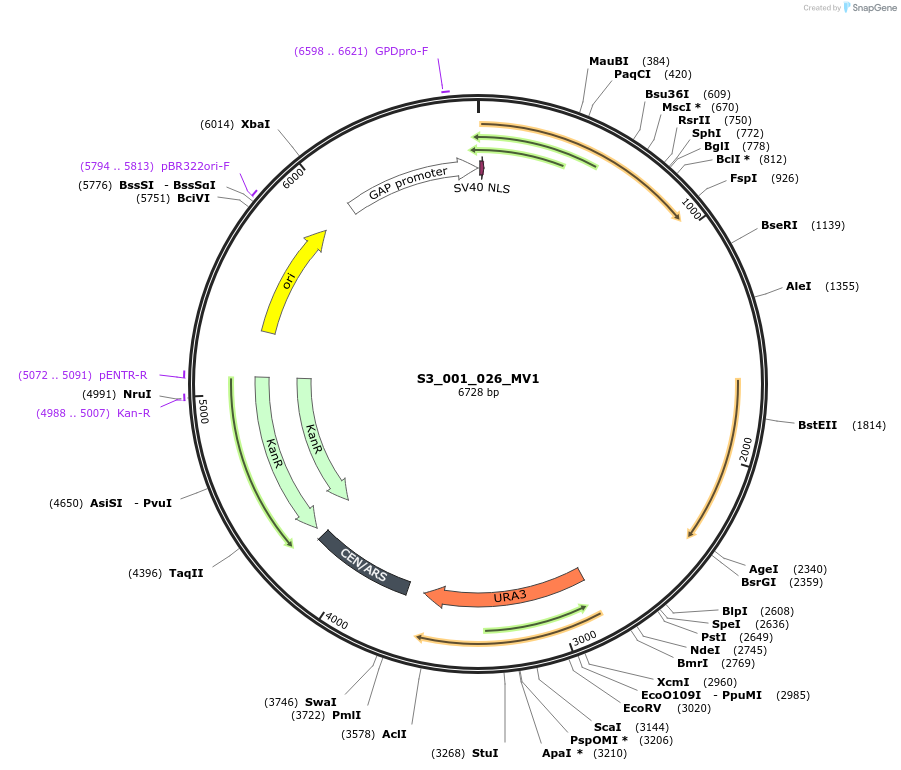
S3_001_026_096
Plasmid#134402PurposepYTK096-pTDH3_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
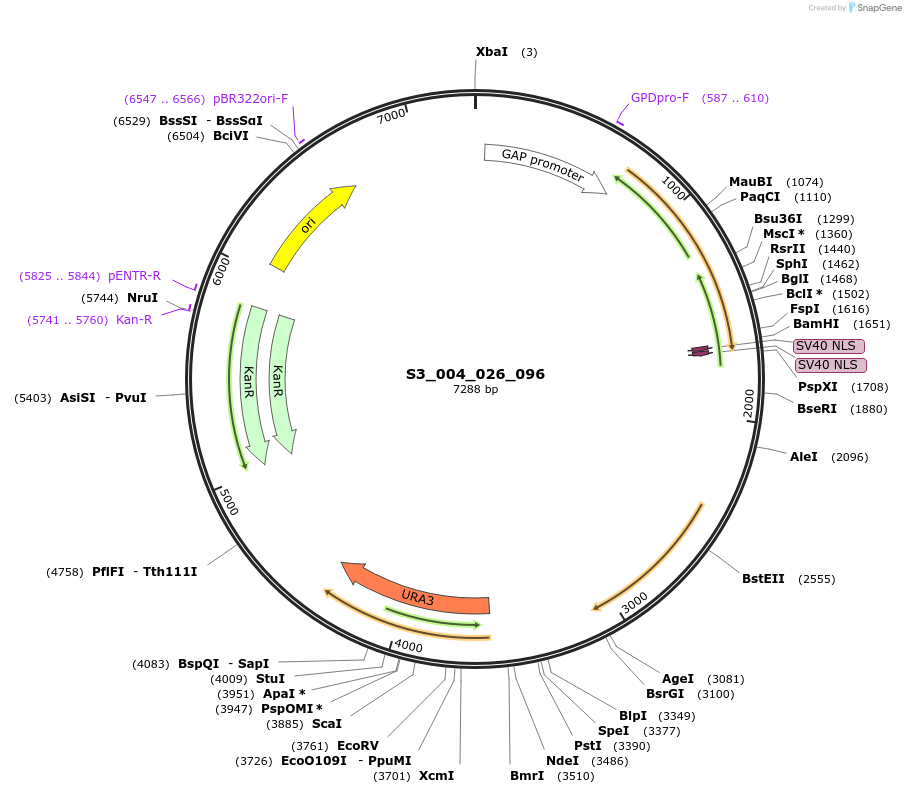
S3_004_026_096
Plasmid#134407PurposepYTK096-pTDH3_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
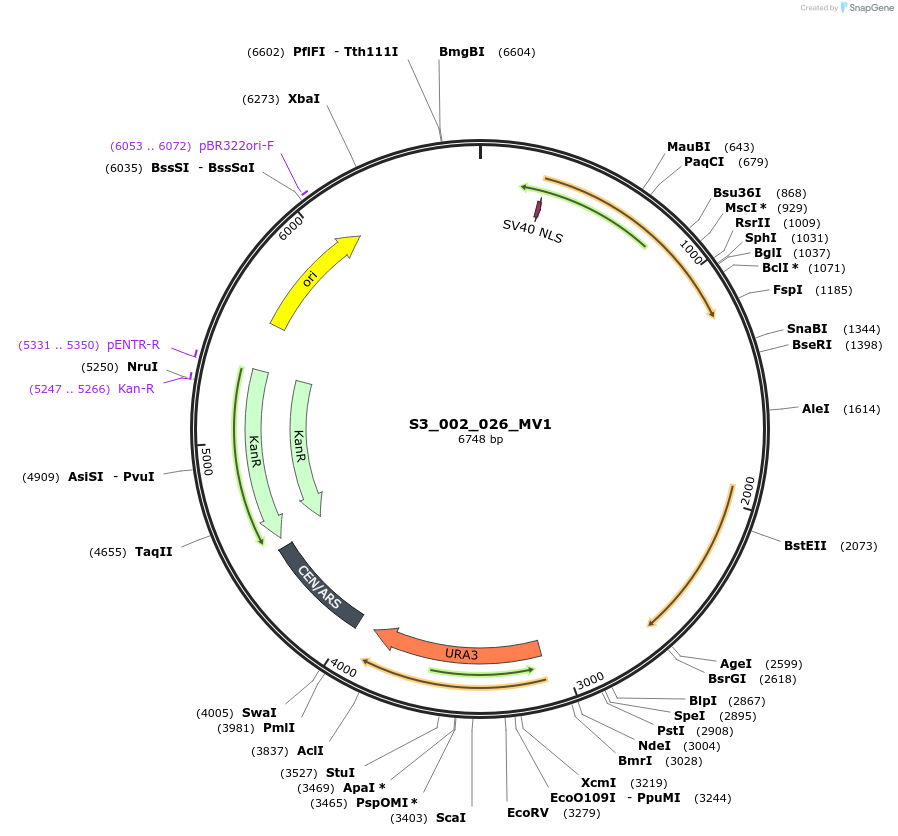
S3_002_026_MV1
Plasmid#134414PurposepYTKMV1-pRPL18B_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pRPL18BAvailable SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
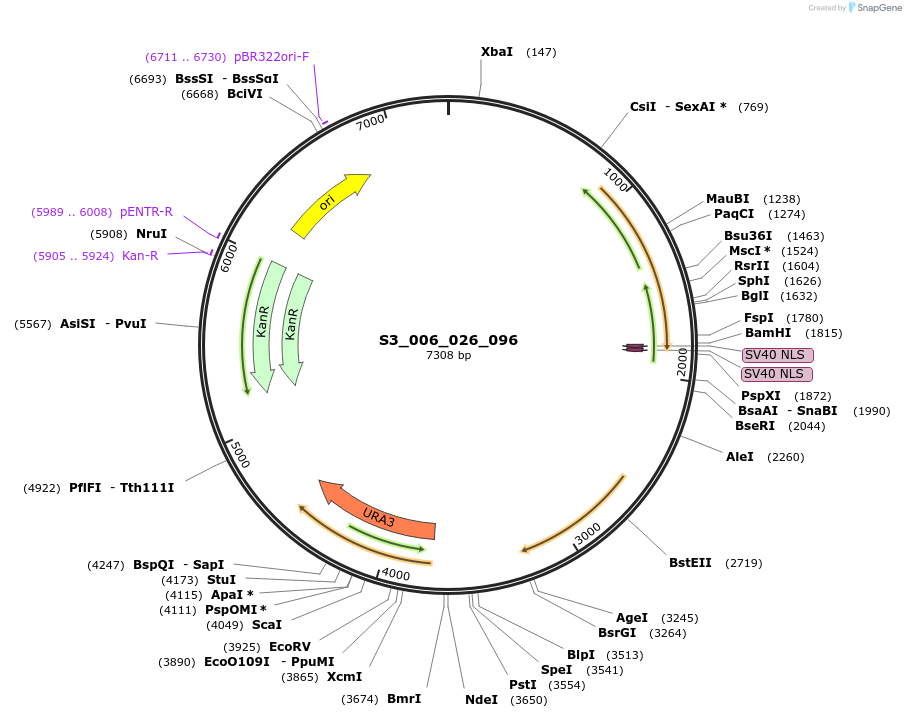
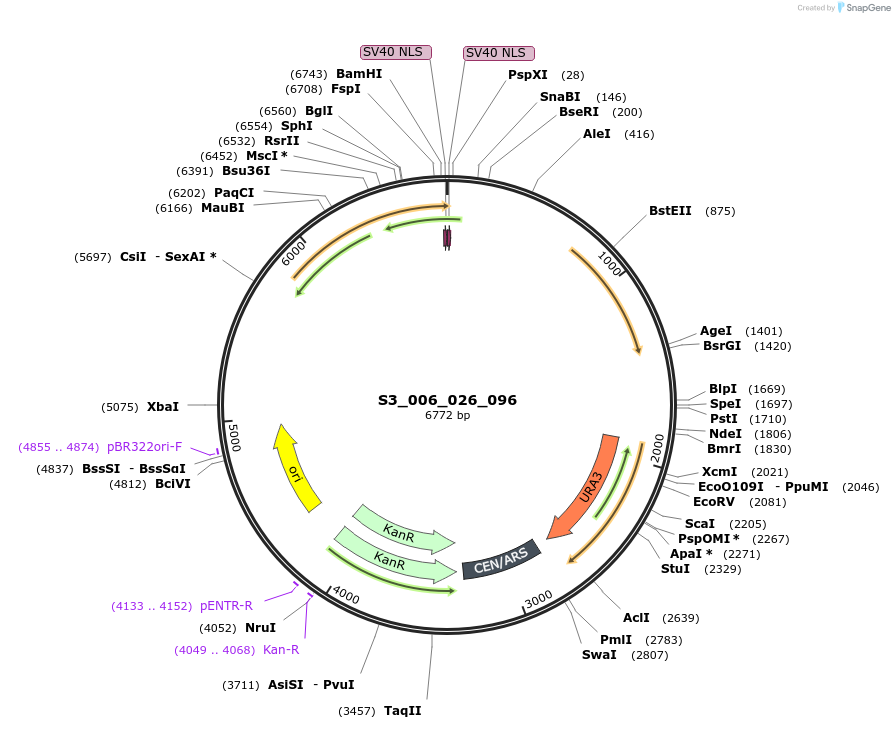
S3_006_026_096
Plasmid#134409PurposepYTK096-pREV1_FdeR-N_2NLS_tPGK1- pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pREV1Available SinceDec. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
S3_006_026_096
Plasmid#134418PurposepYTK096-pREV1_FdeR-N_2NLS_tPGK1- pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
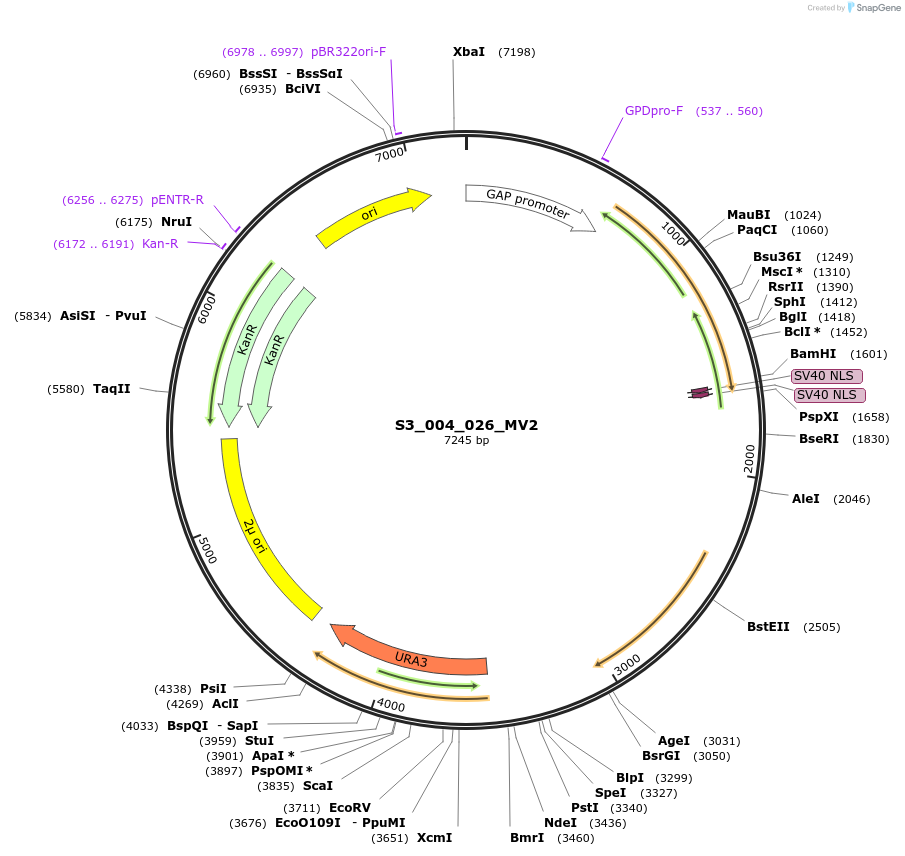
S3_004_026_MV2
Plasmid#134423PurposepYTKMV2-pTDH3_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pTDH3Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
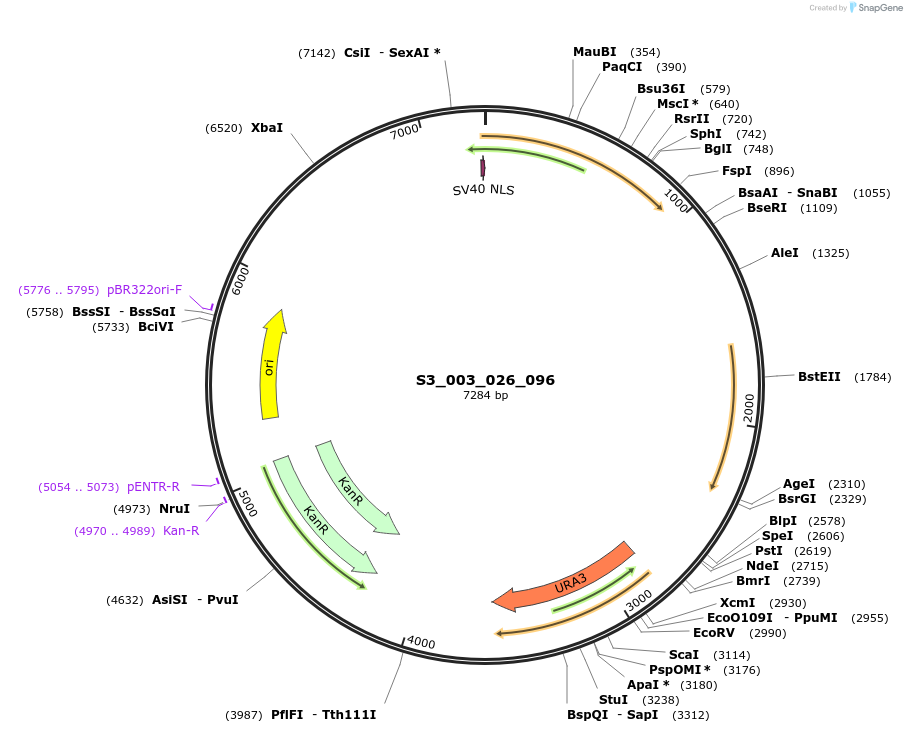
S3_003_026_096
Plasmid#134406PurposepYTK096-pREV1_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pREV1Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
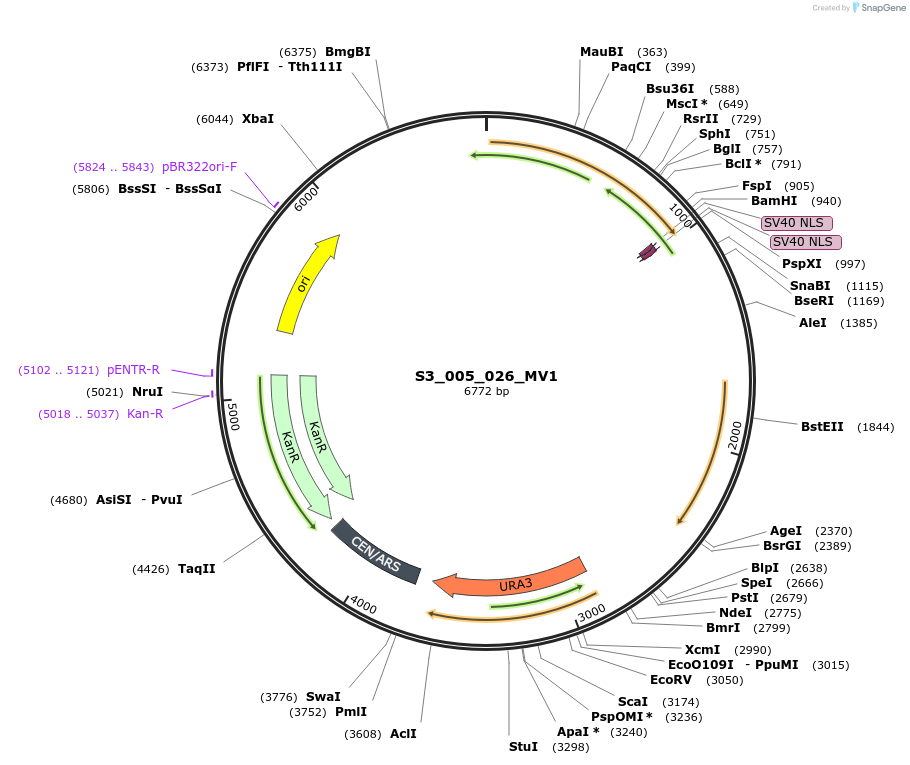
S3_005_026_MV1
Plasmid#134417PurposepYTKMV1-pRPL18B_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pRPL18BAvailable SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
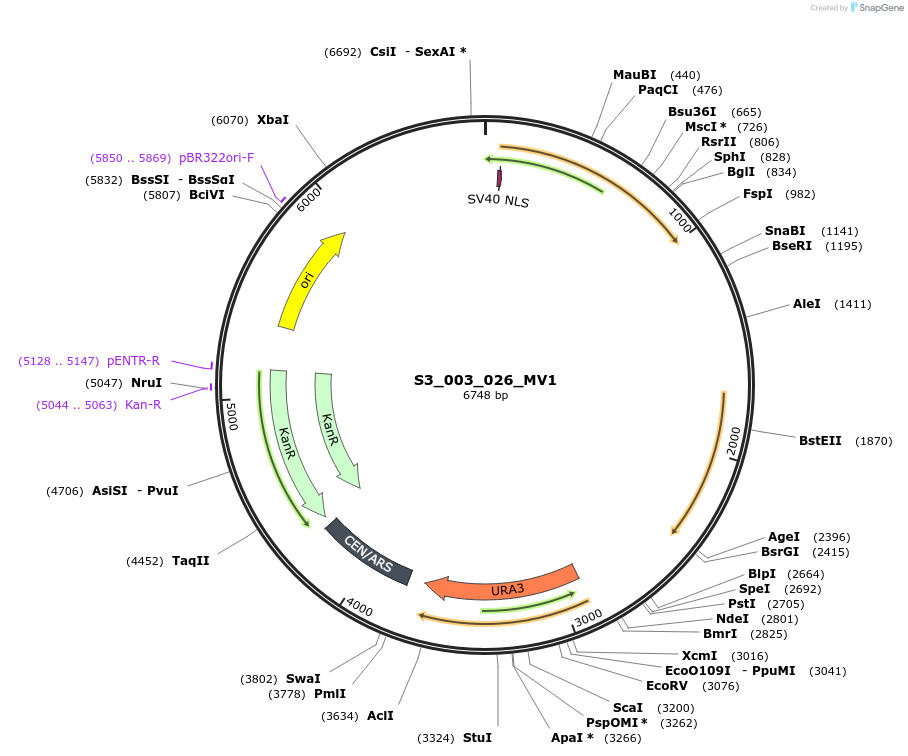
S3_003_026_MV1
Plasmid#134415PurposepYTKMV1-pREV1_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGMP1 and pREV1Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
S3_001_026_MV1
Plasmid#134413PurposepYTKMV1-pTDH3_NLS_FdeR-N_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
NLS_FdeR-N
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pTDH3Available SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
S3_005_026_096
Plasmid#134408PurposepYTK096-pRPL18B_FdeR-N_2NLS_tPGK1-pGPM1-fdeO_mcherry_tTDH1DepositorInsertsmCherry
FdeR-N_2xNLS
TagsnoneExpressionYeastMutationnonePromoterpGPM1 and pRPL18BAvailable SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
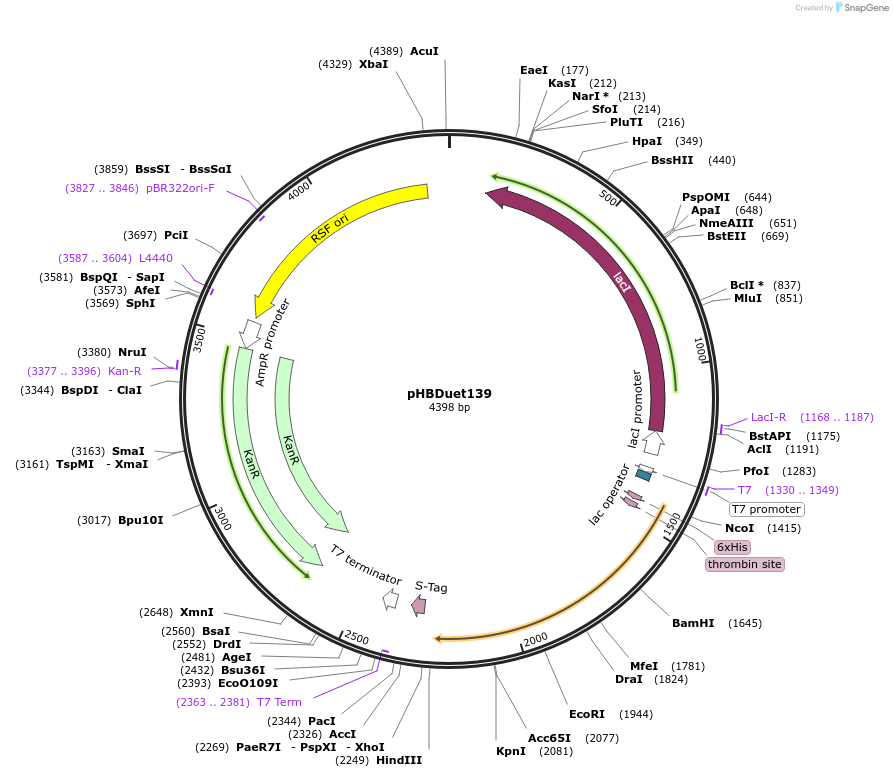
pHBDuet139
Plasmid#121913PurposeCharge-Introduced NpuDnaB mini-inteinDepositorInsertCI-NpuDnaBΔ290 intein
TagsGB1 and His6-GB1ExpressionBacterialMutationDeletion of 290-residue endonuclease domain, I53K…PromoterT7Available SinceDec. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
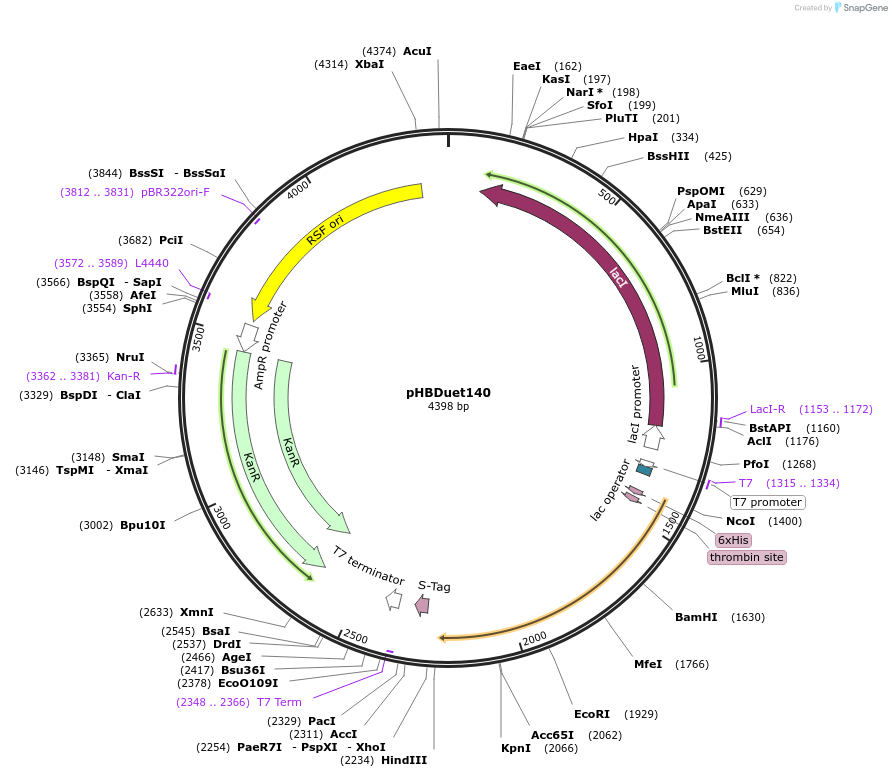
pHBDuet140
Plasmid#121914PurposeOrthogonal NpuDnaB mini-intein inteinDepositorInsertOth-NpuDnaBΔ290 intein
TagsGB1 and His6-GB1ExpressionBacterialMutationDeletion of 290-residue endonuclease domain, I53K…PromoterT7Available SinceDec. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
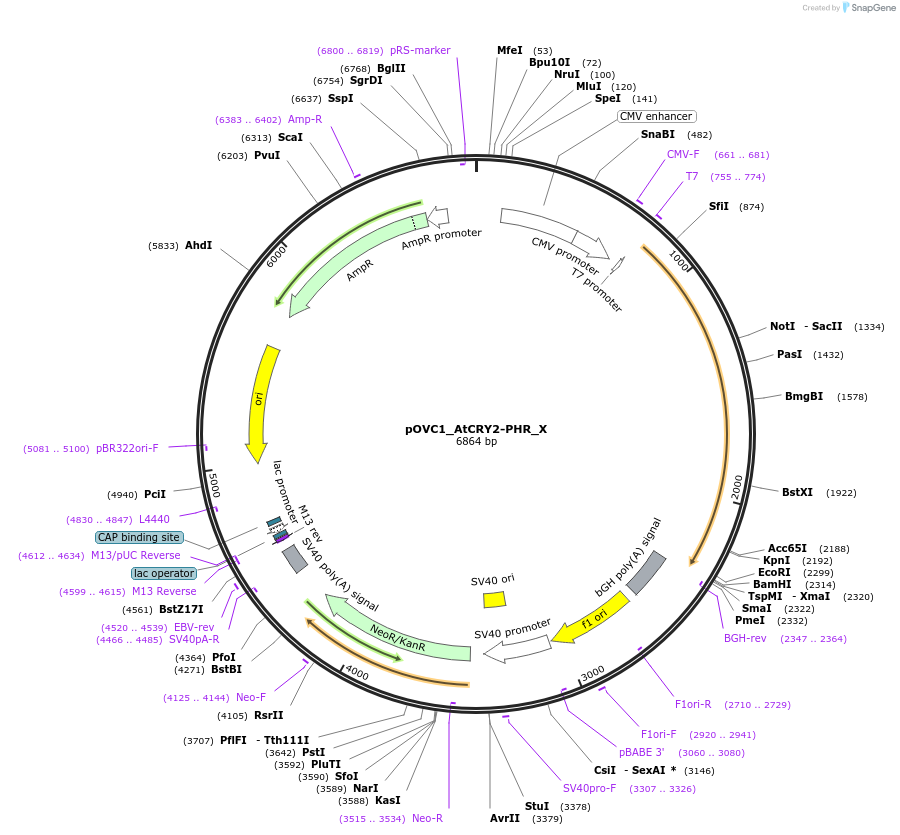
pOVC1_AtCRY2-PHR_X
Plasmid#131180PurposeExpresses AtCRY2-PHR (Arabidopsis thaliana) followed by restriction sitesDepositorInsertAtCRY2-PHR
ExpressionMammalianPromoterCMVAvailable SinceNov. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
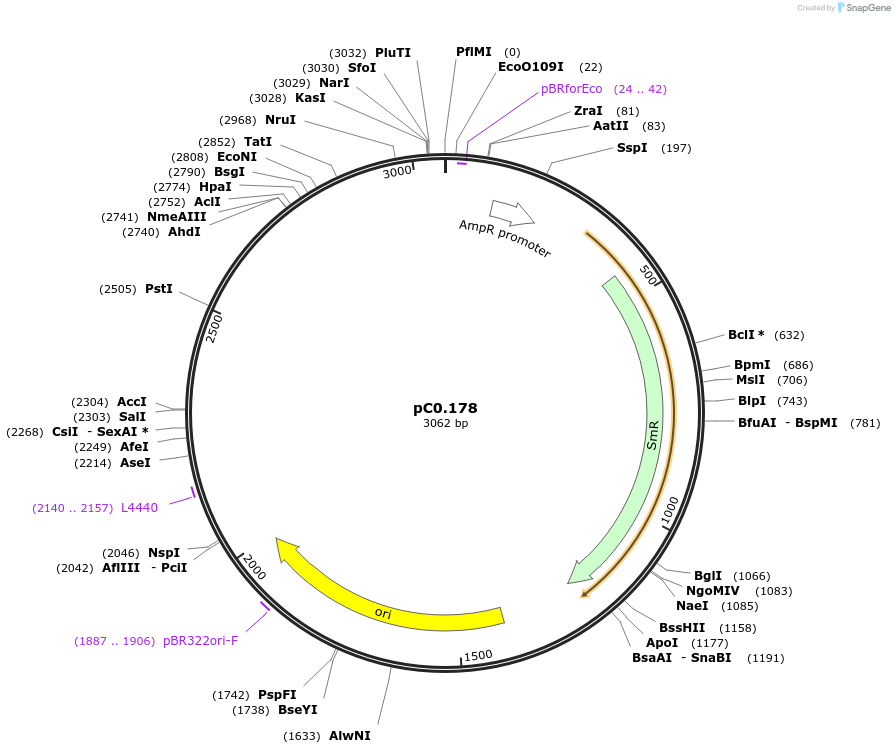
pC0.178
Plasmid#119645PurposeLevel 0 Part. Insertion siteDepositorInsert7942 NS3 Down Flank
UseSynthetic BiologyMutation4 nucleotide insertion between nt 65 and 66 in NS…Available SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
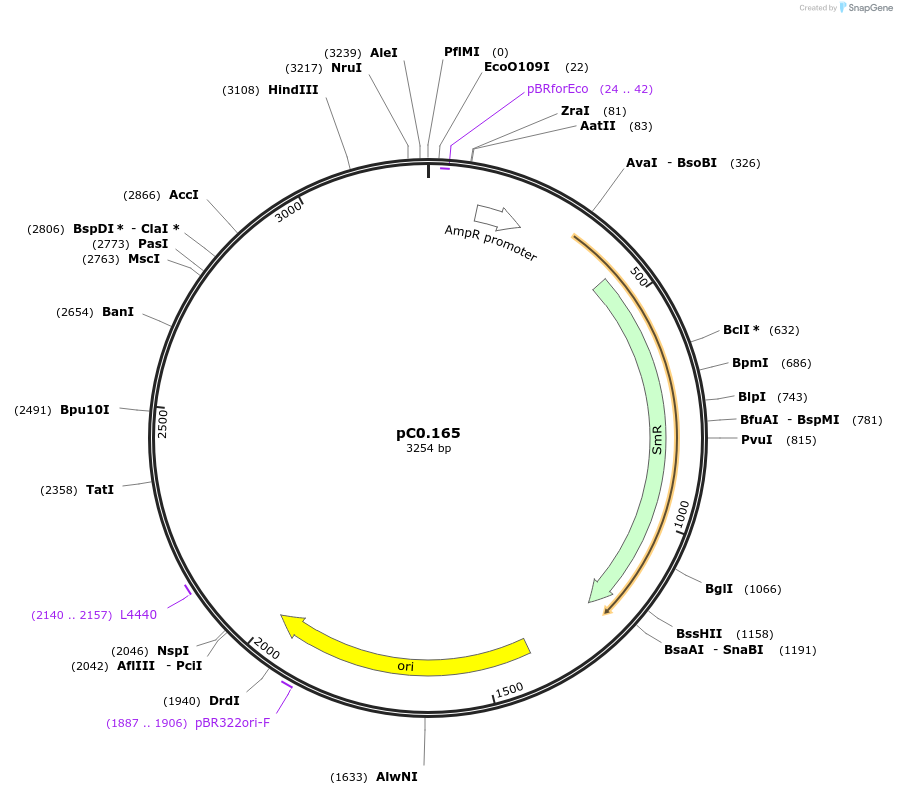
pC0.165
Plasmid#119634PurposeLevel 0 Part. Insertion siteDepositorInsert6803 NS1 Up Flank
UseSynthetic BiologyMutation1 nucleotide insertion between nt 21 and 22 in NS…Available SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
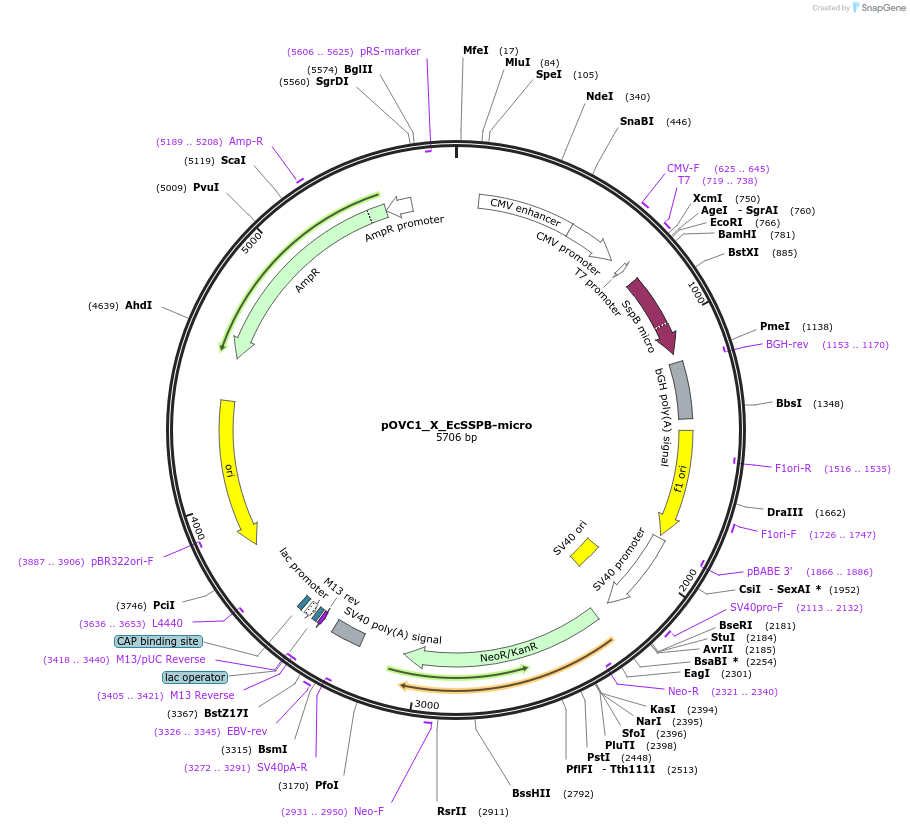
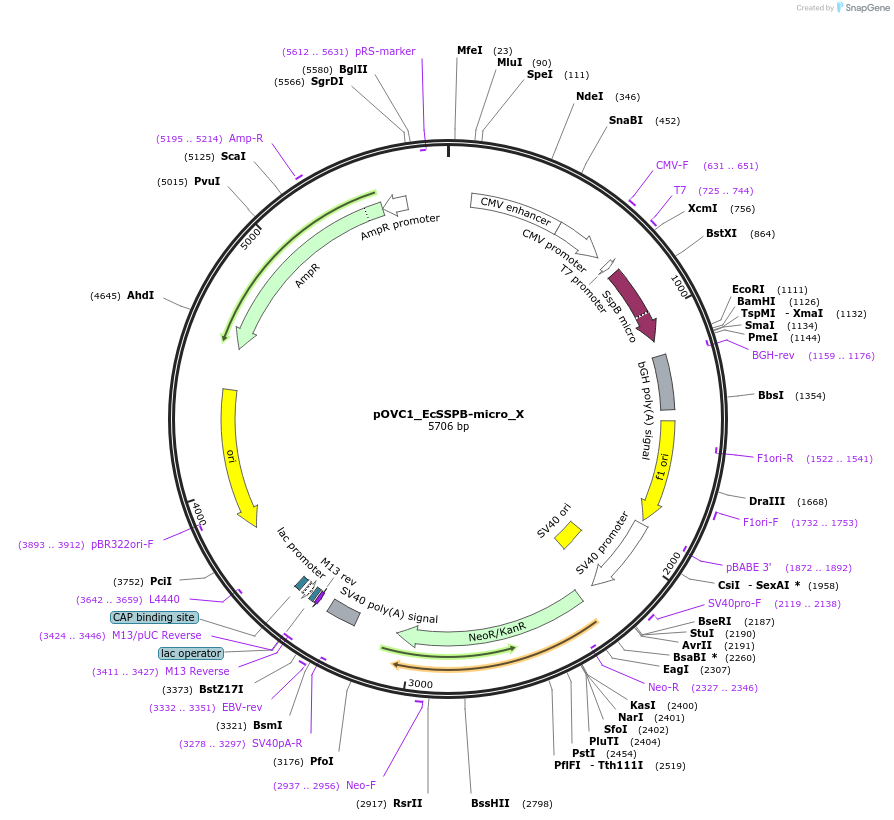
pOVC1_X_EcSSPB-micro
Plasmid#131218PurposeExpresses EcSSPB-nano (Escherichia coli) preceded by restriction sitesDepositorInsertEcSSPB-micro
ExpressionMammalianPromoterCMVAvailable SinceSept. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
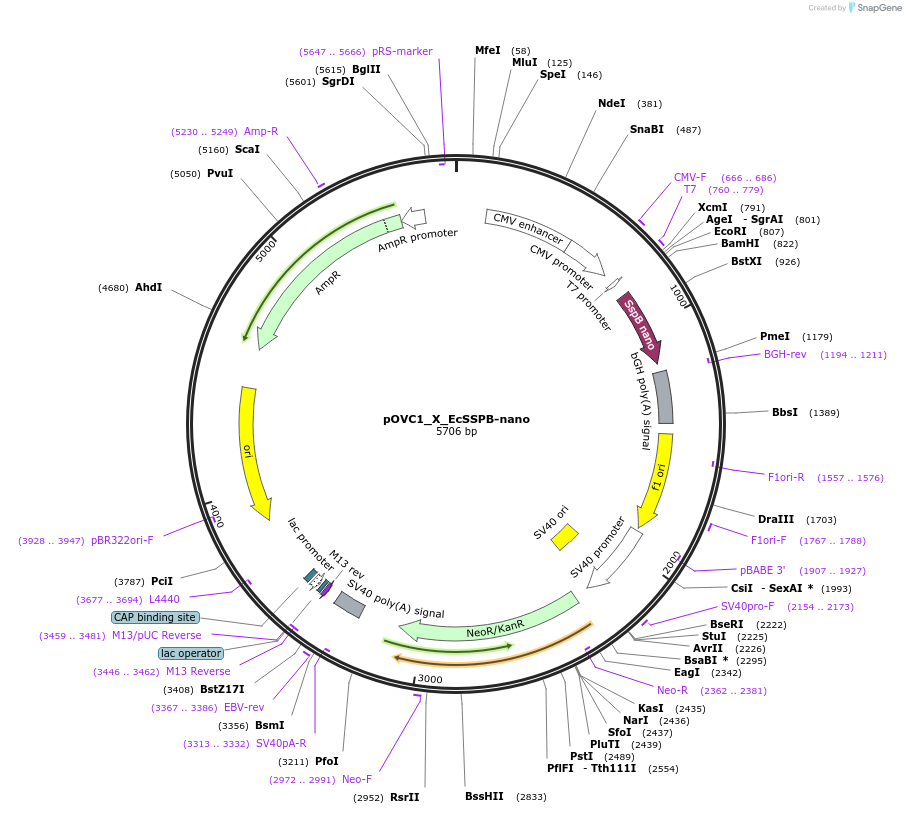
pOVC1_X_EcSSPB-nano
Plasmid#131219PurposeExpresses EcSSPB-micro (Escherichia coli) preceded by restriction sitesDepositorInsertEcSSPB-nano
ExpressionMammalianPromoterCMVAvailable SinceSept. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
pOVC1_EcSSPB-micro_X
Plasmid#131213PurposeExpresses EcSSPB-micro (Escherichia coli) followed by restriction sitesDepositorInsertEcSSPB-micro
ExpressionMammalianPromoterCMVAvailable SinceSept. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
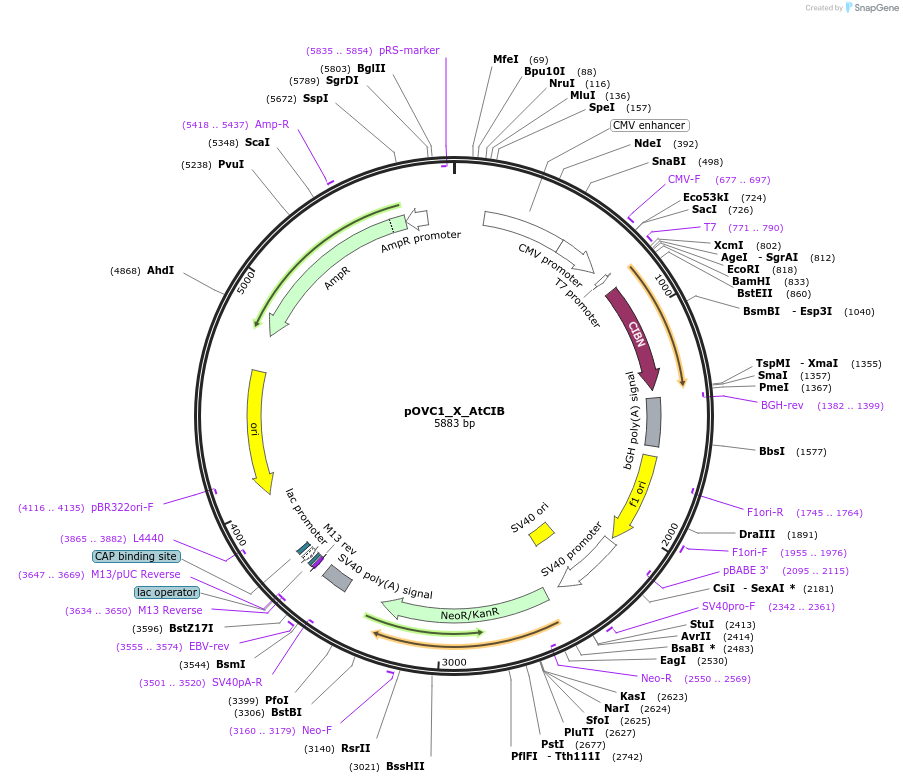
pOVC1_X_AtCIB
Plasmid#131194PurposeExpresses AtCIB (Arabidopsis thaliana) preceded by restriction sitesDepositorInsertAtCIB
ExpressionMammalianPromoterCMVAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
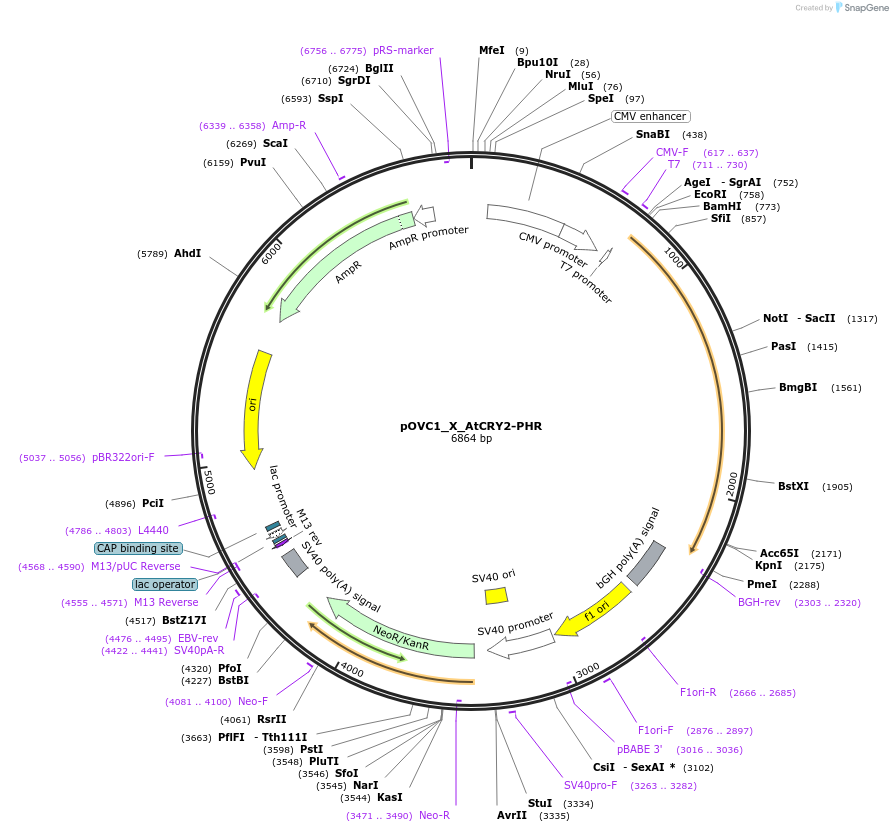
pOVC1_X_AtCRY2-PHR
Plasmid#131191PurposeExpresses AtCRY2-PHR (Arabidopsis thaliana) preceded by restriction sitesDepositorInsertAtCRY2-PHR
ExpressionMammalianPromoterCMVAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
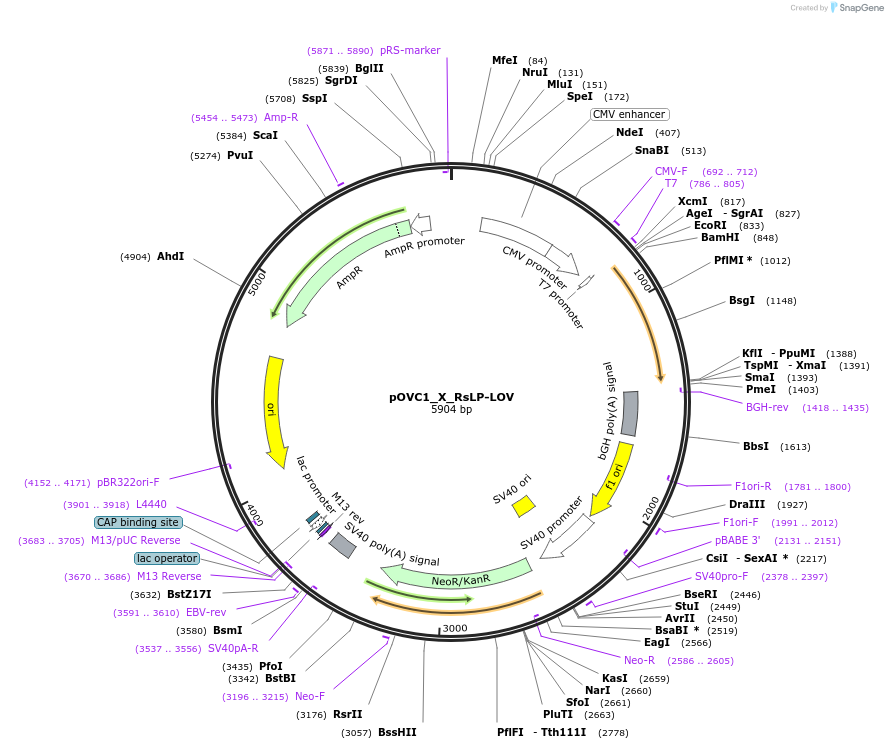
pOVC1_X_RsLP-LOV
Plasmid#131188PurposeExpresses RsLP-LOV (Rhodobacter sphaeroides) preceded by restriction sitesDepositorInsertRsLP-LOV
ExpressionMammalianPromoterCMVAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
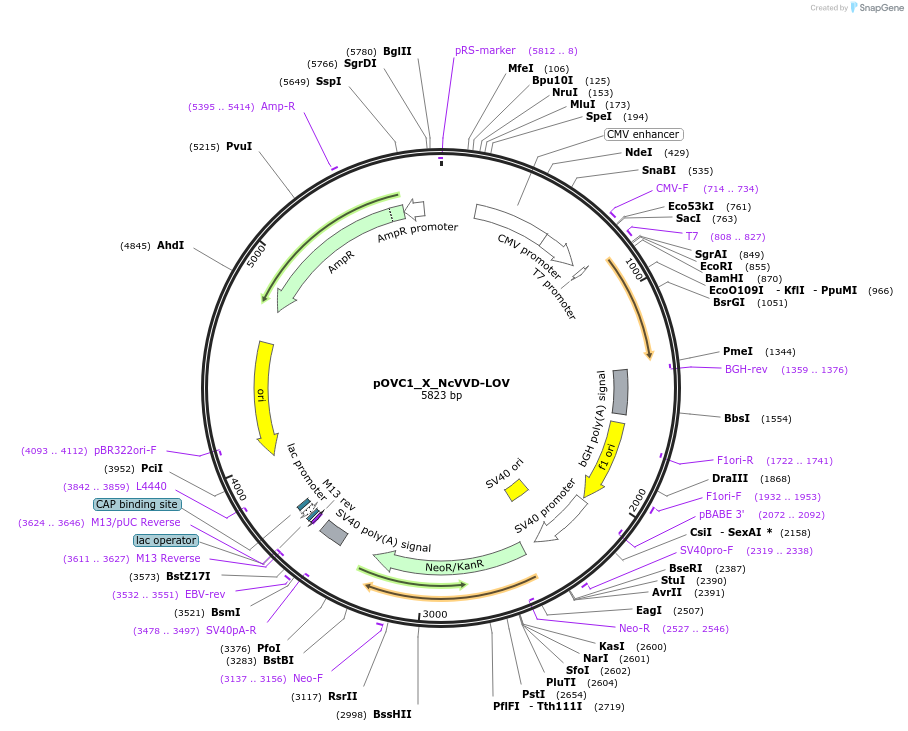
pOVC1_X_NcVVD-LOV
Plasmid#131187PurposeExpresses NcVVD-LOV (Neurosporra crassa) preceded by restriction sitesDepositorInsertNcVVD-LOV
ExpressionMammalianPromoterCMVAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
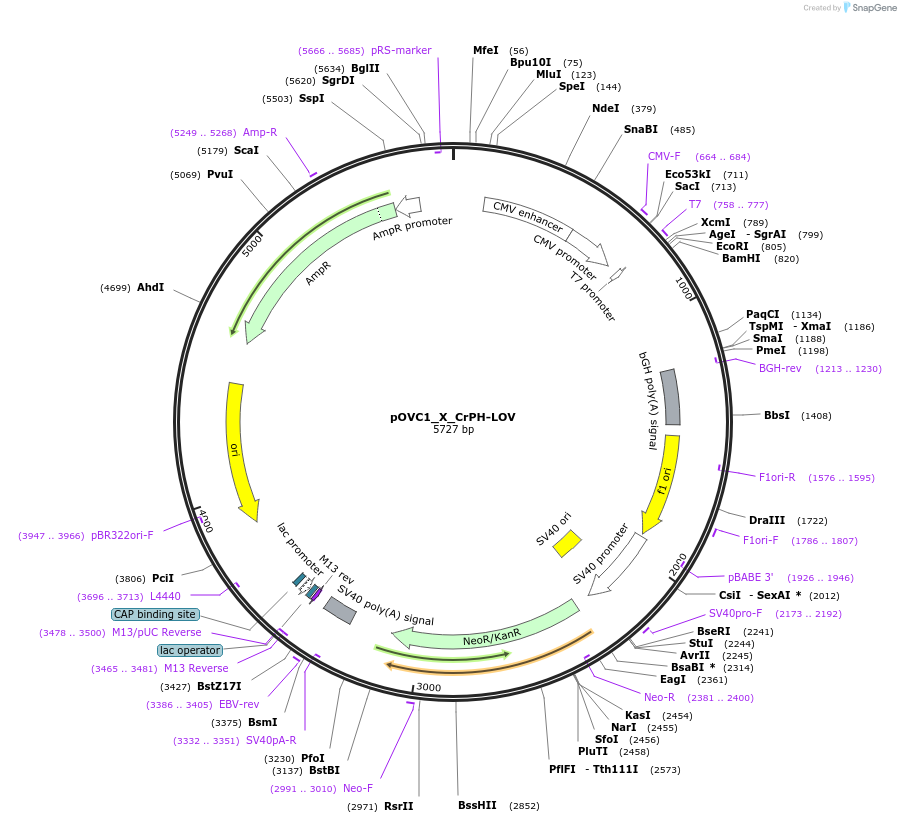
pOVC1_X_CrPH-LOV
Plasmid#131186PurposeExpresses CrPh-LOV domain (Chlamydomonas reinhardtii) preceded by restriction sitesDepositorInsertCrPH-LOV
ExpressionMammalianPromoterCMVAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
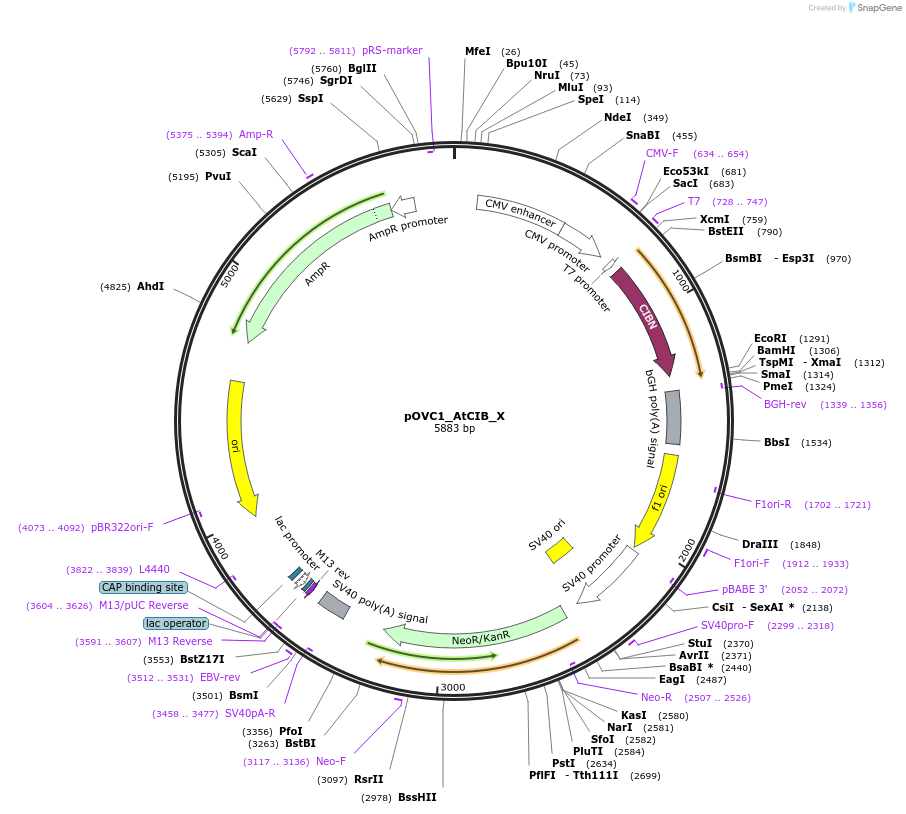
pOVC1_AtCIB_X
Plasmid#131181PurposeExpresses AtCIB (Arabidopsis thaliana) followed by restriction sitesDepositorInsertAtCIB
ExpressionMammalianPromoterCMVAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
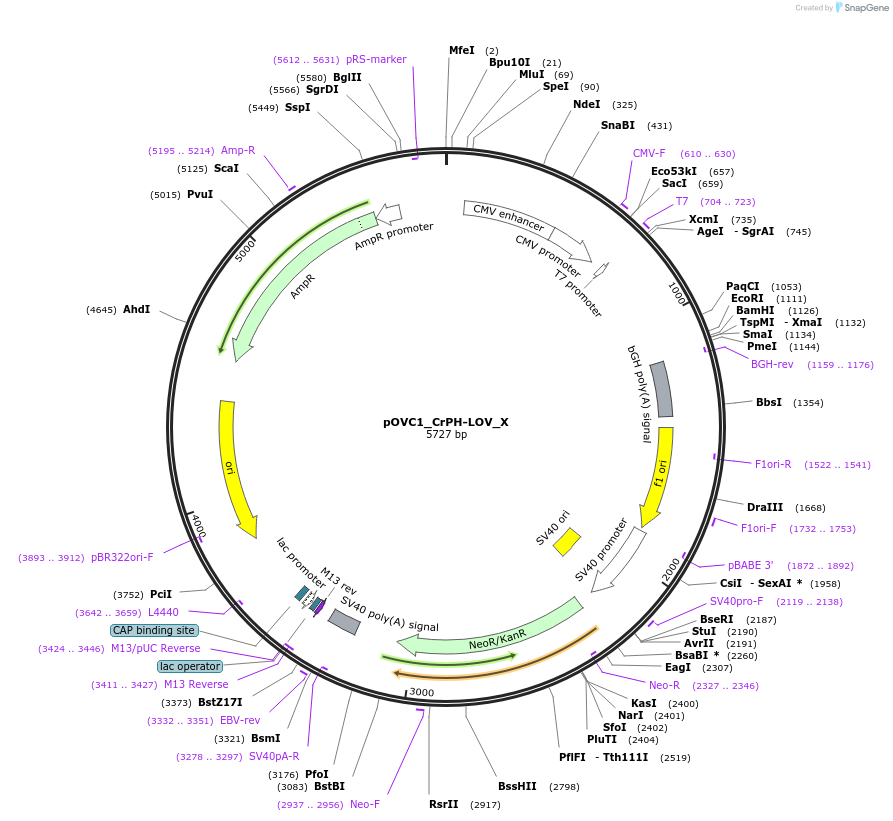
pOVC1_CrPH-LOV_X
Plasmid#131175PurposeExpresses CrPh-LOV domain (Chlamydomonas reinhardtii) followed by restriction sitesDepositorInsertCrPH-LOV
ExpressionMammalianPromoterCMVAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
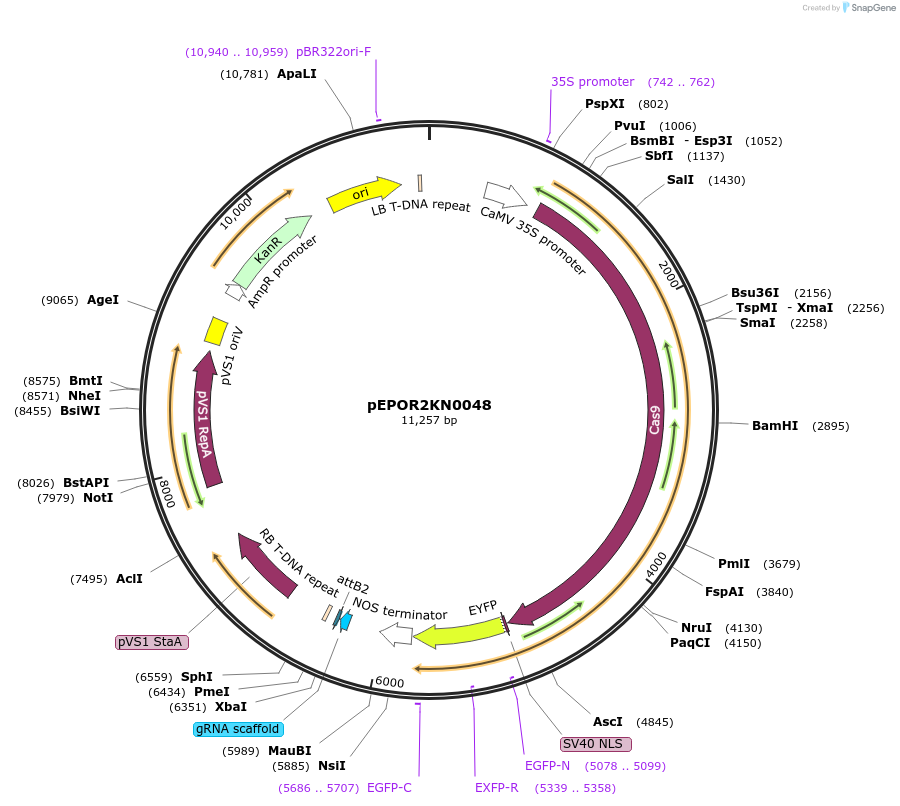
pEPOR2KN0048
Plasmid#117611PurposeLevel2 MoClo construct, containing level1 plant expression cassettes for pEPOR1CB0012(pEPOR1CB0012) and sgRNA_Rpn2/Psmd1 subunit {AT2G32730}(pEPOR1CB0075)DepositorInsert[35S:pEPOR1CB0012(pEPOR1CB0012) ] +[AtU6-26:sgRNA]
ExpressionPlantAvailable SinceSept. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
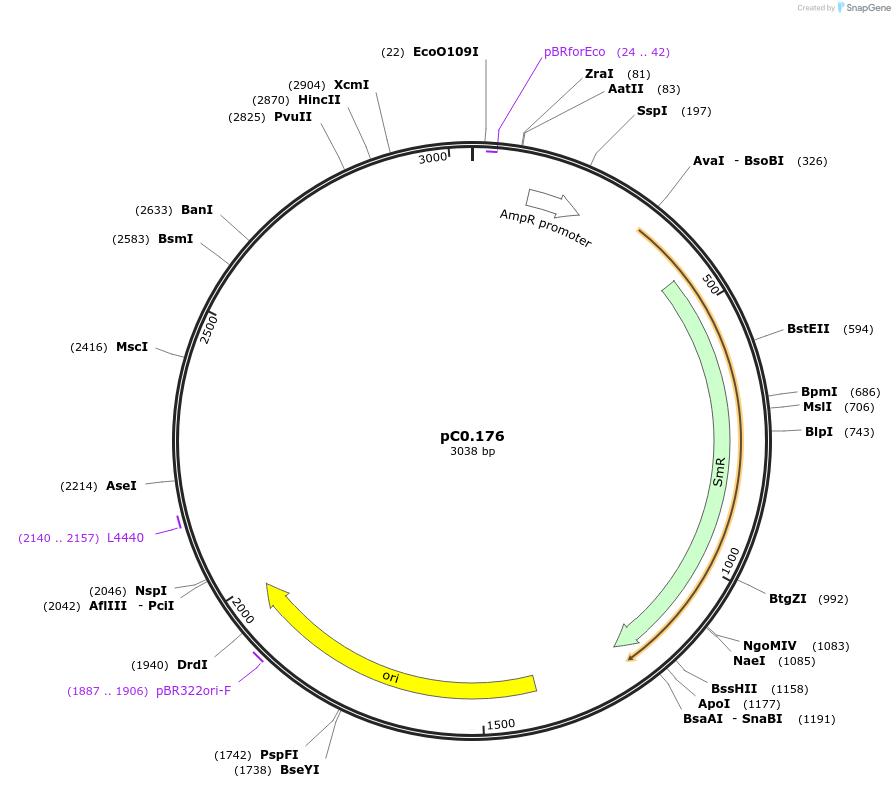
pC0.176
Plasmid#119644PurposeLevel 0 Part. Insertion siteDepositorInsert7942 NS2 Down Flank
UseSynthetic BiologyMutationbp 302 T to CAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
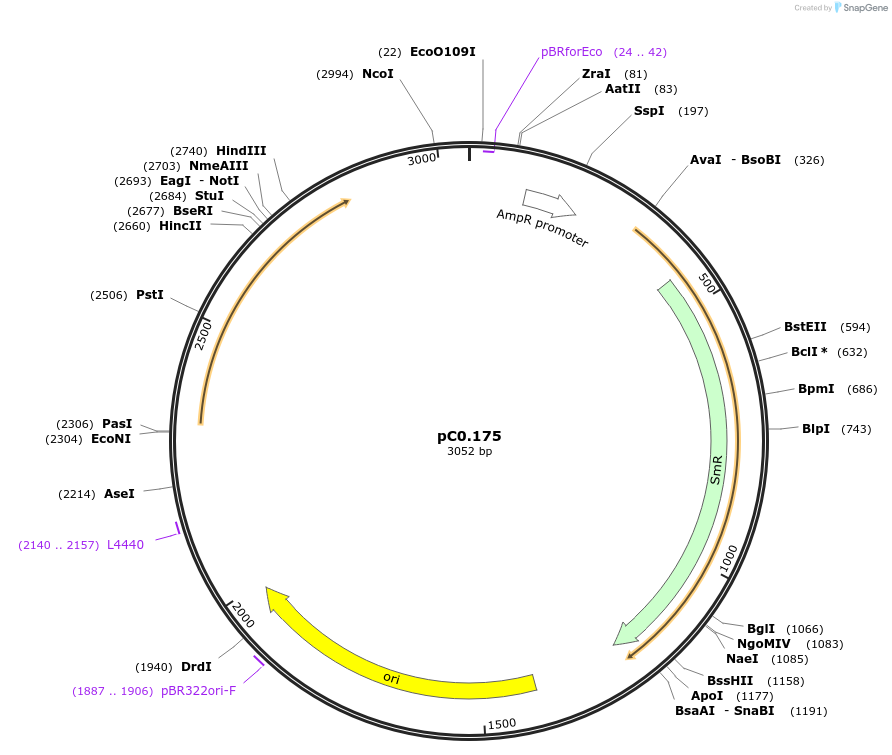
pC0.175
Plasmid#119643PurposeLevel 0 Part. Insertion siteDepositorInsert7942 NS2 Up Flank
UseSynthetic BiologyMutationbp 422 A to T, bp 423 A to T, bp 663 inserted G. …Available SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
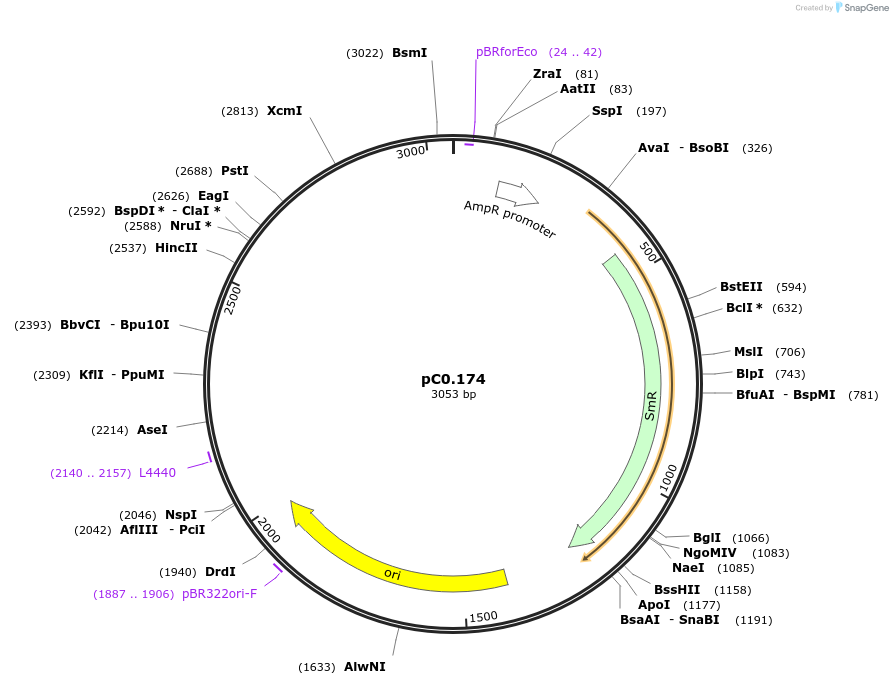
pC0.174
Plasmid#119641PurposeLevel 0 Part. Insertion siteDepositorInsert7942 NS1 Down Flank
UseSynthetic BiologyMutationbp 658 G to AAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
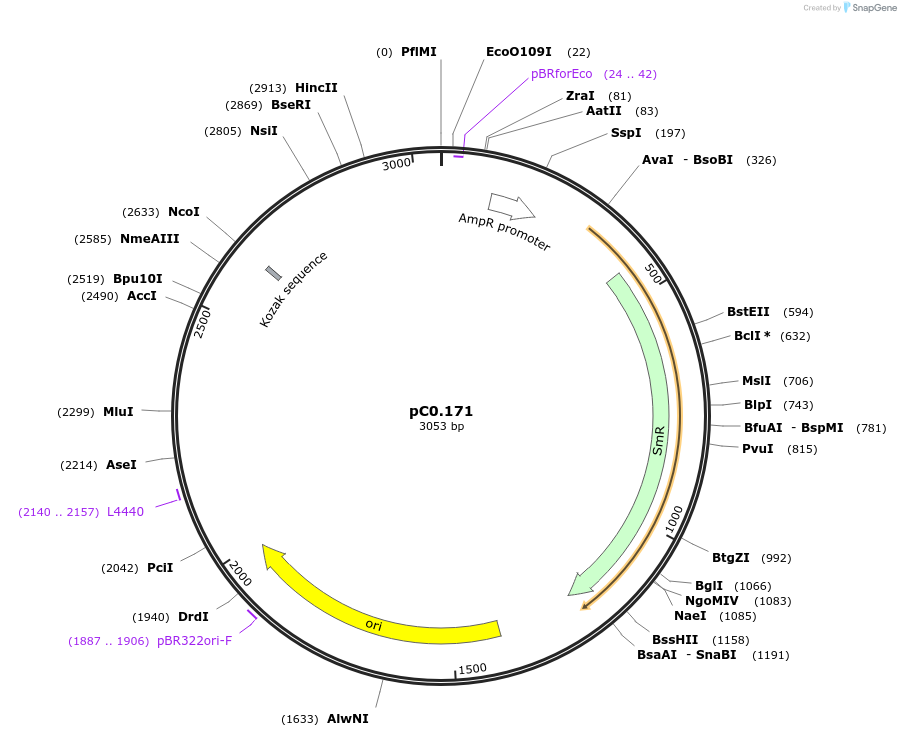
pC0.171
Plasmid#119639PurposeLevel 0 Part. Insertion siteDepositorInsert6803 NS4 Up Flank
UseSynthetic BiologyMutationA691T in NS4 Up FlankAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
pC0.169
Plasmid#119637PurposeLevel 0 Part. Insertion siteDepositorInsert6803 NS3 Up Flank
UseSynthetic BiologyMutationbp 551 T to C, bp 554 T to CAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
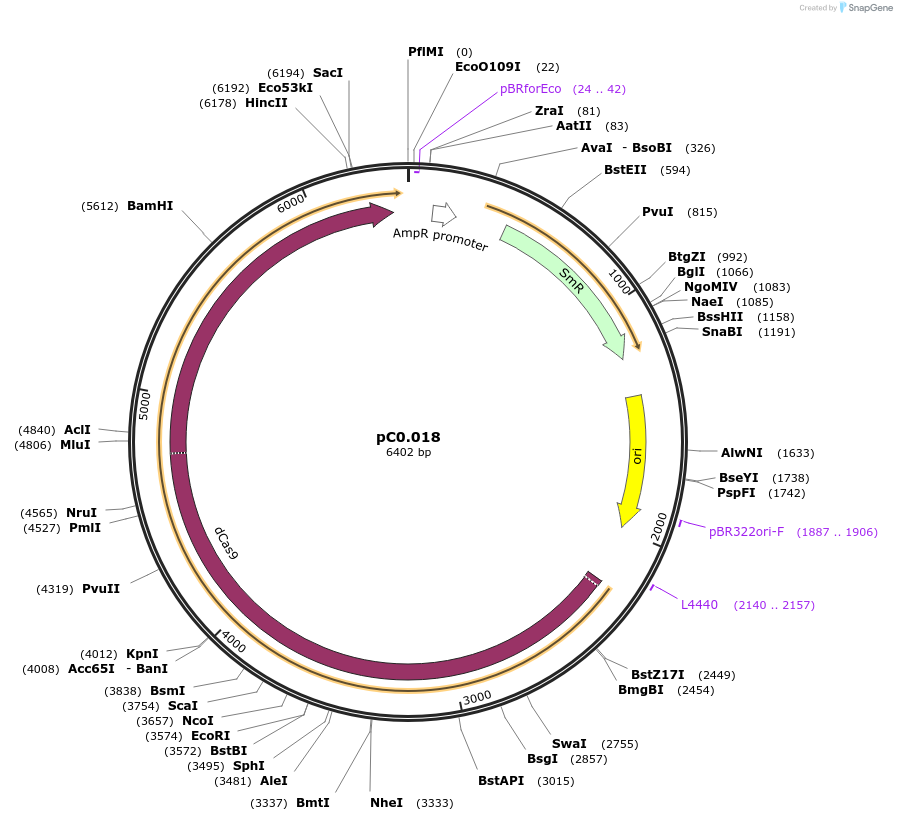
pC0.018
Plasmid#119622PurposeLevel 0 Part. CDSDepositorInsertdCas9+deg-tag
UseSynthetic BiologyMutationbp 331 C to A , 1237 A to C, 1284 C to T, 2121 C …Available SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
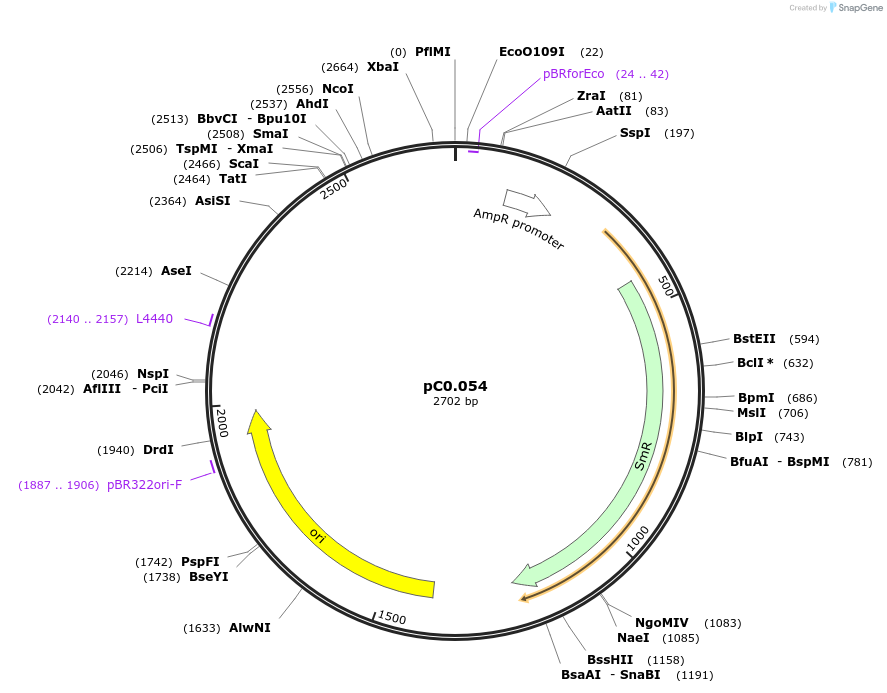
pC0.054
Plasmid#119572PurposeLevel 0 part. PromoterDepositorInsertPpetE
UseSynthetic BiologyMutationRBS* added (22 bp) upstream of ATG. Deletion of n…Available SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
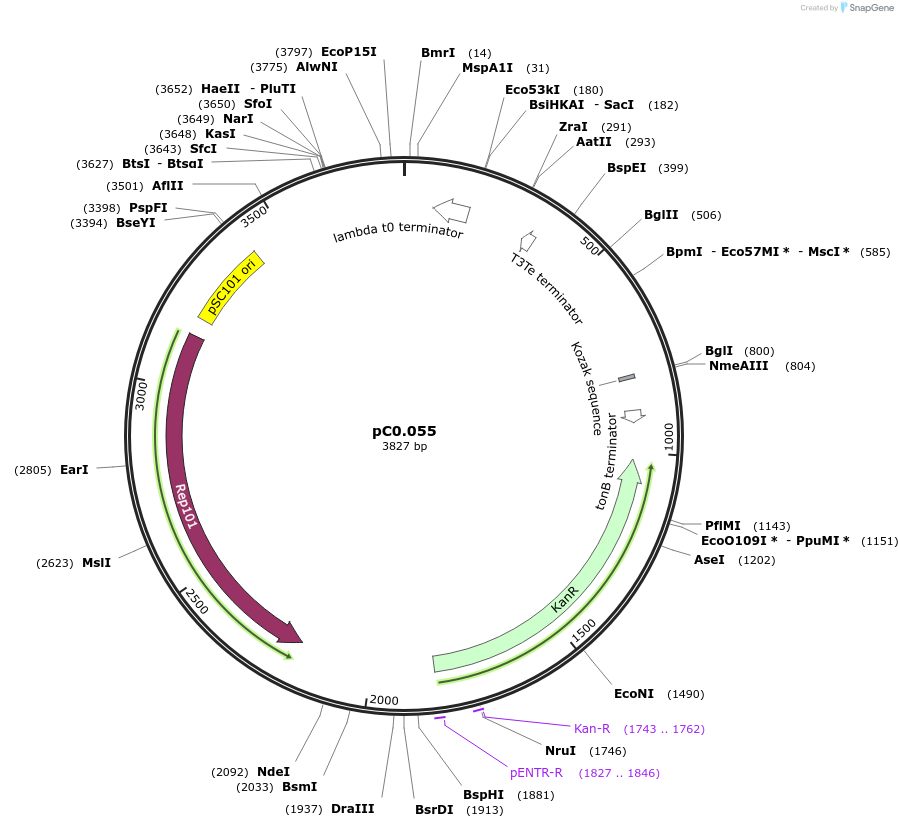
pC0.055
Plasmid#119566PurposeLevel 0 part. PromoterDepositorInsertPisiAB
UseSynthetic BiologyMutationbp 391 G to A, RBS* added (22 bp) upstream of ATG…Available SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
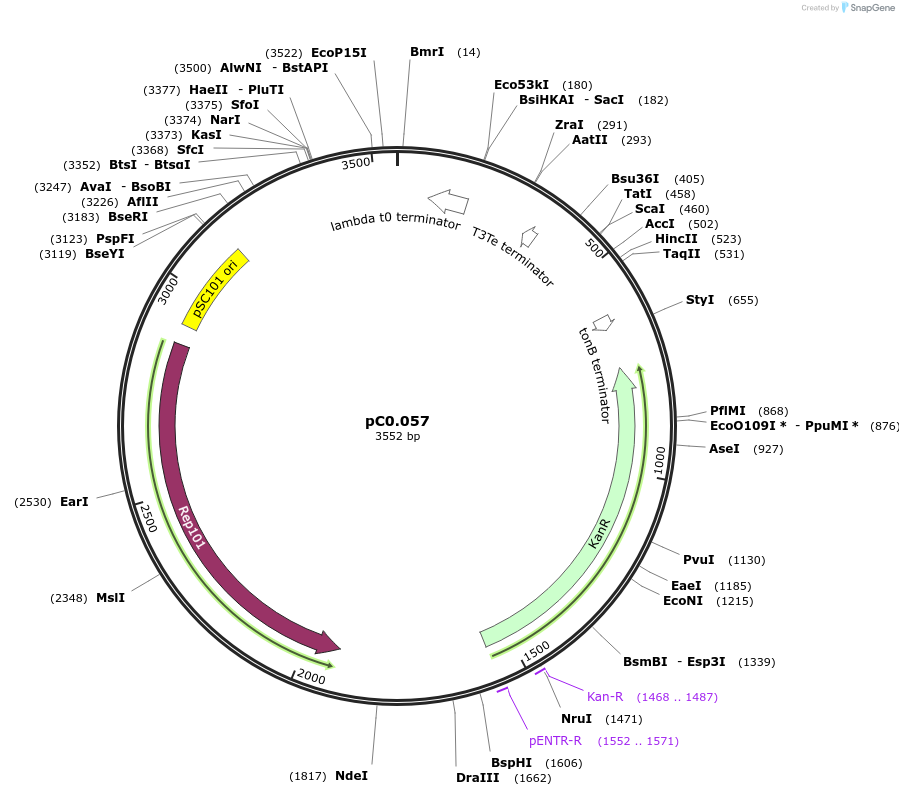
pC0.057
Plasmid#119565PurposeLevel 0 part. PromoterDepositorInsertParsB
UseSynthetic BiologyMutationbp 117 G to A, RBS* added (22 bp) upstream of ATG.Available SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
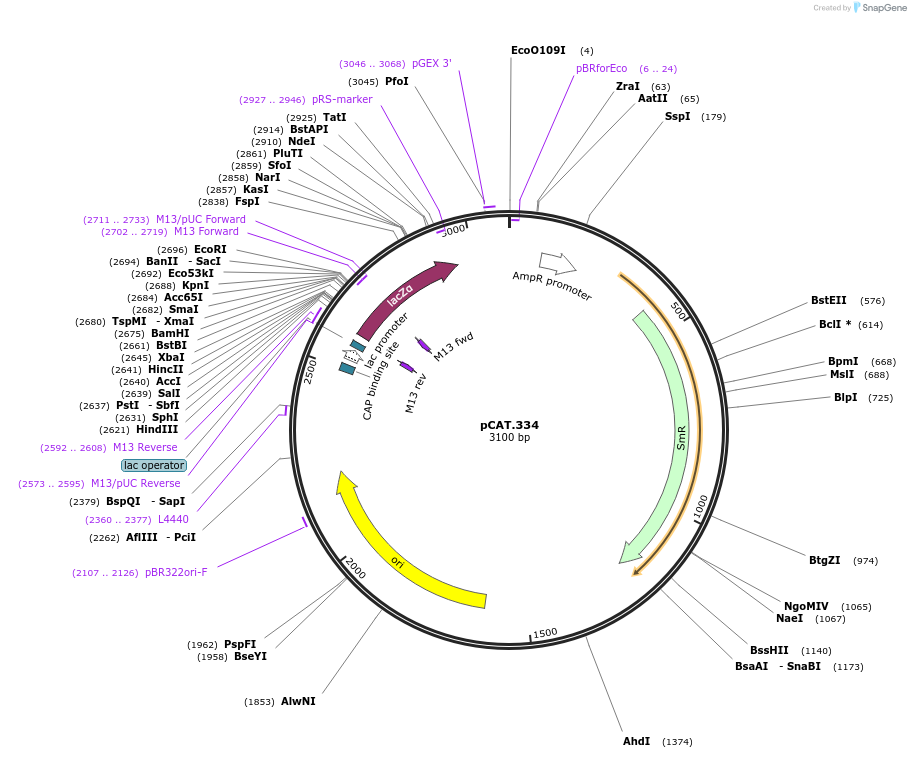
pCAT.334
Plasmid#119556PurposeLevel T acceptor vector for chromosomal integration with lacZ compatible with blue/white screening. Modified pUC19 vector.DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
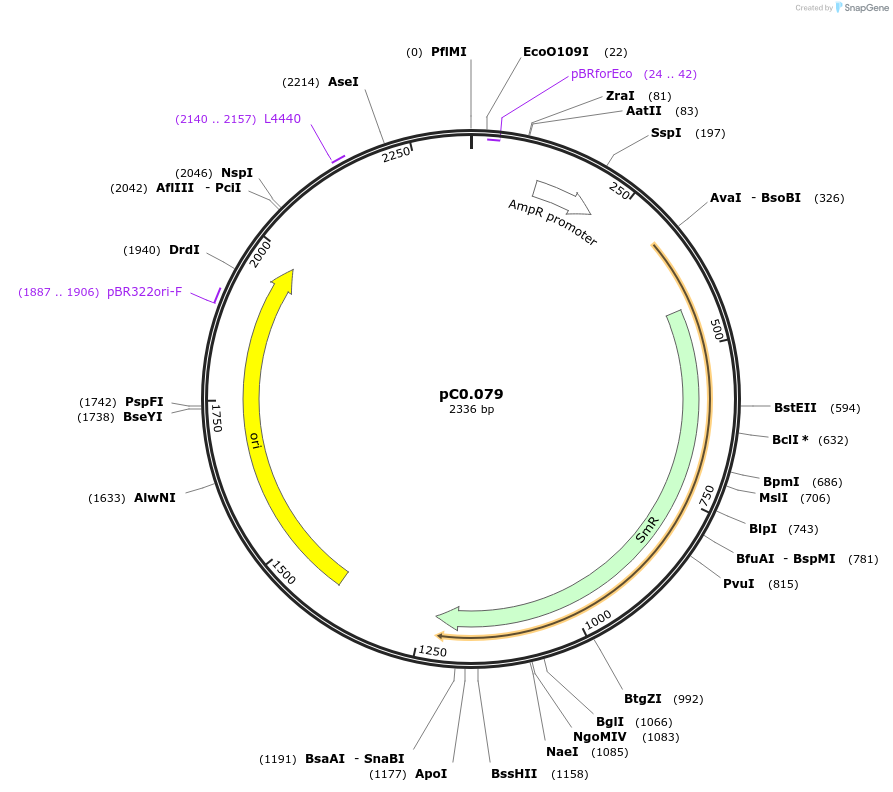
pC0.079
Plasmid#119667PurposeLevel 0 part. Terminator for the psbA2 gene Synechocystis sp. PCC 6803.DepositorInsertTpsbA2
UseSynthetic BiologyAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
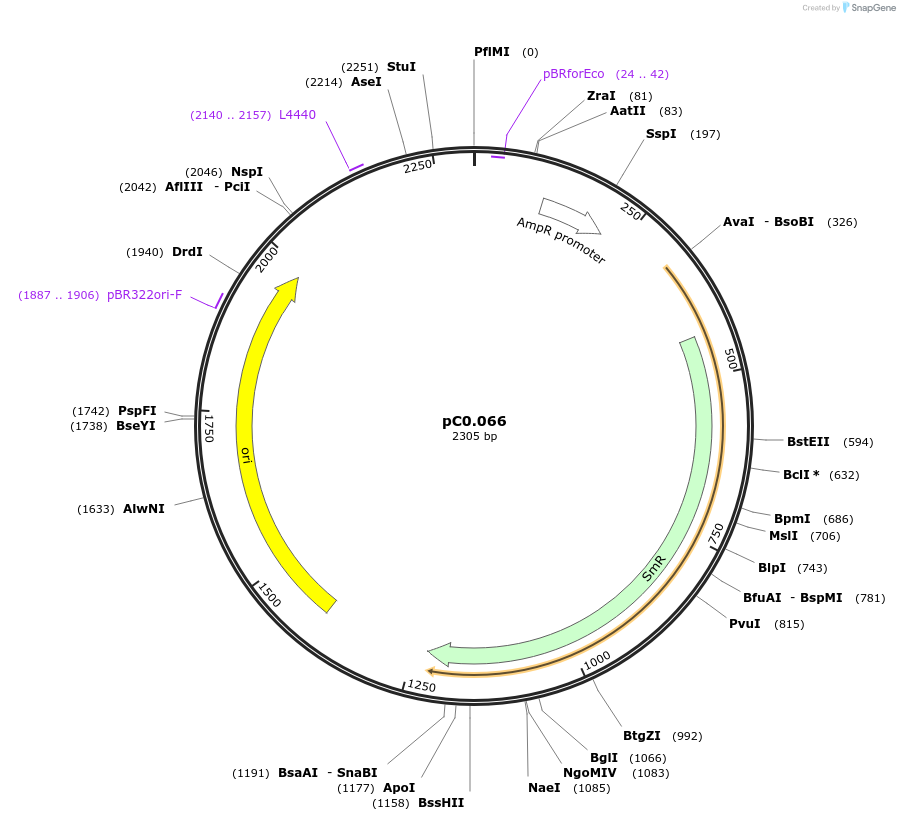
pC0.066
Plasmid#119666PurposeLevel 0 part. Terminator from pheA gene from E. coli.DepositorInsertTpheA-1
UseSynthetic BiologyAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only