We narrowed to 5,921 results for: crispr cas9 expression plasmids
-
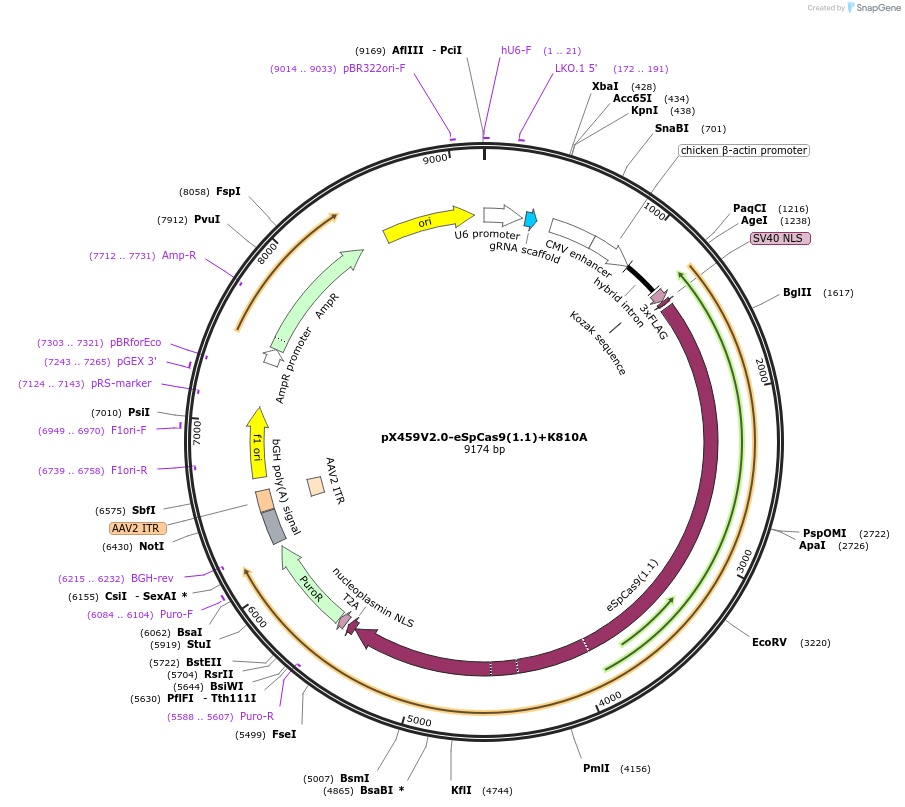
Plasmid#108297PurposepX459 V2.0 (Plasmid #62988) with the K810A, K848A, K1003A, and R1060A mutationsDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceMay 18, 2018AvailabilityAcademic Institutions and Nonprofits only
-
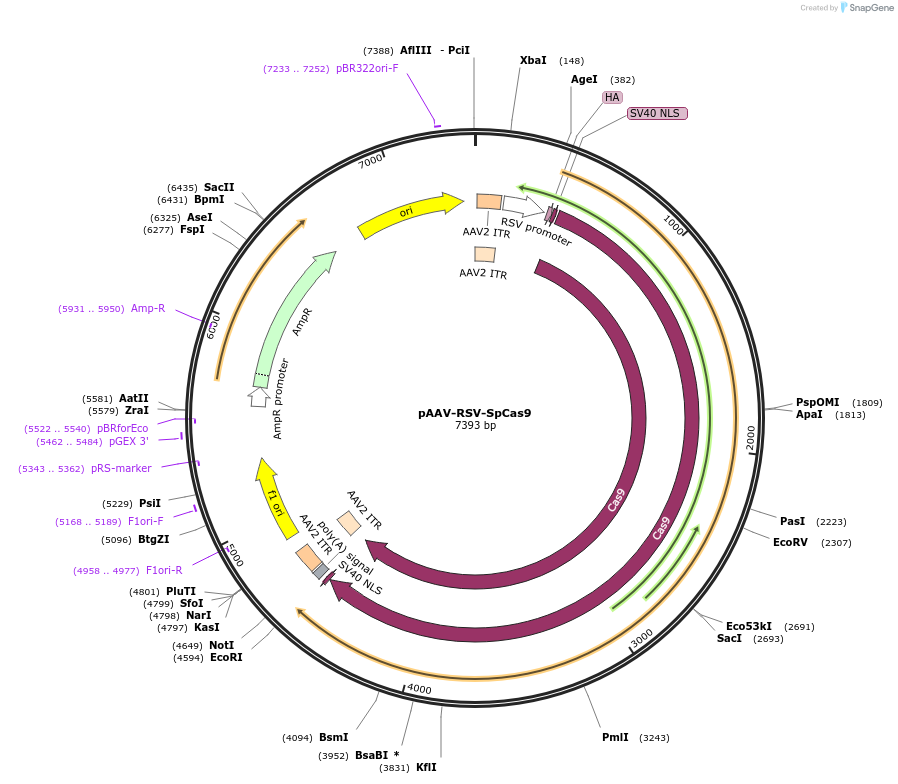
pAAV-RSV-SpCas9
Plasmid#85450PurposeExpress SpCas9 in mammalian cellsDepositorInsertpRSV
UseAAVPromoterpRSVAvailable SinceJan. 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
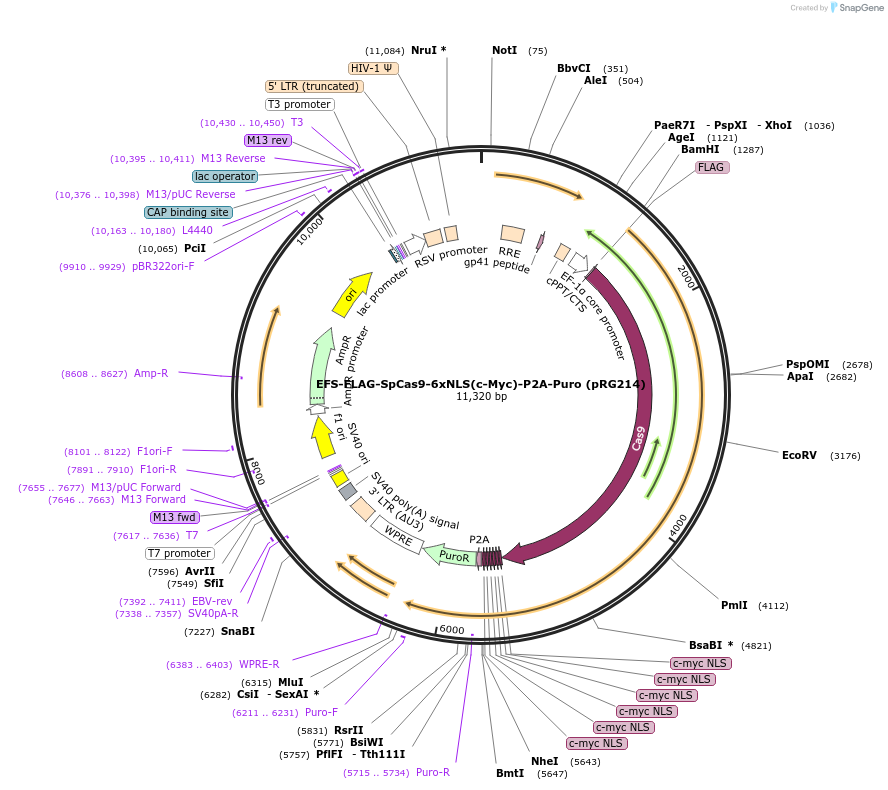
EFS-FLAG-SpCas9-6xNLS(c-Myc)-P2A-Puro (pRG214)
Plasmid#221385PurposeLentiviral expression of SpCas9-6xNLS(c-Myc) with puromycin resistance geneDepositorInsertSpCas9
UseCRISPR and LentiviralTagsFLAGExpressionMammalianAvailable SinceJune 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
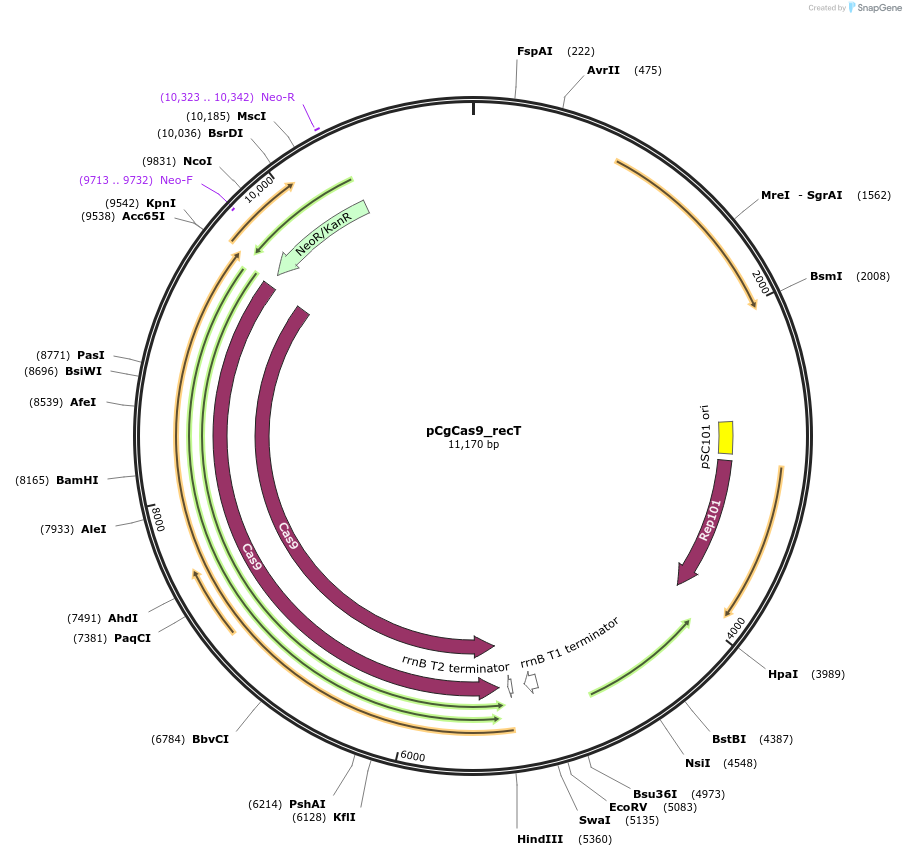
pCgCas9_recT
Plasmid#153337PurposeDouble-plasmid-based CRISPR-Cas9 system in Corynebacterium glutamicum; pBL1ts oriVC. glutamicum; pSC101 oriVE. coli PlacM-SpCas9 Streptomyces coelicolor codon-optimized, Peftu-RecT; KanrDepositorInsertCas9
UseCRISPRExpressionBacterialAvailable SinceJuly 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
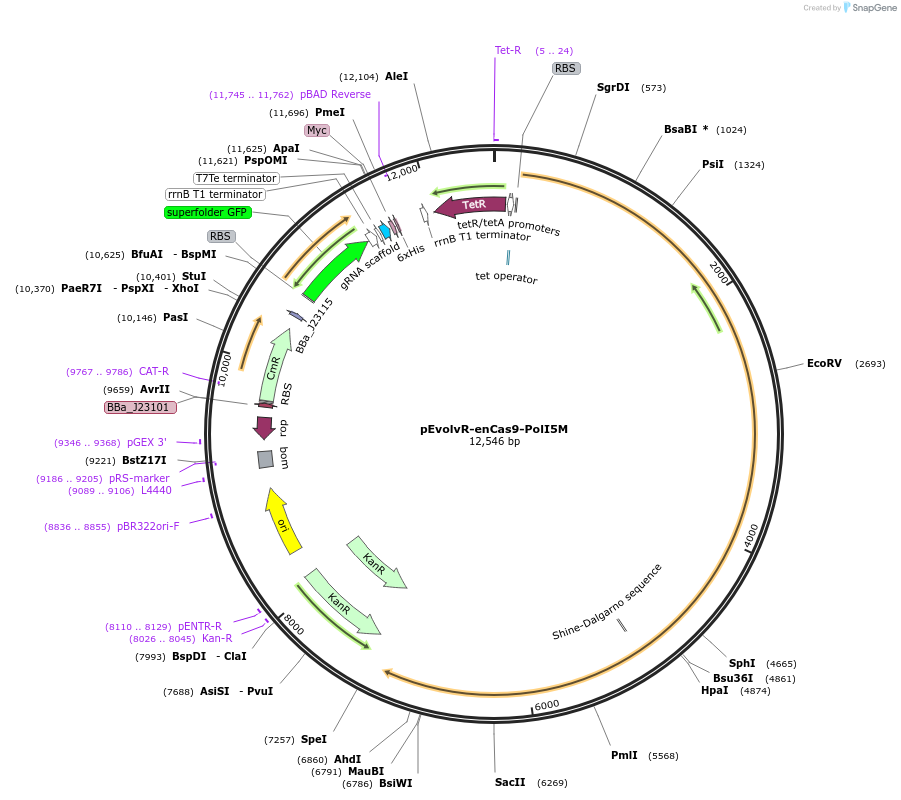
pEvolvR-enCas9-PolI5M
Plasmid#113078PurposeExpresses enCas9 fused to PolI5M in E. coli. contains a BsmbI-flanked GFP cassette for sgRNA cloning.DepositorInsertenCas9-PolI5M
UseCRISPRExpressionBacterialAvailable SinceAug. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
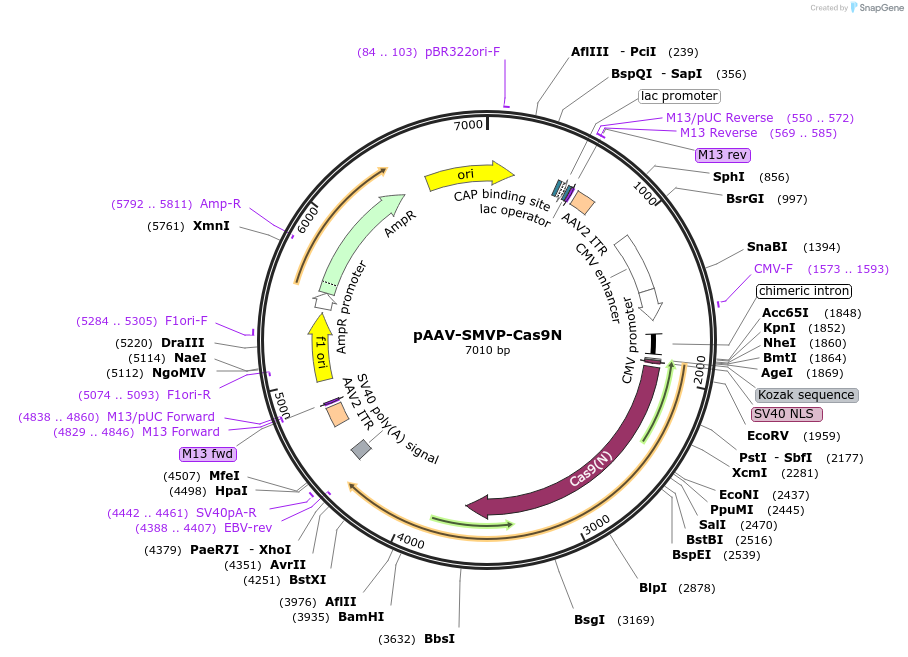
pAAV-SMVP-Cas9N
Plasmid#80930PurposeExpresses Cas9N in mammalian cells.DepositorInsertCas9N
UseAAV, CRISPR, and Synthetic BiologyExpressionMammalianAvailable SinceOct. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
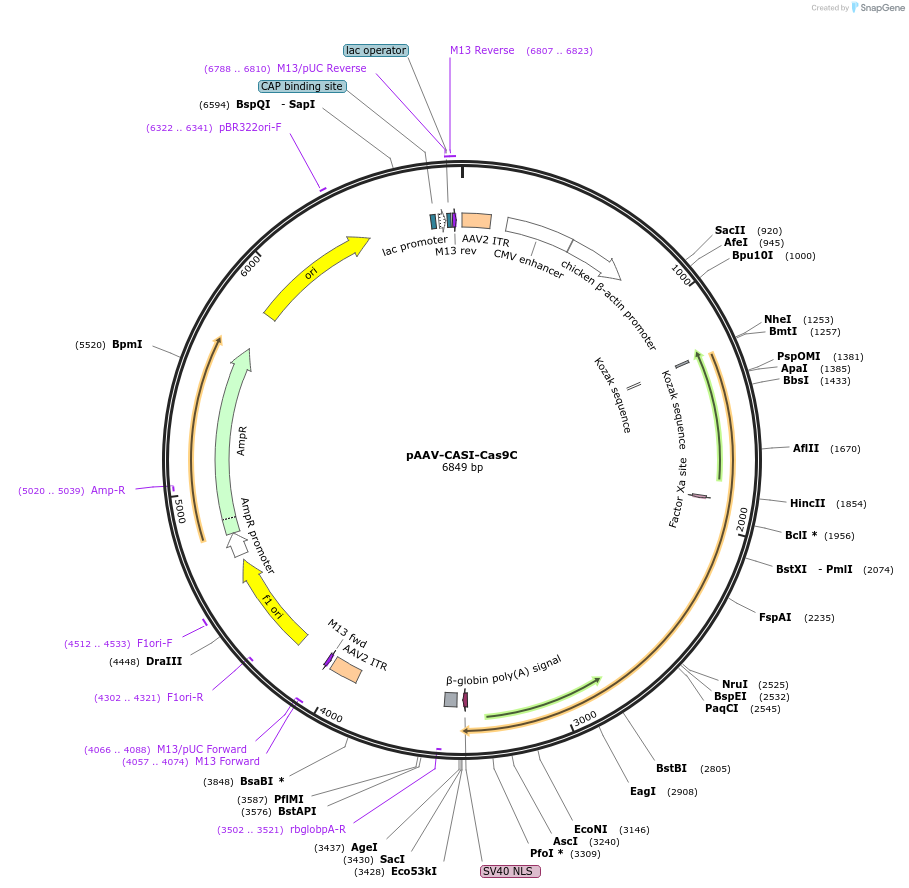
pAAV-CASI-Cas9C
Plasmid#80932PurposeExpresses Cas9C in mammalian cells; derived from pZac2.1 with CASI promoter.DepositorInsertCas9C
UseAAV, CRISPR, and Synthetic BiologyExpressionMammalianAvailable SinceOct. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
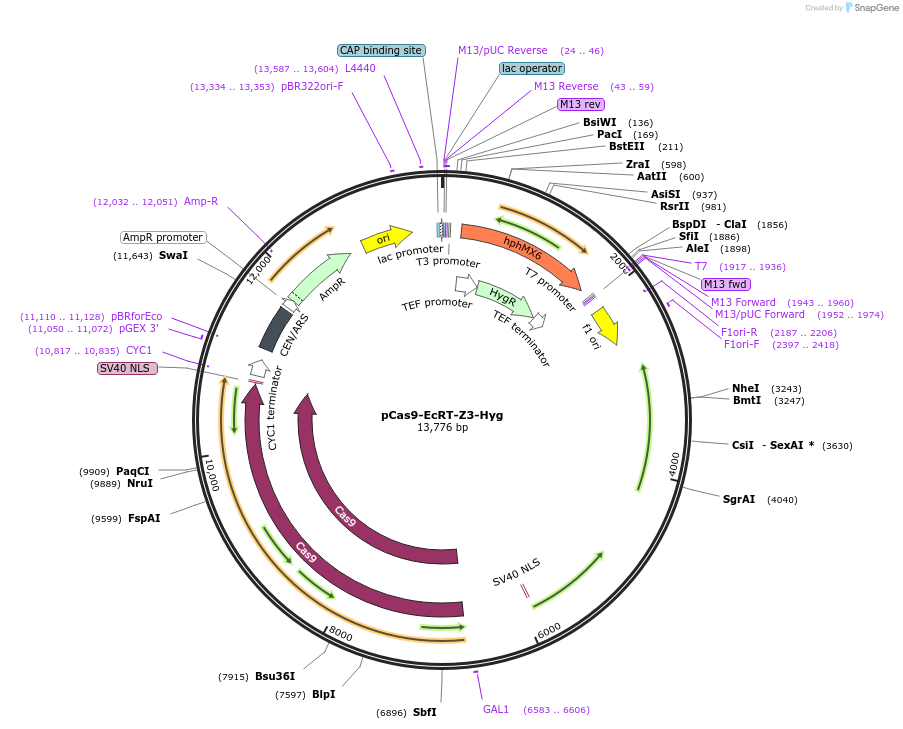
pCas9-EcRT-Z3-Hyg
Plasmid#232095PurposeYeast CEN plasmid for estradiol-inducible expression of spCas9 and Ec86 reverse transcriptase.DepositorInsertZ3 promoter and Z3EV transcription factor
UseCRISPRExpressionYeastPromoterZ3Available SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
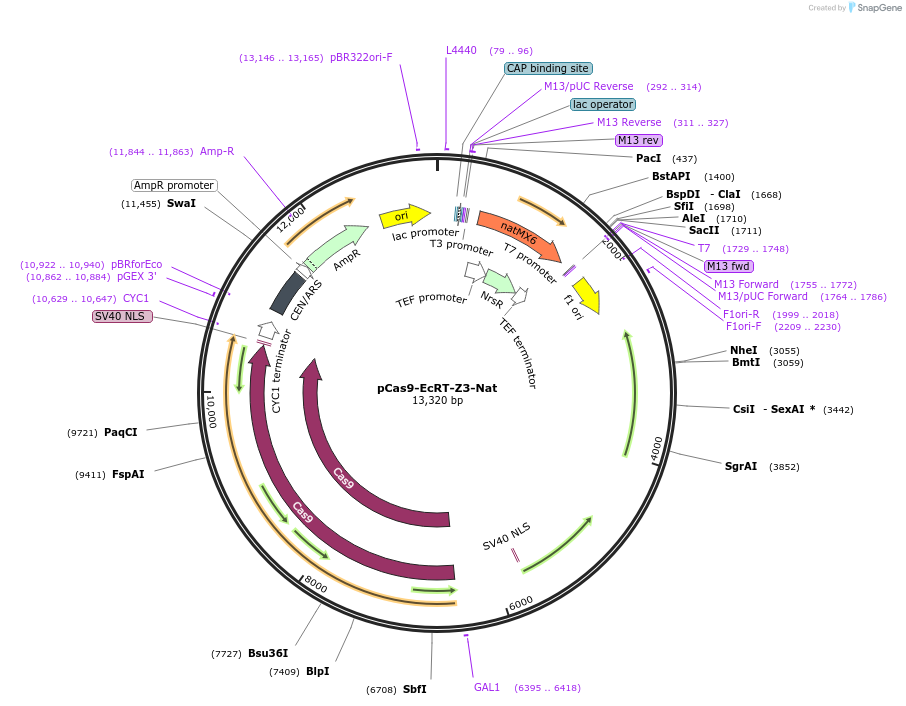
pCas9-EcRT-Z3-Nat
Plasmid#232094PurposeYeast CEN plasmid for estradiol-inducible expression of spCas9 and Ec86 reverse transcriptase.DepositorInsertZ3 promoter and Z3EV transcription factor
UseCRISPRExpressionYeastPromoterZ3Available SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
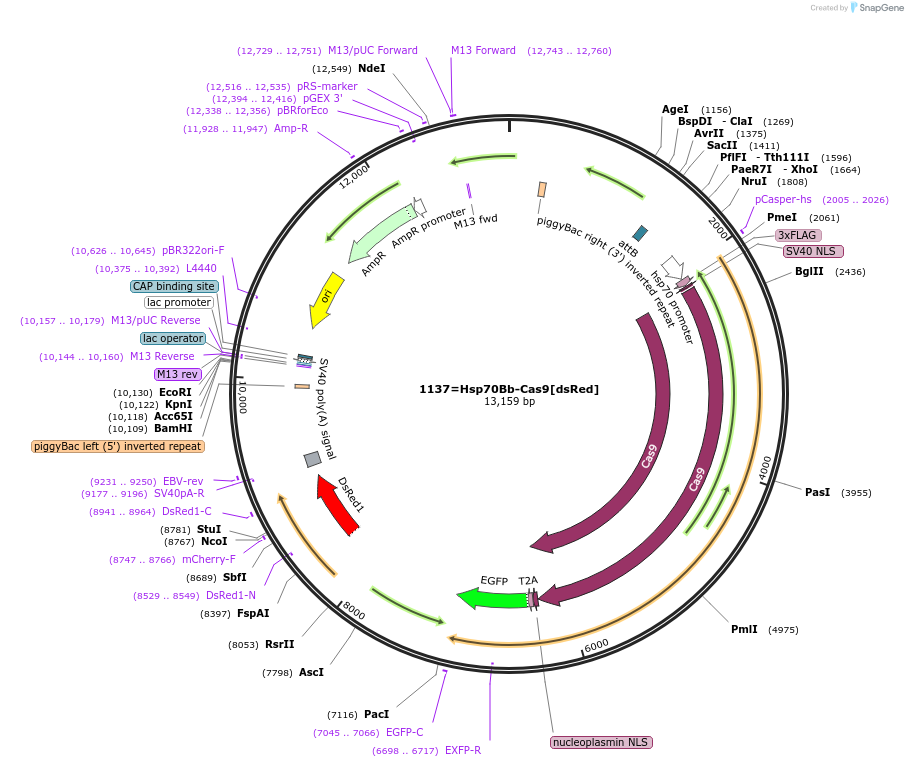
1137=Hsp70Bb-Cas9[dsRed]
Plasmid#149424PurposeThe piggyBac plasmid harboring Hsp70Bb-Cas9-T2A-eGFP-p10 and Opie2-dsRed-SV10 (marker gene)DepositorInsertCas9
UseCRISPRTagseGFPExpressionInsectPromoterDmel Hsp70BbAvailable SinceMarch 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
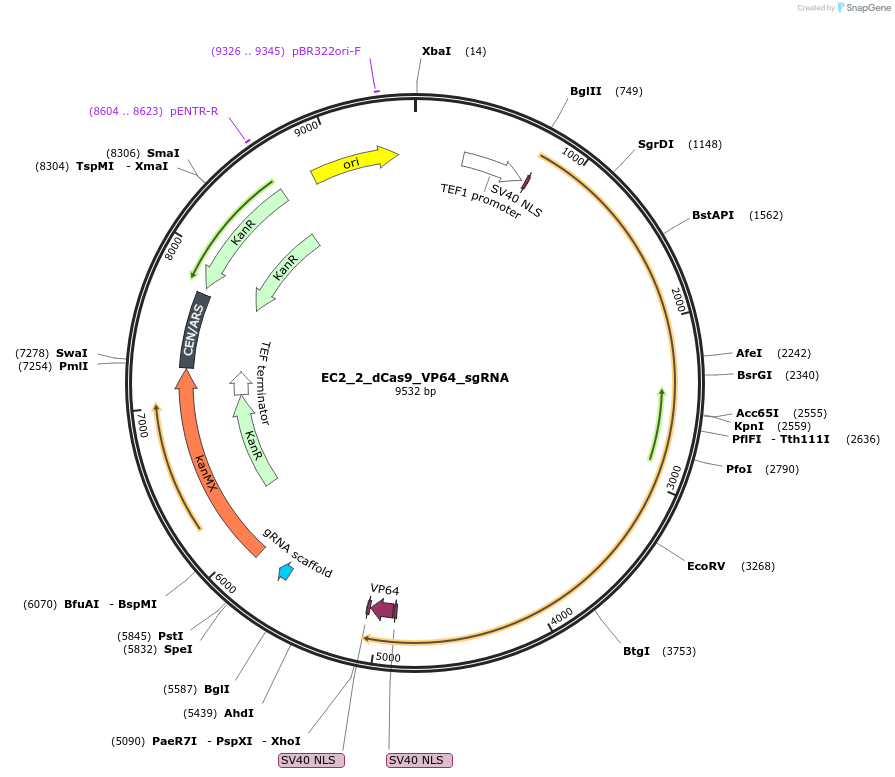
EC2_2_dCas9_VP64_sgRNA
Plasmid#163707PurposeYeast low copy plasmid with dCas9, sgRNA expression cassette and VP64 activation domainDepositorInsertsdCas9
VP64+SV40 NLS
ExpressionYeastMutationPoint mutations D10A and H840APromoterScTEF1Available SinceNov. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
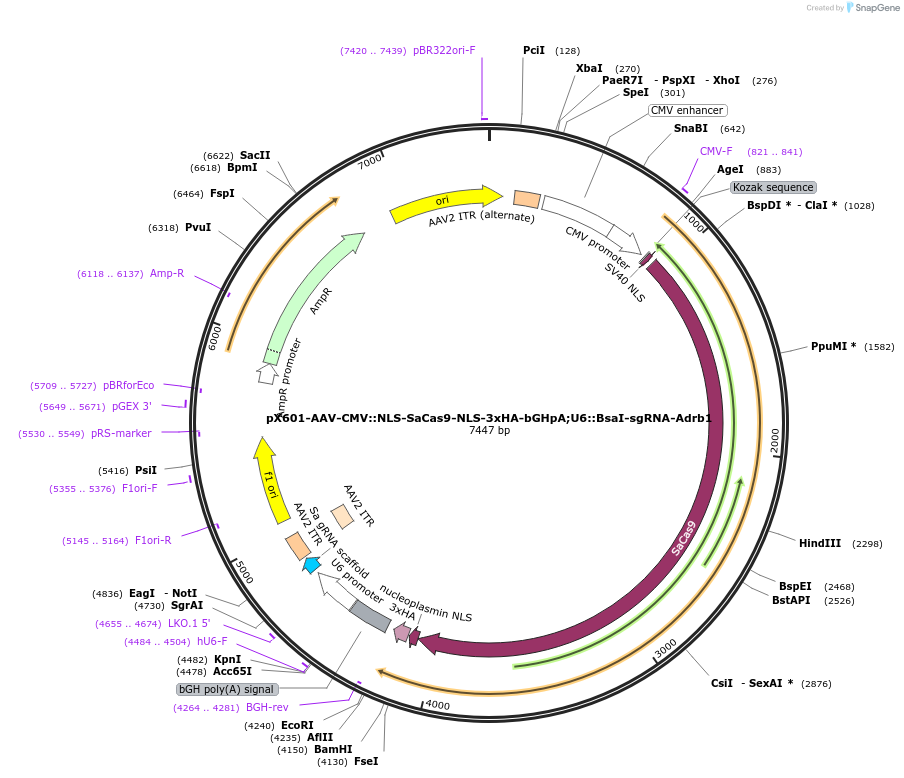
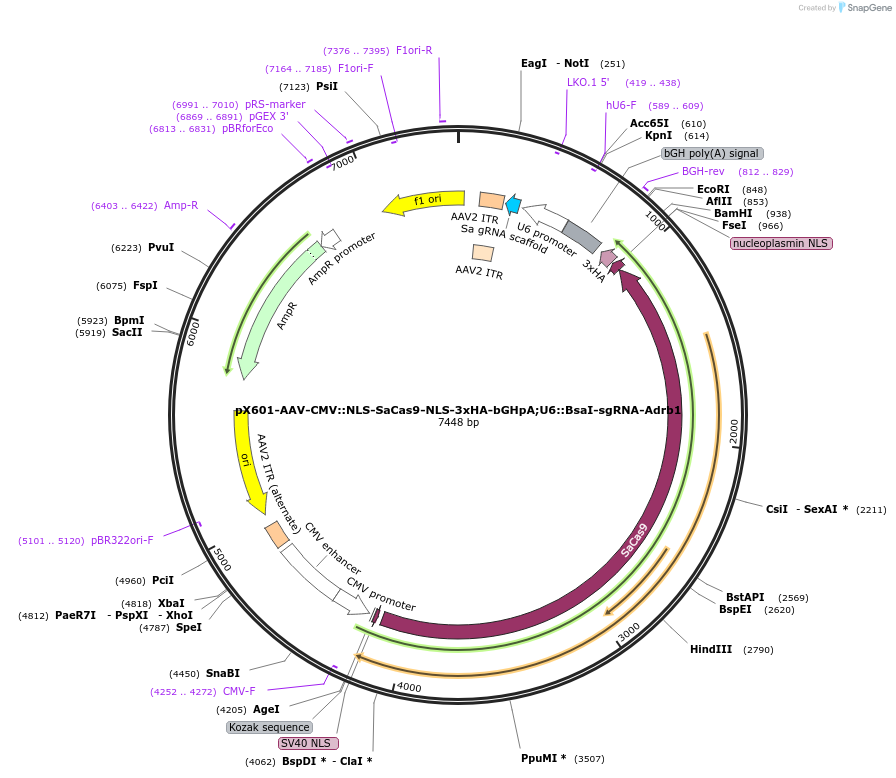
pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA-Adrb1
Plasmid#184294PurposeA single vector containing Cas9 from Staphylococcus aureus (SaCas9) and its sgRNA targeting Adrb1DepositorInsertsgRNA-Adrb1
UseAAV and CRISPRExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
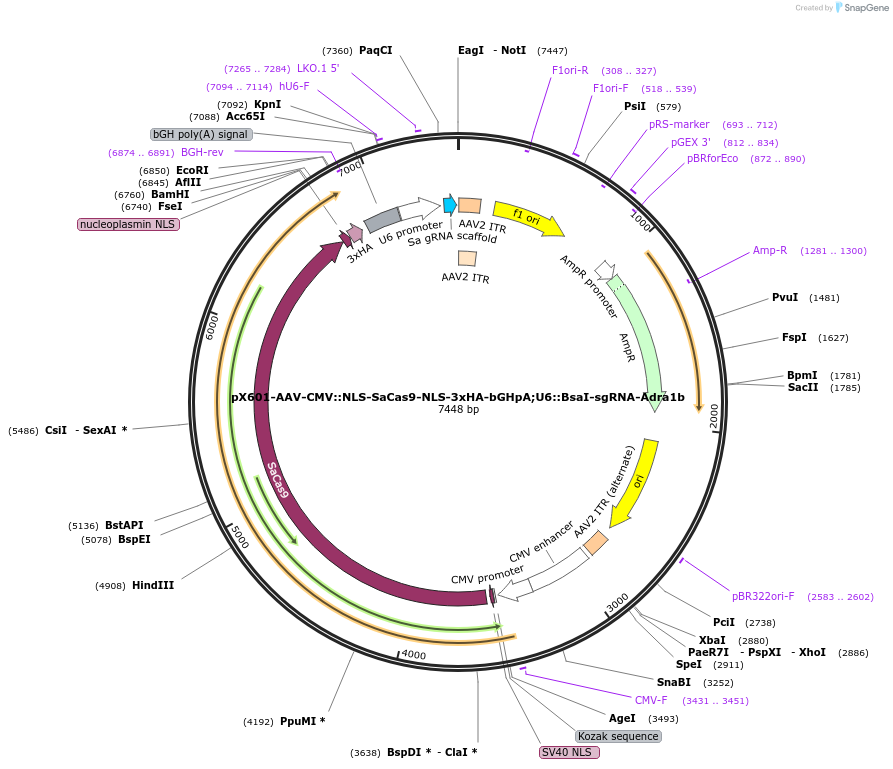
pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA-Adra1b
Plasmid#184293PurposeA single vector containing Cas9 from Staphylococcus aureus (SaCas9) and its sgRNA targeting Adra1bDepositorInsertssgRNA-Adra1b
sgRNA-Adra1b
UseAAV and CRISPRExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
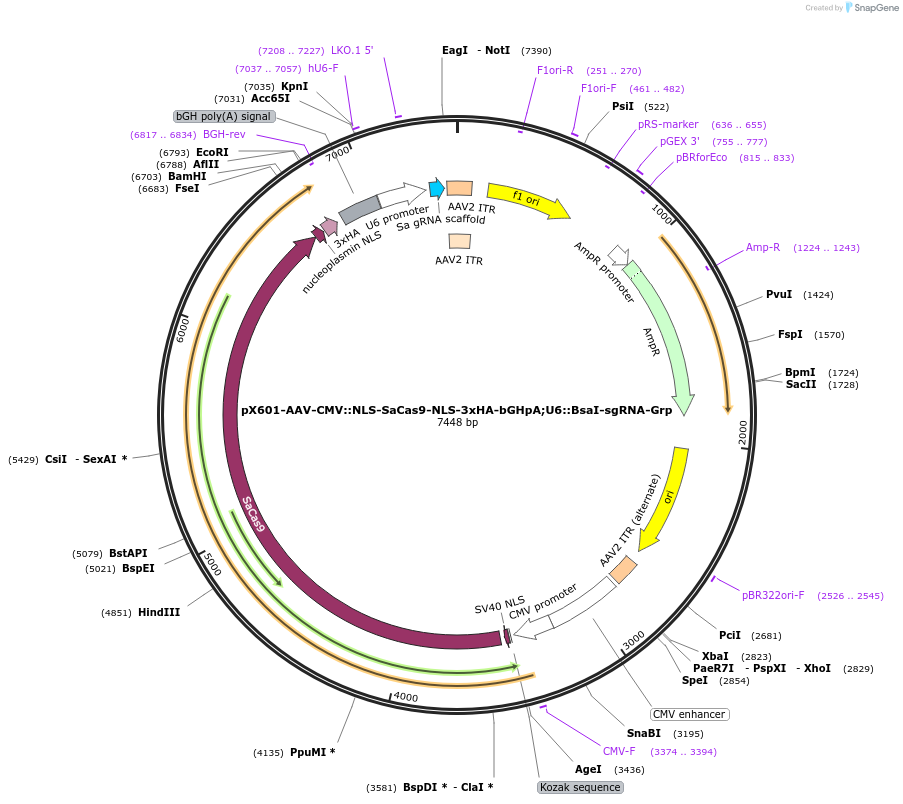
pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA-Grp
Plasmid#184292PurposeA single vector containing Cas9 from Staphylococcus aureus (SaCas9) and its sgRNA targeting GrpDepositorInsertsgRNA-Grp
UseAAV and CRISPRExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
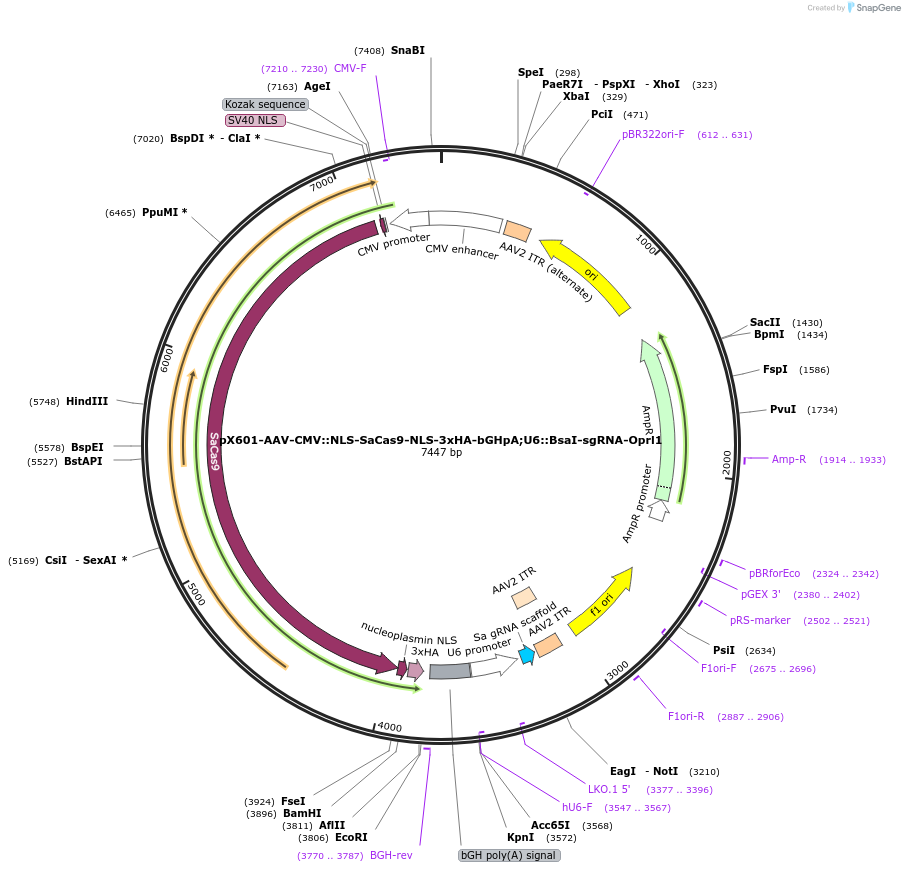
pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA-Oprl1
Plasmid#184291PurposeA single vector containing Cas9 from Staphylococcus aureus (SaCas9) and its sgRNA targeting Oprl1DepositorInsertsgRNA-Oprl1
UseAAV and CRISPRExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA-Rxfp3
Plasmid#184290PurposeA single vector containing Cas9 from Staphylococcus aureus (SaCas9) and its sgRNA targeting Rxfp3DepositorInsertsgRNA-Rxfp3
UseAAV and CRISPRExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA-Adrb1
Plasmid#184295PurposeA single vector containing Cas9 from Staphylococcus aureus (SaCas9) and its sgRNA targeting Adrb1DepositorInsertsgRNA-Adrb1
UseAAV and CRISPRExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
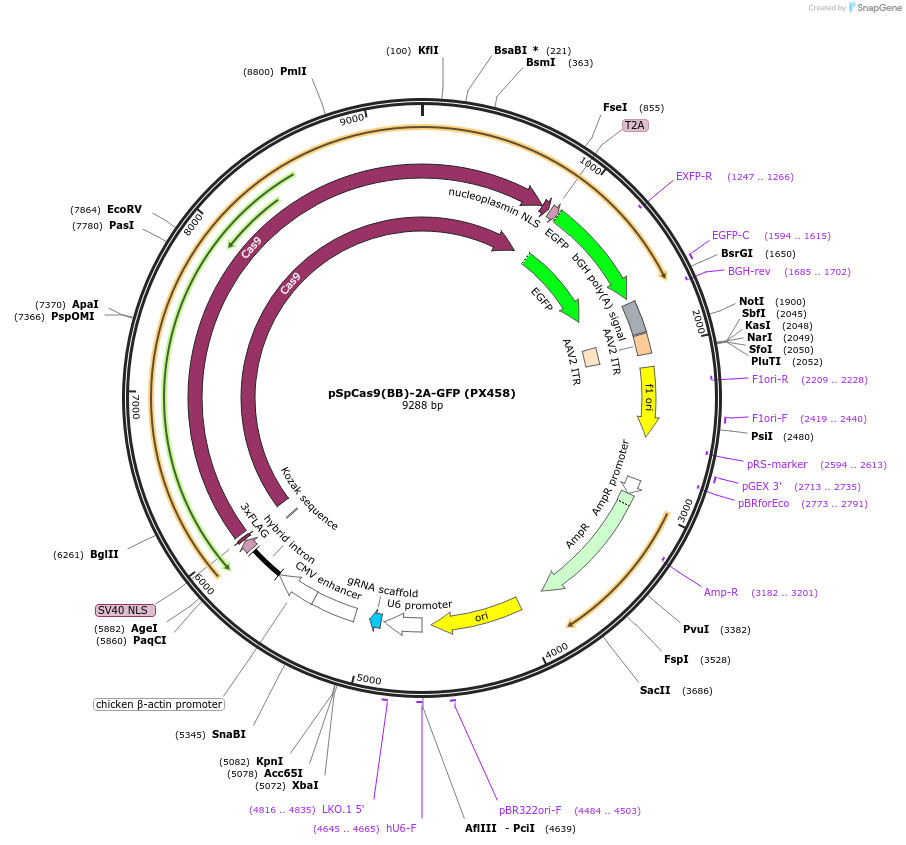
pSpCas9(BB)-2A-GFP (PX458)
Plasmid#48138PurposeCas9 from S. pyogenes with 2A-EGFP, and cloning backbone for sgRNADepositorHas ServiceCloning Grade DNAInserthSpCas9
UseCRISPRTags3XFLAG and GFPExpressionMammalianPromoterCbhAvailable SinceOct. 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
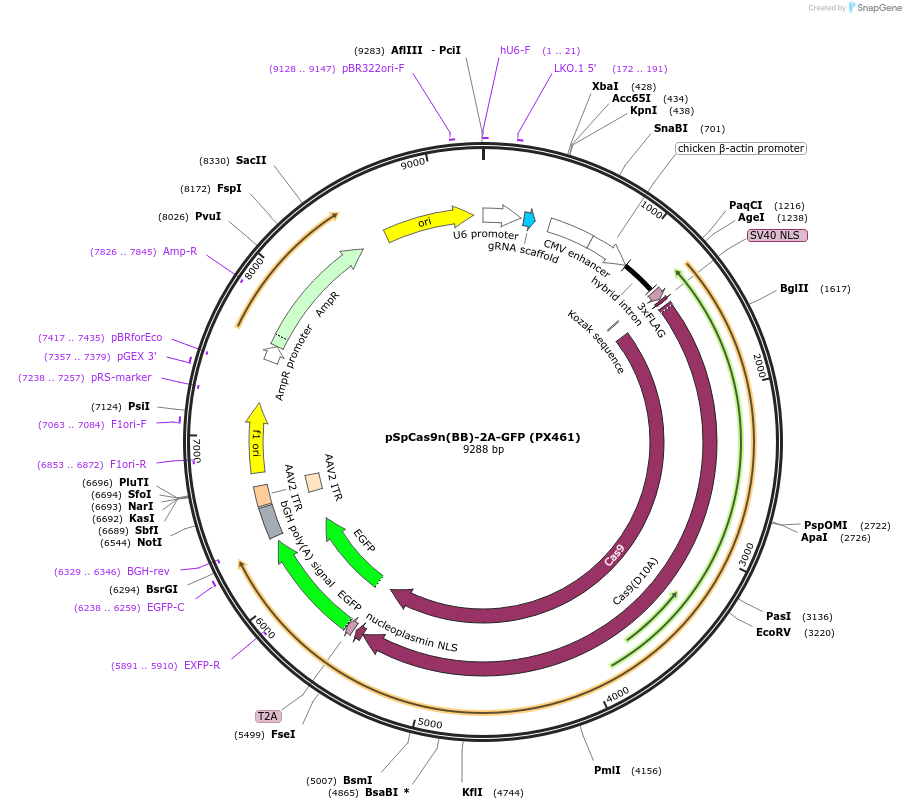
pSpCas9n(BB)-2A-GFP (PX461)
Plasmid#48140PurposeCas9n (D10A nickase mutant) from S. pyogenes with 2A-EGFP, and cloning backbone for sgRNADepositorInserthSpCas9n
UseCRISPRTags3XFLAG and GFPExpressionMammalianMutationCas9 D10A nickase mutantPromoterCbhAvailable SinceOct. 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
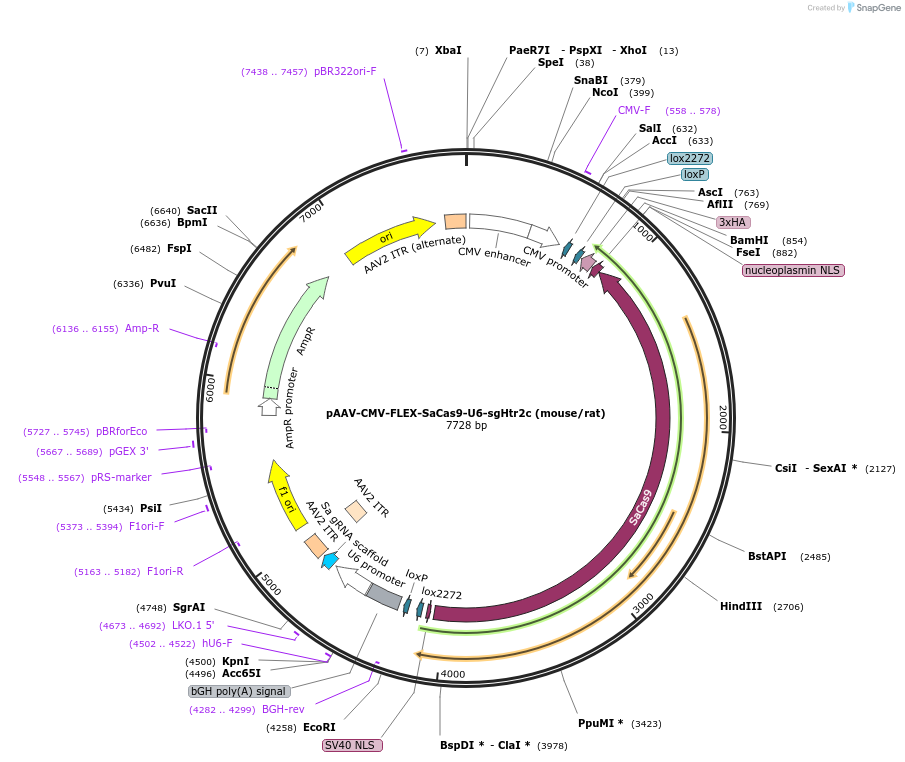
pAAV-CMV-FLEX-SaCas9-U6-sgHtr2c (mouse/rat)
Plasmid#249541PurposeCre-dependent editing of Htr2c with SaCas9DepositorAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only