We narrowed to 3,846 results for: 28
-
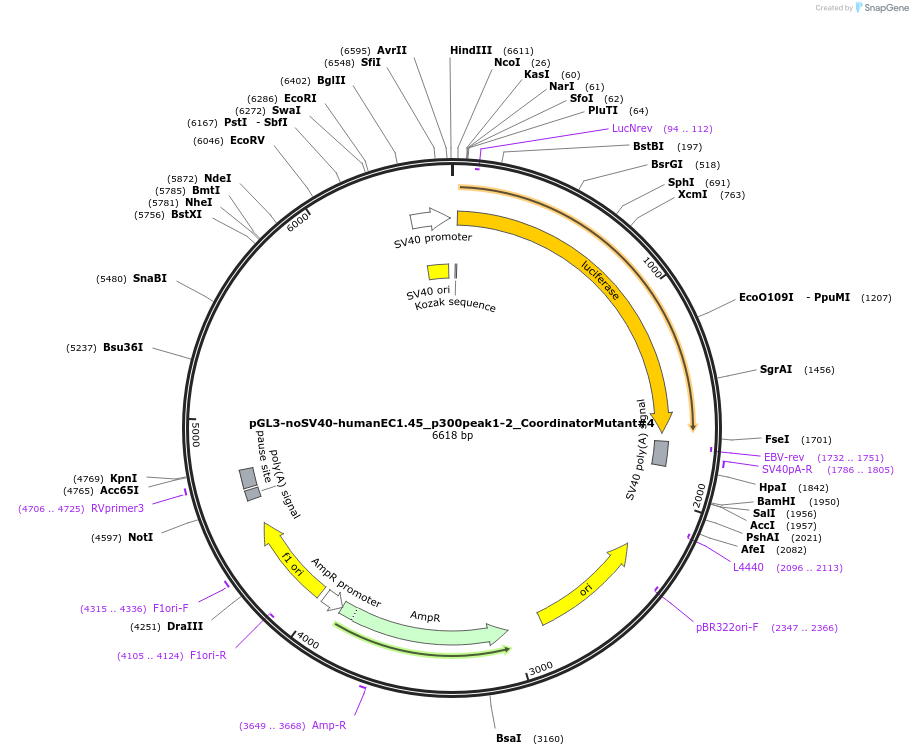
Plasmid#173988PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #4 mutated
UseLuciferaseMutationCoordinator motif mutation #4PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
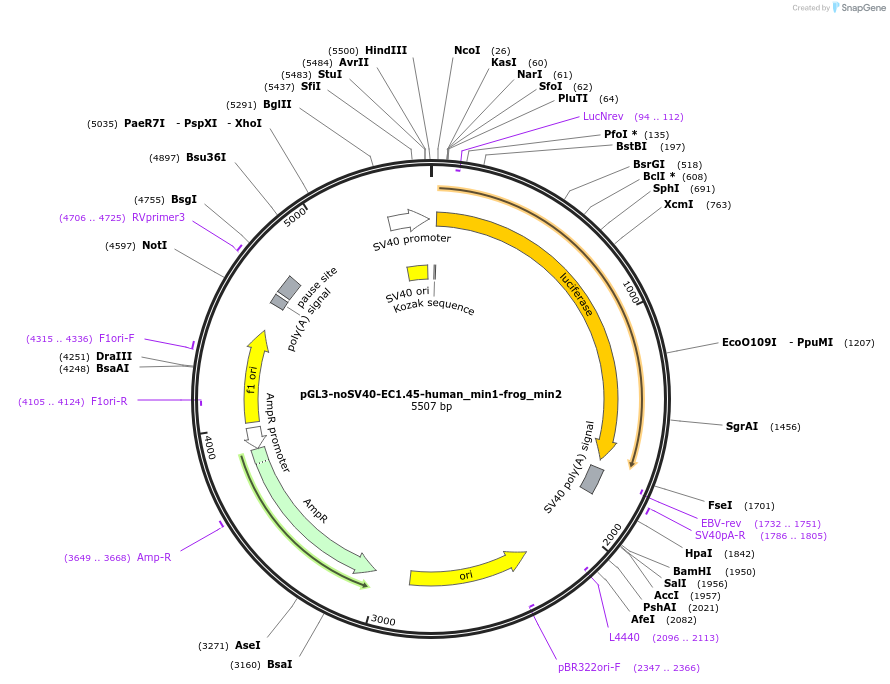
pGL3-noSV40-EC1.45-human_min1-frog_min2
Plasmid#173970PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and frog minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
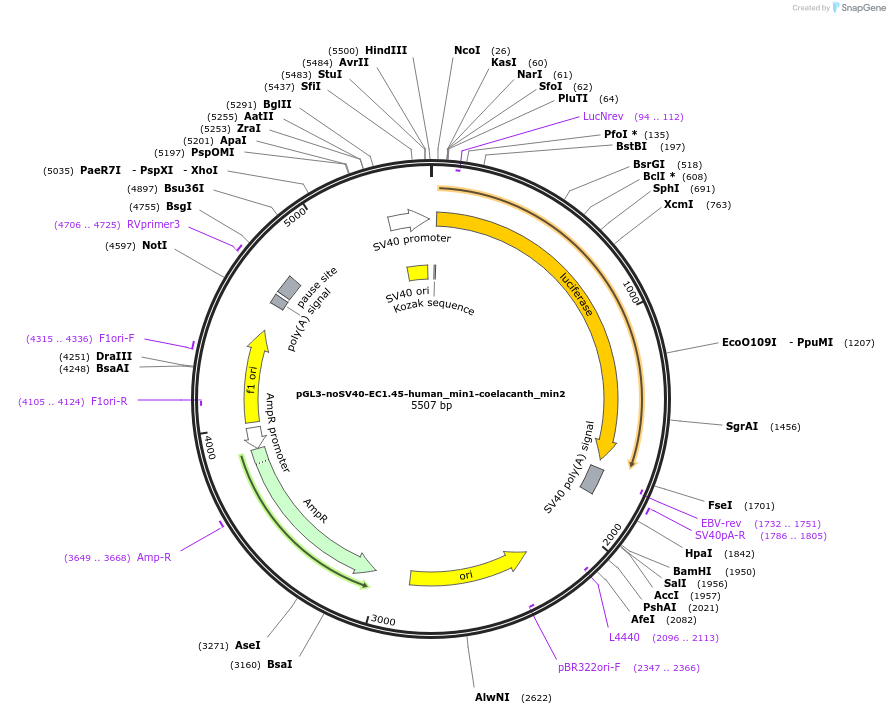
pGL3-noSV40-EC1.45-human_min1-coelacanth_min2
Plasmid#173971PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and coelacanth minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
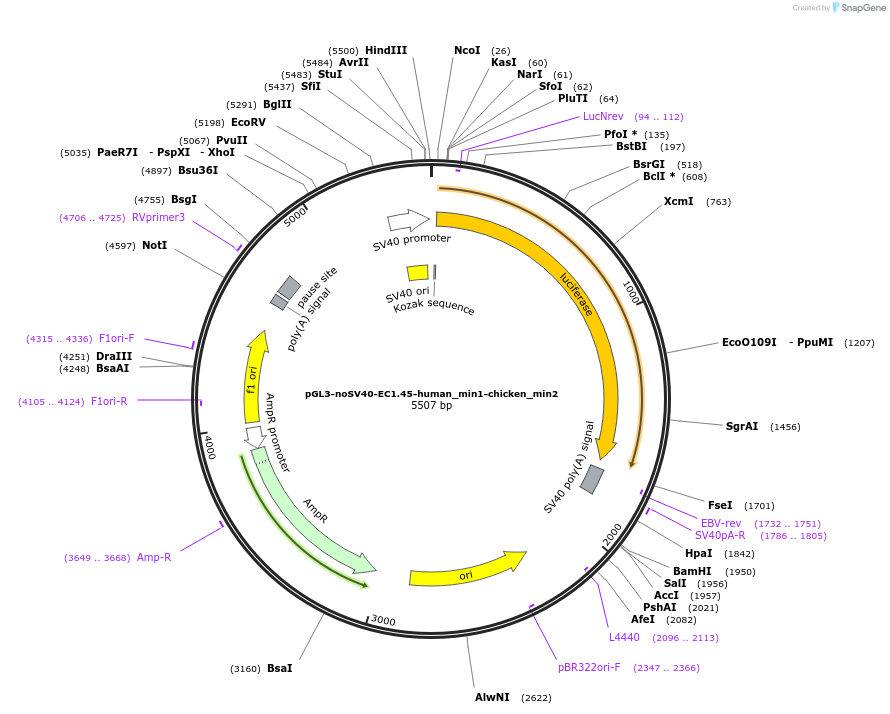
pGL3-noSV40-EC1.45-human_min1-chicken_min2
Plasmid#173968PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and chicken minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
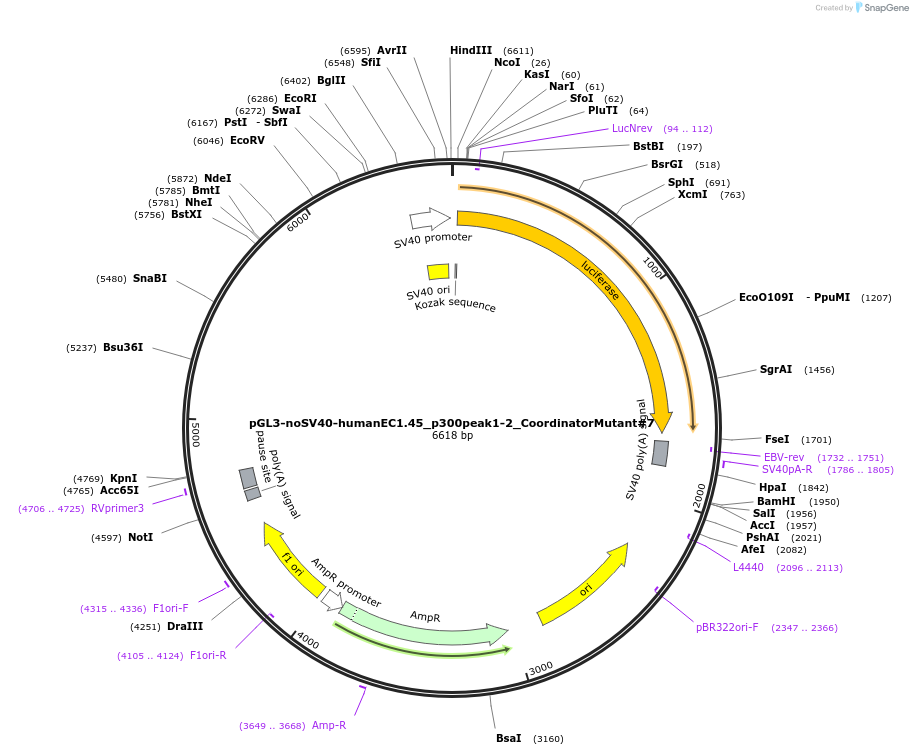
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#7
Plasmid#173991PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #7 mutated
UseLuciferaseMutationCoordinator motif mutation #7PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
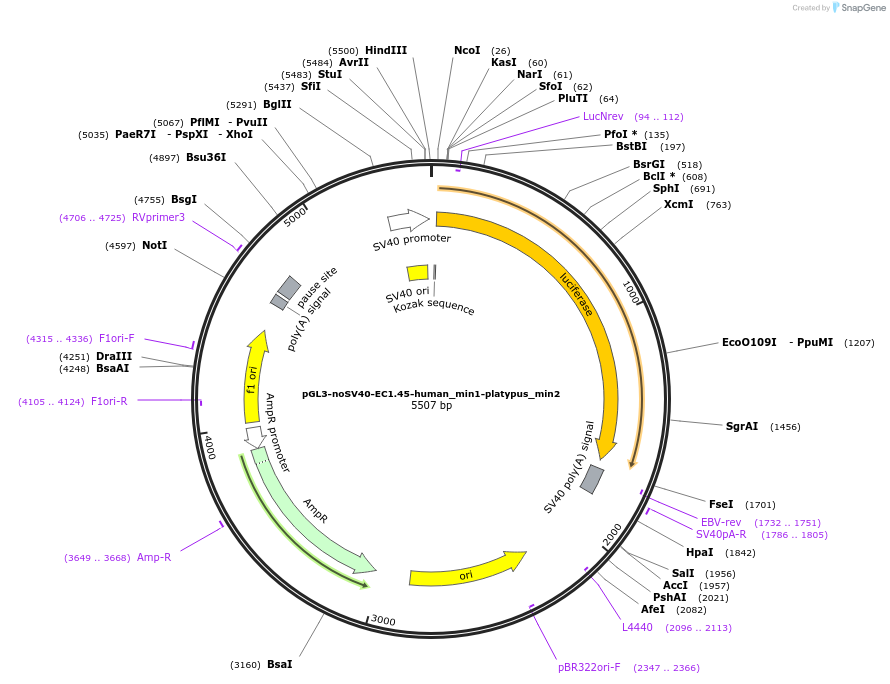
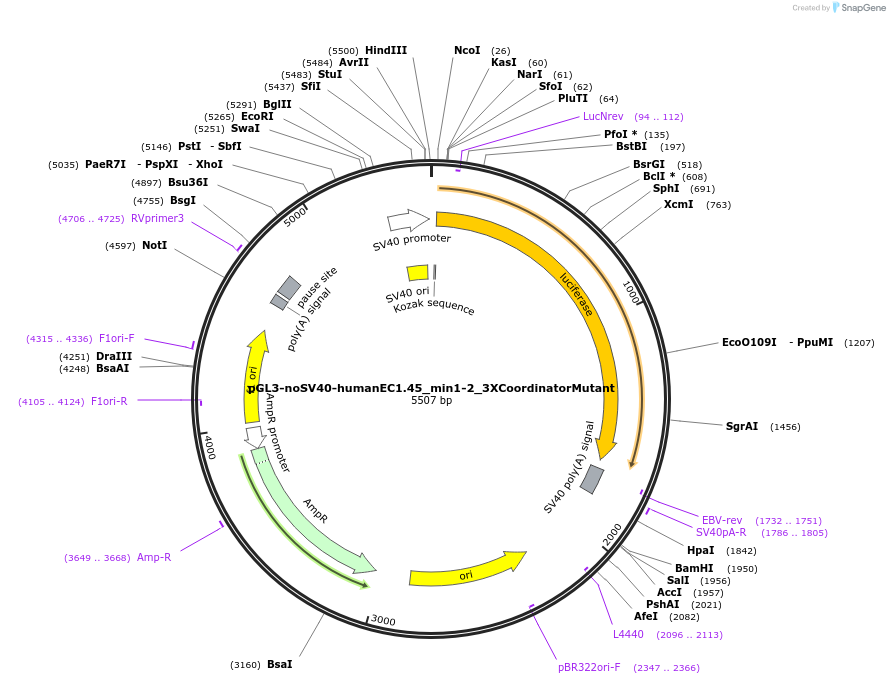
pGL3-noSV40-EC1.45-human_min1-platypus_min2
Plasmid#173967PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and platypus minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
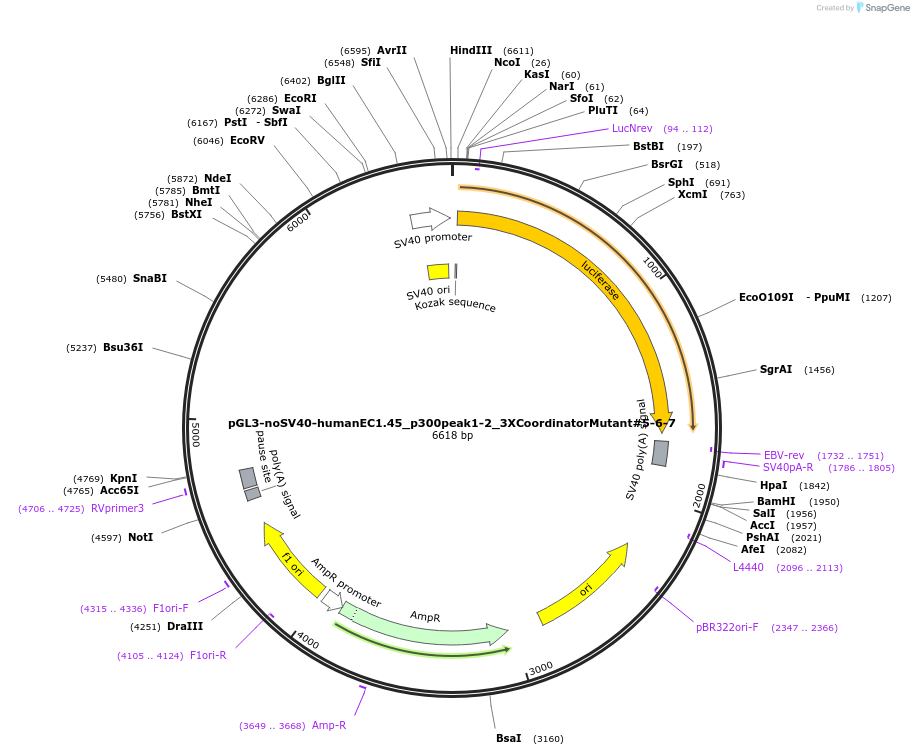
pGL3-noSV40-humanEC1.45_p300peak1-2_3XCoordinatorMutant#5-6-7
Plasmid#173993PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motifs #5, #6 and #7 mutated
UseLuciferaseMutation3X Coordinator motif mutation #5-6-7PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_min1-2_3XCoordinatorMutant
Plasmid#173964PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer regions 1 and 2 (min1 and min2) from SOX9 enhancer cluster EC1.45 with 3 mutated Coordinator motifs
UseLuciferaseMutation3 mutated Coordinator motifsPromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
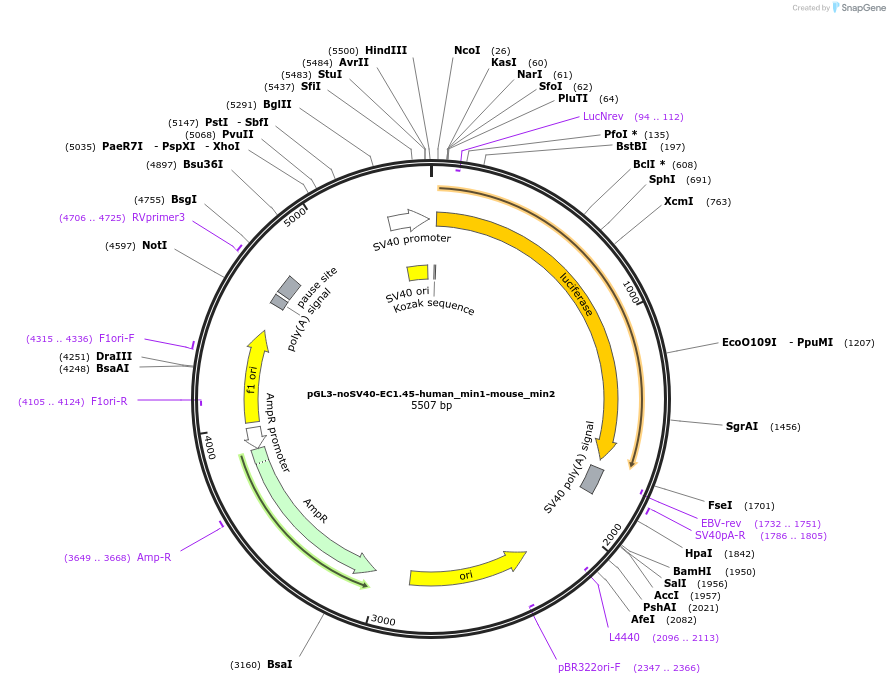
pGL3-noSV40-EC1.45-human_min1-mouse_min2
Plasmid#173965PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and mouse minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
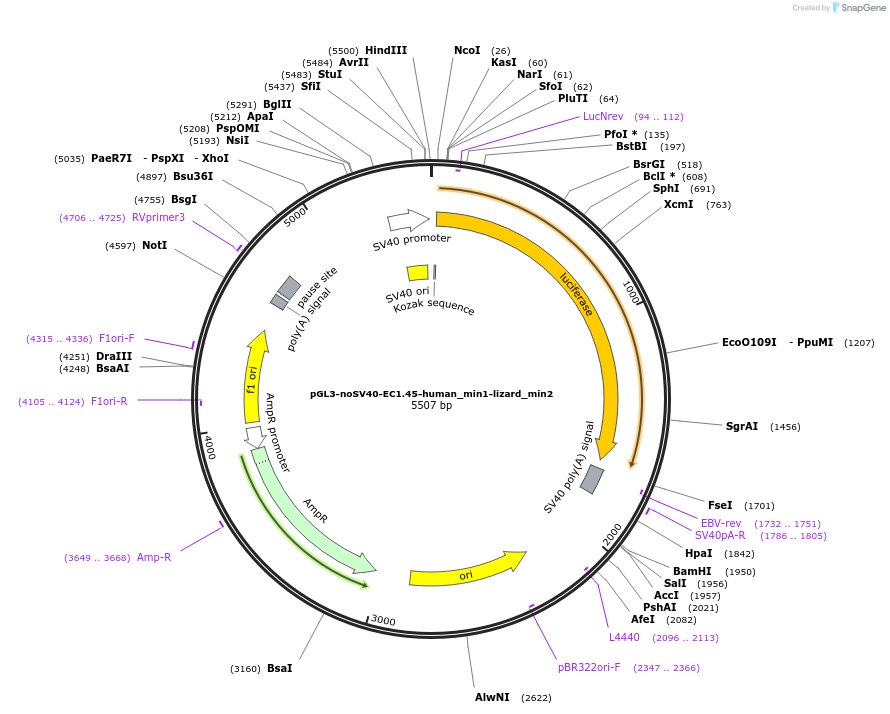
pGL3-noSV40-EC1.45-human_min1-lizard_min2
Plasmid#173969PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and lizard minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
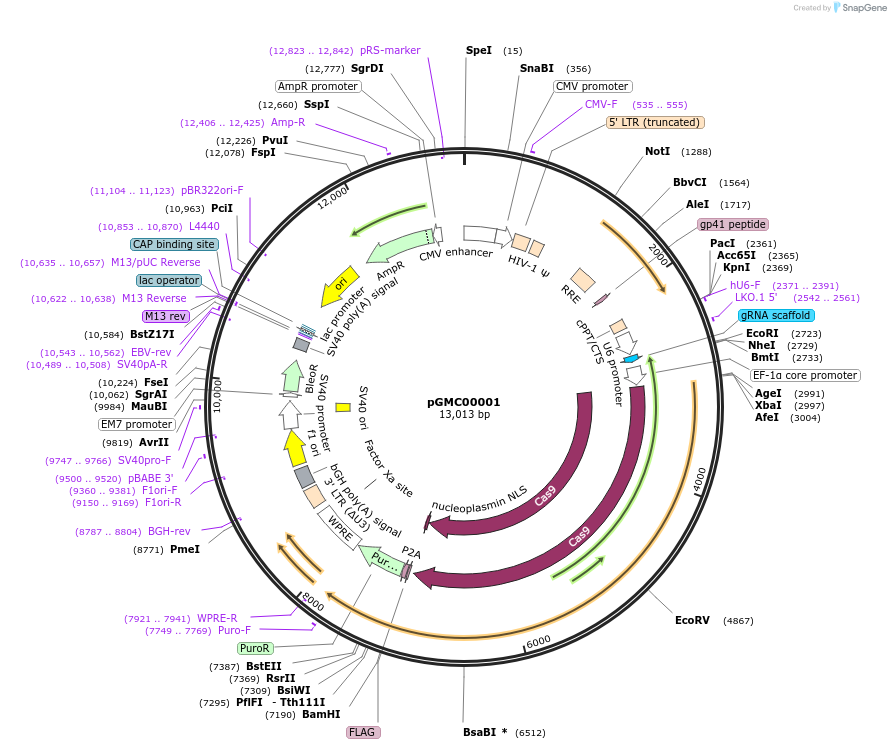
pGMC00001
Plasmid#166718PurposesgRNA against human BAP1DepositorAvailable SinceJune 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
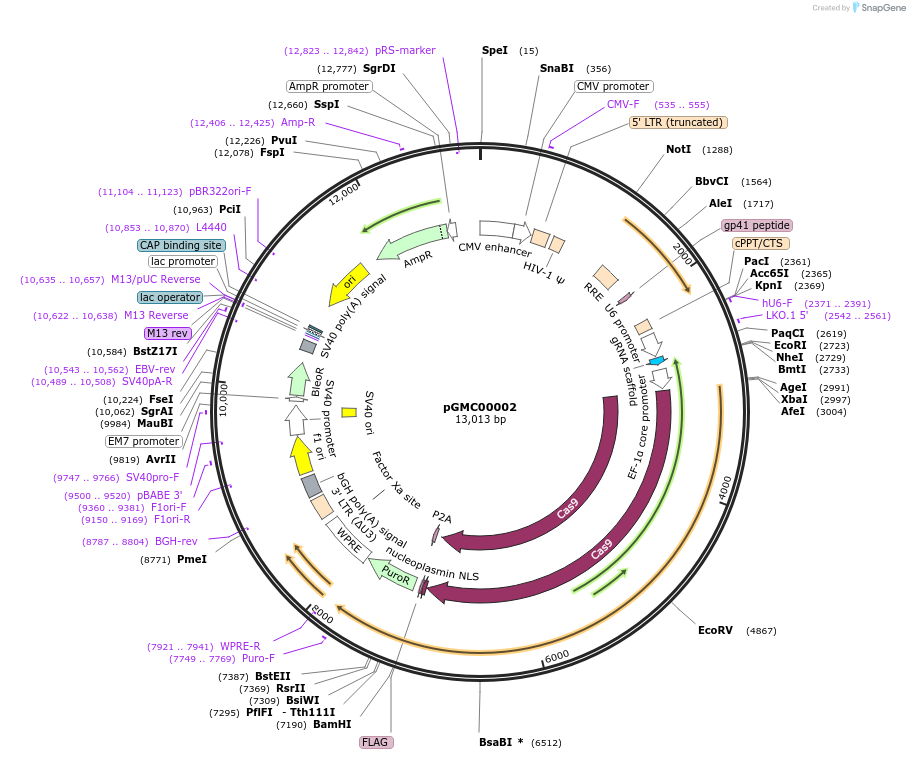
pGMC00002
Plasmid#166719PurposesgRNA against human BAP1DepositorAvailable SinceJune 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
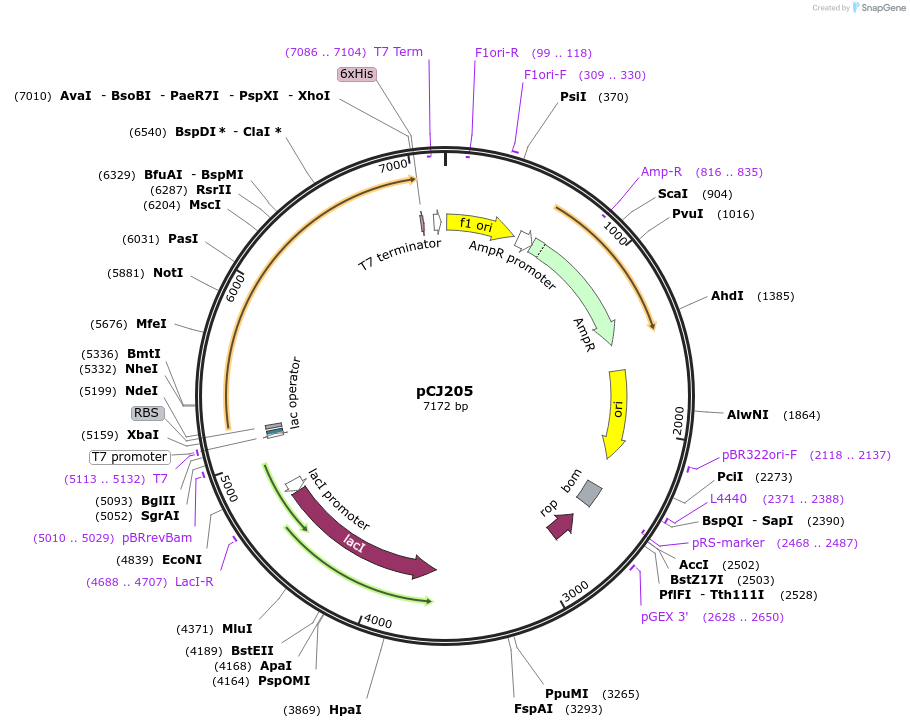
pCJ205
Plasmid#162676PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224A anDepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224A, C529A; codon optimized for expression in E…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCJ198
Plasmid#162671PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating F495I mutation.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationF495I; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
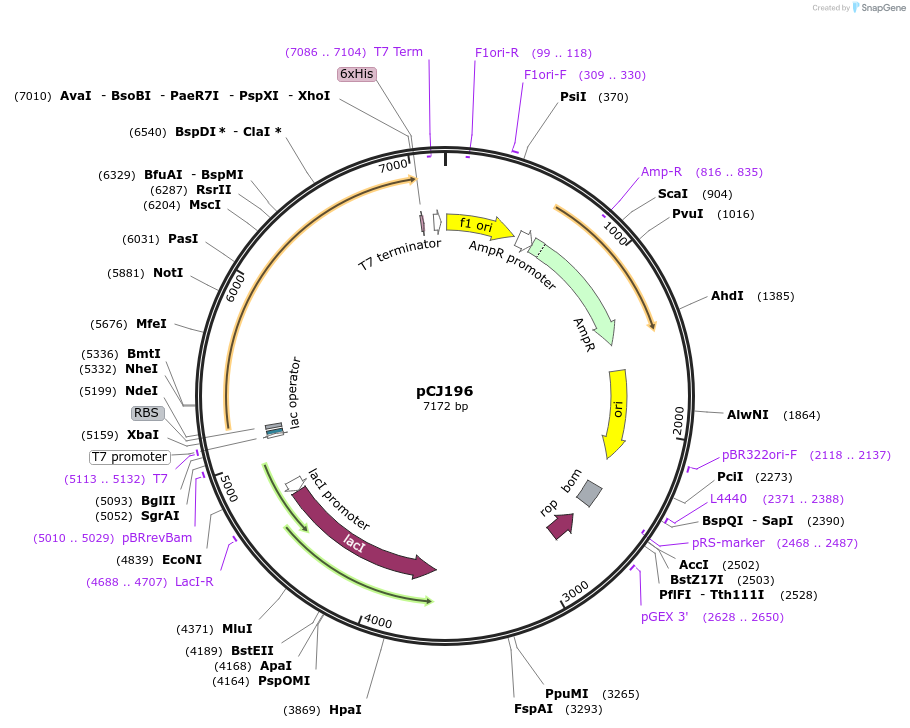
pCJ196
Plasmid#162669PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating catalytic mutation, S225ADepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS225A; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
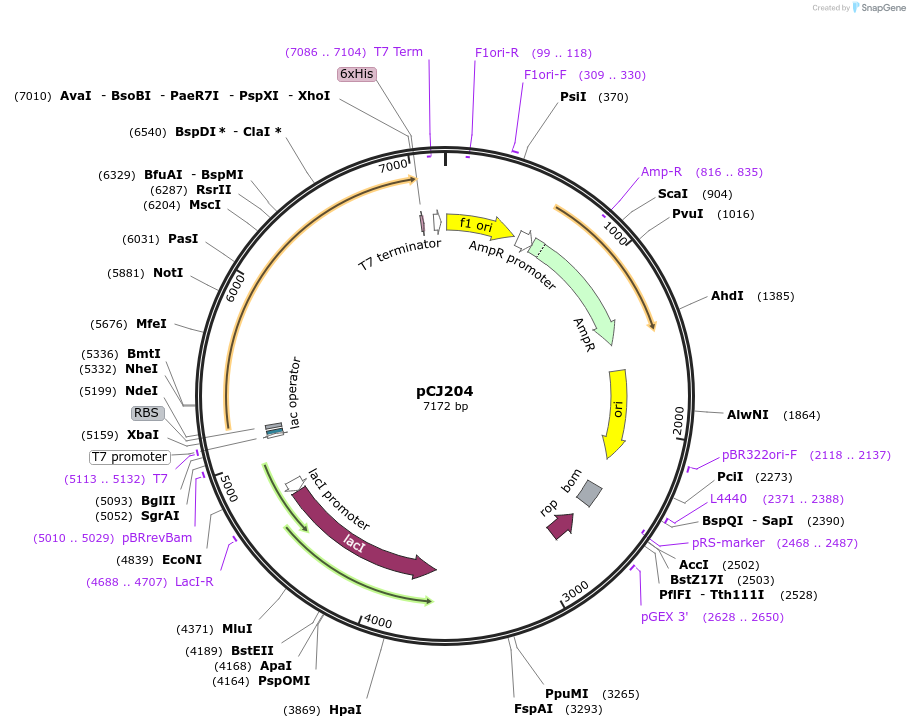
pCJ204
Plasmid#162677PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224H and C529F mutations.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224H, C529F; codon optimized for expression in E…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
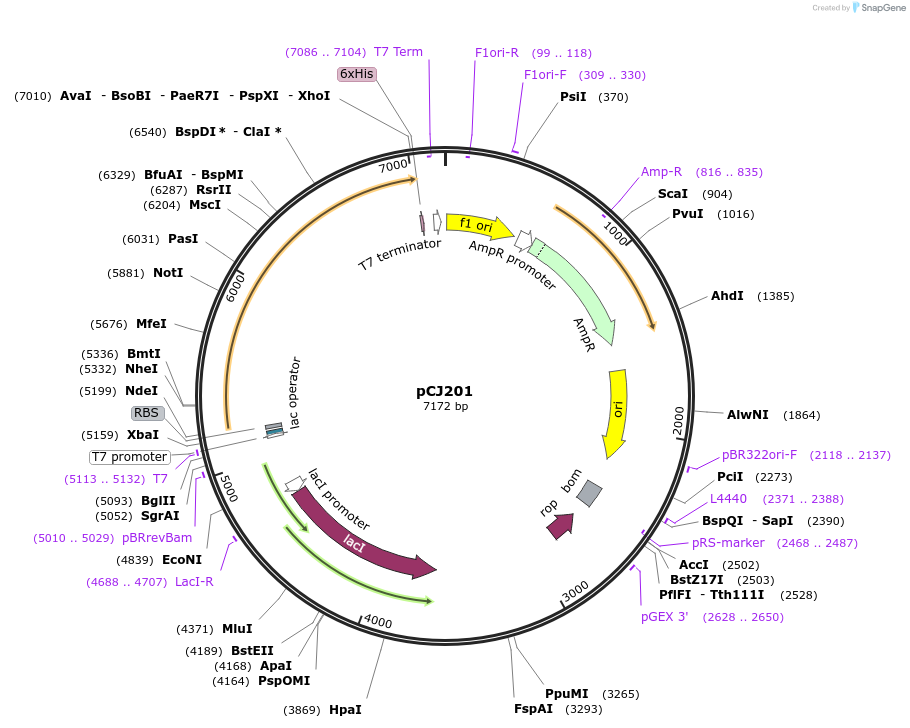
pCJ201
Plasmid#162674PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224W and C529SS mutations.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224W, C529SS; codon optimized for expression in …PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
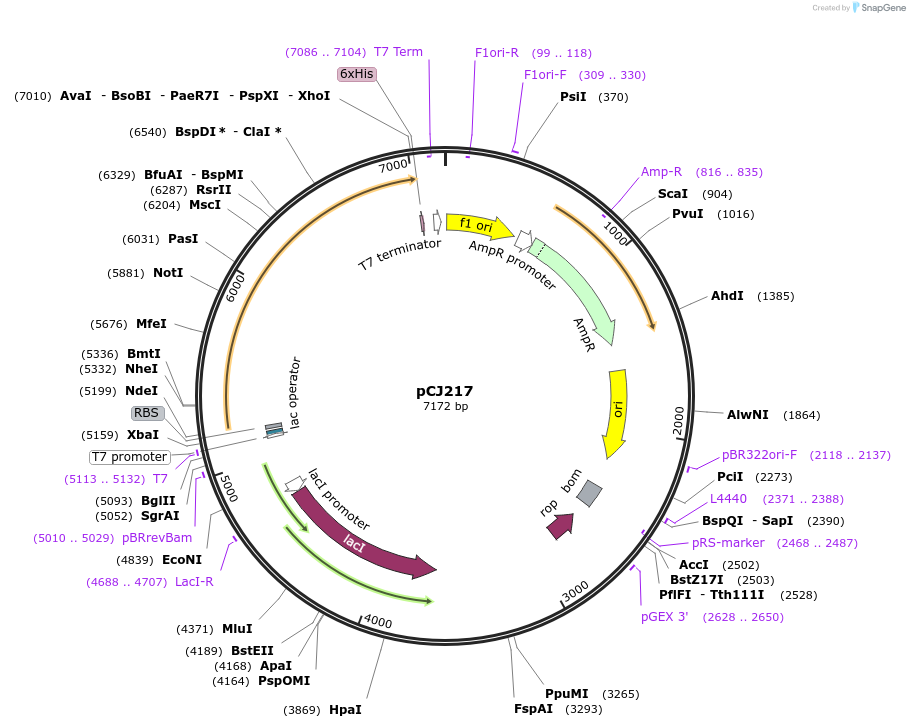
pCJ217
Plasmid#162685PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating two point mutations, G489C and S530C, to introduce a 6th disulfide bond (from PETase).DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with mutations to introduce a 6th disulfide bond from PETase
TagsHisExpressionBacterialMutationG489C, S530C; codon optimized for E. coli K12PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
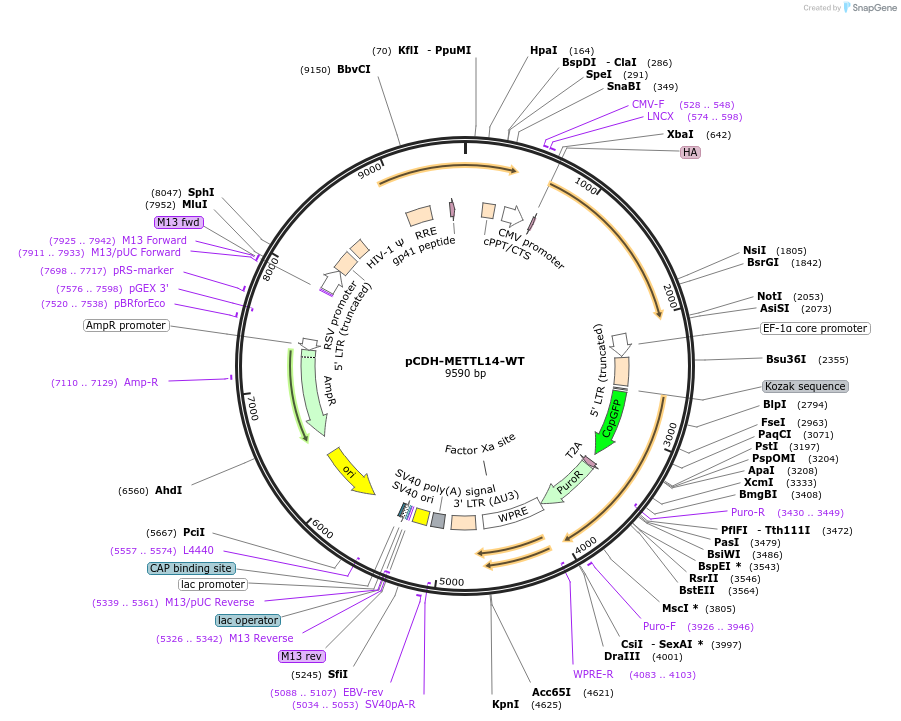
pCDH-METTL14-WT
Plasmid#141112PurposeOverexpression of WT METTL14 in mammalian cells.DepositorInsertHMETTL14 (METTL14 Human)
ExpressionMammalianAvailable SinceJune 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
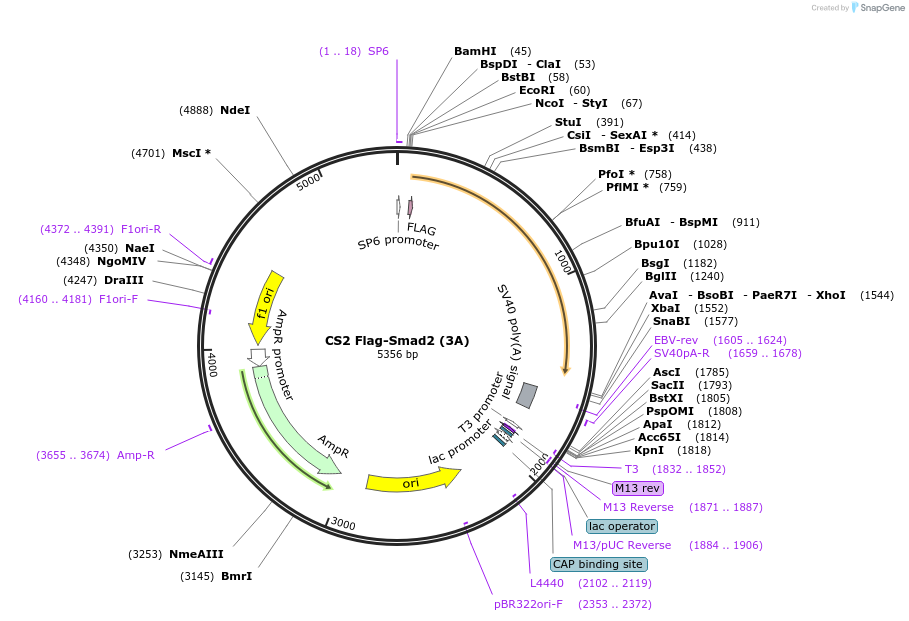
CS2 Flag-Smad2 (3A)
Plasmid#101763PurposeExpresses SMAD2 (3A) in mammalian cellsDepositorInsertSMAD2 (SMAD2 Human)
TagsFlagExpressionMammalianMutationThree Ser to Ala mutations in the phosphorylation…PromoterCMVAvailable SinceOct. 20, 2017AvailabilityAcademic Institutions and Nonprofits only