We narrowed to 13,765 results for: FLU
-
Plasmid#54765PurposeLocalization: N1 Cloning Vector, Excitation: 493, Emission: 509DepositorTypeEmpty backboneTagsmWasabiExpressionMammalianAvailable SinceJune 20, 2014AvailabilityAcademic Institutions and Nonprofits only
-
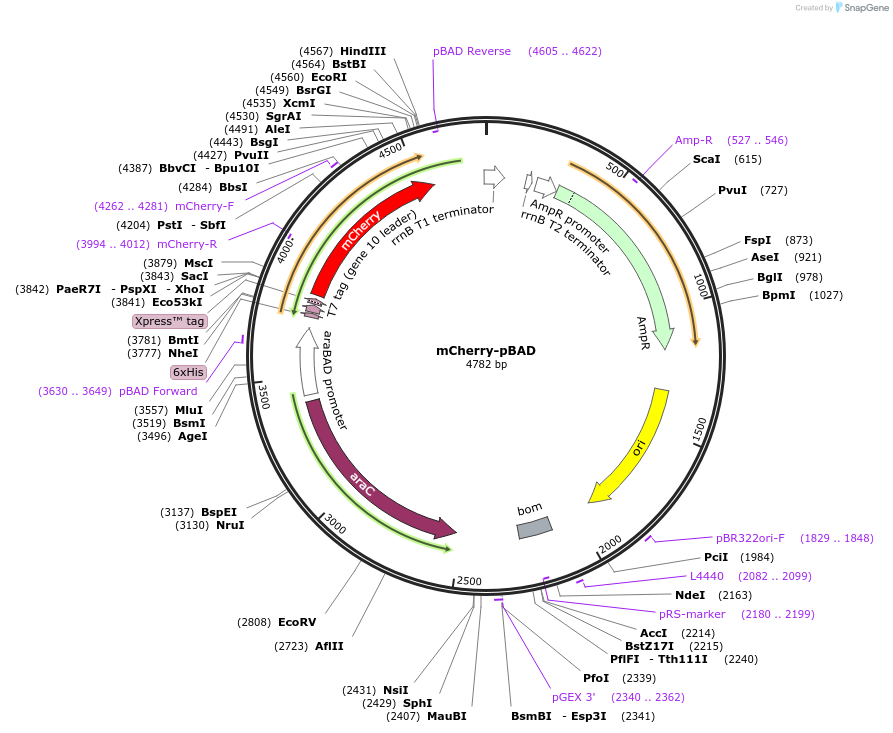
mCherry-pBAD
Plasmid#54630PurposeLocalization: Protein Expression Vector , Excitation: 587, Emission: 610DepositorTypeEmpty backboneTags6xHis and mCherryExpressionBacterialAvailable SinceJuly 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
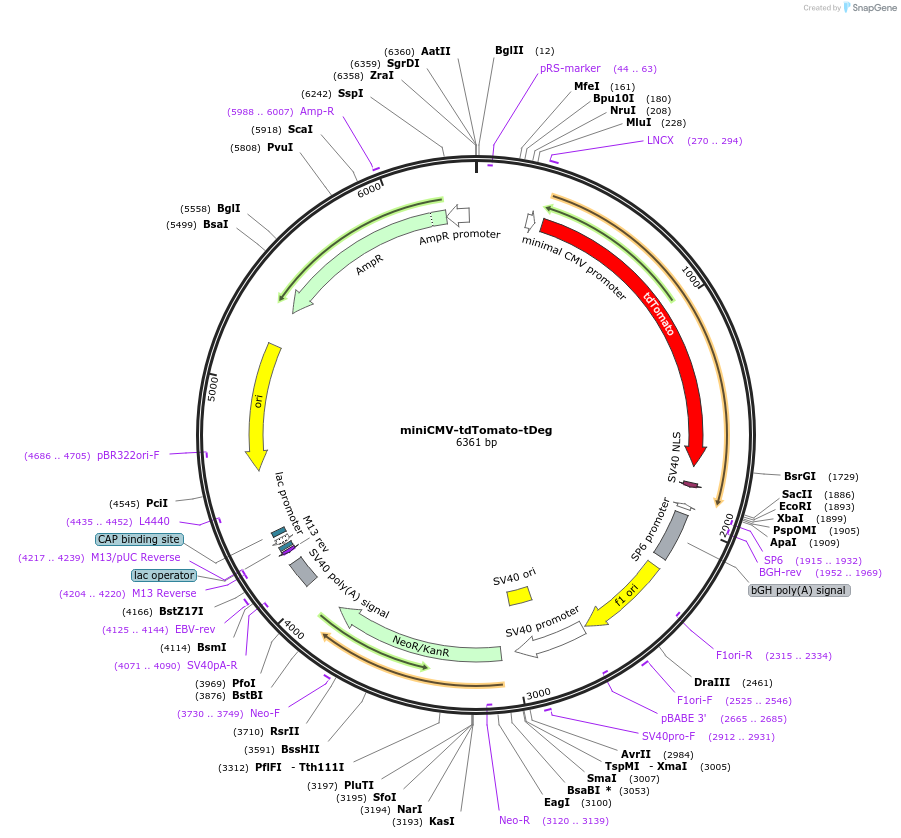
miniCMV-tdTomato-tDeg
Plasmid#217521PurposeExpresses tdTomato-tDeg fluorogenic protein in mammalian cells with miniCMV promoterDepositorInserttdTomato-tDeg fluorogenic protein
ExpressionMammalianPromoterminiCMVAvailable SinceJune 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
mTFP1-N1
Plasmid#54521PurposeLocalization: N1 Cloning Vector, Excitation: 462, Emission: 492DepositorTypeEmpty backboneTagsmTFP1ExpressionMammalianAvailable SinceJune 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
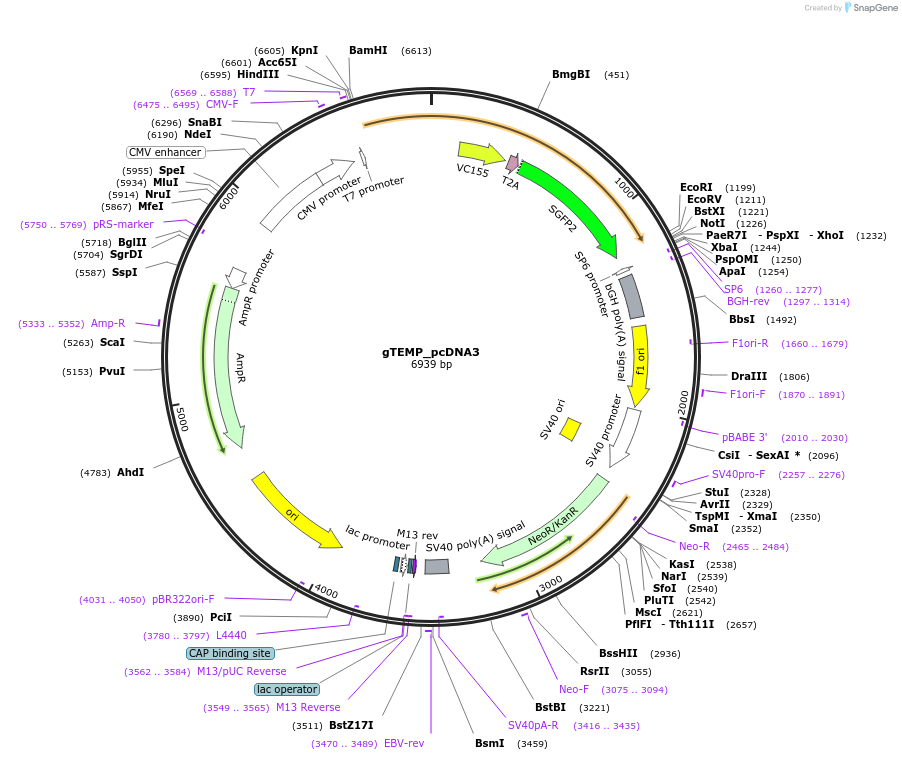
gTEMP_pcDNA3
Plasmid#89583PurposeGenetically encoded ratiometric fluorescent thermometerDepositorInsertgTEMP
ExpressionMammalianPromoterCMVAvailable SinceMay 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
mTagRFP-T-Endosomes-14
Plasmid#58014PurposeLocalization: Endosomes, Excitation: 555, Emission: 584DepositorAvailable SinceJuly 31, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
tdTomato-pBAD
Plasmid#54856PurposeLocalization: Protein Expression Vector , Excitation: 554, Emission: 581DepositorTypeEmpty backboneTags6xHis and tdTomatoExpressionBacterialAvailable SinceJuly 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
mEmerald-Vinexin-C-14
Plasmid#54305PurposeLocalization: Focal Adhesions, Excitation: 487, Emission: 509DepositorAvailable SinceJune 16, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
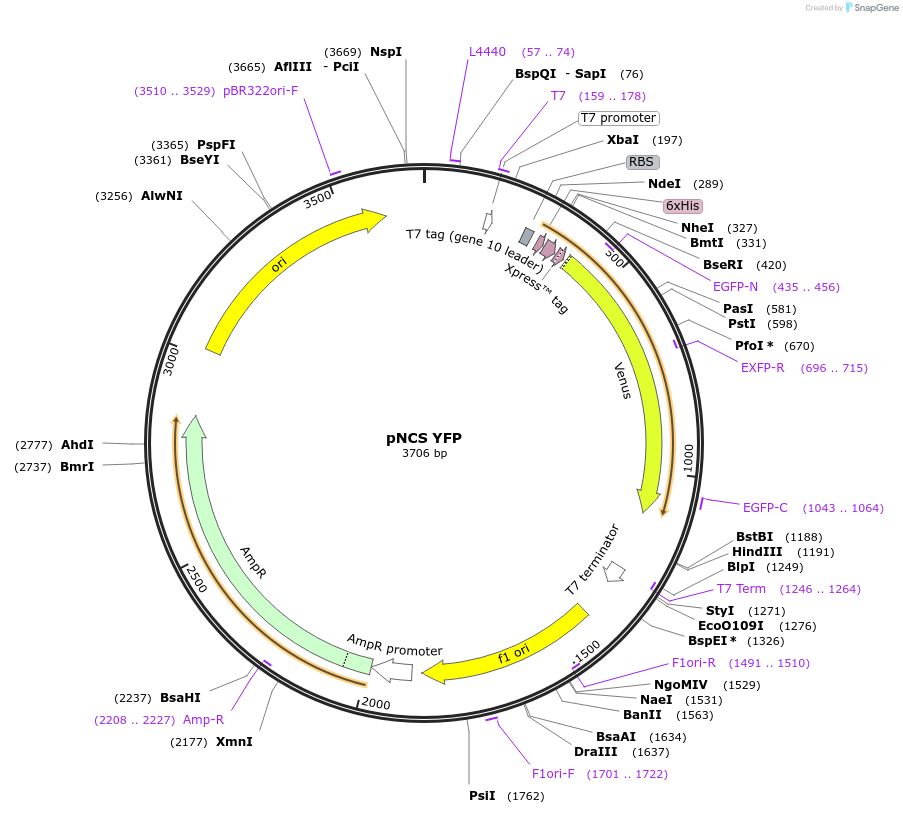
pNCS YFP
Plasmid#91759PurposeBacterial expression of a fluorescent protein, Yellow Fluorescent Protein (YFP)DepositorInsertVenus YFP (A206K)
TagsHexa-Histidine tag, Xpress epitope for detection …ExpressionBacterialPromoterT7Available SinceJune 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
mStrawberry-N1
Plasmid#54644PurposeLocalization: N1 Cloning Vector, Excitation: 574, Emission: 596DepositorTypeEmpty backboneTagsmStrawberryExpressionMammalianAvailable SinceJune 11, 2014AvailabilityAcademic Institutions and Nonprofits only -
mKusabira Orange-C1 (mKO)
Plasmid#54834PurposeLocalization: C1 Cloning Vector, Excitation: 548, Emission: 561DepositorTypeEmpty backboneTagsmKusabira OrangeExpressionMammalianAvailable SinceJune 12, 2014AvailabilityAcademic Institutions and Nonprofits only -
pCD86-mEGFP
Plasmid#190745PurposeExpression of mEGFP-tagged CD86 in mammalian cells.DepositorAvailable SinceSept. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
mTFP1-C1
Plasmid#54613PurposeLocalization: C1 Cloning Vector, Excitation: 462, Emission: 492DepositorTypeEmpty backboneTagsmTFP1ExpressionMammalianAvailable SinceJune 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
MiCy-pBAD
Plasmid#54565PurposeLocalization: Protein Expression Vector , Excitation: 472, Emission: 495DepositorTypeEmpty backboneTags6xHis and MiCyExpressionBacterialAvailable SinceJuly 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
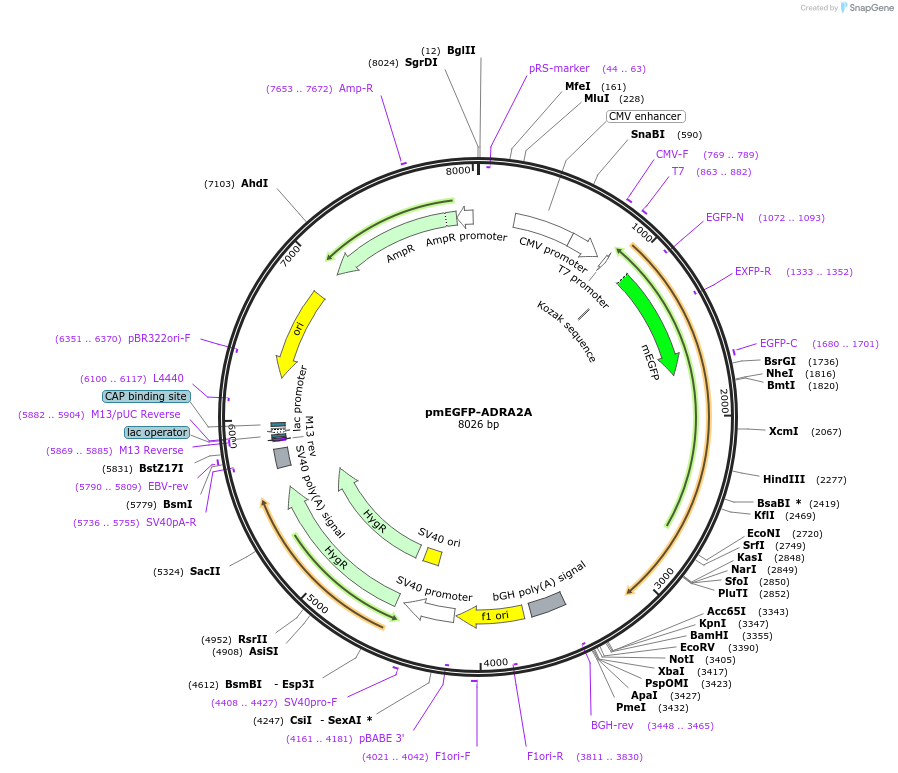
pmEGFP-ADRA2A
Plasmid#190753PurposeExpression of ADRA2A with mEGFP tag on N-terminus.DepositorAvailable SinceSept. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
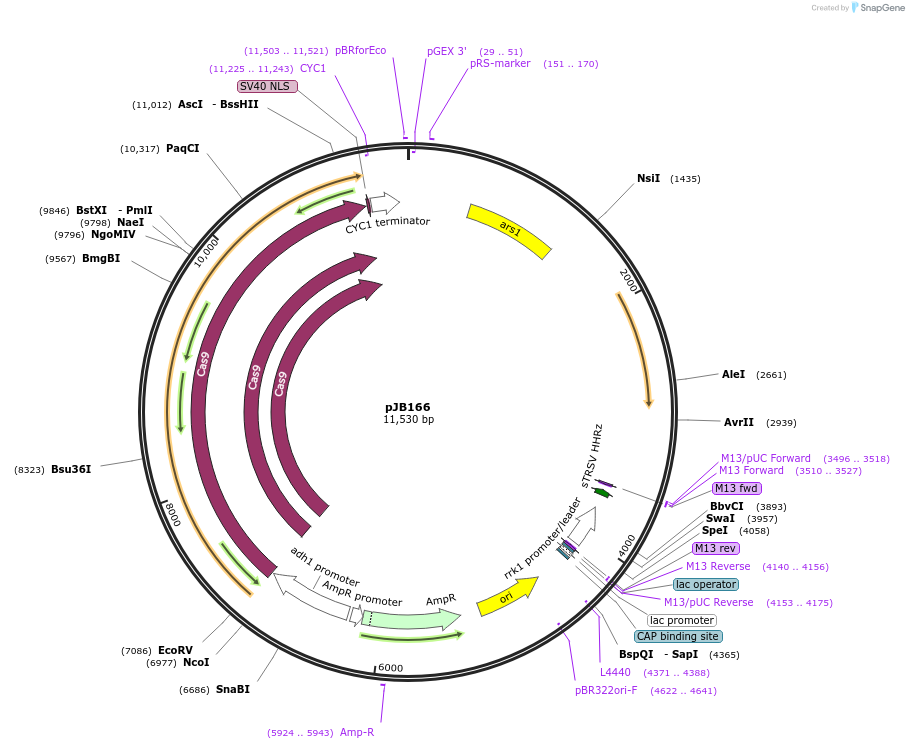
pJB166
Plasmid#86998PurposeCRISPR/Cas9 in fission yeast using fluoride selectionDepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 22, 2017AvailabilityAcademic Institutions and Nonprofits only -
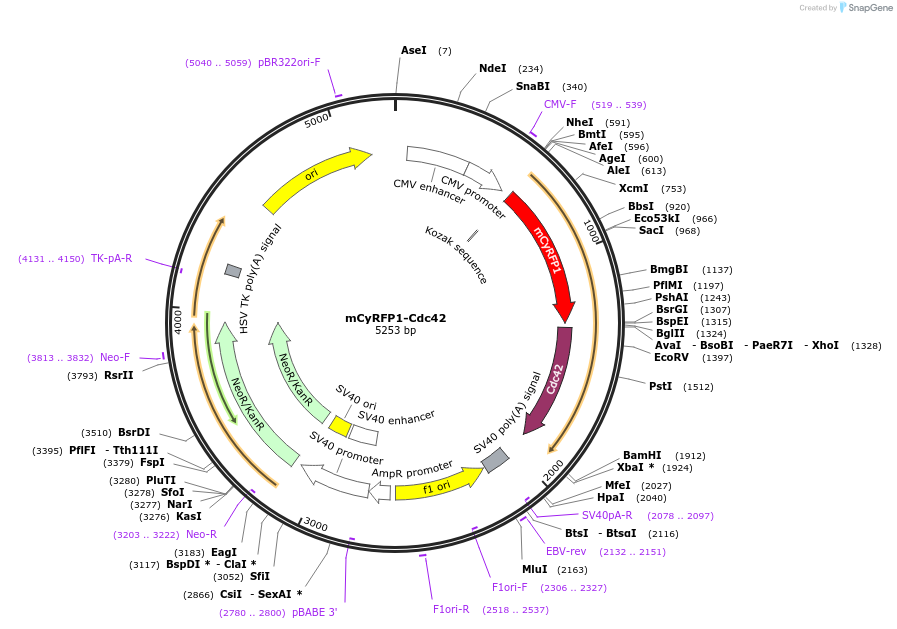
mCyRFP1-Cdc42
Plasmid#84360Purposefusion protein of Cdc42, FRET donorDepositorInsertmCyRFP1-Cdc42
TagsmCyRFP1 N terminal fusion to Cdc42ExpressionMammalianPromoterCMVAvailable SinceNov. 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
pCMV-ECFPEYFP
Plasmid#24520DepositorInsertECFP, EYFP
TagsECFP and EYFPExpressionMammalianAvailable SinceJune 3, 2011AvailabilityAcademic Institutions and Nonprofits only -
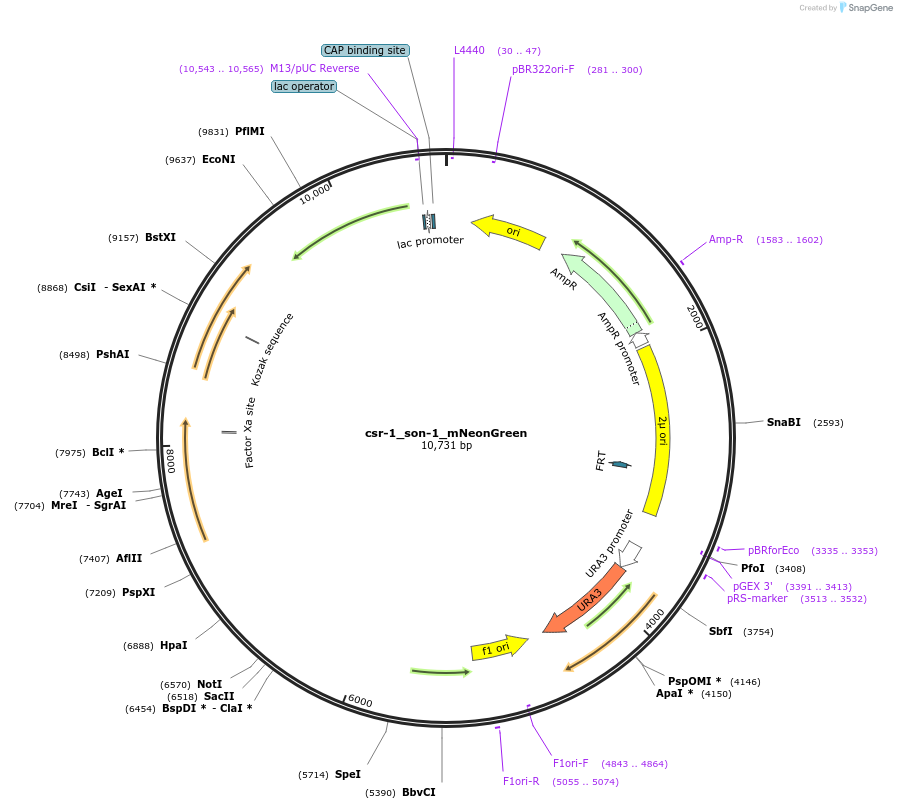
csr-1_son-1_mNeonGreen
Plasmid#191743PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and mNeonGreenExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pJOE7771.1
Plasmid#135077PurposeMannitol, arabitol, and glucitol-inducible mtlR/PmtlE expression vectorDepositorInsertmtlR
ExpressionBacterialAvailable SinceFeb. 5, 2020AvailabilityAcademic Institutions and Nonprofits only