We narrowed to 20,082 results for: ATO
-
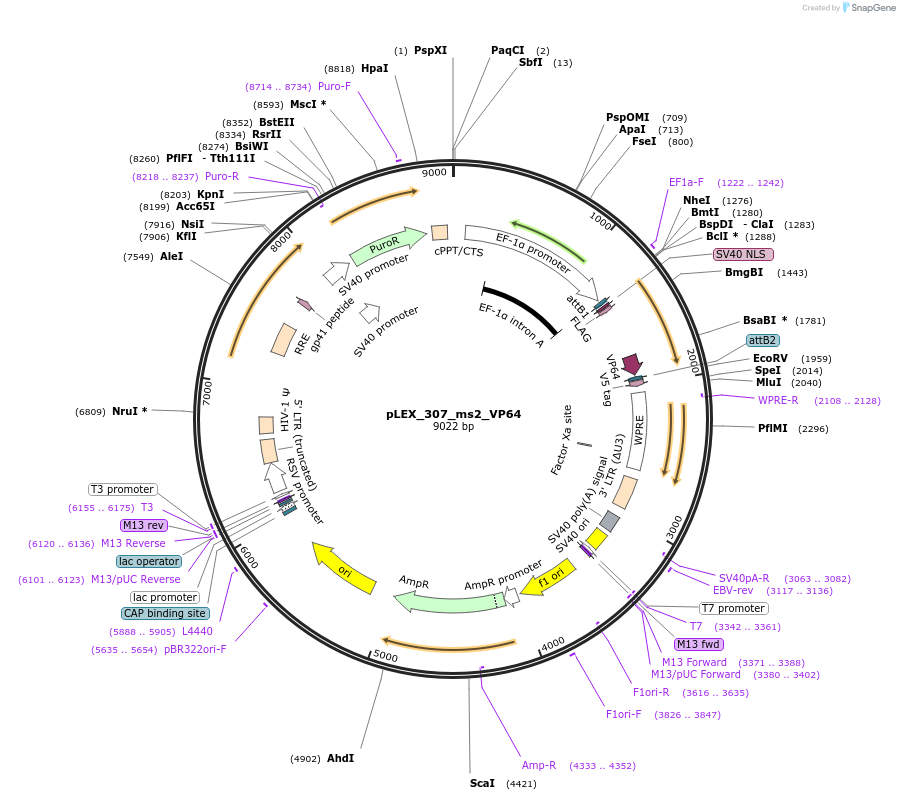
Plasmid#79375PurposeRecruits VP64 to compatible gRNAs for transcriptional activationDepositorInsertms2-VP64
UseCRISPRExpressionMammalianAvailable SinceSept. 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
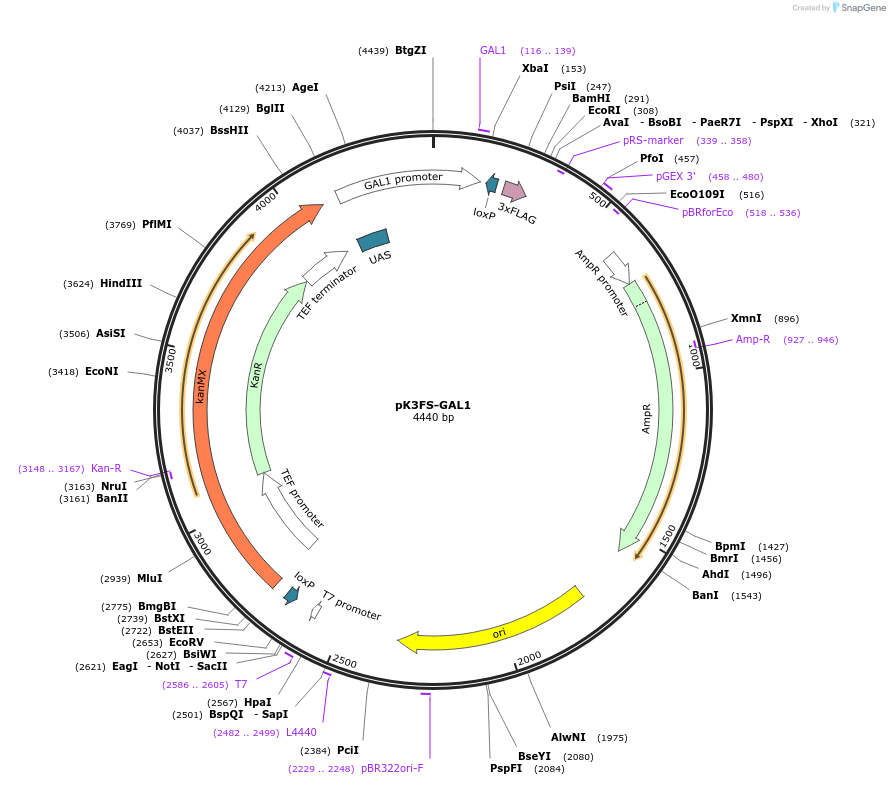
pK3FS-GAL1
Plasmid#85778PurposeN-ICE plasmid pK3FS-GAL1DepositorInsertloxP-KanMX-loxP-GAL1p-3FLAG
UseCre/LoxExpressionYeastAvailable SinceJan. 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
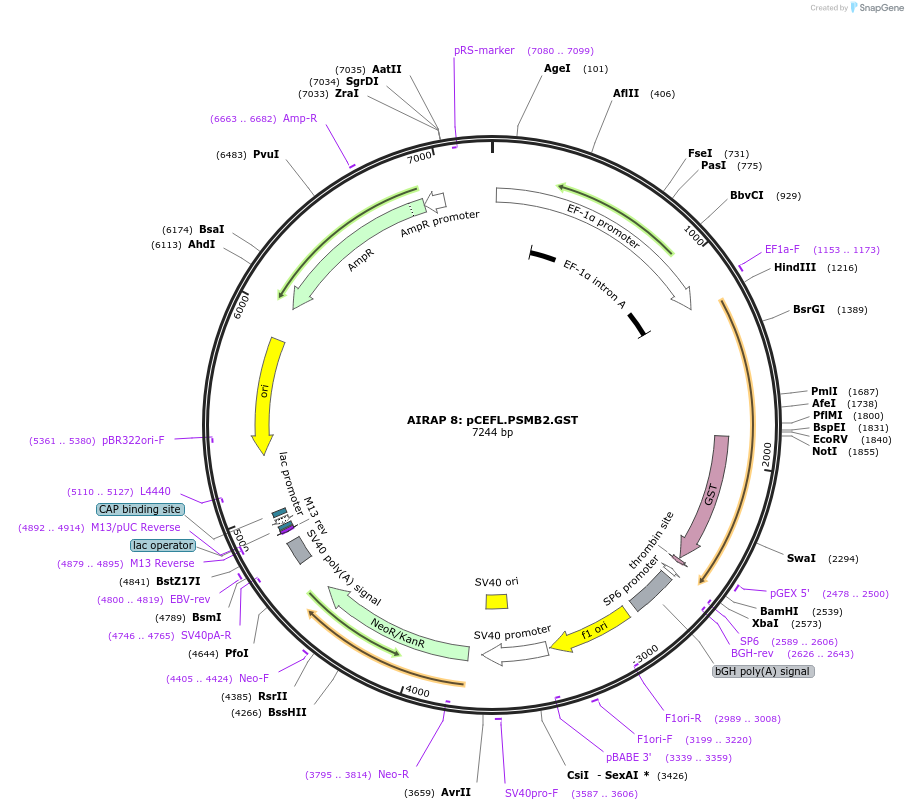
AIRAP 8: pCEFL.PSMB2.GST
Plasmid#21798DepositorAvailable SinceSept. 14, 2009AvailabilityAcademic Institutions and Nonprofits only -
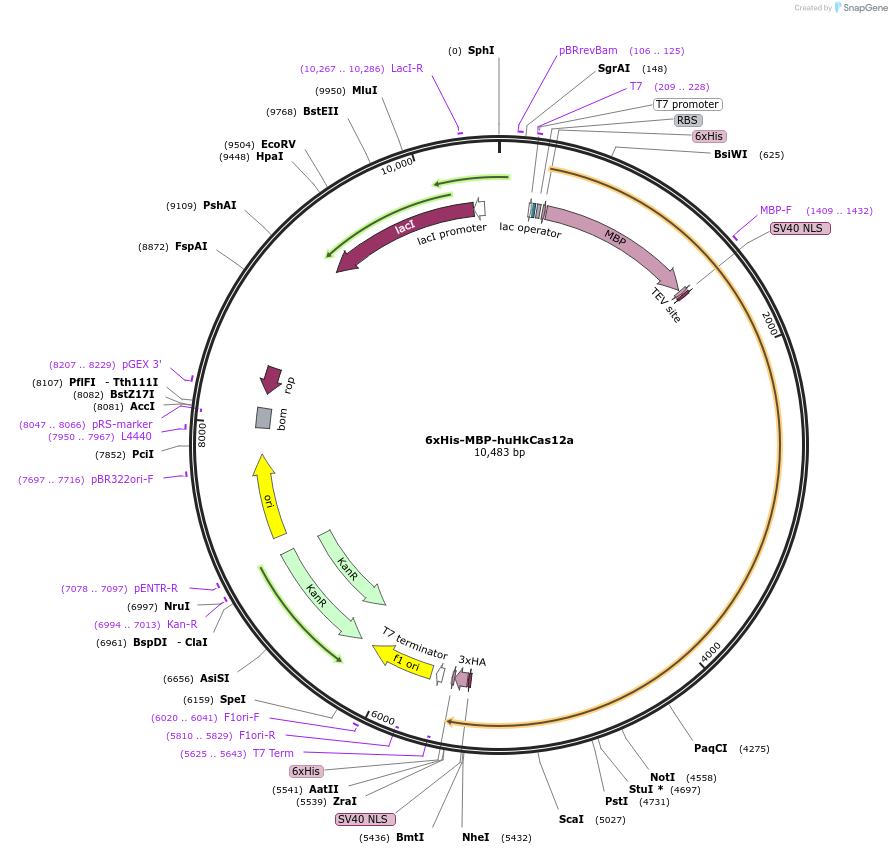
6xHis-MBP-huHkCas12a
Plasmid#174683PurposeExpresses 6xHis-MBP tagged HkCas12aDepositorArticleInserthuHkCas12a
UseCRISPR and Synthetic BiologyTags6xHis, MBP, TEV, NLS and NLS, 6xHis, 3xHAExpressionBacterialPromoterT7Available SinceOct. 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
pSMP-Suv39H2_1
Plasmid#36344DepositorAvailable SinceMay 2, 2012AvailabilityAcademic Institutions and Nonprofits only -
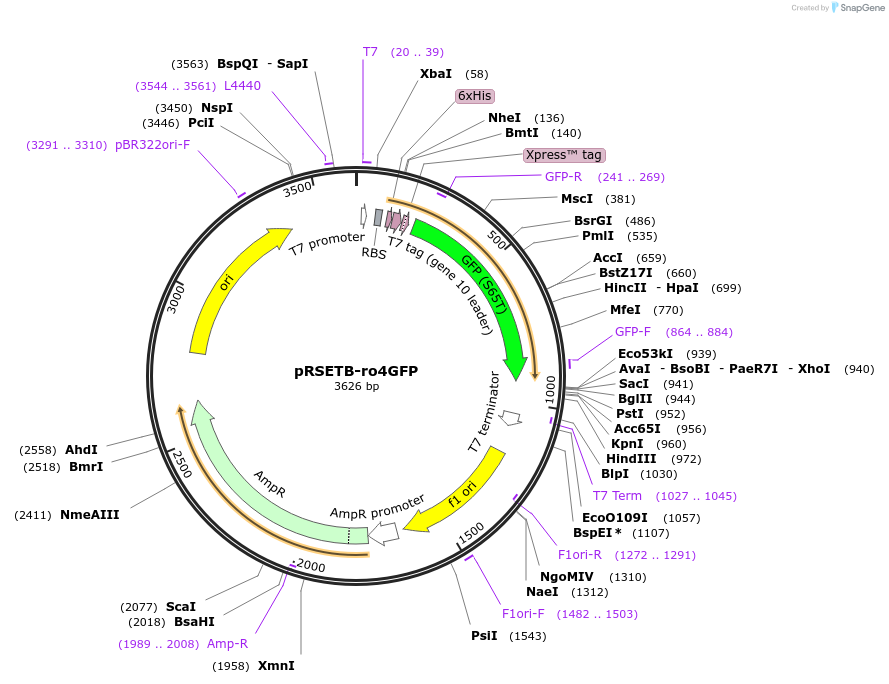
pRSETB-ro4GFP
Plasmid#82368PurposeInducible expression of redox sensitive GFP in bacteriaDepositorInsertro4GFP
Tags6xHISExpressionBacterialMutationC48S/S65T/Q80R/N149C/S202CPromoterT7Available SinceSept. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pSMP-Suv39H2_2
Plasmid#36345DepositorAvailable SinceMay 2, 2012AvailabilityAcademic Institutions and Nonprofits only -
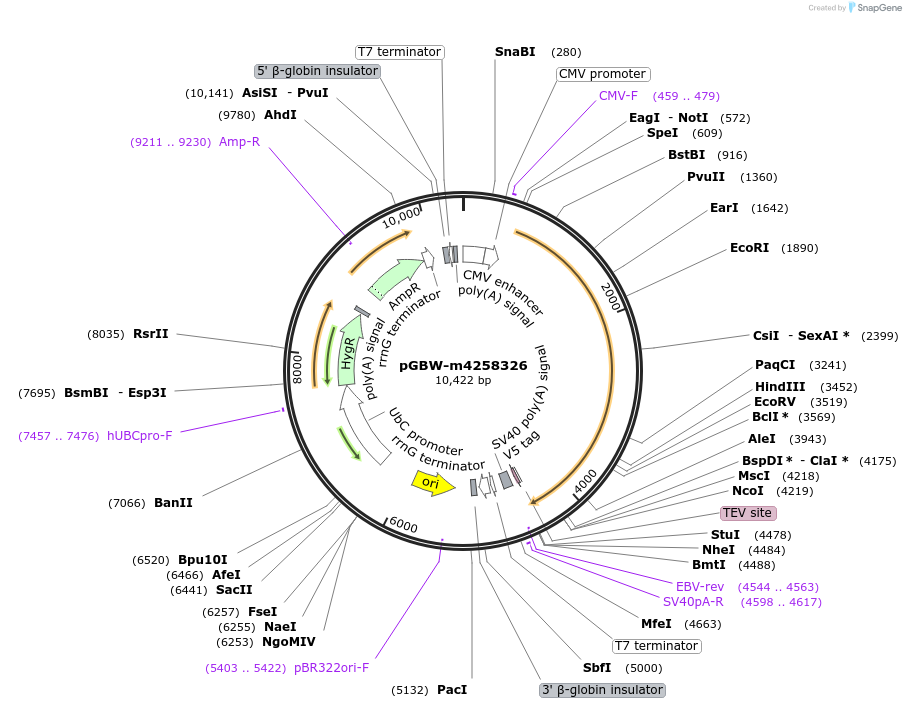
pGBW-m4258326
Plasmid#154793PurposeMammalian expression plasmid for SARS-CoV-2 surface glycoprotein (Spike protein)DepositorInsertSARS-CoV-2 surface glycoprotein (Spike protein) (S Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;V5ExpressionMammalianMutationwtPromoterCMVAvailable SinceJuly 30, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
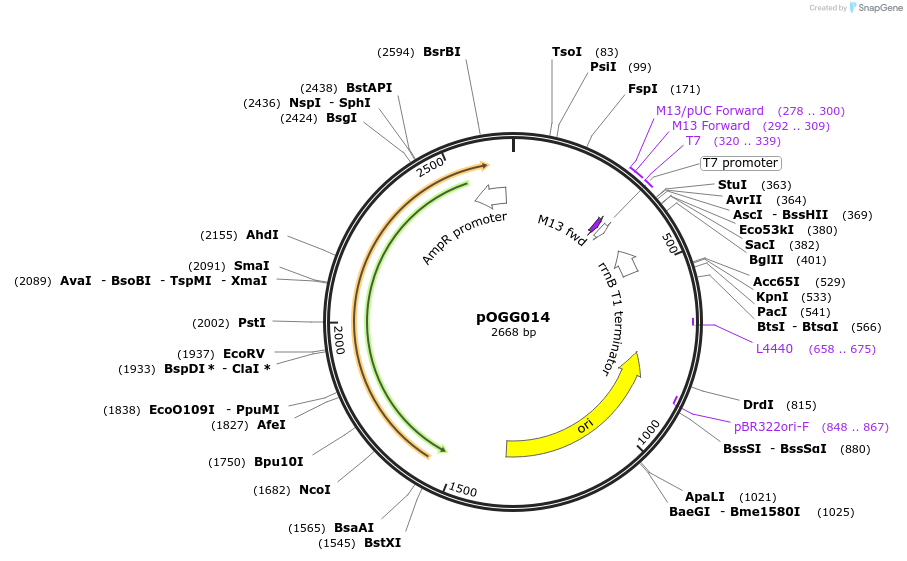
pOGG014
Plasmid#113988PurposeVector construction module. Endlinker/terminator module to circularize plasmid following Part 4DepositorInsertEndlinker/terminator Part 4
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceJan. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
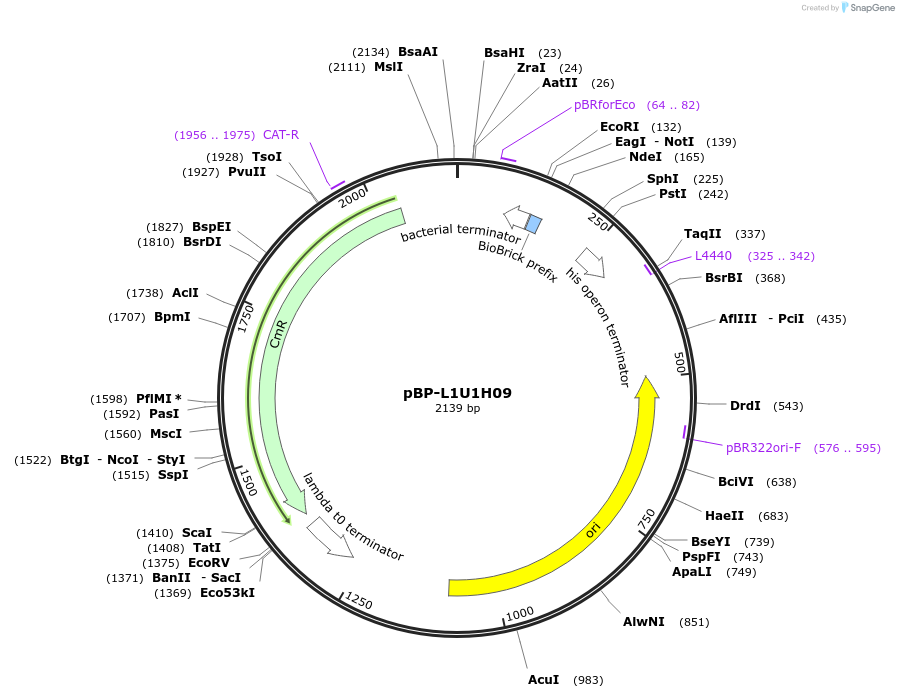
pBP-L1U1H09
Plasmid#73008PurposeL1U1H09 terminatorDepositorInsertL1U1H09 terminator
UseSynthetic BiologyMutationCAT gene - C435G (nucleotide) - silent mutagenesi…Available SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
pBP-mTAZ-NFLAG
Plasmid#31791DepositorAvailable SinceAug. 29, 2011AvailabilityAcademic Institutions and Nonprofits only -
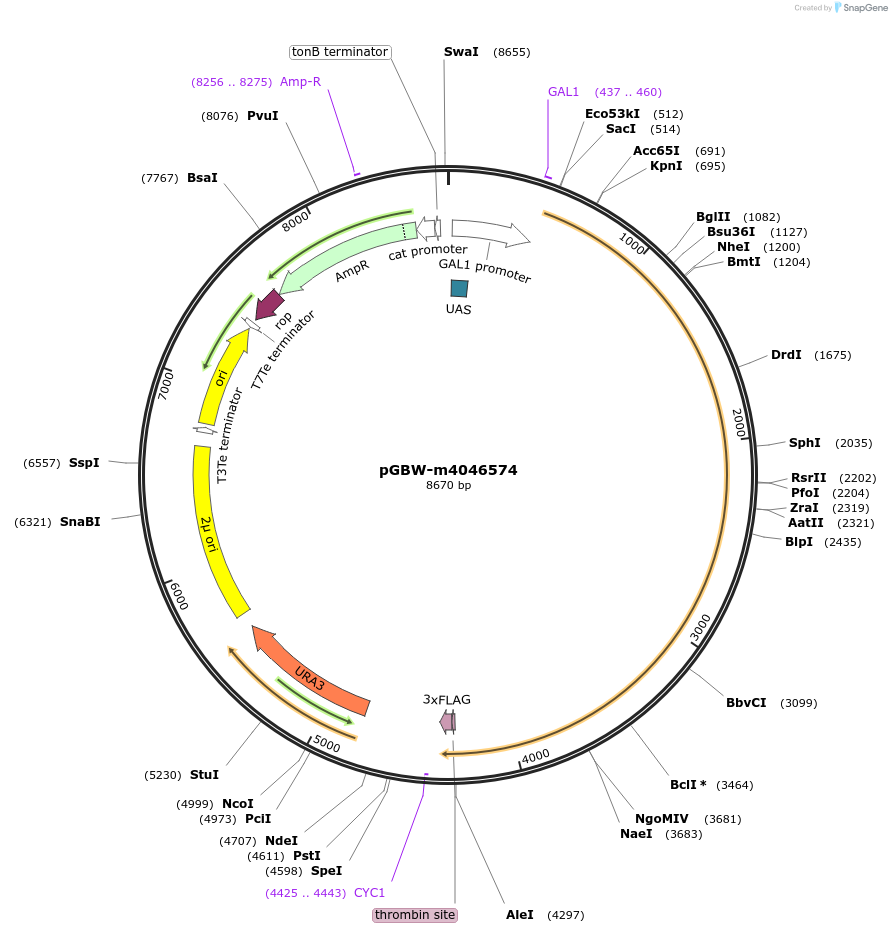
pGBW-m4046574
Plasmid#148976PurposeYeast Expression plasmid for SARS-CoV-2 surface glycoprotein (Spike protein)DepositorInsertSARS-CoV-2 S (surface glycoprotein) (S Severe acute respiratory syndrome coronavirus 2, Budding Yeast)
TagsCleavable thrombin;3xFLAGExpressionYeastMutationSaccharomyces cerevisiae recode 1PromoterpGAL1Available SinceMay 14, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
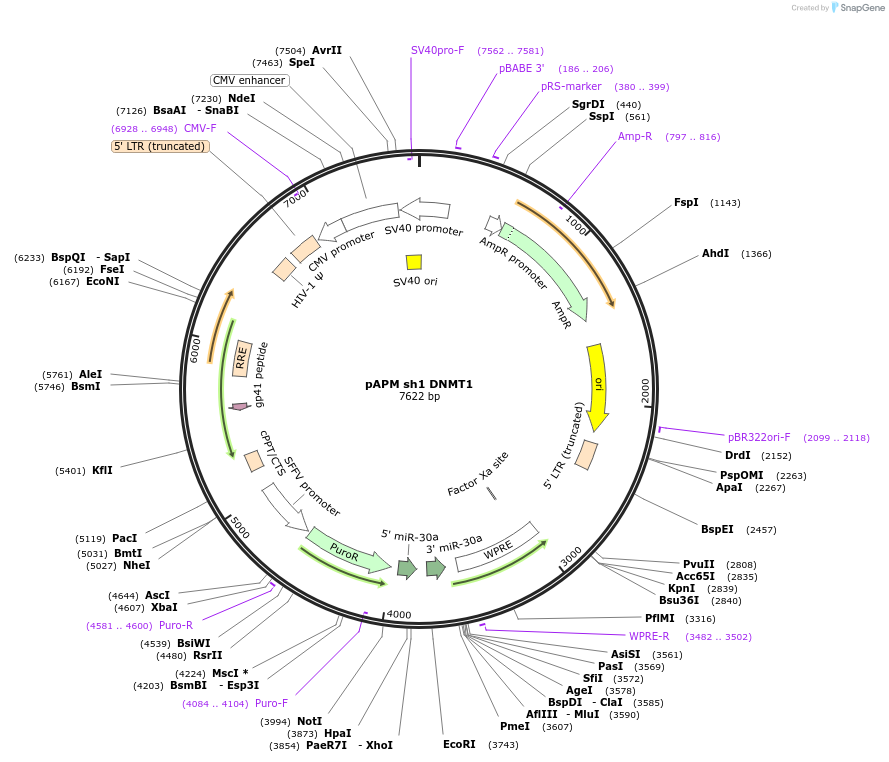
pAPM sh1 DNMT1
Plasmid#88884PurposeLentiviral vector expressing shRNA against murine DNMT1DepositorInsertshRNA no.1 DNMT1
UseLentiviralExpressionMammalianPromoterSFFV (spleen focus forming virus)Available SinceApril 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
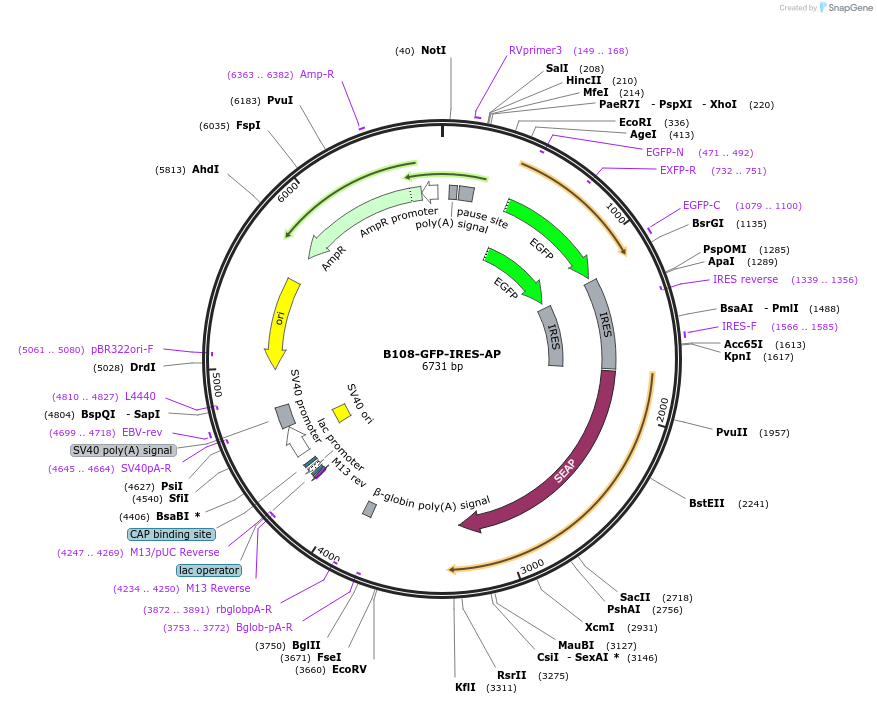
B108-GFP-IRES-AP
Plasmid#73988PurposeB108 enhancer of Blimp1 gene drives EGFP expression.DepositorInsertB108 enhancer
ExpressionMammalianAvailable SinceJune 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
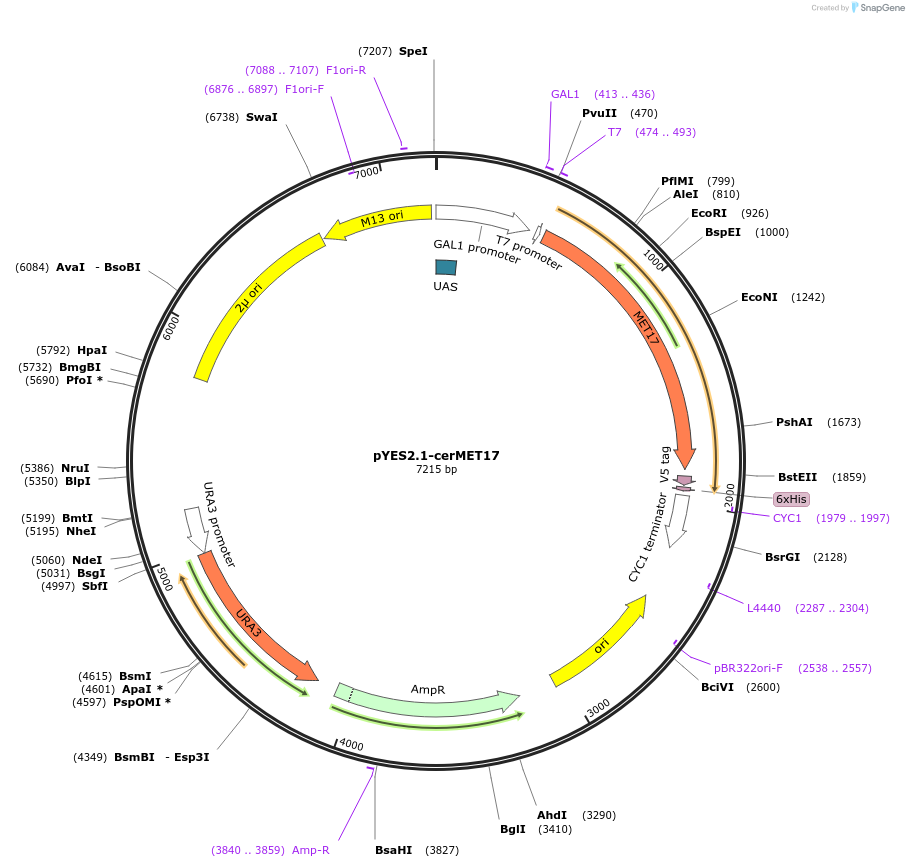
pYES2.1-cerMET17
Plasmid#107443PurposeOverexpression of MET fusion protein in S. cerevisiaeDepositorAvailable SinceApril 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
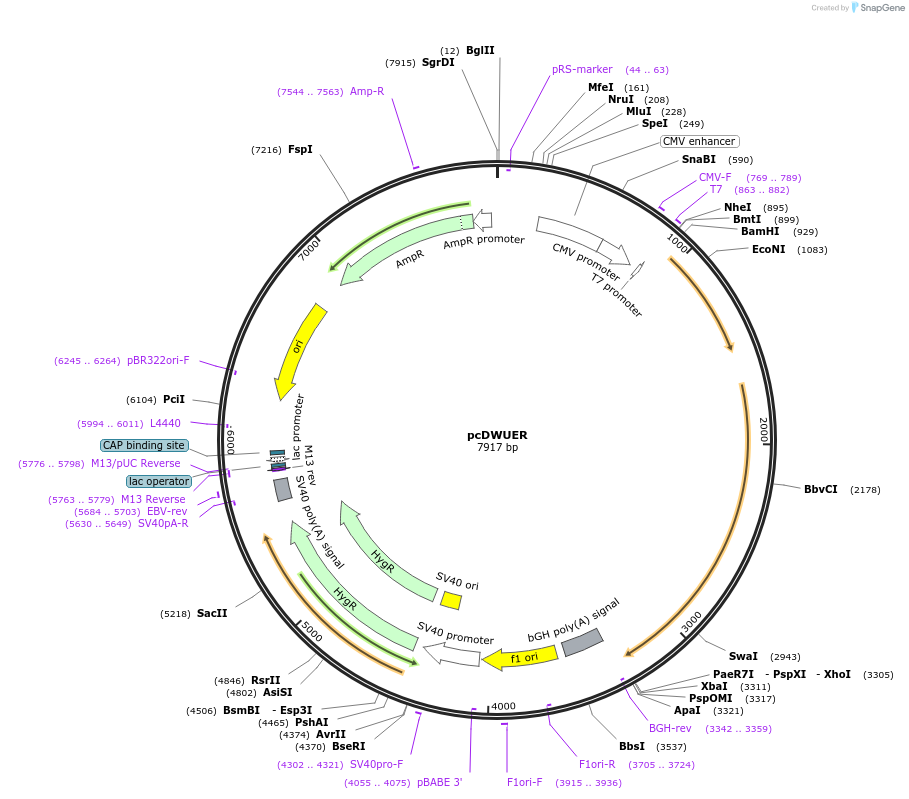
pcDWUER
Plasmid#37094DepositorExpressionMammalianPromoterCMVAvailable SinceJuly 3, 2012AvailabilityAcademic Institutions and Nonprofits only -
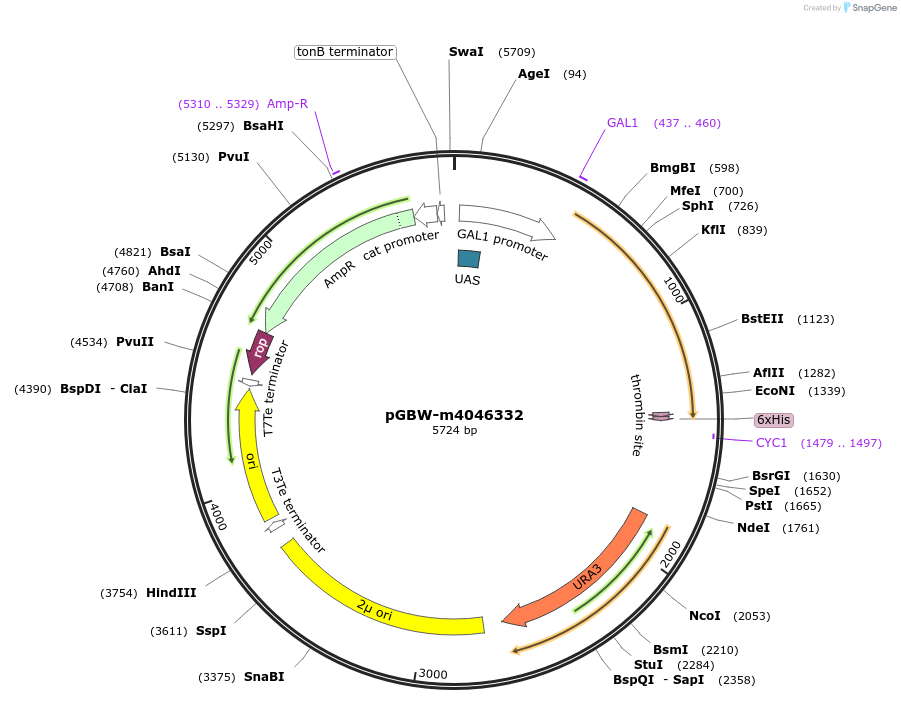
pGBW-m4046332
Plasmid#145413PurposeYeast Expression plasmid for SARS-CoV-2 3C-like proteinaseDepositorInsertSARS-CoV-2 3C-like proteinase (ORF1ab Severe acute respiratory syndrome coronavirus 2, Budding Yeast)
TagsCleavable thrombin;6xHISExpressionYeastMutationSaccharomyces cerevisiae recode 1PromoterpGAL1Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pAdTrackCMV-CDKN2A
Plasmid#37105DepositorAvailable SinceNov. 5, 2012AvailabilityAcademic Institutions and Nonprofits only -
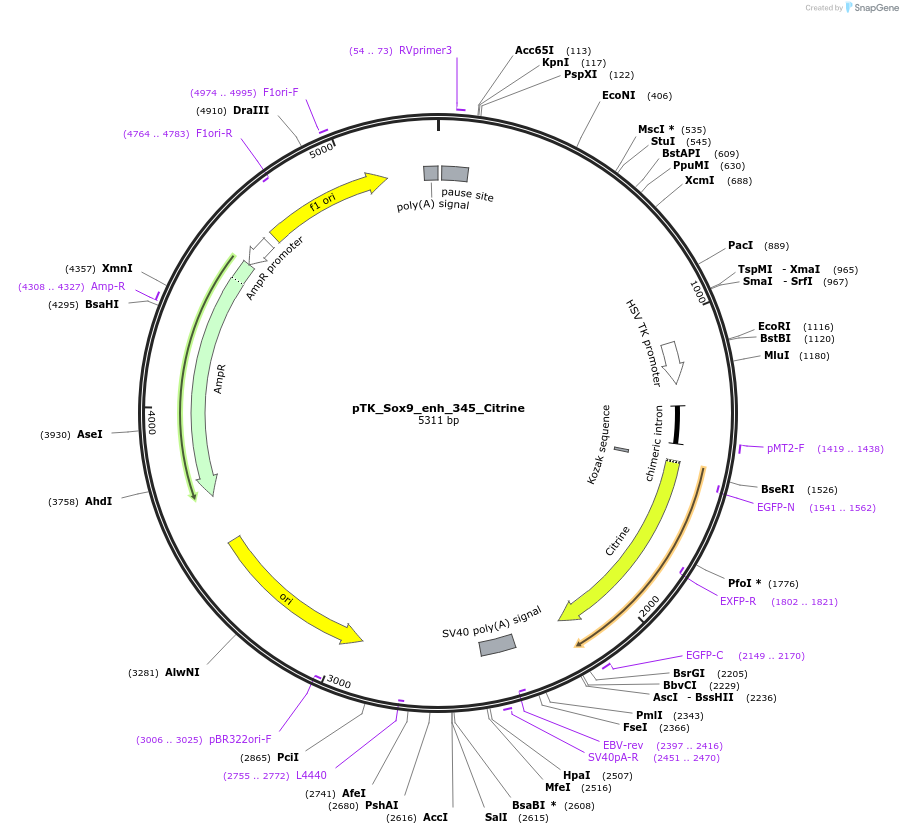
pTK_Sox9_enh_345_Citrine
Plasmid#130592Purposeenhancer reporterDepositorInsertSox9_enh_345
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
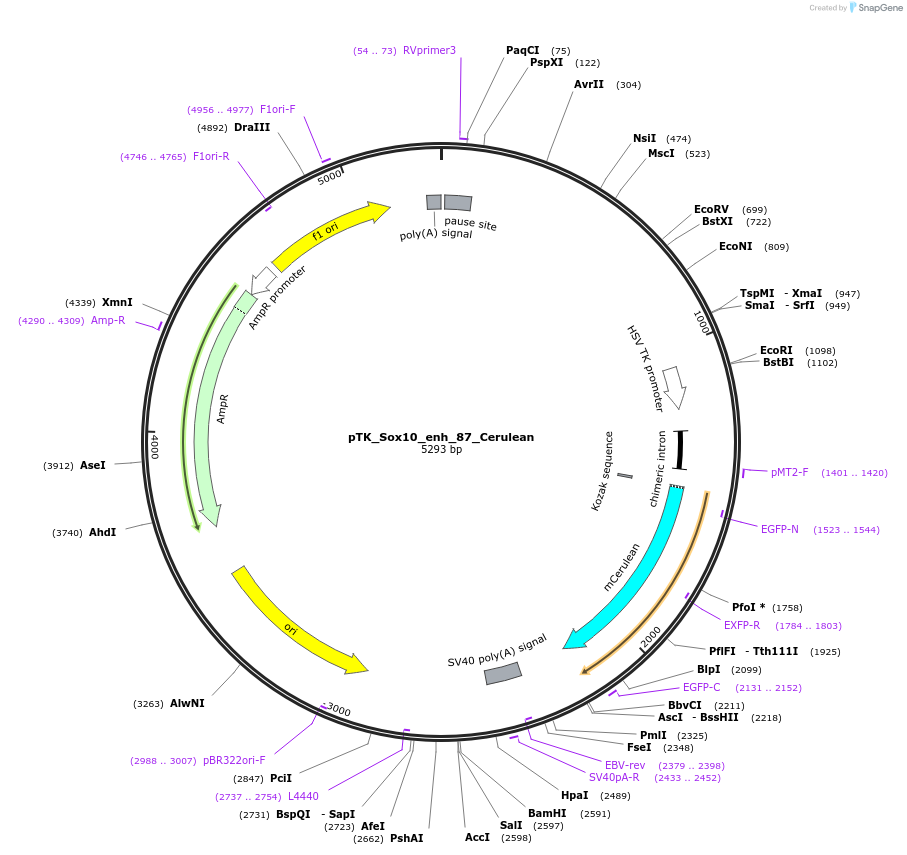
pTK_Sox10_enh_87_Cerulean
Plasmid#130586Purposeenhancer reporterDepositorInsertSox10_enh_87
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only