We narrowed to 14,145 results for: Lor;
-
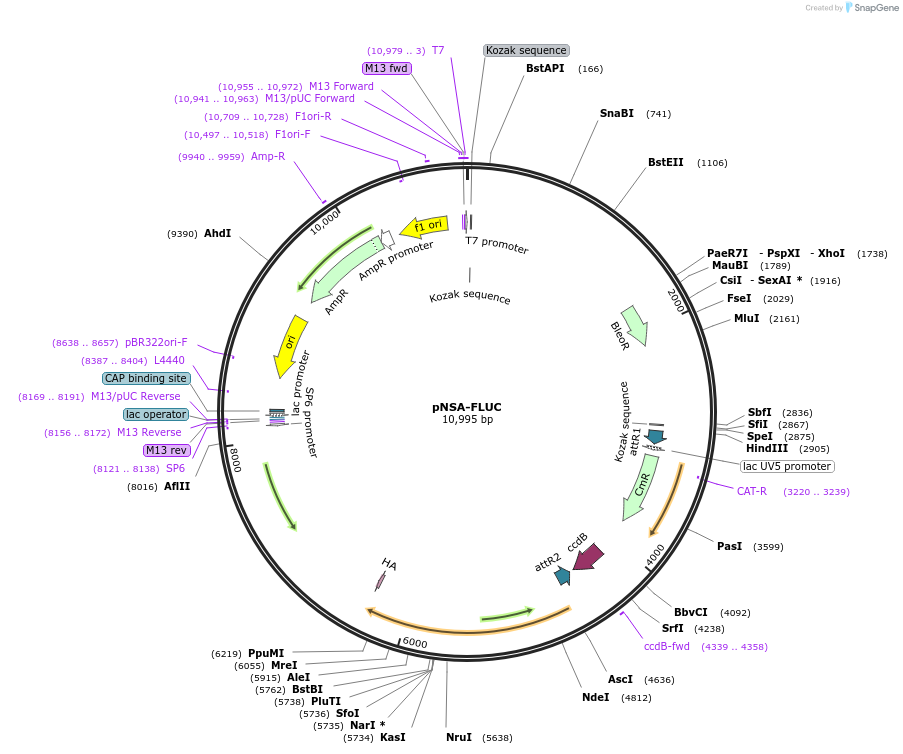
Plasmid#98143PurposeGateway compatible construct for N' terminal fusion or promoter driven firefly luciferase reporter, zeocin resistance cassetteDepositorTypeEmpty backboneUseAlgae, nannochloropsisTagsHAAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only
-
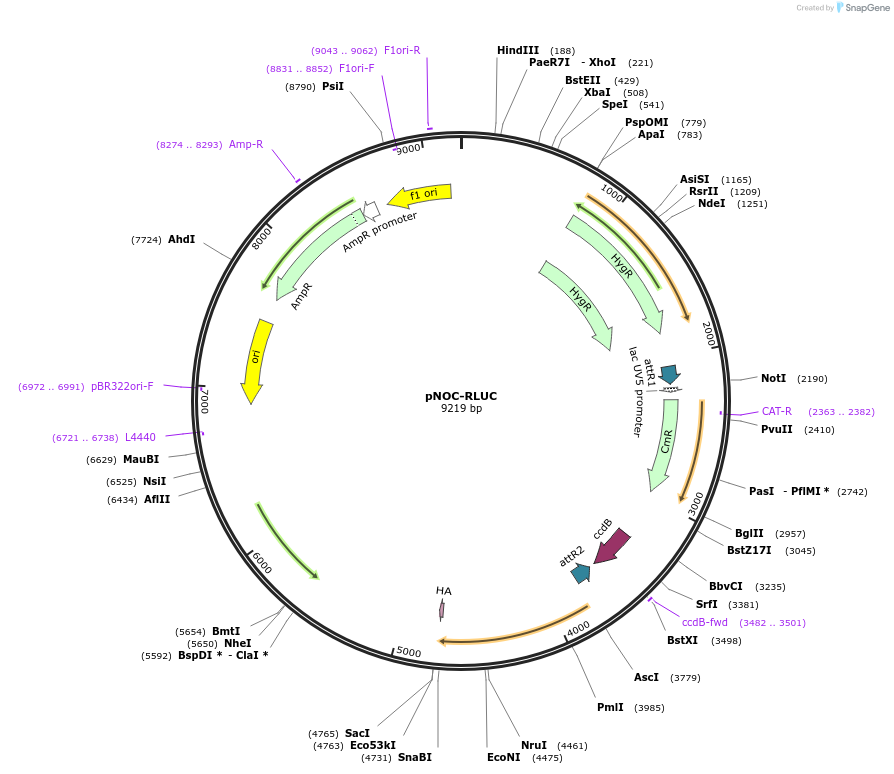
pNOC-RLUC
Plasmid#98142PurposeGateway compatible construct for N' terminal fusion or promoter driven renilla luciferase reporter, hygromycin resistance cassetteDepositorTypeEmpty backboneUseAlgae, nannochloropsisTagsHAAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
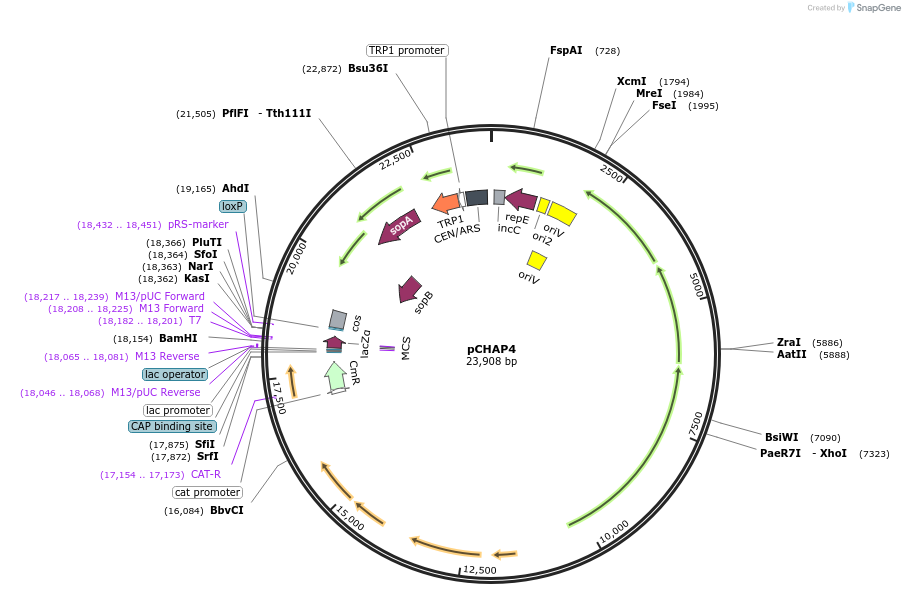
pCHAP4
Plasmid#206850PurposeA plasmid containing a 16,121 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
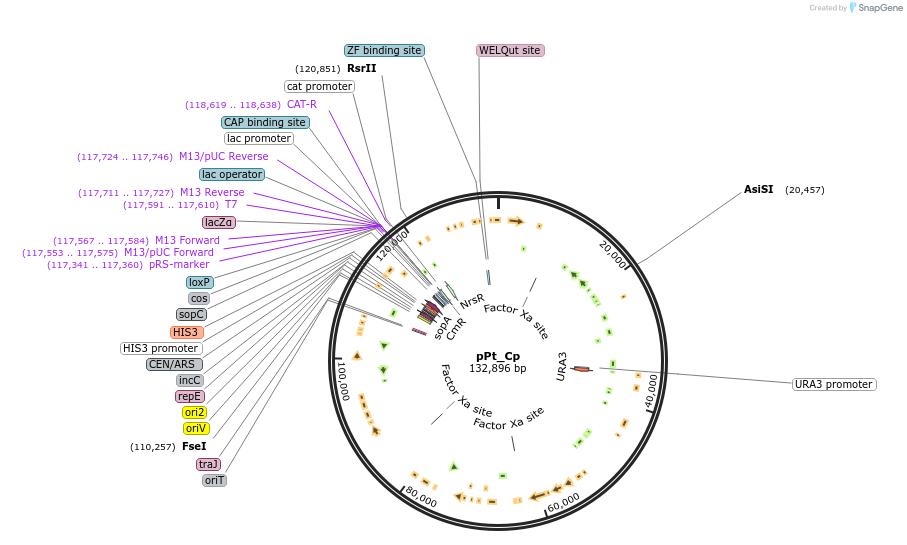
pPt_Cp
Plasmid#206855PurposeA plasmid containing the whole P. tricornutum chloroplast genome, cloned with a pCC1BAC-based backbone. The marker URA3 was integrated into the genome.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
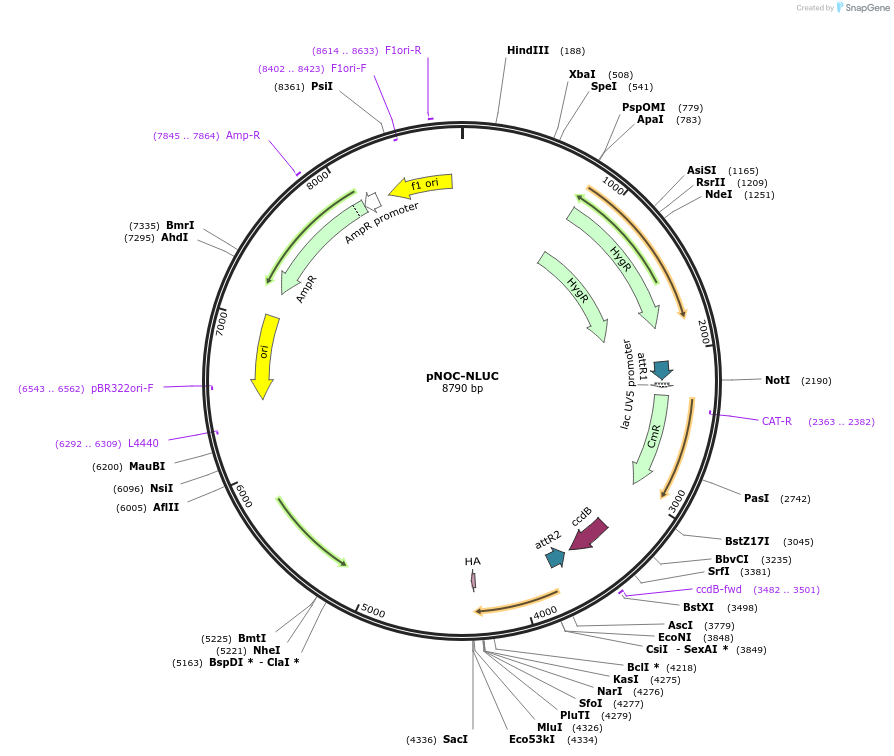
pNOC-NLUC
Plasmid#98141PurposeGateway compatible construct for N' terminal fusion or promoter driven NanoLuciferase reporter, hygromycin resistance cassetteDepositorTypeEmpty backboneUseAlgae, nannochloropsisTagsHAAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
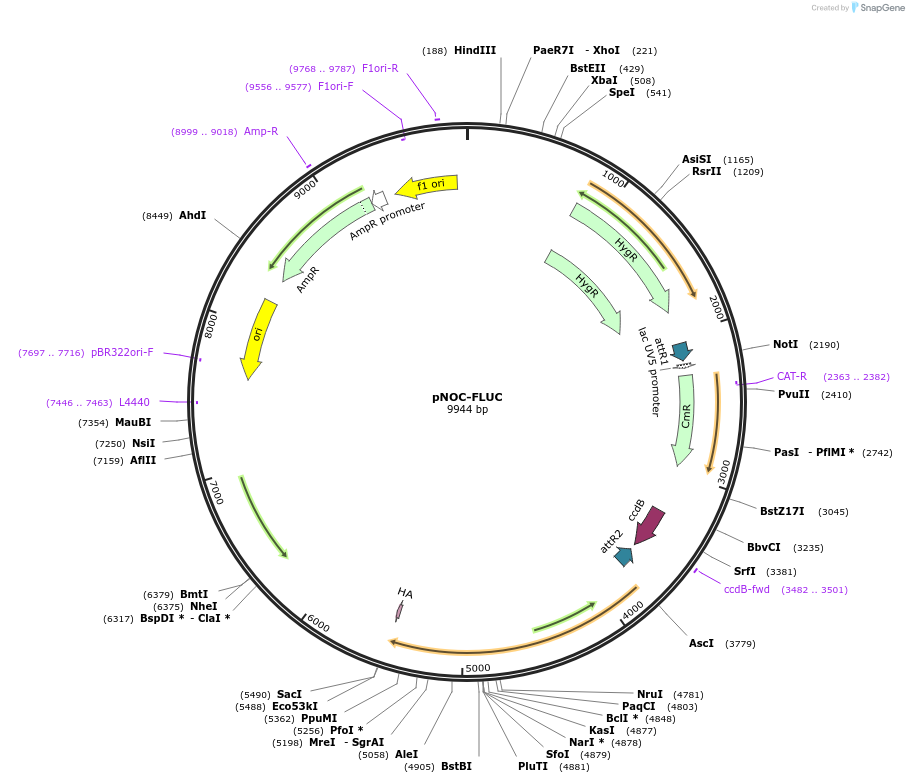
pNOC-FLUC
Plasmid#98140PurposeGateway compatible construct for N' terminal fusion or promoter driven firefly luciferase reporter, hygromycin resistance cassetteDepositorTypeEmpty backboneUseAlgae, nannochloropsisTagsHAAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
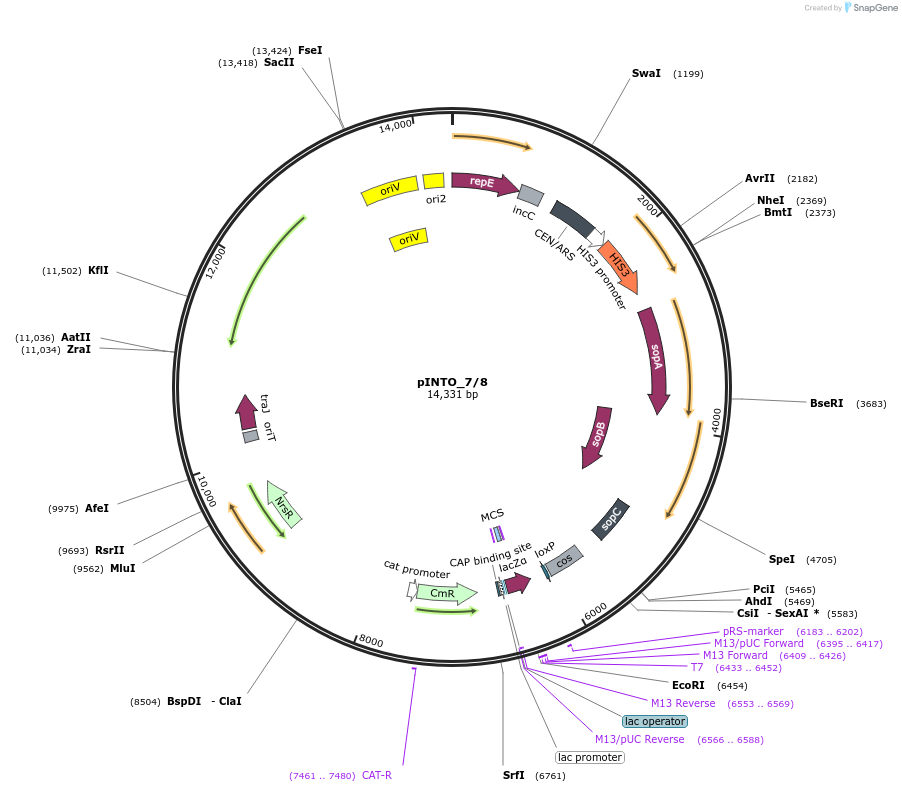
pINTO_7/8
Plasmid#206431PurposeA vector backbone (pCC1BAC-based) containing a single SapI recognition site. When digested, it exposes overhangs that allow integration of the vector into the P. tricornutum chloroplast genome.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
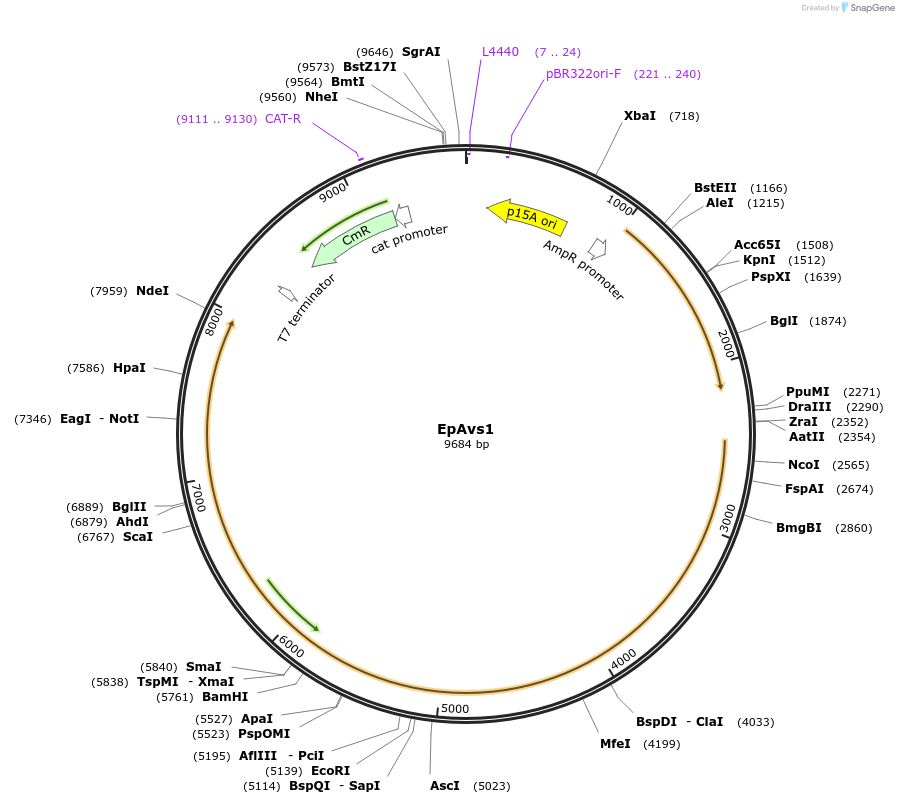
EpAvs1
Plasmid#188788PurposeAvs1 from Erwinia piriflorinigrans CFBP5888DepositorInsertEpAvs1
ExpressionBacterialPromoterblaAvailable SinceAug. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
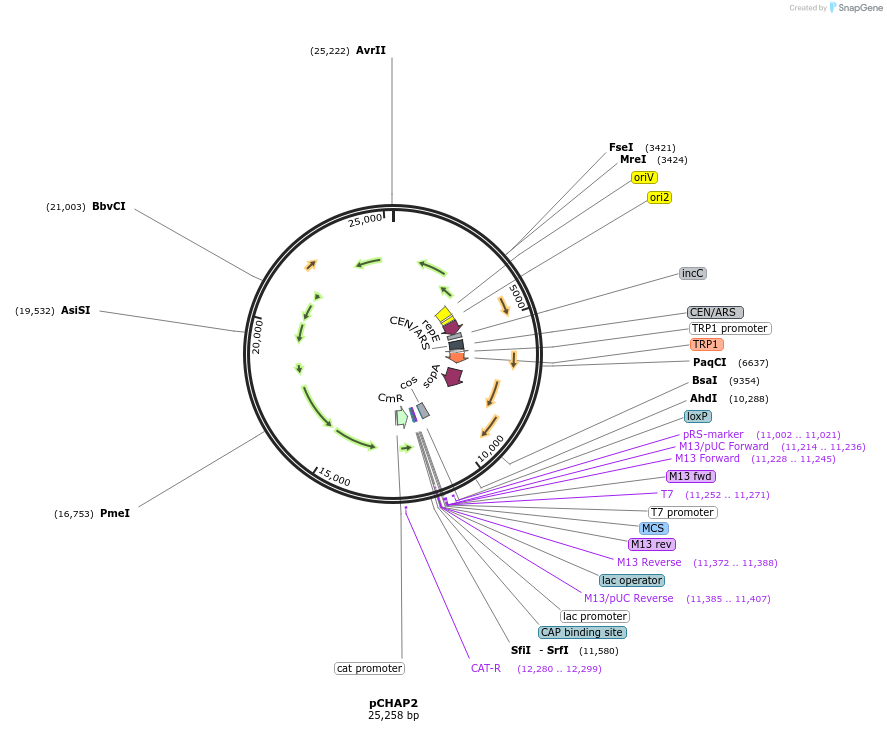
pCHAP2
Plasmid#206847PurposeA plasmid containing a 15,249 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector pSAP1.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
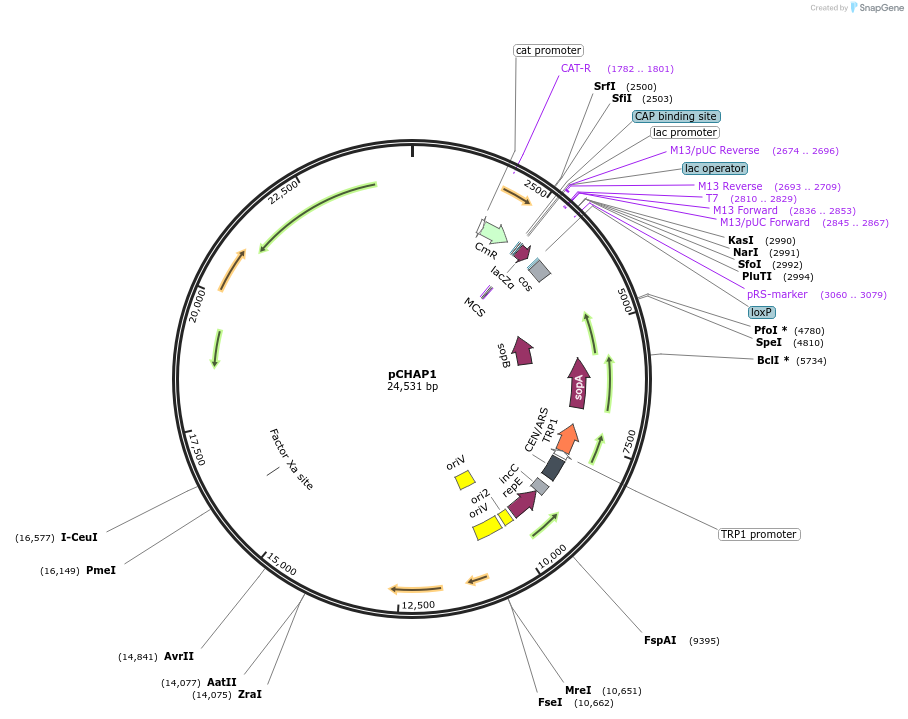
pCHAP1
Plasmid#206846PurposeA plasmid containing a 14,519 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector pSAP1.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
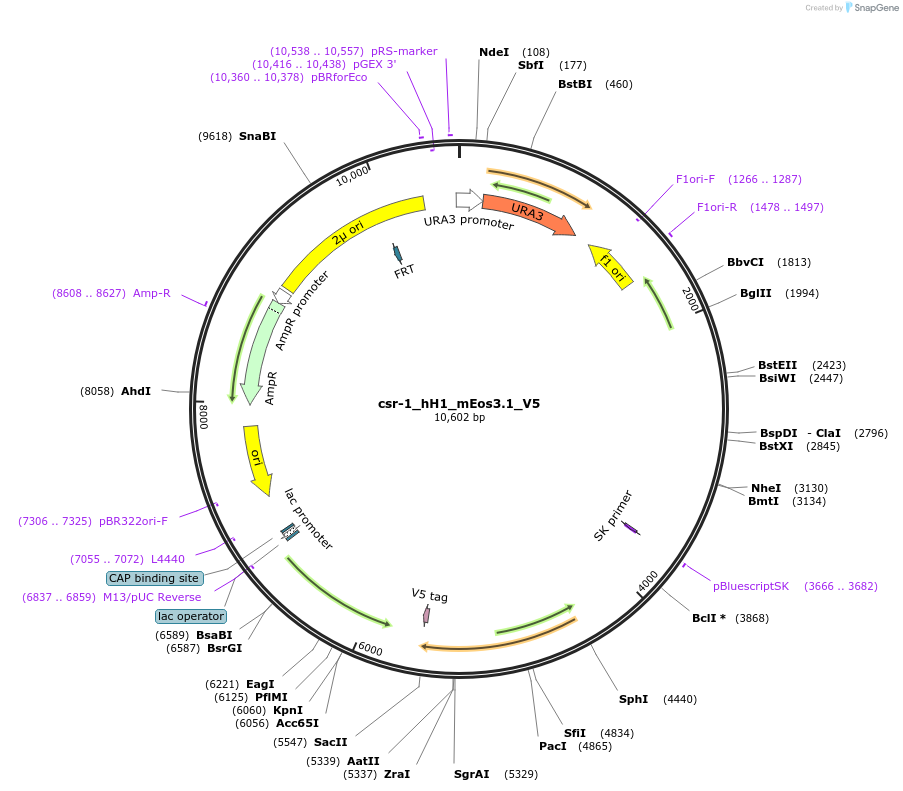
csr-1_hH1_mEos3.1_V5
Plasmid#191750PurposePhotoconvertible fluorescent nuclear reporter.DepositorAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
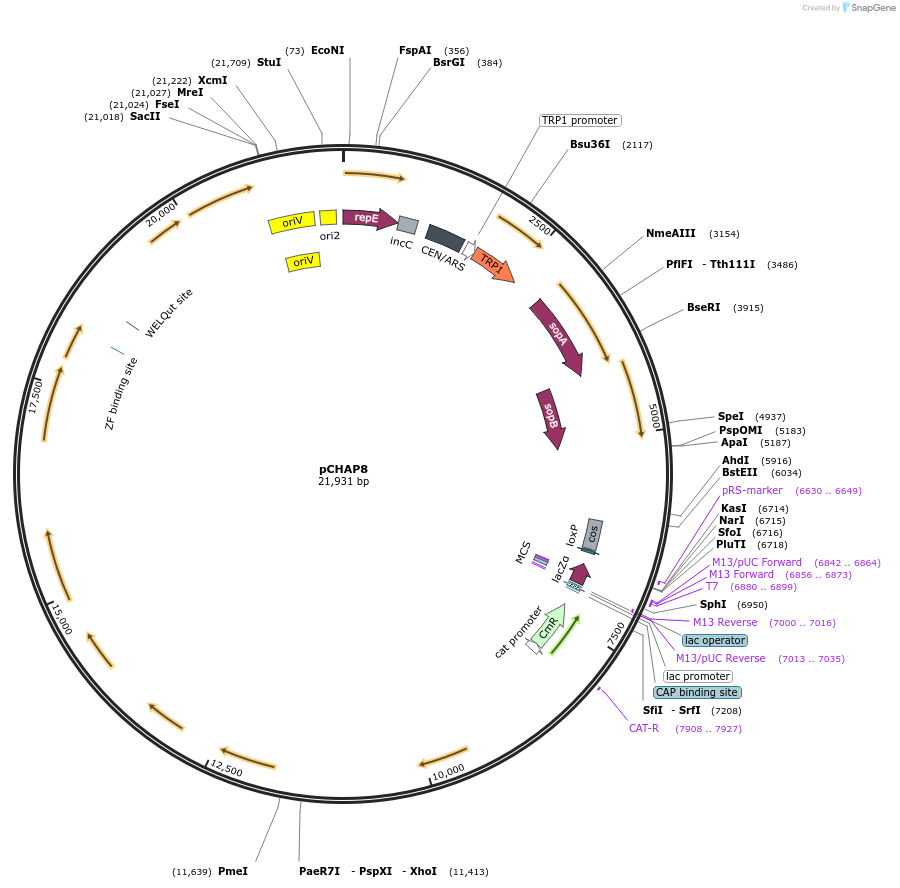
pCHAP8
Plasmid#206857PurposeA plasmid containing a 11,944 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector pSAP1.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
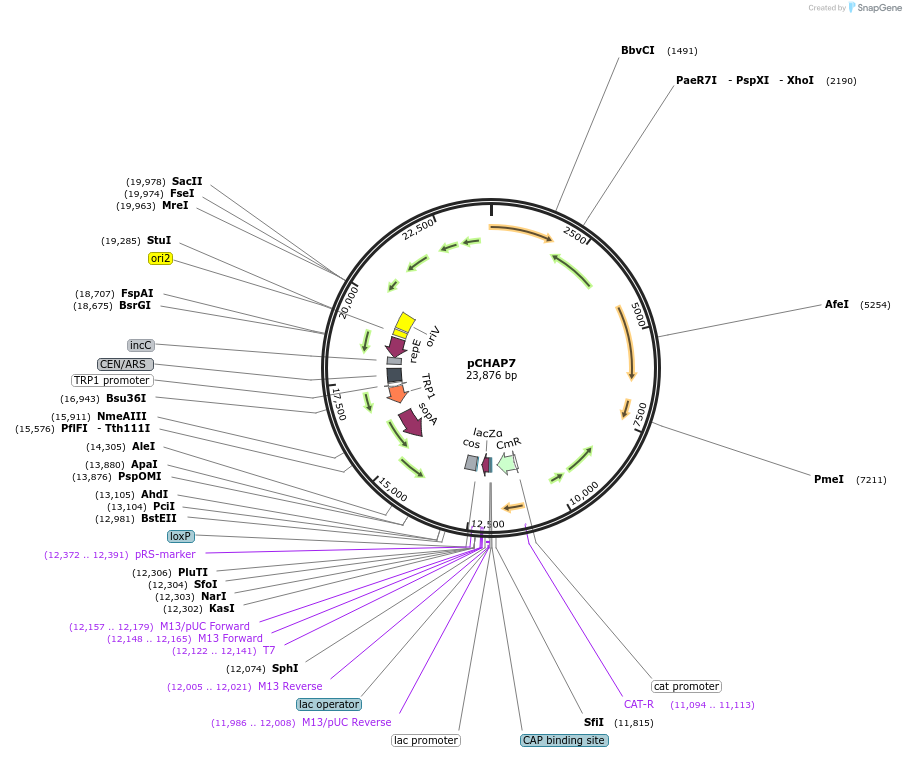
pCHAP7
Plasmid#206853PurposeA plasmid containing a 13,748 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector pSAP1.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
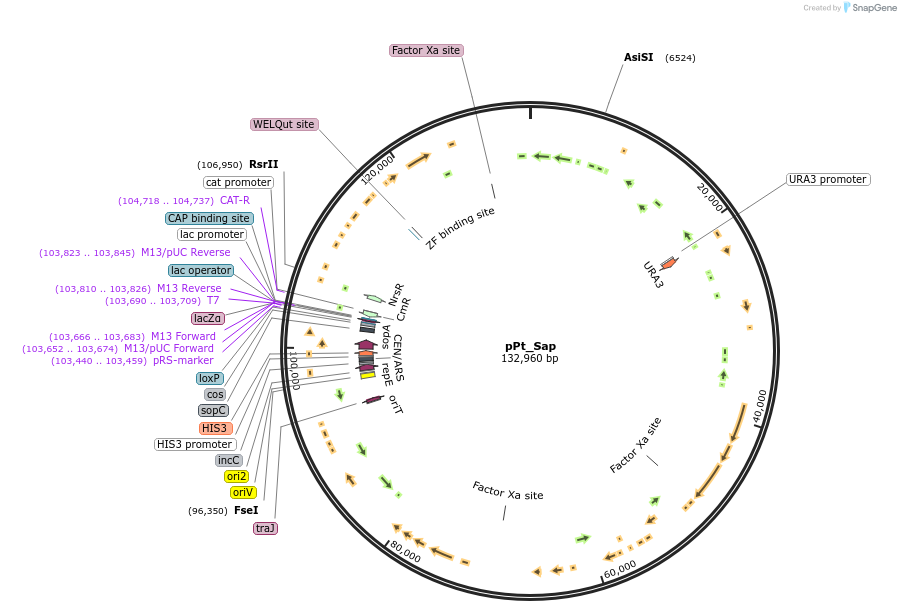
pPt_Sap
Plasmid#206856PurposeA plasmid containing the whole P. tricornutum chloroplast genome, cloned with a pCC1BAC-based backbone. It contains a single SapI site located in the URA3 marker, which was integrated into the genome.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
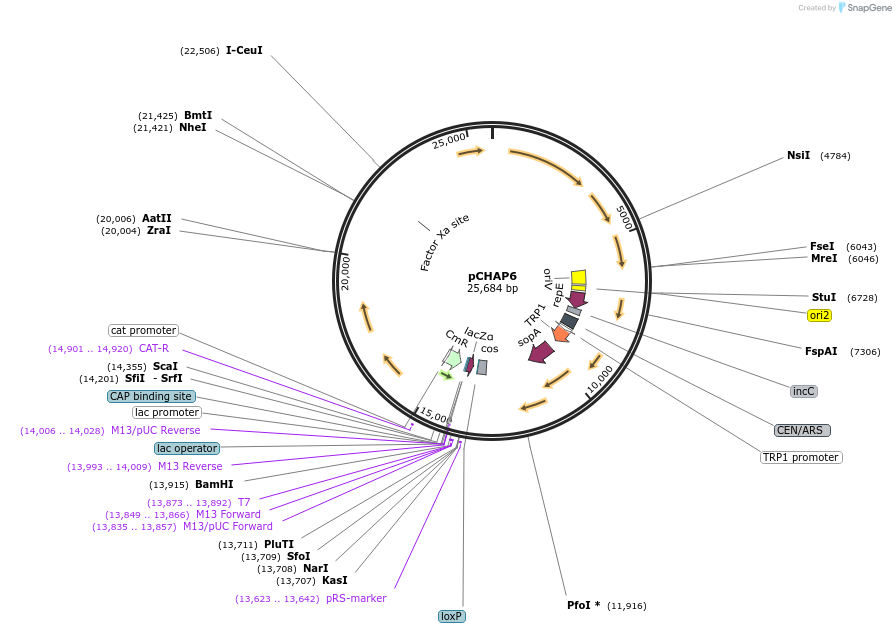
pCHAP6
Plasmid#206852PurposeA plasmid containing a 15,671 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector pSAP1.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
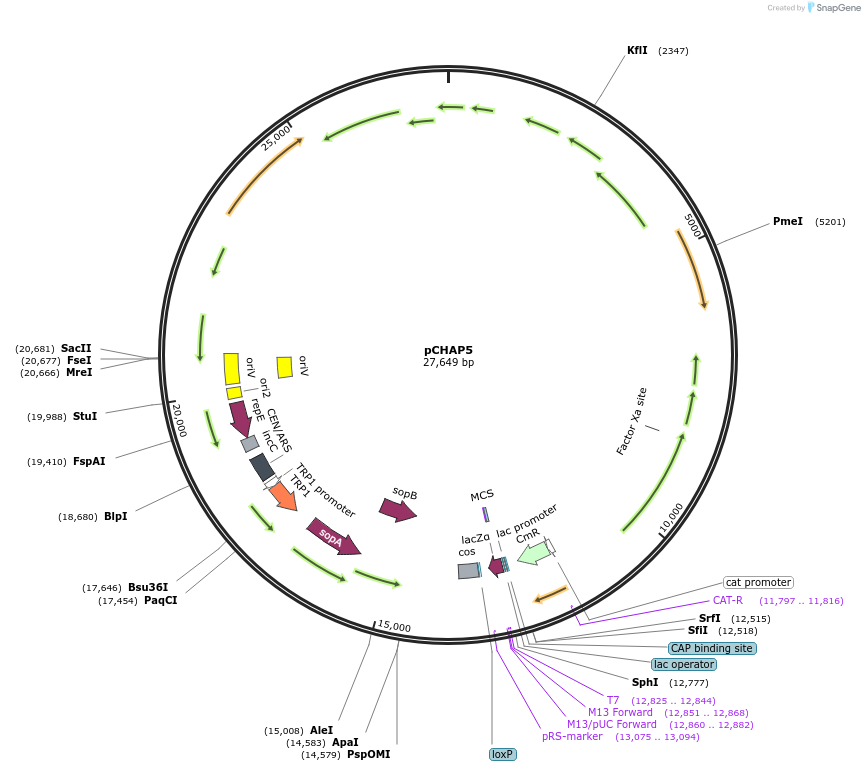
pCHAP5
Plasmid#206851PurposeA plasmid containing a 17,639 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector pSAP1.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
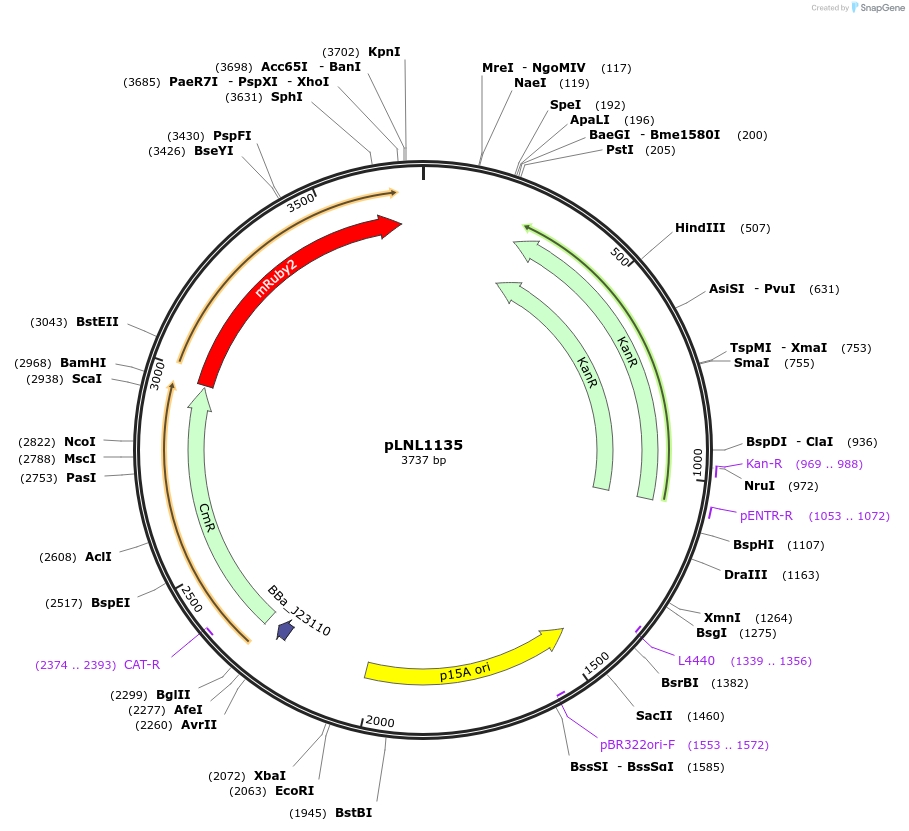
pLNL1135
Plasmid#160866PurposePJ23110-RBS-CAT-taa-mRuby2-tL3S2P24_KanR_p15ADepositorInsertChloramphenicol Acetyltransferase
UseSynthetic BiologyExpressionBacterialPromoterPJ23110_BBa_B0029Available SinceJan. 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
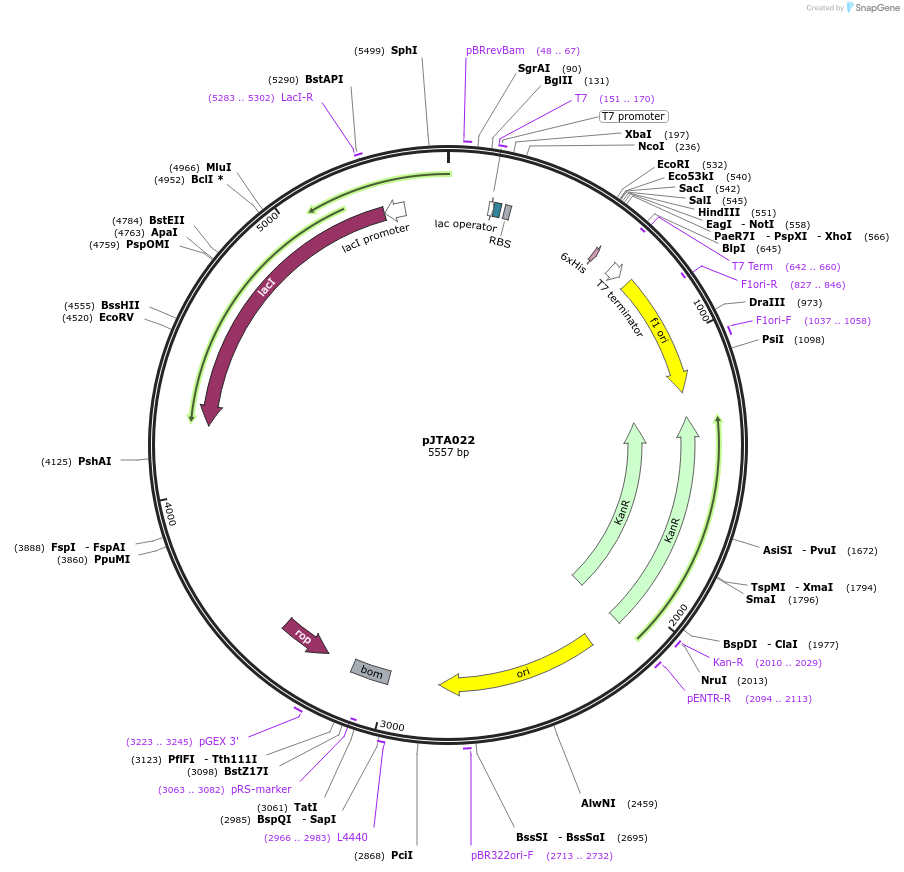
pJTA022
Plasmid#137975PurposeInucible expression of Prochlorococcus phage P-SSM2 phage Fd for purificationDepositorInsertProchlorococcus phage P-SSM2 ferredoxin
UseSynthetic BiologyAvailable SinceJune 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
pET21a(+)-HpDsbG C162A
Plasmid#39330DepositorInsertoxidoreductase
TagsHis-tagExpressionBacterialMutationchanged Cysteine 162 to AlaninePromoterT7Available SinceSept. 18, 2012AvailabilityAcademic Institutions and Nonprofits only -
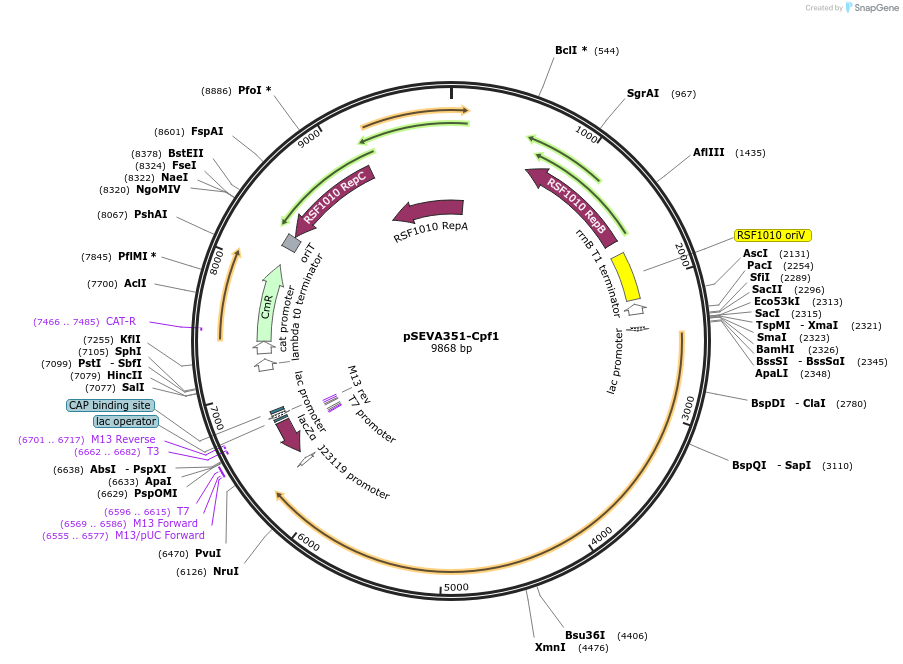
pSEVA351-Cpf1
Plasmid#192361PurposeCRISPR-based vector for genome editingDepositorInsertCas12a, gRNA scaffold
UseCRISPRAvailable SinceSept. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
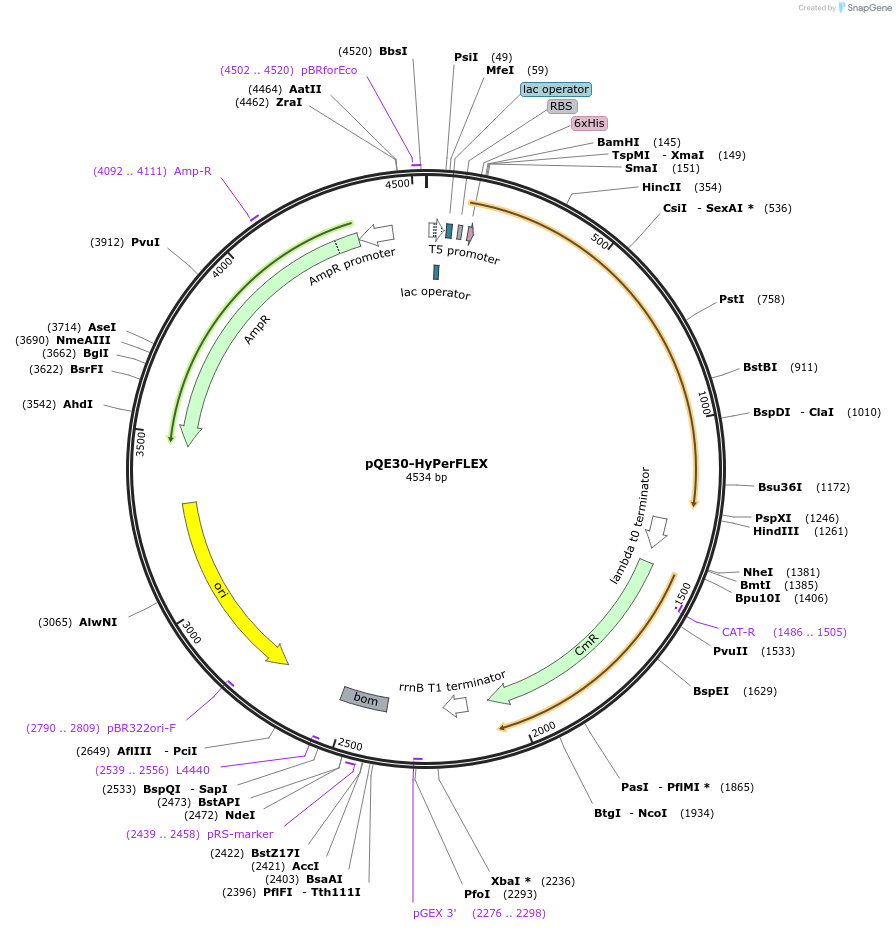
pQE30-HyPerFLEX
Plasmid#217569PurposeBacterial expression of HyPerFLEXDepositorInsertHyPerFLEX
Tags6xHisExpressionBacterialPromoterT5Available SinceOct. 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
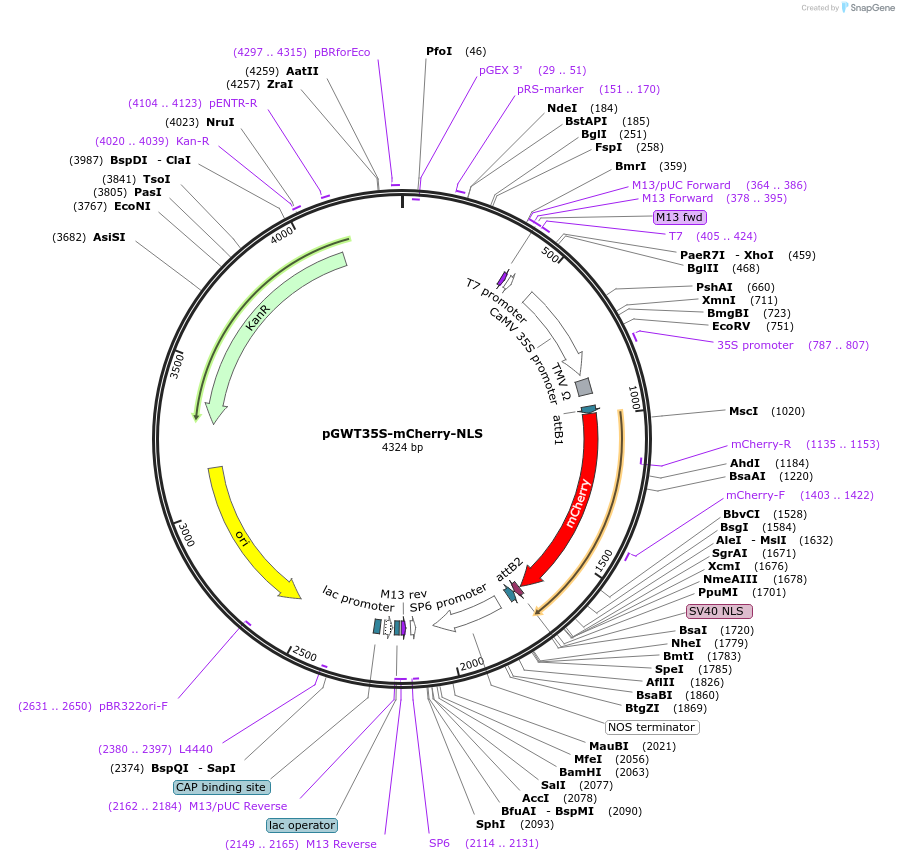
pGWT35S-mCherry-NLS
Plasmid#194049PurposeTransient expression of mCherry-NLS in plant cell (Nucleus)DepositorInsertmCherry
TagsSV40 (Nuclear localization signal)ExpressionPlantPromoterCaMV 35S promoterAvailable SinceJan. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
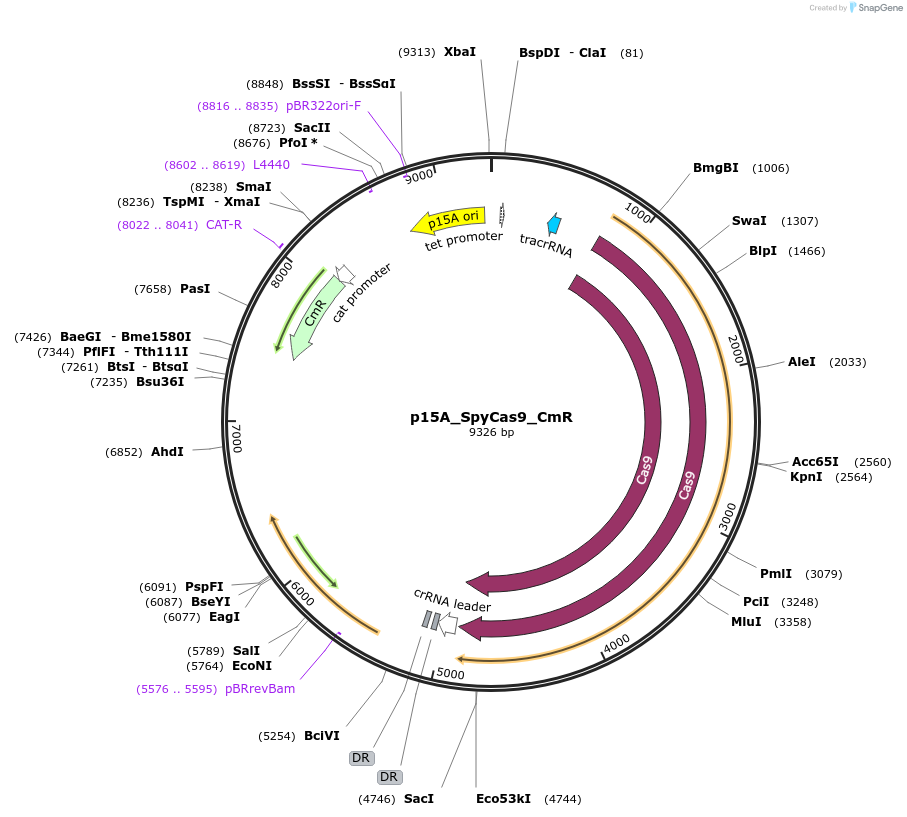
p15A_SpyCas9_CmR
Plasmid#119162PurposeExpresses S. pyogenes Cas9DepositorInsertCas9 from S. pyogenes (NEWENTRY )
UseCrisprAvailable SinceDec. 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
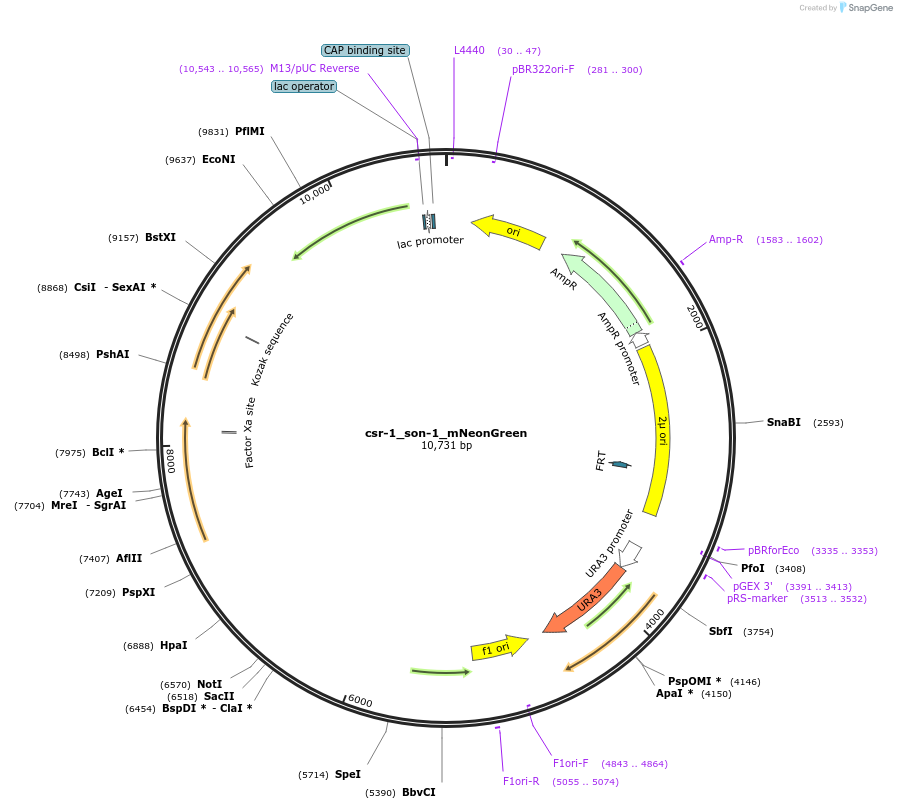
csr-1_son-1_mNeonGreen
Plasmid#191743PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and mNeonGreenExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
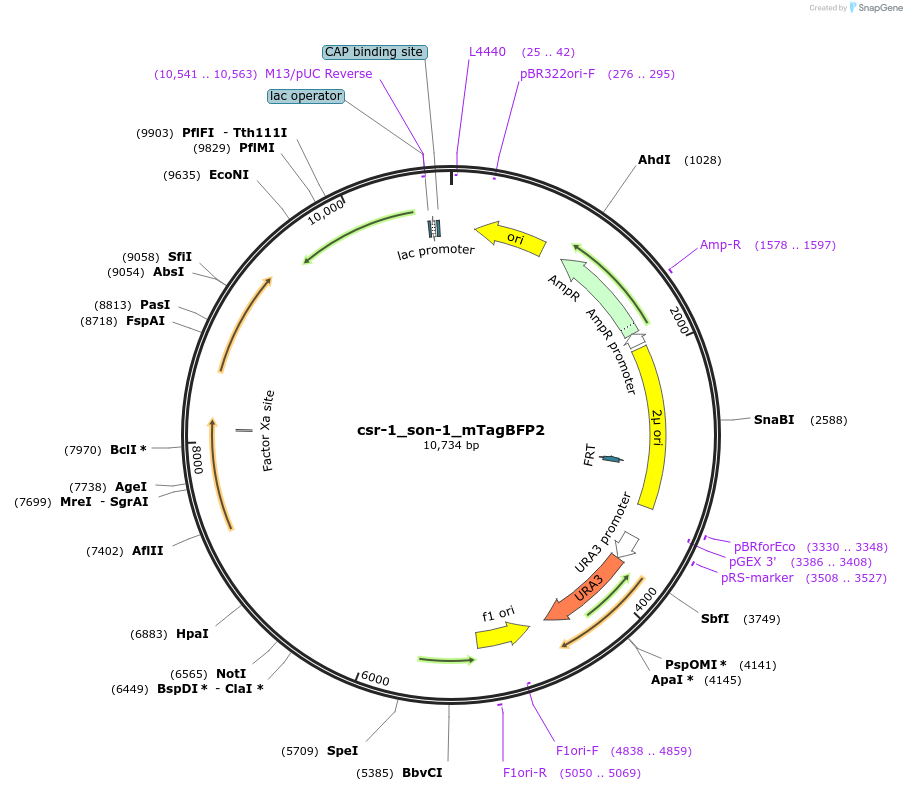
csr-1_son-1_mTagBFP2
Plasmid#191746PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and mTagBFP2ExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
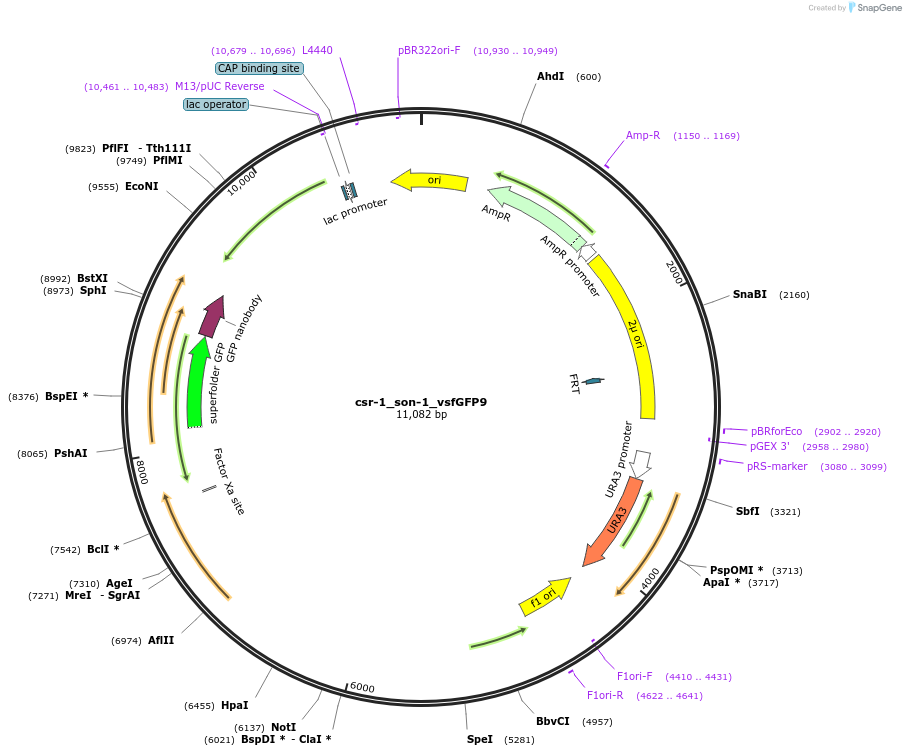
csr-1_son-1_vsfGFP9
Plasmid#191747PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and vsfGFP-9ExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
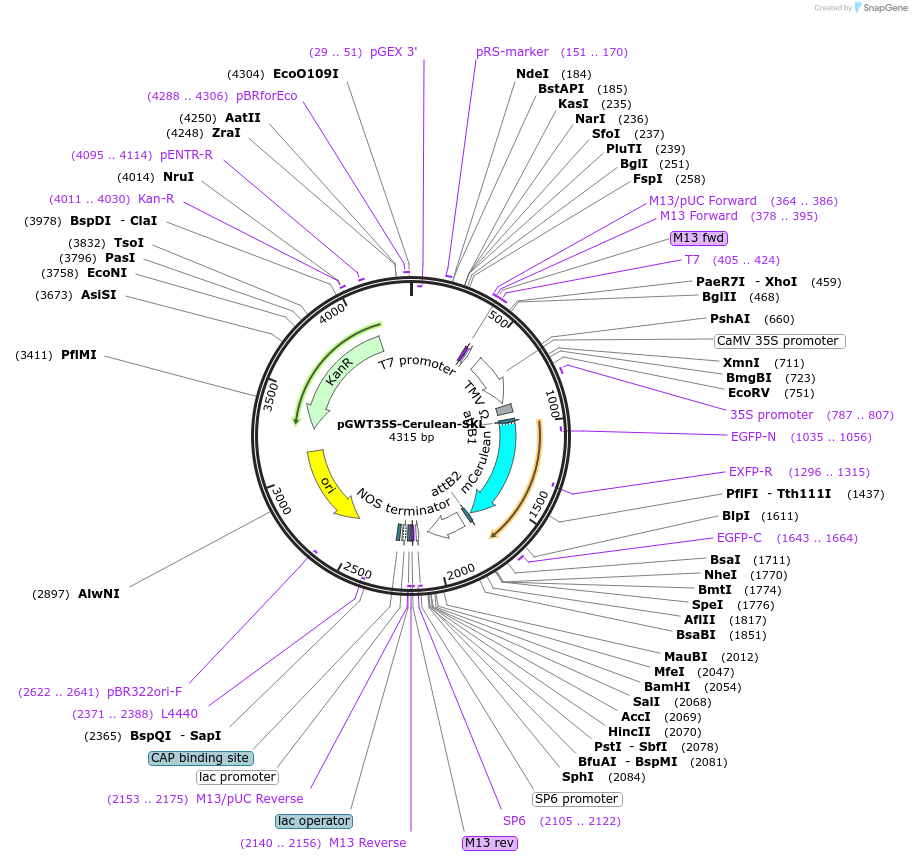
pGWT35S-Cerulean-SKL
Plasmid#194048PurposeTransient expression of Cerulean-SKL in plant cell (Peroxisome)DepositorInsertCerulean
TagsSer-Lsy-LeuExpressionPlantPromoterCaMV 35S promoterAvailable SinceJan. 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
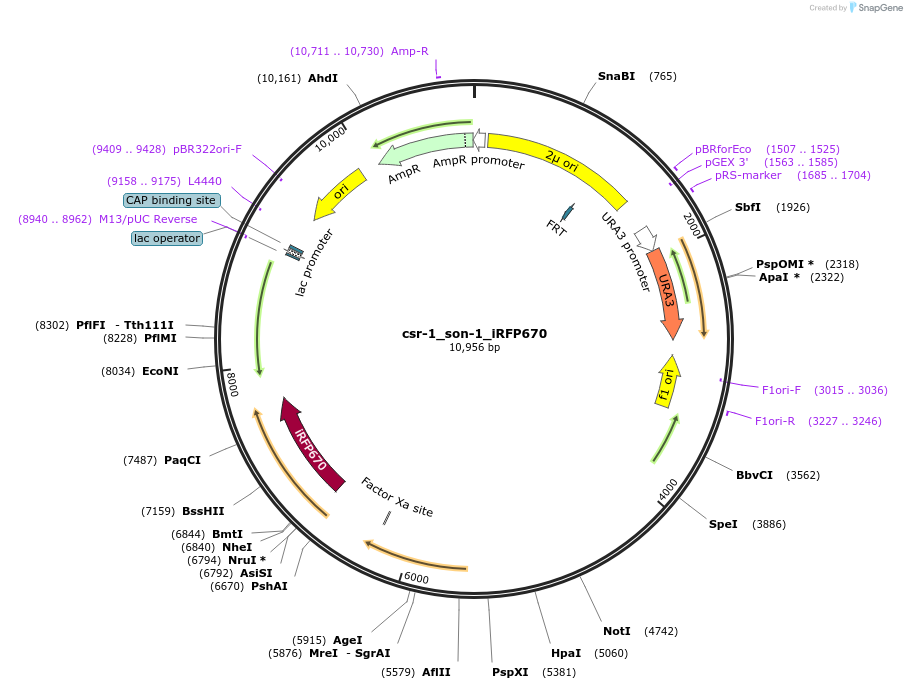
csr-1_son-1_iRFP670
Plasmid#191749PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and iRFP670ExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
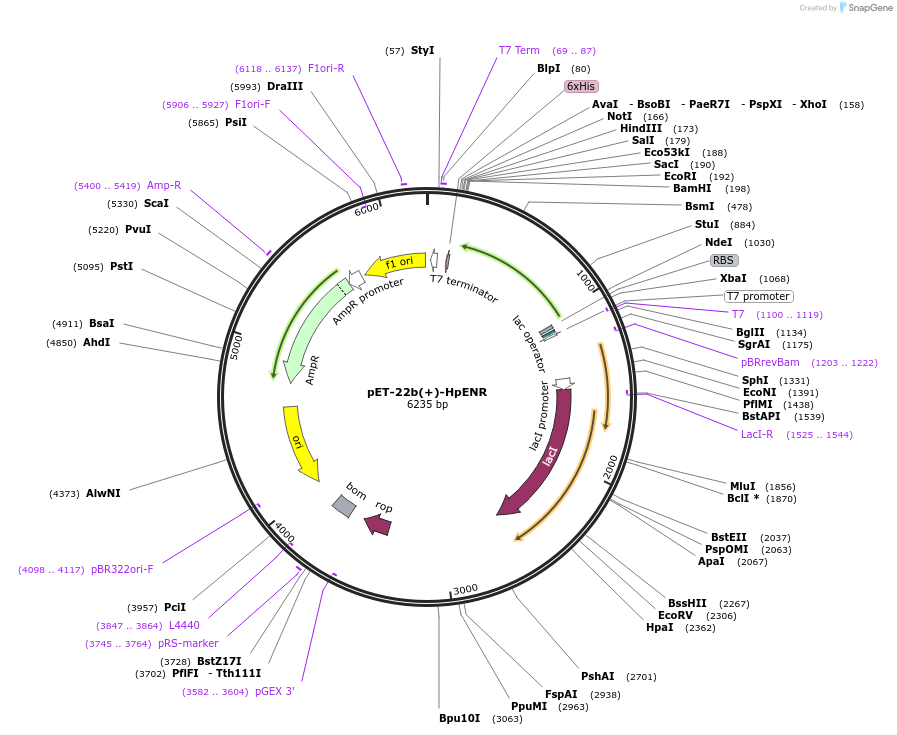
pET-22b(+)-HpENR
Plasmid#12651DepositorInsertnoyl-acyl carrier protein reductase (fabI Helicobacter pylori)
ExpressionBacterialAvailable SinceJan. 5, 2007AvailabilityAcademic Institutions and Nonprofits only -
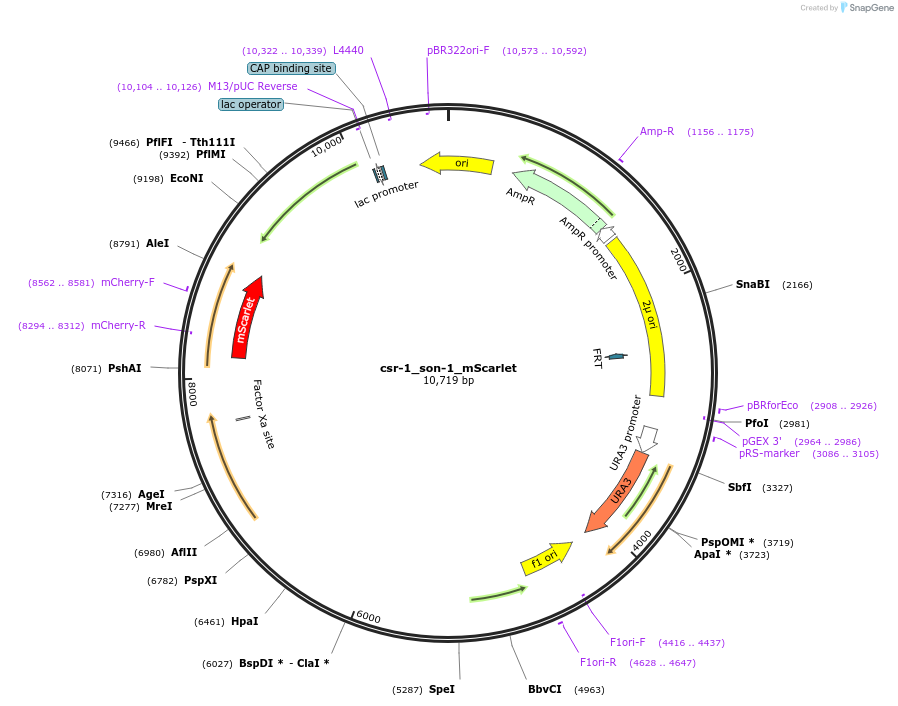
csr-1_son-1_mScarlet
Plasmid#191745PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and mScarletExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
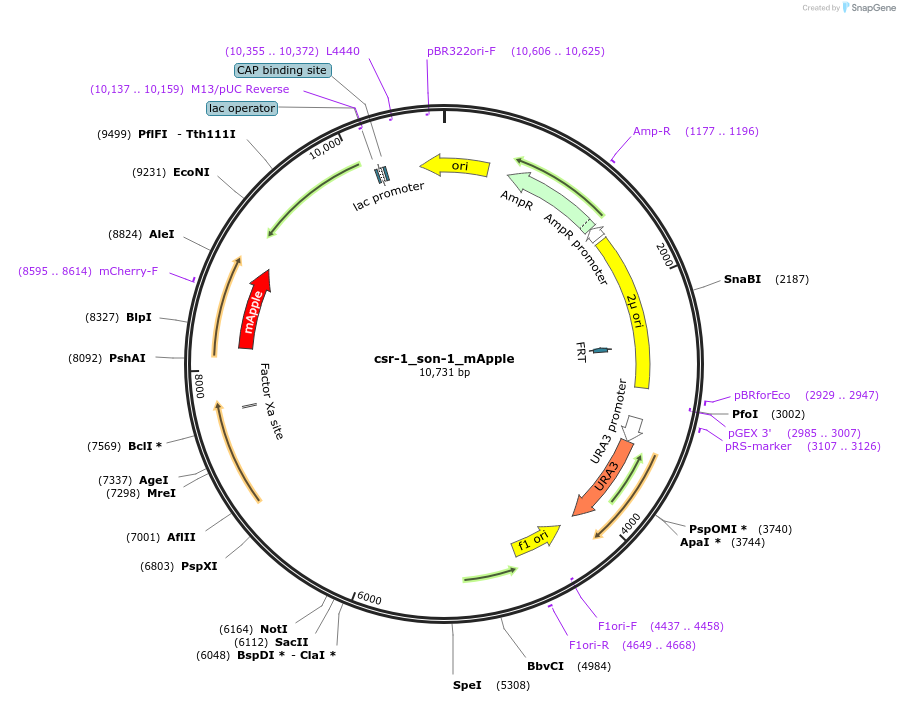
csr-1_son-1_mApple
Plasmid#191741PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
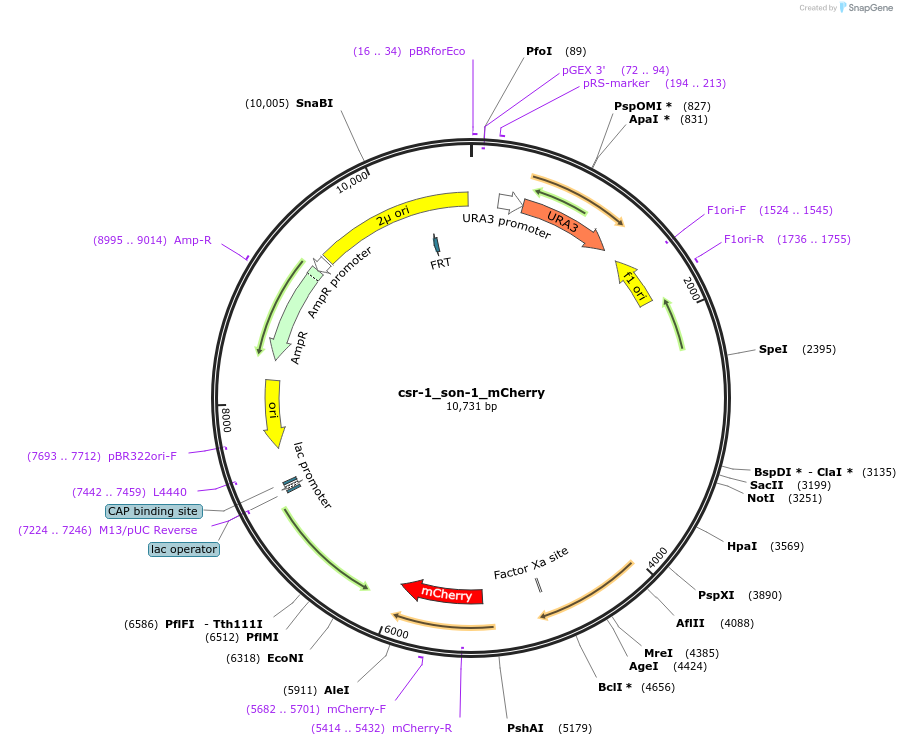
csr-1_son-1_mCherry
Plasmid#191742PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and mCherryExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
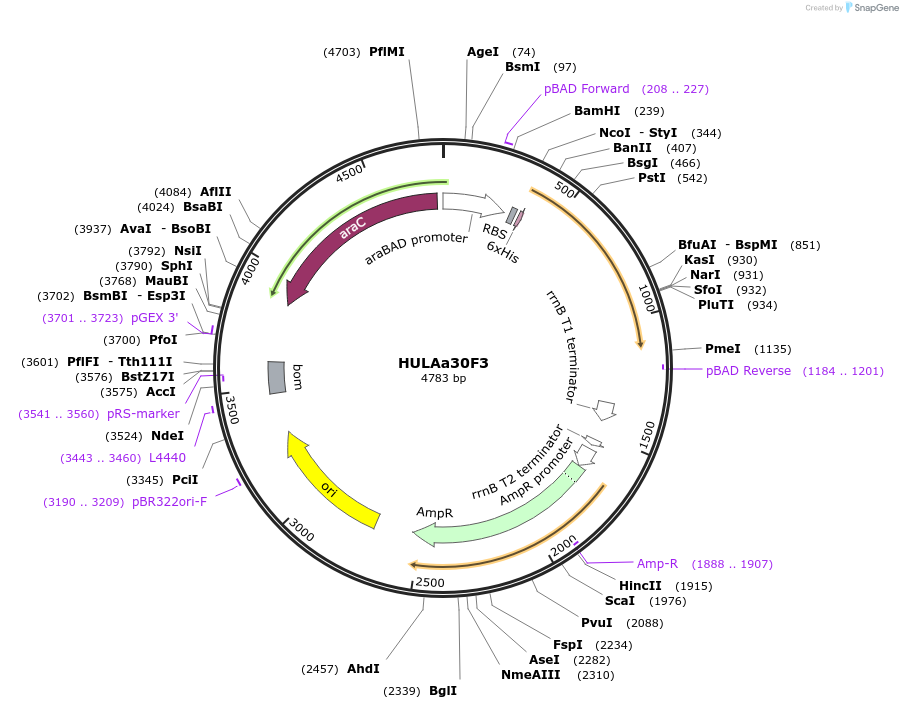
HULAa30F3
Plasmid#179620PurposeExpresses heliorhodopsin gene in E.coli cellsDepositorInsertHULAa30F3
Tags6xHisExpressionBacterialPromoteraraBADAvailable SinceOct. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
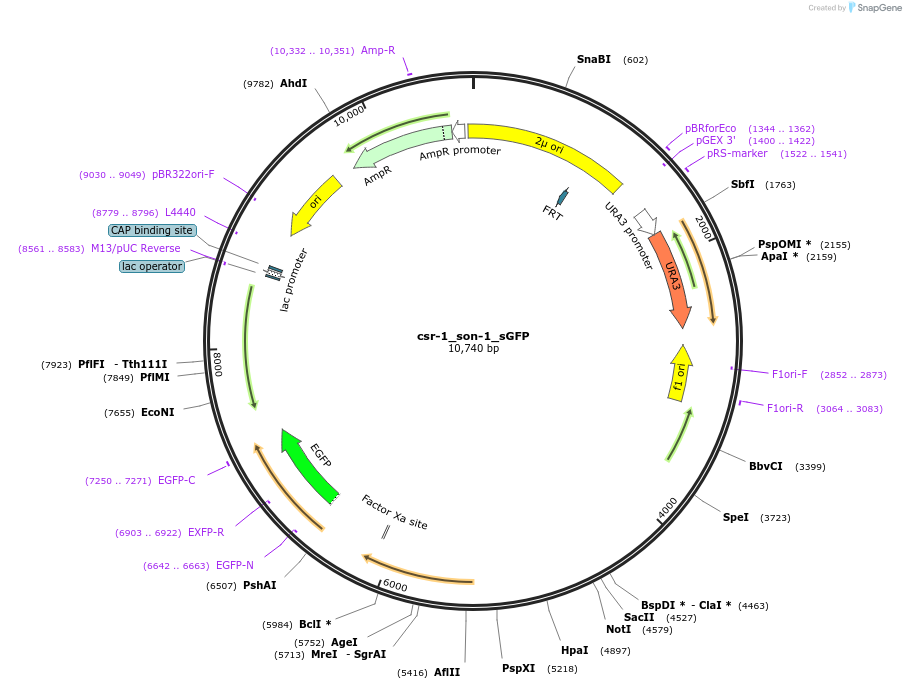
csr-1_son-1_sGFP
Plasmid#191561PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorAvailable SinceJan. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
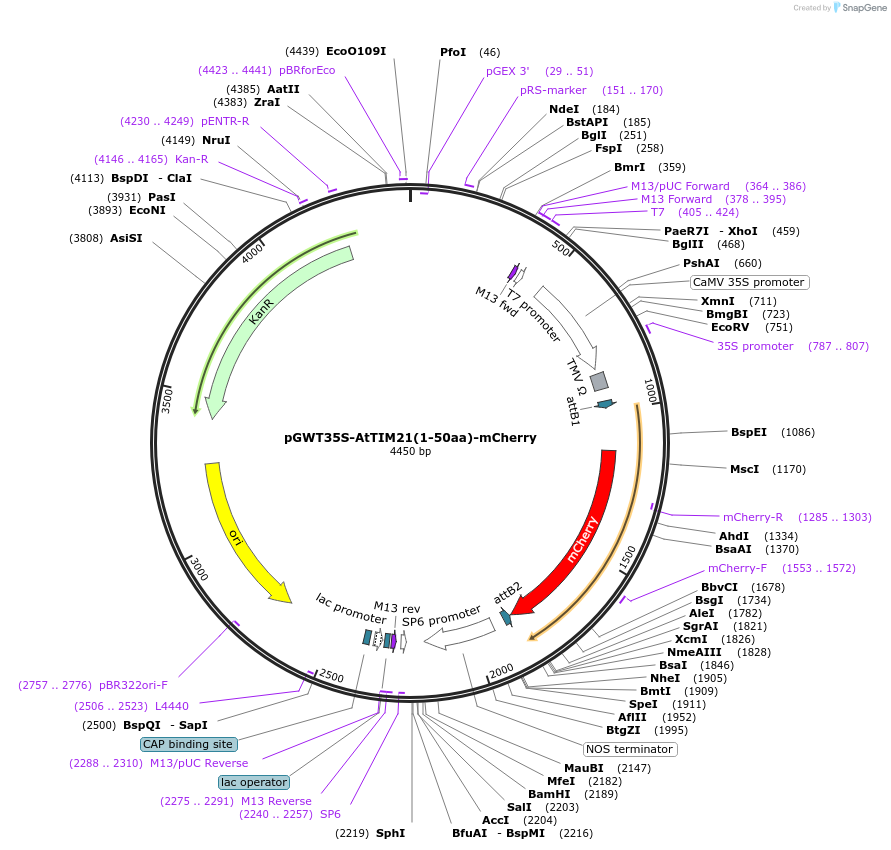
pGWT35S-AtTIM21(1-50aa)-mCherry
Plasmid#194047PurposeTransient expression of AtTIM21-mCherry in plant cell (Mitochondria)DepositorInsertmCherry
TagsAtTIM21 1–50aaExpressionPlantPromoterCaMV 35S promoterAvailable SinceJan. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
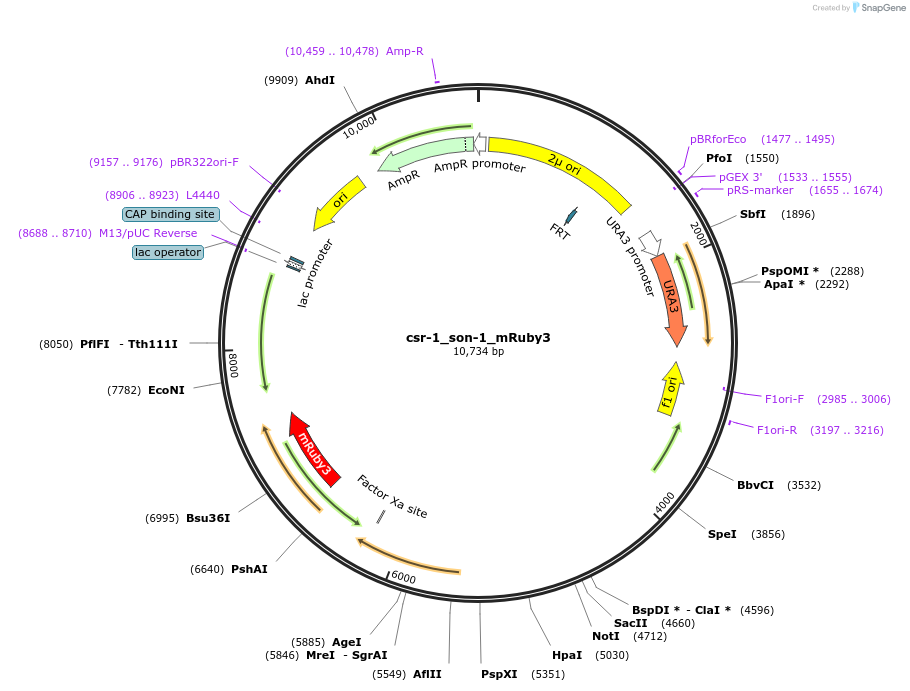
csr-1_son-1_mRuby3
Plasmid#191744PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
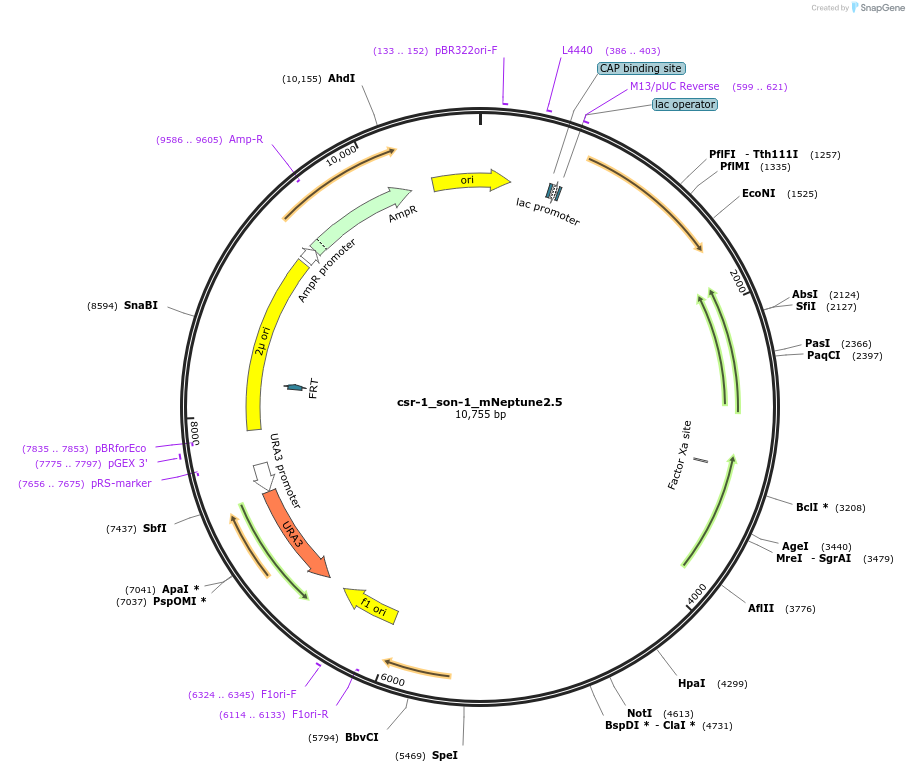
csr-1_son-1_mNeptune2.5
Plasmid#191748PurposeTest fluorescent properties in Neurospora crassa. Nuclear envelope fluorescent reporter.DepositorInsertNCU04288 (NCU04288 Neurospora crassa)
TagsNCU04502 promoter and mNeptune2.5ExpressionBacterialAvailable SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
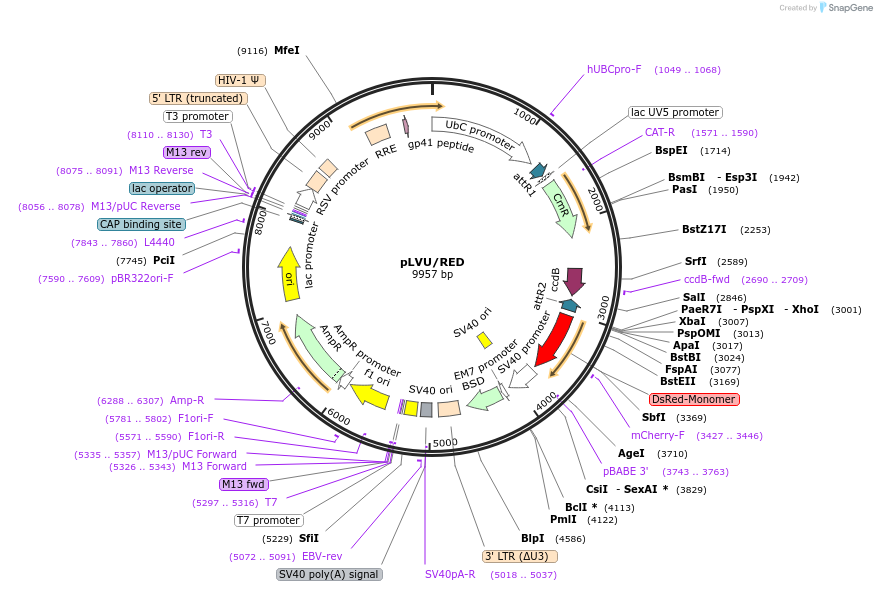
pLVU/RED
Plasmid#24178DepositorInsertChloramphenicol resistance gene (CmR) - ccdB gene
UseLentiviral; GatewayTagsDsREDExpressionMammalianAvailable SinceSept. 23, 2010AvailabilityAcademic Institutions and Nonprofits only -
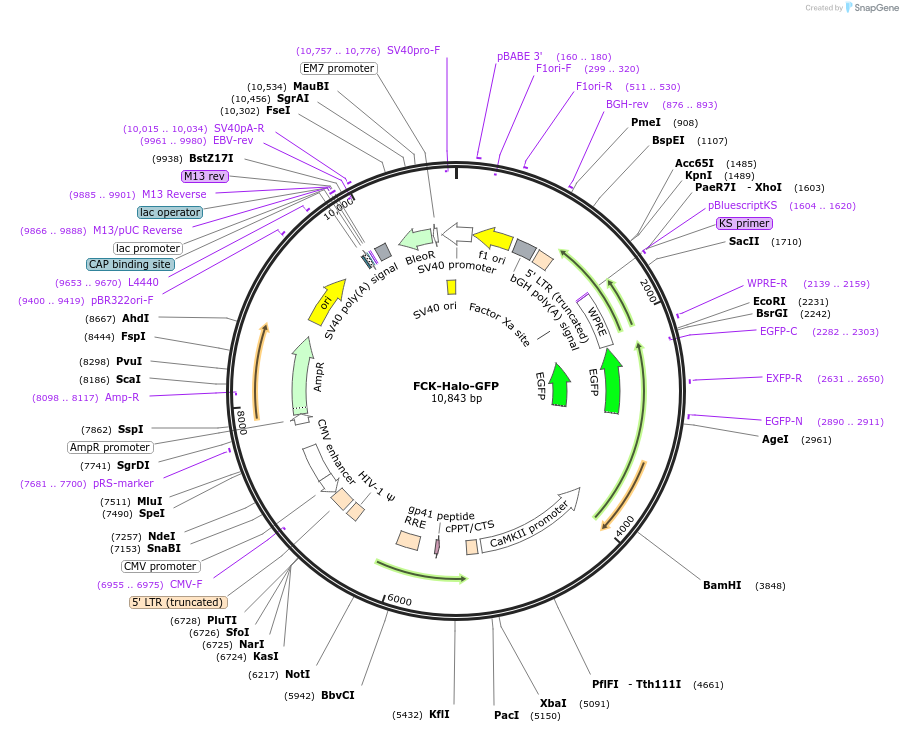
FCK-Halo-GFP
Plasmid#14750PurposeLentiviral expression of Halo-GFP for neuronal inhibitionDepositorInsertMammalian codon-optimized halorhodopsin, from Natronomas pharaonis (N. pharaonis), fused with GFP at the C-terminus
UseLentiviralTagsGFPExpressionMammalianMutationN. pharaonis halorhodopsin is mammalian codon-opt…PromoterCaMKIIaAvailable SinceApril 5, 2007AvailabilityAcademic Institutions and Nonprofits only -
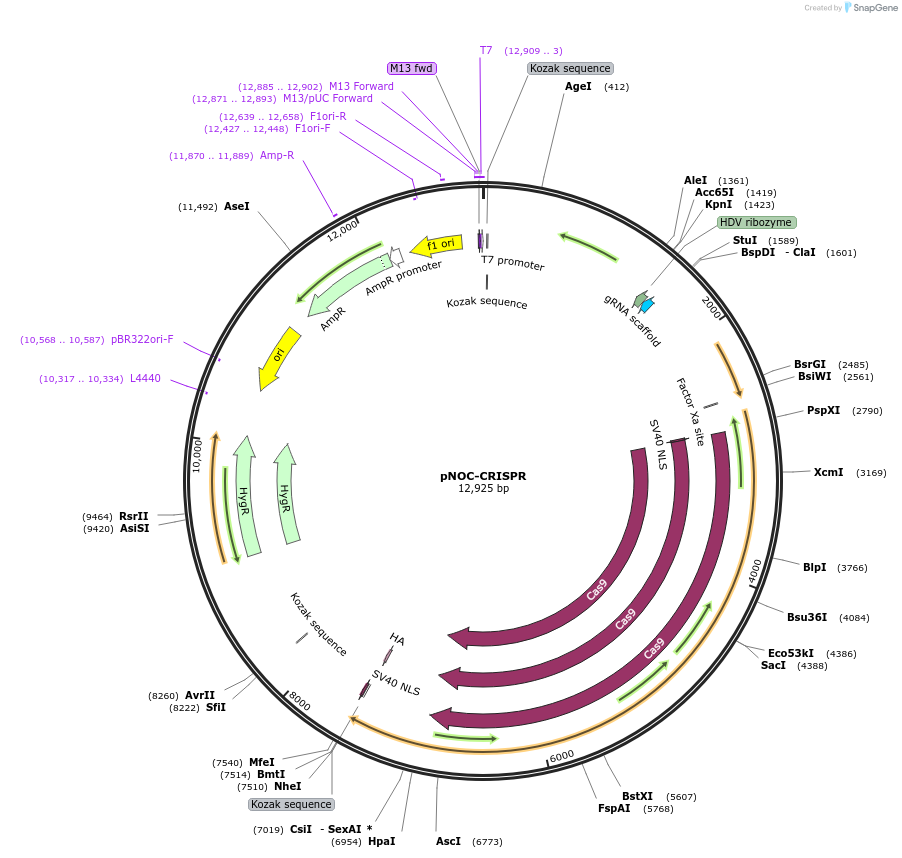
pNOC-CRISPR
Plasmid#100008PurposeExpresses Cas9-Nlux and sgRNA in Nannochloropsis oceanica, contains hygromycin resistance cassette, for integration into genomeDepositorTypeEmpty backboneUseAlgae, nannochloropsis expressionTagsCas9-NluxPromoterRibi promoterAvailable SinceFeb. 11, 2019AvailabilityAcademic Institutions and Nonprofits only