We narrowed to 23,118 results for: Sis
-
Plasmid#72872PurposeThe retroviral GOT1 vector was generated by cloning an sgRNA resistant human GOT1 gene block into the pMXS-ires-blast vector by Gibson Assembly.DepositorInsertGOT1 (GOT1 Human)
UseRetroviralExpressionMammalianMutationsgRNA resistant human GOT1, 7 silent mutations c1…Available SinceFeb. 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
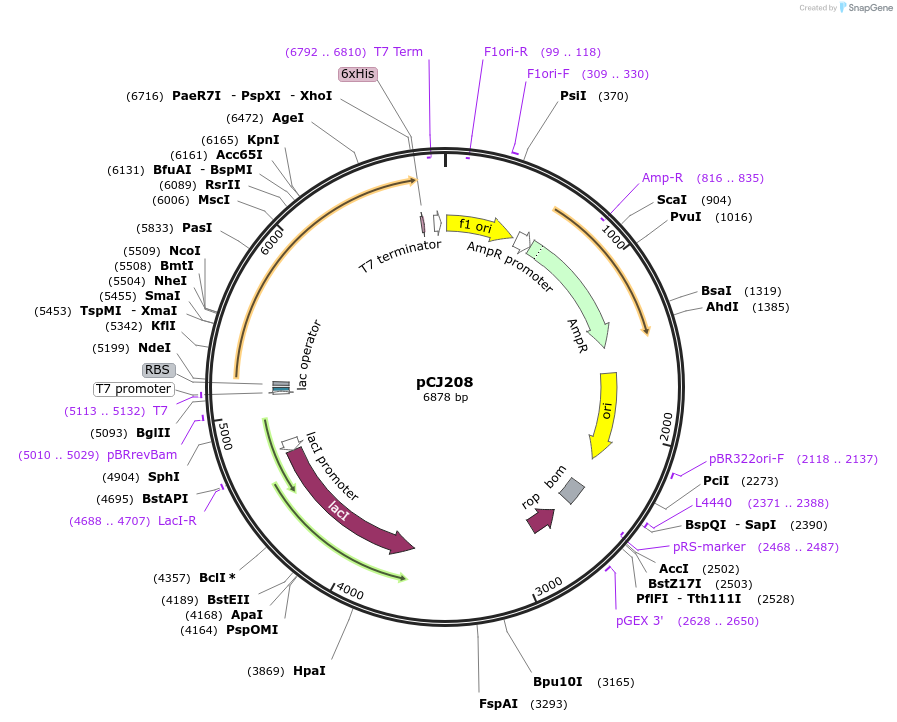
pCJ208
Plasmid#162681PurposeLidded variant of PETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1) removing a seven-residue loop of PETase (Trp185:Phe191, PETase numbering) and replacing it with Gly251:Thr472 from MHETase; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertPETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1) incorporating the MHETase lid
TagsHisExpressionBacterialMutationReplacement of W185-F191 with G251-T472 from MHET…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
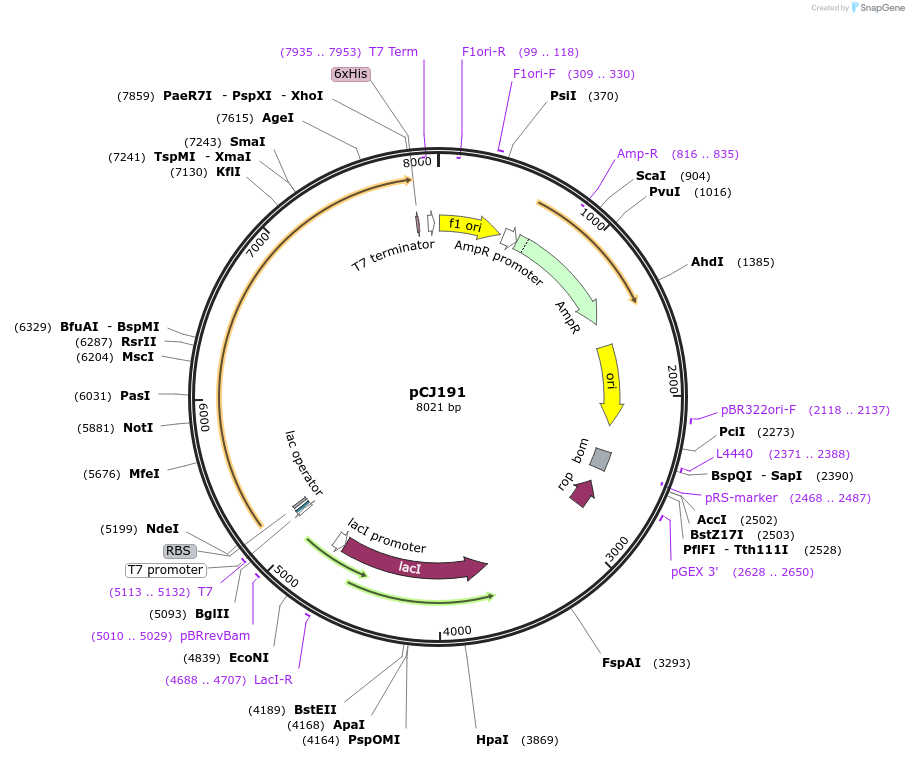
pCJ191
Plasmid#162668PurposepET-21b(+) based plasmid for expresion of MHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 20 residue Gly/Ser linker; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 20 residue Gly/Ser linker
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
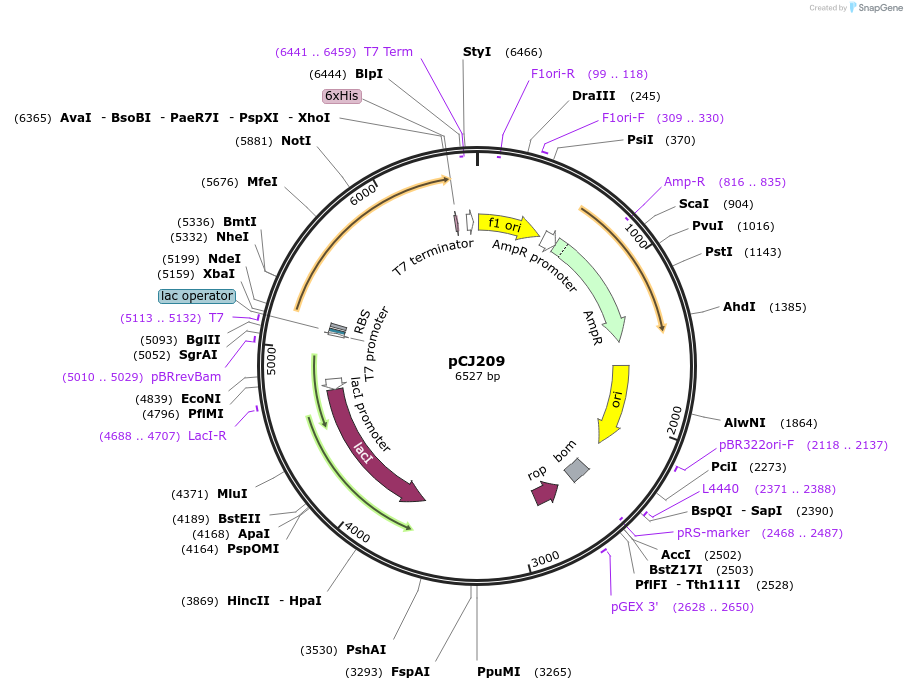
pCJ209
Plasmid#162682PurposeLidless variant of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) replacing Gly251:Thr472 of MHETase with the 7-residue loop (Trp185:Phe191, PETase numbering) of PETase , codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) with the lid removed
TagsHisExpressionBacterialMutationReplacement of G251-T472 with W185-F191 from PETa…PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
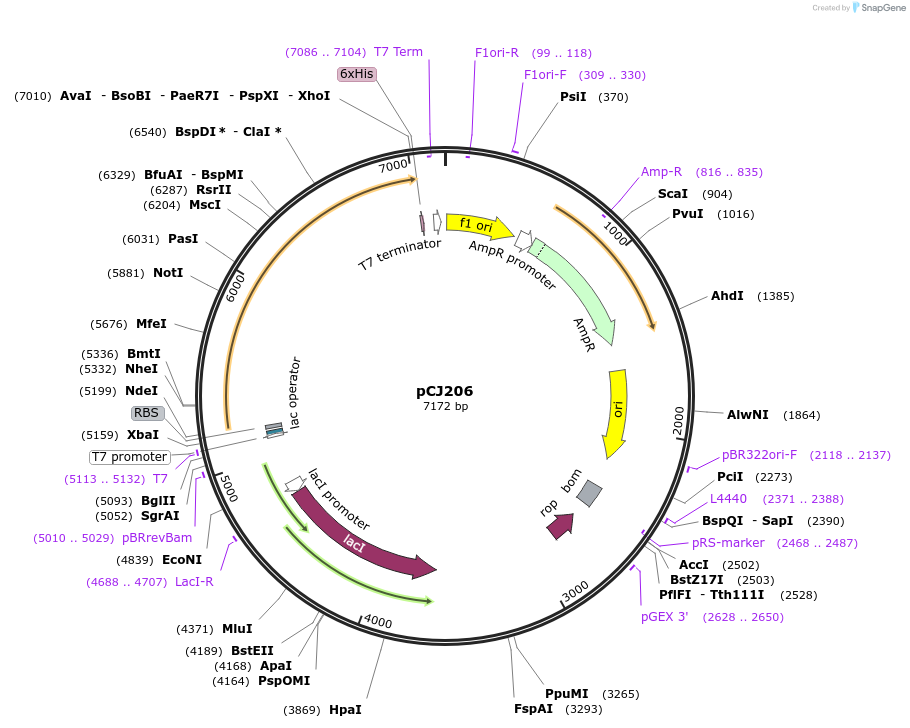
pCJ206
Plasmid#162679PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating the E226T mutation to the putative lipase box.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), incorporating the E226T mutation to the putative lipase box
TagsHisExpressionBacterialMutationE226T; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pABD1275
Plasmid#68017PurposePIF1 protein overproduction for purification from bacterial cellsDepositorInsertPHY-INTERACTING FACTOR 1 (PIF1) (PIL5 AT2G20180, Mustard Weed)
Tags6x-HisExpressionBacterialMutationCodon Optimized for bacterial expression (GenScri…PromoterT7Available SinceDec. 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
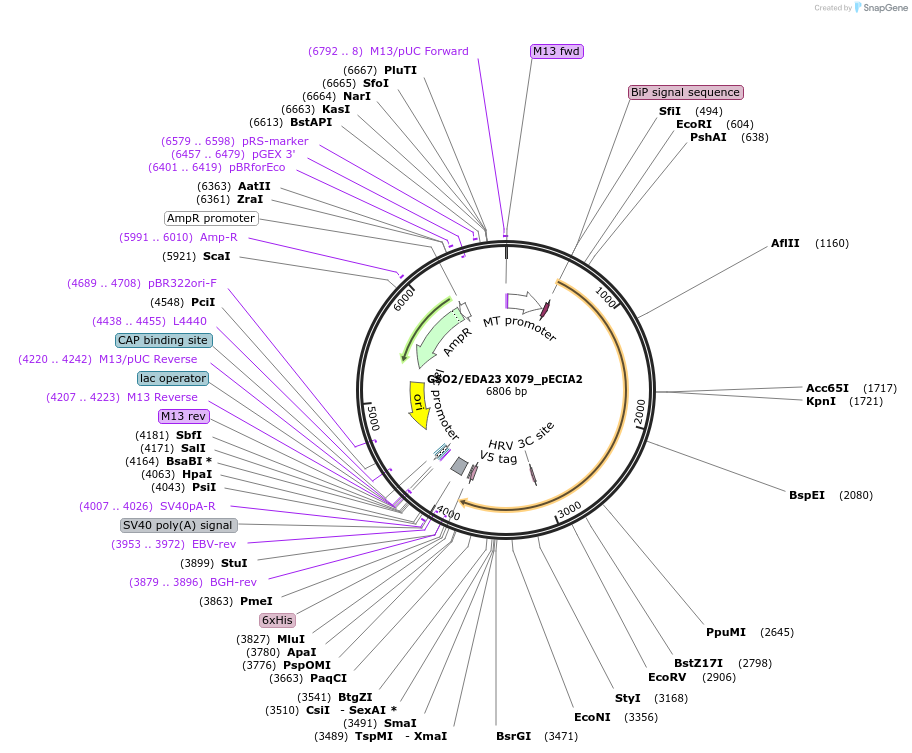
GSO2/EDA23 X079_pECIA2
Plasmid#115111PurposeBait vector GSO2/EDA23 X079_pECIA2 should be used with prey vector GSO2/EDA23 X079_pECIA14.DepositorInsertAT5G44700 (GSO2 Mustard Weed)
TagsFc, V5 and hexahistidine tagsExpressionInsectPromoterMetallothionein (Copper-Inducible)Available SinceMarch 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
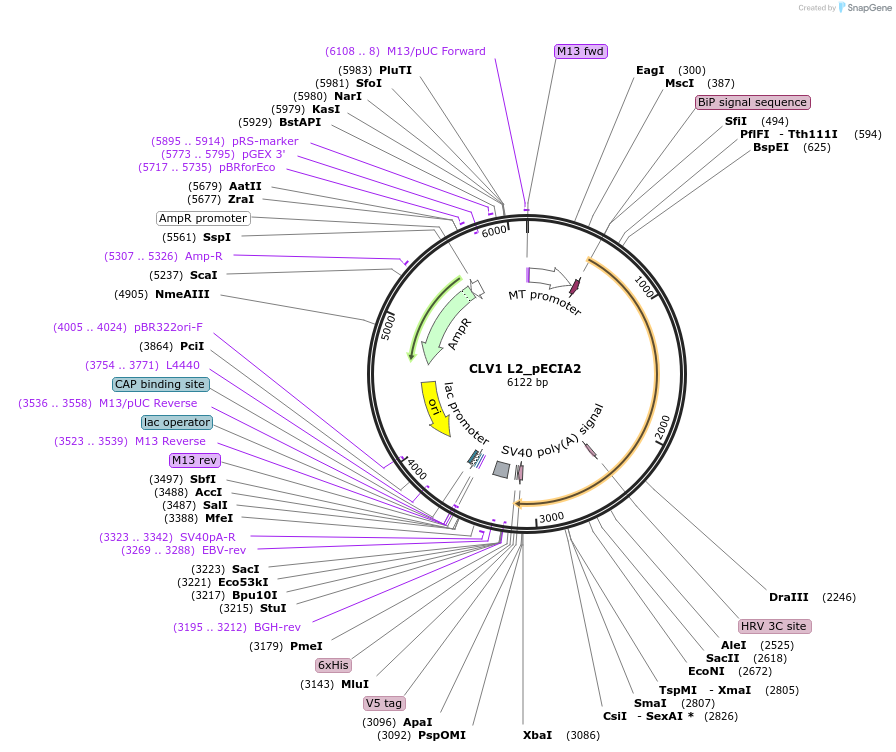
CLV1 L2_pECIA2
Plasmid#115100PurposeBait vector CLV1 L2_pECIA2 should be used with prey vector CLV1 L2_pECIA14.DepositorInsertAT1G75820 (CLV1 Mustard Weed)
TagsFc, V5 and hexahistidine tagsExpressionInsectPromoterMetallothionein (Copper-Inducible)Available SinceMarch 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
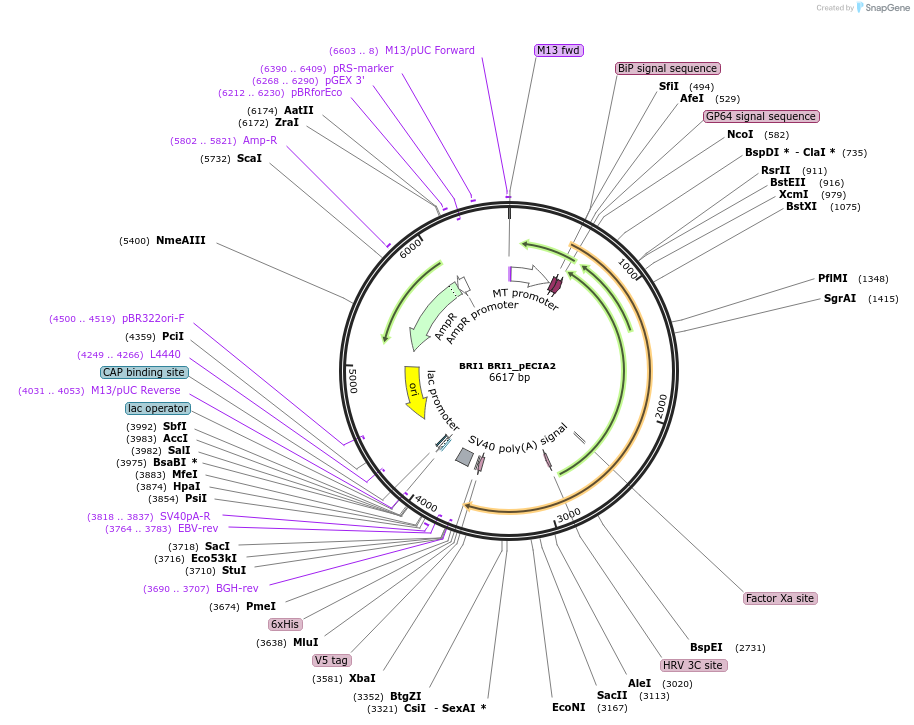
BRI1 BRI1_pECIA2
Plasmid#115086PurposeBait vector BRI1 BRI1_pECIA2 should be used with prey vector BRI1 BRI1_pECIA14.DepositorInsertAT4G39400 (BRI1 Mustard Weed)
TagsFc, V5 and hexahistidine tagsExpressionInsectPromoterMetallothionein (Copper-Inducible)Available SinceMarch 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
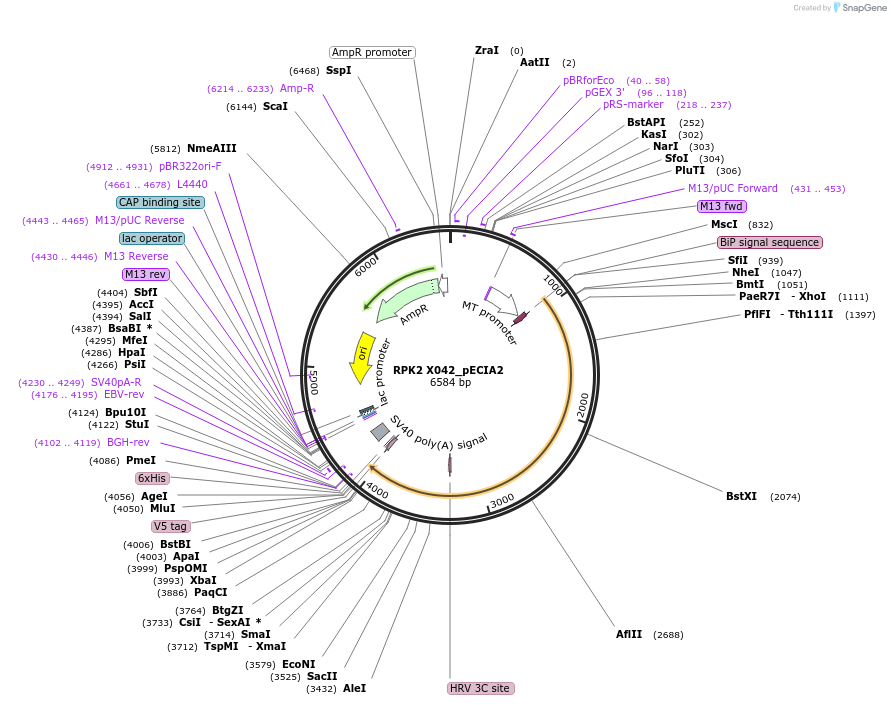
RPK2 X042_pECIA2
Plasmid#115135PurposeBait vector RPK2 X042_pECIA2 should be used with prey vector RPK2 X042_pECIA14.DepositorInsertAT3G02130 (RPK2 Mustard Weed)
TagsFc, V5 and hexahistidine tagsExpressionInsectPromoterMetallothionein (Copper-Inducible)Available SinceMarch 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
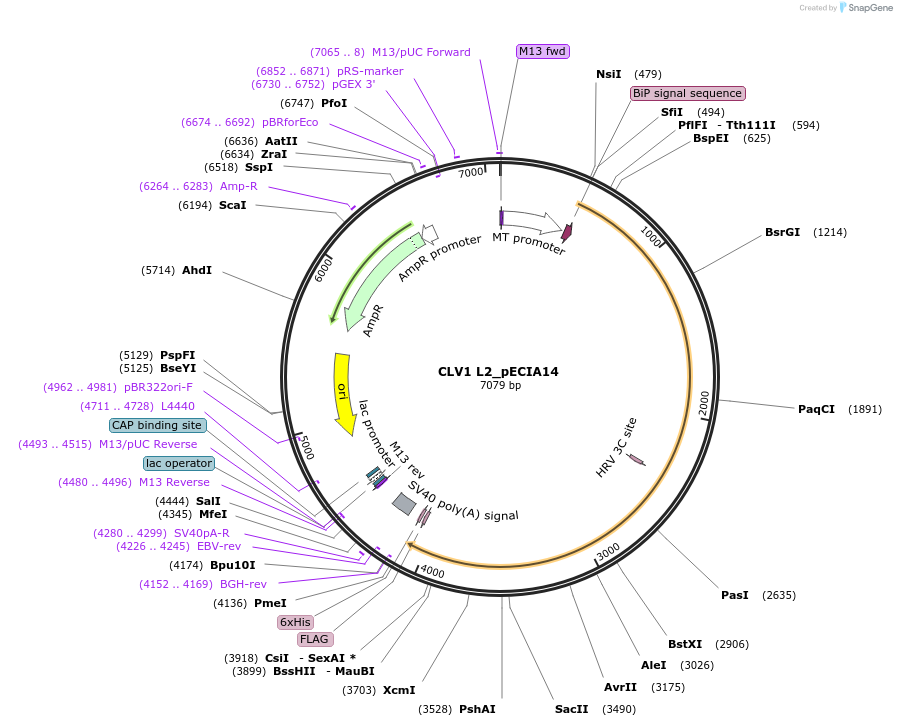
CLV1 L2_pECIA14
Plasmid#114900PurposePrey vector CLV1 L2_pECIA14 should be used with bait vector CLV1 L2_pECIA2.DepositorAvailable SinceMarch 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
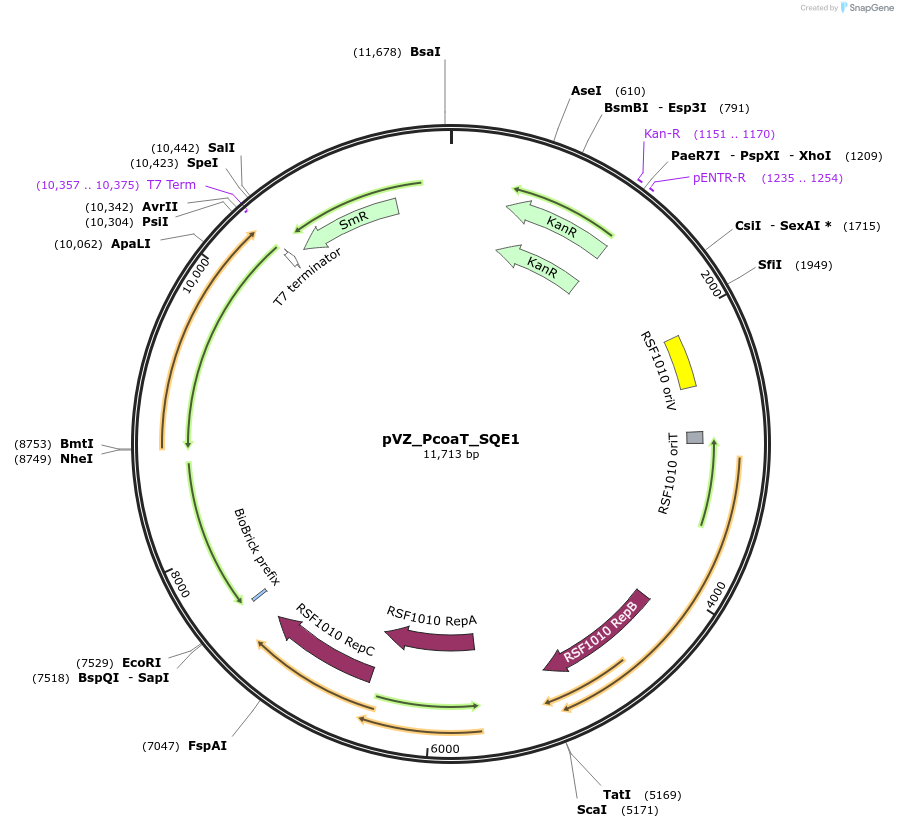
pVZ_PcoaT_SQE1
Plasmid#103860PurposeHeterologous, cobalt-inducible expression of SQE1 from A. thaliana in Synechocystis sp. PCC6803DepositorInsertsqualene epoxidase 1 (XF1 Mustard Weed)
UseSynthetic BiologyExpressionBacterialMutationinserted gene is codon optimized. 2nd amino acid …PromoterPcoaT (from Synechocystis sp. PCC 6803)Available SinceJan. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
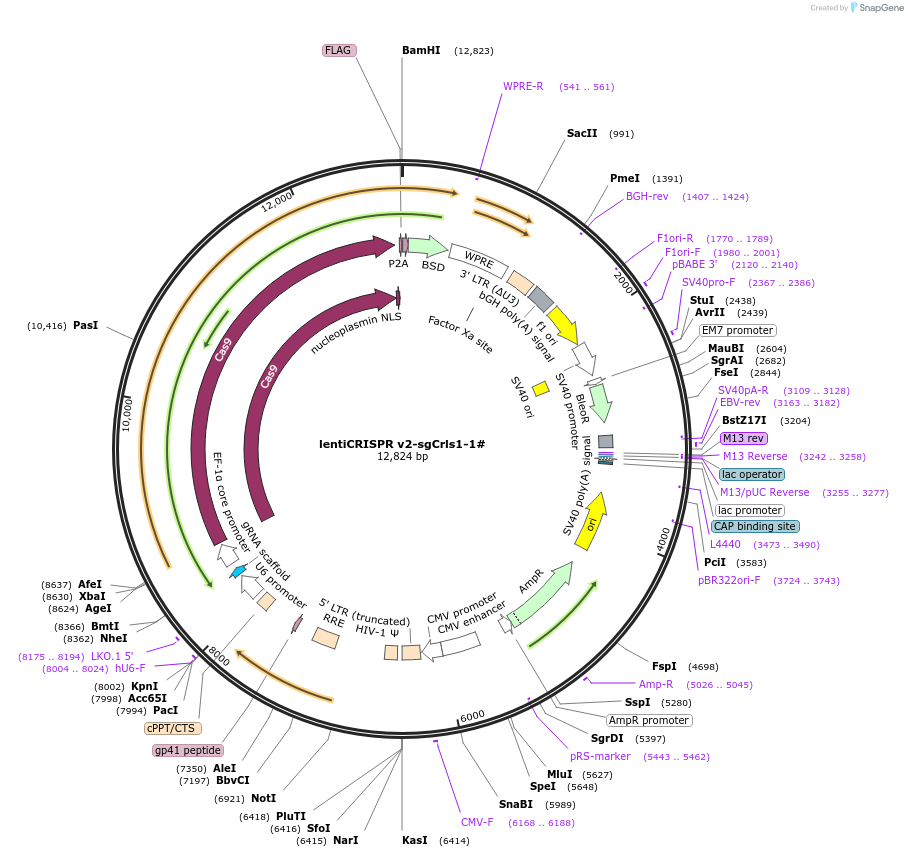
lentiCRISPR v2-sgCrls1-1#
Plasmid#234765Purposeknockout Crls1DepositorInsertCrls1 (Crls1 Mouse)
UseLentiviralAvailable SinceMarch 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
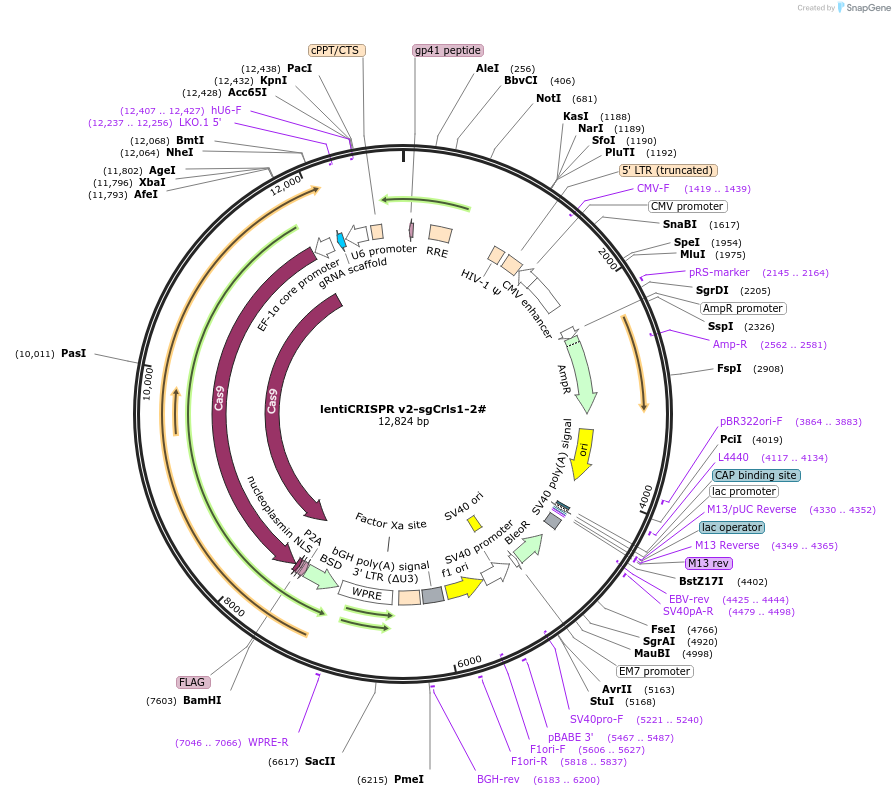
lentiCRISPR v2-sgCrls1-2#
Plasmid#234766Purposeknockout CrlsDepositorInsertCrls1 (Crls1 Mouse)
UseLentiviralAvailable SinceMarch 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
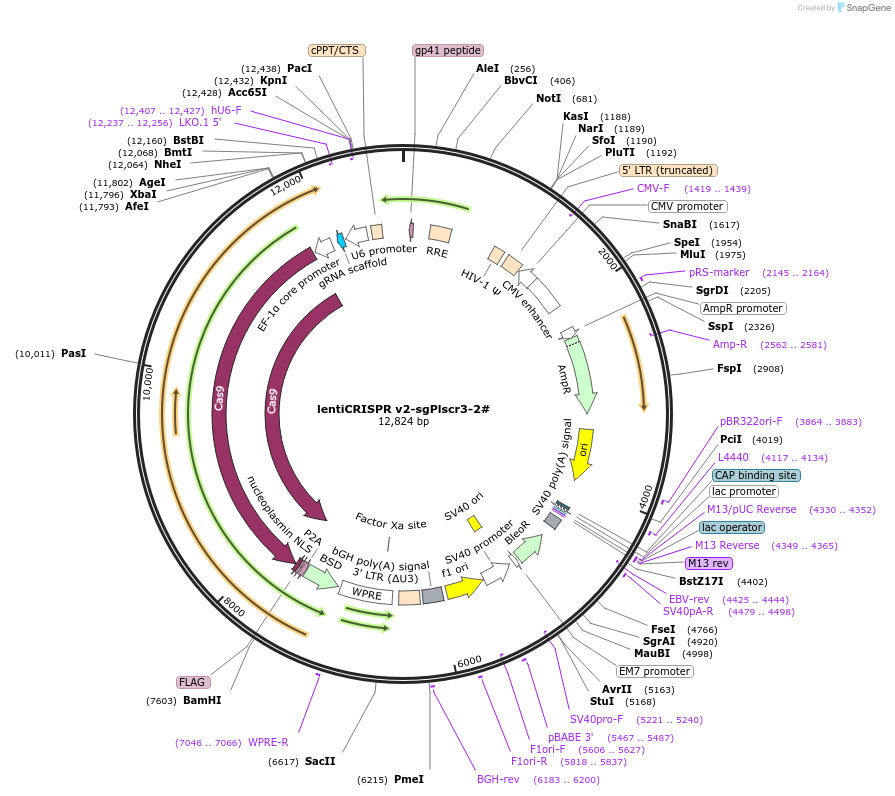
lentiCRISPR v2-sgPlscr3-2#
Plasmid#234772Purposeknockout Plscr3DepositorInsertPlscr3 (Plscr3 Mouse)
UseLentiviralAvailable SinceMarch 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
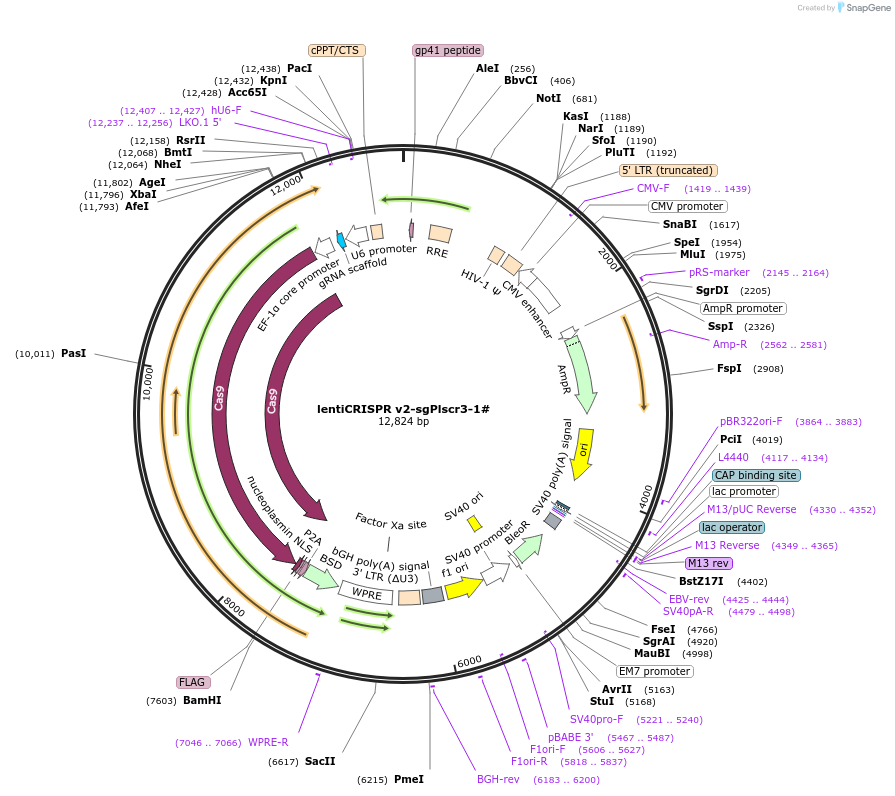
lentiCRISPR v2-sgPlscr3-1#
Plasmid#234767Purposeknockout Plscr3DepositorInsertPlscr3 (Plscr3 Mouse)
UseLentiviralAvailable SinceMarch 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
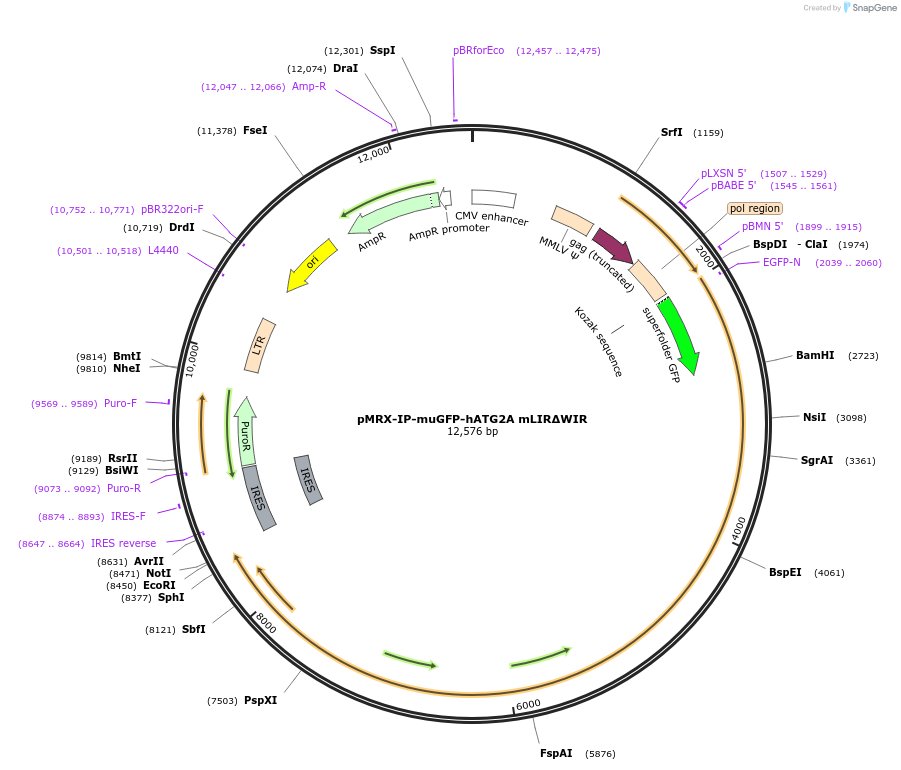
pMRX-IP-muGFP-hATG2A mLIRΔWIR
Plasmid#215508PurposeExpresses GFP tagged human ATG2A which has the mutation in LC3 interacting region and lacks WIPI interacting region.DepositorInsertAutophagy related 2A (ATG2A Human)
UseRetroviralTagsmuGFPExpressionMammalianMutationP656R (natural variant with no functional relevan…Available SinceJune 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
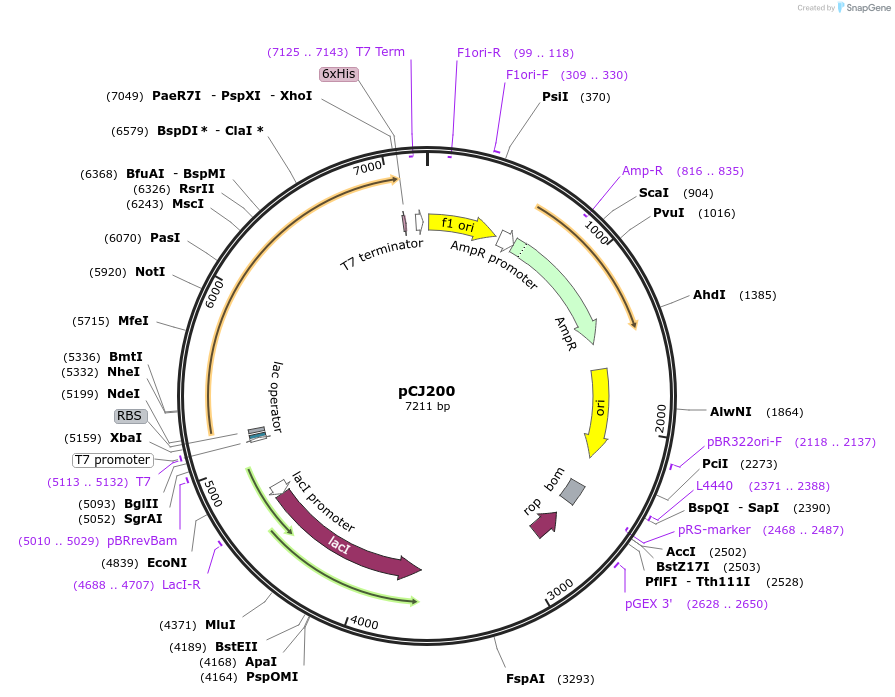
pCJ200
Plasmid#162673PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C and a sequence Tyr75:Phe104 to introduce a 6th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C and a sequence Y75-F104 to introduce a 6th…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
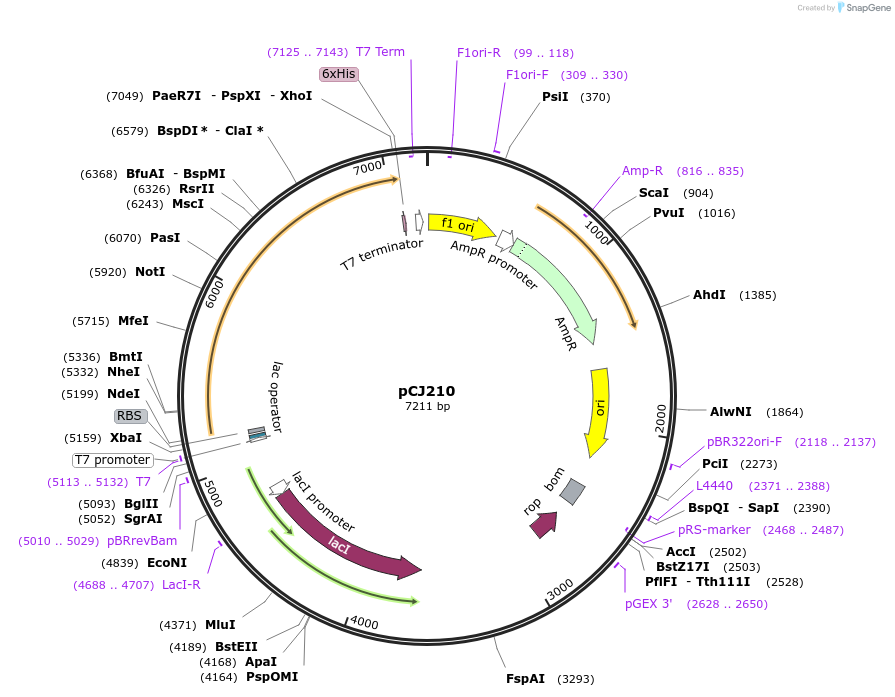
pCJ210
Plasmid#162683PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C, G489C, S530C, and a sequence Tyr75:Phe104 to introduce a 6th and 7th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C, G489C, S530C, and a sequence Y75-F104 to i…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
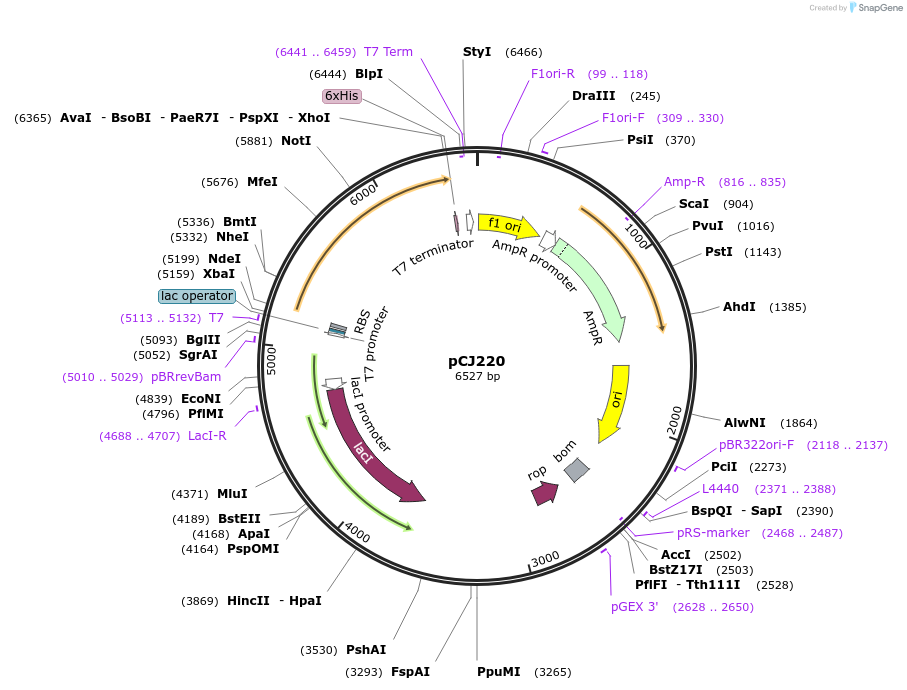
pCJ220
Plasmid#162686PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224W and C529S mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) with lid deletion and C224W and C529S mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224W, C529S; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
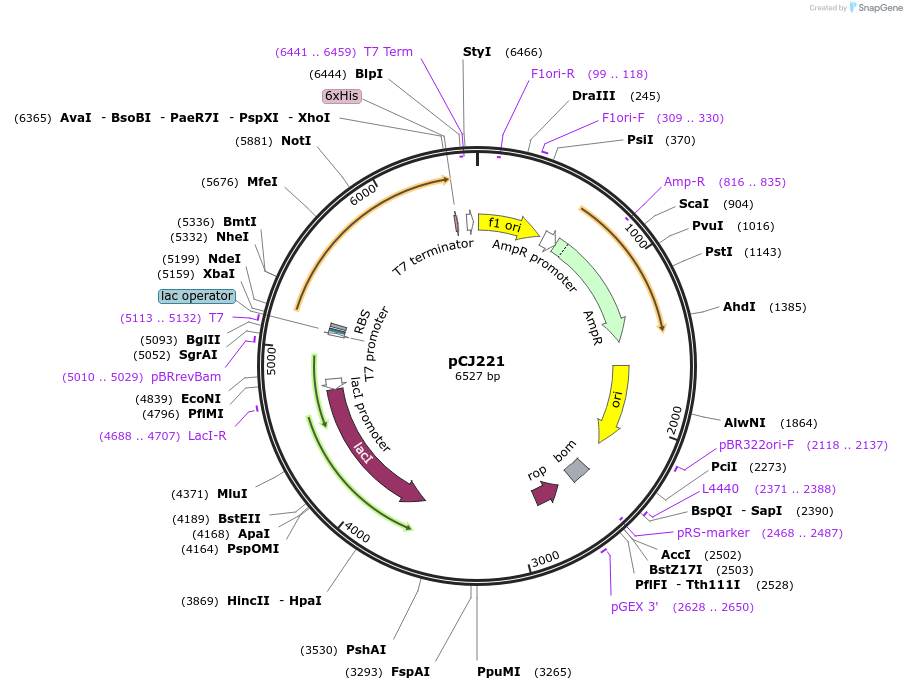
pCJ221
Plasmid#162687PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224H and C529F mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with lid deletion and C224H and C529F mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224H, C529F; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
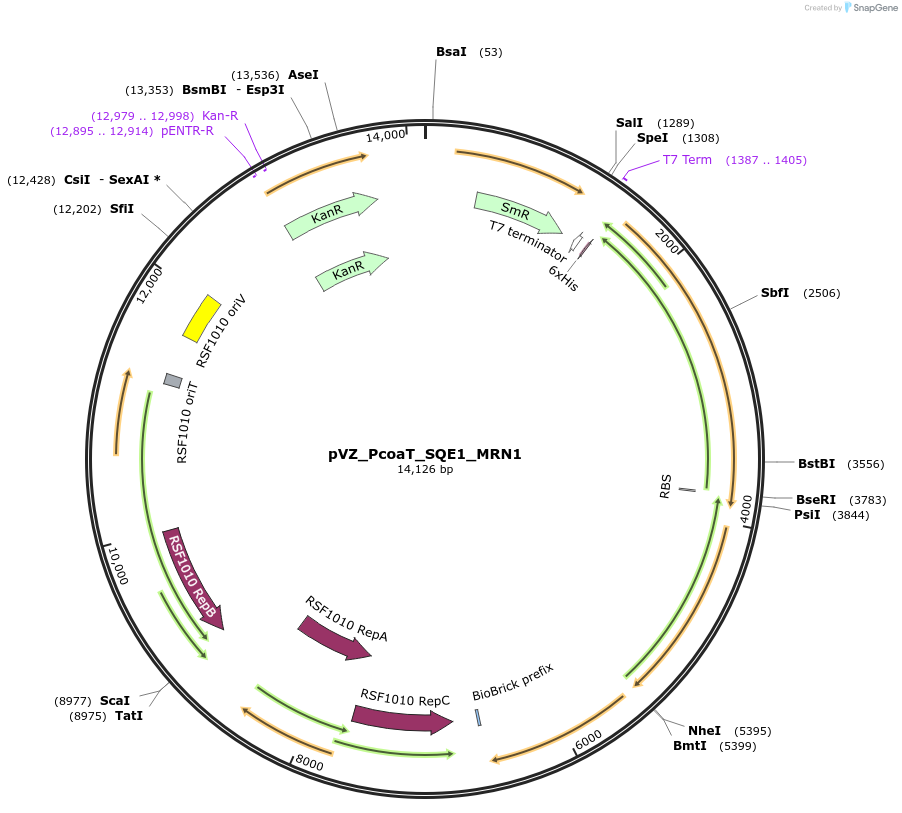
pVZ_PcoaT_SQE1_MRN1
Plasmid#103858PurposeHeterologous, cobalt-inducible expression of SQE1 and MRN1 from A. thaliana in Synechocystis sp. PCC6803DepositorInsertmarneral synthase 1 (MRN1 Mustard Weed)
UseSynthetic BiologyExpressionBacterialMutationinserted genes are codon optimized2nd amino acid …PromoterPcoaT (from Synechocystis sp. PCC 6803)Available SinceFeb. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
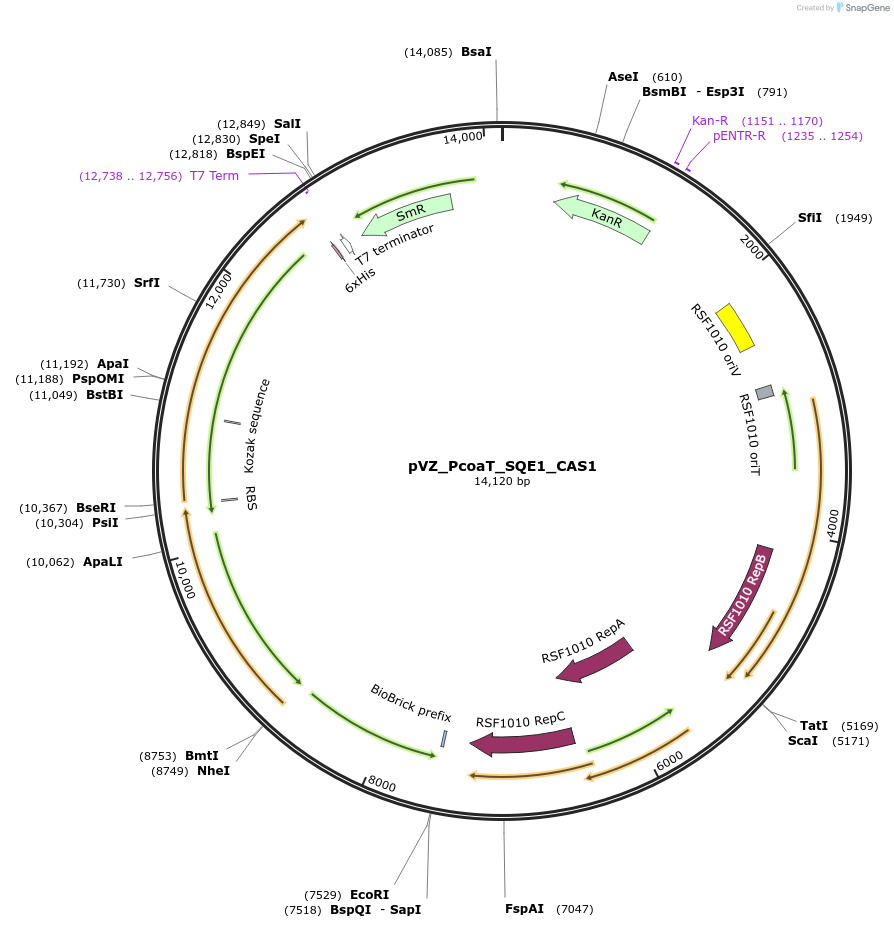
pVZ_PcoaT_SQE1_CAS1
Plasmid#103856PurposeHeterologous, cobalt-inducible expression of SQE1 and CAS1 from A. thaliana in Synechocystis sp. PCC6803DepositorInsertcycloartenol synthase 1 (CAS1 Mustard Weed)
UseSynthetic BiologyExpressionBacterialMutationinserted genes are codon optimized. 2nd amino aci…PromoterPcoaT (from Synechocystis sp. PCC 6803)Available SinceJan. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
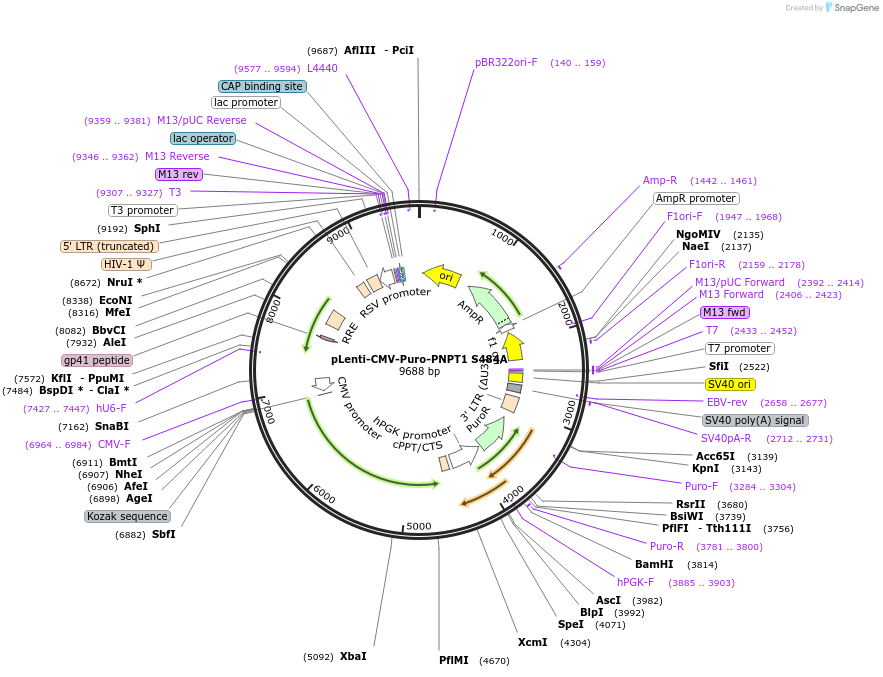
pLenti-CMV-Puro-PNPT1 S484A
Plasmid#223316PurposeLentiviral vector expressing human PNPT1 S484A variantDepositorInsertPolyribonucleotide Nucleotidyltransferase 1 (PNPT1 Human)
UseLentiviralExpressionMammalianMutationChanged serine 484 to alanineAvailable SinceOct. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
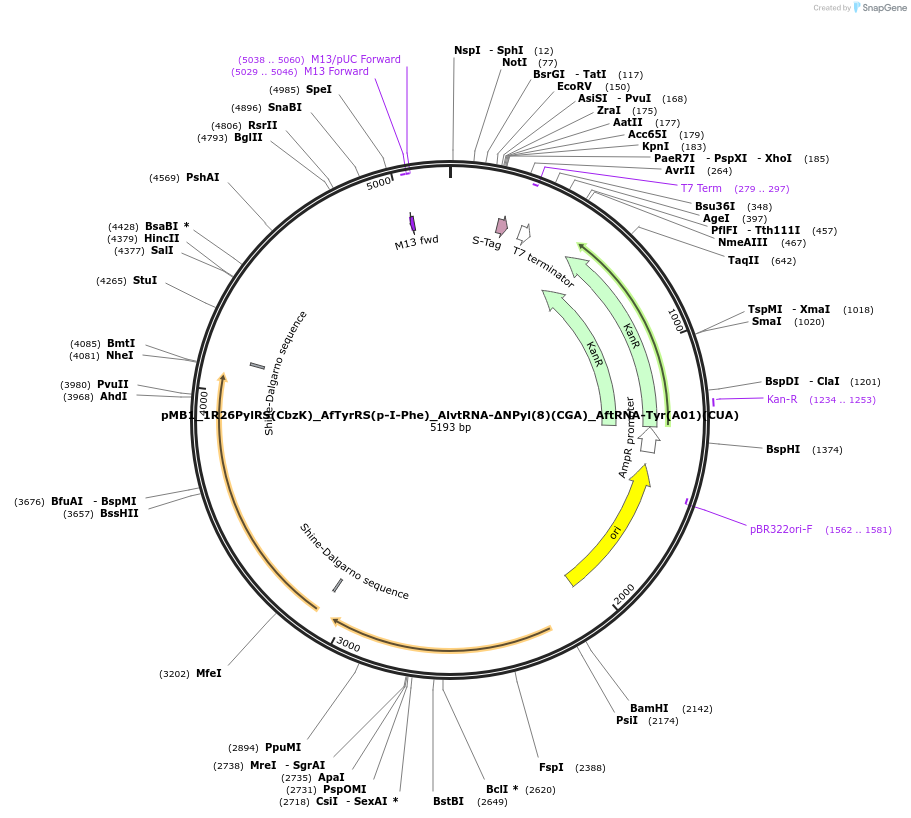
pMB1_1R26PylRS(CbzK)_AfTyrRS(p-I-Phe)_AlvtRNA-ΔNPyl(8)(CGA)_AftRNA-Tyr(A01)(CUA)
Plasmid#174516PurposeRecoded double aaRS/tRNA plasmid with 1R26PylRS/AlvtRNA-ΔNPyl(8)(CGA) & AfTyrRS/AftRNA-Tyr(A01)(CUA) to incorporate CbzK into the TCG codon & p-I-Phe into the TAG codon.DepositorInsertsPylS_1R26 - CBZK
Af_TyrRS - p-I-Phe
Ma tRNA pylT(8) with CGA anticodon
A.fulgidus tRNA with CUA anticodon
ExpressionBacterialMutationCUA anticodon, M.alvus Pyl tRNA with CGA anticodo…PromoterGlnS and lppAvailable SinceOct. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
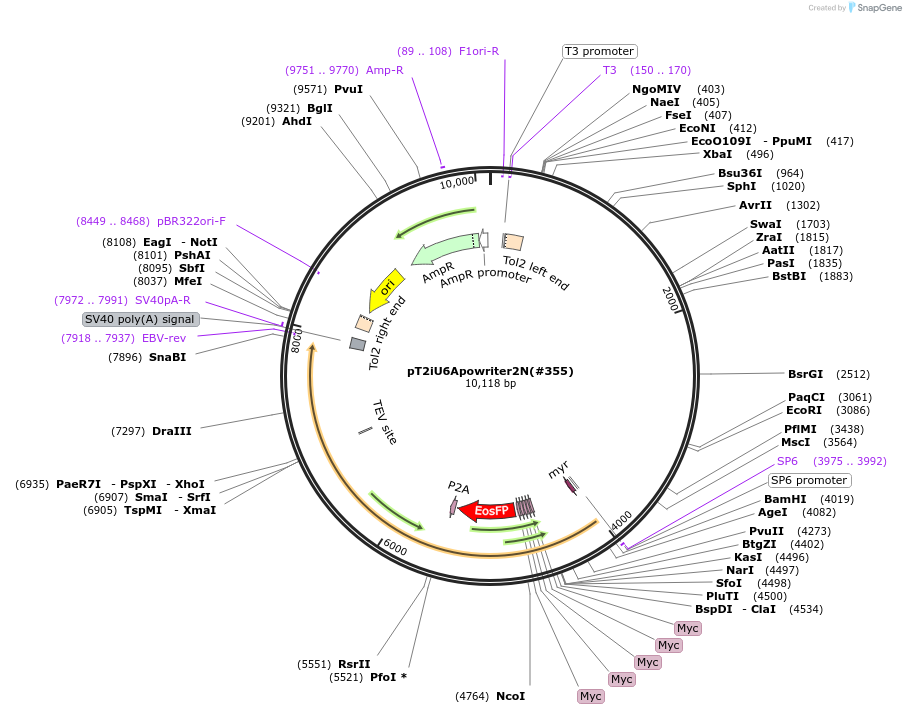
pT2iU6Apowriter2N(#355)
Plasmid#184106Purposecyclofen-inducible apoptosis activation and imaging (tEosFP) by zebrafish transient transgenesis or mRNA synthesisDepositorInsertmyr-DIAP1-5myc-tEosFP-nls'-P2A-Casp9-ERT2
UseZebrafish transient transgenesis + in vitro trans…Tagsinternal tag: 5mycMutationDIAP1: deleted 1-2, N19G, N20V, deleted 146-438; …PromoterZf-Ubi+SP6Available SinceJune 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
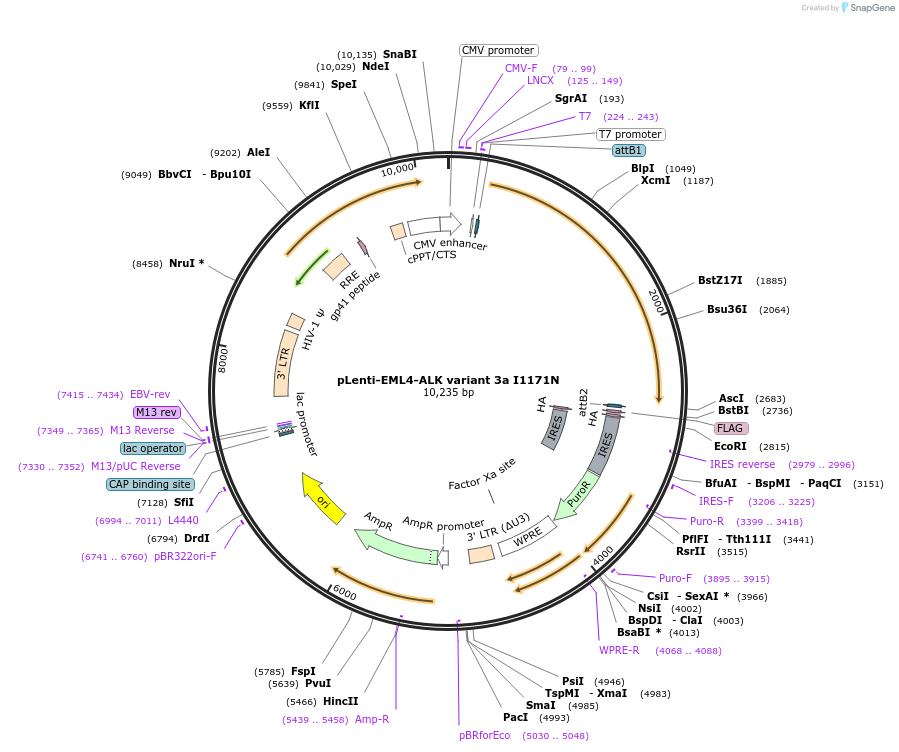
pLenti-EML4-ALK variant 3a I1171N
Plasmid#183832PurposeExpress EML4-ALK fusion variant 3a (E6a;A20) harboring ALK I1171N mutation in mammalian cellsDepositorUseLentiviralTagsThe fusion protein of EML4 exon 1-6a and ALK exon…ExpressionMammalianMutationI1171NPromoterCMVAvailable SinceJune 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
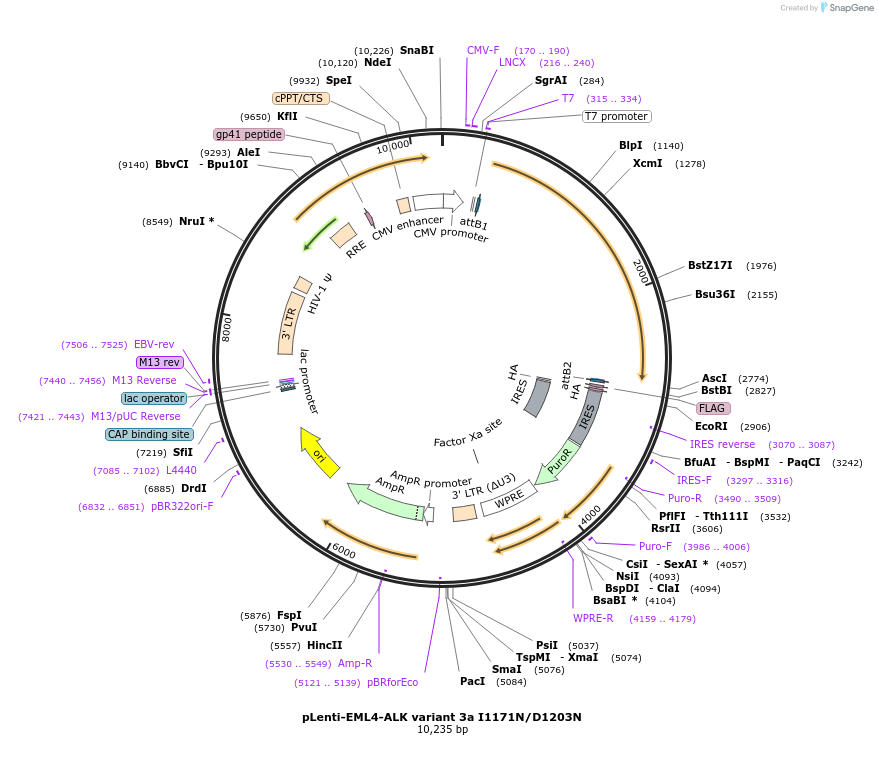
pLenti-EML4-ALK variant 3a I1171N/D1203N
Plasmid#183833PurposeExpress EML4-ALK fusion variant 3a (E6a;A20) harboring ALK I1171N/D1203N mutation in mammalian cellsDepositorUseLentiviralTagsThe fusion protein of EML4 exon 1-6a and ALK exon…ExpressionMammalianMutationI1171N/D1203NPromoterCMVAvailable SinceJune 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
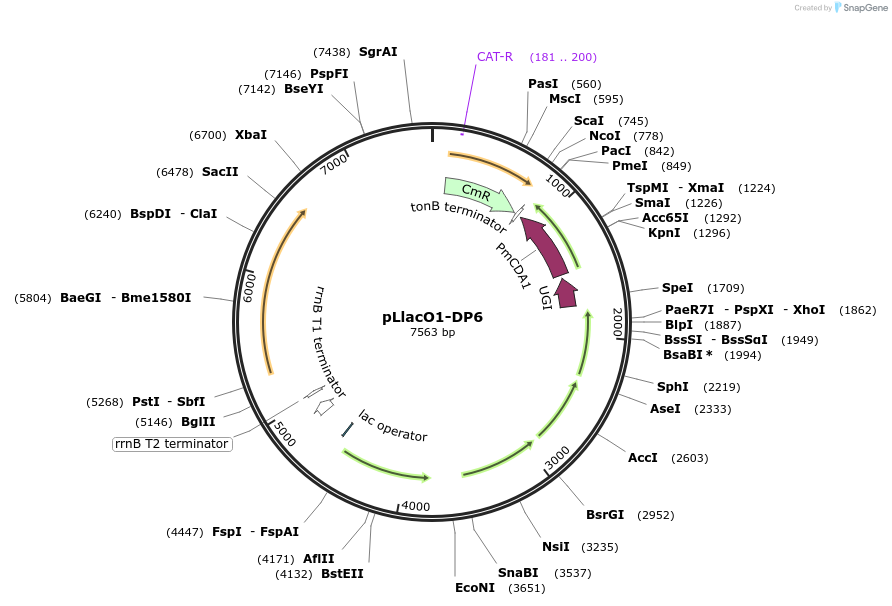
pLlacO1-DP6
Plasmid#242138PurposeAraC compatible Mutagenesis Circuit (drift plasmid)DepositorInsertGene III
ExpressionBacterialAvailable SinceSept. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
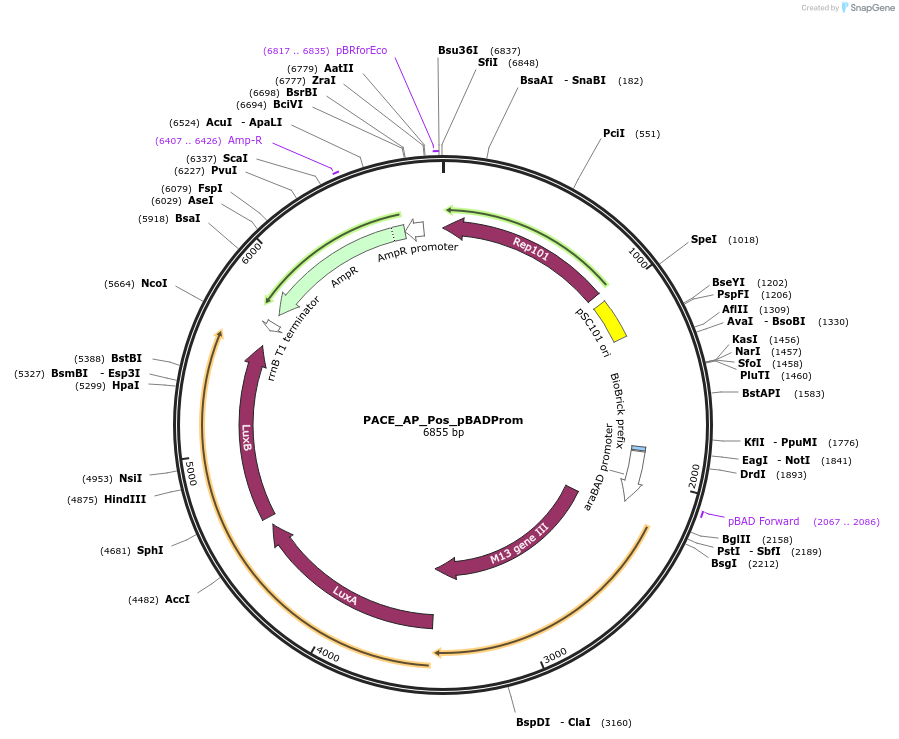
PACE_AP_Pos_pBADProm
Plasmid#242139PurposepBAD-sd8-GIII Cassette; AP Pos Circuit for AraC phage-assisted evolutionDepositorInsertGene III
ExpressionBacterialAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
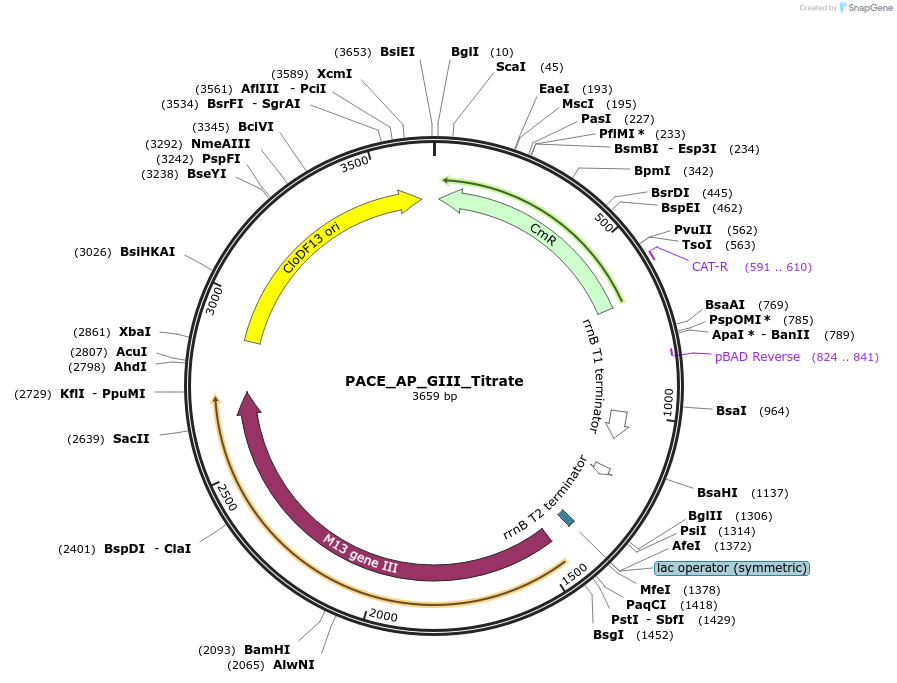
PACE_AP_GIII_Titrate
Plasmid#242140PurposePSP-LacO Cassette; AP Prop Circuit for AraC phage-assisted evolutionDepositorInsertGene III
ExpressionBacterialAvailable SinceAug. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
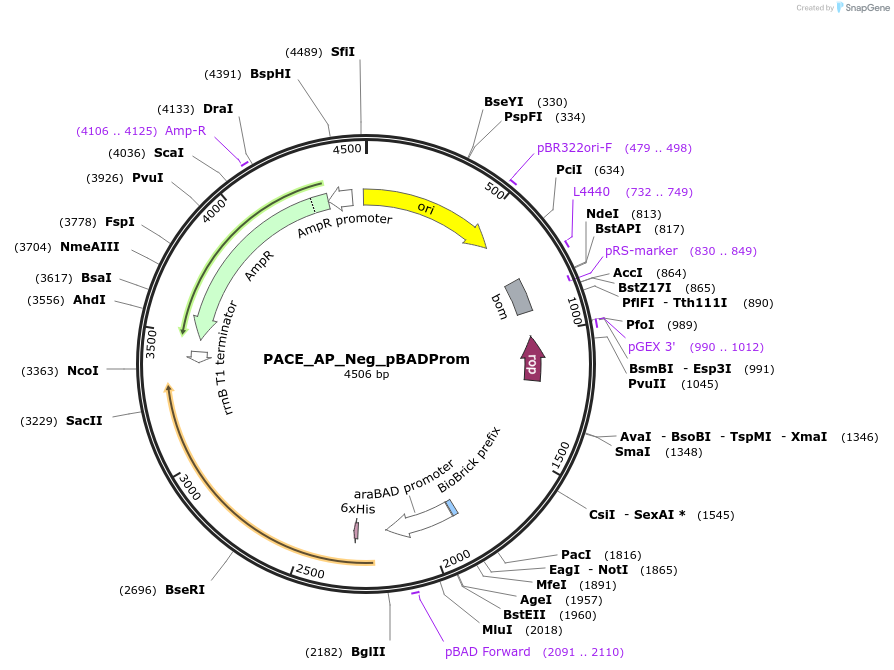
PACE_AP_Neg_pBADProm
Plasmid#242141PurposepBAD-SD8-GIII Neg; AP Neg Circuit for AraC phage-assisted evolutionDepositorInsertGene III Negative
ExpressionBacterialAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
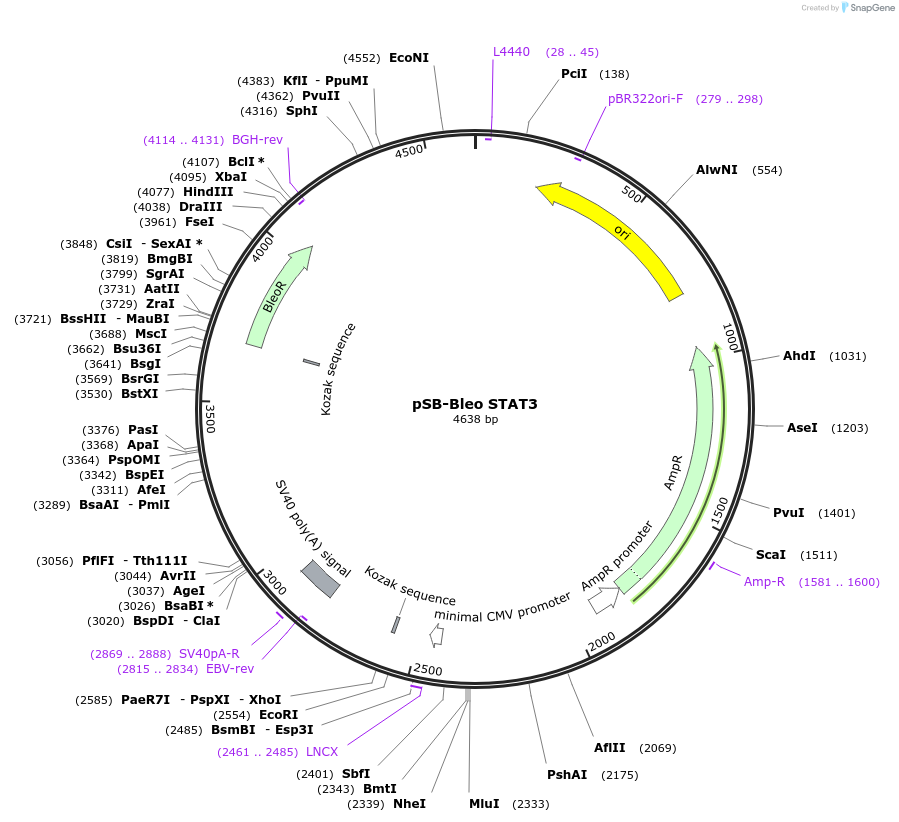
pSB-Bleo STAT3
Plasmid#139184PurposeEmpty backbone encoding constitutive bleomycin resistance with an inducible STAT3 promoter to allow for an insert in a Sleeping Beauty backbone.DepositorTypeEmpty backboneUseTransposonExpressionMammalianPromoterSTAT3Available SinceJuly 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
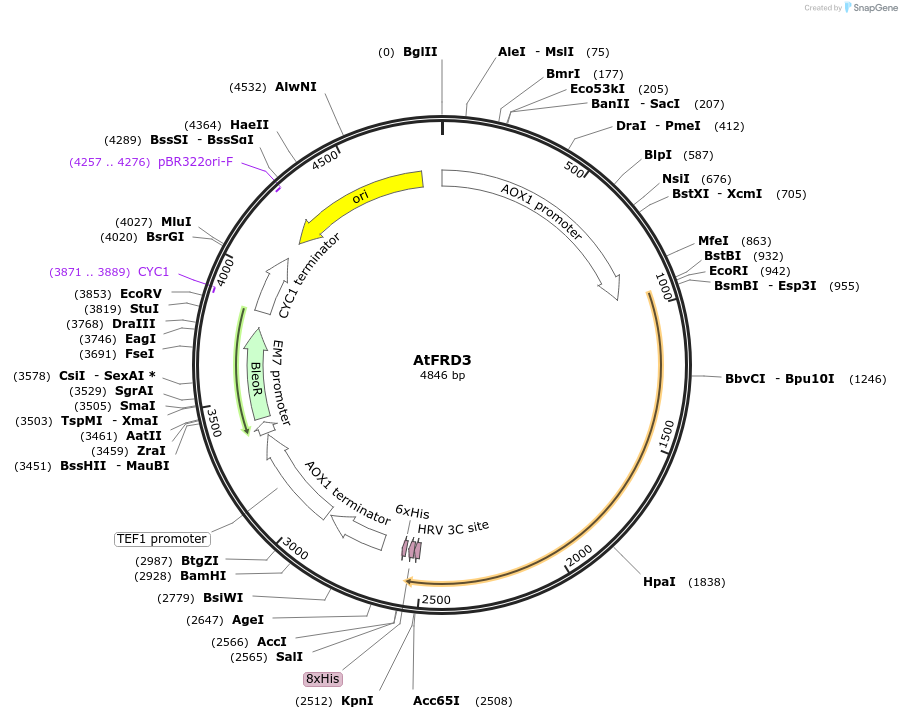
AtFRD3
Plasmid#117086Purposeprotein expression in yeastDepositorInsertAtFRD3
TagsHisExpressionYeastPromoterAOXAvailable SinceNov. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
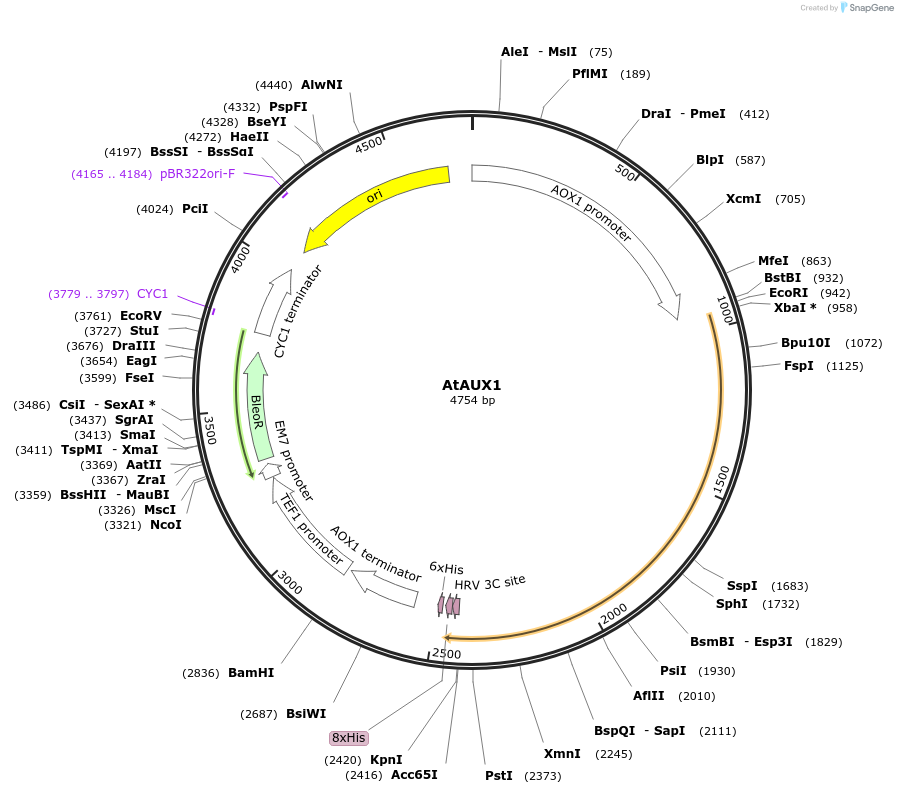
AtAUX1
Plasmid#117087Purposeprotein expression in yeastDepositorInsertAtAUX1
TagsHisExpressionYeastPromoterAOXAvailable SinceNov. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
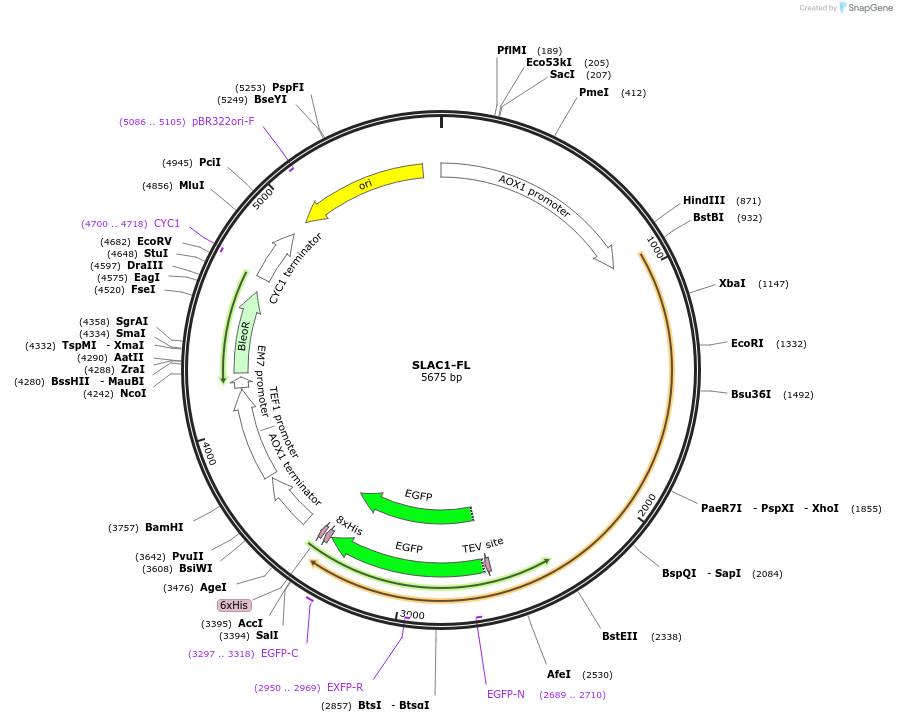
SLAC1-FL
Plasmid#117100Purposeprotein expression in yeastDepositorInsertSLAC1-FL
TagsHisExpressionYeastPromoterAOXAvailable SinceNov. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
pLVU/myc
Plasmid#24179DepositorInsertChloramphenicol resistance gene (CmR) - ccdB gene
UseLentiviral; GatewayTagsMycExpressionMammalianAvailable SinceNov. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
-
-
-
-
-
-
-
-
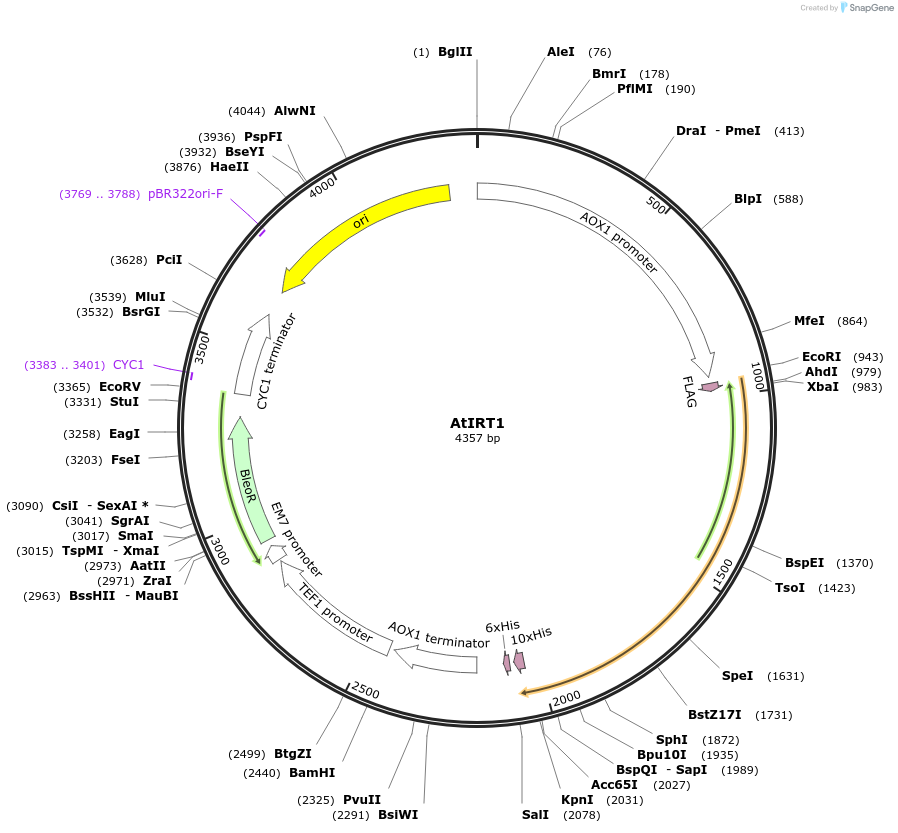
AtIRT1
Plasmid#117884Purposeprotein expressionDepositorAvailable SinceDec. 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
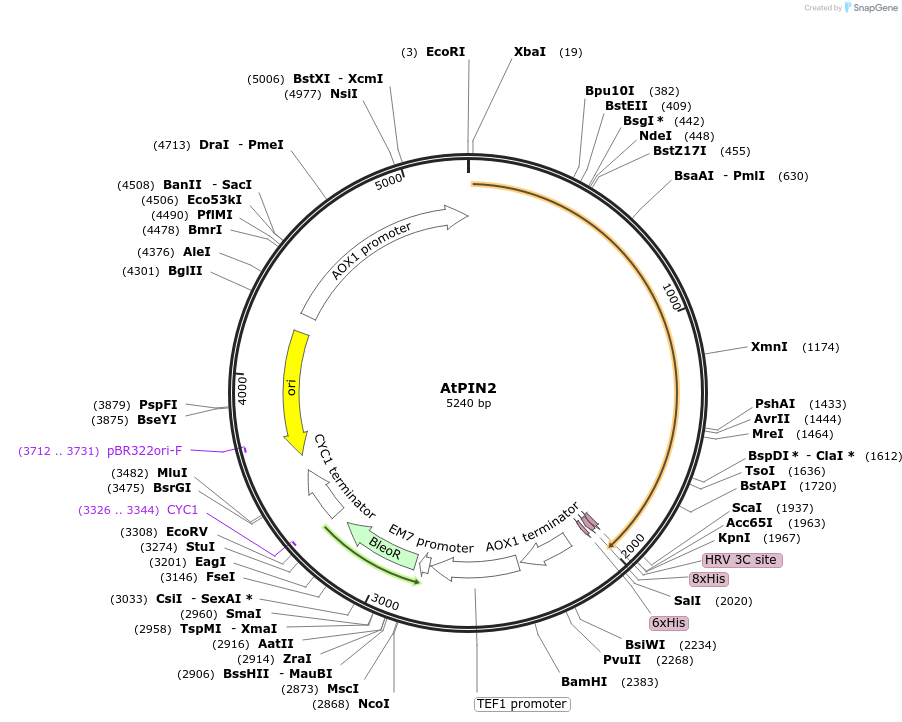
AtPIN2
Plasmid#117089Purposeprotein expression in yeastDepositorInsertAtPIN2
TagsHisExpressionYeastPromoterAOXAvailable SinceNov. 6, 2018AvailabilityAcademic Institutions and Nonprofits only