We narrowed to 89,717 results for: MAL
-
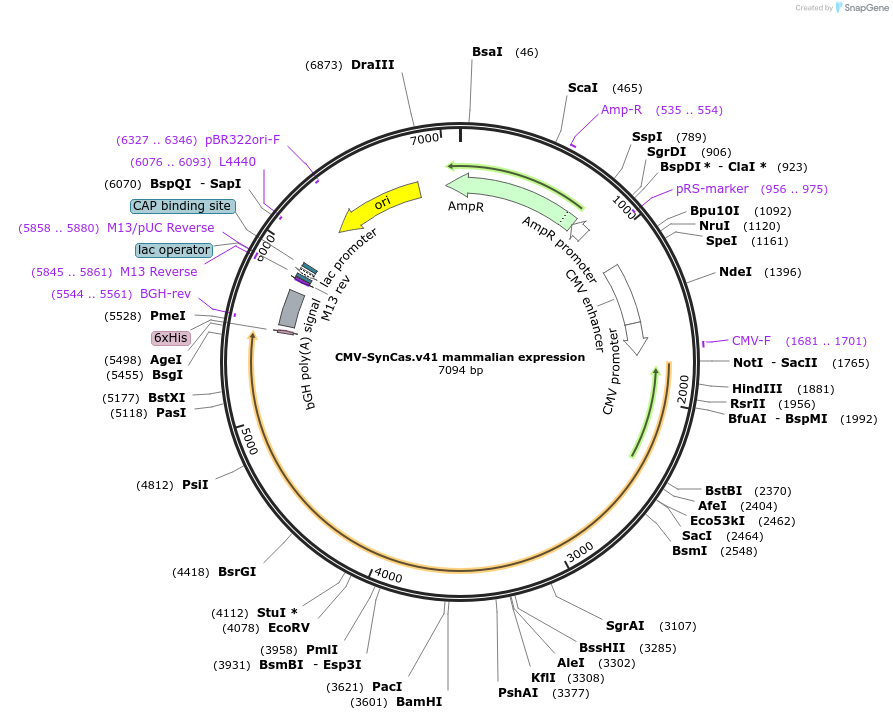
Plasmid#208593PurposeExpression of SynCas.v41 for analysis of RNA knockdownDepositorInsertSynCas.v41
ExpressionMammalianPromoterCMVAvailable SinceApril 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
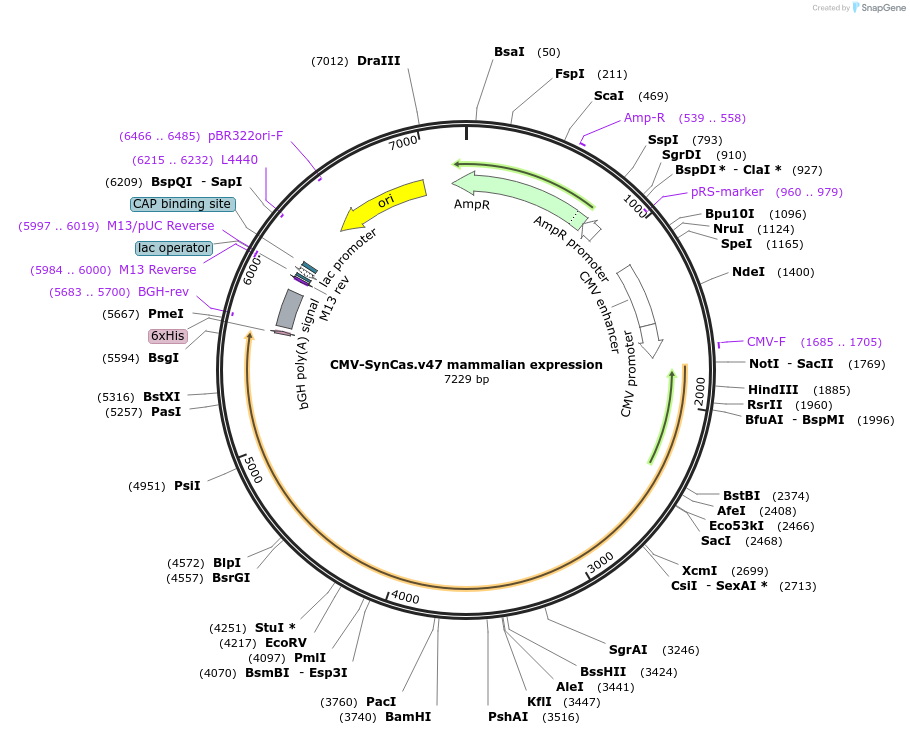
CMV-SynCas.v47 mammalian expression
Plasmid#209003PurposeExpression of SynCas.v47 for analysis of RNA knockdownDepositorInsertSynCas.v47
ExpressionMammalianPromoterCMVAvailable SinceApril 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
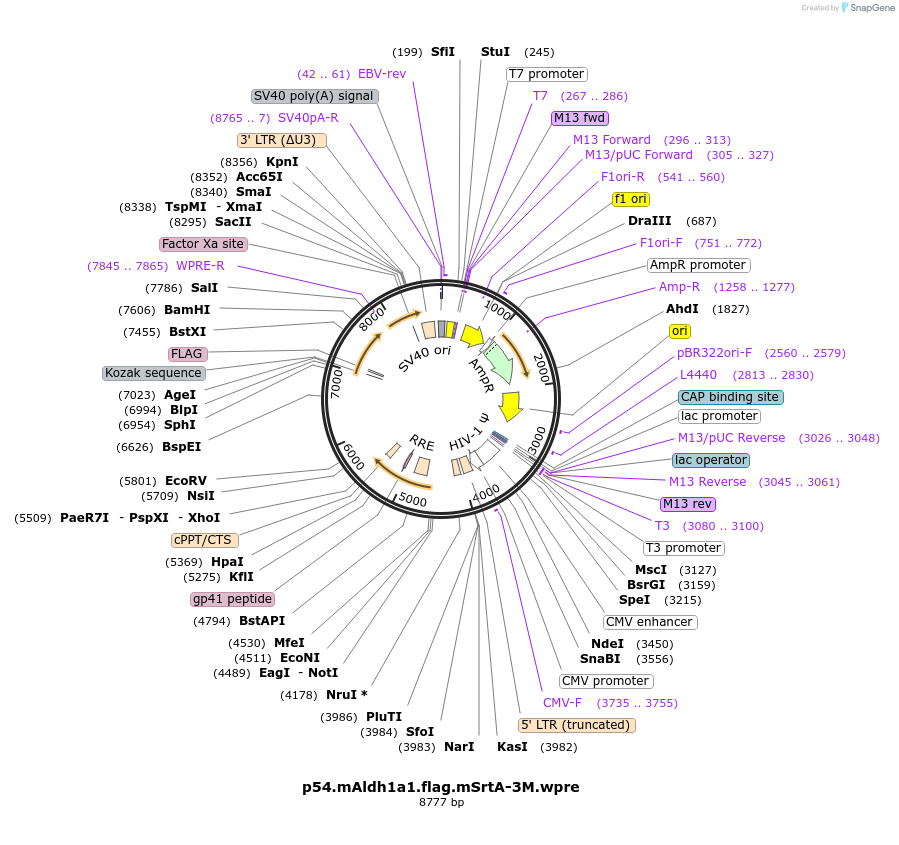
p54.mAldh1a1.flag.mSrtA-3M.wpre
Plasmid#226771Purposesecretory SortaseA in p52 (mAldh1a backbone)DepositorInsertsortase A (srtA )
ExpressionMammalianAvailable SinceNov. 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
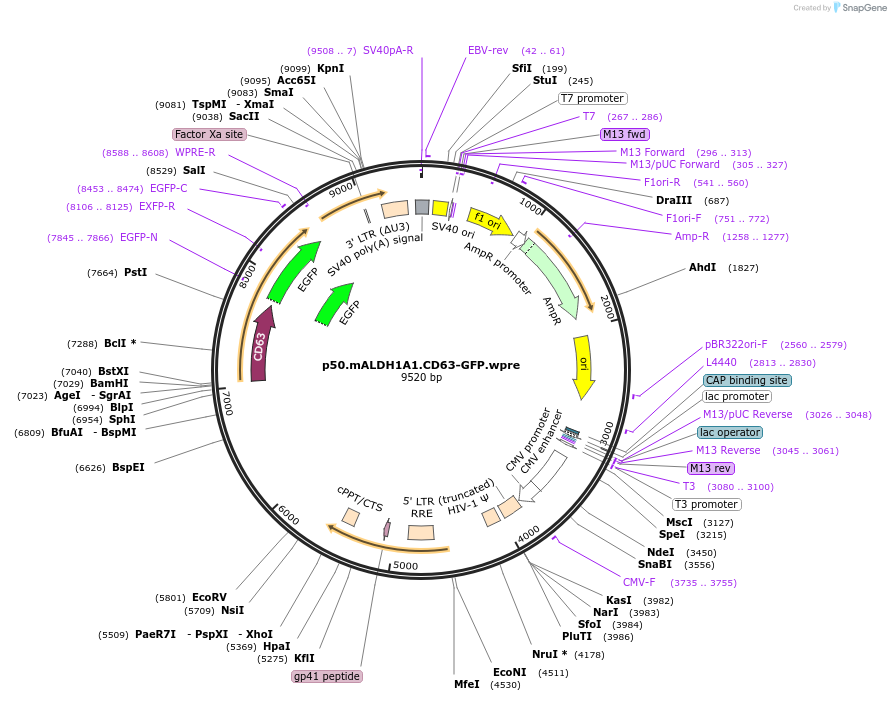
p50.mALDH1A1.CD63-GFP.wpre
Plasmid#226770PurposeCD63-GFP in p52 (mAldh1a backbone)DepositorInsertCD63 (CD63 )
ExpressionMammalianAvailable SinceNov. 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
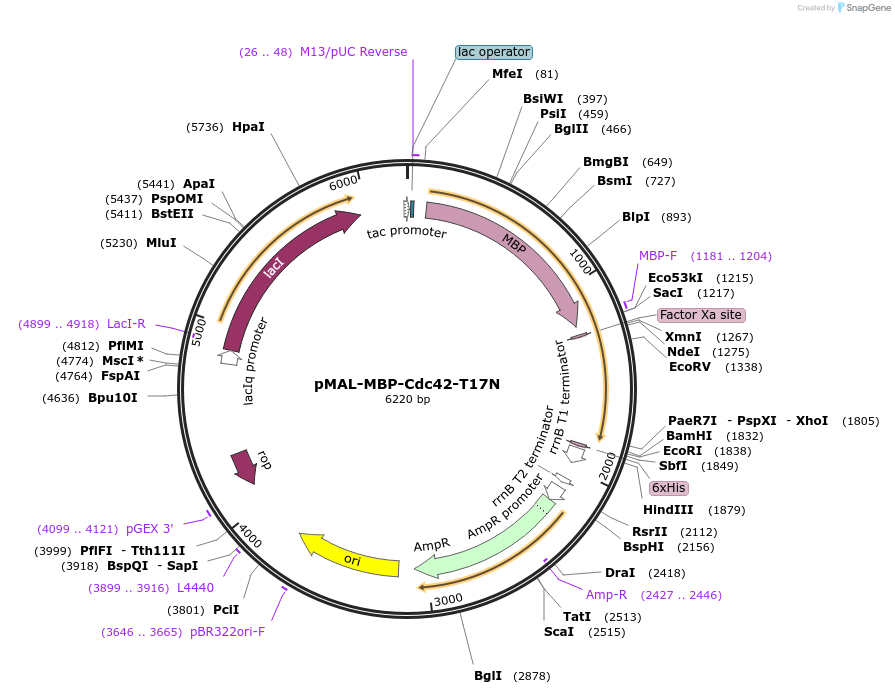
pMAL-MBP-Cdc42-T17N
Plasmid#214289PurposeExpress dominant negative Cdc42 fused with MBPDepositorInsertDominant negative Cdc42 (CDC42 Human)
TagsMaltose-binding protein (MBP)ExpressionBacterialMutationCdc42-T17NPromoterTacAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
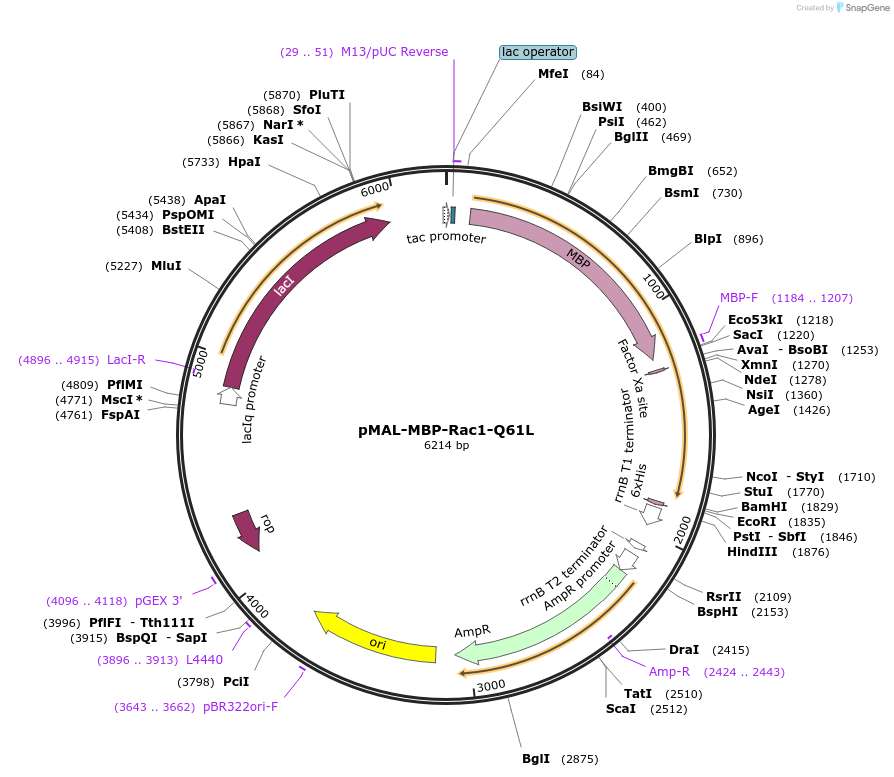
pMAL-MBP-Rac1-Q61L
Plasmid#214292PurposeExpress constitutively active Rac1 fused with MBPDepositorInsertConstitutively active Rac1 (RAC1 Human)
TagsMaltose-binding protein (MBP)ExpressionBacterialMutationRac1-Q61LPromoterTacAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
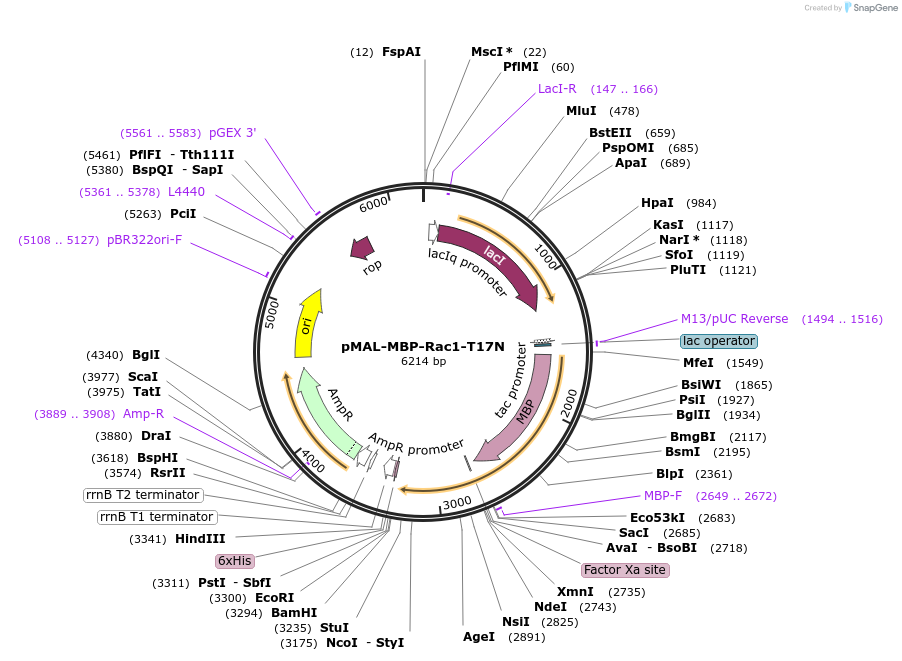
pMAL-MBP-Rac1-T17N
Plasmid#214293PurposeExpress dominant negative Rac1 fused with MBPDepositorInsertDominant negative Rac1 (RAC1 Human)
TagsMaltose-binding protein (MBP)ExpressionBacterialMutationRac1-T17NPromoterTacAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
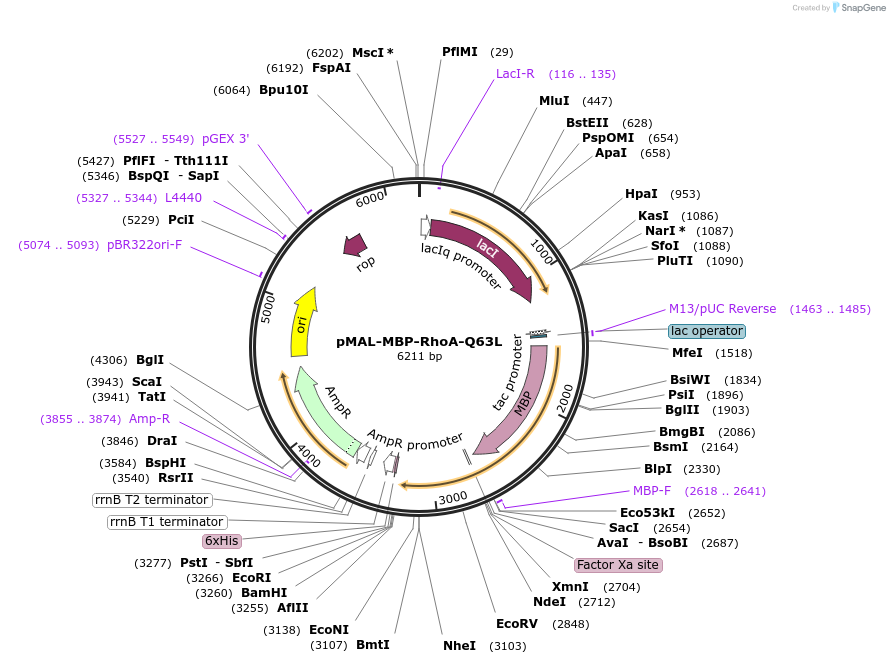
pMAL-MBP-RhoA-Q63L
Plasmid#214296PurposeExpress constitutively active RhoA fused with MBPDepositorInsertConstitutively active RhoA (RHOA Human)
TagsMaltose-binding protein (MBP)ExpressionBacterialMutationRhoA-Q63LPromoterTacAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
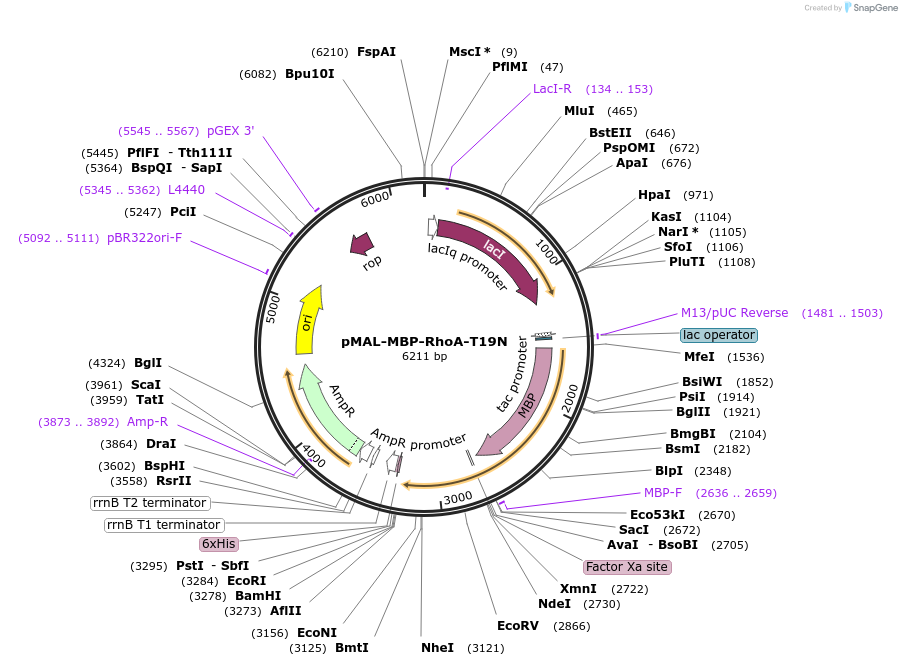
pMAL-MBP-RhoA-T19N
Plasmid#214297PurposeExpress dominant negative RhoA fused with MBPDepositorInsertDominant negative RhoA (RHOA Human)
TagsMaltose-binding protein (MBP)ExpressionBacterialMutationRhoA-T19NPromoterTacAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
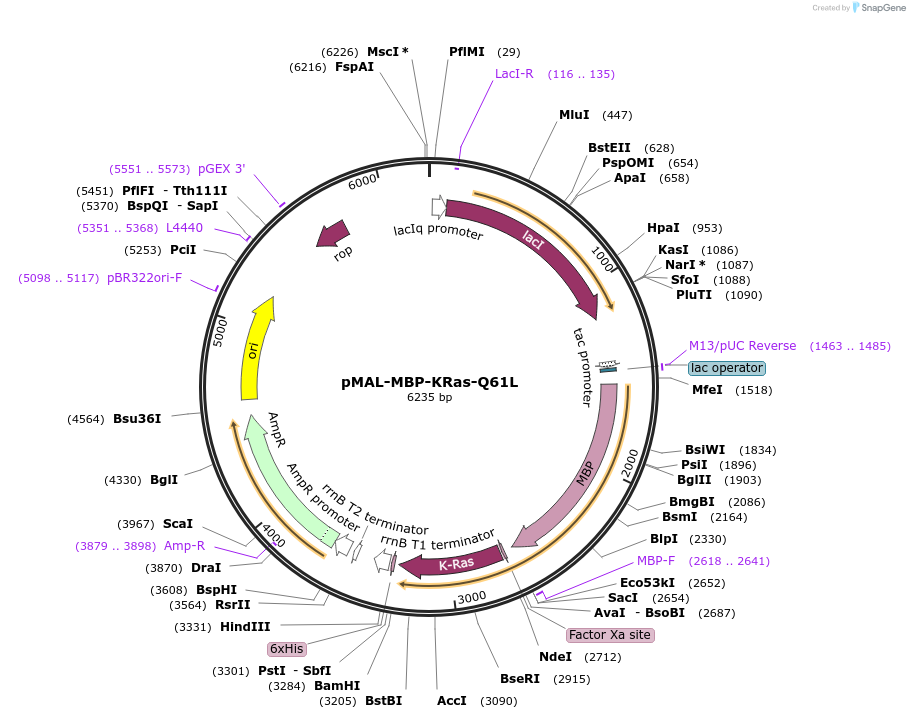
pMAL-MBP-KRas-Q61L
Plasmid#214300PurposeExpress constitutively active KRas fused with MBPDepositorInsertConstitutively active KRas (KRAS Human)
TagsMaltose-binding protein (MBP)ExpressionBacterialMutationKRas-Q61LPromoterTacAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
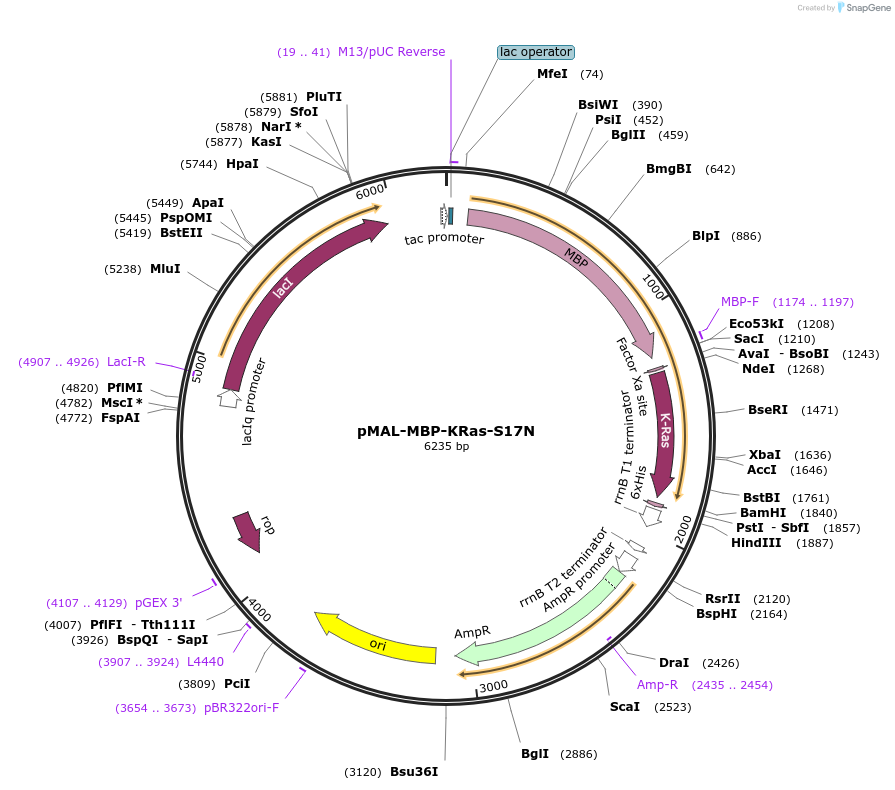
pMAL-MBP-KRas-S17N
Plasmid#214301PurposeExpress dominant negative KRas fused with MBPDepositorInsertDominant negative KRas (KRAS Human)
TagsMaltose-binding protein (MBP)ExpressionBacterialMutationKRas-S17NPromoterTacAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
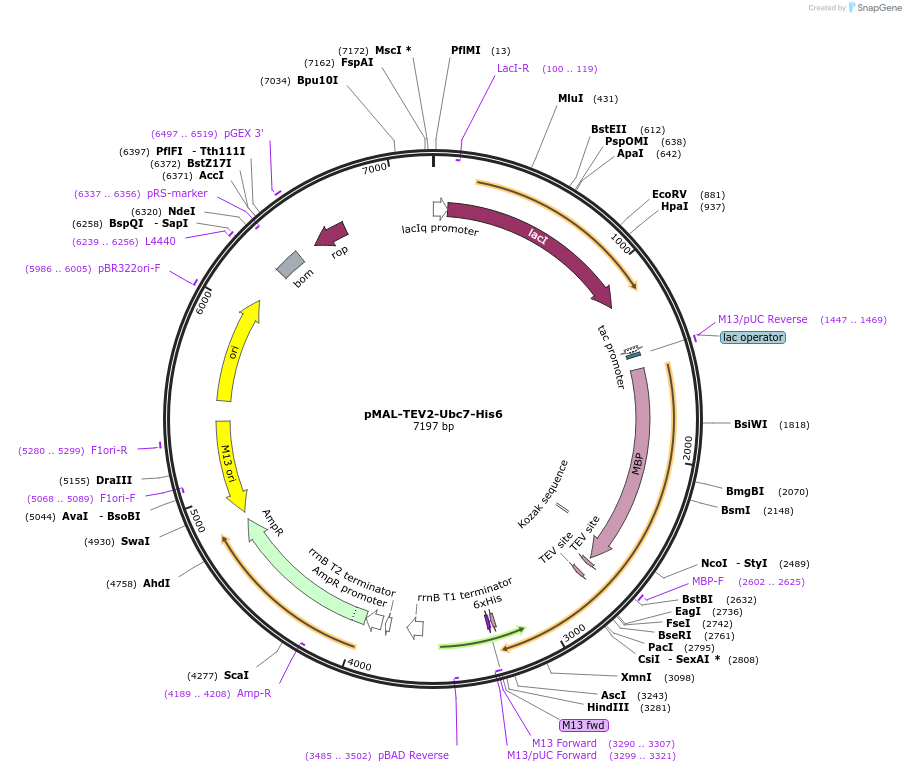
pMAL-TEV2-Ubc7-His6
Plasmid#212713PurposeBacterial expression of MBP-Ubc7-His6, MBP-tag cleavable with TEV-proteaseDepositorInsertUbc7
Tags6xHis and MBP-TEV2ExpressionBacterialAvailable SinceJan. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
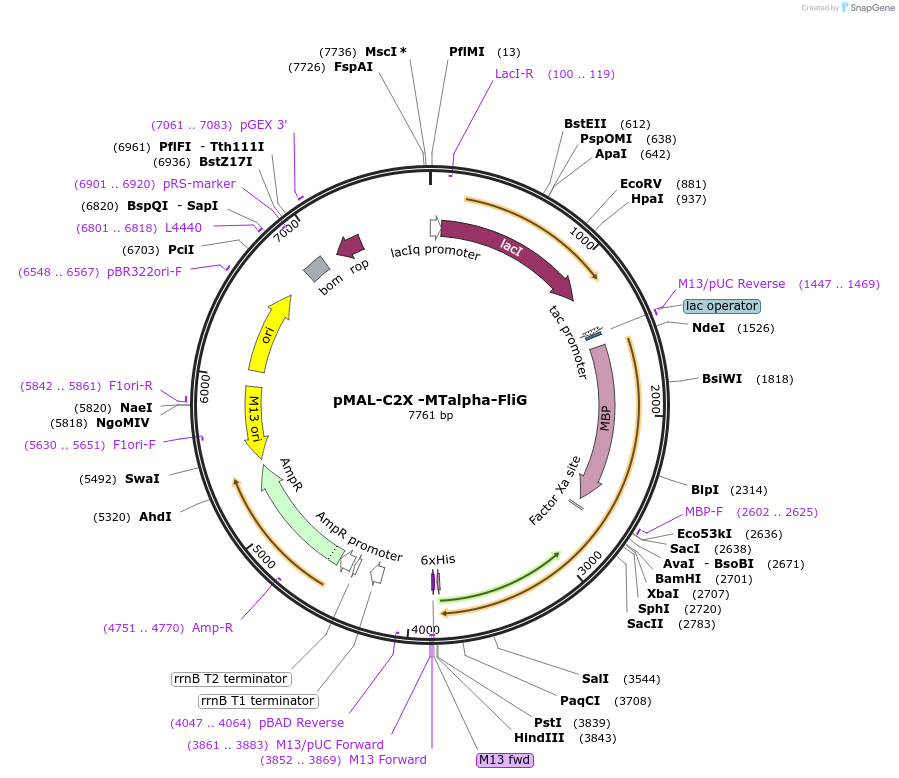
pMAL-C2X -MTalpha-FliG
Plasmid#154361PurposeBacterial expression of MtalphaDepositorInsertMtalpha
ExpressionBacterialAvailable SinceNov. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
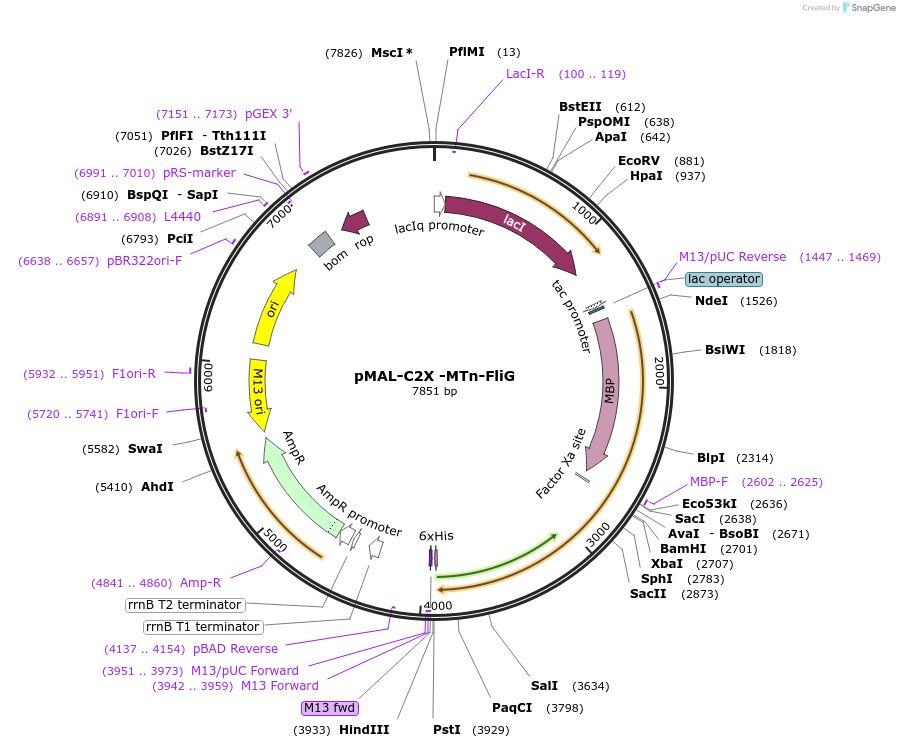
pMAL-C2X -MTn-FliG
Plasmid#154360PurposeBacterial expression of MTnDepositorInsertMTn
ExpressionBacterialAvailable SinceOct. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
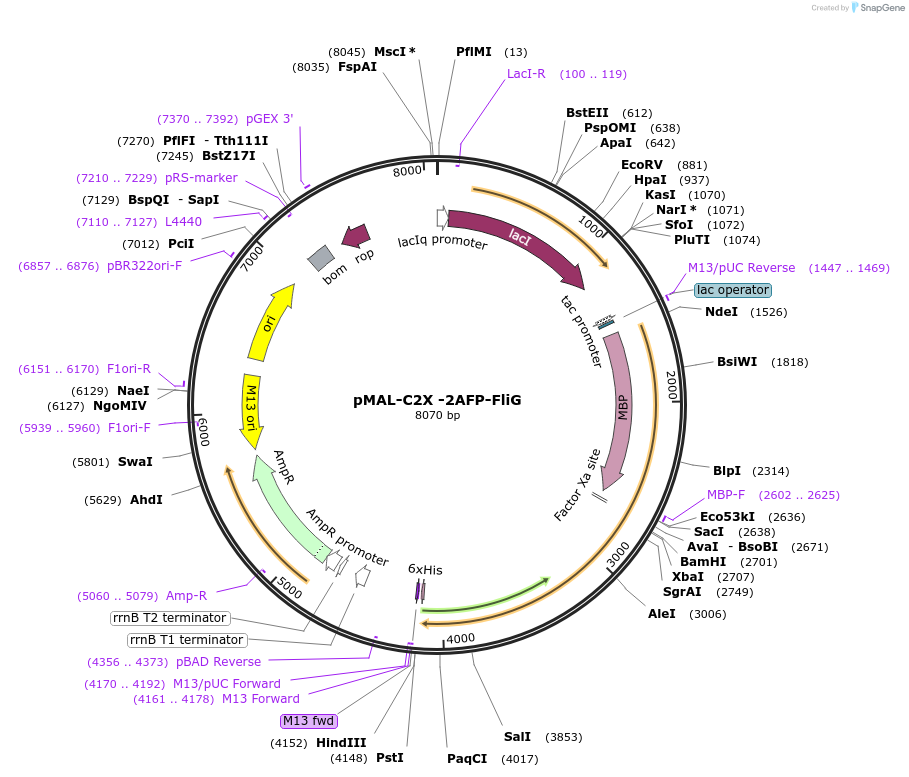
pMAL-C2X -2AFP-FliG
Plasmid#154362PurposeBacterial expression of 2AFPDepositorInsert2AFP
ExpressionBacterialAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
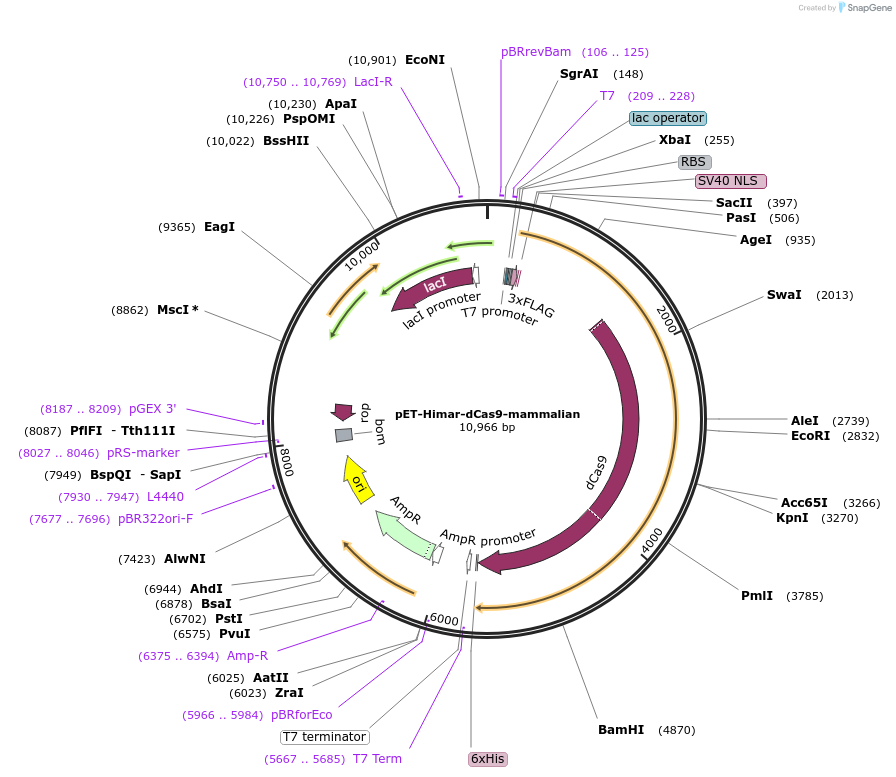
pET-Himar-dCas9-mammalian
Plasmid#138414PurposeProtein purification of Himar-dCas9 with 3x-FLAG, SV40 NLS, and 6x-His tagDepositorInsertHimar-dCas9
Tags3xFLAG, 6xHis, and SV40 NLSExpressionBacterialPromoterT7Available SinceMarch 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
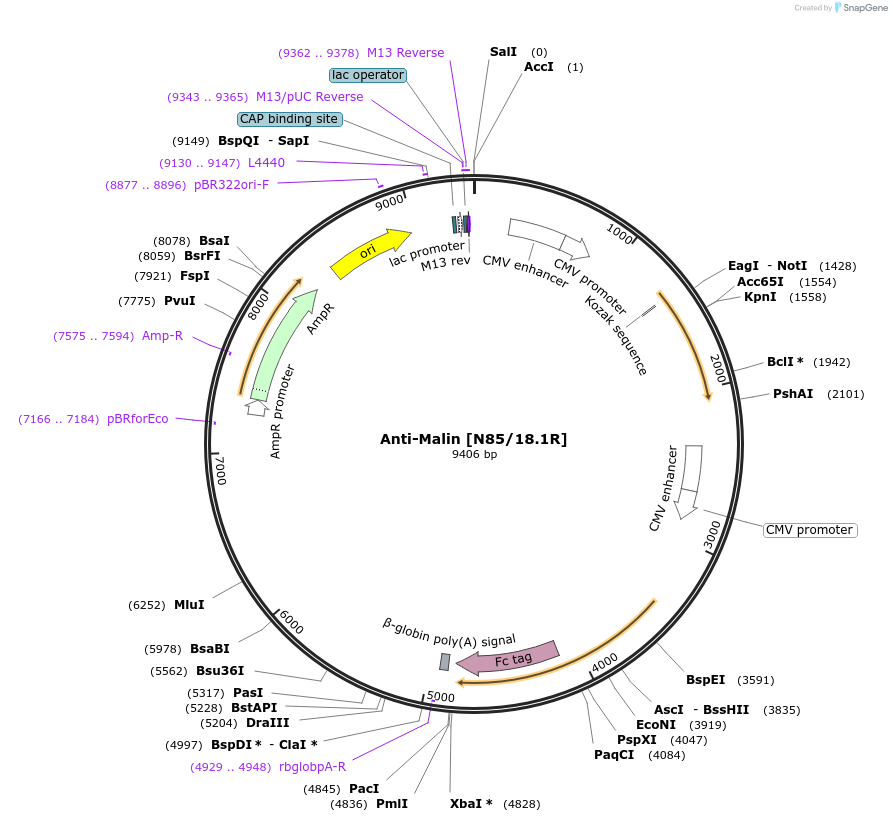
Anti-Malin [N85/18.1R]
Plasmid#114513PurposeMammalian Expression Plasmid of anti-Malin (Human). Derived from hybridoma N85/18.1.DepositorInsertanti-Malin (Homo sapiens) recombinant mouse monoclonal antibody (NHLRC1 Mouse)
ExpressionMammalianPromoterdual CMVAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
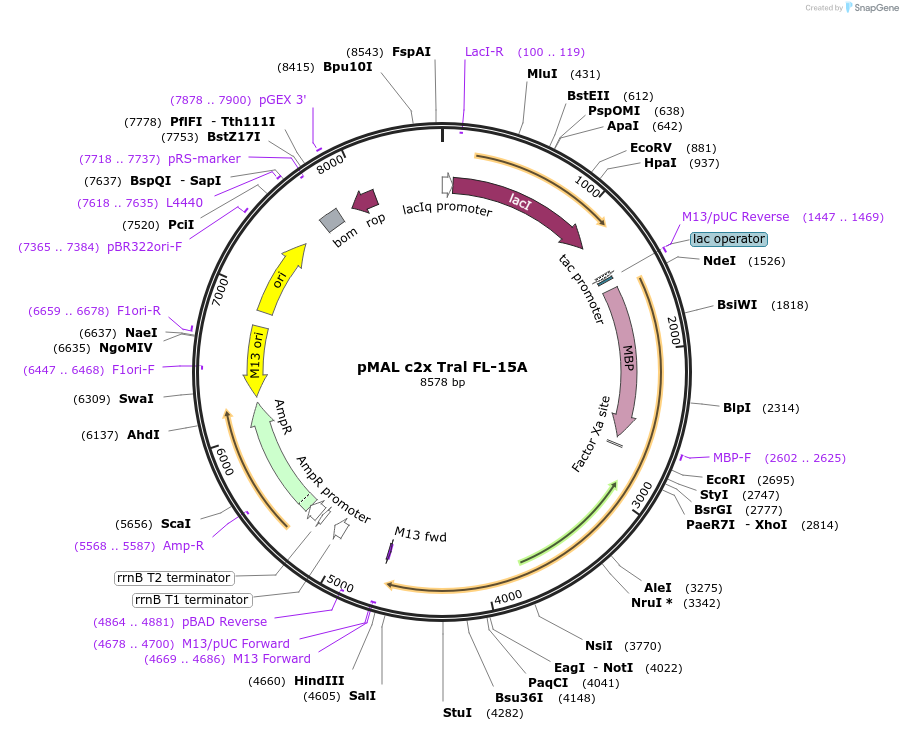
pMAL c2x Tral FL-15A
Plasmid#113011PurposeFor bacterial expression of MBP fusion of full-length Drosophila Tral with phosphomutations (15A)DepositorInsertfull length tral (tral Fly)
ExpressionBacterialMutation15 PNG phosphorylation sites mutated to AAvailable SinceAug. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
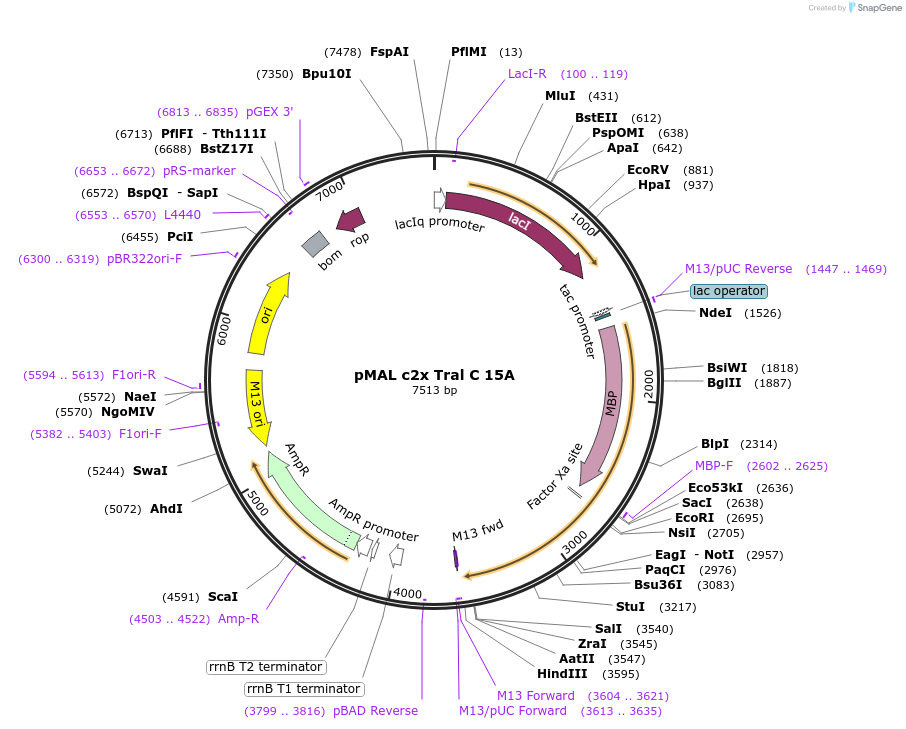
pMAL c2x Tral C 15A
Plasmid#113007PurposeFor bacterial expression of MBP fusion of C terminal region of Drosophila Tral phosphomutant (15A)DepositorInsertC terminus of tral (tral Fly)
ExpressionBacterialMutationPNG phosphorylation sites mutated to AAvailable SinceAug. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
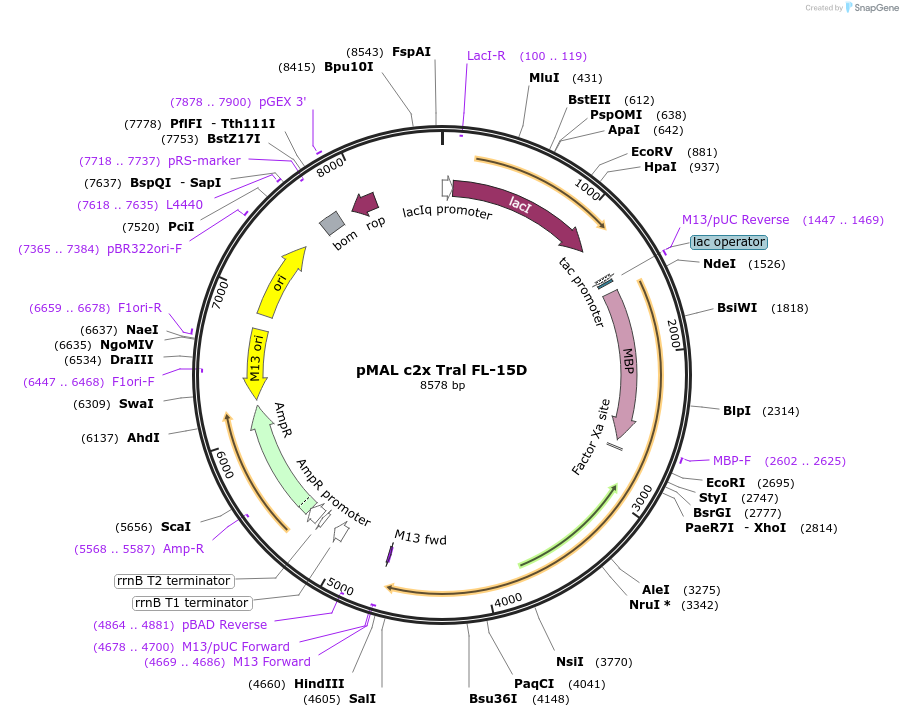
pMAL c2x Tral FL-15D
Plasmid#113010PurposeFor bacterial expression of MBP fusion of full-length Drosophila Tral with Phos-mimic mutationsDepositorInsertfull length tral (tral Fly)
ExpressionBacterialMutation15 PNG phosphorylation sites mutated to DAvailable SinceJuly 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
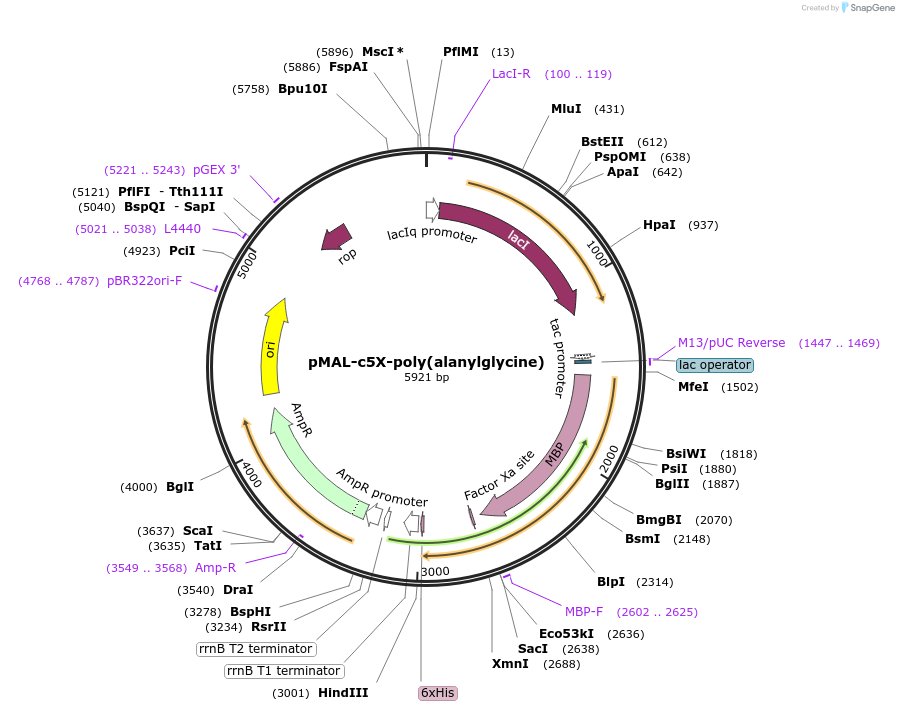
pMAL-c5X-poly(alanylglycine)
Plasmid#67003PurposeExpresses malE-poly(alanylglycine) in E. coliDepositorInsertmalE-poly(alanylglycine)
TagsHis6 and malE-Factor XaExpressionBacterialPromotertacAvailable SinceJan. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
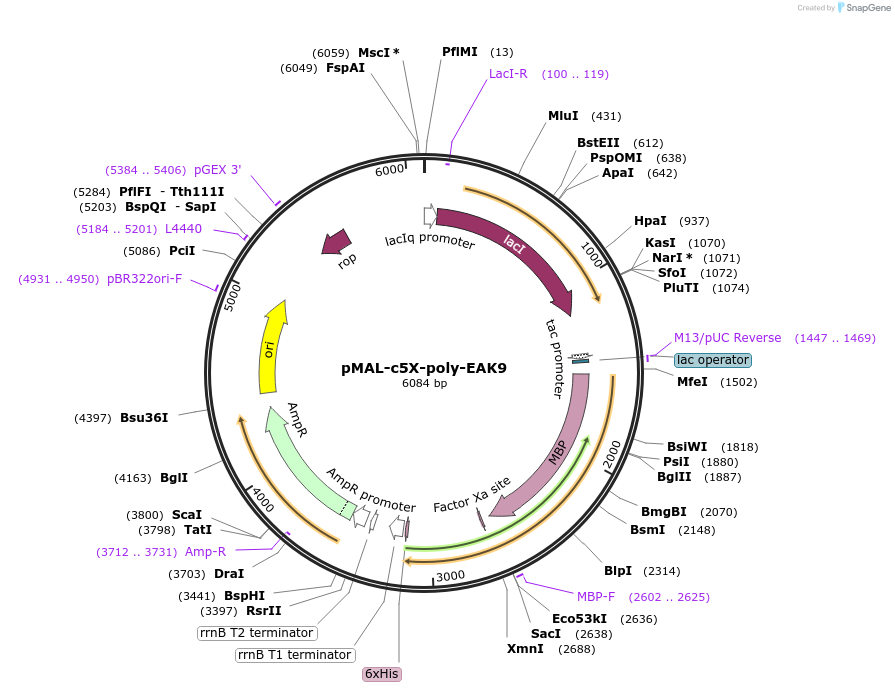
pMAL-c5X-poly-EAK9
Plasmid#67010PurposeExpresses malE-poly-EAK9 in E. coliDepositorInsertmalE-poly-EAK9
TagsHis6 and malE-Factor XaExpressionBacterialPromotertacAvailable SinceJan. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
LV-CMV-Cer-FL- Bmal L95E
Plasmid#47374PurposeBmal full length with cer tag used for mutationDepositorAvailable SinceJan. 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
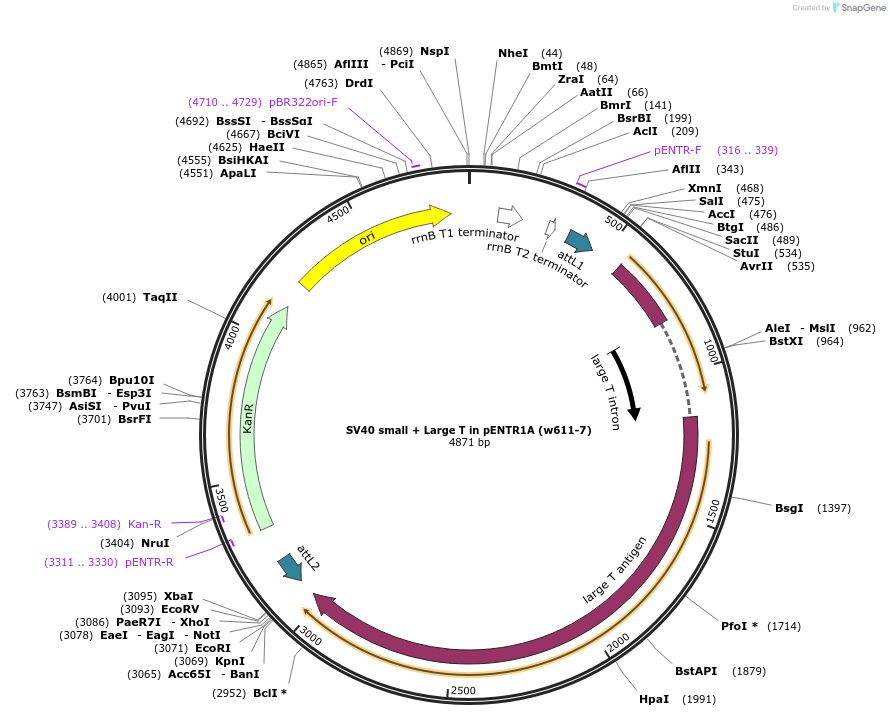
SV40 small + Large T in pENTR1A (w611-7)
Plasmid#22297DepositorInsertSV40 small + Large T antigen
UseEntry vectorAvailable SinceJan. 21, 2010AvailabilityAcademic Institutions and Nonprofits only -
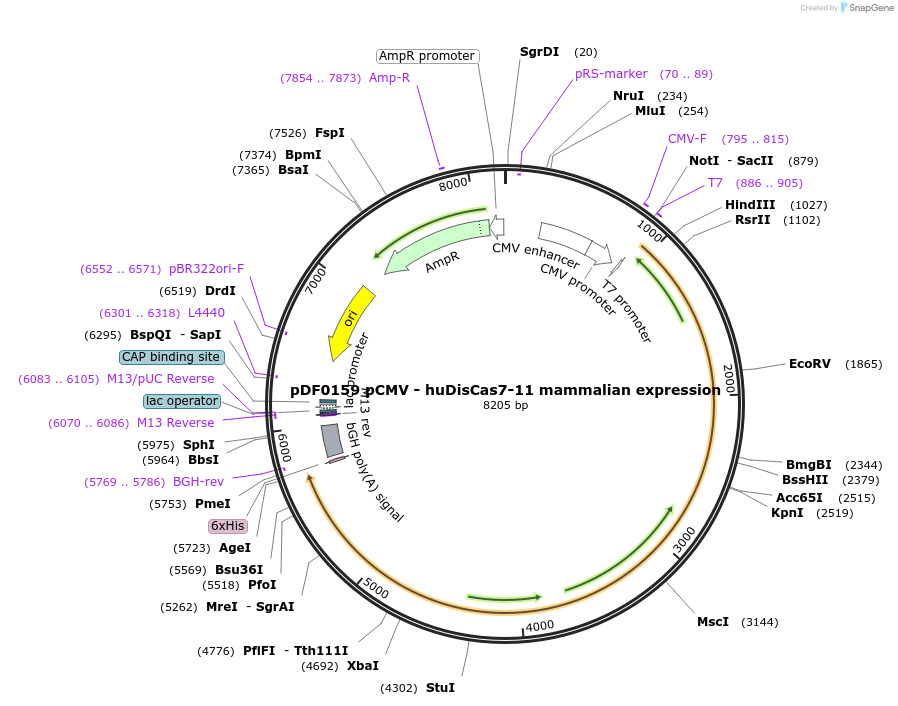
pDF0159 pCMV - huDisCas7-11 mammalian expression
Plasmid#172507PurposeEncodes human codon optimized DisCas7-11 protein alone for expression in mammalian cellsDepositorInsertpCMV - huDisCas7-11 mammalian expression
ExpressionMammalianAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
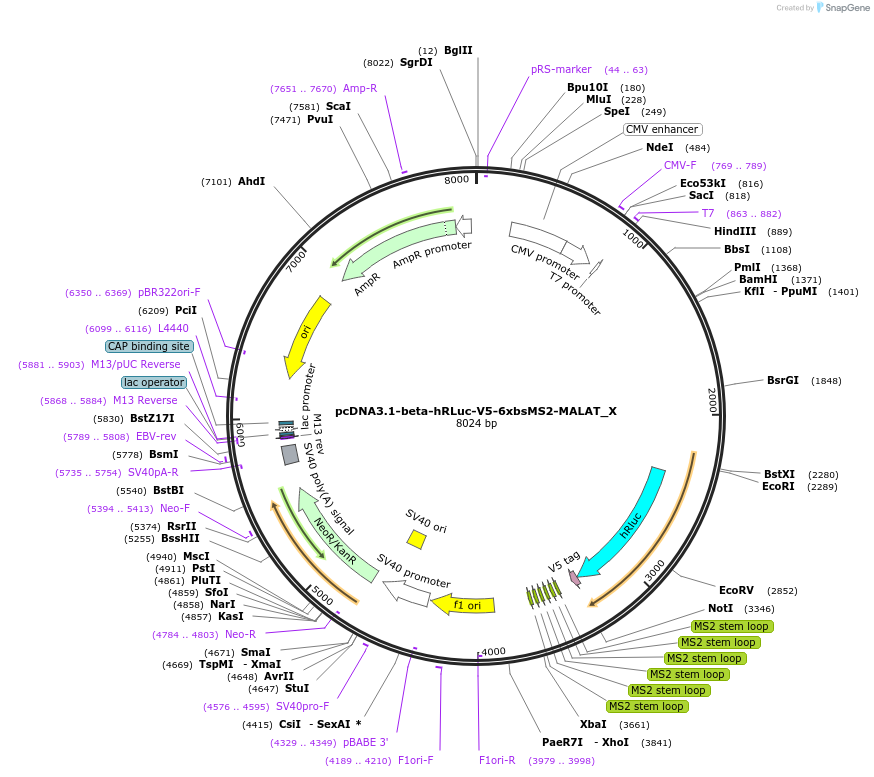
pcDNA3.1-beta-hRLuc-V5-6xbsMS2-MALAT_X
Plasmid#147924PurposeMammalian Expression of 6xbsMS2-MalatDepositorInsert6xbsMS2-Malat (Malat1 Mouse)
ExpressionMammalianAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pLpC-Reg-L-IFP-F[2]+Venus (mammalian)
Plasmid#52903PurposeBinary retroviral pl. (puromycin). Reg:Linker (GGGGS)X2+IFP-F[2] (PacI/EcoRI). In the second MCS, the Venus fluorescent protein is cloned in between PmeI/SalI.Confers ampicillin resistance to DH5α.DepositorInsertReg-L-IFP-F[2]+Venus
UseRetroviralTagsL-IFP-F[2]ExpressionMammalianPromoterUnknownAvailable SinceJune 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pLpC-Cat-L-IFP-F[1]+Plum (mammalian)
Plasmid#52905PurposeBinary retroviral pl. (puromycin). Cat(PacI/ClaI):Linker (GGGGS)X2+IFP-F[2]. In the second MCS, the Plum fluorescent protein is cloned in between PmeI/SalI.Confers ampicillin resistance to DH5α.DepositorInsertCat-L-IFP-F[1]+Plum
UseRetroviralTagsL-IFP-F[1]ExpressionMammalianPromoterUnknownAvailable SinceJune 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
FL MALT1 Catalytic Mutant in pET29b-Strep
Plasmid#48969PurposeExpresses Full length catalytic mutant MALT1 in e.coliDepositorInsertMALT1 isoform A (MALT1 Human)
TagsHis and StrepExpressionBacterialMutationCatalytic Mutant C464APromoterT7Available SinceNov. 6, 2013AvailabilityAcademic Institutions and Nonprofits only -
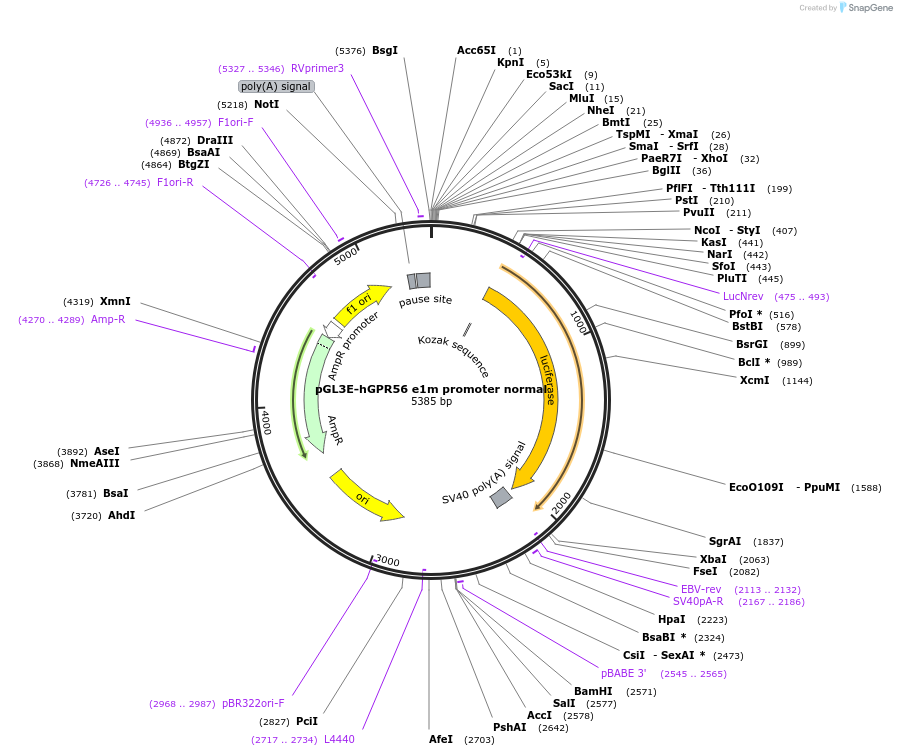
pGL3E-hGPR56 e1m promoter normal
Plasmid#52298PurposeReporter for normal human GPR56 exon 1m promoter activity. Human GPR56 e1m promoter (chr16:56,230,630-56,230,943 (hg18)) was inserted into pGL3E.DepositorInsertADGRG1 adhesion G protein-coupled receptor G1 (ADGRG1 Human)
UseLuciferaseAvailable SinceApril 15, 2014AvailabilityAcademic Institutions and Nonprofits only -
pLpC-Zipp.-L-IFP-F[2]+Venus (mammalian)
Plasmid#52906PurposeBinary retroviral pl. (puromycin). Zipper (PacI/ClaI):Linker (GGGGS)X2+IFP-F[2]. In the second MCS, the Venus fluorescent protein is cloned in between PmeI/SalI.Confers ampicillin resistance to DH5α.DepositorInsertZipp.-L-IFP-F[2]+Venus
UseRetroviralTagsL-IFP-F[2]ExpressionMammalianPromoterUnknownAvailable SinceJune 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pLpC-Zipp.-L-IFP-F[1]+Plum (mammalian)
Plasmid#52907PurposeBinary retroviral pl. (puromycin). Zipper (PacI/ClaI):Linker (GGGGS)X2+IFP-F[1]. In the second MCS, the Plum fluorescent protein is cloned in between PmeI/SalI. Confers ampicillin resistance to DH5α.DepositorInsertZipp.-L-IFP-F[1]+Plum
UseRetroviralTagsL-IFP-F[1]ExpressionMammalianPromoterUnknownAvailable SinceJune 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
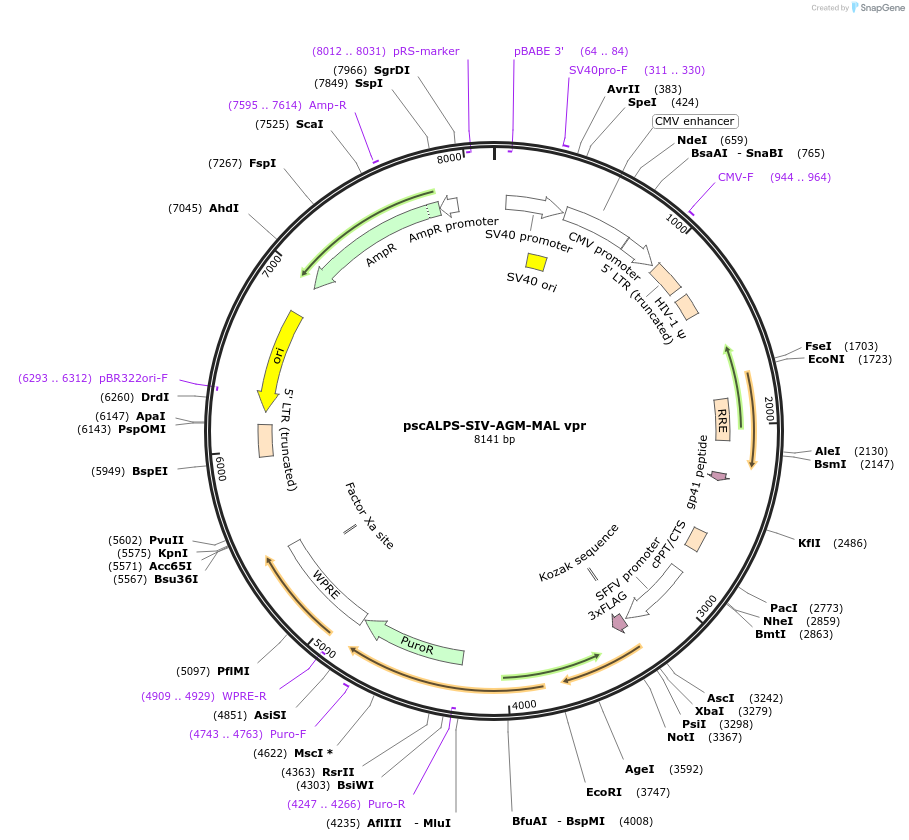
pscALPS-SIV-AGM-MAL vpr
Plasmid#115828PurposeEncodes codon optimized 3xFLAG-SIVAGM Vpr (MAL_ZMB) (LC114462) and puromycin resistance proteinDepositorInsertSIVAGM Vpr (MAL_ZMB) (LC114462)
UseLentiviralTags3x-FLAGPromoterSFFVAvailable SinceJan. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
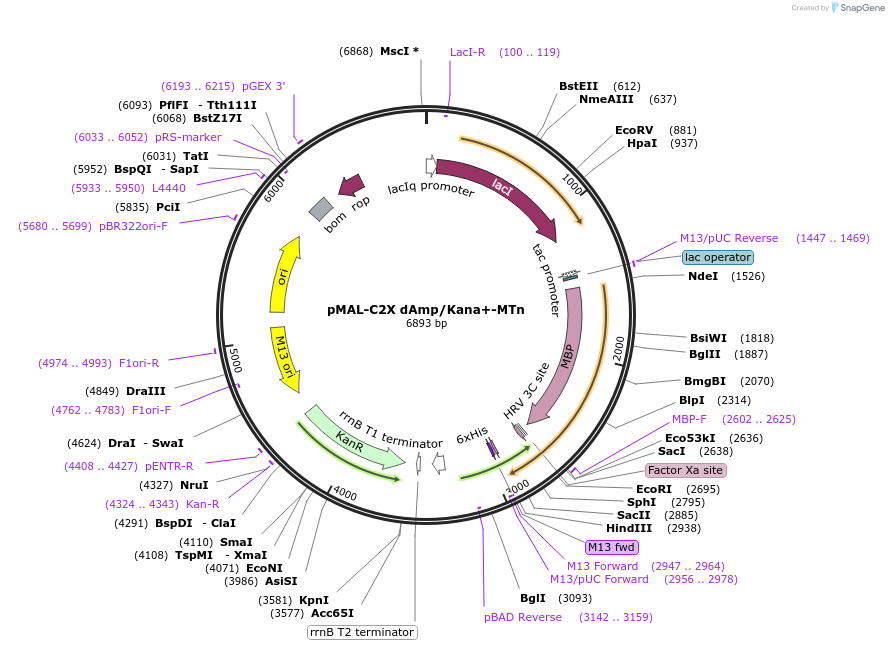
pMAL-C2X dAmp/Kana+-MTn
Plasmid#154355PurposeBacterial expression of MTnDepositorInsertMTn
TagsHisExpressionBacterialAvailable SinceJuly 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
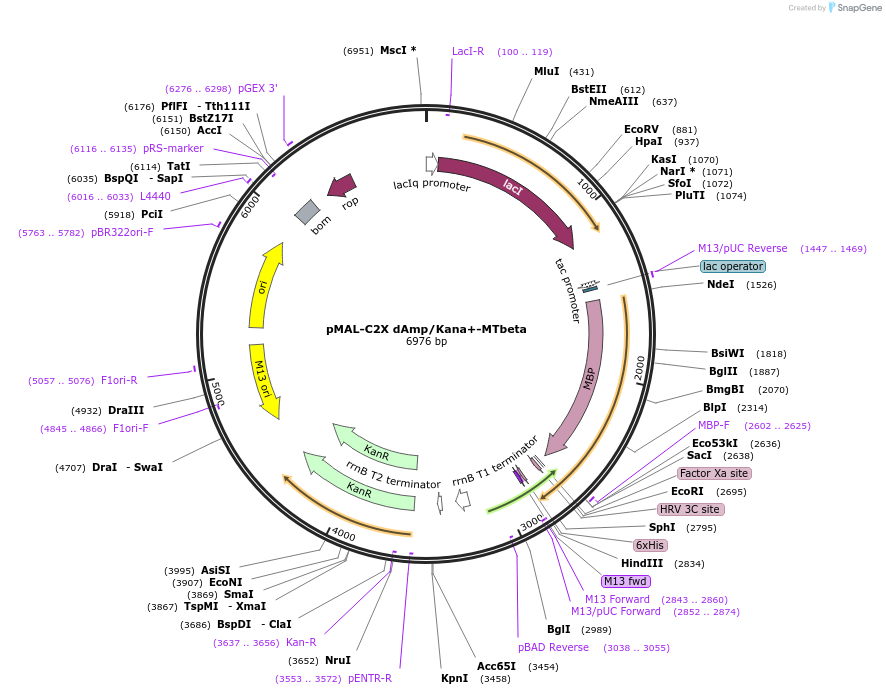
pMAL-C2X dAmp/Kana+-MTbeta
Plasmid#154357PurposeBacterial expression of MTbetaDepositorInsertMTbeta
TagsHisExpressionBacterialAvailable SinceJuly 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
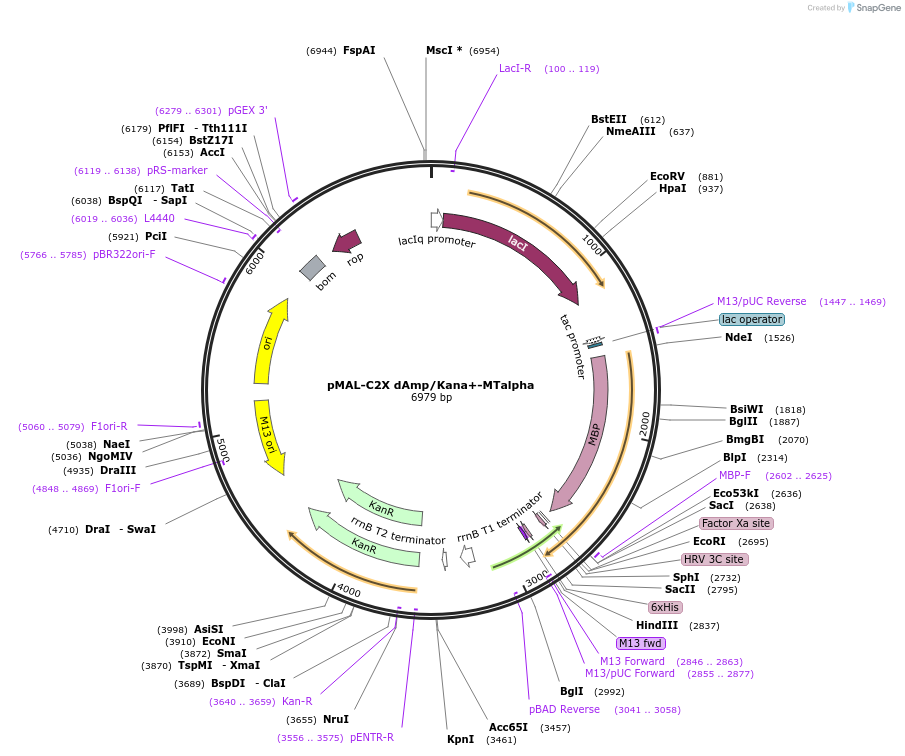
pMAL-C2X dAmp/Kana+-MTalpha
Plasmid#154356PurposeBacterial expression of MTalphaDepositorInsertMTalpha
TagsHisExpressionBacterialAvailable SinceJuly 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
pCDNA3-HA/Bmal L95E/L150E/W427A
Plasmid#47352PurposePcHA-Bmal backbone used for the above mutation. Ref.Genes & Dev 17:1921-1932 2003DepositorAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
HLH Bmal Pas 1-465-VenC
Plasmid#47364PurposeBmal fragment cloned with Venus tag and used for BiFCDepositorAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
pBABE-zeo small T C97S E102Q
Plasmid#9064DepositorInsertSV40 small T Antigen 97, 102 (SV40gp7 simian virus)
UseRetroviralExpressionMammalianMutationC97S, E102QAvailable SinceSept. 23, 2005AvailabilityAcademic Institutions and Nonprofits only -
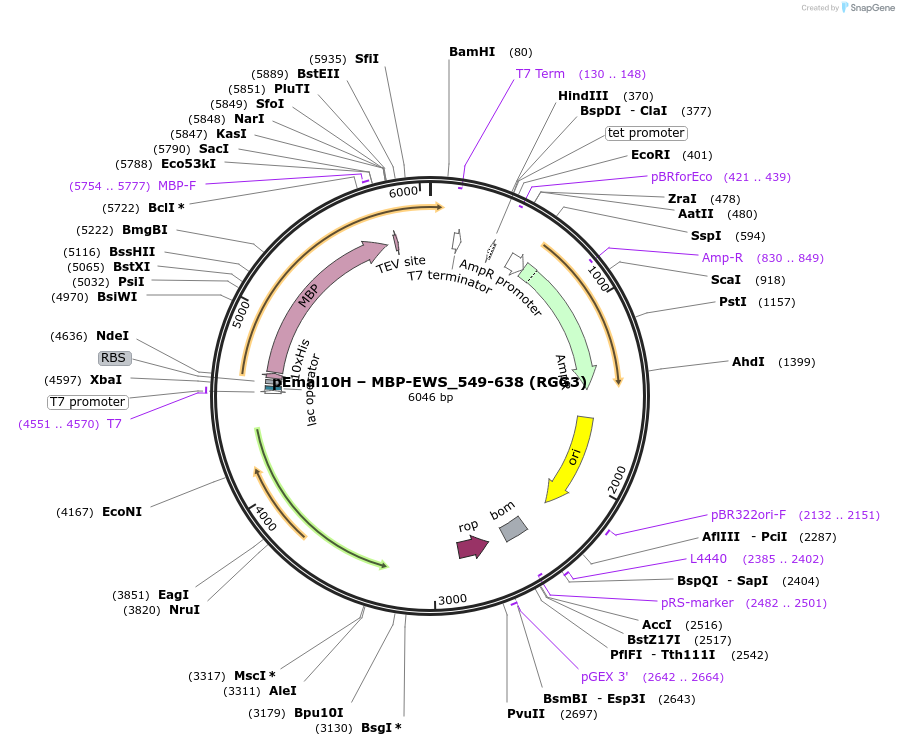
pEmal10H – MBP-EWS_549-638 (RGG3)
Plasmid#238370PurposeExpresses the RGG3 (549-638) region from the RNA-binding protein EWSDepositorInsertEWSR1 (EWSR1 Human)
Tags10xHis-MBPExpressionBacterialMutationdeleted amino acids 1-548PromoterT7Available SinceSept. 2, 2025AvailabilityAcademic Institutions and Nonprofits only