We narrowed to 17,800 results for: JUN
-
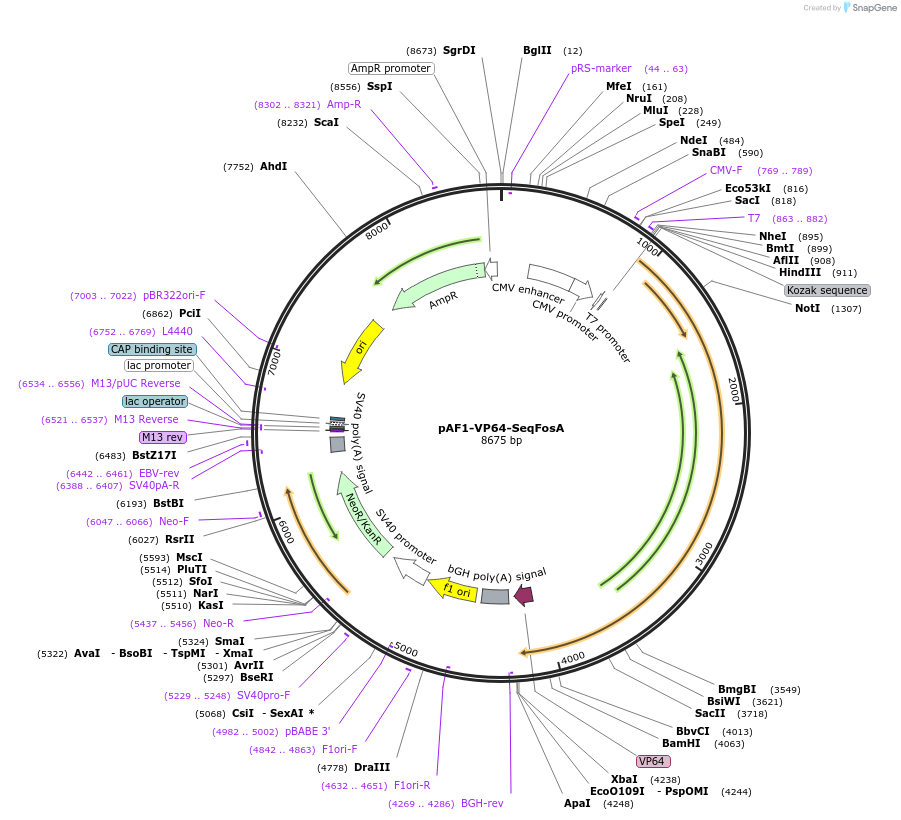
Plasmid#51430PurposeArtificial transcription factor with VP64 targeting c-Fos (TALE A)DepositorInsertSeqFosA
ExpressionMammalianPromoterCMVAvailable SinceJuly 2, 2014AvailabilityAcademic Institutions and Nonprofits only -
Kohinoor-Golgi/pcDNA3
Plasmid#67779PurposeGolgi-targetted Kohinoor for mammalian cell expressionDepositorInsertKohinoor
TagsGTExpressionMammalianPromoterCMVAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
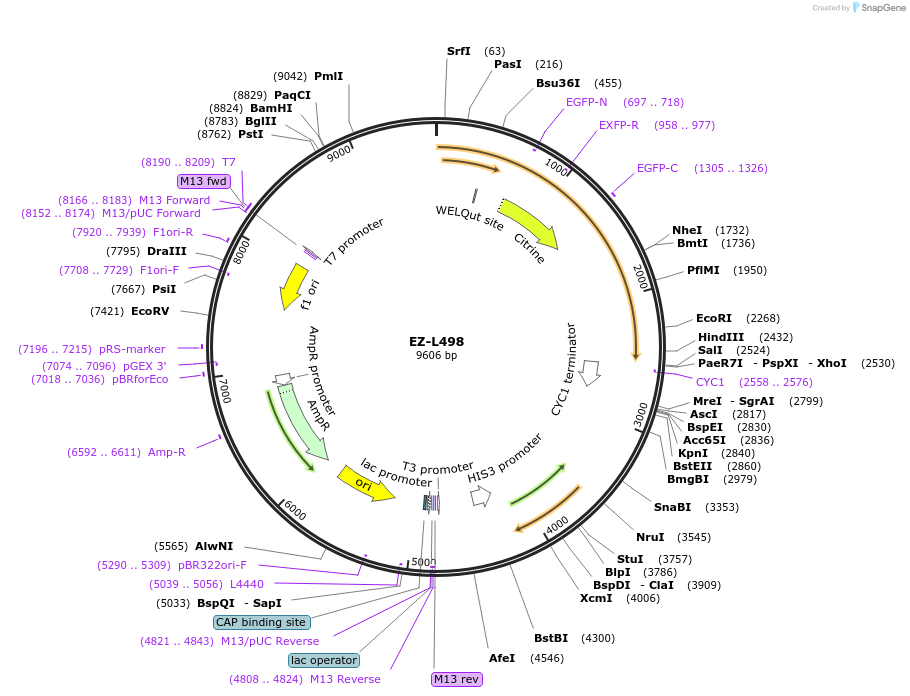
EZ-L498
Plasmid#131771PurposePPGK1_FUSN_Citrine _PixE_TCYC1DepositorInsertPPGK1_FUSN_Citrine _PixE_TCYC1
UseIntegration into his3 locusExpressionYeastPromoterPGKAvailable SinceSept. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
pAW-GFP-2xmiR-34 (pAW-AS-25)
Plasmid#40833PurposemiRNA sensor used in Drosophila cell linesDepositorInsertTwo target sites perfectly complementary to miR-34
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
Kohinoor-Zyxin/pcDNA3
Plasmid#67777PurposeZyxin-targetted Kohinoor for mammalian cell expressionDepositorInsertKohinoor
TagsZyxinExpressionMammalianPromoterCMVAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
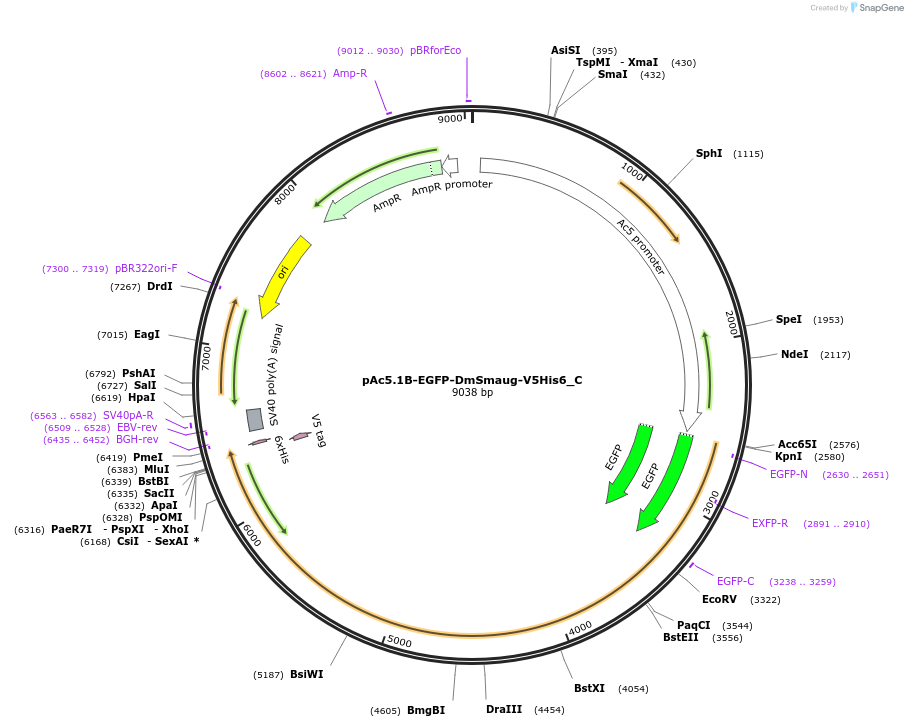
pAc5.1B-EGFP-DmSmaug-V5His6_C
Plasmid#146040PurposeInsect Expression of DmSmaugDepositorInsertDmSmaug (smg Fly)
ExpressionInsectMutation10 silent mutations and a deletion of the AA 961-…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F31
Plasmid#23067DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed lysine 14 to Glutamine K14QAvailable SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
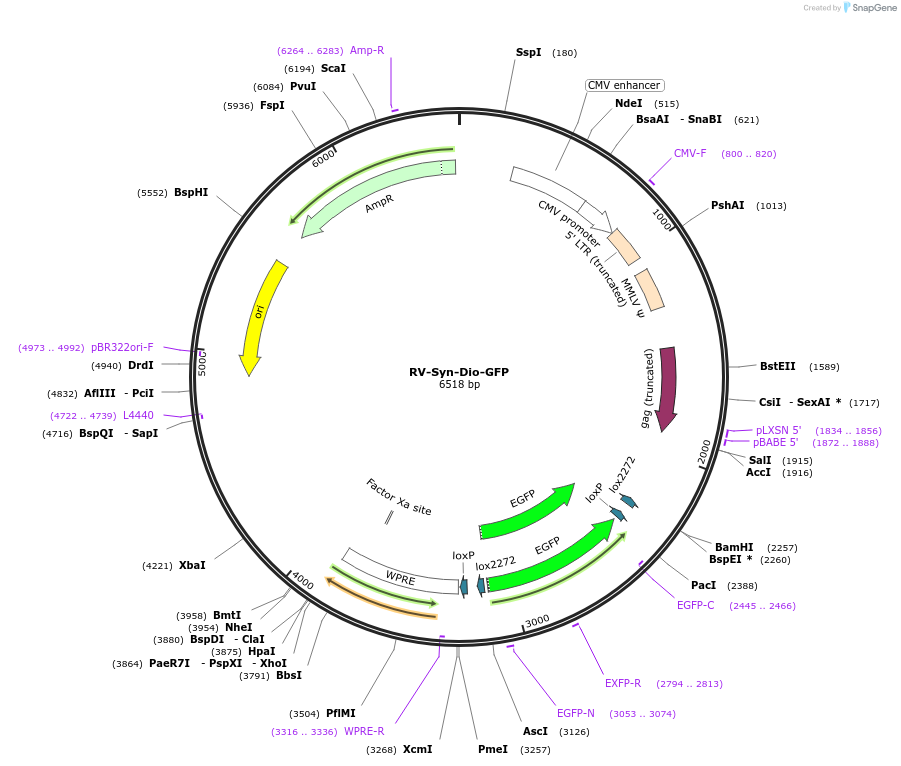
RV-Syn-Dio-GFP
Plasmid#87673Purposeretroviral vector encoding cre dependent expression of GFPDepositorInserteGFP
UseRetroviralPromoterSynapsinAvailable SinceMay 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
pYes2-AtPCS1
Plasmid#49766PurposeExpresses Arabidopsis phytochelatin synthase (AtPCS1) in Saccharomyces CerevisiaeDepositorAvailable SinceJan. 2, 2014AvailabilityAcademic Institutions and Nonprofits only -
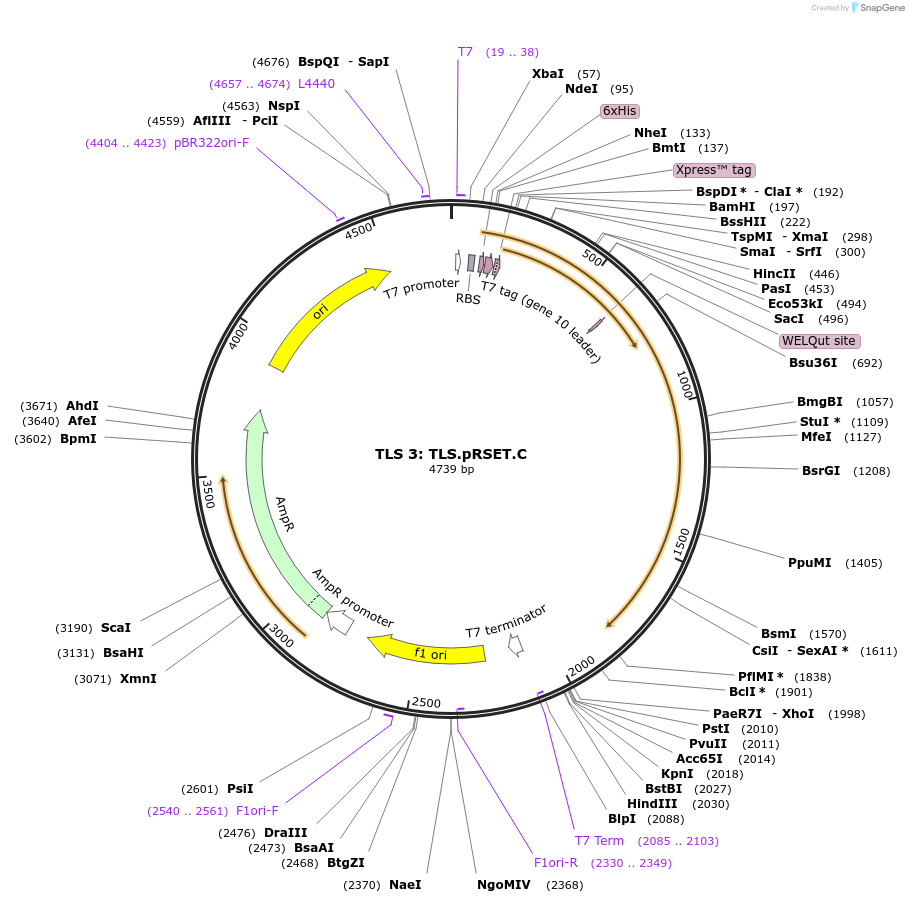
TLS 3: TLS.pRSET.C
Plasmid#21829DepositorAvailable SinceSept. 10, 2009AvailabilityAcademic Institutions and Nonprofits only -
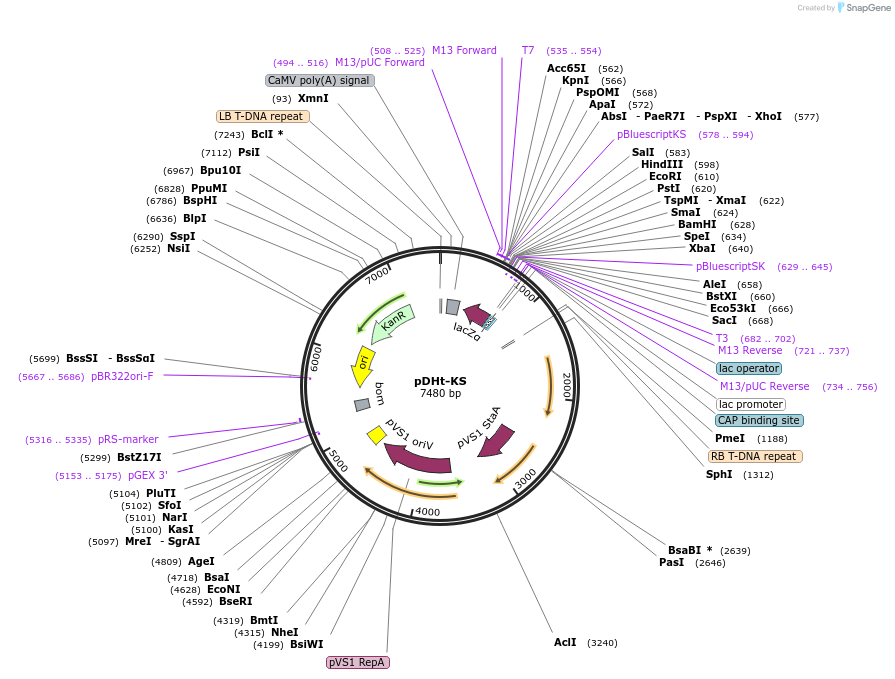
pDHt-KS
Plasmid#63616PurposeThe multiple cloning site of pGreenII0000 cloned into binary vector pDHtDepositorTypeEmpty backboneUseFungal expressionAvailable SinceJuly 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
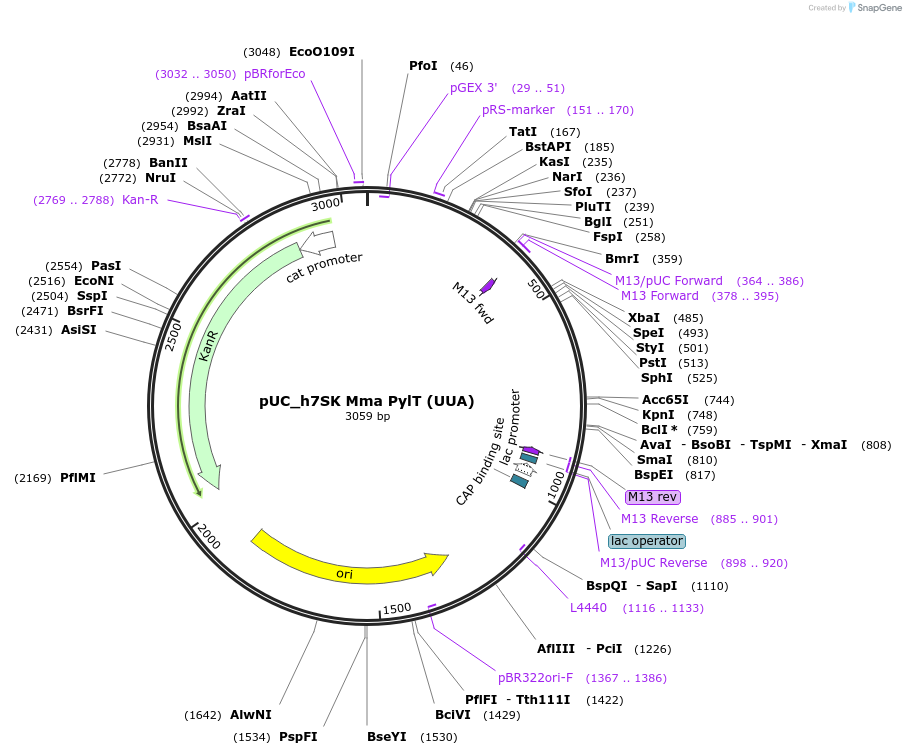
pUC_h7SK Mma PylT (UUA)
Plasmid#154783Purposeplasmid with single copy of Mma PylT (UUA anticodon)DepositorInsertMma PylT
ExpressionMammalianMutationU25C and UUA anticodonPromoterh7SKAvailable SinceSept. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
pAW-EGFP-2xmiR-1 (pAW-AS-21)
Plasmid#40832PurposemiRNA sensor used in Drosophila cell linesDepositorInsertTwo target sites perfectly complementary to miR-1
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
XE40 pSP64T-PABP (Clone 63)
Plasmid#17019DepositorInsertPoly(A) binding protein (pabpc1.S Frog)
UseXenopus expressionAvailable SinceApril 15, 2008AvailabilityAcademic Institutions and Nonprofits only -
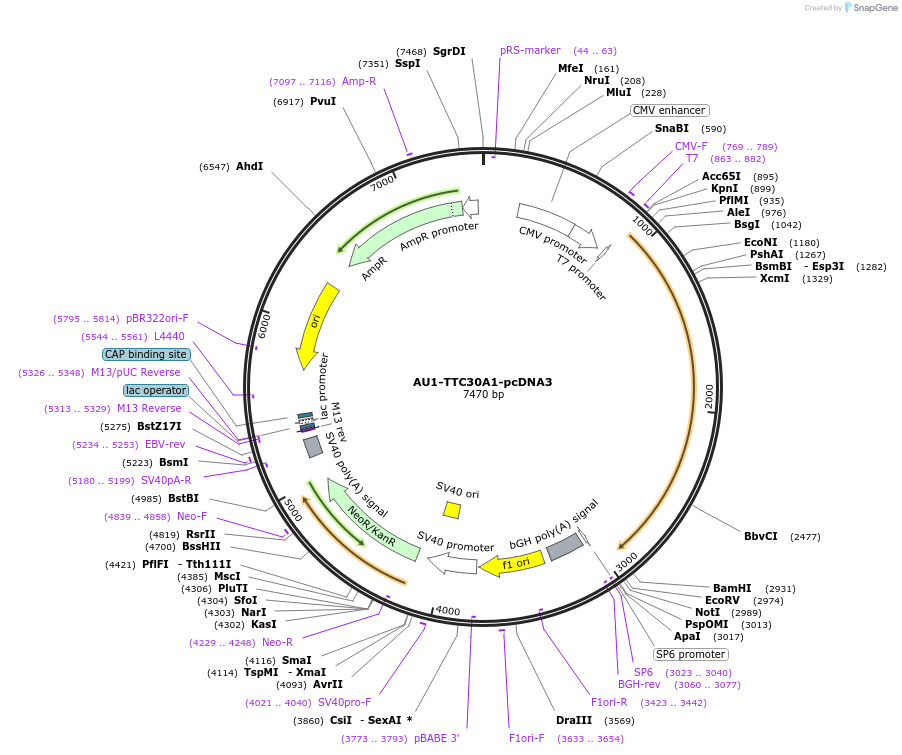
AU1-TTC30A1-pcDNA3
Plasmid#47323PurposeExpresses AU1 epitope-tagged TTC30A1 in mammalian cellsDepositorAvailable SinceSept. 3, 2013AvailabilityAcademic Institutions and Nonprofits only -
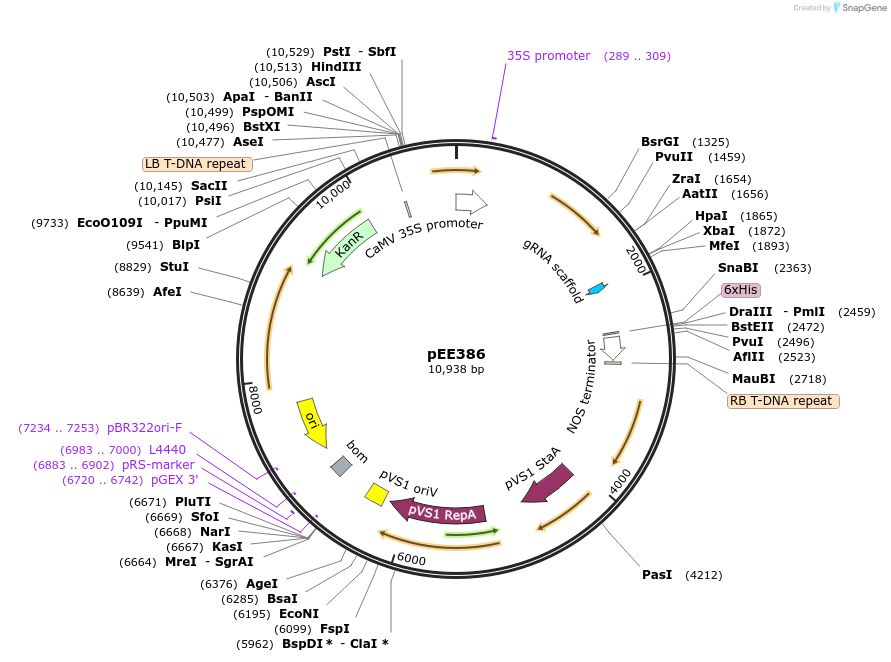
pEE386
Plasmid#149284PurposeT-DNA encoding TRV2 with gRNA targeting NbAGDepositorInsertp35S:TRV2_NbAGsgRNA1:tNos
ExpressionPlantPromoter35SAvailable SinceJuly 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
pEE388
Plasmid#149286PurposeT-DNA encoding TRV2 with mFT augmented gRNA targeting NbAGDepositorInsertp35S:TRV2_NbAGsgRNA1_mFT:tNos
ExpressionPlantPromoter35SAvailable SinceJuly 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
pfMDH_H303A
Plasmid#67730PurposeExpresses H303A mutant of mannitol 2-dehydrogenase from Pseudomonas fluorescens, codon optimized for expression in E. coliDepositorInsertMannitol 2-Dehydrogenase from Pseudomonas fluorescens
Tags6x-His tagExpressionBacterialMutationChanged histidine 303 to alaninePromoterT7Available SinceSept. 11, 2015AvailabilityAcademic Institutions and Nonprofits only -
pfMDH_D230A
Plasmid#67725PurposeExpresses D230A mutant of mannitol 2-dehydrogenase from Pseudomonas fluorescens, codon optimized for expression in E. coliDepositorInsertMannitol 2-Dehydrogenase from Pseudomonas fluorescens
Tags6x-His tagExpressionBacterialMutationChanged aspartate 230 to alaninePromoterT7Available SinceSept. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pfMDH_N191A
Plasmid#67724PurposeExpresses N191A mutant of mannitol 2-dehydrogenase from Pseudomonas fluorescens, codon optimized for expression in E. coliDepositorInsertMannitol 2-Dehydrogenase from Pseudomonas fluorescens
Tags6x-His tagExpressionBacterialMutationChanged asparagine 191 to alaninePromoterT7Available SinceSept. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pfMDH_K295A
Plasmid#67726PurposeExpresses K295A mutant of mannitol 2-dehydrogenase from Pseudomonas fluorescens, codon optimized for expression in E. coliDepositorInsertMannitol 2-Dehydrogenase from Pseudomonas fluorescens
Tags6x-His tagExpressionBacterialMutationChanged lysine 295 to alaninePromoterT7Available SinceSept. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
p224 pCMV-RFP/CREM
Plasmid#8401PurposeFloxed RFP inserted within the coding sequence of cre recombinase.DepositorInsertRFP/CREM
UseCre/LoxExpressionMammalianMutationmodified Cre, LOX 511 sites surrounding RFP withi…PromoterCMVAvailable SinceAug. 4, 2006AvailabilityAcademic Institutions and Nonprofits only -
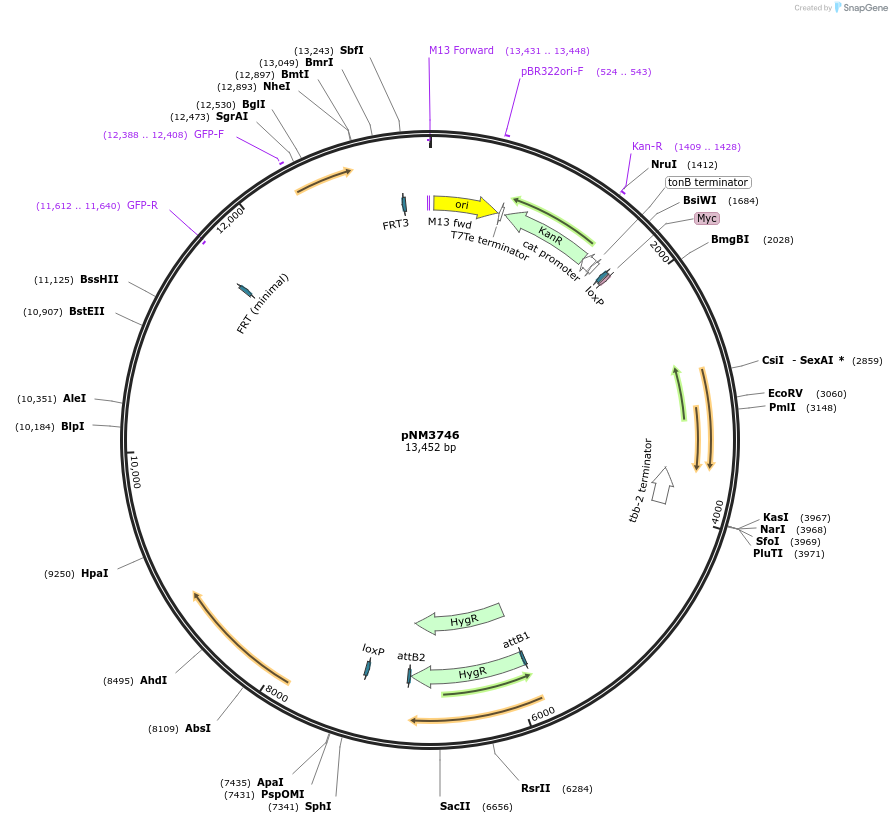
pNM3746
Plasmid#154109PurposeloxP FLP FRT FRT3 landing site in a Golden Gate SapI GCG ACG assembly vectorDepositorInsertsLoxP-SEC-LoxP
FRT::rpl-28p::GFP::His-58::FRT3
mex-5p::FLP D5::sl2::mNG::glh2 3'
UseCre/Lox; RmceExpressionWormPromoternot applicableAvailable SinceJuly 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
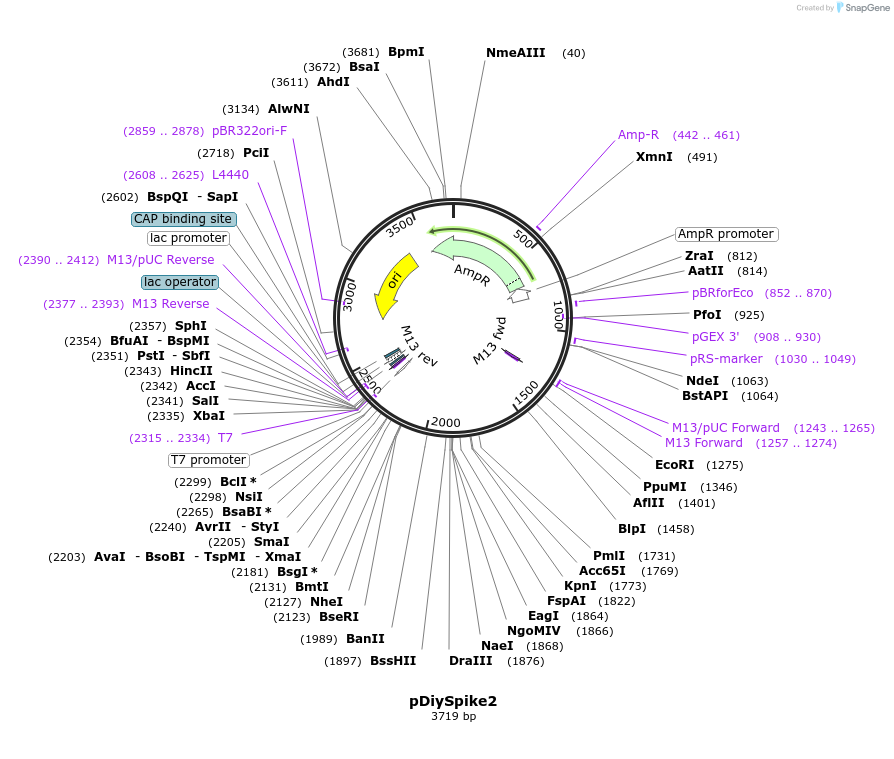
pDiySpike2
Plasmid#136949PurposePlasmid for the In Vitro transcription of DiySpike2DepositorInsertDiySpike2
UseSynthetic BiologyPromoterT7Available SinceApril 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
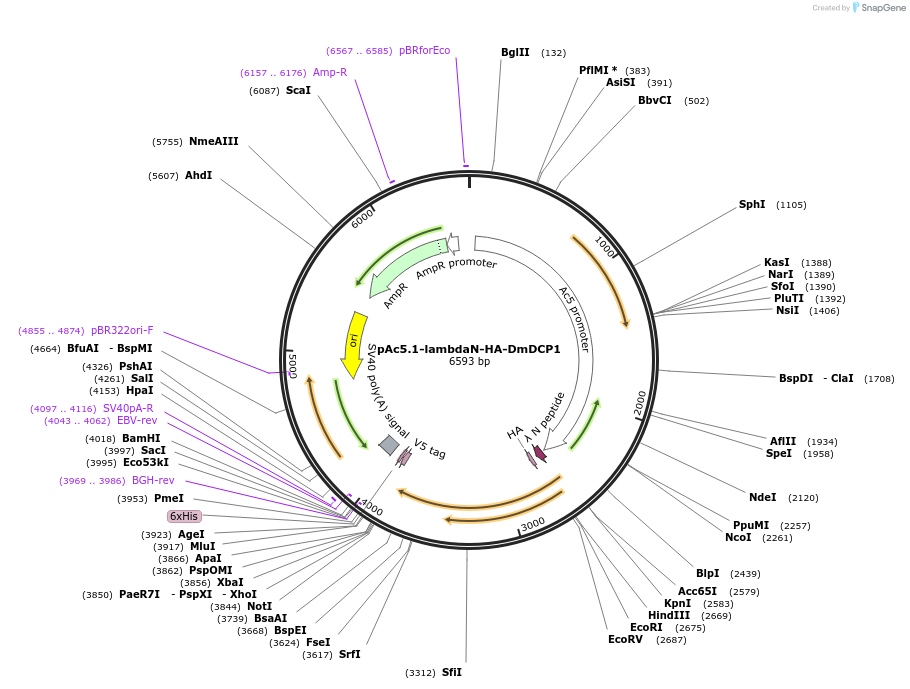
pAc5.1-lambdaN-HA-DmDCP1
Plasmid#21685DepositorAvailable SinceAug. 5, 2009AvailabilityAcademic Institutions and Nonprofits only -
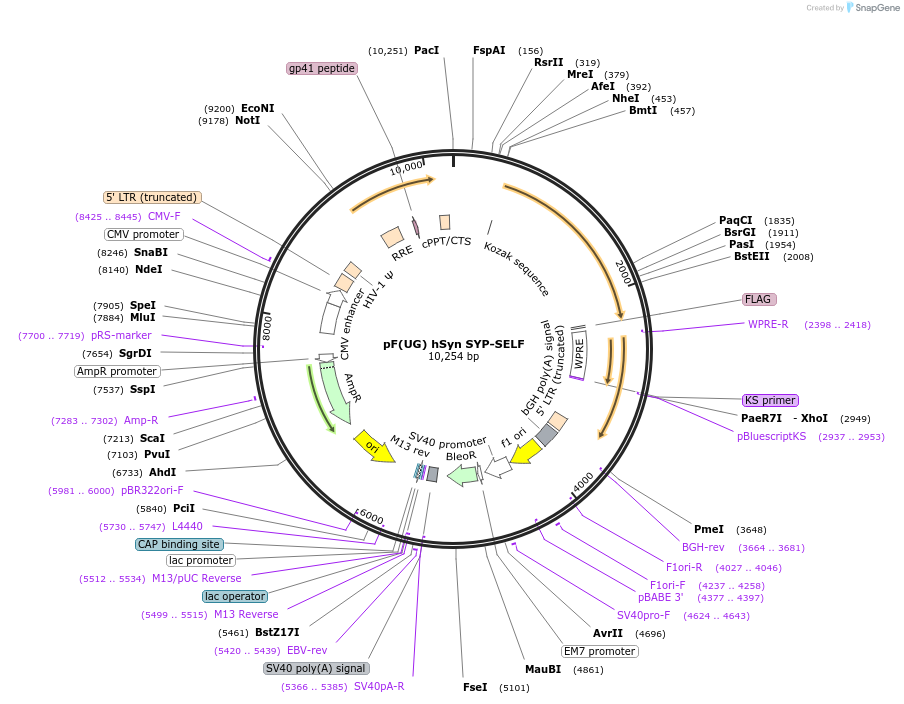
pF(UG) hSyn SYP-SELF
Plasmid#158770PurposeExpression of synaptophysin 1 variant with self-cleaving c-terminal tail.DepositorInsertSynaptophysin 1-NS3/4a (Syt1 Mouse)
UseLentiviralMutationFlexible linkers, ADVVCC/QMSY cleavage siteAvailable SinceMarch 31, 2021AvailabilityAcademic Institutions and Nonprofits only -
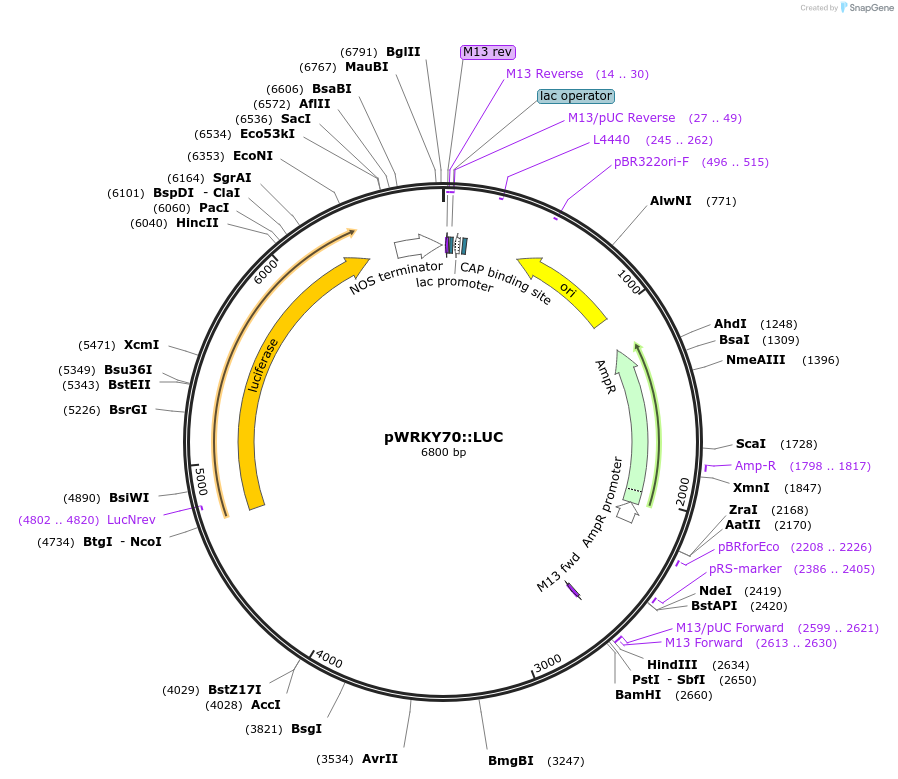
pWRKY70::LUC
Plasmid#141171PurposeLuminescent reporter for salicylic acid signalingDepositorInsertAtWRKY70 promoter
UseLuciferaseExpressionPlantPromoterAtWRKY70, AT3G56400Available SinceAug. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
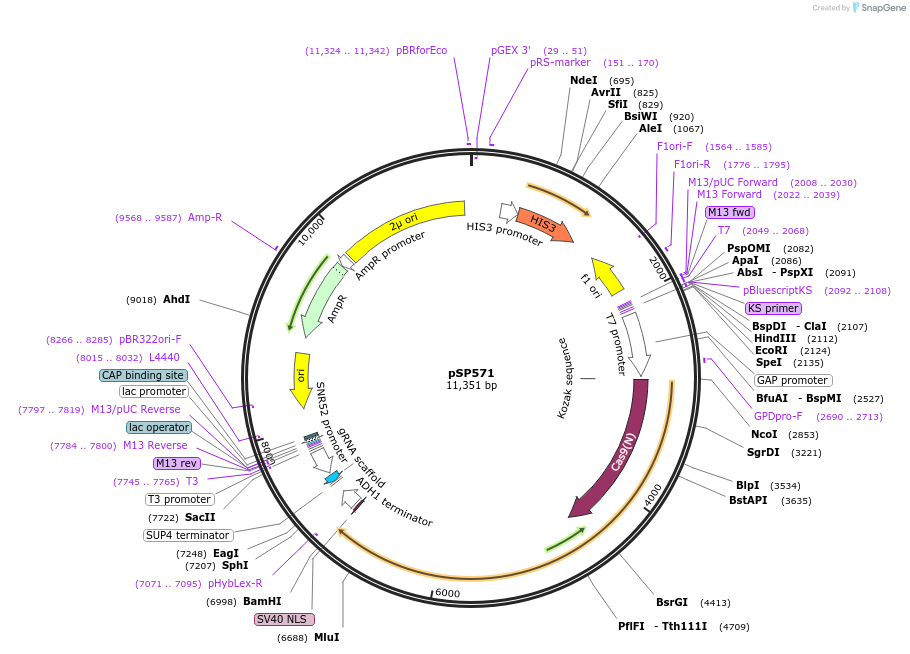
pSP571
Plasmid#139498PurposeCRISPR-Cas9 plasmid to generate double strand break in STL1 locus in S. cerevisiae. Expresses both Cas9 and STL1 sgRNADepositorInsertpGPD Cas9 / sgRNA (STL162)
ExpressionYeastMutationWTPromoterpGPDAvailable SinceJune 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
pIS1-wt Fzd3 3'UTR
Plasmid#25024DepositorInsertwt Fzd3 3'UTR
UseLuciferaseExpressionMammalianAvailable SinceJuly 1, 2010AvailabilityAcademic Institutions and Nonprofits only -
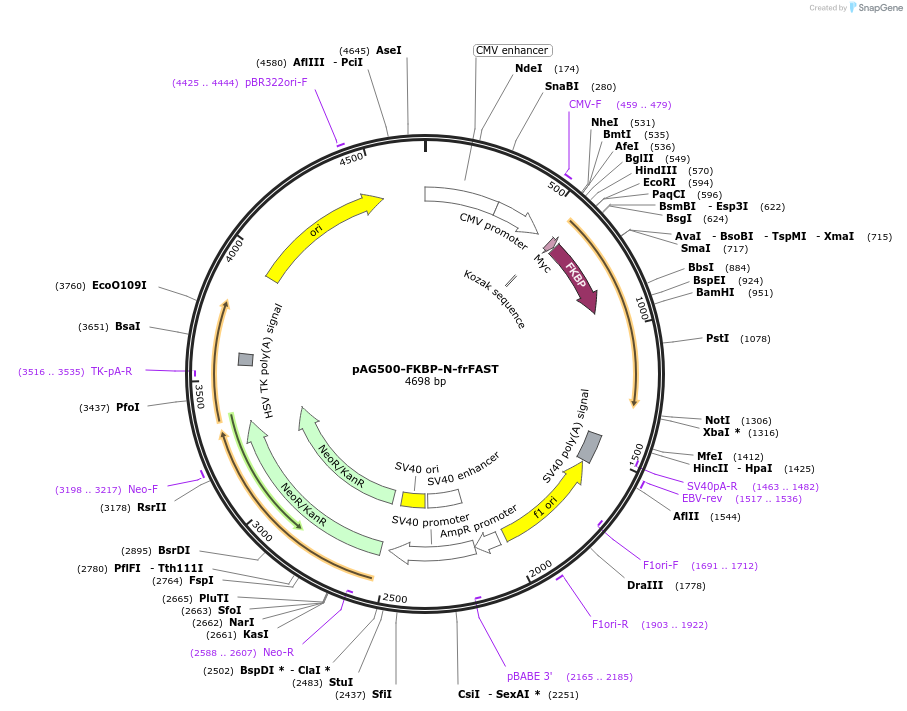
pAG500-FKBP-N-frFAST
Plasmid#160060PurposeExpresses FKBP-N-frFAST in mammalian cellsDepositorInsertFKBP-N-frFAST
ExpressionMammalianAvailable SinceOct. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
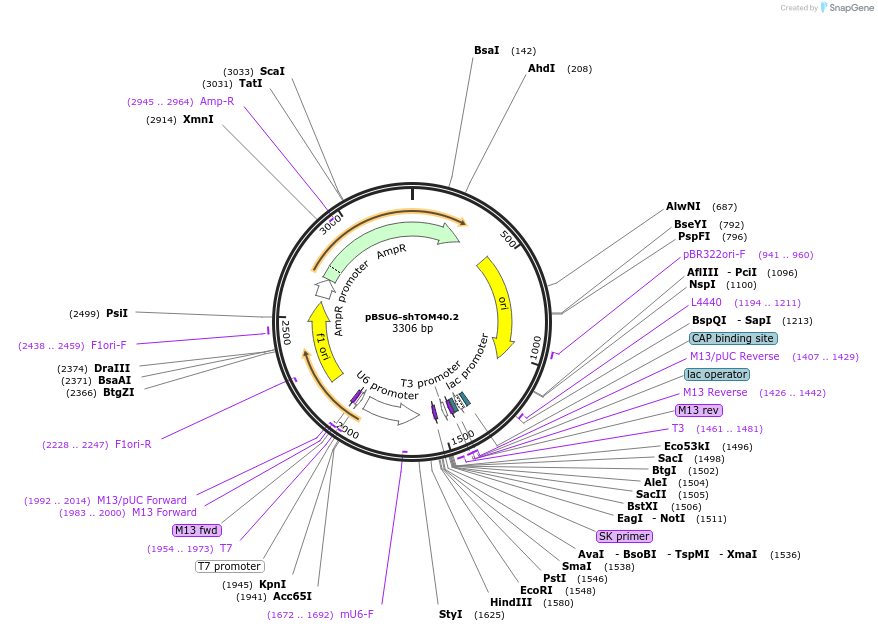
pBSU6-shTOM40.2
Plasmid#89882PurposeExpresses shRNA against TOM40DepositorInsertshRNA Tom40
TagsnoneExpressionMammalianAvailable SinceJune 13, 2017AvailabilityAcademic Institutions and Nonprofits only -
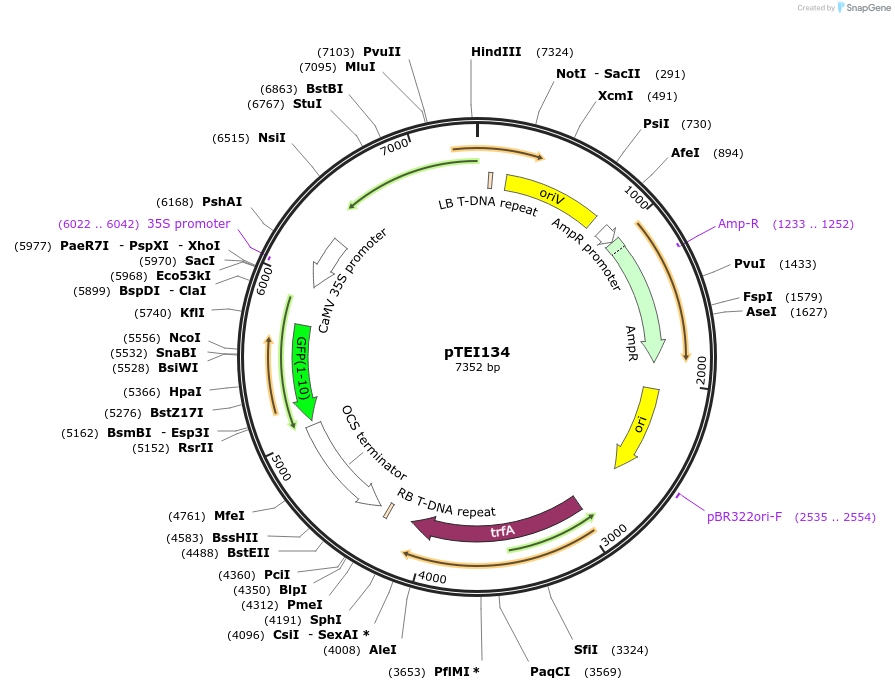
pTEI134
Plasmid#126071PurposeVector for expression of plastid targeted YFP1-10 (T203Y) in plant cellDepositorInsertChloroplastic FNR transit peptide
UseBinaryTagsYFP1-10ExpressionPlantAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
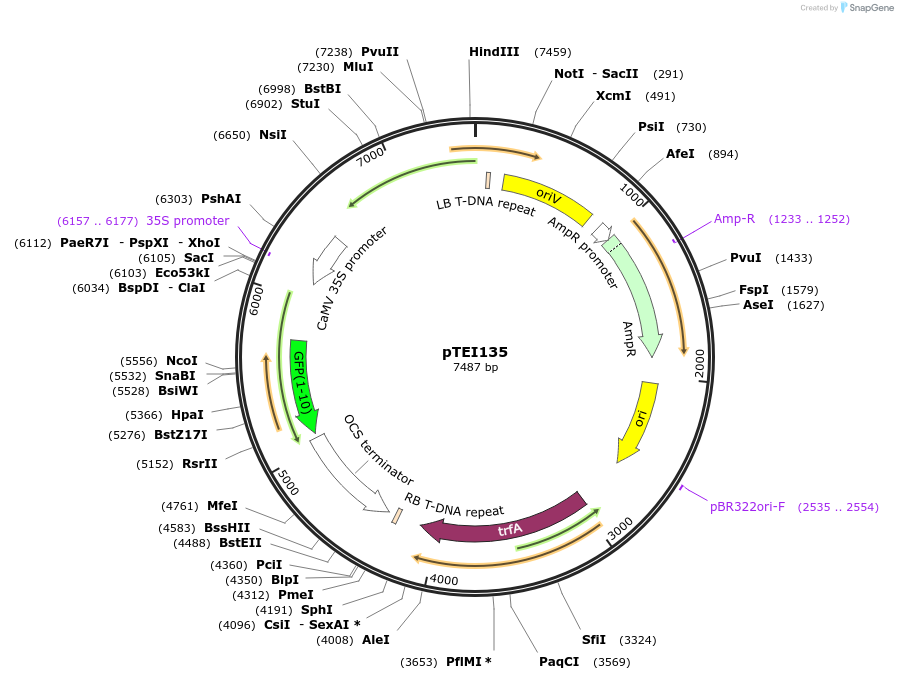
pTEI135
Plasmid#126073PurposeVector for expression of mitochondria targeted YFP1-10 (T203Y) in plant cellDepositorInsertmitochondria Rieske iron sulphur protein (1-100 aa)
UseBinaryTagsYFP1-10ExpressionPlantAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSuperRetro-puro-RalB 45i
Plasmid#11122DepositorAvailable SinceMay 25, 2006AvailabilityAcademic Institutions and Nonprofits only -
SI15-CD276
Plasmid#46305DepositorAvailable SinceFeb. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
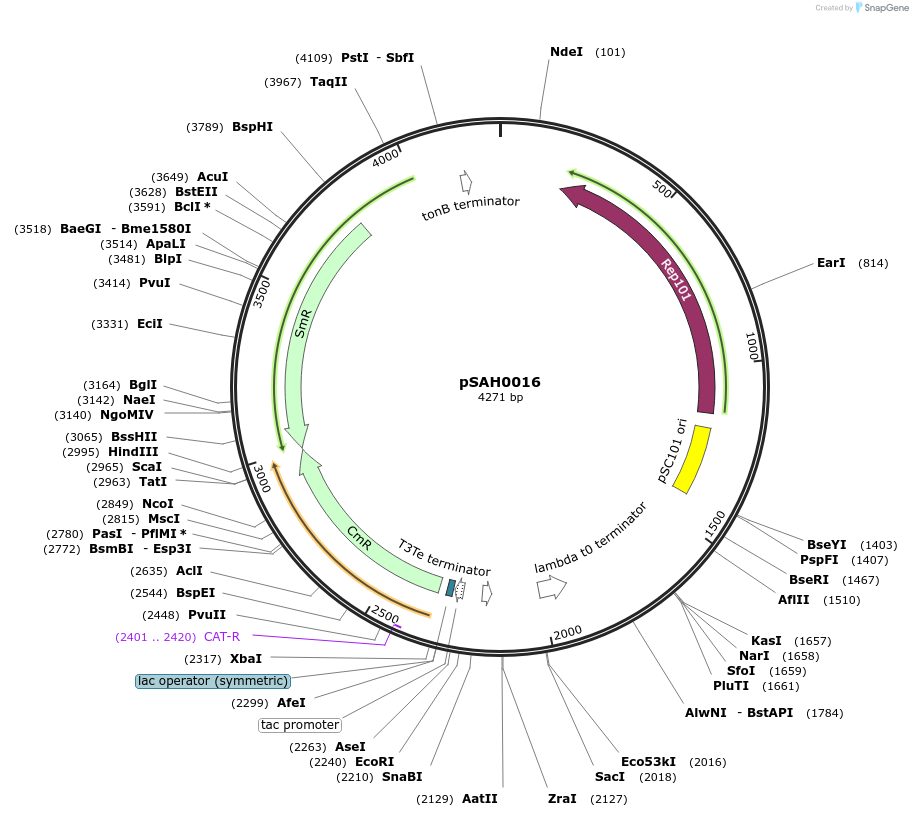
pSAH0016
Plasmid#79824PurposeDual selection plasmid with lac operator, selection for binding with spectinomycin, for non-binding with chloramphenicolDepositorInsertscat
aadA
PromoterAATTCAAAAAAATGCTTGACAATTAATCATCGGCTCGTATAATGTGTGG…Available SinceAug. 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
SI6-IGSF21a
Plasmid#36141DepositorAvailable SinceOct. 16, 2012AvailabilityAcademic Institutions and Nonprofits only -
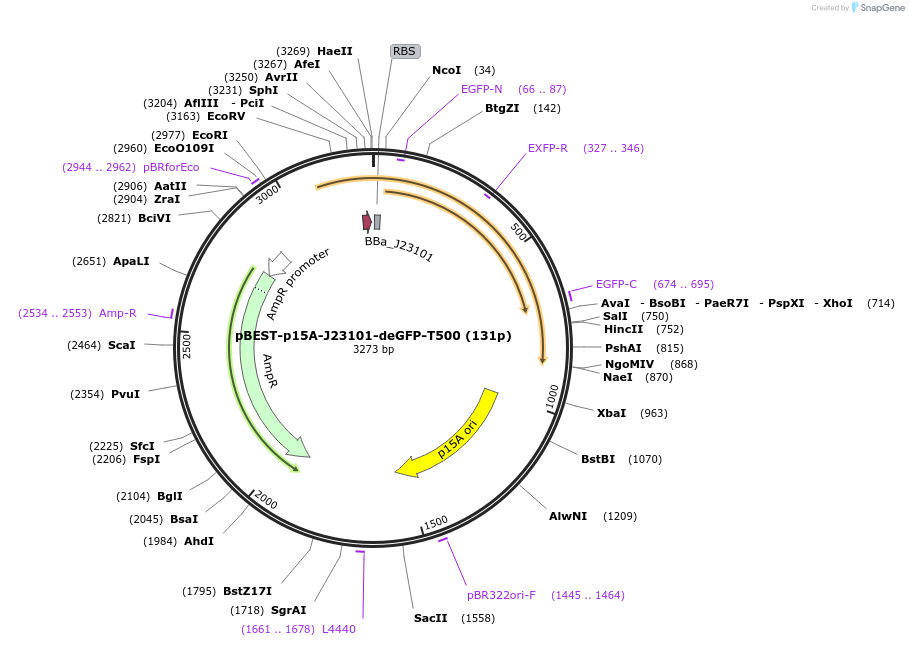
pBEST-p15A-J23101-deGFP-T500 (131p)
Plasmid#70207PurposePromoter panelDepositorInsertdeGFP
Available SinceFeb. 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
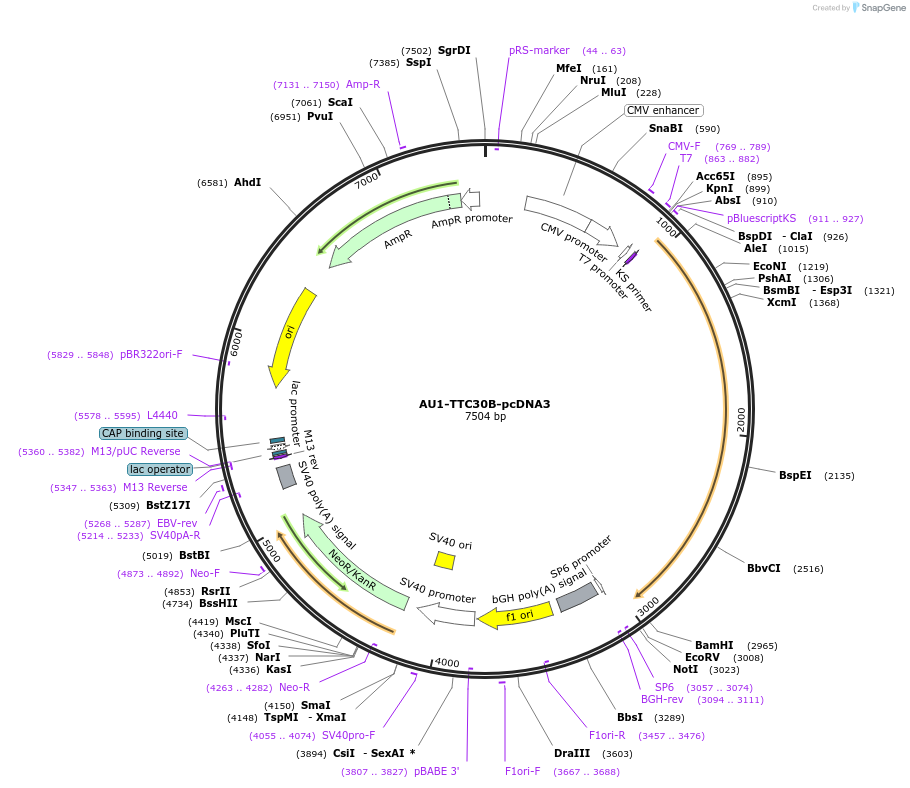
AU1-TTC30B-pcDNA3
Plasmid#47325PurposeExpresses AU1 epitope-tagged TTC30B in mammalian cellsDepositorAvailable SinceSept. 3, 2013AvailabilityAcademic Institutions and Nonprofits only -
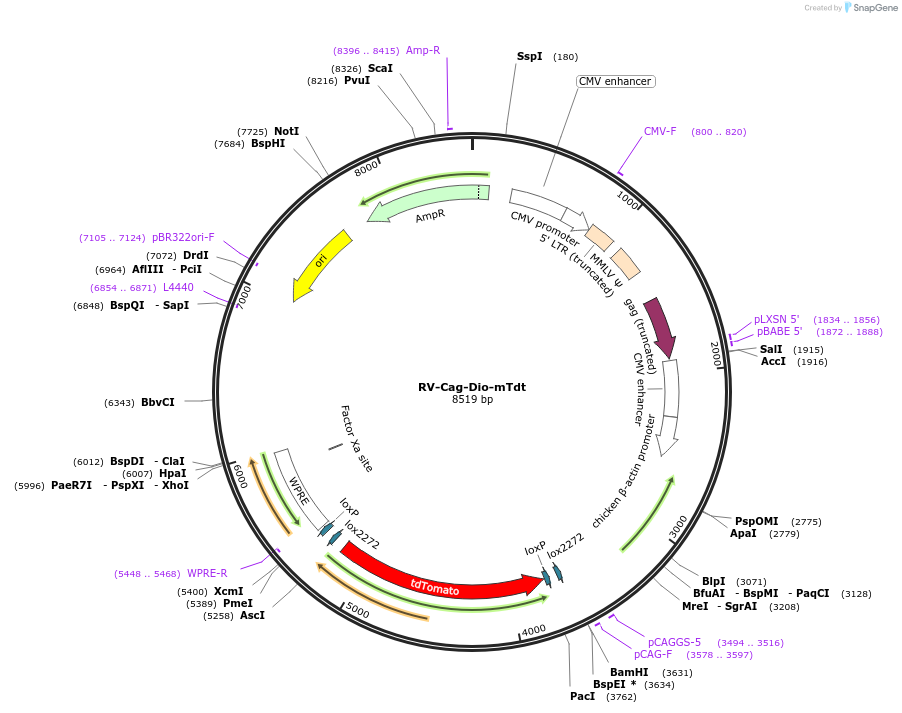
RV-Cag-Dio-mTdt
Plasmid#87665Purposeretroviral vector encoding cre dependent expression of membrane TdtomatoDepositorInsertMem-Tdtomato
UseRetroviralPromoterCAGAvailable SinceMay 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
p236 pPr-CREM
Plasmid#8404PurposeExpression of Cre from tissue-specific, androgen-inducible probasin promoter, with chimeric/β-globin intron within the Cre coding sequence.DepositorInsertprobasin driven CREM
UseCre/LoxExpressionMammalianMutationmodified CrePromoterProbasin promoterAvailable SinceApril 28, 2006AvailabilityAcademic Institutions and Nonprofits only -
pGL3-p107 1*
Plasmid#32389DepositorInsertp107 promoter (Rbl1 Mouse)
UseLuciferaseTagsLuciferaseExpressionMammalianMutation1st E2F binding site mutation: GCG-->AAAAvailable SinceSept. 27, 2011AvailabilityAcademic Institutions and Nonprofits only -
pIS1-miR-126 site
Plasmid#25026DepositorInsertsynthetic miR-126 site
UseLuciferaseExpressionMammalianAvailable SinceJune 18, 2010AvailabilityAcademic Institutions and Nonprofits only -
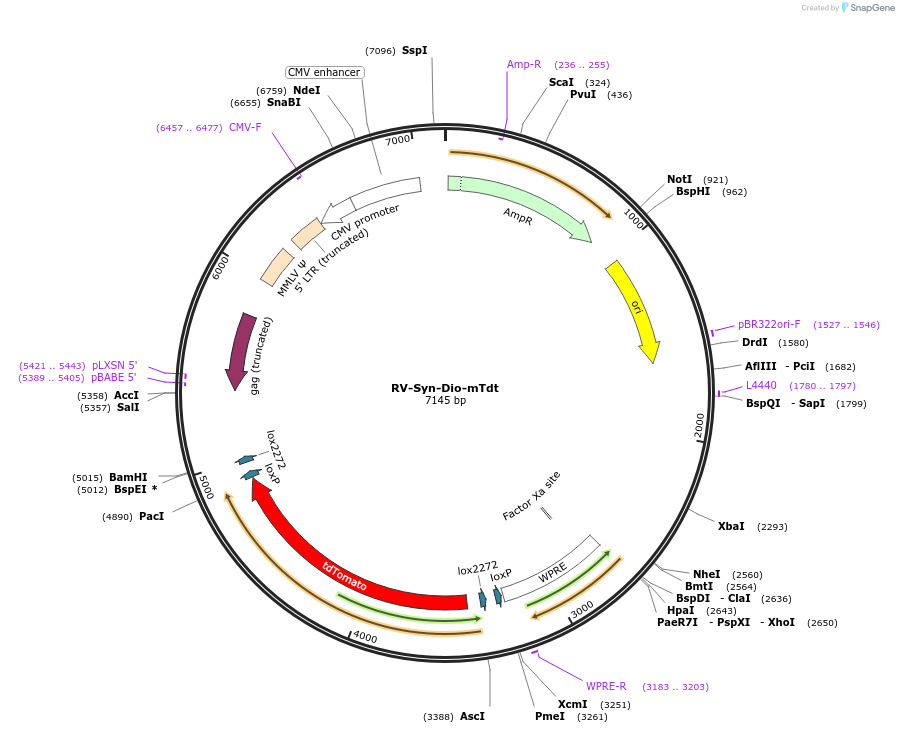
RV-Syn-Dio-mTdt
Plasmid#87667Purposeretroviral vector encoding cre dependent expression of membrane TdtomatoDepositorInsertMem-Tdtomato
UseRetroviralTagspalmitylationPromoterSynapsinAvailable SinceMay 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
pENTR-fishRab8A[T22N]
Plasmid#24906DepositorInsertfishRab8A
UseGateway entry vectorMutationChanged Threonine 22 to AsparagineAvailable SinceJuly 7, 2010AvailabilityAcademic Institutions and Nonprofits only -
pENTR-fishRab8A
Plasmid#24904DepositorInsertfishRab8A
UseGateway entry vectorAvailable SinceJune 28, 2010AvailabilityAcademic Institutions and Nonprofits only -
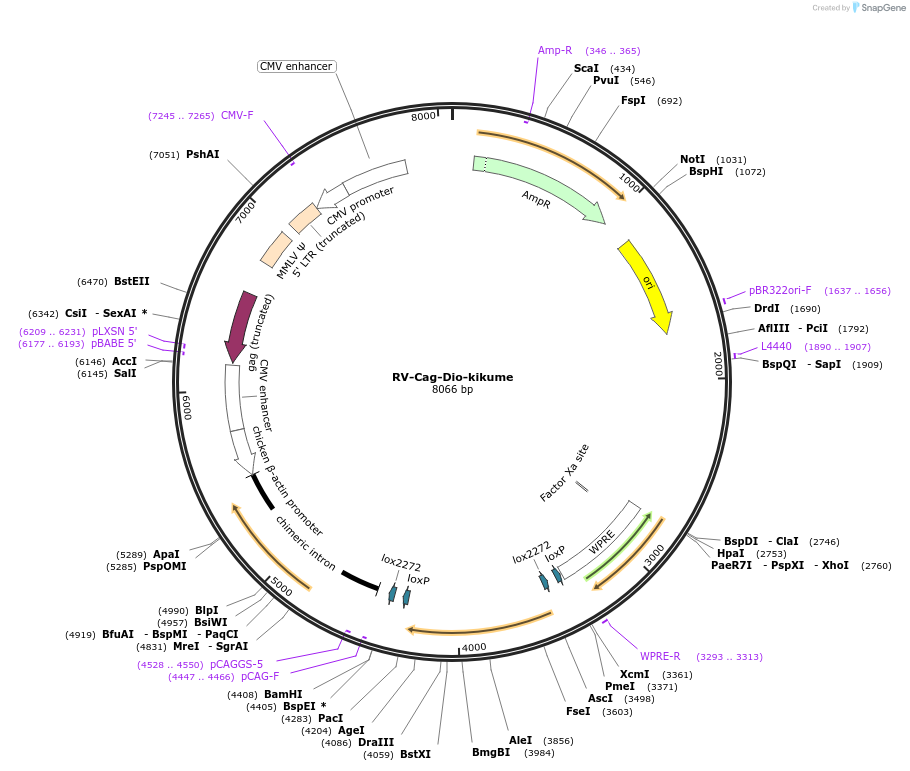
RV-Cag-Dio-kikume
Plasmid#87663Purposeretroviral vector encoding cre-dependent expression of kikumeDepositorInsertkikume
UseRetroviralPromoterCAGAvailable SinceMay 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
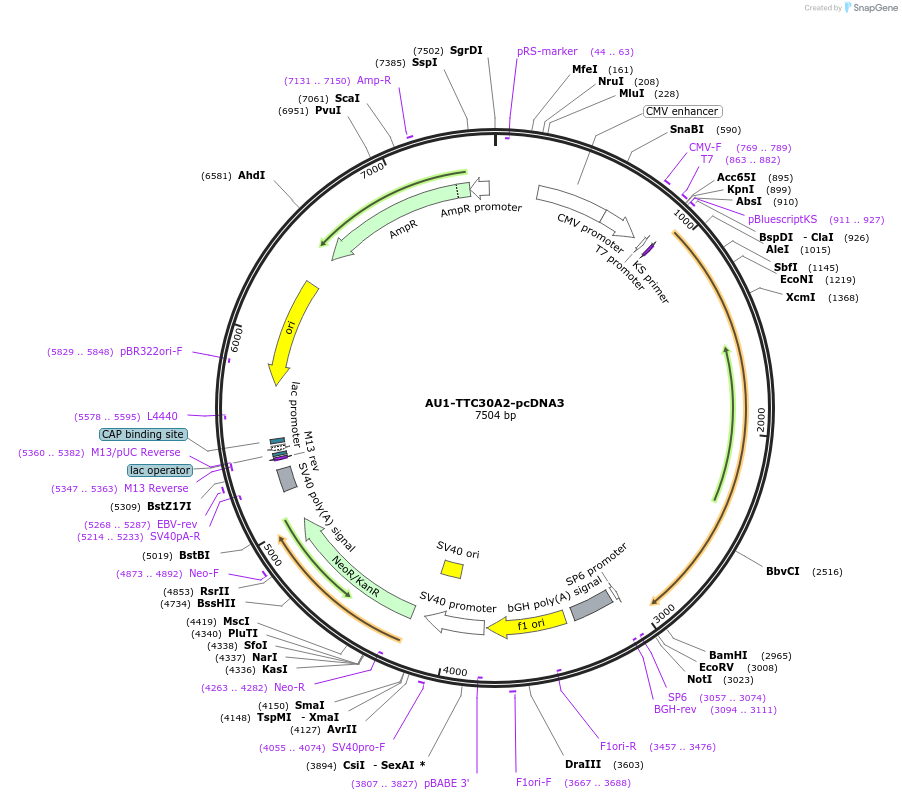
AU1-TTC30A2-pcDNA3
Plasmid#47324PurposeExpresses AU1 epitope-tagged TTC30A2 in mammalian cellsDepositorAvailable SinceSept. 3, 2013AvailabilityAcademic Institutions and Nonprofits only -
pBSU6-shTOM40.3
Plasmid#89796PurposeExpresses shRNA against TOM40.DepositorInsertshRNA Tom40
TagsnoneExpressionMammalianPromoterU6Available SinceMay 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
SI5-IGSF21a
Plasmid#36115DepositorAvailable SinceOct. 16, 2012AvailabilityAcademic Institutions and Nonprofits only