We narrowed to 4,220 results for: PRS
-
Plasmid#207444Purpose6xHis-SS-TEV-ErkB-Y178A bacterial expression vectorDepositorInsertErkB
UseUnspecifiedMutationY178AAvailable SinceSept. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAF9D94 (pAF10C6)
Plasmid#185848PurposeExpressing artificial trans-repressor in Saccharomyces cerevisiaeDepositorInsertPHAC1>TetR>CYC8>PABF1
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
KBB441
Plasmid#185131PurposeSwi6 full length with L->A mutated in nuclear localization signalDepositorInsertSWI6
TagsGFPExpressionYeastMutationSWI6 deletion of amino acids 450-458Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
KBB556
Plasmid#185108Purpose1st 200 amino acids (N-term + transmembrane domain) of Pom152 fused to HA and GFP with S158A, F159A, F160ADepositorInsertPOM152
TagsGFPExpressionYeastMutation1st 200 amino acids (N-term + transmembrane domai…Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
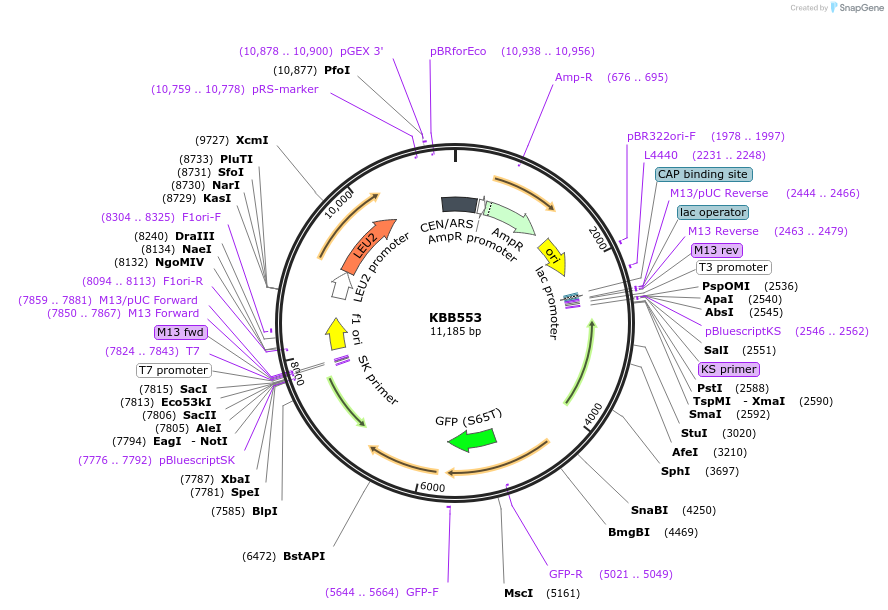
KBB553
Plasmid#185107Purpose1st 200 amino acids (N-term + transmembrane domain) of Pom152 fused to HA and GFP with Y180L, Q182LDepositorInsertPOM152
TagsGFPExpressionYeastMutation1st 200 amino acids (N-term + transmembrane domai…Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
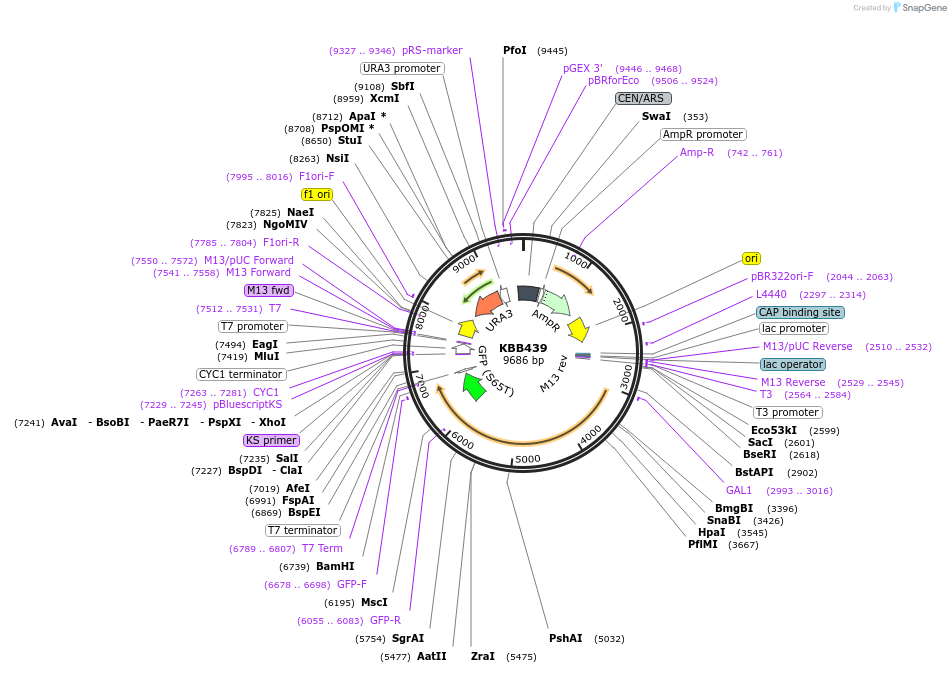
KBB439
Plasmid#185093PurposeNMA111-GFP wild type under control of GAL1 promoter (complements nma111 deletion)DepositorInsertNMA111
TagsGFPExpressionYeastMutationGAL1::NMA111-GFPPromoterGAL1Available SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
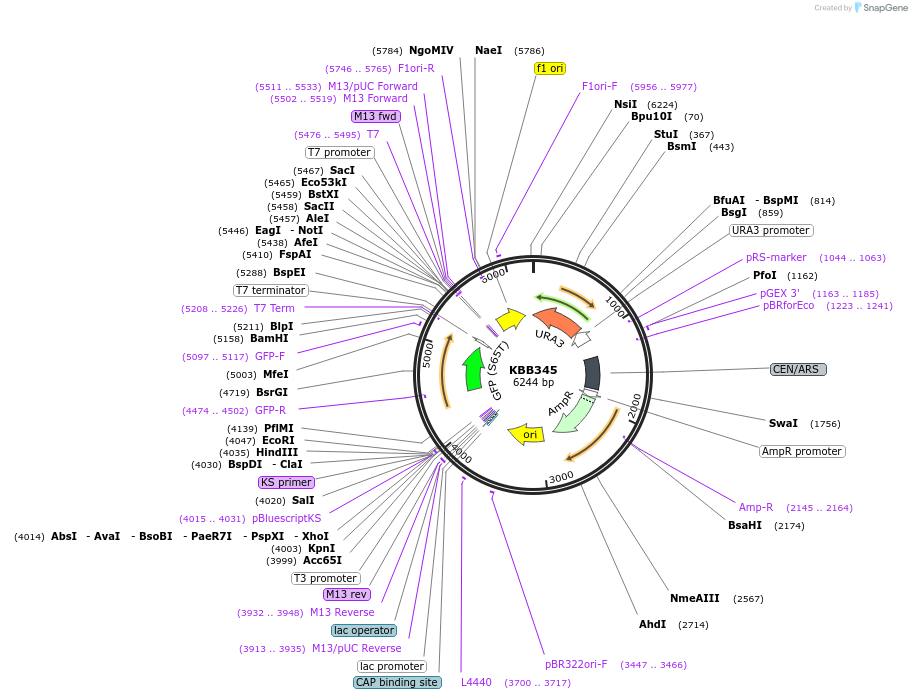
KBB345
Plasmid#185078Purposerfa3D46 truncation fused to GFPDepositorInsertRFA3
TagsGFPExpressionYeastMutationRfa3 truncation aa 1-46Available SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
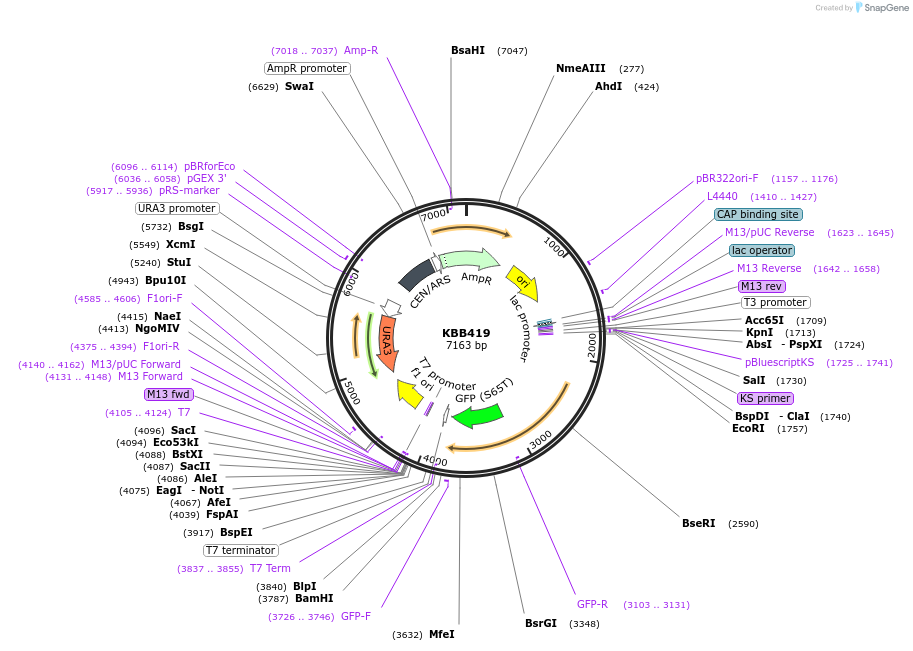
KBB419
Plasmid#185089PurposeSwi6L-GFP expressing N-term 273 amino acids of Swi6DepositorInsertSWI6
TagsGFPExpressionYeastMutationSwi6 truncation at aa 273Available SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
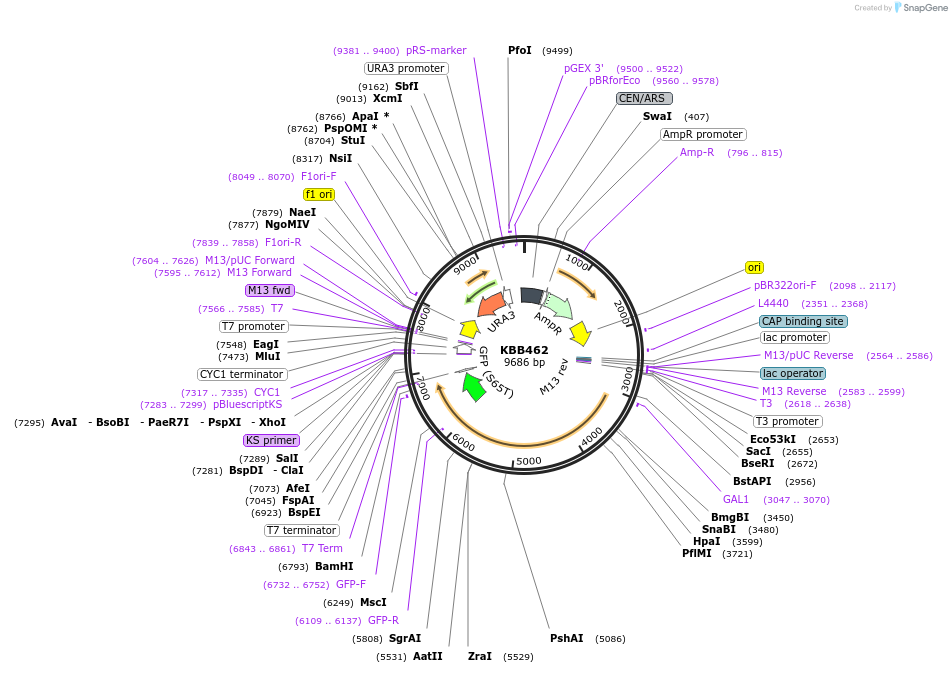
KBB462
Plasmid#185095PurposeNMA111-GFP with NLS1 and NLS2 mutated under control of GAL1 promoterDepositorInsertNMA111
TagsGFPExpressionYeastMutationGAL1::nma111Dnls1Dnls2-GFPPromoterGAL1Available SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
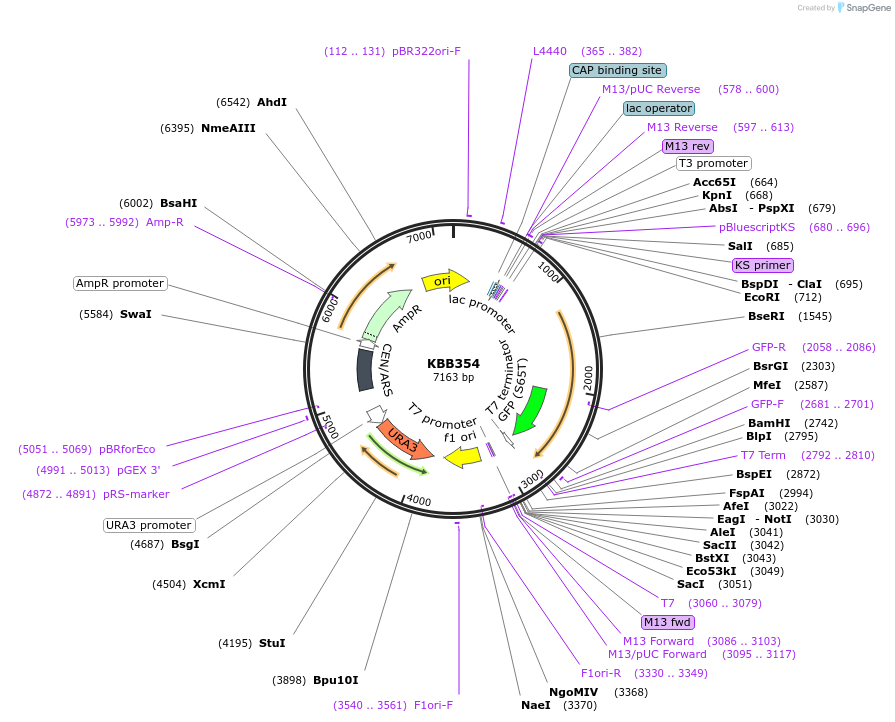
KBB354
Plasmid#185082PurposeSwi6L-GFP expressin N-term 273 amino acids of Swi6DepositorInsertSWI6
TagsGFPExpressionYeastMutationSwi6 truncation aa 1-273Available SinceJuly 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
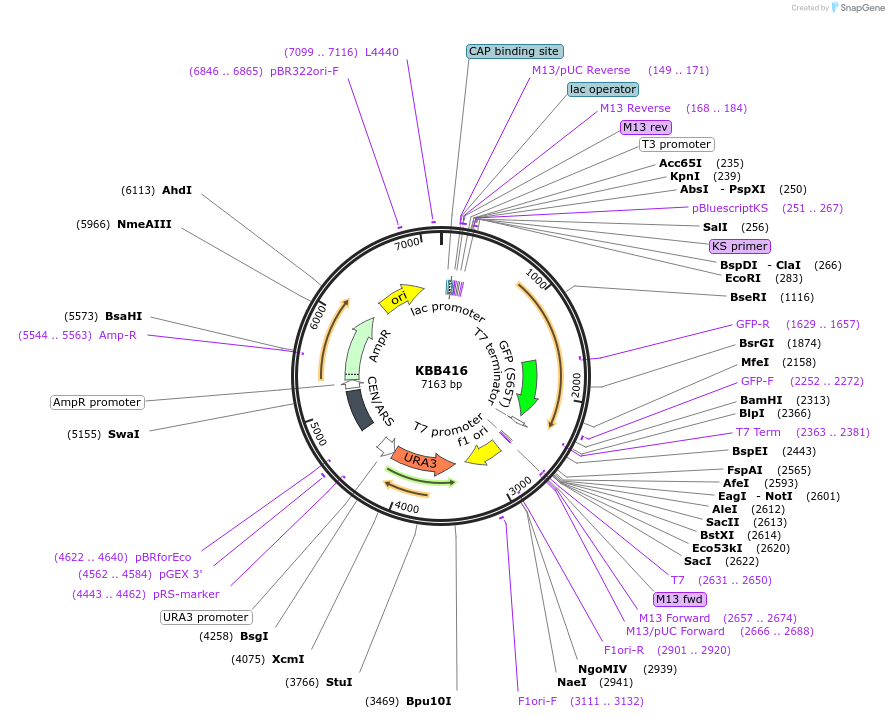
KBB416
Plasmid#185087PurposeSwi6L-GFP with nuclear export signal mutated to alaninesDepositorInsertSWI6
TagsGFPExpressionYeastMutationSwi6 nuclear export signal L to A mutationsAvailable SinceJuly 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
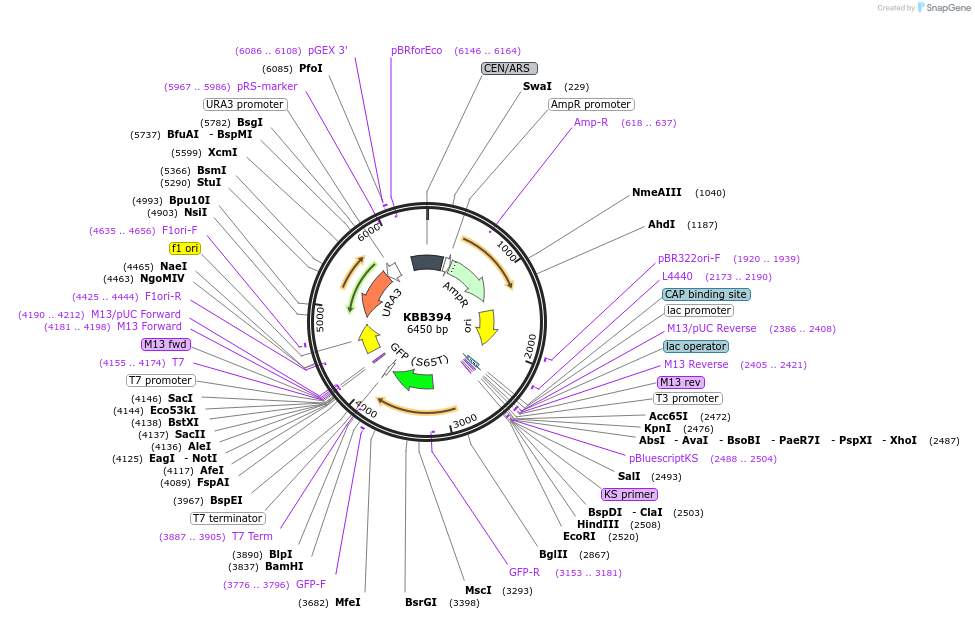
KBB394
Plasmid#185086PurposeNMA111 with both nuclear localization signals mutated to alanines fused to GFP (mutagenesis of KBB280)DepositorInsertNMA111
TagsGFPExpressionYeastMutationNma111 KKR 9-11 AAA; KRK 28-30 AAAAvailable SinceJuly 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
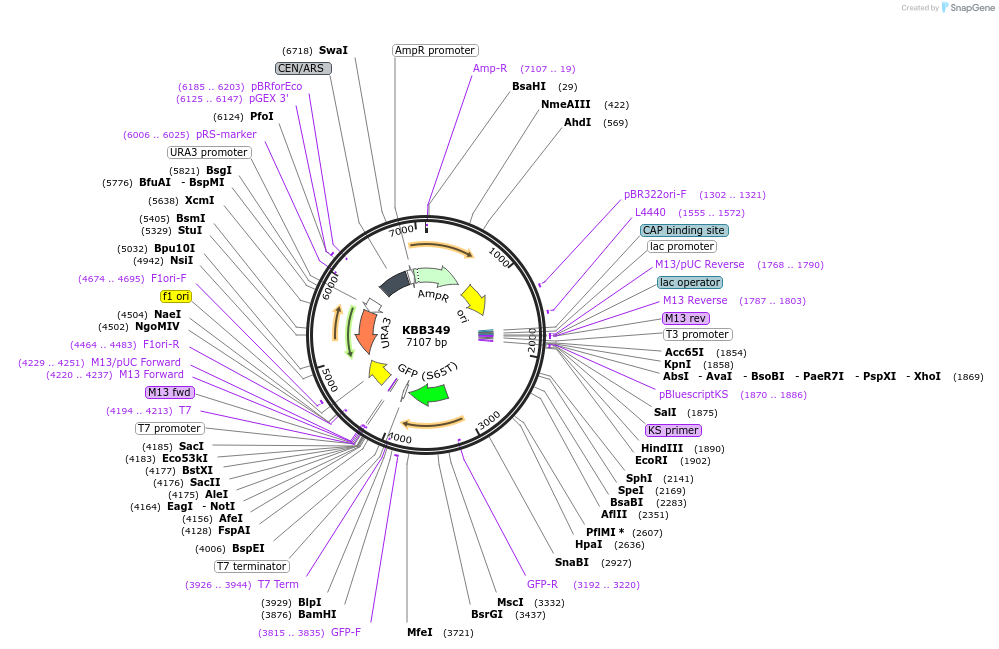
KBB349
Plasmid#185080Purposerfa2D248 truncation fused to GFPDepositorInsertRFA2
TagsGFPExpressionYeastMutationRfa2 truncation aa 1-247Available SinceJuly 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
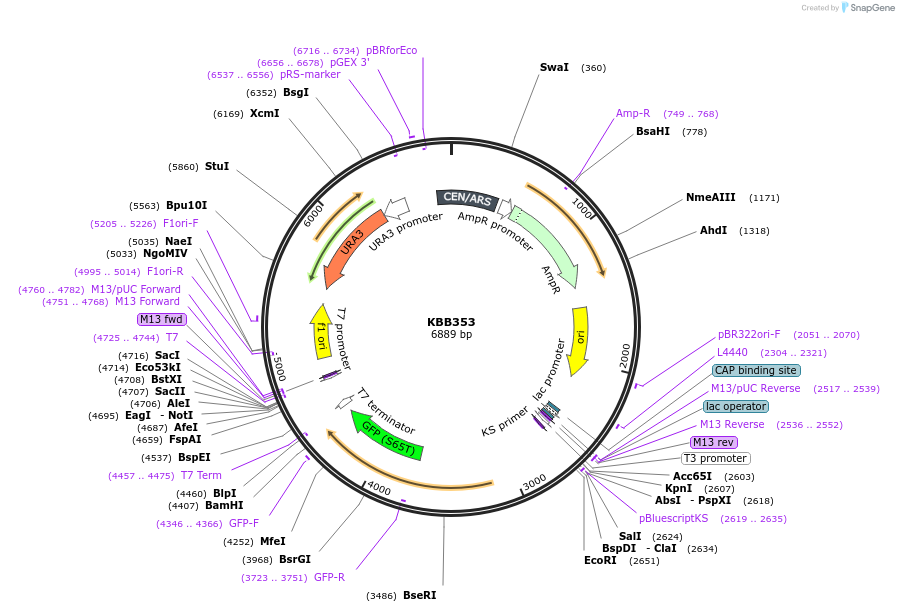
KBB353
Plasmid#185081PurposeSwi6M-GFP expressing N-term 181 amino acids of Swi6DepositorInsertSWI6
TagsGFPExpressionYeastMutationSwi6 truncation aa 1-181Available SinceJuly 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
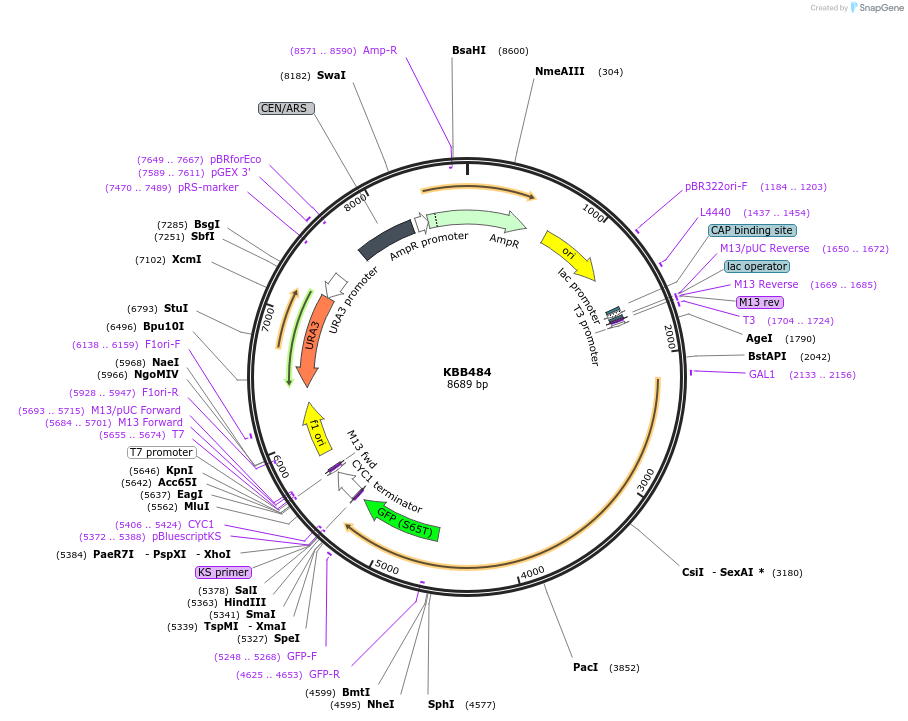
KBB484
Plasmid#185132PurposeSWI6-GFP full length under control of GAL1 promotionDepositorInsertSWI6
TagsGFPExpressionYeastAvailable SinceJuly 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
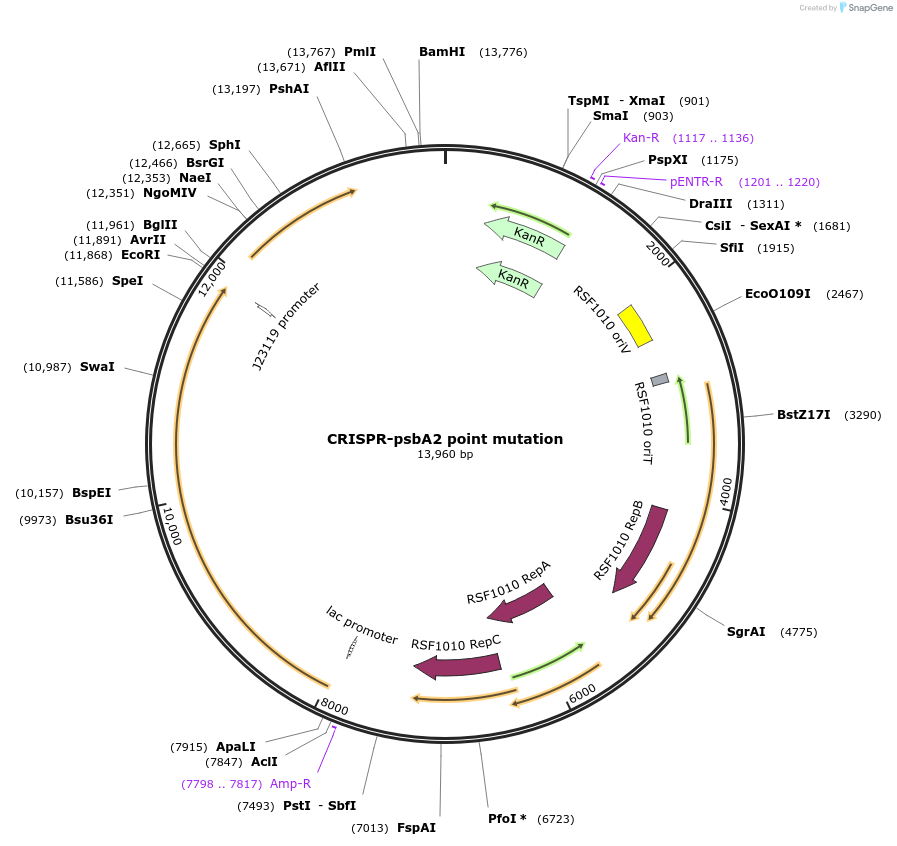
CRISPR-psbA2 point mutation
Plasmid#182929Purposepoint mutation on psbA2 by CRISPR in Synechocystis 6803DepositorInsertsddcpf1
gRNA targeting psbA2 in Synechocystis 6803: gatcttcggtcgcttgatctttc
psbA
UseCRISPRMutationS264A, and silent mutation to remove PAM siteAvailable SinceApril 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
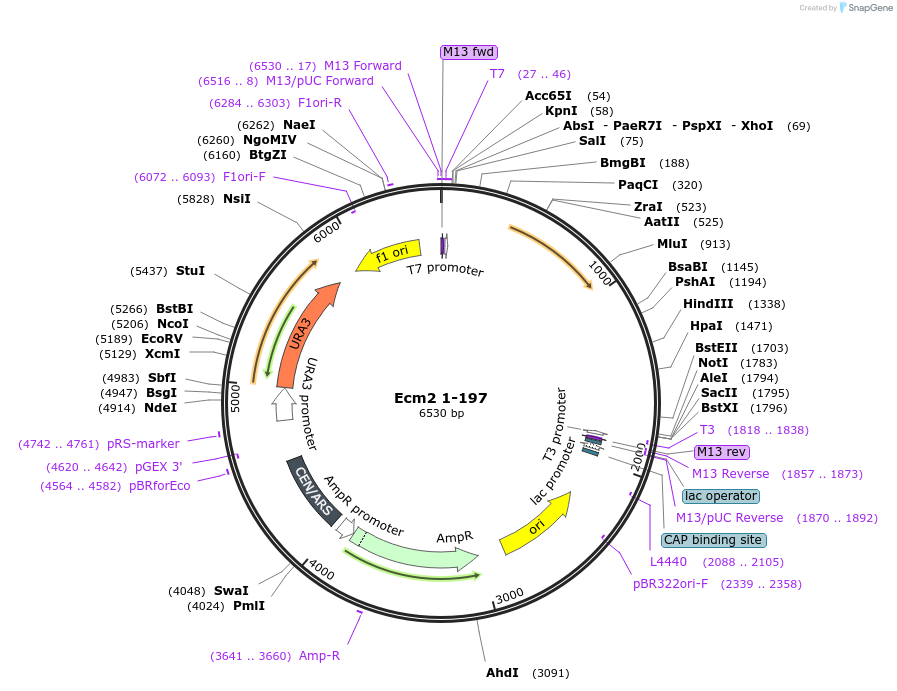
Ecm2 1-197
Plasmid#169924PurposeWT Ecm2 mutated to introduce stop codon after AA 197 of Ecm2DepositorInsertEcm2 1-197
ExpressionBacterial and YeastAvailable SinceOct. 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
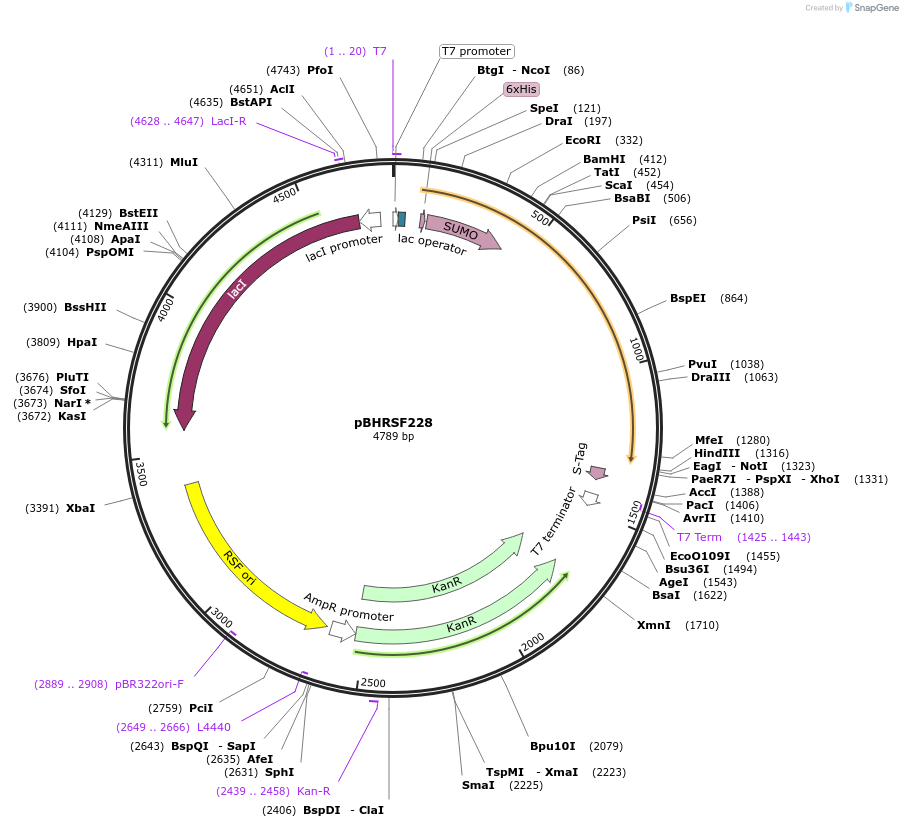
pBHRSF228
Plasmid#135007PurposeMusF2 prenyltransferases from Nostoc sp. UHCC 0398DepositorInsertMusF2 prenyltransferase
ExpressionBacterialAvailable SinceSept. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
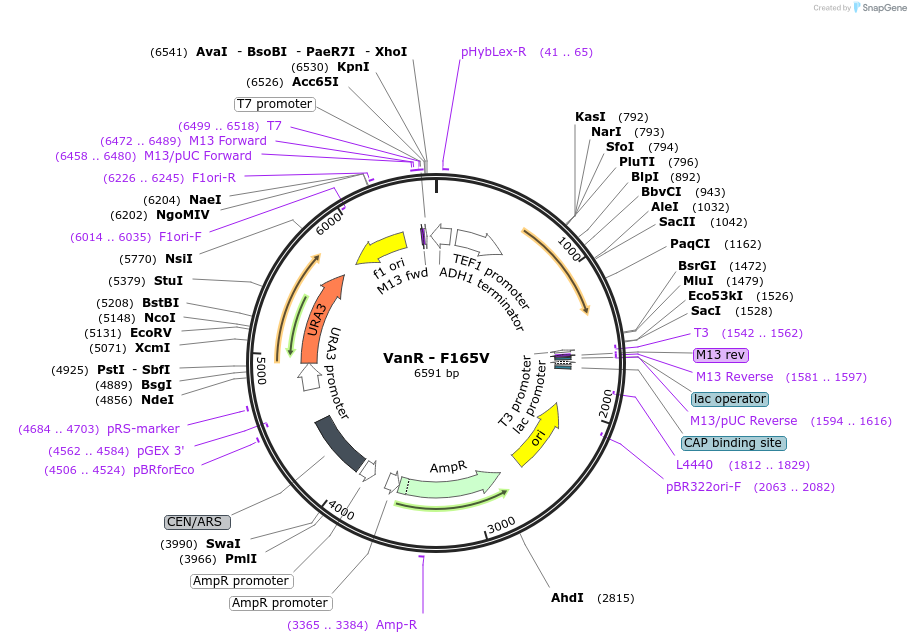
VanR - F165V
Plasmid#158965PurposeWild type VanR where the residue in position 165 was mutated from Phenylalanine to Valine. This mutation abolished the response to vanillic acid. The gene is under the control of the TEF1 promoter.DepositorInsertVanR - F165V
ExpressionYeastMutationWild type VanR from Caulobacter crescentus with m…PromoterTEF1Available SinceJan. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
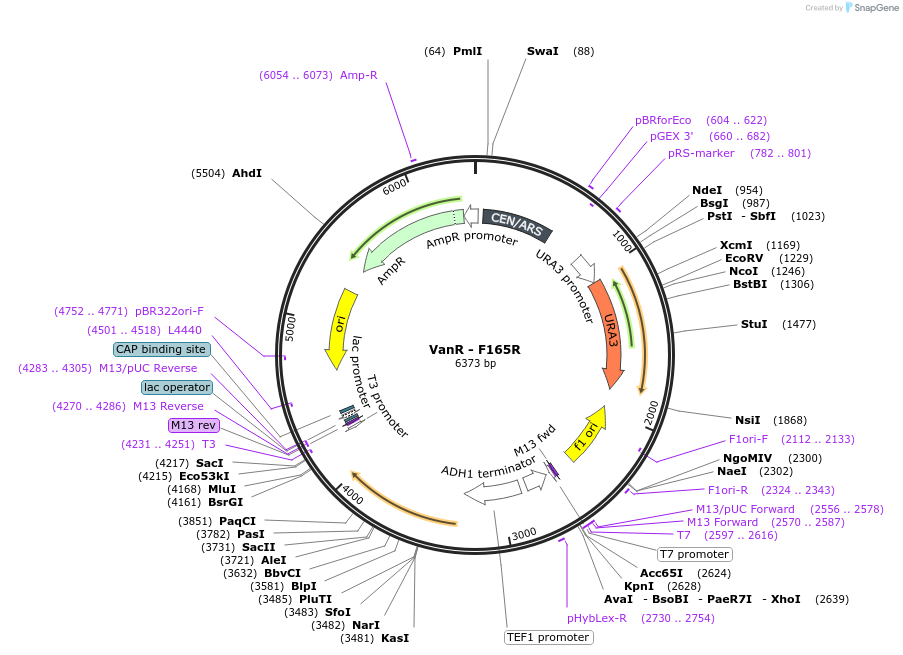
VanR - F165R
Plasmid#158966PurposeWild type VanR where the residue in position 165 was mutated from Phenylalanine to Arginine. This mutation abolished the response to vanillic acid. The gene is under the control of the TEF1 promoter.DepositorInsertVanR - F165R
ExpressionYeastMutationWild type VanR from Caulobacter crescentus with m…PromoterTEF1Available SinceJan. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
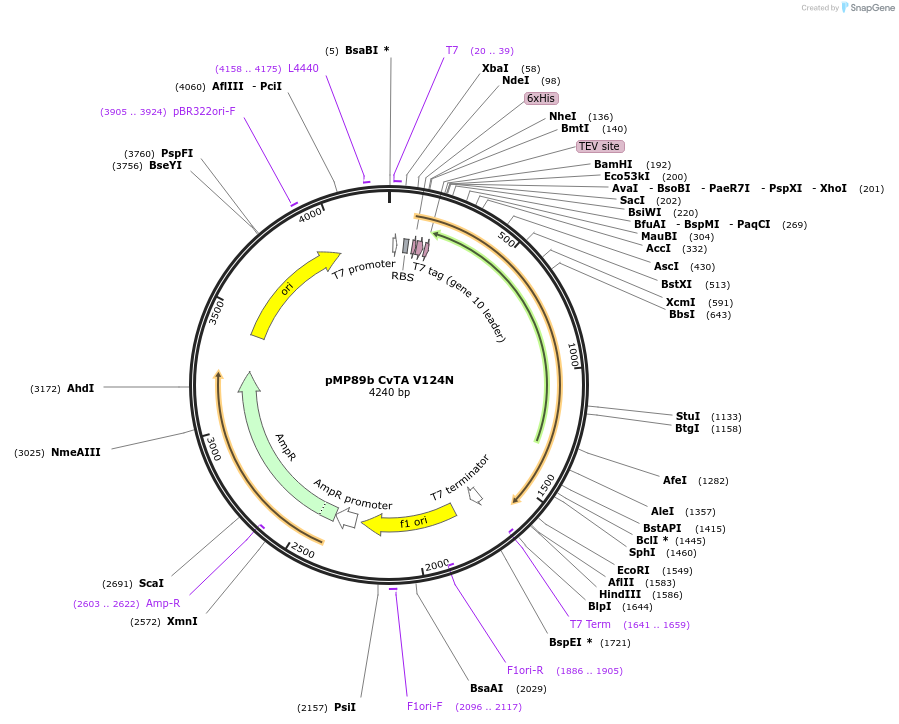
pMP89b CvTA V124N
Plasmid#141207PurposeChromobacterium violaceum transaminase with enhances affinity for PLP. TEV cleaving site after the N-terminal Histag.DepositorInsertCvTA
ExpressionBacterialMutationV124NAvailable SinceJune 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
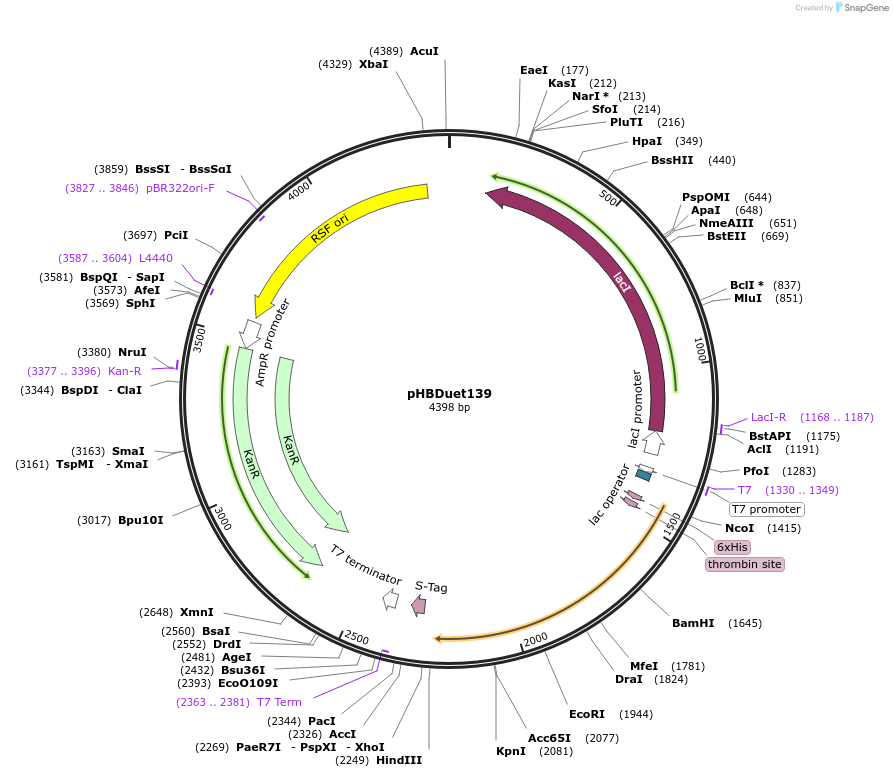
pHBDuet139
Plasmid#121913PurposeCharge-Introduced NpuDnaB mini-inteinDepositorInsertCI-NpuDnaBΔ290 intein
TagsGB1 and His6-GB1ExpressionBacterialMutationDeletion of 290-residue endonuclease domain, I53K…PromoterT7Available SinceDec. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
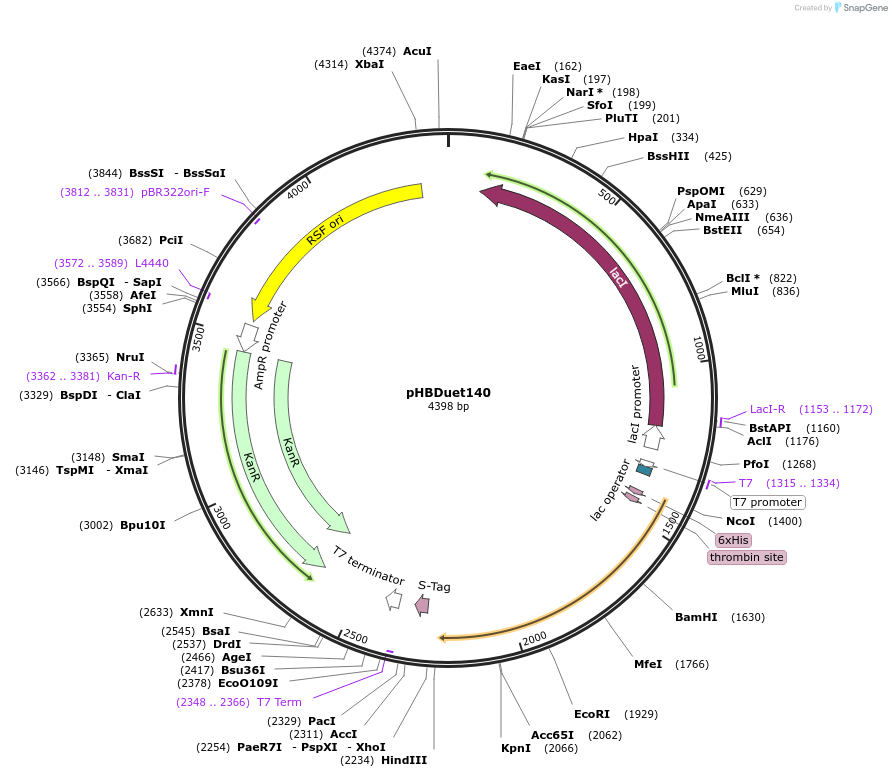
pHBDuet140
Plasmid#121914PurposeOrthogonal NpuDnaB mini-intein inteinDepositorInsertOth-NpuDnaBΔ290 intein
TagsGB1 and His6-GB1ExpressionBacterialMutationDeletion of 290-residue endonuclease domain, I53K…PromoterT7Available SinceDec. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
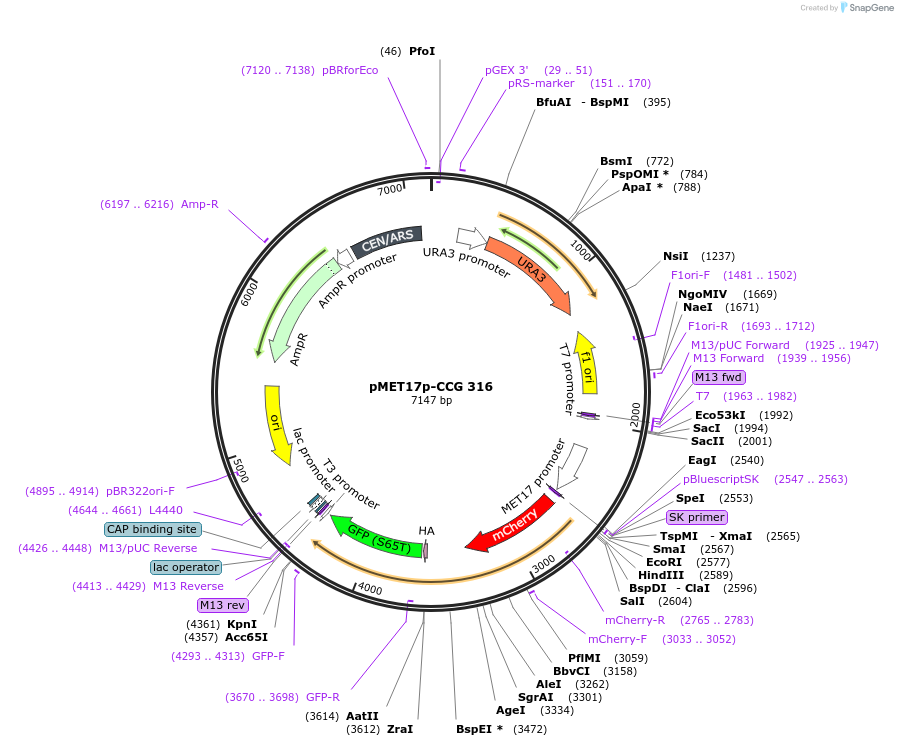
pMET17p-CCG 316
Plasmid#131166PurposeFor ScMET17 promoter regulated expression of mCherry-Cub-R-GFP (CCG) fused bait proteins, centromeric ARS plasmid, URA3 complements the uracil auxotrophy of S. cerevisiae ura mutantDepositorTypeEmpty backboneTagsCCGExpressionYeastAvailable SinceSept. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
Spt2-K166R-GFP
Plasmid#115573PurposeExpresses yeast Spt2-GFP fusion protein mutated from lysine to arginine at site 166DepositorInsertSPT2
TagsGFPExpressionBacterial and YeastMutationLysine 166 mutated to ArginineAvailable SinceOct. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
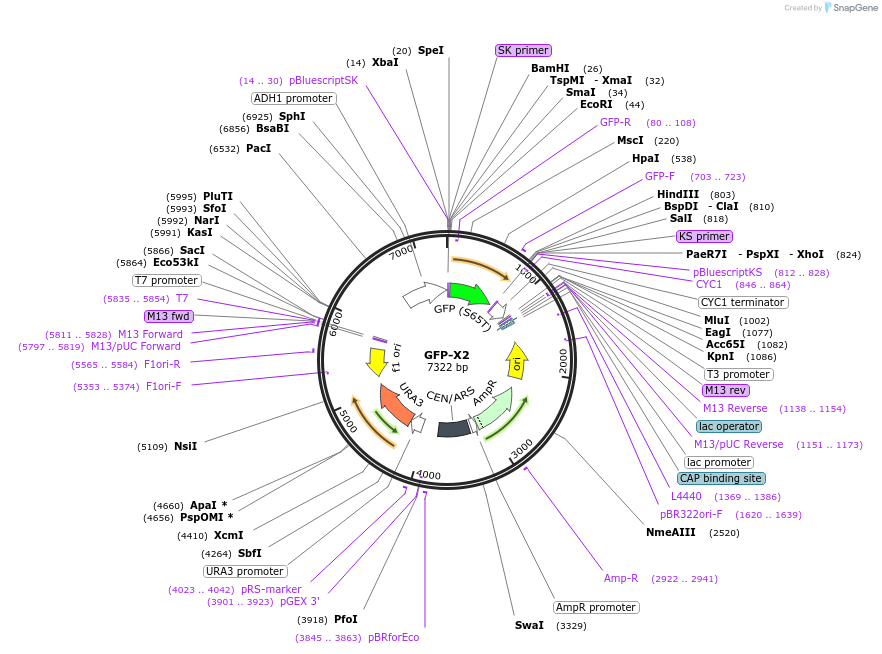
GFP-X2
Plasmid#115567PurposeGFP tethered to two repeats of the Gcn5 consensus motifDepositorInsertGreen Fluorescent Protein
ExpressionBacterial and YeastMutationTwo repeats of the Gcn5 consensus motif are tagge…PromoterADH1Available SinceOct. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
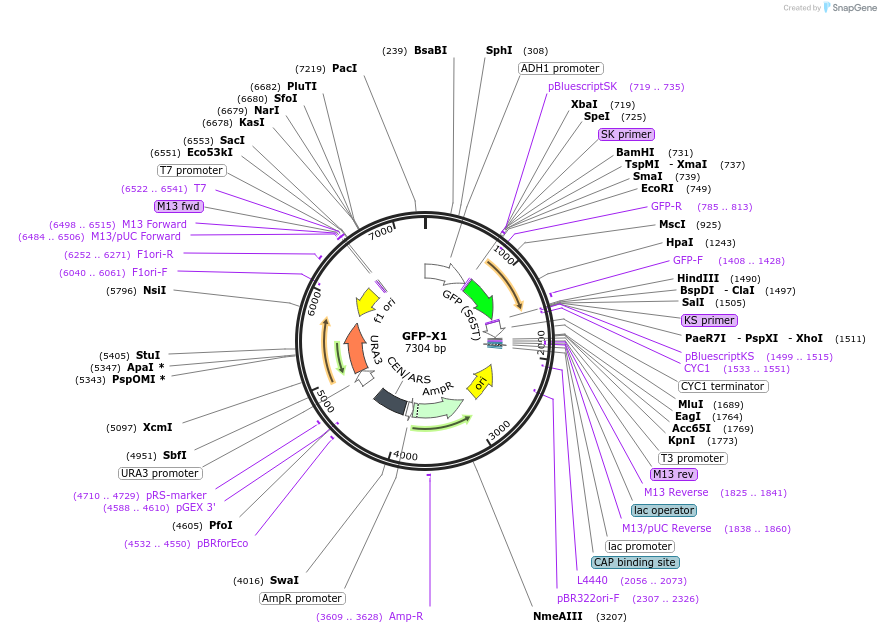
GFP-X1
Plasmid#115566PurposeGFP tethered to a single repeat of the Gcn5 consensus motifDepositorInsertGreen Fluorescent Protein
ExpressionBacterial and YeastMutationSingle Gcn5 consensus motif repeat tethered to C-…PromoterADH1Available SinceOct. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
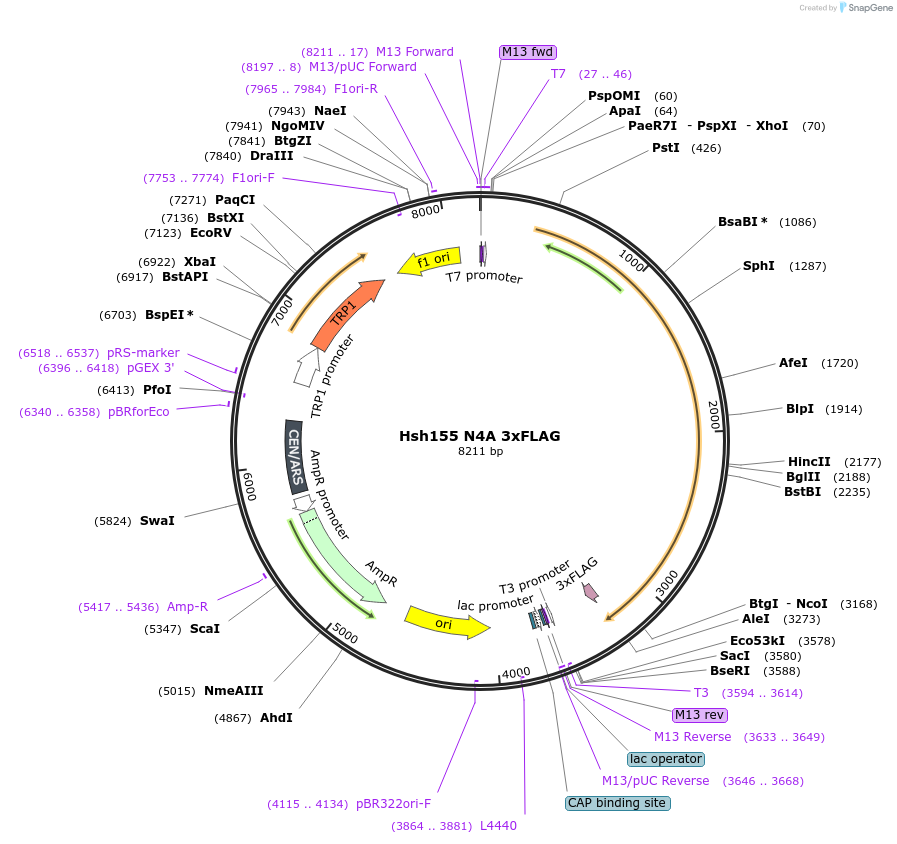
Hsh155 N4A 3xFLAG
Plasmid#111975PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker. It has 3xFLAG at the C-terminus. Contains the N4A mutation.DepositorInsertHsh155 N4A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationT178A R181A R182A R186AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
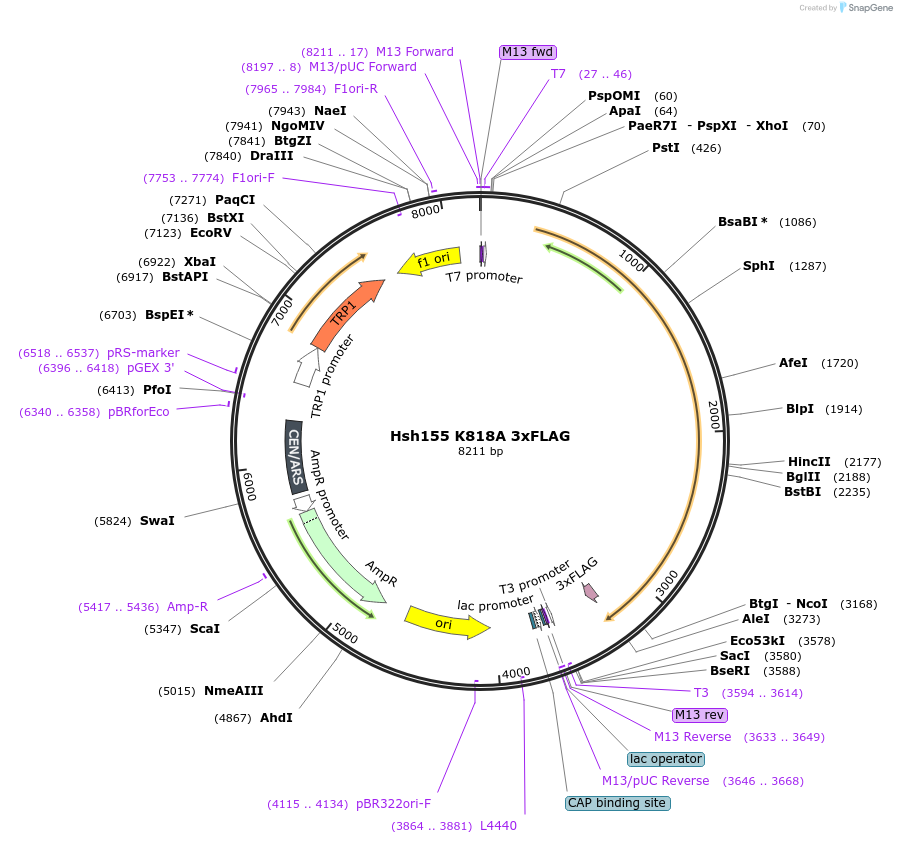
Hsh155 K818A 3xFLAG
Plasmid#111982PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker for shuffle into a HSH155 deletion strain. It has 3xFLAG at the C-terminus. Contains K818A mutation. LethalDepositorInsertHsh155 K818A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationK818AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
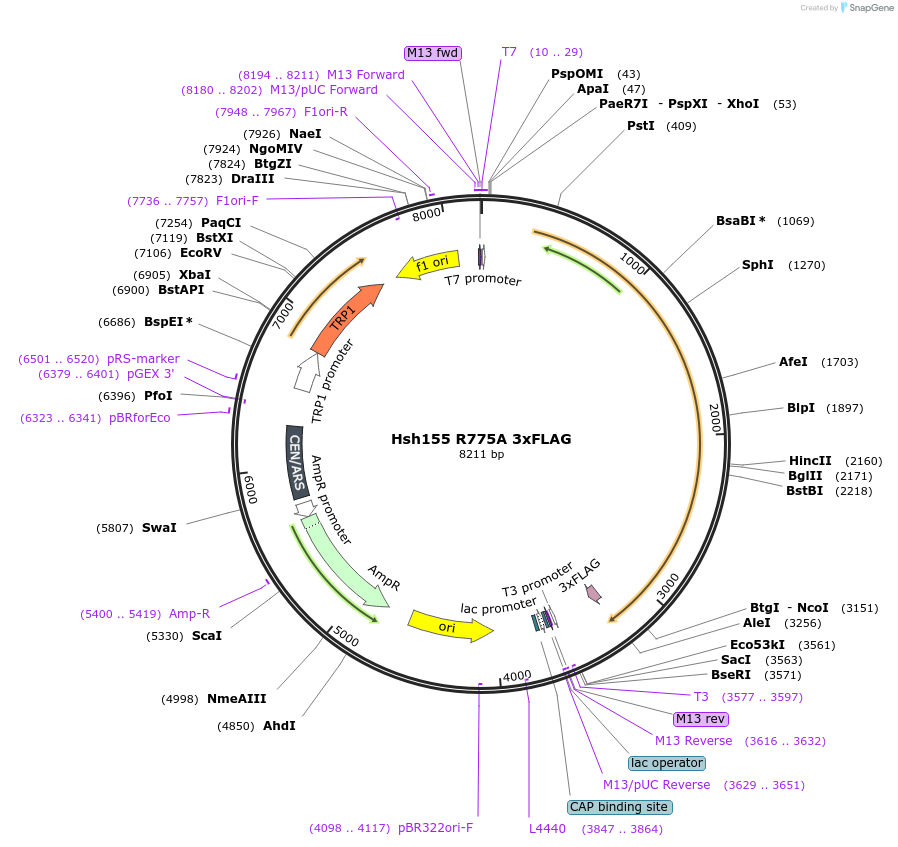
Hsh155 R775A 3xFLAG
Plasmid#111977PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker for shuffle into a HSH155 deletion strain. It has 3xFLAG at the C-terminus. Contains R775A mutation. LethalDepositorInsertHsh155 R775A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationR775AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
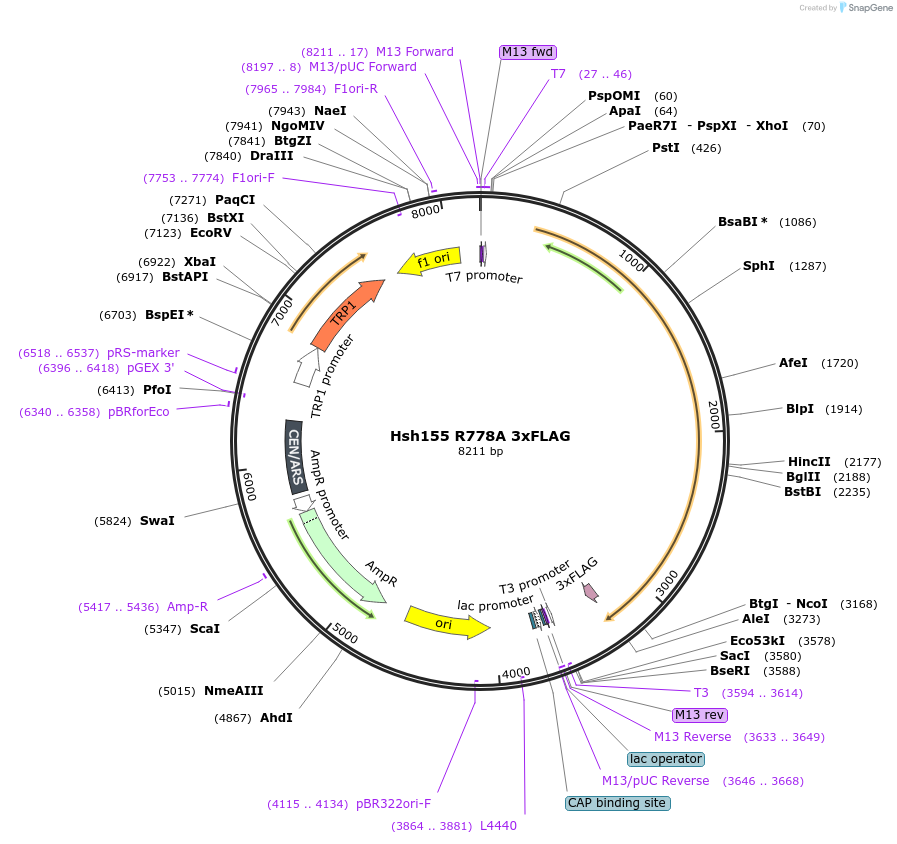
Hsh155 R778A 3xFLAG
Plasmid#111979PurposeShuffle plasmid containing the HSH155 gene and surrounding regions. It bears a TRP marker for shuffle into a HSH155 deletion strain. It has 3xFLAG at the C-terminus. Contains R778A mutation. LethalDepositorInsertHsh155 R778A 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationR778AAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
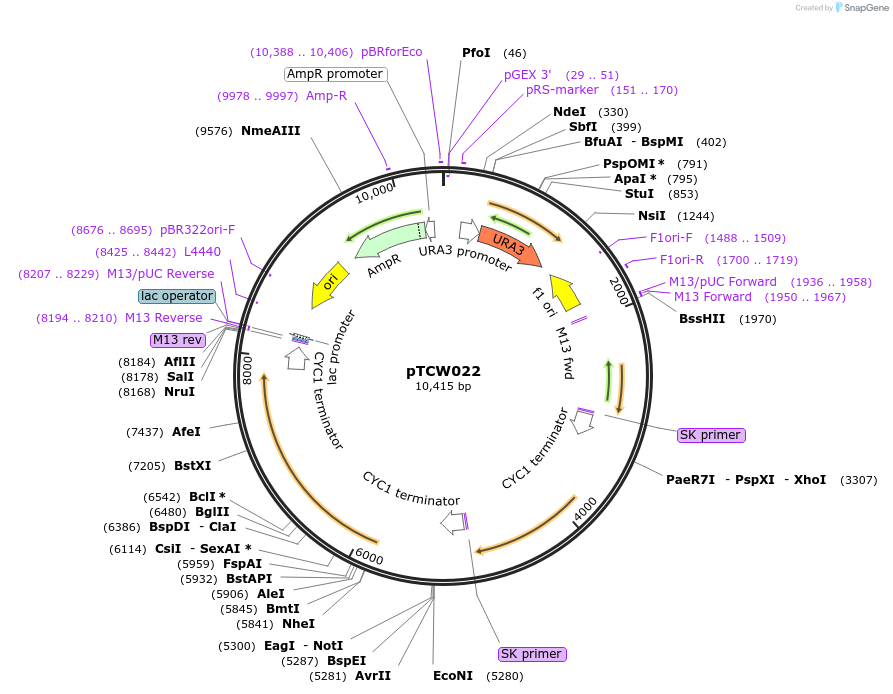
pTCW022
Plasmid#83532Purposepheromone mediated expression of UBiC, ARO4, and TKL1DepositorInsertpFUS1J2-UBiC-CYC1t-pFUS1J2-ARO4*-CYC1t-pFUS1J2-TKL1-CYC1t
ExpressionYeastPromoterpFUS1J2Available SinceDec. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
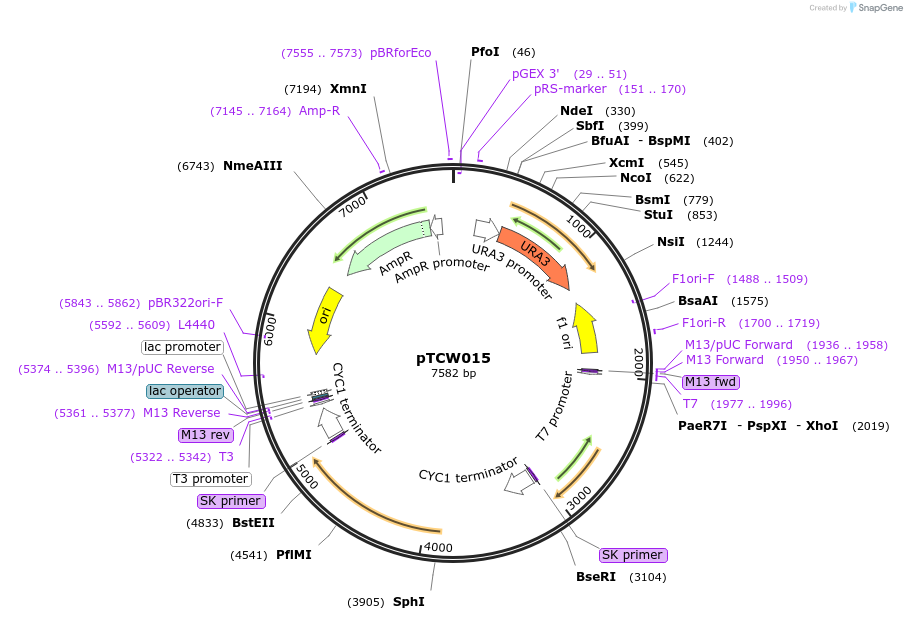
pTCW015
Plasmid#83530PurposeUBiC and feedback resistant ARO4 expression from pheromone inducible FUS1J2 promoterDepositorInsertpFUS1J2-UBiC-CYC1t-pFUS1J2-ARO4*-CYC1t
ExpressionYeastPromoterpFUS1J2Available SinceDec. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
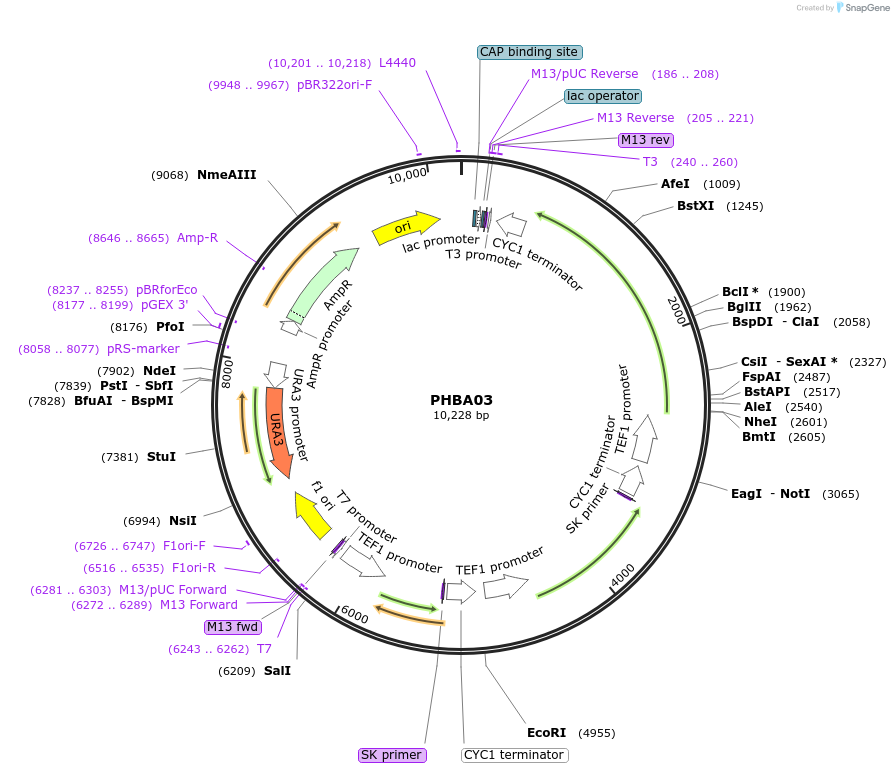
PHBA03
Plasmid#83537PurposeConstitutive TEF1 promoter mediated expression of the UBiC, ARO4*, and TKL1 genesDepositorInsertpTEF1-UBiC-CYC1t-pTEF1-ARO4*-CYC1t-pTEF1-TKL1-CYC1t
ExpressionYeastPromoterpTEF1Available SinceDec. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
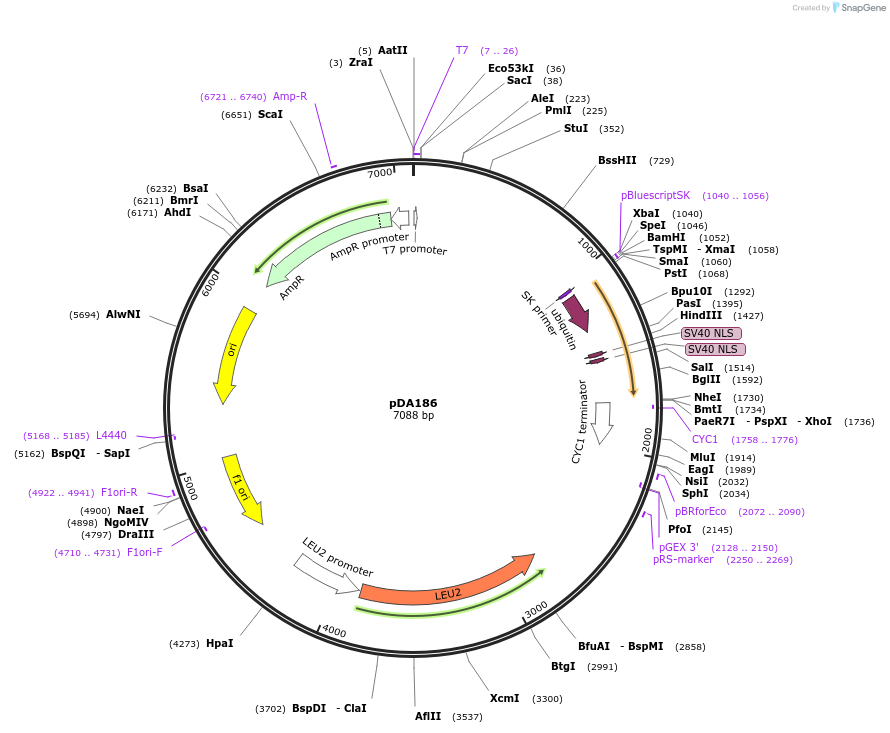
pDA186
Plasmid#74250PurposeIntermediate cloning vector, with MCS2 containing inducible unstable peptide driven by pGPD1 (UbiY 2xNLSs SZ3).DepositorInsertInducible peptide of the dPSTR
ExpressionBacterial and YeastPromoterpGPD1Available SinceMay 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
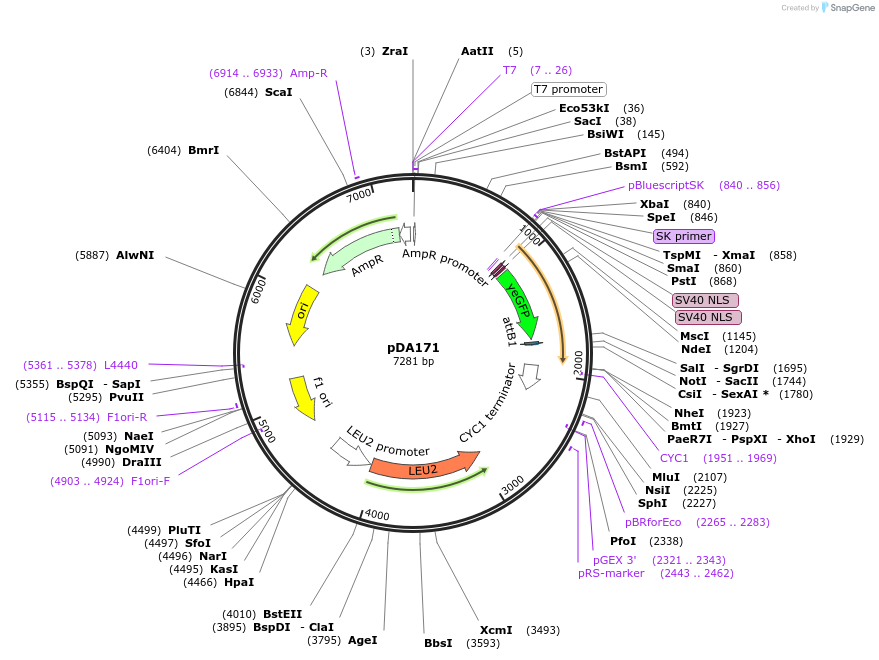
pDA171
Plasmid#74251PurposeIntermediate cloning vector, with MCS2 containing inducible stable peptide driven by pSTL1 (Venus 2xNLSs SZ1).DepositorInsertInducible peptide of the dPSTR
TagsVenusExpressionBacterial and YeastPromoterpSTL1Available SinceMay 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
-
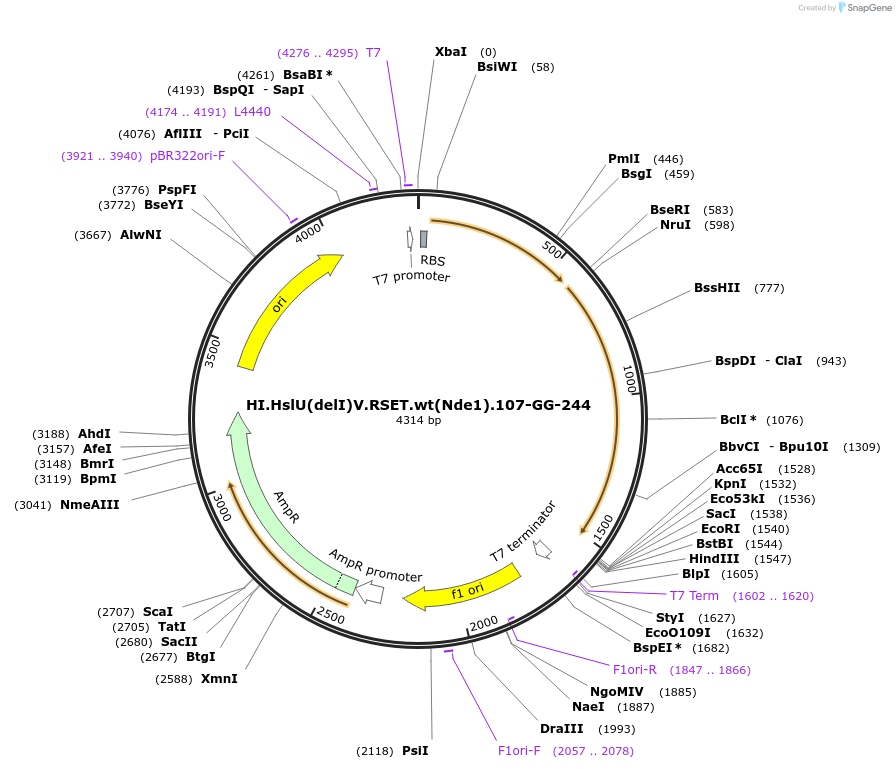
HI.HslU(delI)V.RSET.wt(Nde1).107-GG-244
Plasmid#17859DepositorInsertHaemophilus influenzae heat shock loci U and V
ExpressionBacterialMutationcoding sequence for residues 108-243 of HslU (the…Available SinceMay 29, 2008AvailabilityAcademic Institutions and Nonprofits only -
pJLI-Sup35N-KDG6-C
Plasmid#1238DepositorInsertSUP35 (SUP35 Budding Yeast)
UseYeast integrative plasmidMutationSUP35 middle region (aa124-253) was replaced with…Available SinceJan. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
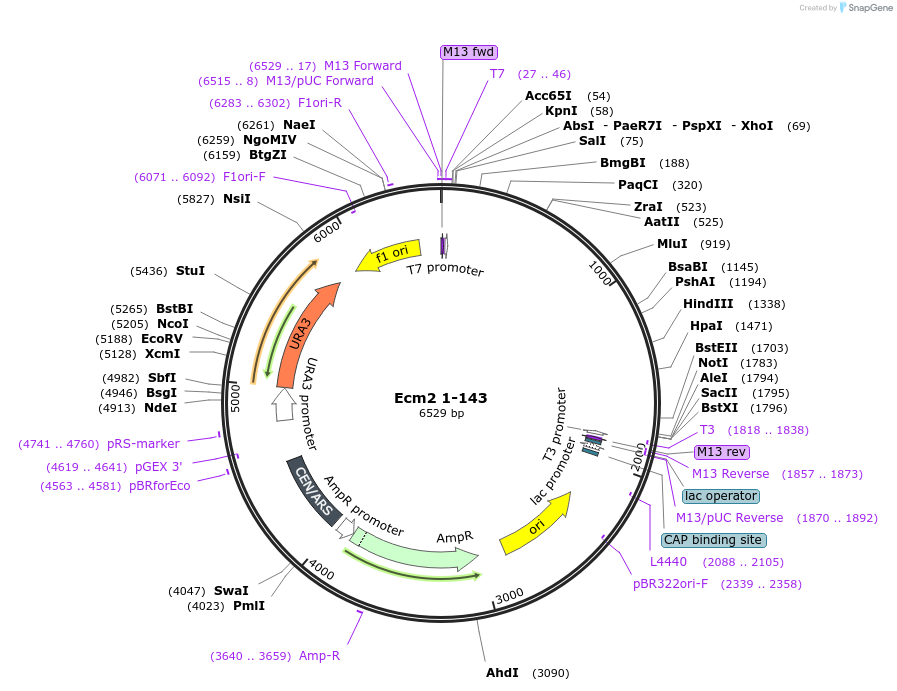
Ecm2 1-143
Plasmid#169923PurposeWT Ecm2 mutated to introduce stop codon after AA 143 of Ecm2DepositorInsertEcm2 1-143
ExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
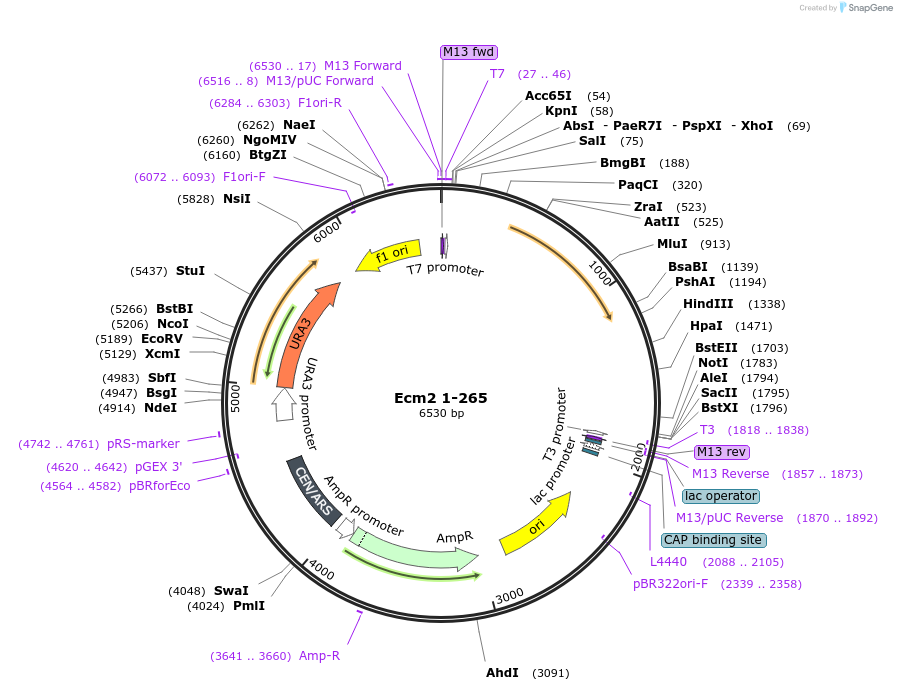
Ecm2 1-265
Plasmid#169925PurposeWT Ecm2 mutated to introduce stop codon after AA 265 of Ecm2DepositorInsertEcm2 1-265
ExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
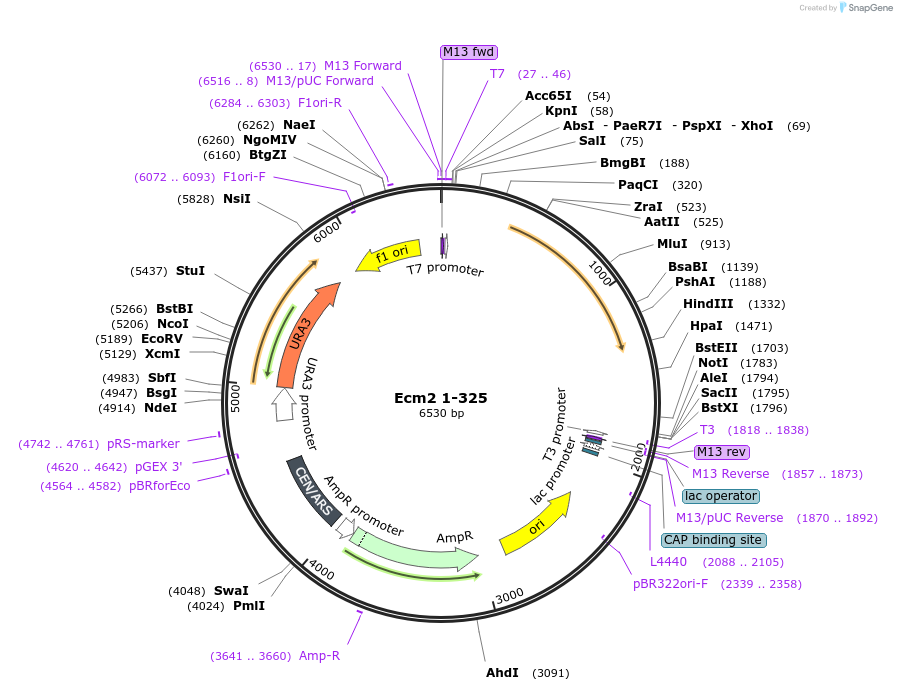
Ecm2 1-325
Plasmid#169926PurposeWT Ecm2 mutated to introduce stop codon after AA 325 of Ecm2DepositorInsertEcm2 1-325
ExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
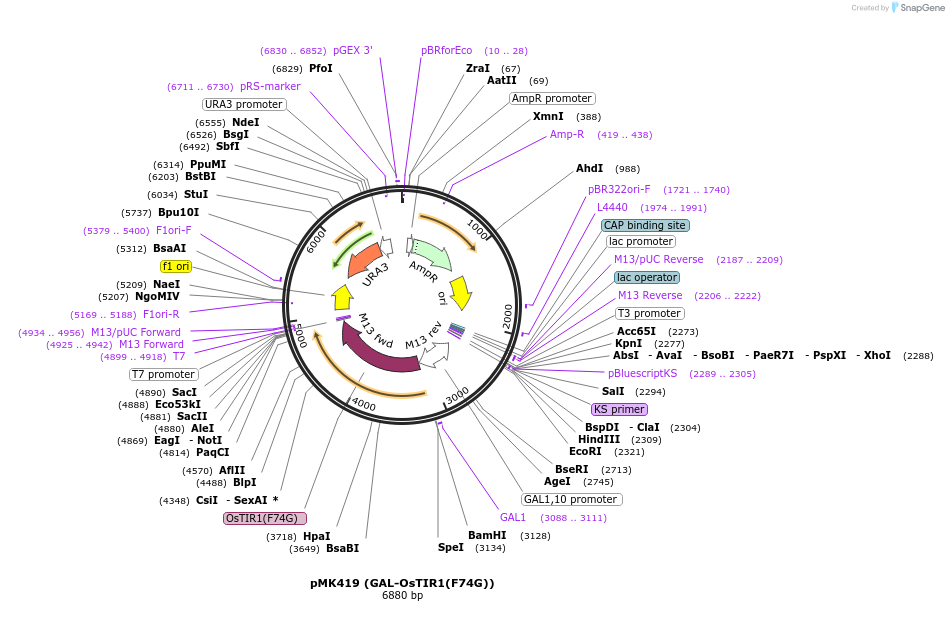
pMK419 (GAL-OsTIR1(F74G))
Plasmid#140656PurposeGAL-OsTIR1(F74G)DepositorInsertGAL-OsTIR1(F74G)
ExpressionYeastAvailable SinceNov. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
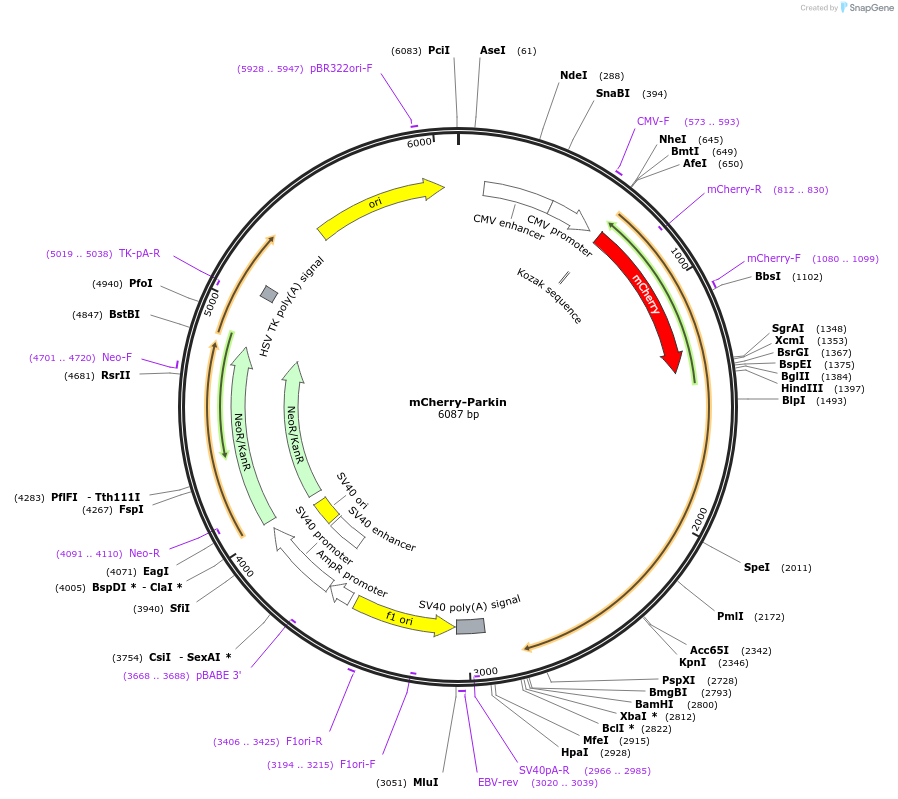
mCherry-Parkin
Plasmid#23956PurposeMammalian expression of human Park2 fused to mCherryDepositorAvailable SinceMarch 24, 2010AvailabilityAcademic Institutions and Nonprofits only -
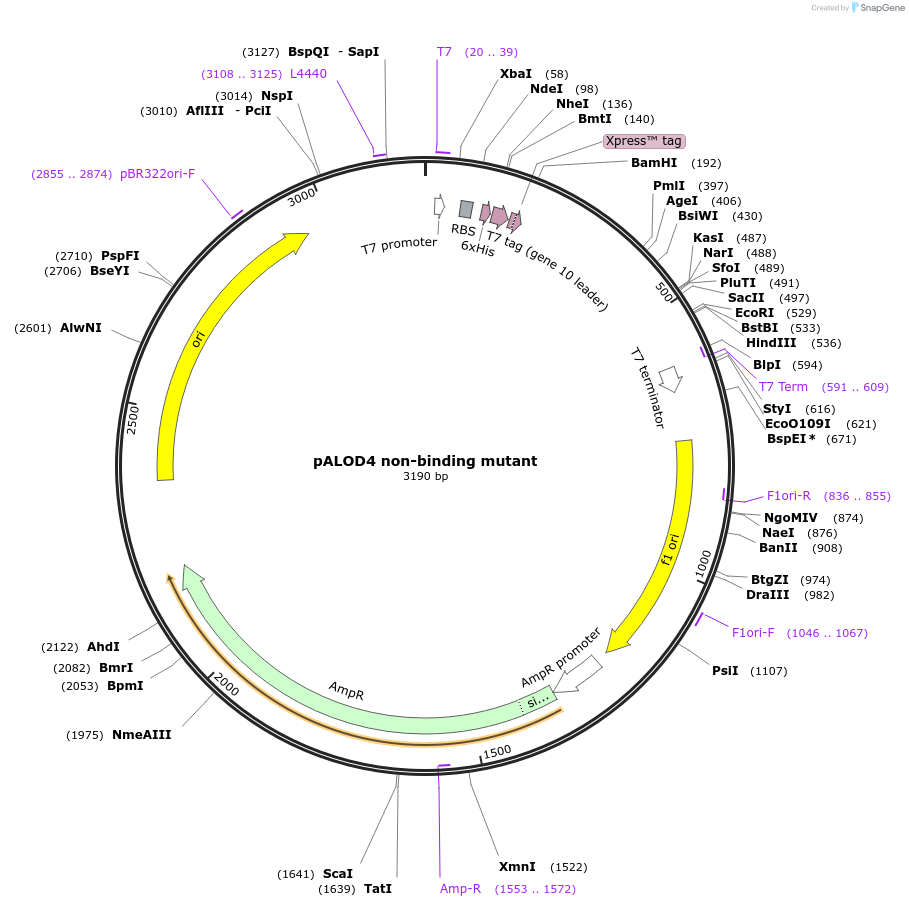
pALOD4 non-binding mutant
Plasmid#111027PurposeExpresses non-binding mutant version of p-ALOD4, has six inactivating mutations (G501A, T502A, T503A, L504A, Y505A, and P506A)DepositorInsertDomain 4 of Anthrolysin O
TagsHis6 tagExpressionBacterialMutationMutations: S404C, C472A, G501A, T502A, T503A, L5…PromoterT7Available SinceJune 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
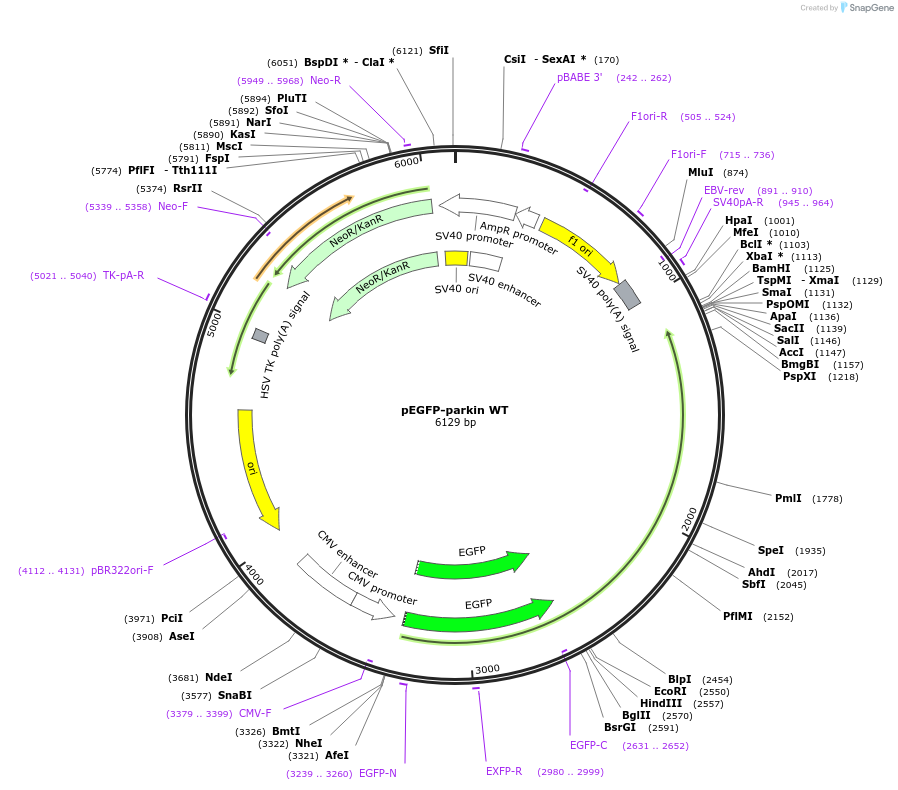
pEGFP-parkin WT
Plasmid#45875DepositorAvailable SinceNov. 14, 2013AvailabilityAcademic Institutions and Nonprofits only -
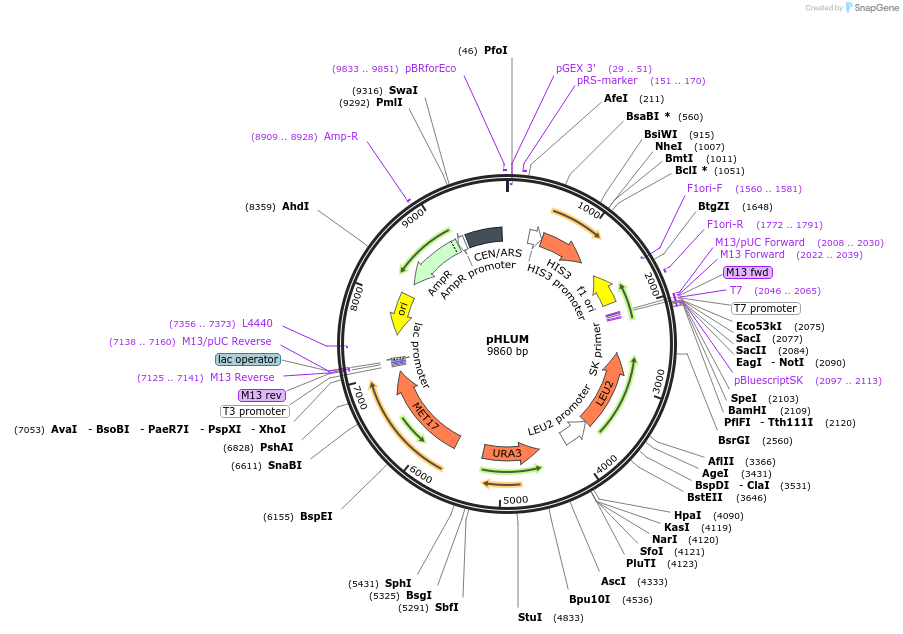
pHLUM
Plasmid#40276DepositorExpressionYeastPromoterendogenous LEU2, endogenous MET15/17, and endogen…Available SinceNov. 8, 2012AvailabilityAcademic Institutions and Nonprofits only -
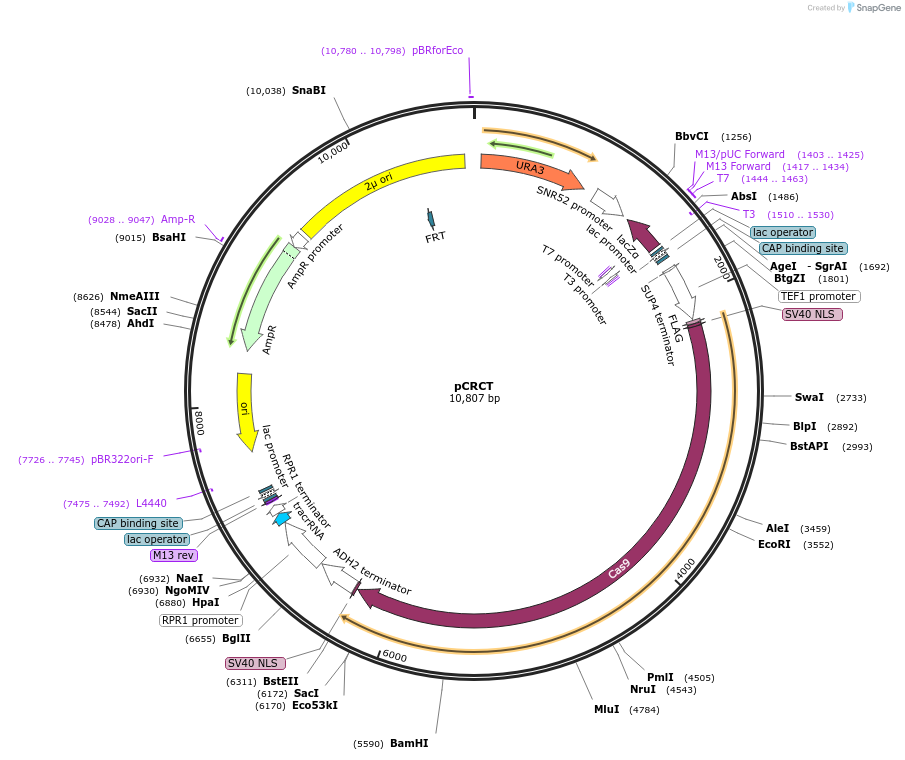
pCRCT
Plasmid#60621PurposePlasmid encoding iCas9, tracrRNA and crRNAsDepositorInsertsiCas9
tracrRNA
UseCRISPRTagsFLAG and SV40 NLSExpressionYeastMutationchanged Aspartate 147 to Tyrosine, Proline 411 to…PromoterRPR1p and TEF1pAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
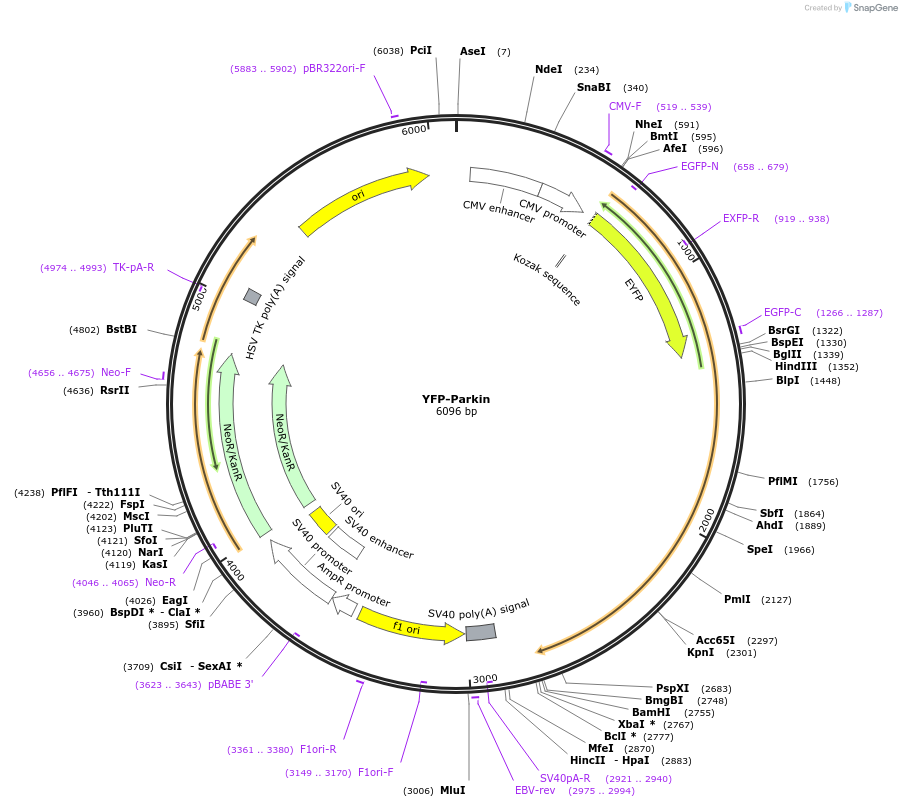
YFP-Parkin
Plasmid#23955PurposeMammalian expression of human Park2 fused to EYFPDepositorAvailable SinceApril 9, 2010AvailabilityAcademic Institutions and Nonprofits only -
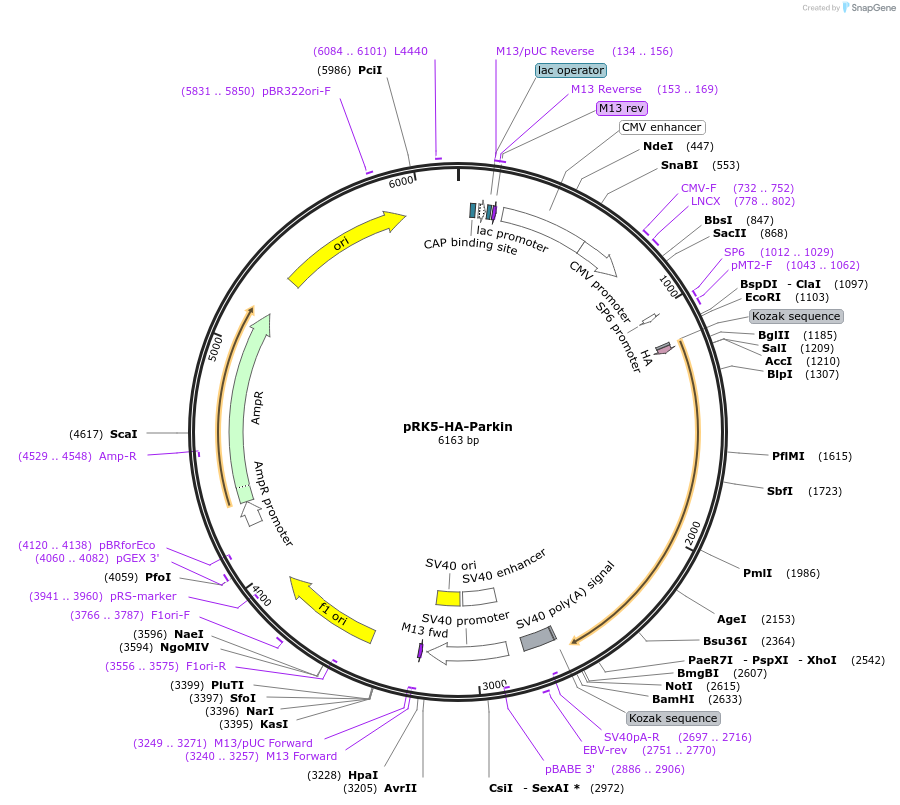
pRK5-HA-Parkin
Plasmid#17613DepositorAvailable SinceApril 3, 2008AvailabilityAcademic Institutions and Nonprofits only