We narrowed to 1,778 results for: GAD
-
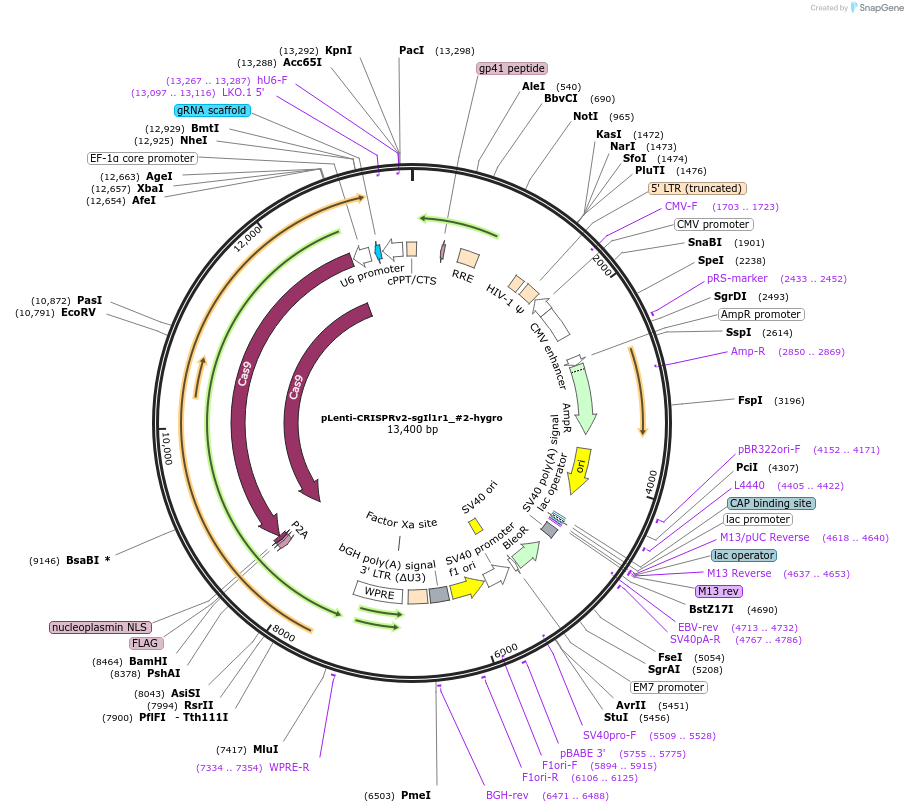
Plasmid#231990PurposeKnockout mouse Il1r1DepositorInsertsgRNA with Cas9 and hygromycin resistance (Il1r1 Mouse)
UseCRISPR and LentiviralAvailable SinceFeb. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
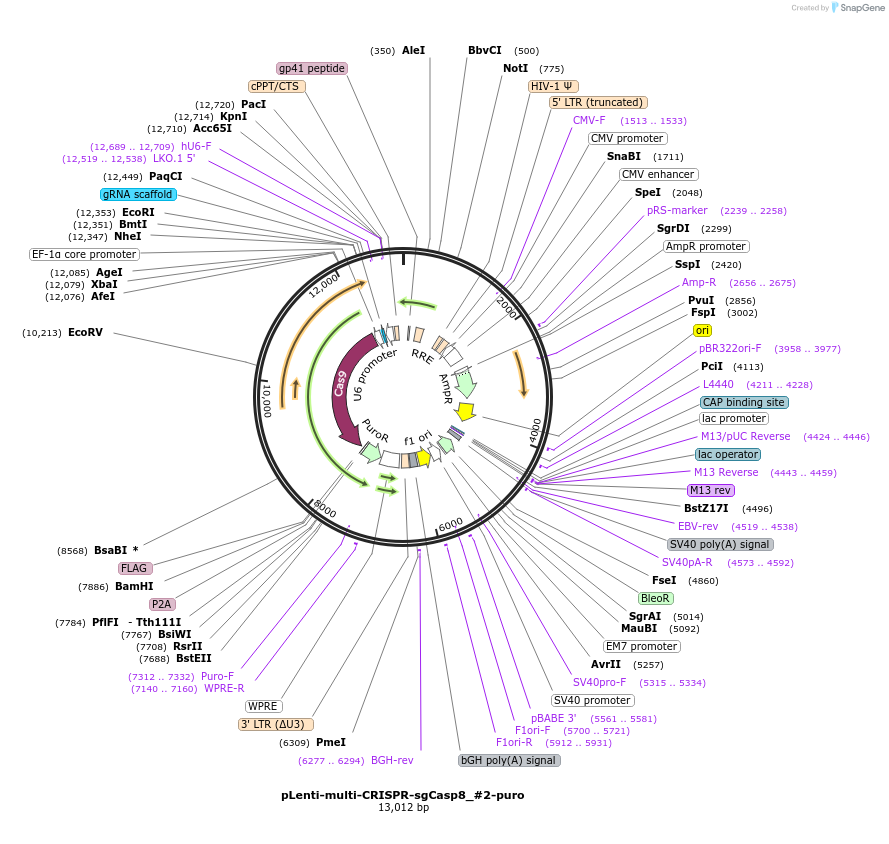
pLenti-multi-CRISPR-sgCasp8_#2-puro
Plasmid#231978PurposeKnockout mouse Casp8DepositorInsertsgRNA with Cas9 with puromycin resistance (Casp8 Mouse)
UseCRISPR and LentiviralAvailable SinceFeb. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
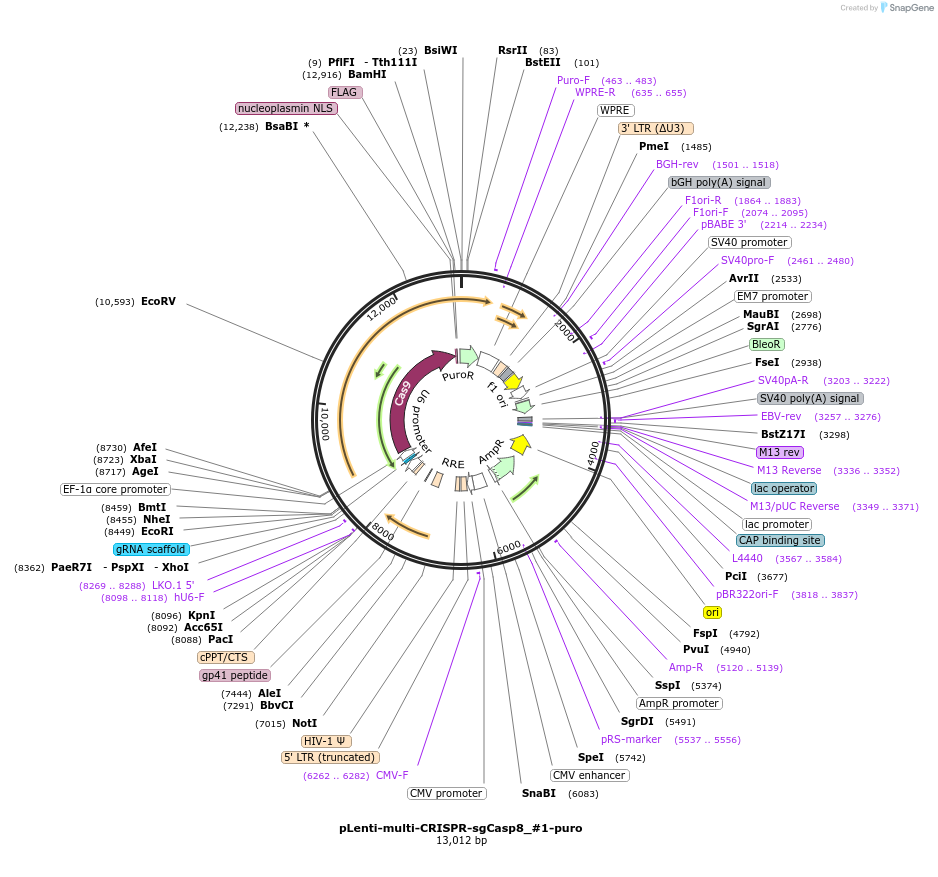
pLenti-multi-CRISPR-sgCasp8_#1-puro
Plasmid#231977PurposeKnockout mouse Casp8DepositorInsertsgRNA with Cas9 with puromycin resistance (Casp8 Mouse)
UseCRISPR and LentiviralAvailable SinceFeb. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
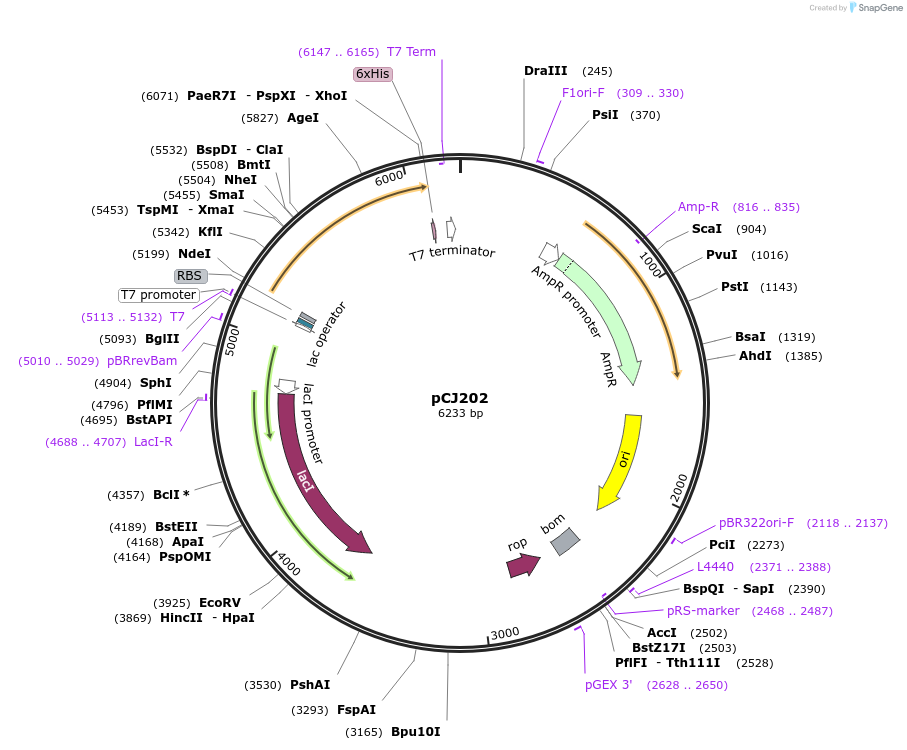
pCJ202
Plasmid#162675PurposepET-21b(+) based plasmid for expression of PETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating W159C and S238C mutations.DepositorInsertPETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1)
TagsHisExpressionBacterialMutationW159C, S238C; codon optimized for expression in E…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
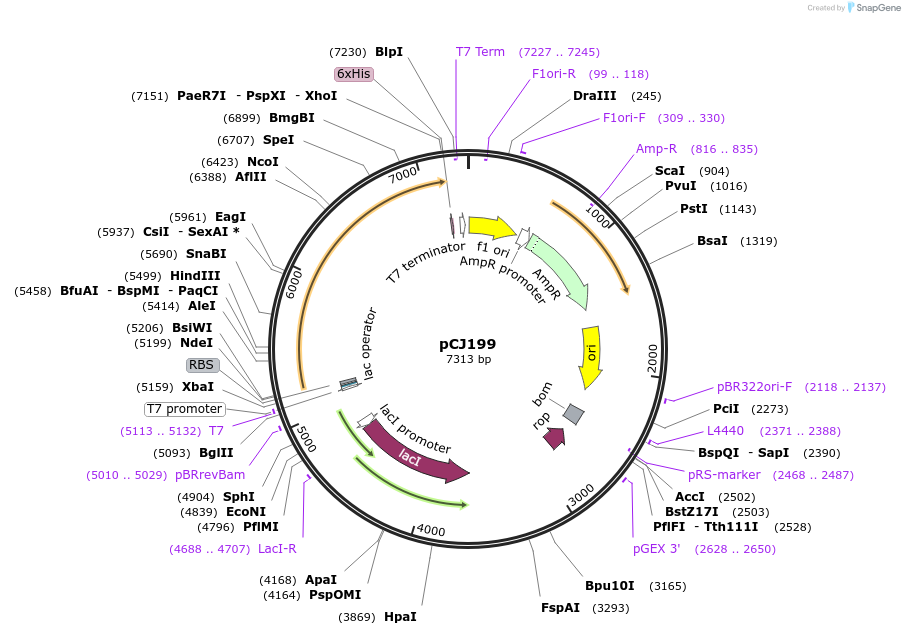
pCJ199
Plasmid#162672PurposepET-21b(+) based plasmid for expression of the putative MHET hydrolase from Comamonas thiooxydans (Genbank WP_080747404.1) with C-terminal His tag, codon optimized for expression in E. coli K12.DepositorInsertPutative MHET hydrolase from Comamonas thiooxydans (Genbank WP_080747404.1) with signal peptide
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
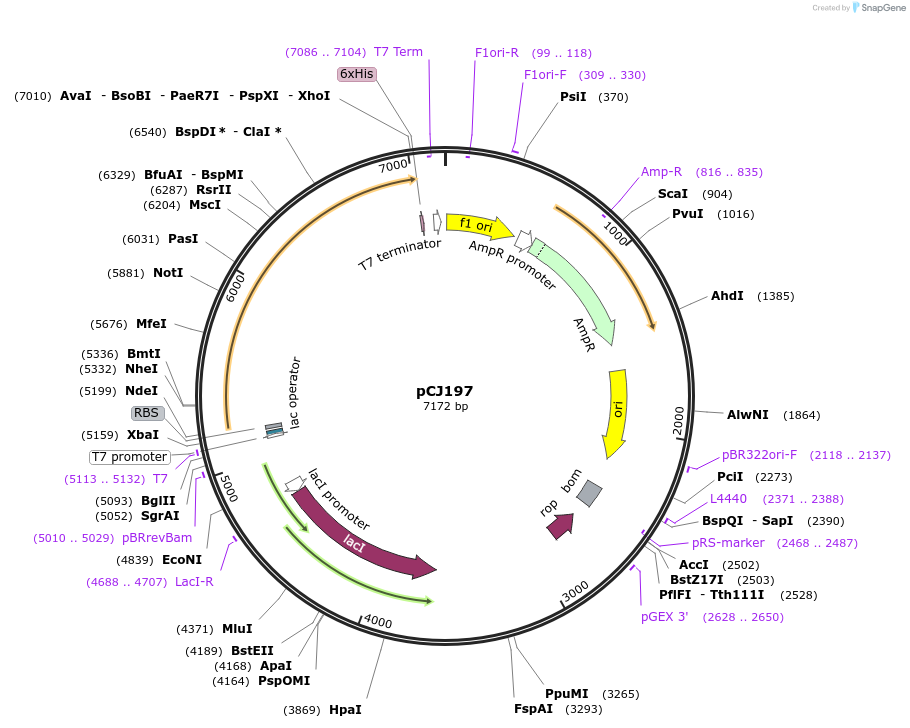
pCJ197
Plasmid#162670PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S131G mutation.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1
TagsHisExpressionBacterialMutationS131G; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
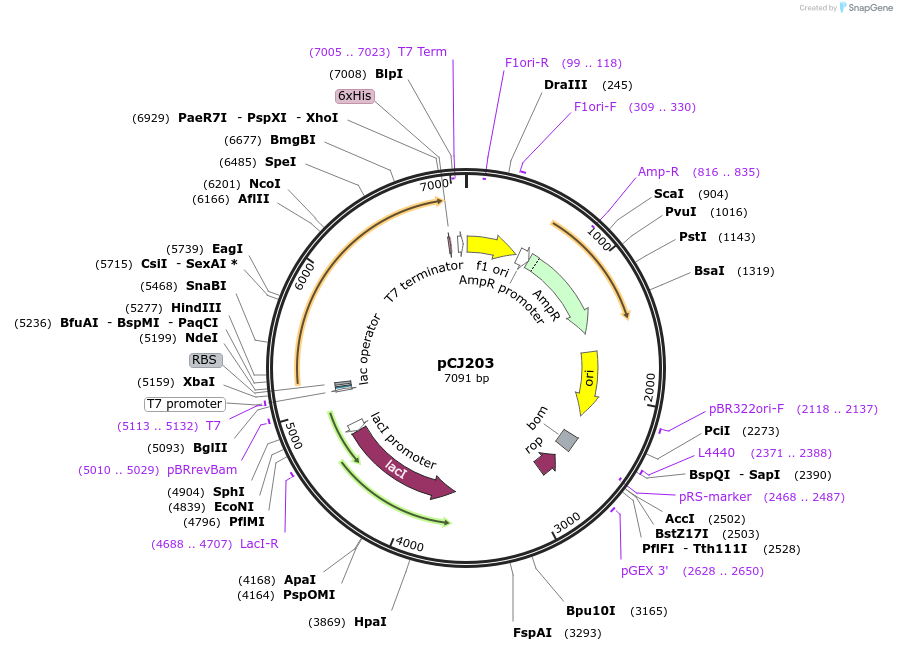
pCJ203
Plasmid#162678PurposepET-21b(+) based plasmid for expression of the putative MHET hydrolase from Comomonas thiooxydans (Genbank WP_080747404.1) with deletion of the predicted signal peptide (Phe2:Ala75)DepositorInsertPutative MHETase from Comomonas thiooxydans (Genbank WP_080747404.1) without 75 residue signal peptide
TagsHisExpressionBacterialMutationdeltaF2-A75; codon optimized for expression in E.…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
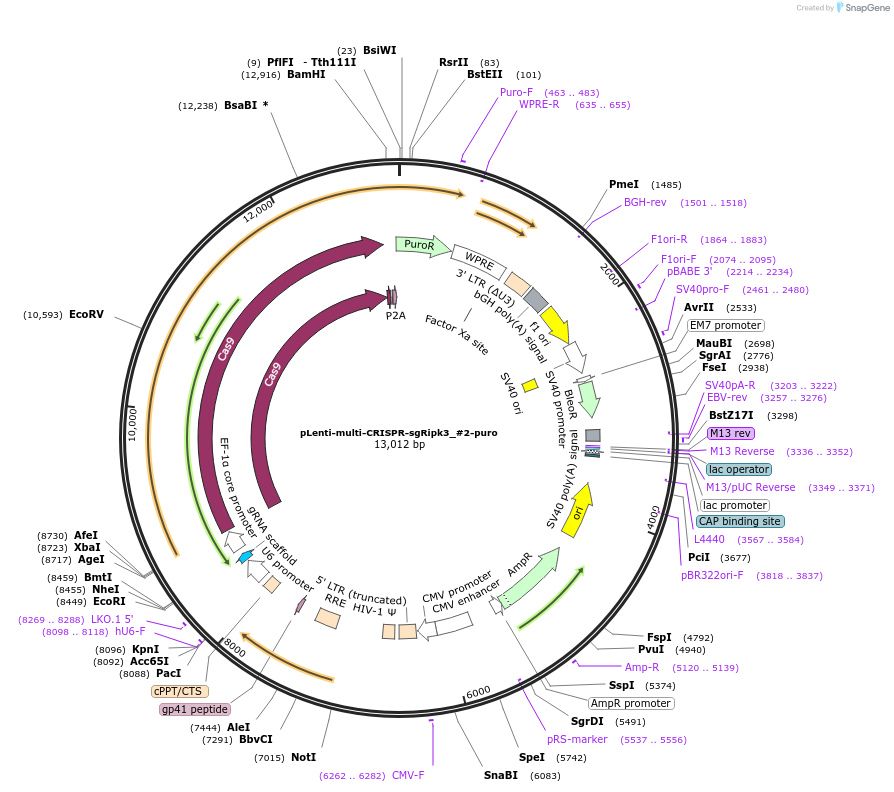
pLenti-multi-CRISPR-sgRipk3_#2-puro
Plasmid#231982PurposeKnockout mouse Ripk3DepositorInsertsgRNA with Cas9 with puromycin resistance (Ripk3 Mouse)
UseCRISPR and LentiviralAvailable SinceFeb. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
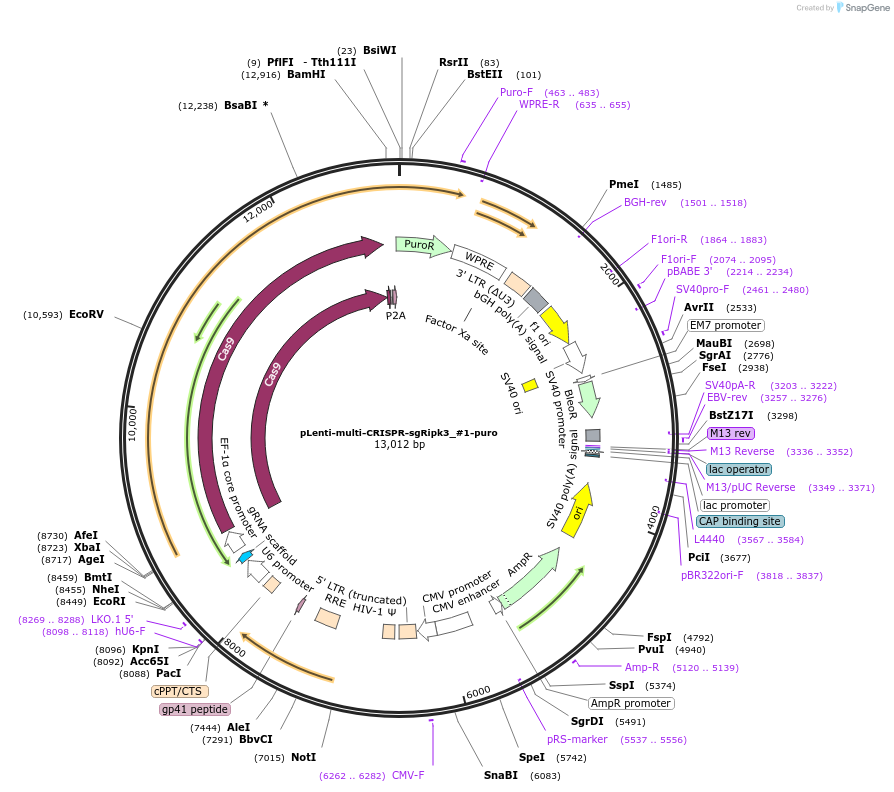
pLenti-multi-CRISPR-sgRipk3_#1-puro
Plasmid#231981PurposeKnockout mouse Ripk3DepositorInsertsgRNA with Cas9 with puromycin resistance (Ripk3 Mouse)
UseCRISPR and LentiviralAvailable SinceFeb. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
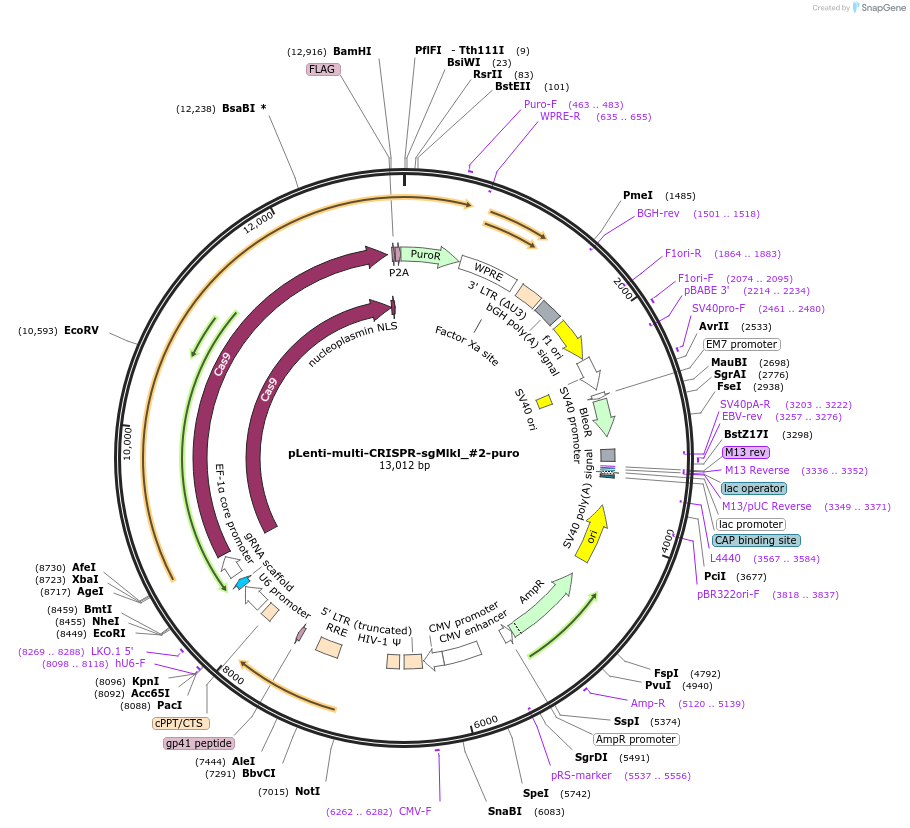
pLenti-multi-CRISPR-sgMlkl_#2-puro
Plasmid#231980PurposeKnockout mouse MlklDepositorInsertsgRNA with Cas9 with puromycin resistance (Mlkl Mouse)
UseCRISPR and LentiviralAvailable SinceFeb. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
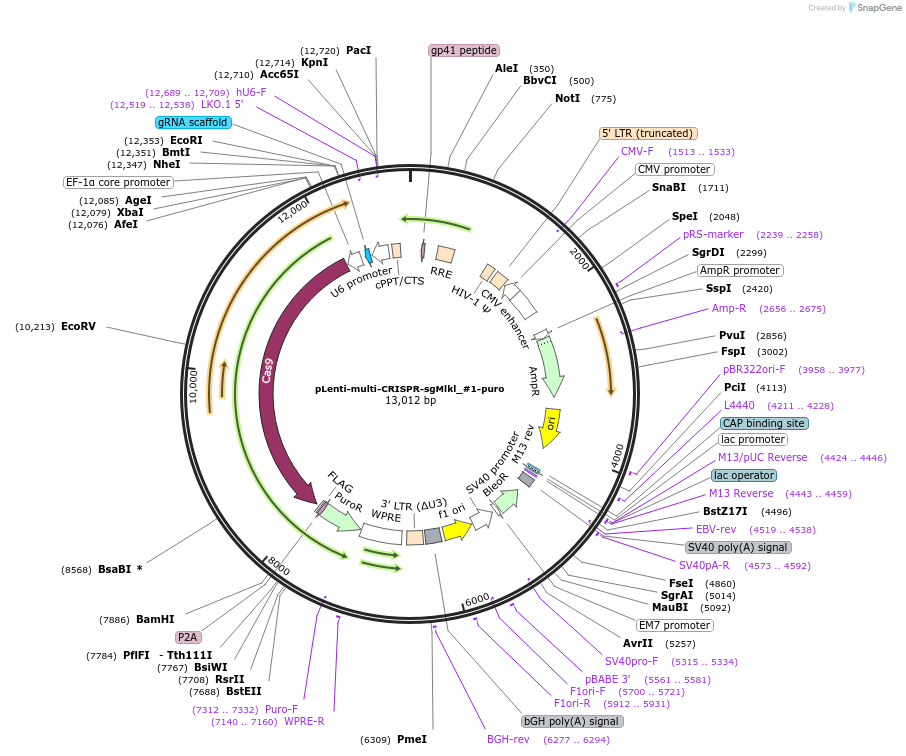
pLenti-multi-CRISPR-sgMlkl_#1-puro
Plasmid#231979PurposeKnockout mouse MlklDepositorInsertsgRNA with Cas9 with puromycin resistance (Mlkl Mouse)
UseCRISPR and LentiviralAvailable SinceFeb. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
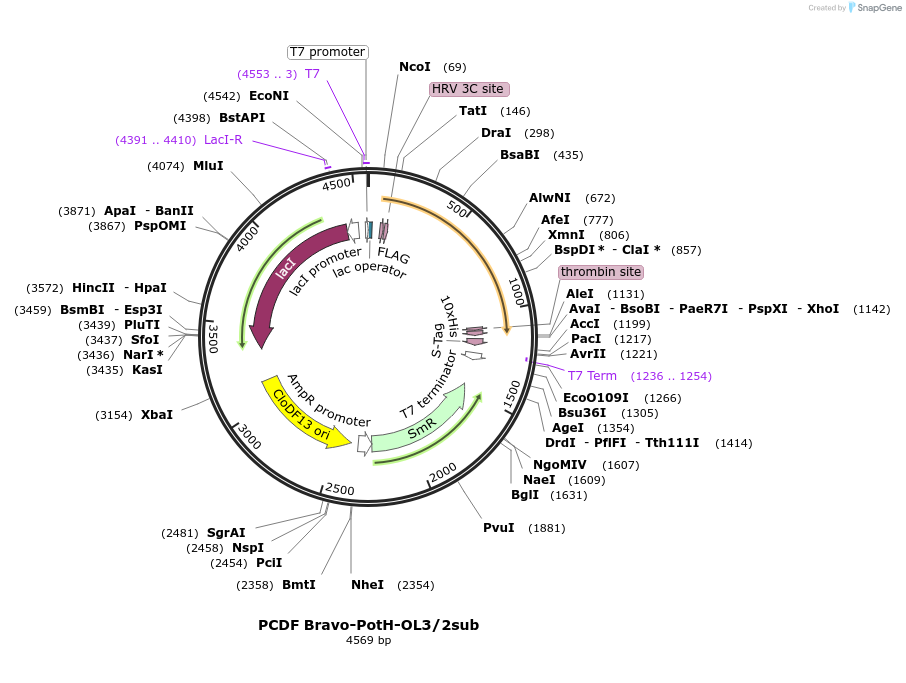
PCDF Bravo-PotH-OL3/2sub
Plasmid#216752PurposeStudy the interaction of PotH in E. coli, specifically focusing on the variant in which outer loop 2 (OL2) is substituted with OL3 in its interaction with exogenous peptides.DepositorInsertpotH (potH E. coli (Bacteria))
Tags10x HisExpressionBacterialMutationOuter loop 2 (OL2) with the sequence of [WMGILKNN…Available SinceMay 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
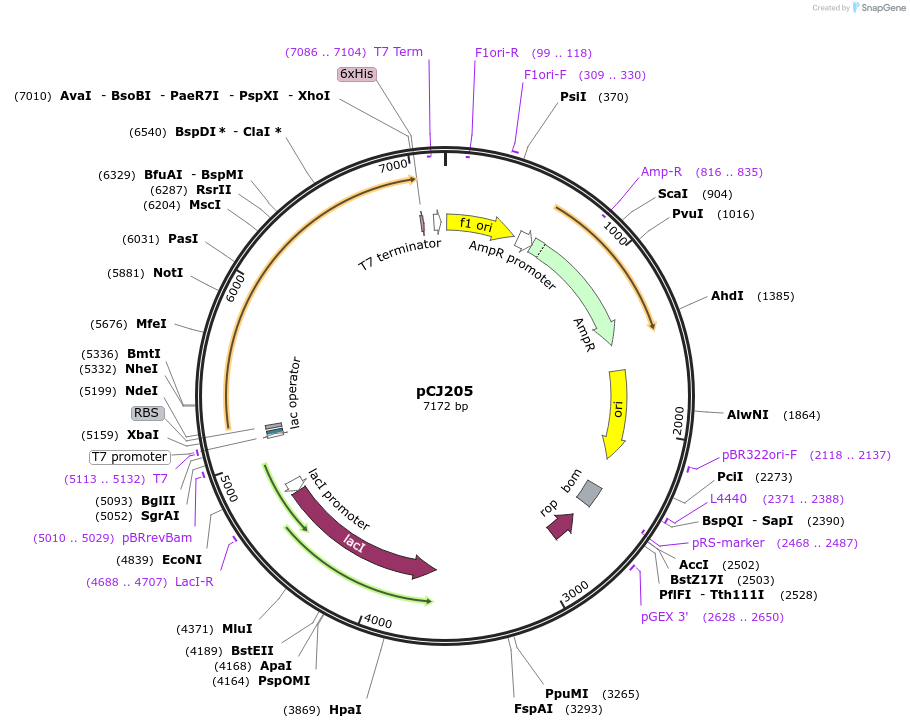
pCJ205
Plasmid#162676PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224A anDepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224A, C529A; codon optimized for expression in E…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCJ198
Plasmid#162671PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating F495I mutation.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationF495I; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
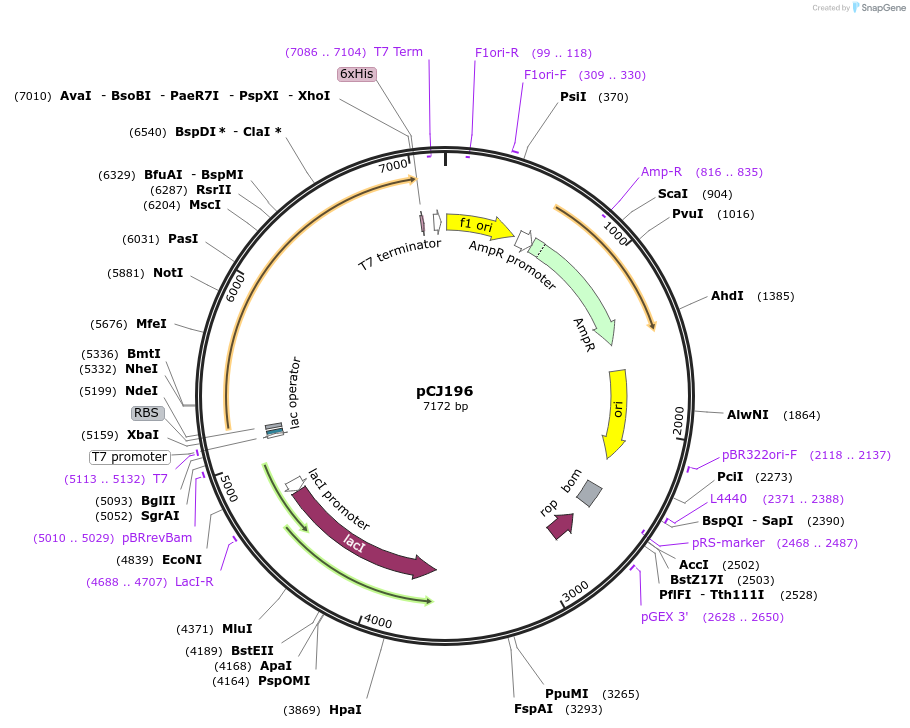
pCJ196
Plasmid#162669PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating catalytic mutation, S225ADepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS225A; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
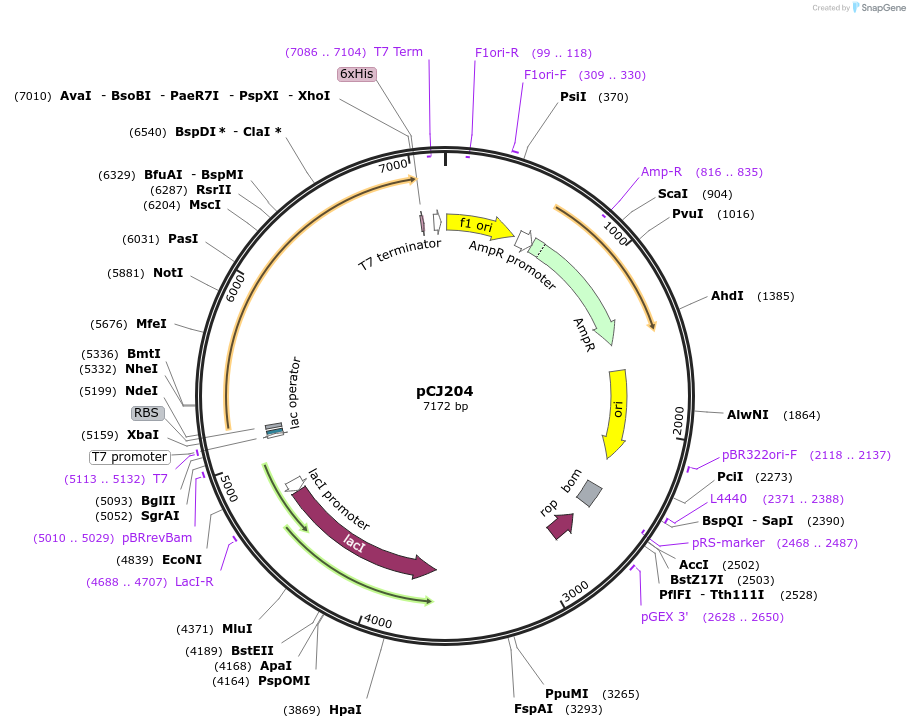
pCJ204
Plasmid#162677PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224H and C529F mutations.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224H, C529F; codon optimized for expression in E…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
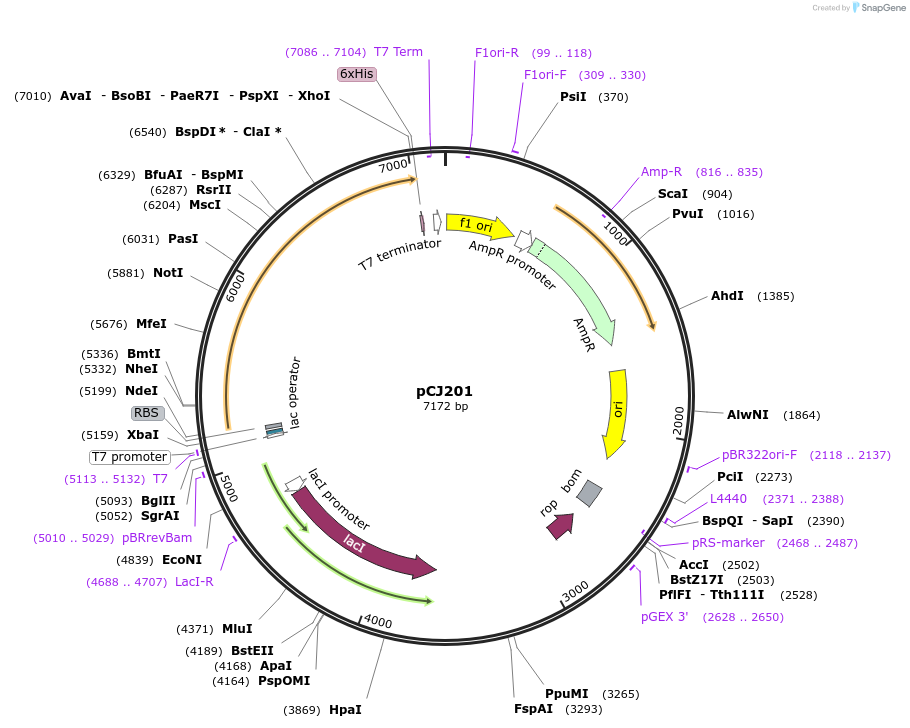
pCJ201
Plasmid#162674PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224W and C529SS mutations.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224W, C529SS; codon optimized for expression in …PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
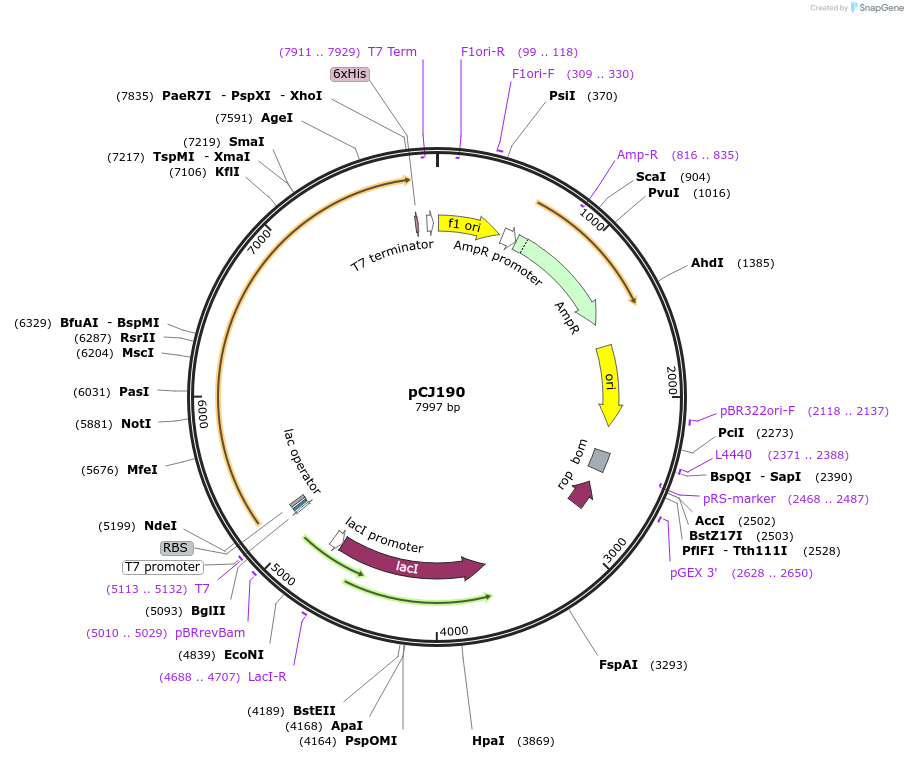
pCJ190
Plasmid#162667PurposepET-21b(+) based plasmid for expresion of MHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 12 residue Gly/Ser linker; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 12 residue Gly/Ser linker
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
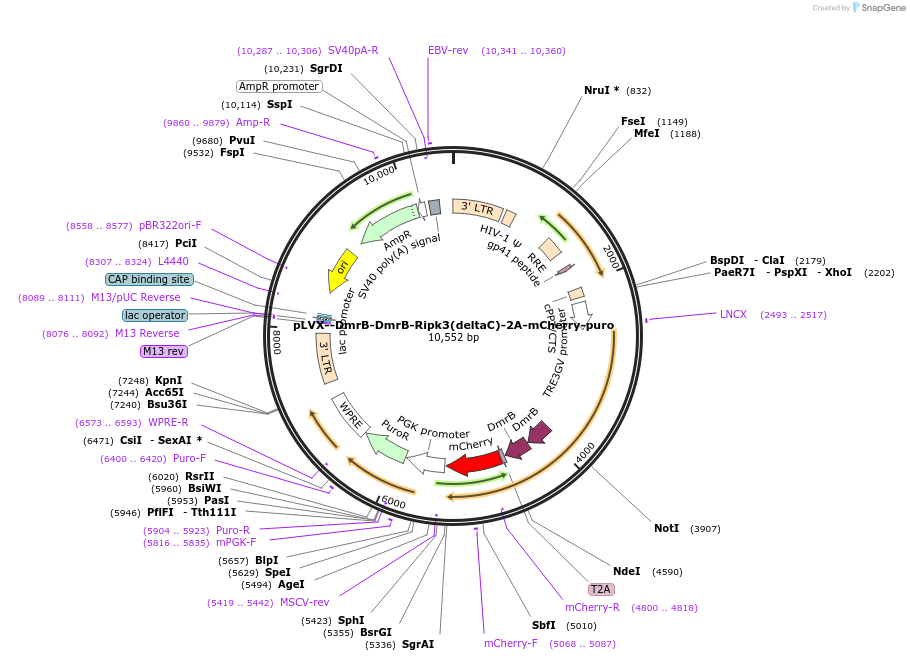
pLVX--DmrB-DmrB-Ripk3(deltaC)-2A-mCherry-puro
Plasmid#231973PurposeTet inducible expression of DmrB-Ripk3 with mCherry to induce Ripk3 necroptosisDepositorInsertRipk3-deltaC dimerizing construct with bi-cistronic mCherry (Ripk3 Mouse)
UseLentiviralAvailable SinceFeb. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
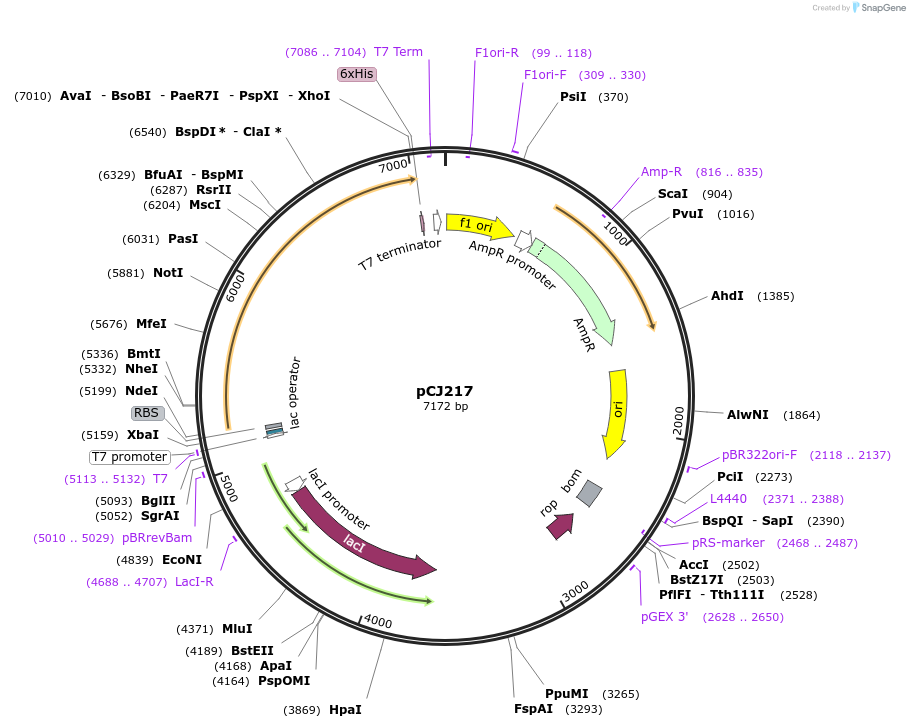
pCJ217
Plasmid#162685PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating two point mutations, G489C and S530C, to introduce a 6th disulfide bond (from PETase).DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with mutations to introduce a 6th disulfide bond from PETase
TagsHisExpressionBacterialMutationG489C, S530C; codon optimized for E. coli K12PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only