We narrowed to 38,511 results for: NAM
-
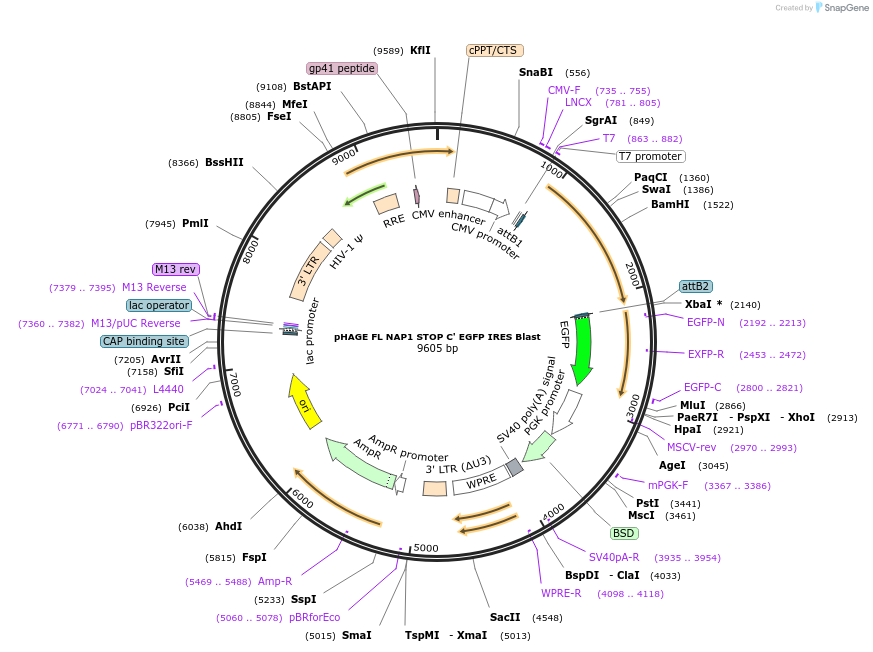
Plasmid#179102PurposeExpresses untagged full length NAP1DepositorAvailable SinceOct. 31, 2023AvailabilityAcademic Institutions and Nonprofits only
-
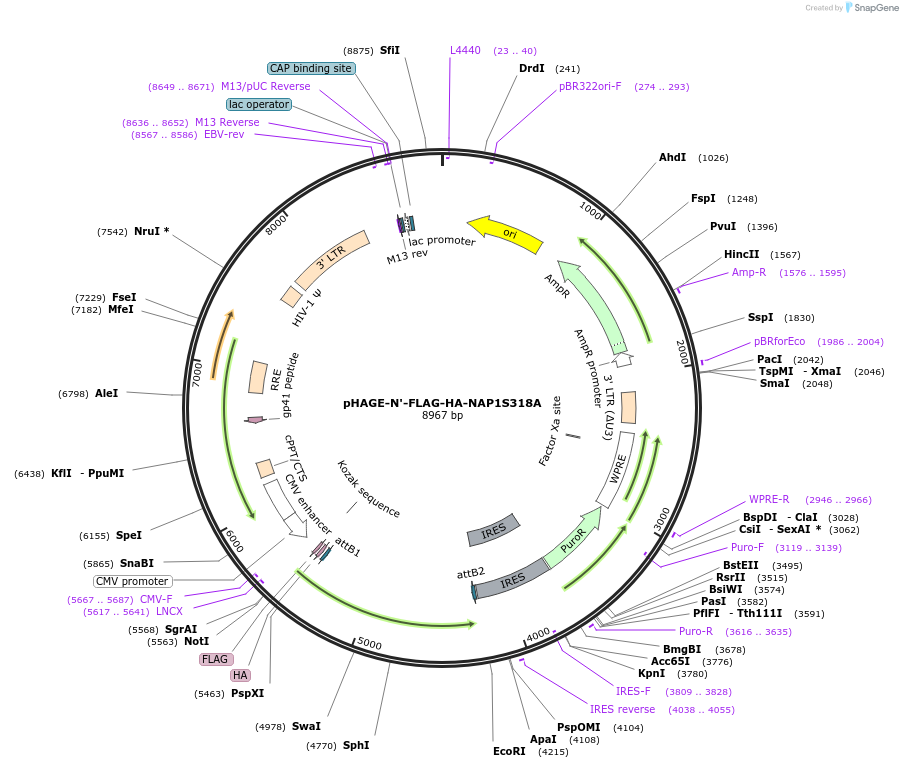
pHAGE-N'-FLAG-HA-NAP1S318A
Plasmid#195410PurposeExpresses FLAG-HA tagged human NAP1 with S318A point mutationDepositorInsertNak associated protein 1 (AZI2 Human)
UseLentiviralTagsFLAG-HAExpressionMammalianMutationSerine 318 turned to AlanineAvailable SinceOct. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
HR700_TP53Exon5mut
Pooled Library#229137PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 5 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon6mut
Pooled Library#229138PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 6 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon7mut
Pooled Library#229139PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 7 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon8mut
Pooled Library#229140PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 8 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 1 (positions 333-436)
Pooled Library#174293PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
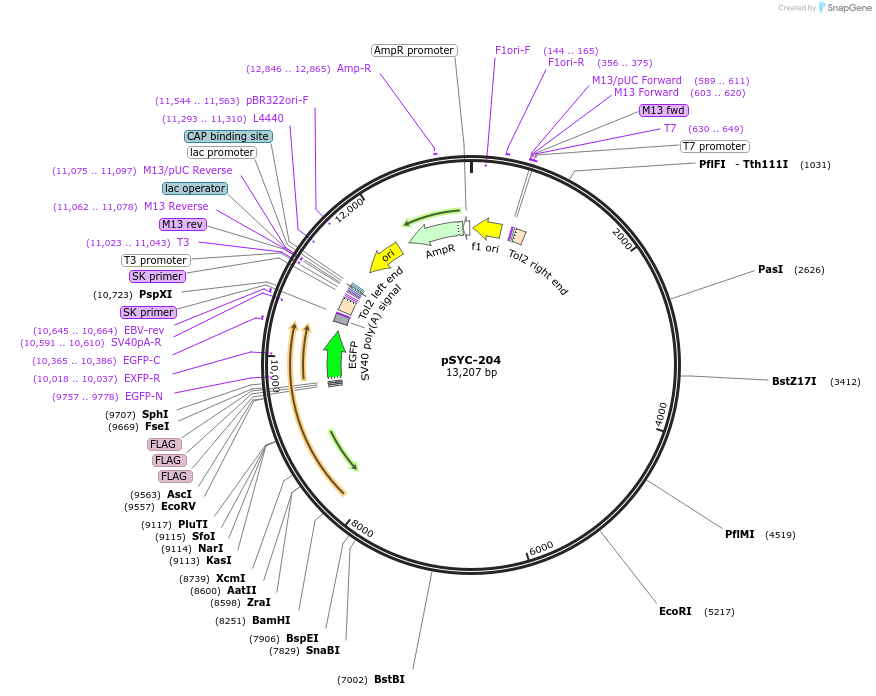
pSYC-204
Plasmid#178964PurposeHuman GFAP (R79C) mutant fused to 3xFLAG tag and EGFP driven by zebrafish gfap promoter for Tol2 mediated recombinationDepositorInsertsUseTol2 mediated recombinationTags3xFLAGMutationR79CPromoterZebrafish gfap promoterAvailable SinceMarch 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
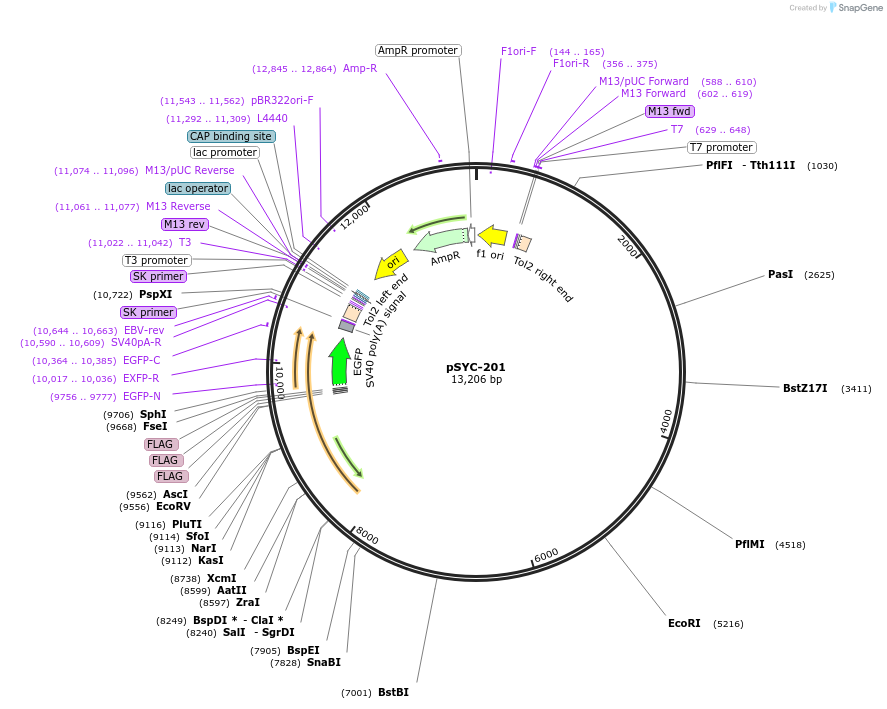
pSYC-201
Plasmid#178963PurposeHuman GFAP fused to 3xFLAG tag and EGFP driven by zebrafish gfap promoter for Tol2 mediated recombinationDepositorInsertsUseTol2 mediated recombinationTags3xFLAGPromoterzebrafish gfap promoterAvailable SinceMarch 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
pTA7001-DEST
Plasmid#71745PurposeGATEWAY compatible derivative of plant binary vector pTA7001 from Nam Chua's laboratory at The Rockefeller University, Dexamethasone inducibleDepositorTypeEmpty backboneExpressionPlantPromoterGAL4Available SinceFeb. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Guzit
Bacterial Strain#196902PurposeGuzit is a RNase HI(rnhA) His-tagged E. coli strain made for a cell-free expression system.DepositorBacterial ResistanceKanamycinAvailable SinceAug. 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
RNase III-His
Bacterial Strain#196903PurposeRNase III-His is an E. coli strain made for a cell-free expression system.DepositorBacterial ResistanceKanamycinAvailable SinceAug. 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
pTA7002-avrPto
Plasmid#49156PurposePlant transformation binary vector for Dexamethasone inducible expression of the type III effector gene avrPtoDepositorInsertavrPto
UsePlant transformation binary vectorPromoterdexamethasone inducibleAvailable SinceApril 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNR2.dnaG.Q576A
Bacterial Strain#41700DepositorBacterial ResistanceAmpicillinAvailable SinceApril 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
Nuc5-.dnaG.Q576A.tolC-
Bacterial Strain#41699DepositorBacterial ResistanceAmpicillinAvailable SinceApril 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
HL 2729 (=HL 716 + rhyB knockout_KanR)
Bacterial Strain#30030DepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceAug. 20, 2012AvailabilityAcademic Institutions and Nonprofits only -
HL 849 (=HL 716 + micC knockout_KanR)
Bacterial Strain#30009DepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceJuly 5, 2012AvailabilityAcademic Institutions and Nonprofits only