We narrowed to 5,107 results for: CAPS;
-
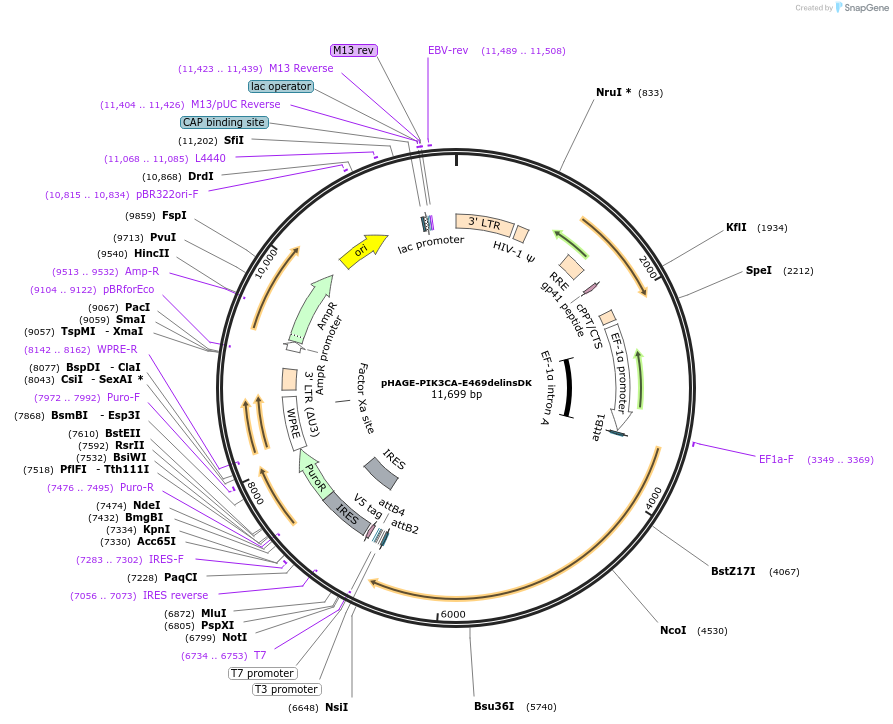
Plasmid#116476PurposeLentiviral expression of PIK3CA E469delinsDKDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only
-
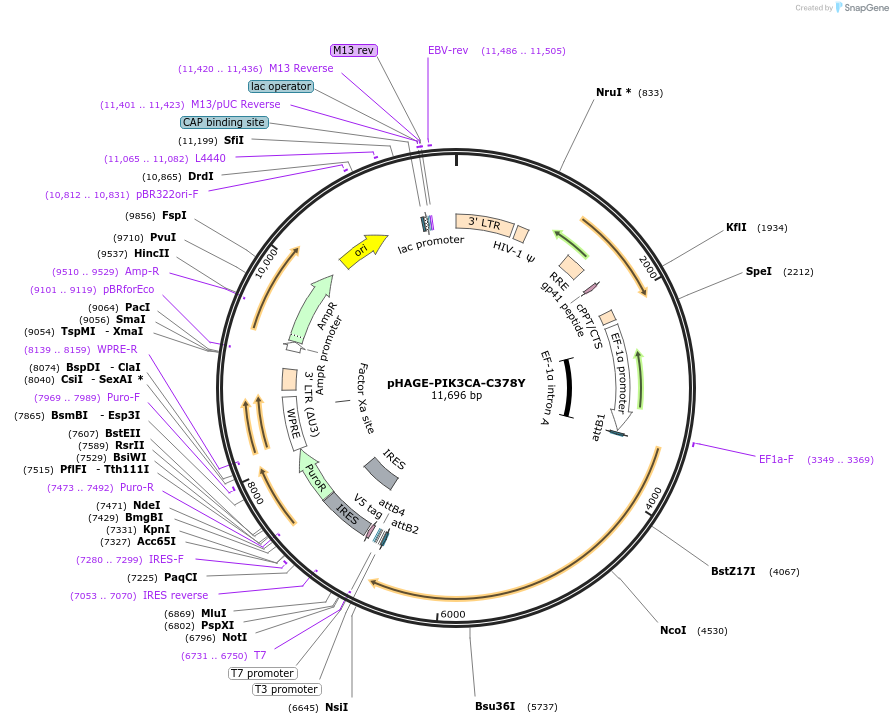
pHAGE-PIK3CA-C378Y
Plasmid#116456PurposeLentiviral expression of PIK3CA C378YDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
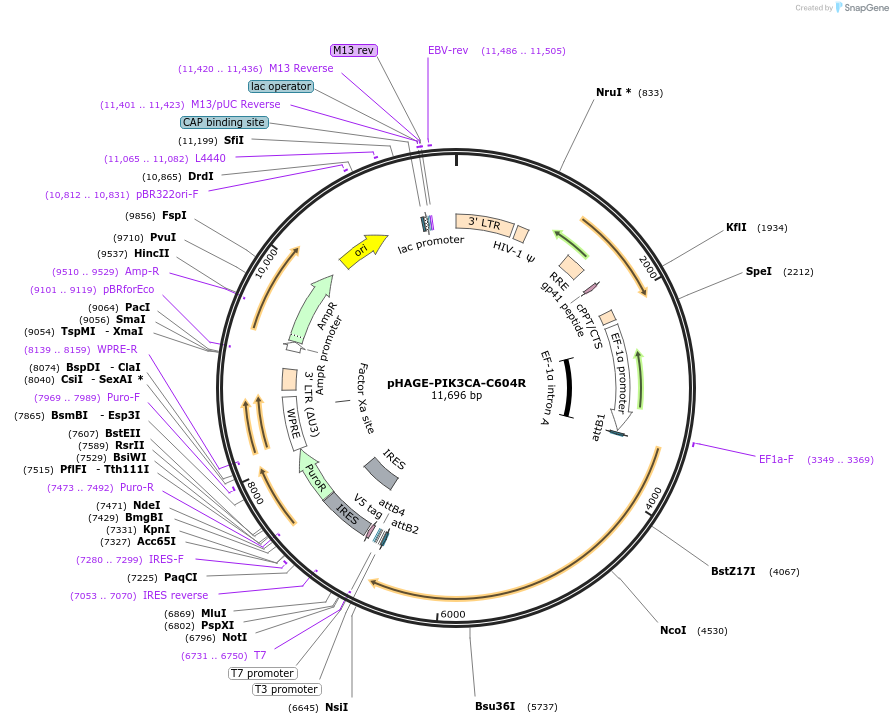
pHAGE-PIK3CA-C604R
Plasmid#116458PurposeLentiviral expression of PIK3CA C604RDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
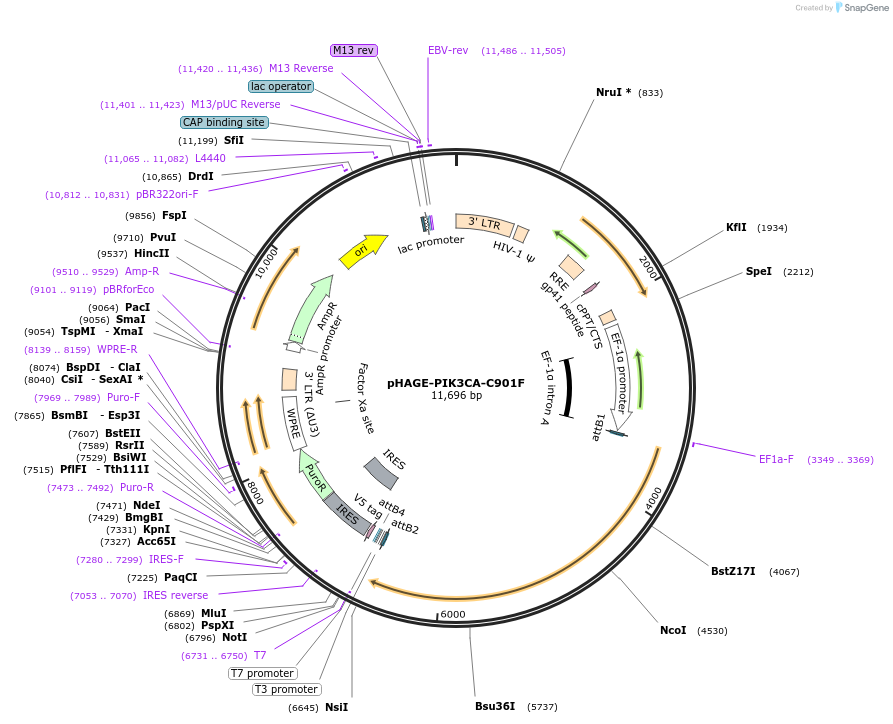
pHAGE-PIK3CA-C901F
Plasmid#116459PurposeLentiviral expression of PIK3CA C901FDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
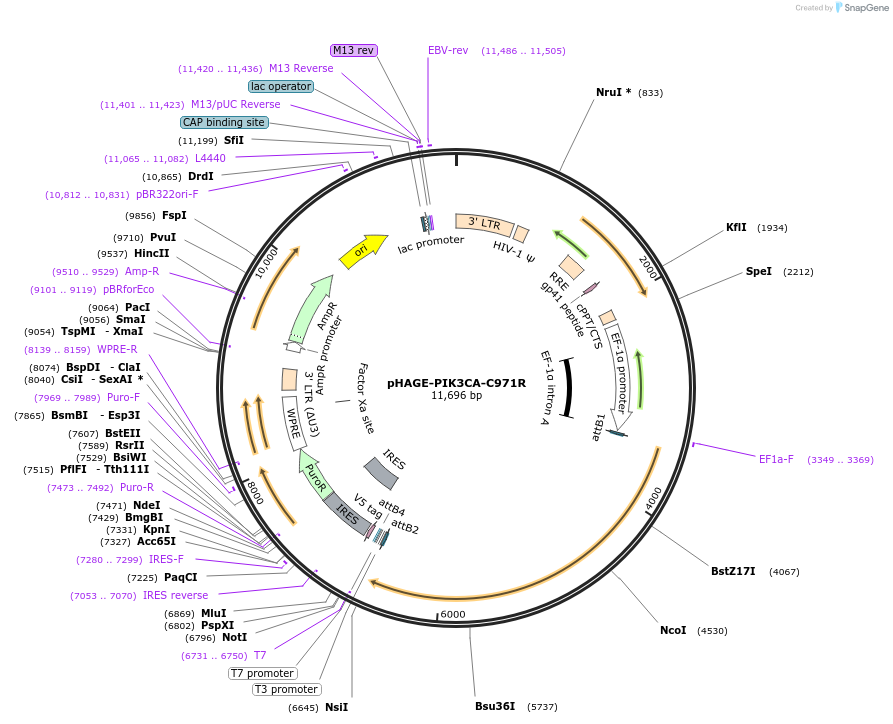
pHAGE-PIK3CA-C971R
Plasmid#116460PurposeLentiviral expression of PIK3CA C971RDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
pHAGE-PIK3CA-D350G
Plasmid#116461PurposeLentiviral expression of PIK3CA D350GDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
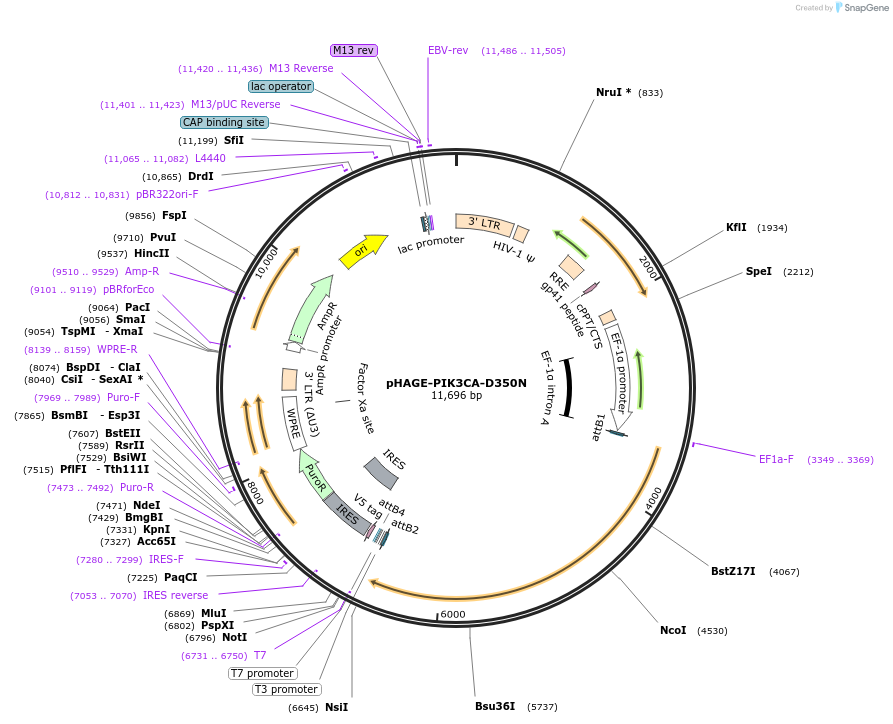
pHAGE-PIK3CA-D350N
Plasmid#116462PurposeLentiviral expression of PIK3CA D350NDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
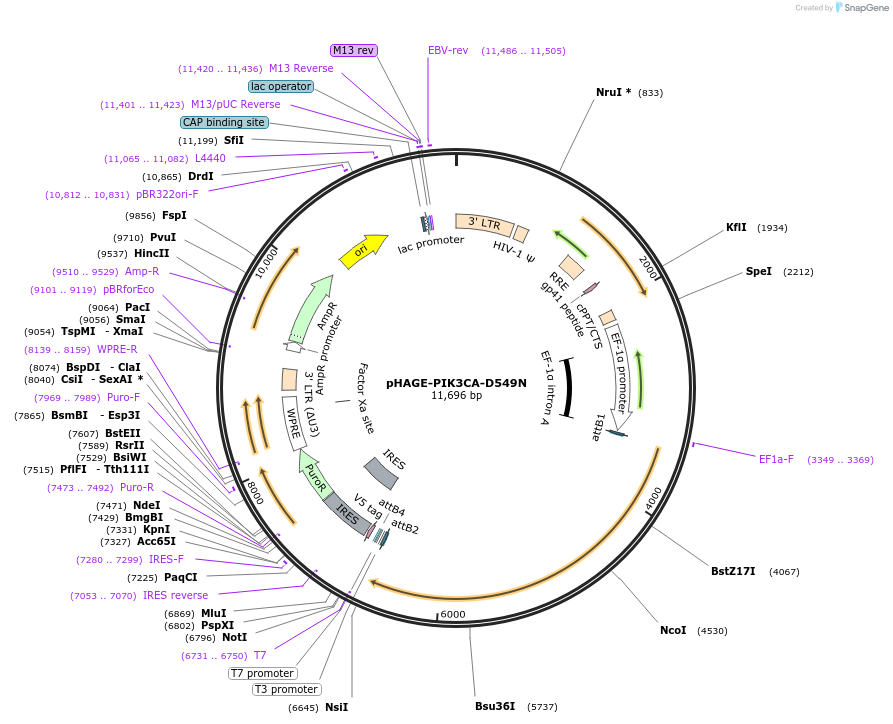
pHAGE-PIK3CA-D549N
Plasmid#116463PurposeLentiviral expression of PIK3CA D549NDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
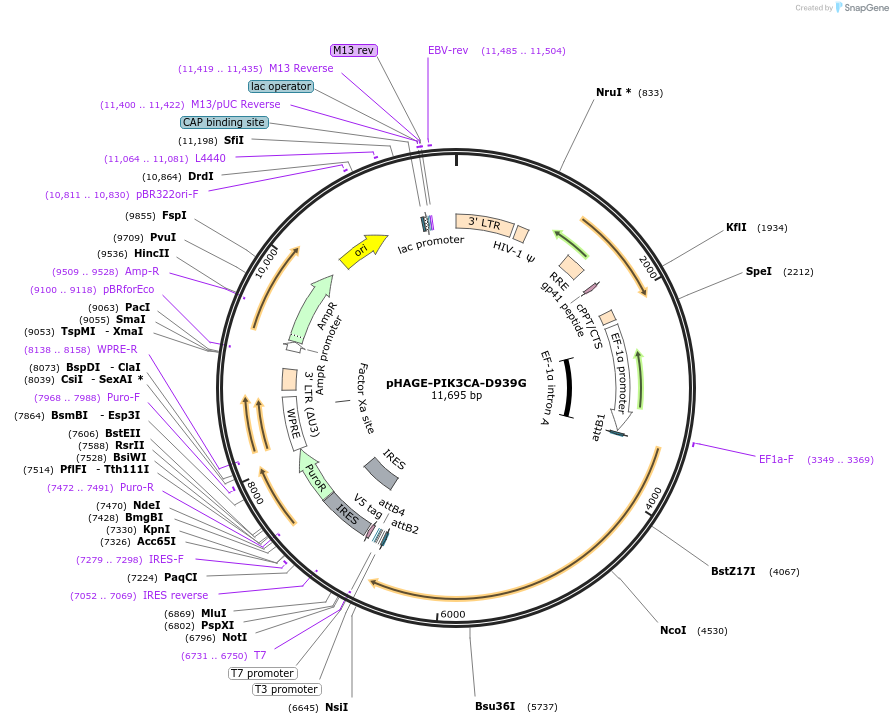
pHAGE-PIK3CA-D939G
Plasmid#116464PurposeLentiviral expression of PIK3CA D939GDepositorAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
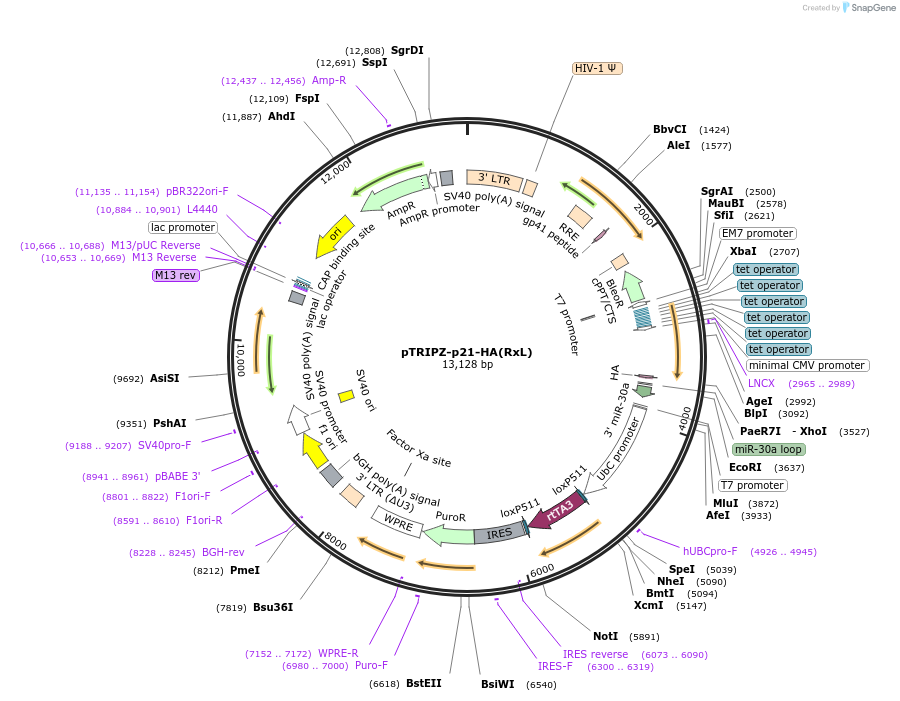
pTRIPZ-p21-HA(RxL)
Plasmid#215111PurposeExpresses p21-HA(RxL) in mammalian cells from an inducible lentiviral vector.DepositorInsertCDKN1A (CDKN1A Human)
UseLentiviralTagsHAExpressionMammalianMutationp21(RxL) denotes mutations: R19A, R20A, L21A, F22…PromoterCMV (Tet inducible)Available SinceApril 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
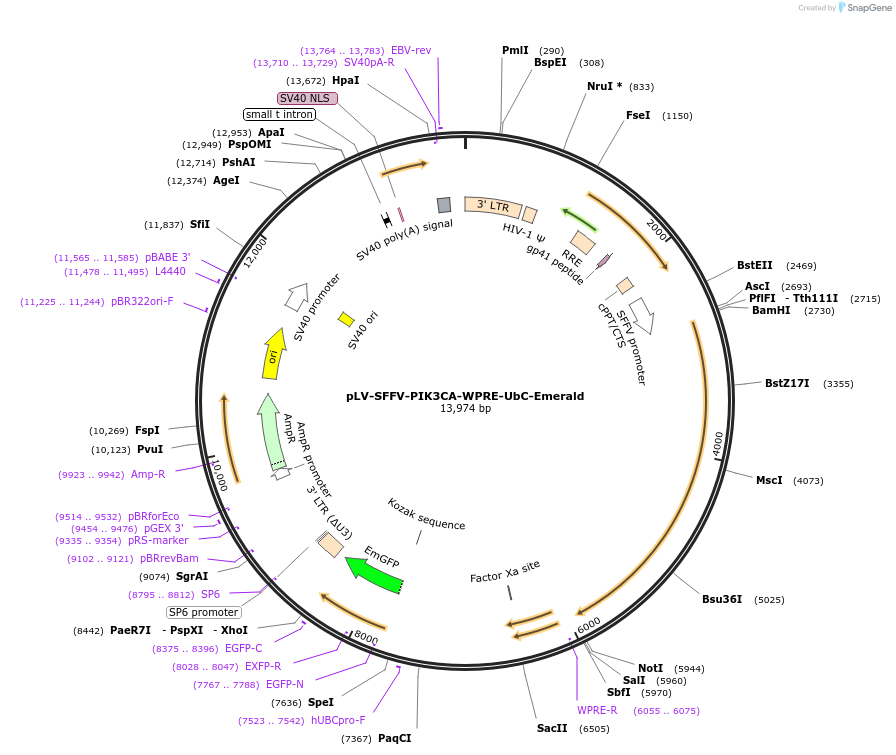
pLV-SFFV-PIK3CA-WPRE-UbC-Emerald
Plasmid#203797PurposeLentiviral vector expressing human phosphoinositide 3-kinase catalytic subunit alpha (PIK3CA) under the spleen focus-forming virus (SFFV) promoter and Emerald under the Ubiquitin C (UbC) promoterDepositorAvailable SinceJuly 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
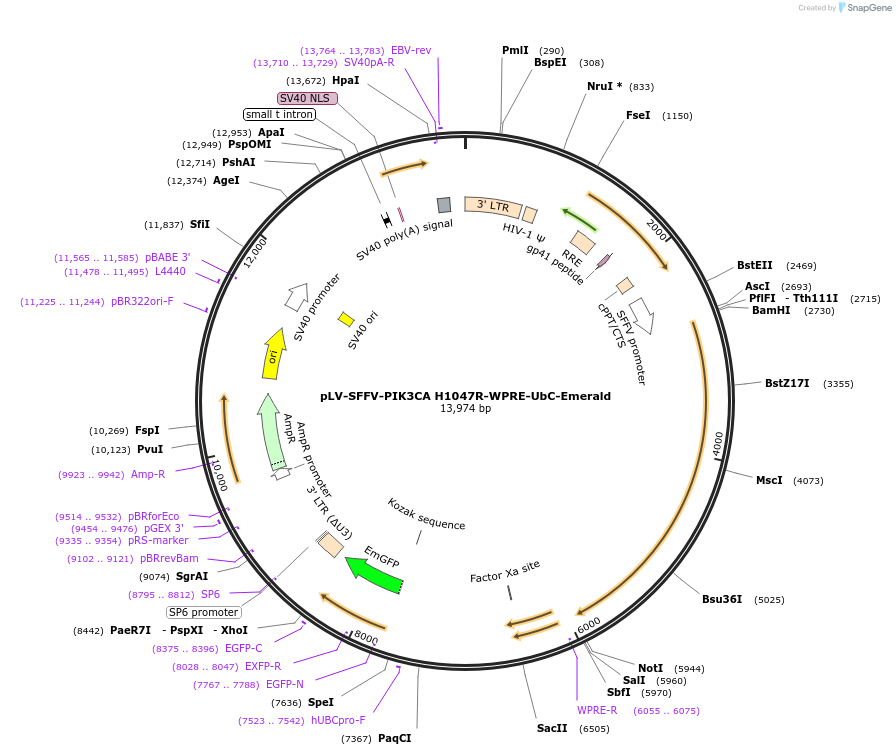
pLV-SFFV-PIK3CA H1047R-WPRE-UbC-Emerald
Plasmid#203798PurposeLentiviral vector expressing human PIK3CA H1047R under the spleen focus-forming virus (SFFV) promoter and Emerald under the Ubiquitin C (UbC) promoterDepositorAvailable SinceJuly 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
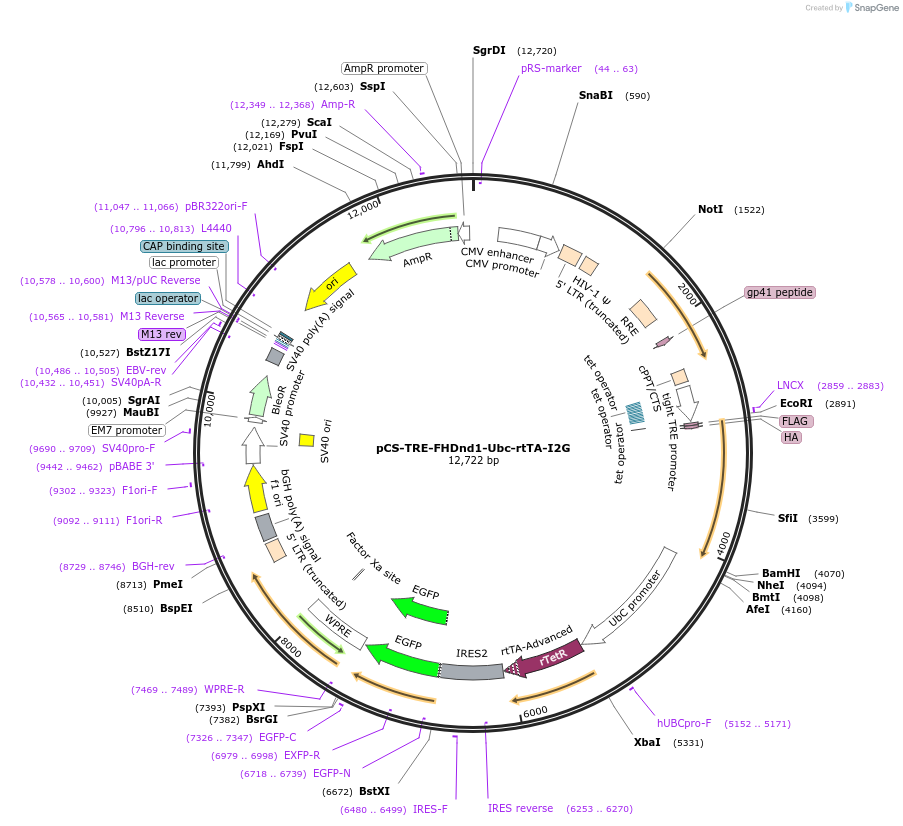
pCS-TRE-FHDnd1-Ubc-rtTA-I2G
Plasmid#70076Purpose2nd gen lentiviral vector. tet/dox controllable expression of FLAG-HA-tagged murine Dnd1 in mammalian cellsDepositorInsertDnd1 (Dnd1 Mouse)
UseLentiviralTagsFLAG and HAExpressionMammalianMutationSilent mutations are introduced to escape from RN…PromoterTet responsible promoterAvailable SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
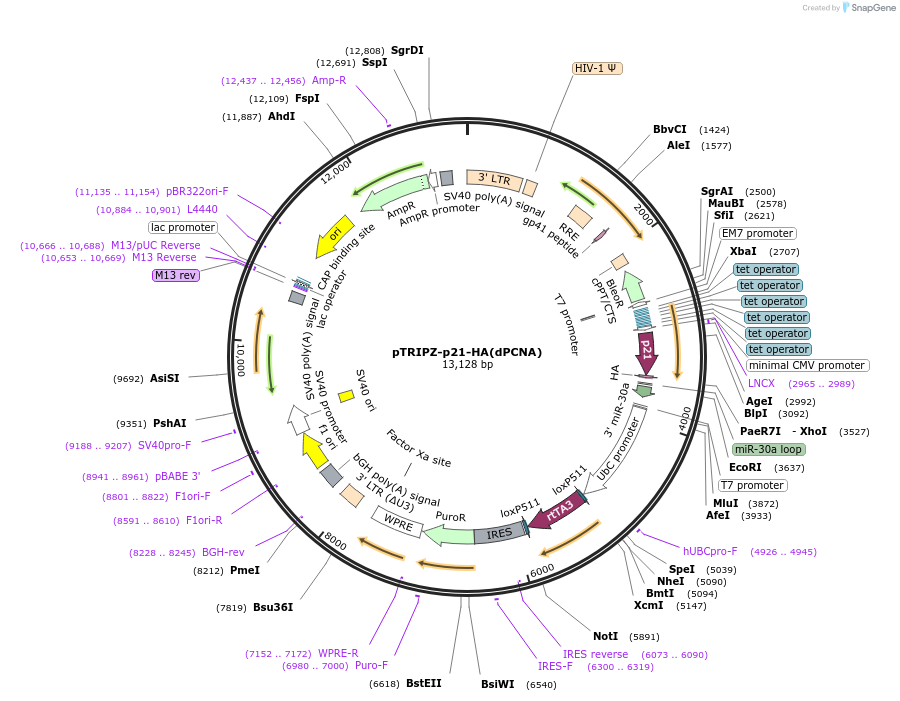
pTRIPZ-p21-HA(dPCNA)
Plasmid#215112PurposeExpresses p21-HA(dPCNA) in mammalian cells from an inducible lentiviral vector.DepositorInsertCDKN1A (CDKN1A Human)
UseLentiviralTagsHAExpressionMammalianMutationp21(dPCNA) denotes mutations M147A, D149A, F150A.PromoterCMV (Tet inducible)Available SinceApril 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
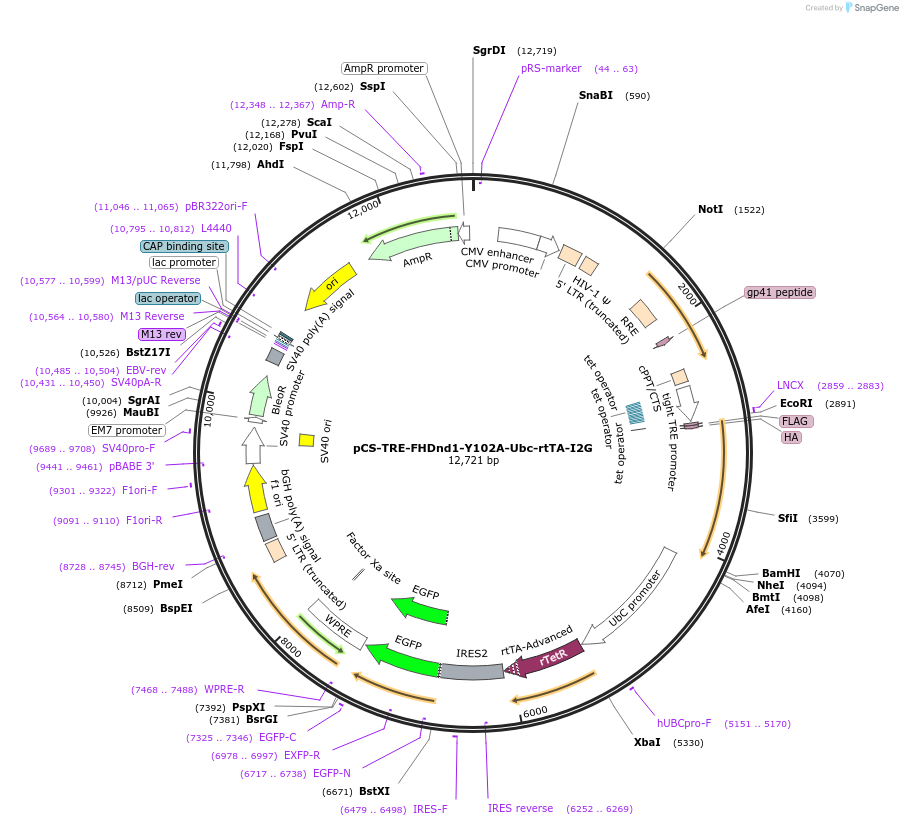
pCS-TRE-FHDnd1-Y102A-Ubc-rtTA-I2G
Plasmid#70075Purpose2nd gen lentiviral vector. tet/dox controllable expression of FLAG-HA-tagged murine Dnd1 (RRM mutant) in mammalian cellsDepositorInsertDnd1 (Dnd1 Mouse)
UseLentiviralTagsFLAG and HAExpressionMammalianMutationY102A. Silent mutations are introduced to escape …PromoterTet responsible promoterAvailable SinceNov. 22, 2017AvailabilityAcademic Institutions and Nonprofits only -
pDONR223_CASP8_WT_V5
Plasmid#82999PurposeGateway Donor vector containing CASP8, used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorInsertCASP8 (CASP8 Human)
UseGateway entry vectorMutationAA 184_191delERSSSLEGinsGEELCGVMTISD; D194R; 196_…PromoterNoneAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
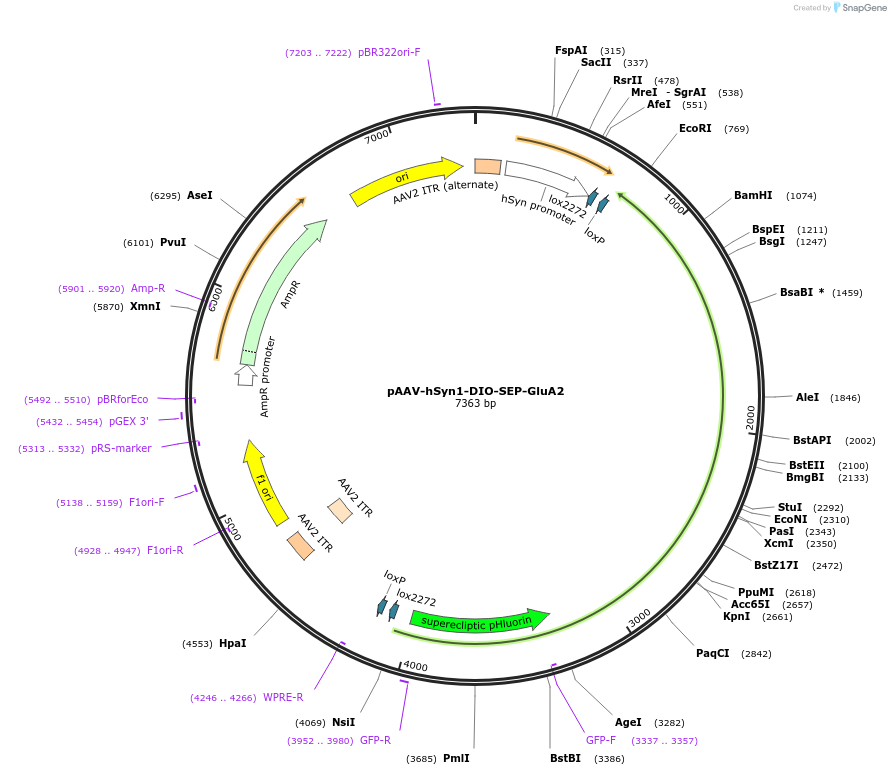
pAAV-hSyn1-DIO-SEP-GluA2
Plasmid#227233PurposeLarge cargo capacity AAV that expresses SEP tagged AMPA receptor subunit GluA2 in neurons expressing Cre recombinaseDepositorInsertSEP-GluA2 (Gria2 Rat)
UseAAVTagssuperecliptic pHluorinExpressionMammalianPromoterhSyn1Available SinceOct. 15, 2025AvailabilityAcademic Institutions and Nonprofits only -
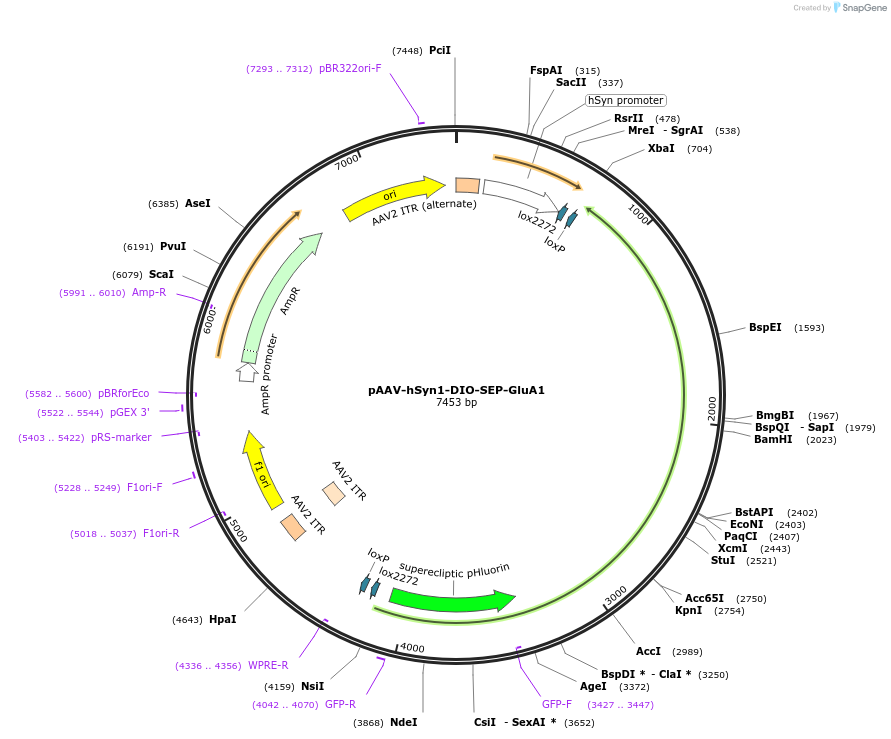
pAAV-hSyn1-DIO-SEP-GluA1
Plasmid#227235PurposeLarge cargo capacity AAV that expresses SEP tagged AMPA receptor subunit GluA1 in neurons expressing Cre recombinaseDepositorInsertSEP-GluA1 (Gria1 Rat)
UseAAVTagssuperecliptic pHluorinExpressionMammalianPromoterhSyn1Available SinceOct. 15, 2025AvailabilityAcademic Institutions and Nonprofits only -
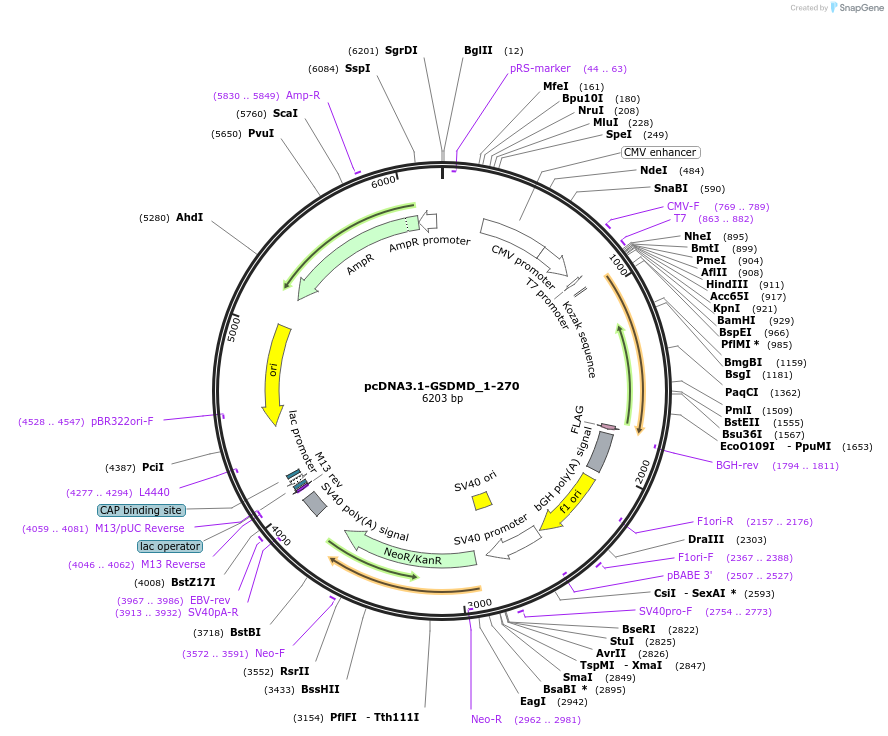
pcDNA3.1-GSDMD_1-270
Plasmid#218229PurposeGasdermin1-270 is a pore-forming GSDMD formed by SARS-CoV-2 3CLpro cleavage. Transfection in HEK293 cells showed GSDMD 1-270 pores secrete 3CLpro, nucleocapsid, and in excess, kill cells by pyroptosisDepositorInsertGSDMD (GSDMD Human)
TagsflagExpressionMammalianMutationconstruct only codes for amino acids 1-270Available SinceJuly 1, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
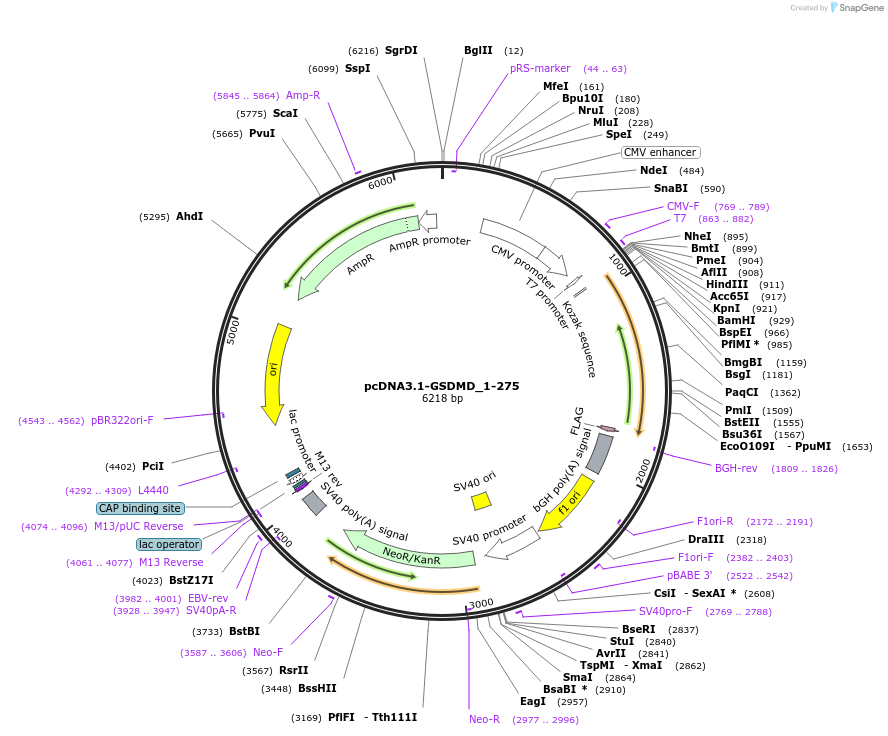
pcDNA3.1-GSDMD_1-275
Plasmid#218230PurposeGasdermin-D_1-275 is a pore-forming GSDMD formed by cleavage by host caspases. Transfection in HEK293 cells showed GSDMD 1-275 pores secrete 3CLpro, nucleocapsid; in excess, kills cells by pyroptosisDepositorInsertGSDMD (GSDMD Human)
TagsflagExpressionMammalianMutationconstruct only codes for amino acids 1-275Available SinceJuly 1, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits