We narrowed to 54,805 results for: Eng;
-
Plasmid#117846PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses aeBlue chromoprotein in E. coliDepositorInsertpromoter, RBS, aeBlue
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
amajLime chromoprotein
Plasmid#117843PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amajLime chromoprotein in E. coliDepositorInsertpromoter, RBS, amajLime
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
fwYellow chromoprotein
Plasmid#117841PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses fwYellow chromoprotein in E. coliDepositorInsertpromoter, RBS, fwYellow
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
tsPurple chromoprotein
Plasmid#117848PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses tsPurple chromoprotein in E. coliDepositorInsertpromoter, RBS, tsPurple
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
pJBEI-6409
Plasmid#47048PurposeBglBrick plasmid (=pBbA5c-MTSAe-T1f-MBI(f)-T1002i-Ptrc-trGPPS(co)-LS) coding for MEV pathway enzymes to produce limonene from glucose in E. coliDepositorInsertCodon-optimized sequences for MEV pathway expression in E. coli to produce limonene
UseSynthetic Biology; BglbrickExpressionBacterialPromoterPlacUV5Available SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pBS3Clux
Plasmid#55172Purposelux-reporter vector, integration at sacA, ampr, cmrDepositorTypeEmpty backboneUseSynthetic Biology; Bacillus biobrick boxPromoternoneAvailable SinceJuly 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
amilCP_Pink chromoprotein
Plasmid#117851PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilCP_Pink chromoprotein in E. coliDepositorInsertpromoter, RBS, amilCP_Pink
UseSynthetic Biology; Escherichia coliMutation64A,65C, BioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
amilCP_Orange chromoprotein
Plasmid#117850PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilCP_Orange chromoprotein in E. coliDepositorInsertpromoter, RBS, amilCP_Orange
UseSynthetic Biology; Escherichia coliMutation64V,65G, BioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
gfasPurple chromoprotein
Plasmid#117849PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses gfasPurple chromoprotein in E. coliDepositorInsertpromoter, RBS, gfasPurple
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
CMV-SauriABE8e
Plasmid#189927PurposePlasmid encoding SauriABE8e driven by the CMV promoter for plasmid transfection.DepositorInsertSauriABE8e
ExpressionMammalianMutationNme2Cas9 D16APromoterCMVAvailable SinceSept. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
spisPink chromoprotein
Plasmid#117839PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses spisPink chromoprotein in E. coliDepositorInsertpromoter, RBS, spisPink
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
asPink chromoprotein
Plasmid#117838PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses asPink chromoprotein in E. coliDepositorInsertpromoter, RBS, asPink
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
scOrange chromoprotein
Plasmid#117840PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses scOrange chromoprotein in E. coliDepositorInsertpromoter, RBS, scOrange
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
pBS2E
Plasmid#55169PurposeEmpty vector, integration at lacA, ampr, mlsrDepositorTypeEmpty backboneUseSynthetic Biology; Bacillus biobrick boxPromoternoneAvailable SinceJuly 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
eforRed chromoprotein
Plasmid#117837PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses eforRed chromoprotein in E. coliDepositorInsertpromoter, RBS, eforRed
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
pJBEI-6410
Plasmid#47049PurposeBglBrick plasmid (=pBbA5a-MTSAe-T1f-MBI(f)-T1002i-Ptrc-trGPPS(co)-LS) coding for MEV pathway enzymes to produce limonene from glucose in E. coliDepositorInsertCodon-optimized sequences for MEV pathway expression in E. coli to produce limonene
UseSynthetic Biology; BglbrickExpressionBacterialPromoterPlacUV5Available SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pBS4S
Plasmid#55170PurposeEmpty vector, integration at thrC, ampr, specrDepositorTypeEmpty backboneUseSynthetic Biology; Bacillus biobrick boxPromoternoneAvailable SinceAug. 1, 2014AvailabilityAcademic Institutions and Nonprofits only -
pCA24N-ligase-NLS-cTF
Plasmid#87746PurposeIPTG-inducible expression of ligase-NLS-cTF fusion for protein purificationDepositorInsertT4 DNA ligase (30 )
Tags6xHis and NLS linker - cTF (chimeric transcriptio…ExpressionBacterialPromoterT5-lacAvailable SinceMarch 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
meffBlue chromoprotein
Plasmid#117845PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses meffBlue chromoprotein in E. coliDepositorInsertpromoter, RBS, meffBlue
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
pLenti-NG-PE2-BSD
Plasmid#176933PurposeExpresses PE2-NG variant in mammalian cellsDepositorInsertPE2-NG variant
UseLentiviralTagsSV40 NLSExpressionMammalianPromoterEFSAvailable SinceDec. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
amilGFP chromoprotein
Plasmid#117842PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilGFP chromoprotein in E. coliDepositorInsertpromoter, RBS, amilGFP
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
pBS1ClacZ
Plasmid#55171PurposelacZ-reporter vector, integration at amyE, ampr, cmrDepositorTypeEmpty backboneUseSynthetic Biology; Bacillus biobrick boxPromoternoneAvailable SinceAug. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
cjBlue chromoprotein
Plasmid#117844PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses cjBlue chromoprotein in E. coliDepositorInsertpromoter, RBS, cjBlue
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
meffRed chromoprotein
Plasmid#117836PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses meffRed chromoprotein in E. coliDepositorInsertpromoter, RBS, meffRed
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
CMV_Npu-BE3.9max C-terminal
Plasmid#137172PurposeExpresses the C-terminal split-intein fragment of BE3.9max from the CMV promoterDepositorInsertNpu-BE3.9max_C-terminal
ExpressionMammalianPromoterCMVAvailable SinceJan. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
CMV_Npu-ABEmax C-terminal
Plasmid#137174PurposeExpresses the C-terminal split-intein fragment of ABEmax from the CMV promoterDepositorInsertNpu-ABEmax_C-terminal
ExpressionMammalianPromoterCMVAvailable SinceJan. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
amilGFP chromoprotein promoter-less
Plasmid#118006PurposeFor course in "Synthetic Biology: A Lab Manual"DepositorInsertRBS, amilGFP
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPPC011
Plasmid#171143PurposeExpression of Sp.pCas9-dCas9 and BBa_J23107-MCP-SoxS(R93A/S101A) on pRK2-KmR plasmidDepositorInsertSp.pCas9-dCas9_BBa_J23107-MCP-SoxS(R93A, S101A)
UseCRISPRExpressionBacterialMutationSoxS has R93A and S101A mutationsPromoterSp.pCas9, BBa_J23107Available SinceOct. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
tsPurple chromoprotein promoter-less
Plasmid#118007PurposeFor course in "Synthetic Biology: A Lab Manual"DepositorInsertRBS, tsPurple
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
ku70 insUP+pPGI+NAT+tNOS+ku70 insD (SBE217)
Plasmid#195049Purposeintegration cassette for ku70 deletion and expression of NAT gene under pPGI for characterizationDepositorInsertNAT
ExpressionBacterialPromoterpPGIAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
promoter J23110 (upstream of RBS, mRFP1)
Plasmid#117852PurposeFor course in "Synthetic Biology: A Lab Manual"DepositorInsertmedium-high promoter, RBS, mRFP1
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
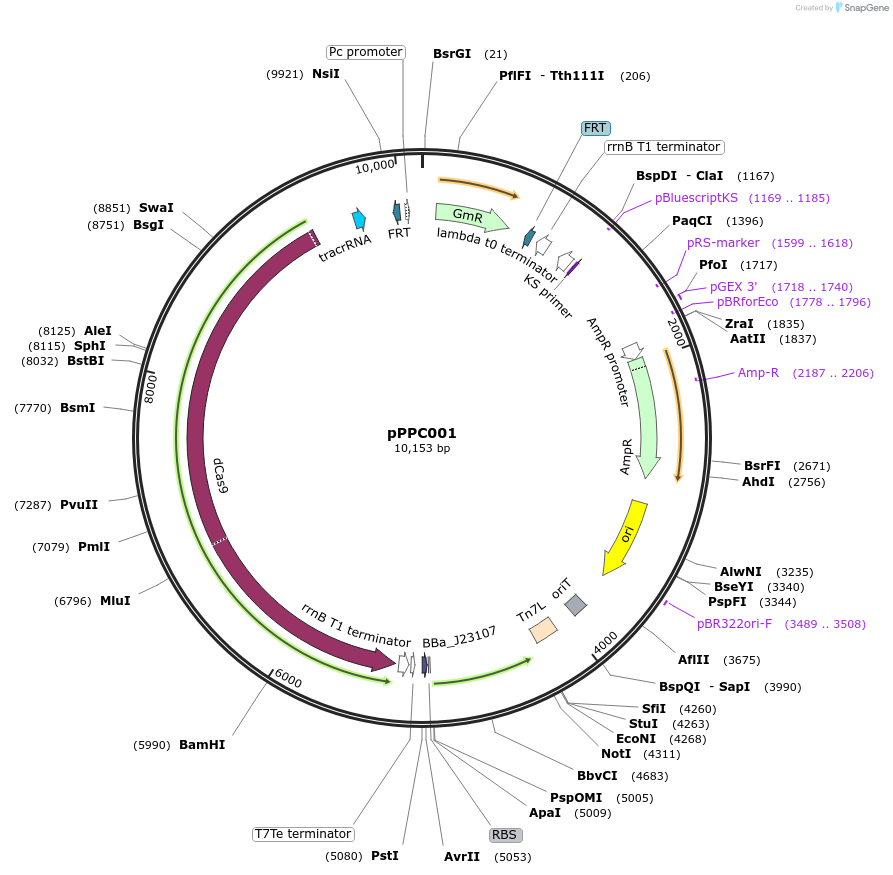
pPPC001
Plasmid#171138PurposeFor integration of Sp.pCas9-dCas9 and BBa_J23107-MCP-SoxS(R93A/S101A) with miniTn7T methodDepositorInsertSp.pCas9-dCas9_BBa_J23107-MCP-SoxS(R93A, S101A)
UseCRISPRExpressionBacterialMutationSoxS has R93A and S101A mutationsPromoterSp.pCas9, BBa_J23107Available SinceOct. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
H446 EF1a RA-ABE6.3 p2A Puro
Plasmid#139104PurposeLentiviral expression of ABE6.3.DepositorInsertABE RA6.3
UseCRISPR and LentiviralExpressionMammalianPromoterEF1aAvailable SinceMarch 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
(K) tNOS (S)_T3 (SBE120)
Plasmid#195010Purposeterminator from Agrobacterium tumefaciens nopaline synthase (tNOS, MF116010) for T3 position (K/S)DepositorInsert(K) tNOS (S)
ExpressionBacterialAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
(H) tNOS (I) _T2 (SBE119)
Plasmid#195006Purposeterminator from Agrobacterium tumefaciens nopaline synthase (tNOS, MF116010) for T2 position (H/I)DepositorInsert(H) tNOS (I)
ExpressionBacterialAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
(B) HYG marker(R) (SBE264)
Plasmid#195013Purposehygromycin (HYG) resistance genes codon-optimized for R. toruloides and expressed under pXYL and tGPDDepositorInsertpXYL+HYG+tGPD
ExpressionBacterialPromoterpXYLAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
(E) tNOS (F) _T1 (SBE118)
Plasmid#194997Purposeterminator from Agrobacterium tumefaciens nopaline synthase (tNOS, MF116010) for T1 position (E/F)DepositorInsert(E) tNOS (F)
ExpressionBacterialAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
(B) BLE marker(R) (SBE262)
Plasmid#195012Purposebleomycin (BLE) resistance genes codon-optimized for R. toruloides and expressed under pXYL and tGPDDepositorInsertpXYL+BLE+tGPD
ExpressionBacterialPromoterpXYLAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
pZ_lacO-cP-AOX1-eGFP
Plasmid#126720Purposeyeast plasmid overexpressing eGFPDepositorInsertegfp
Tags6xHis,MycExpressionYeastAvailable SinceJune 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
p1585588
Plasmid#62620PurposeePathBrick pETM6 vector encoding CsF3H, CsDFR, and DuLAR (1:3:3 Ratio)DepositorInsertsflavaone 3B-hydroxylase
dihydroflavonol 4-reductase
leucoanthocyanidin reductase
dihydroflavonol 4-reductase
dihydroflavonol 4-reductase
leucoanthocyanidin reductase
leucoanthocyanidin reductase
UseSynthetic BiologyTagsGBD, PDZ, and SH3ExpressionBacterialPromoterT7Available SinceJuly 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCHC025 (∆CBBp)
Plasmid#226203PurposeElectroporation knockout plasmid (pK18sB) to delete the C. necator megaplasmid copy of the CBB operon (deltaCBBp).DepositorInsertHomology arms for megaplasmid copy of the CBB operonthe CBB operon (deltaCBBp).
UseSynthetic BiologyMutation∆cbbR ∆cbbLpSpXpYpEpFpPpTpZpGpKpApAvailable SinceNov. 18, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCHC024 (∆SH)
Plasmid#226202PurposeElectroporation knockout plasmid (pK18sB) to delete the C. necator soluble hydrogenase operon (deltaSH).DepositorInsertHomology arms for soluble hydrogenase operon
UseSynthetic BiologyMutation∆hoxFUYHWI ∆hypA2B2F2Available SinceNov. 18, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCHC023 (∆MBH)
Plasmid#226201PurposeElectroporation knockout plasmid (pK18sB) to delete the C. necator membrane-bound hydrogenase operon (deltaMBH).DepositorInsertHomology arms for membrane bound hydrogenase operon
UseSynthetic BiologyMutation∆hoxKGZMLOQRTV ∆hypA1B1F1CDEX ∆hoxABCJAvailable SinceNov. 18, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCHC027 (∆pemK)
Plasmid#226204PurposeConjugation knockout plasmid (pK18msB) to delete the C. necator addiction system toxin PemK, to enable deletion of the pHG1 megaplasmid.DepositorInsertUpstream/downstream homology arms for addiction system toxinPemK.
UseSynthetic BiologyAvailable SinceNov. 14, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pACB108
Plasmid#222207PurposeFor recombinant expression of fghA (PP_1617 from Pseudomonas putida) with a C-terminal His tagDepositorInsertfghA
Tags6x HisExpressionBacterialPromoterT7Available SinceAug. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACB109
Plasmid#222208PurposeFor recombinant expression of fghA (PP_1617 from Pseudomonas putida) with the K10N mutation and a C-terminal His tagDepositorInsertfghA (K10N)
Tags6x HisExpressionBacterialMutationChanged Lys10 to AsnPromoterT7Available SinceAug. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACB110
Plasmid#222209PurposeFor recombinant expression of fghA (PP_1617 from Pseudomonas putida) with the G258D mutation and a C-terminal His tagDepositorInsertfghA (G258D)
Tags6x HisExpressionBacterialMutationChanged Gly258 to AspartatePromoterT7Available SinceAug. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
(R)pGPD1(D)_P1(SBE109)
Plasmid#194993Purposepromoter of the gene glyceraldehyde-3-phosphate dehydrogenase from R. toruloides for a P1 position (overhangs R/D)DepositorInsert(R) pGPD (D)
ExpressionBacterialAvailable SinceFeb. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
WT_IFL_A28C_R
Plasmid#200184PurposeWT PA14 I-F Leader and A28C RepeatDepositorInsertWildtype I-F Leader and a mutant I-F Locus (A28C)
ExpressionBacterialMutationA28C in the I-F repeatAvailable SinceDec. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
(F)pGPD(G)_P2 (SBE110)
Plasmid#194992Purposepromoter of the gene glyceraldehyde-3-phosphate dehydrogenase from R. toruloides for a P2 position (overhangs F/G)DepositorInsert(F) pGPD (G)
ExpressionBacterialAvailable SinceDec. 7, 2023AvailabilityAcademic Institutions and Nonprofits only