We narrowed to 18,970 results for: REV
-
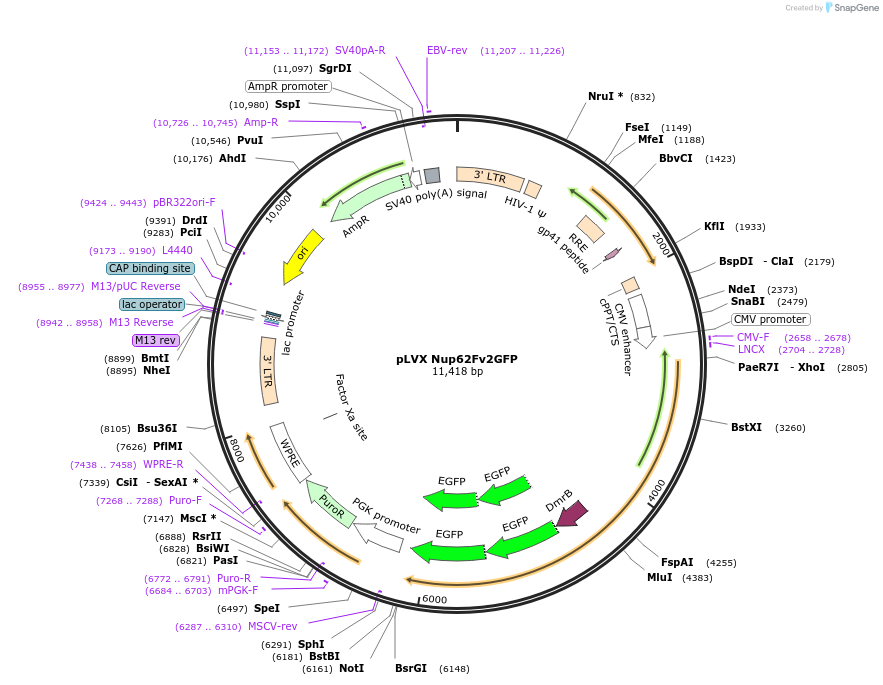
Plasmid#139683PurposeNup62 dimerization construct fused to 2 copies of GFP under an CMV promoterDepositorInsertNup62 (NUP62 Human)
UseLentiviralTags2 copies of GFP and Dimerization domain FKBPExpressionMammalianPromoterCMVAvailable SinceJune 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
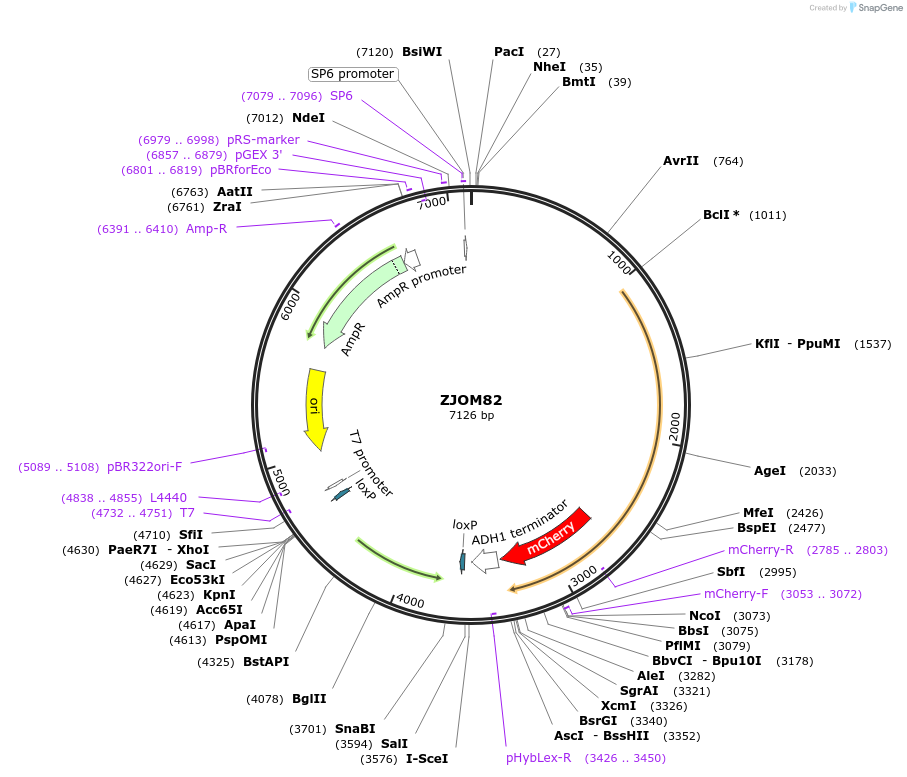
ZJOM82
Plasmid#133648PurposeIntegration of NAB2-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceJan. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
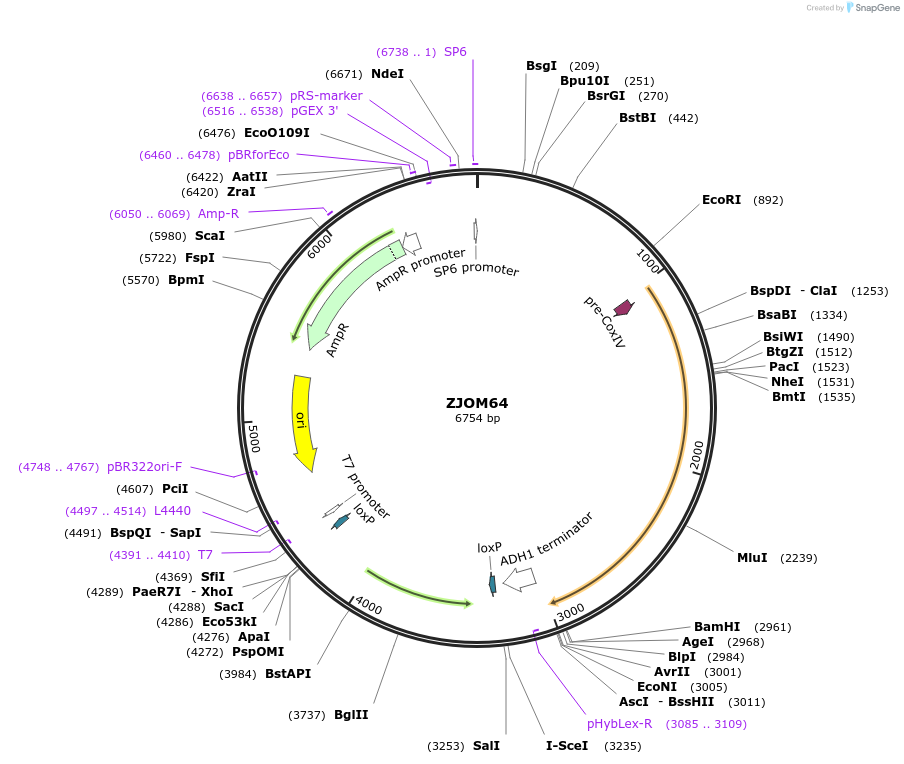
ZJOM64
Plasmid#133655PurposeIntegration of COX4-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
ZJOM135
Plasmid#133659PurposeIntegration of SEC63-2mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
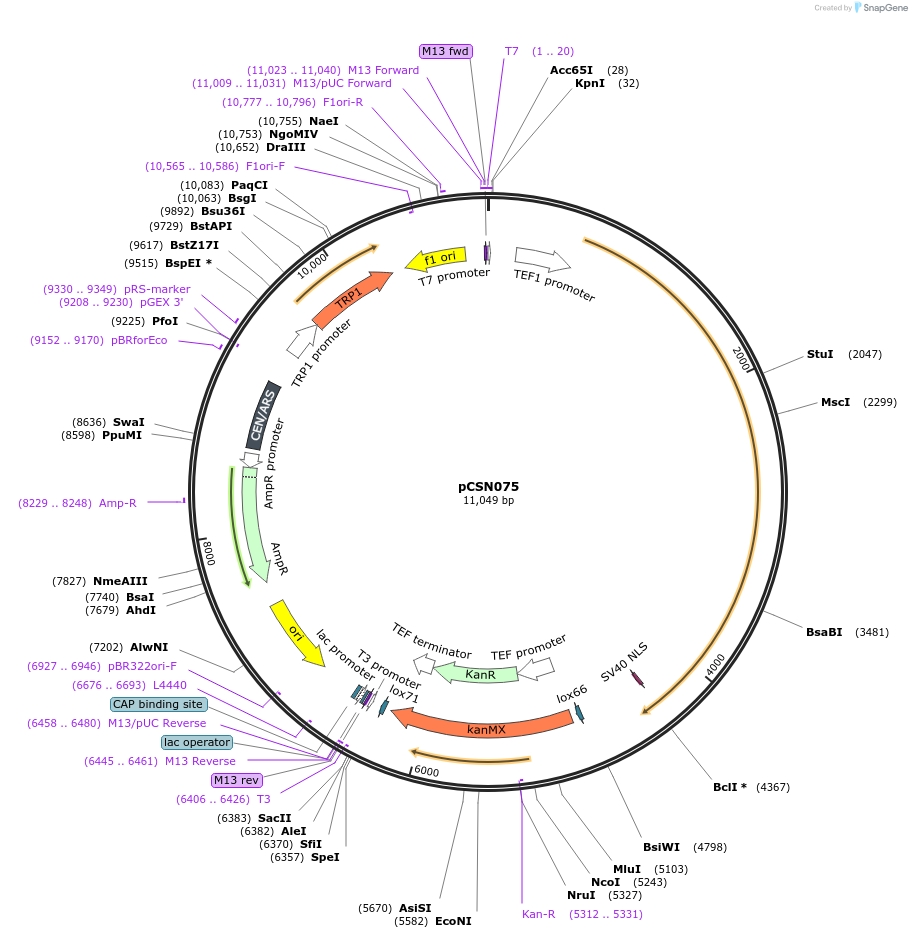
pCSN075
Plasmid#166732PurposeSingle copy yeast plasmid expressing DNase-dead dLbCas12a E925A fused to NLS and Mxi, codon optimized for Saccharomyces cerevisiaeDepositorInsertdCas12a-NLS-Mxi1
UseCRISPRTagsMxi1 and SV40 NLSExpressionYeastMutationE925APromoterS. cerevisiae TEF1Available SinceJune 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
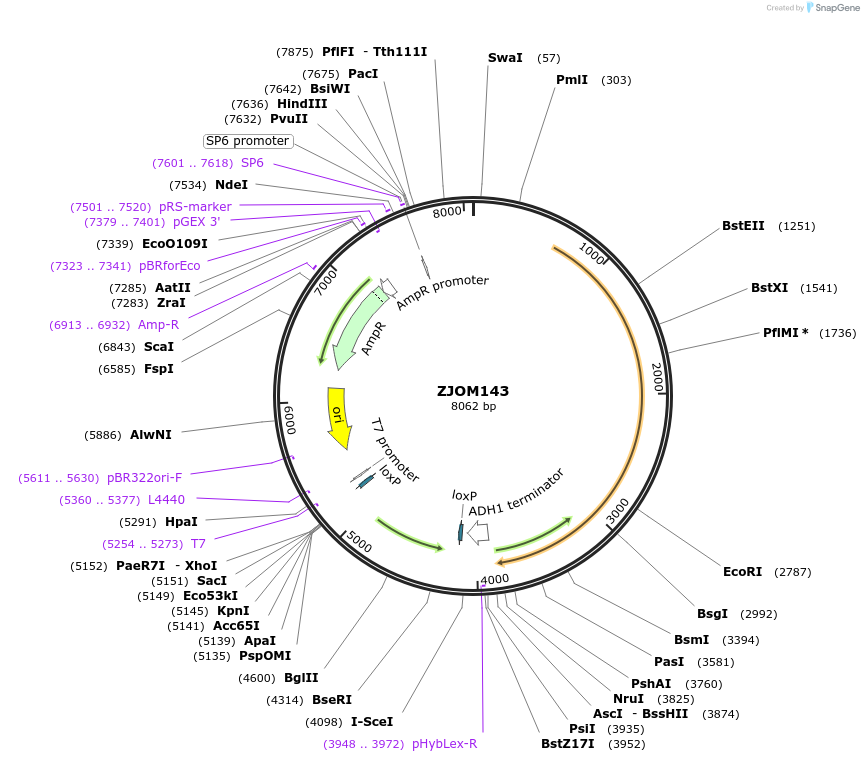
ZJOM143
Plasmid#133667PurposeIntegration of VPH1-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3-AKAR4-NLS
Plasmid#138217PurposeNuclear-targeted FRET-based PKA activity reporter.DepositorInsertAKAR4-NLS
Tags6xHIS - T7 tag (gene 10 leader) - Xpress (TM) tag…ExpressionMammalianPromoterCMVAvailable SinceJan. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
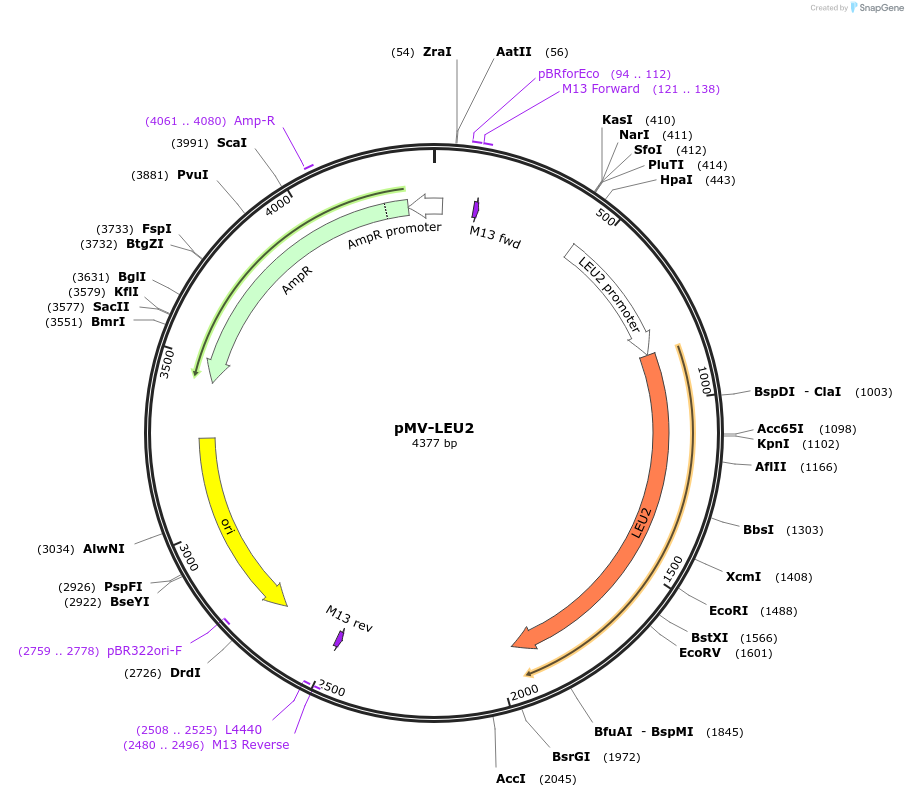
pMV-LEU2
Plasmid#65334Purposestandard plasmids for exogenus pathway assembly haboring yeast auxotrophic marker LEU2 which could be released and ligated to series of TUsDepositorInsertLEU2
UseSynthetic BiologyAvailable SinceFeb. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
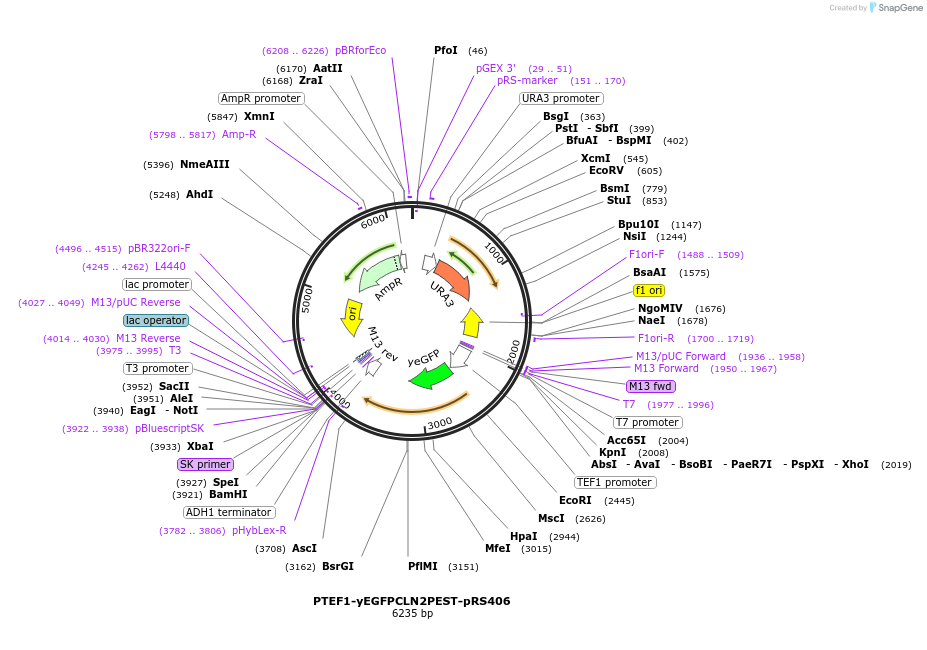
PTEF1-yEGFPCLN2PEST-pRS406
Plasmid#64406PurposeTEF promoter driven GFP expressionDepositorInsertTEF1 promoter (TEF1 Budding Yeast)
TagsyEGFPCLN2PEST (destabilized yeast EGFP)ExpressionYeastPromoterTEF1 promoterAvailable SinceMay 19, 2015AvailabilityAcademic Institutions and Nonprofits only -
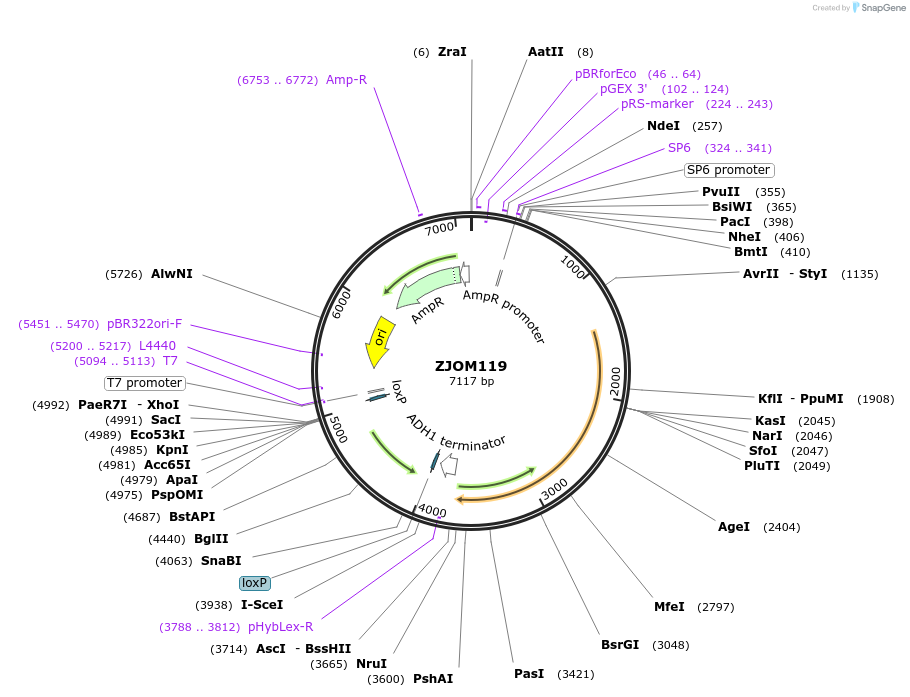
ZJOM119
Plasmid#133661PurposeIntegration of NAB2-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceDec. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
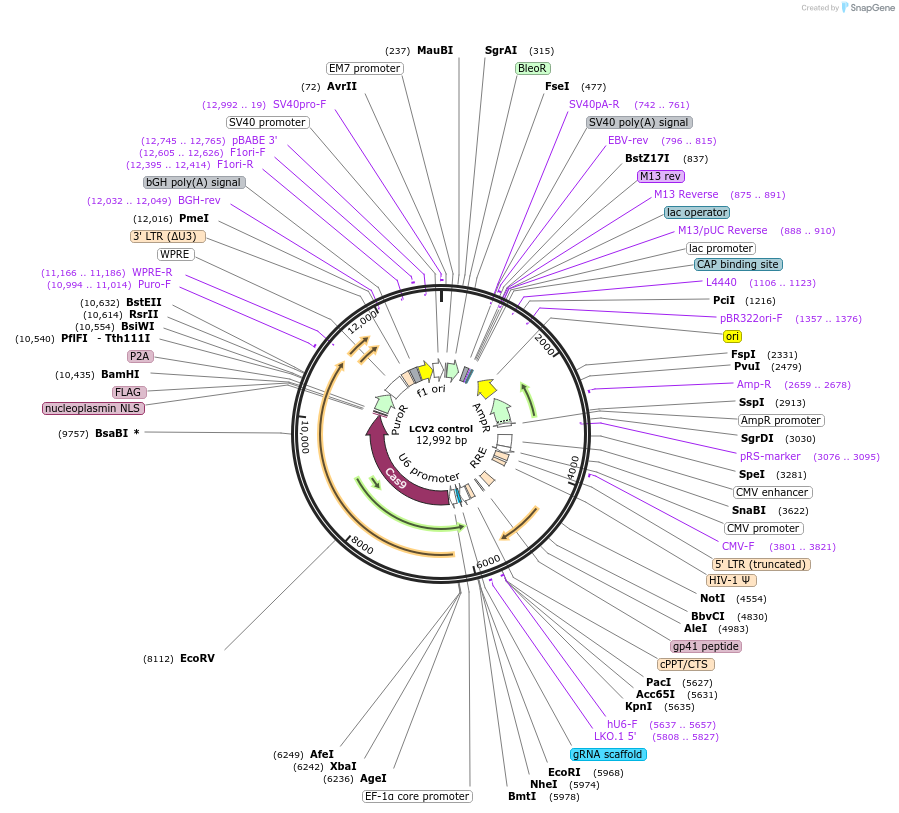
LCV2 control
Plasmid#217443PurposeLentiviral vector expressing Cas9 without a targeting sgRNADepositorTypeEmpty backboneUseCRISPR and LentiviralExpressionMammalianAvailable SinceJan. 28, 2025AvailabilityAcademic Institutions and Nonprofits only -
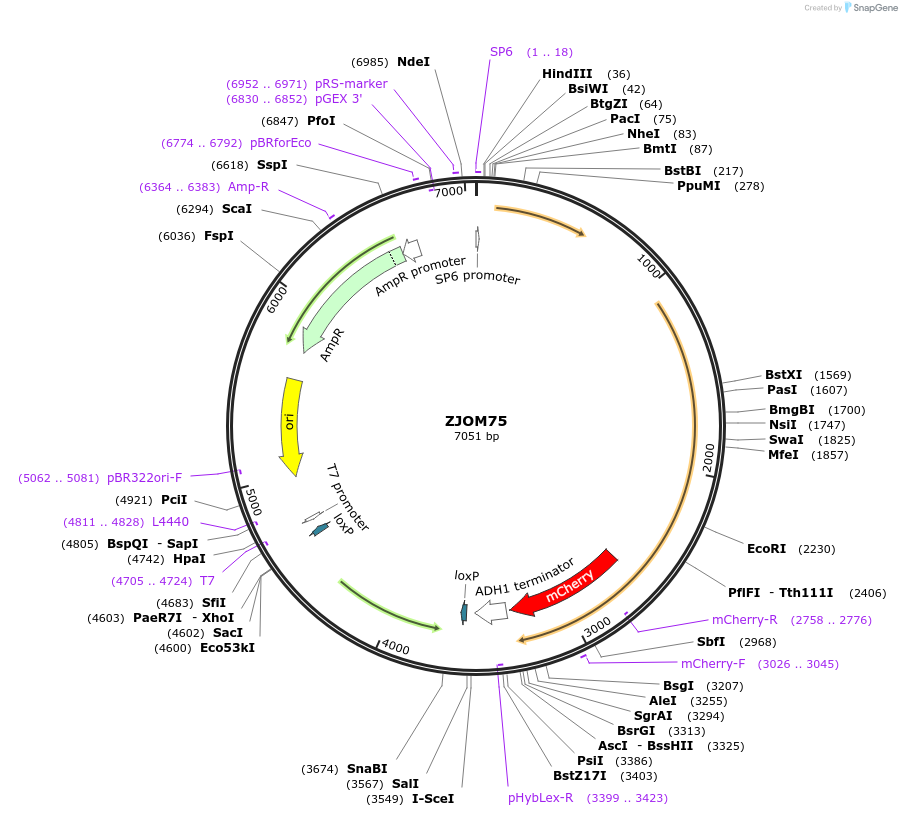
ZJOM75
Plasmid#133649PurposeIntegration of ANP1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceJan. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
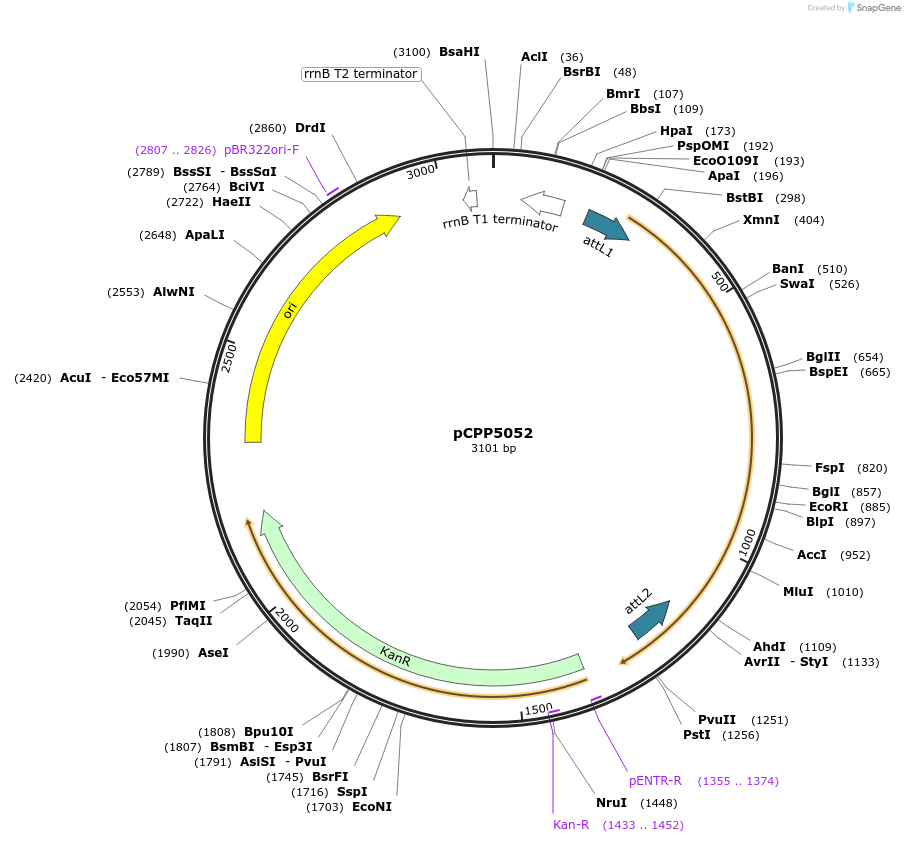
pCPP5052
Plasmid#128729PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopAM1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
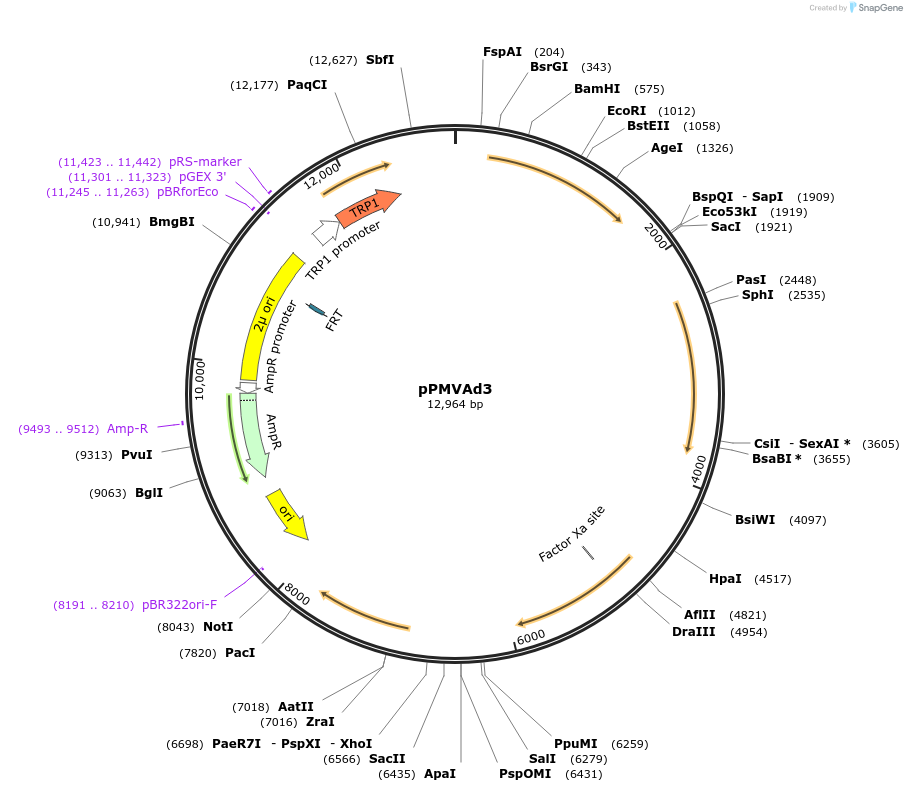
pPMVAd3
Plasmid#98298PurposeEnhancing the lower-part mevalonate pathway; overexpressing yeast mevalonate pathway genes under the control of consitutive promoters.DepositorInsertPRPL8B-ERG12-TNAT5-PSSB1-ERG8-TIDP1-PRPL3-MVD1-TIDP1-PYEF3-IDI1-TRPL15A
ExpressionYeastAvailable SinceAug. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
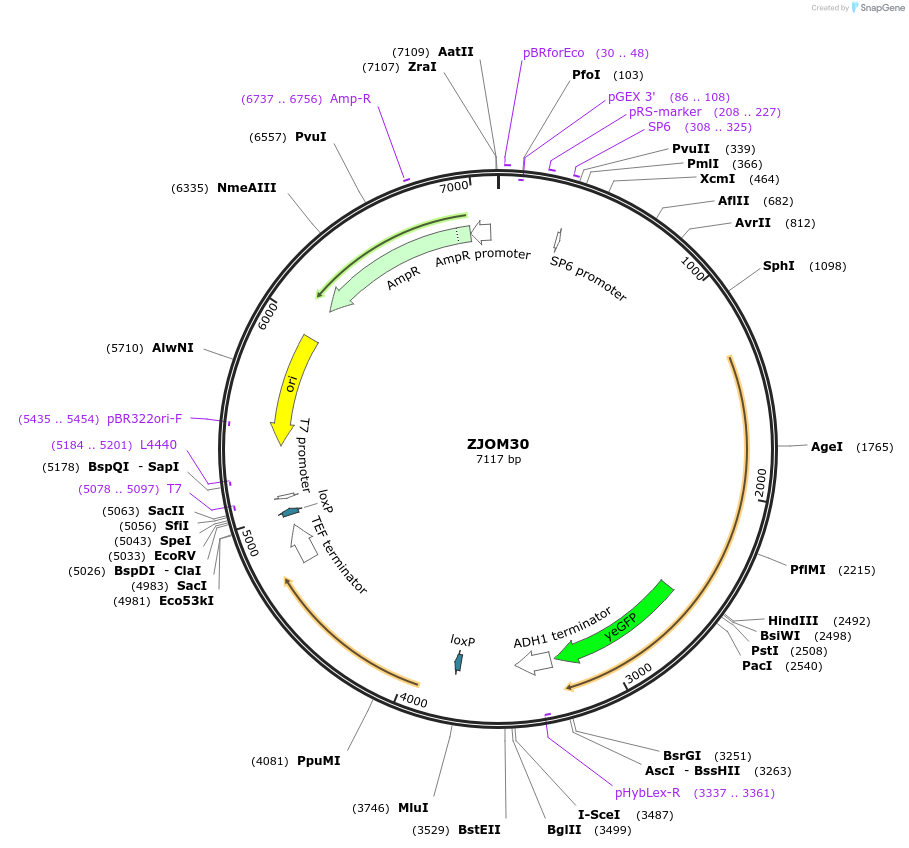
ZJOM30
Plasmid#133673PurposeIntegration of ERG6-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Present on ER and lipid droplets.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
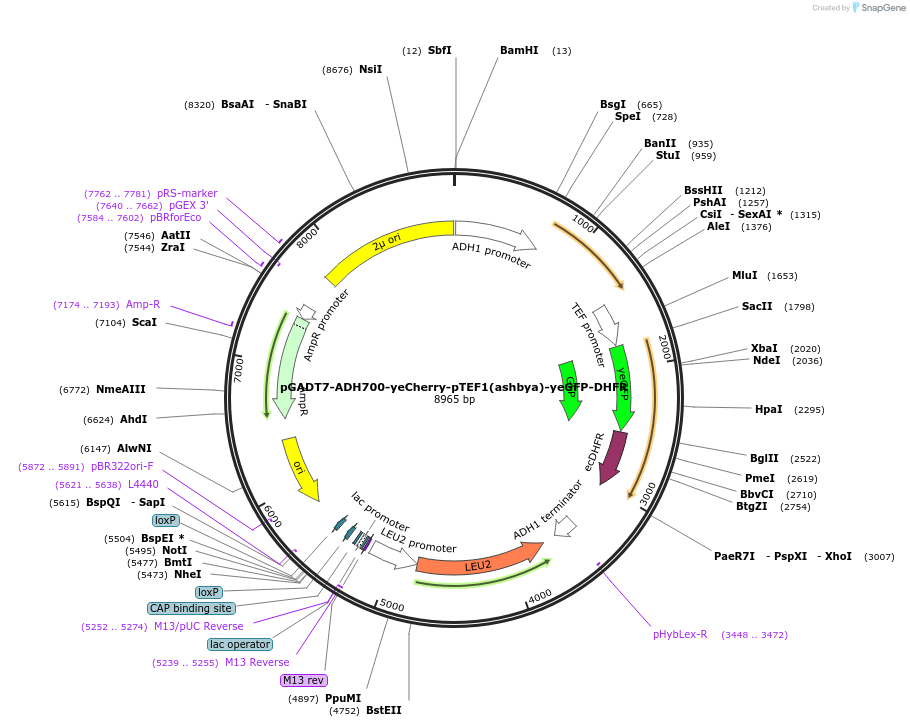
pGADT7-ADH700-yeCherry-pTEF1(ashbya)-yeGFP-DHFR
Plasmid#24585DepositorInsertpADH1 yeCherry::pTEF1 (Ashbya)yeGFP-DHFR
ExpressionYeastMutation1. Truncated version of ADH700 promoter (SbfI/Spe…Available SinceMay 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
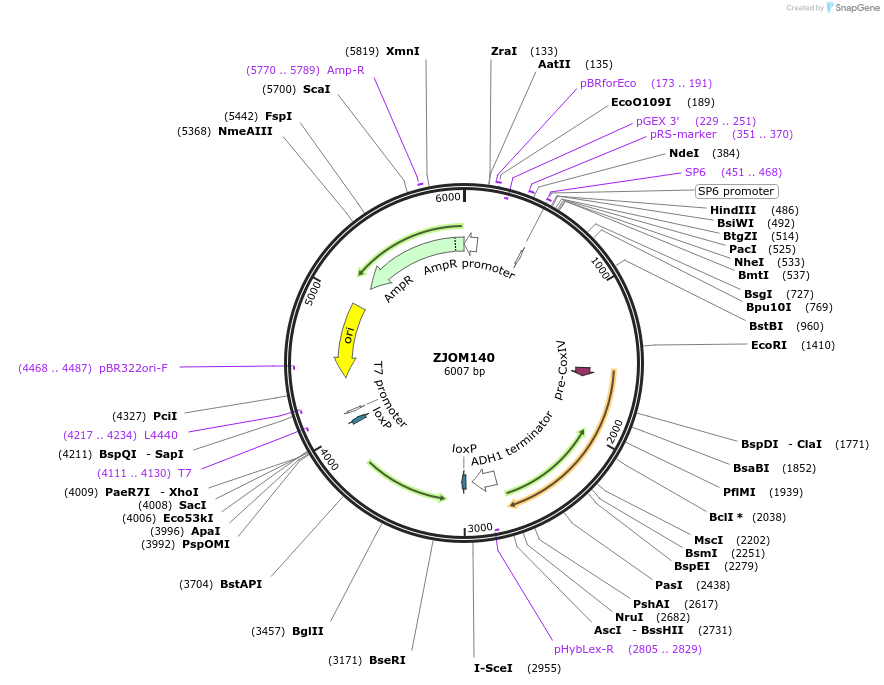
ZJOM140
Plasmid#133668PurposeIntegration of COX4-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
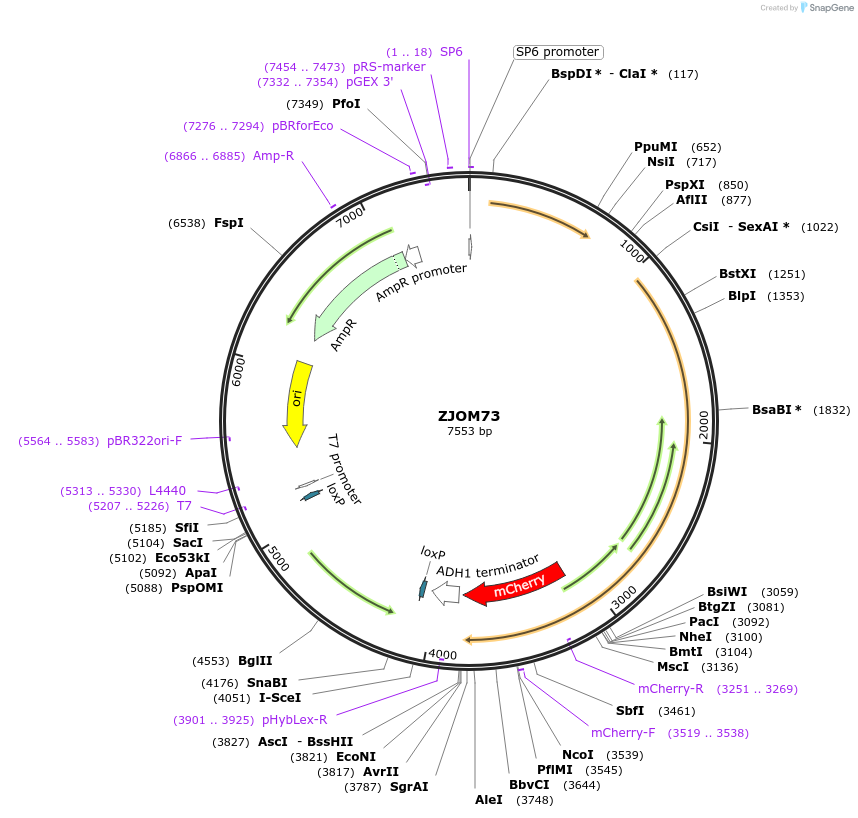
ZJOM73
Plasmid#133651PurposeIntegration of CHS5-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
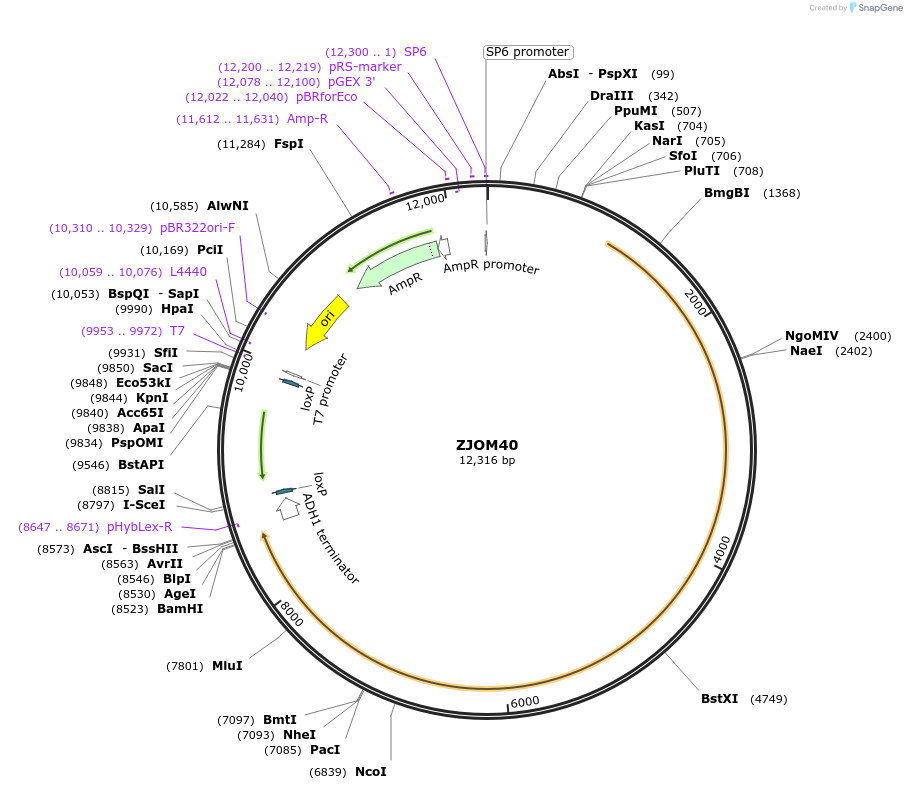
ZJOM40
Plasmid#133650PurposeIntegration of SEC7-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
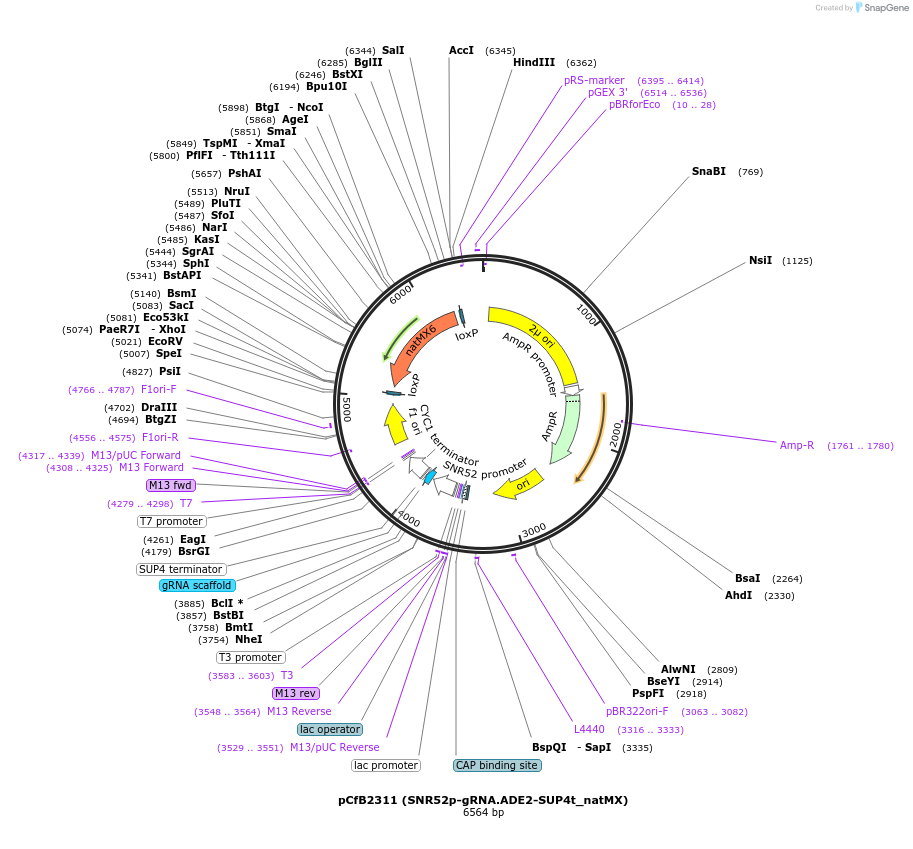
pCfB2311 (SNR52p-gRNA.ADE2-SUP4t_natMX)
Plasmid#83947PurposegRNA cassette-carrying vector with natMX markerDepositorInsertgRNA targeting ADE2 gene
ExpressionYeastAvailable SinceOct. 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
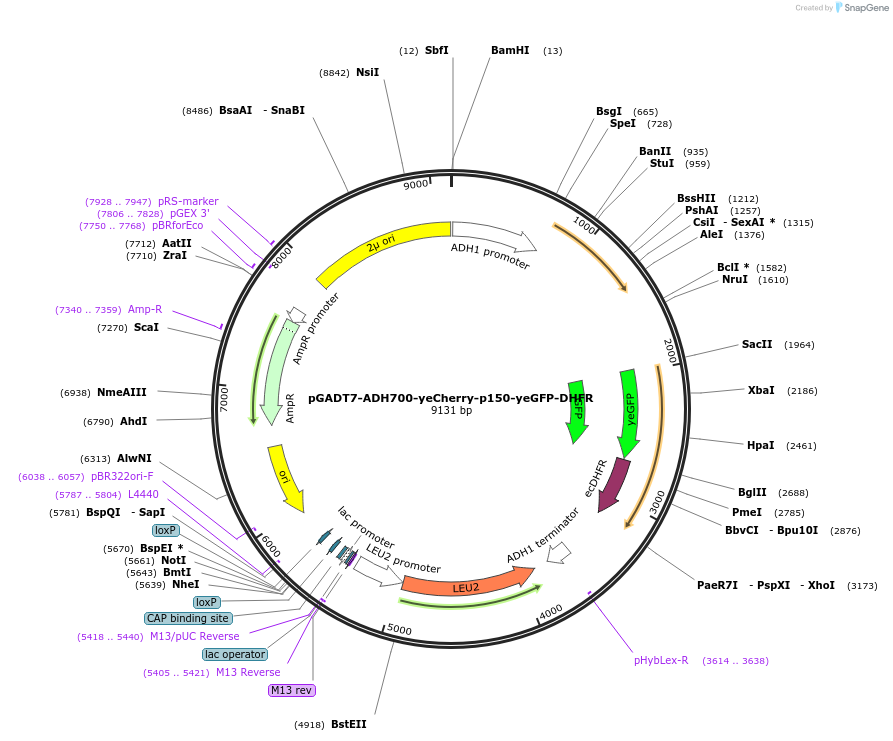
pGADT7-ADH700-yeCherry-p150-yeGFP-DHFR
Plasmid#24378DepositorInsertpADH1 yeCherry::p150yeGFP-DHFR
ExpressionYeastMutation1. Truncated version of ADH700 promoter (SbfI/Spe…Available SinceMay 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
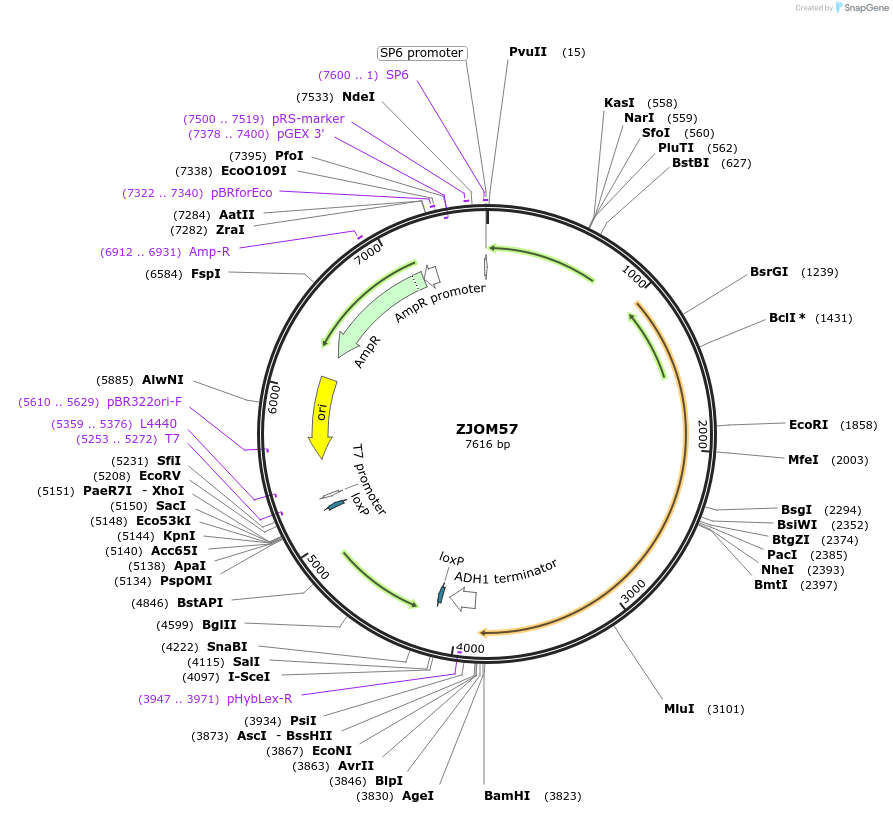
ZJOM57
Plasmid#133656PurposeIntegration of PEX3-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast peroxisome.DepositorAvailable SinceNov. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
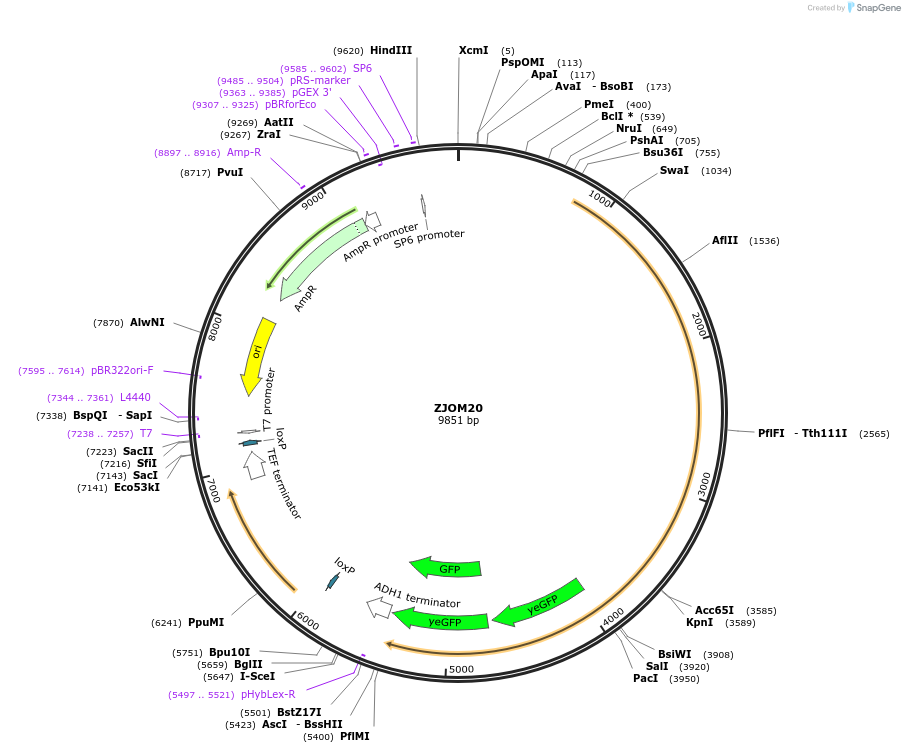
ZJOM20
Plasmid#133644PurposeIntegration of PEX1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast peroxisome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
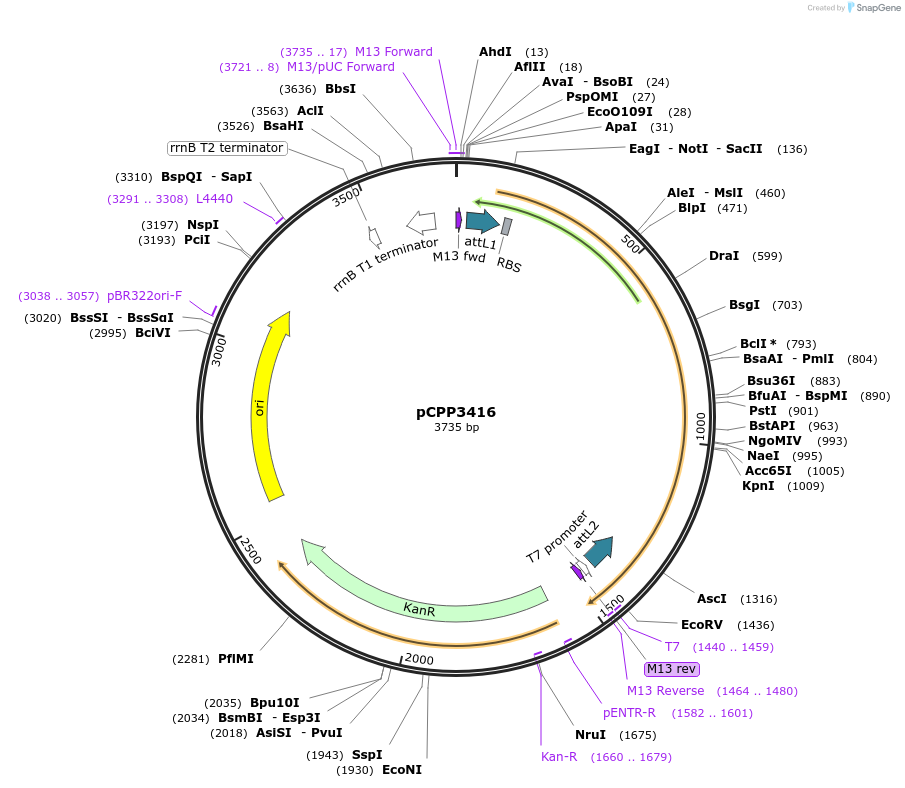
pCPP3416
Plasmid#128720PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopT1-1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
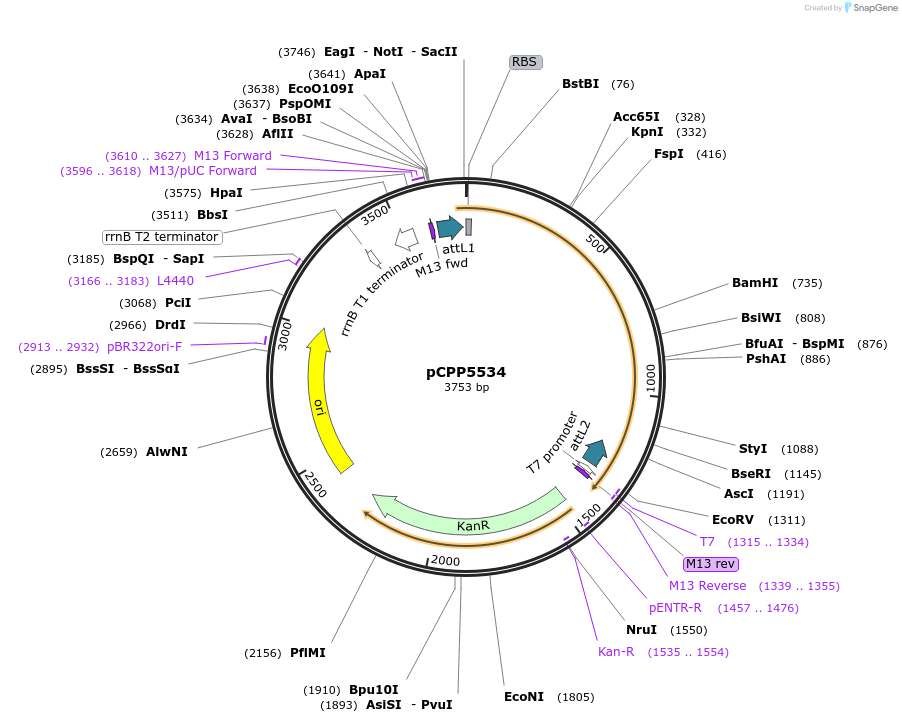
pCPP5534
Plasmid#128723PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopX1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
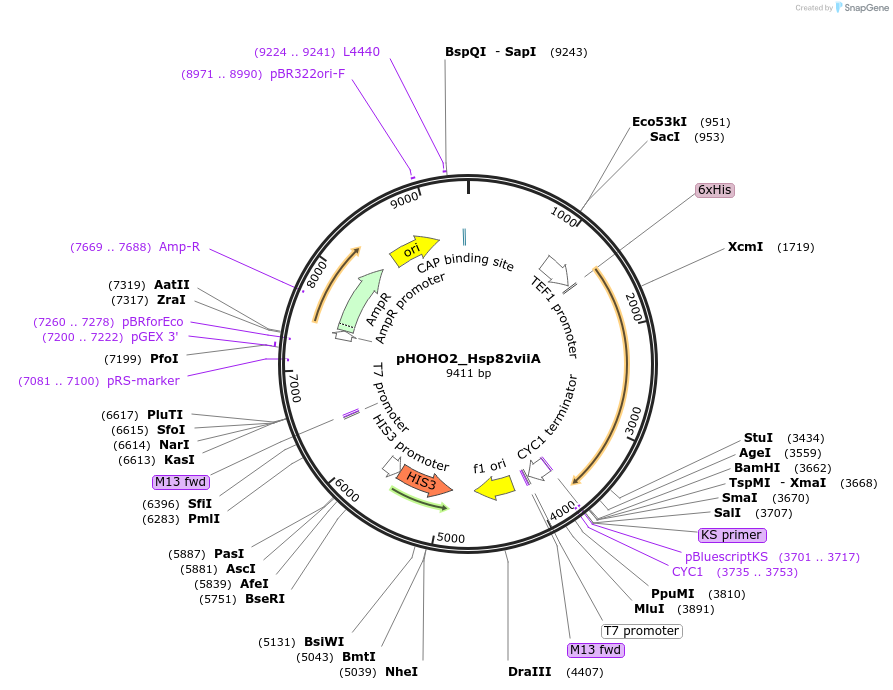
pHOHO2_Hsp82viiA
Plasmid#61708PurposeYeast integration plasmid targeted to the HO loci with heterodimeric Hsp82. Digest with NotI prior to addition to yeast.DepositorInsertHsp82viiA (HSP82 Budding Yeast)
UseYeast integration vector targeting inserts to the…Tags6xHisExpressionYeastMutationHeteromeric coiled coil inserted after amino acid…PromoterTEF1Available SinceFeb. 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
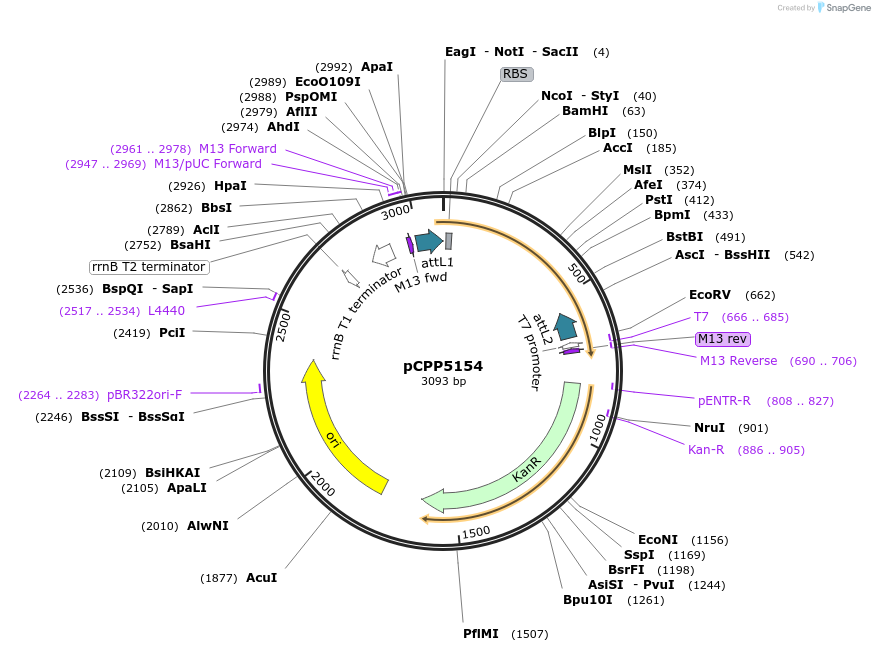
pCPP5154
Plasmid#128704PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInsertavrPto
ExpressionBacterialAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
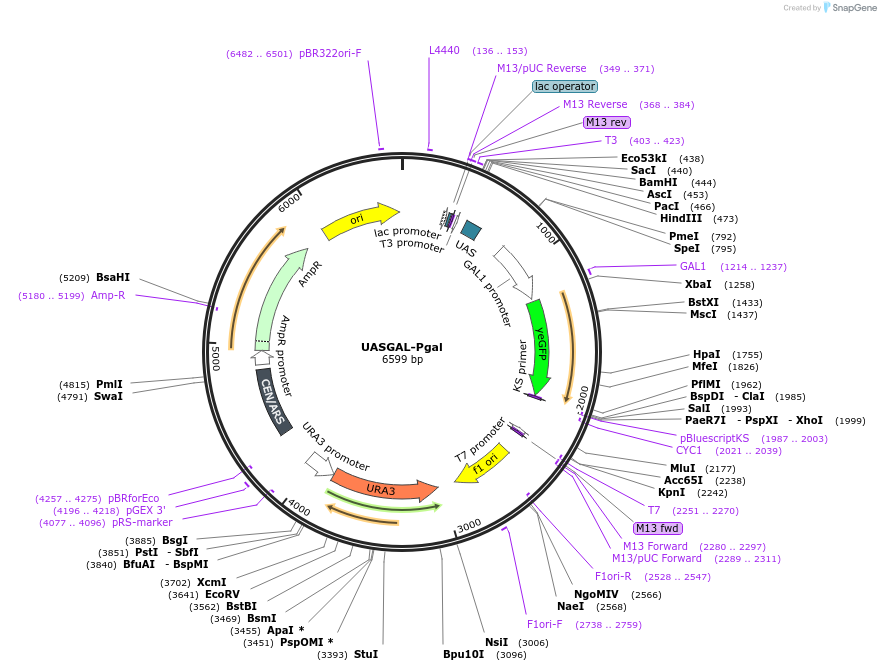
UASGAL-Pgal
Plasmid#60338PurposeContains a promoter of activating sequence derived from native galactose promoter coupled with full length native galactose promoterDepositorInsertpromoter
UseSynthetic Biology; Yeast synthetic hybrid promote…ExpressionYeastAvailable SinceNov. 6, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
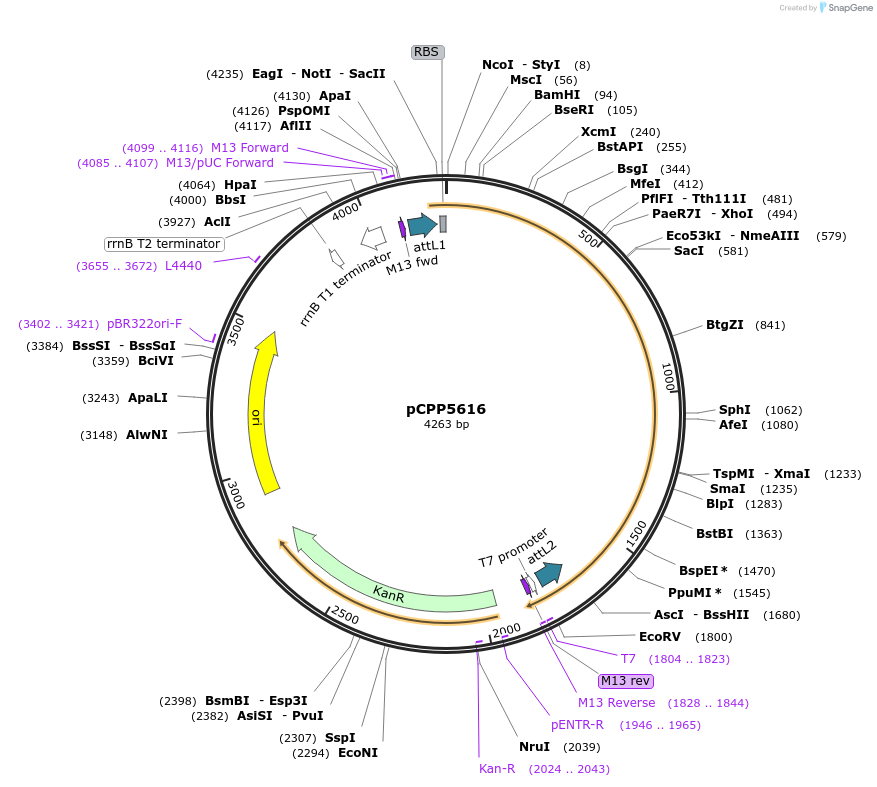
pCPP5616
Plasmid#128727PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInsertavrPtoB
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
GFP-ATG8(414)/GFP-AUT7(414)
Plasmid#49424PurposeExpresses ATG8 fused at the N terminus to GFP under the control of the endogenous promoter in the pRS414 vector.DepositorInsertautophagy-related 8 (ATG8 Budding Yeast)
TagsGFPExpressionBacterial and YeastPromoterATG8Available SinceJan. 2, 2014AvailabilityAcademic Institutions and Nonprofits only -
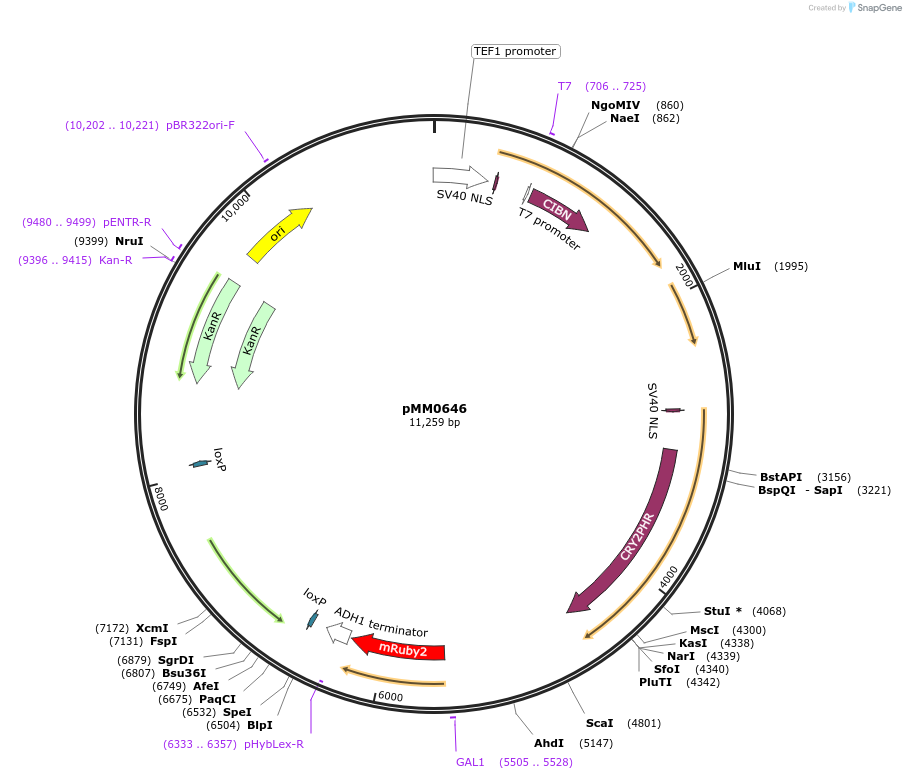
pMM0646
Plasmid#129007PurposeIntegration vector for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) and pZF-mRUBY2 with loxable KIURA3 markerDepositorInsertKanR-ColE1 URA 5'-pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pZF(BS)-mRUBY2-tADH1-loxP-KIURA3-loxP-URA3 3'
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
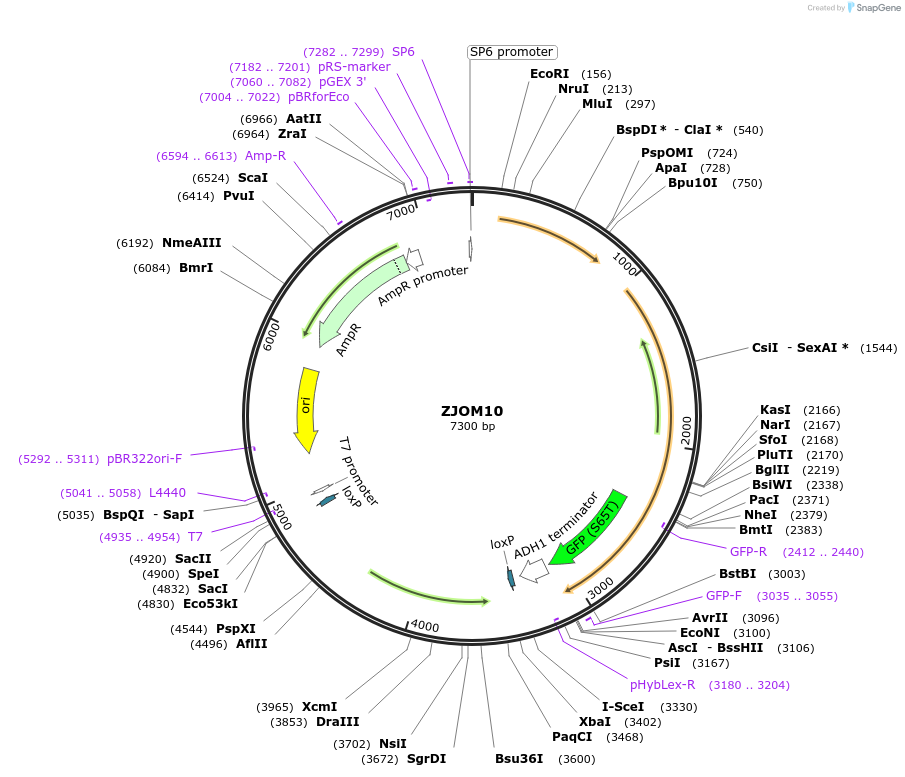
ZJOM10
Plasmid#133639PurposeIntegration of VPS4-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast late endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
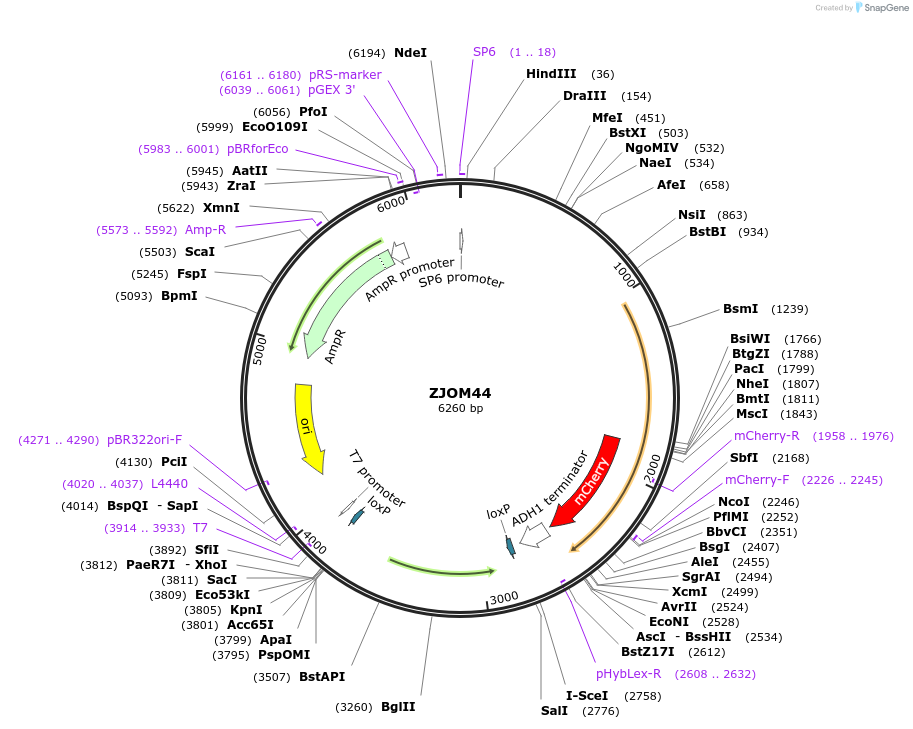
ZJOM44
Plasmid#133653PurposeIntegration of SNF7-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
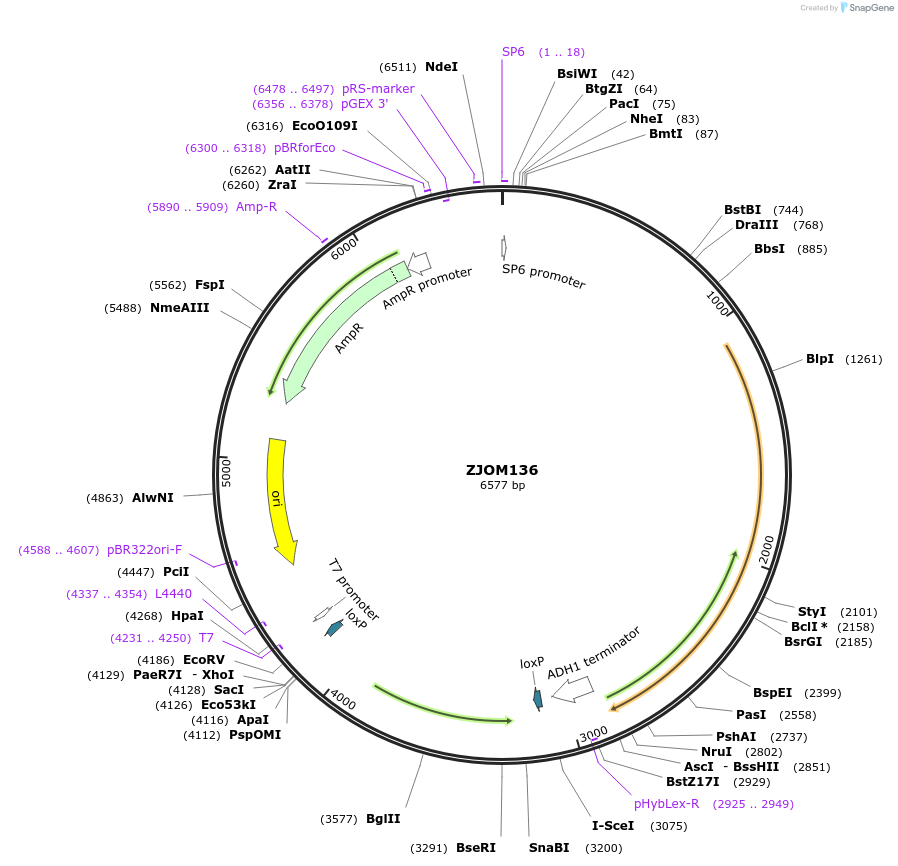
ZJOM136
Plasmid#133660PurposeIntegration of ELO3-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
ZJOM5
Plasmid#133635PurposeIntegration of CHS5-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
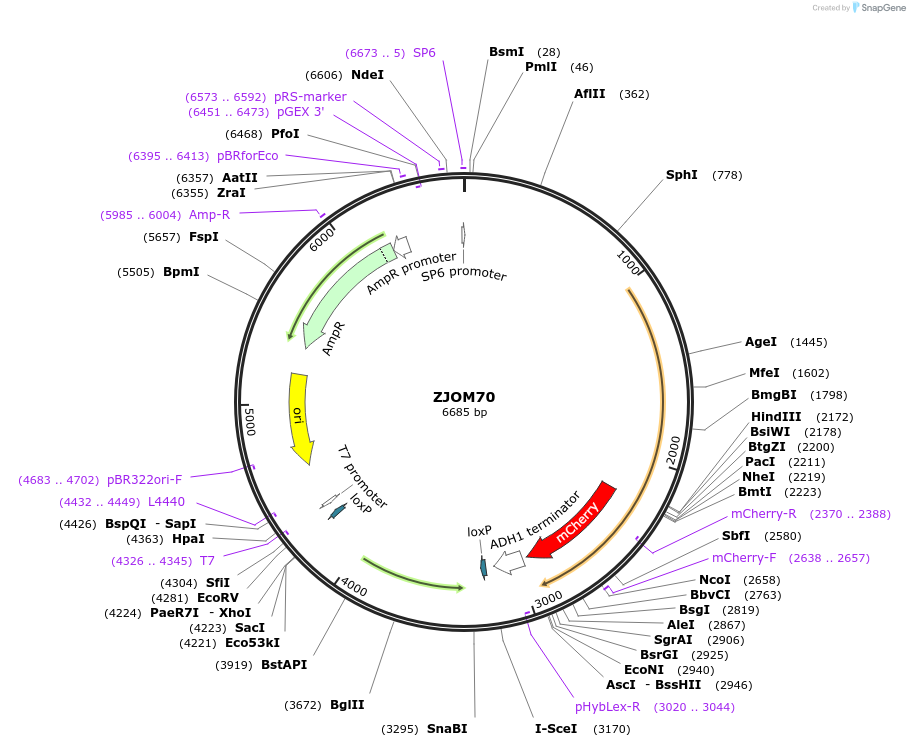
ZJOM70
Plasmid#133658PurposeIntegration of ERG6-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast lipid droplet.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
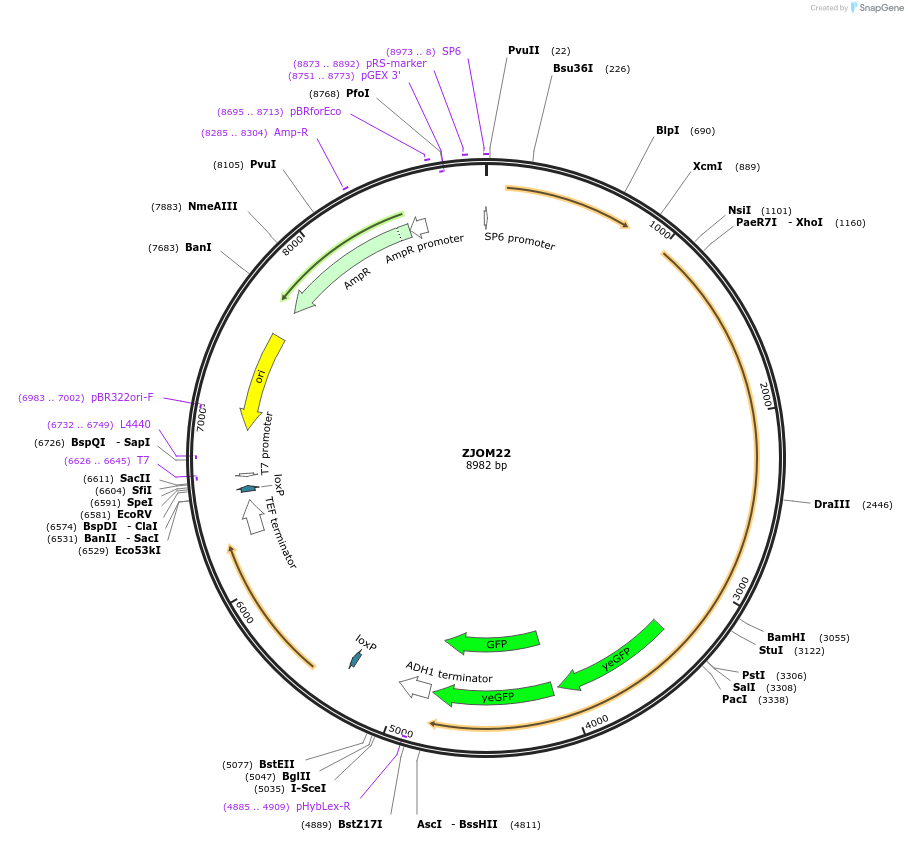
ZJOM22
Plasmid#133629PurposeIntegration of EMC1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
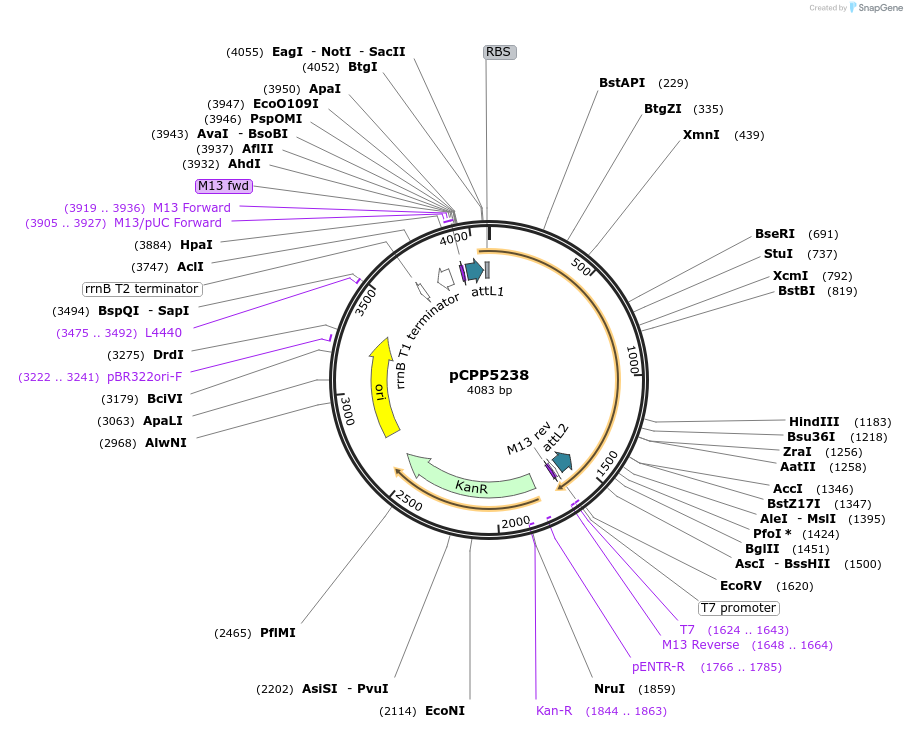
pCPP5238
Plasmid#128711PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopG1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
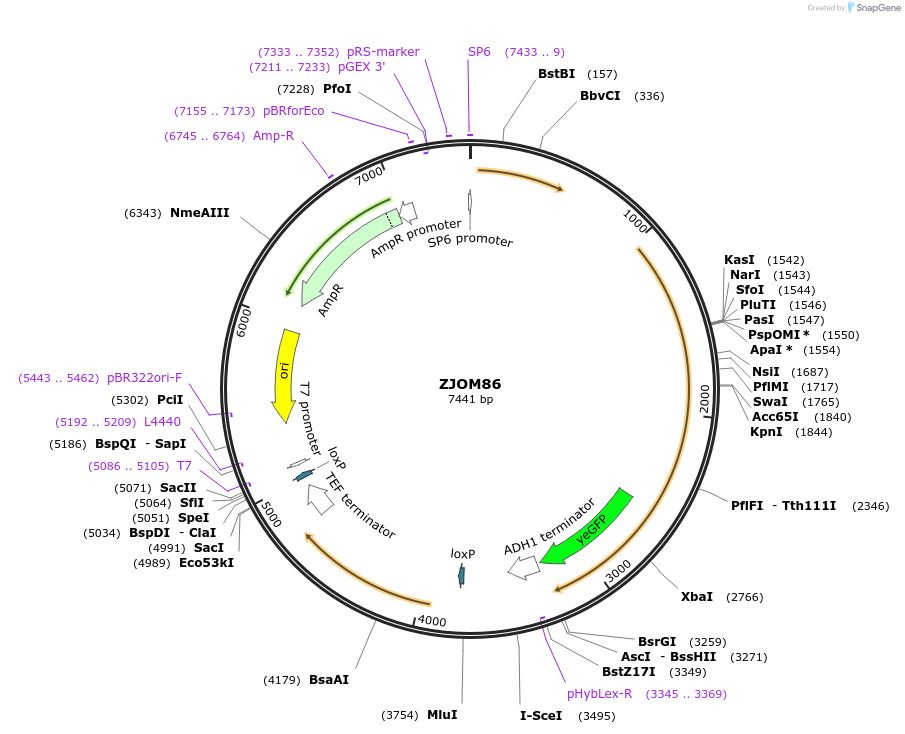
ZJOM86
Plasmid#133634PurposeIntegration of ANP1-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast early golgi.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
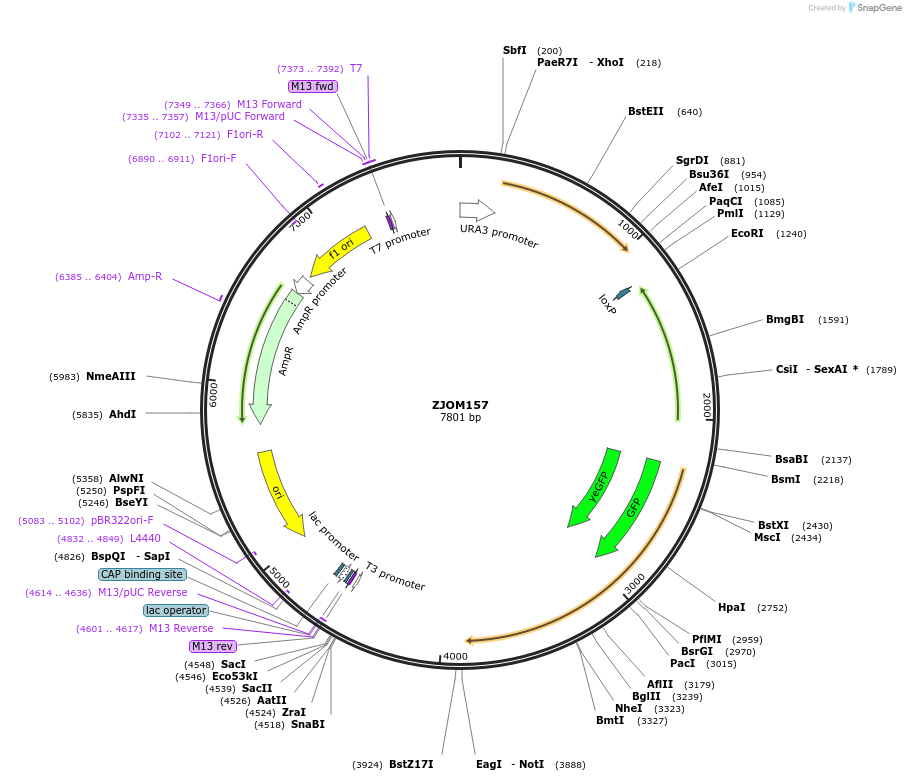
ZJOM157
Plasmid#133640PurposeIntegration of GFP-PEP12 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis).Marker for yeast late endosome.DepositorAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
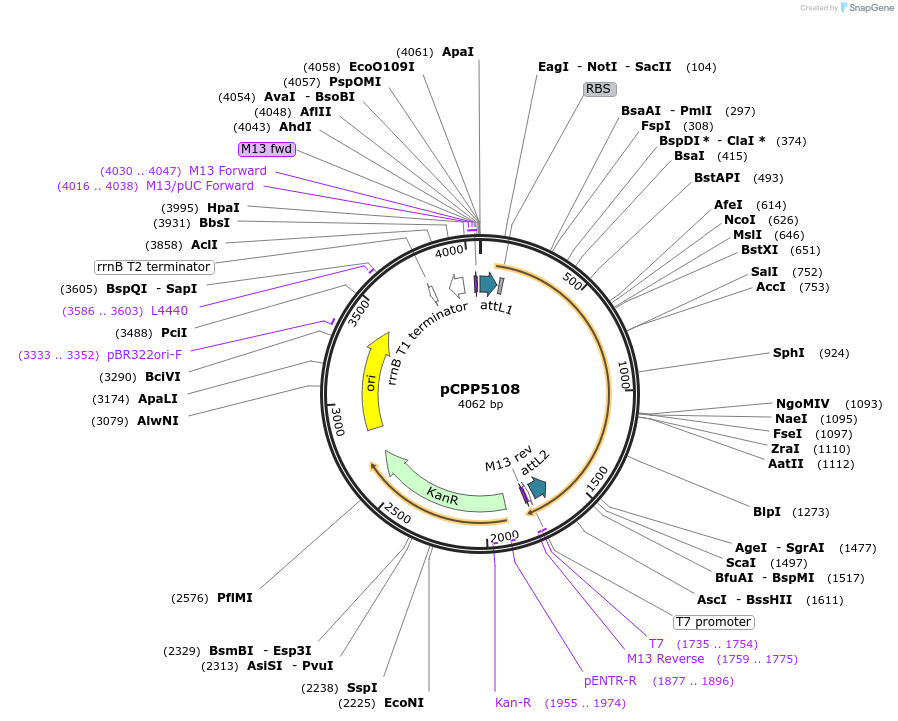
pCPP5108
Plasmid#128726PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopAA1-2
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
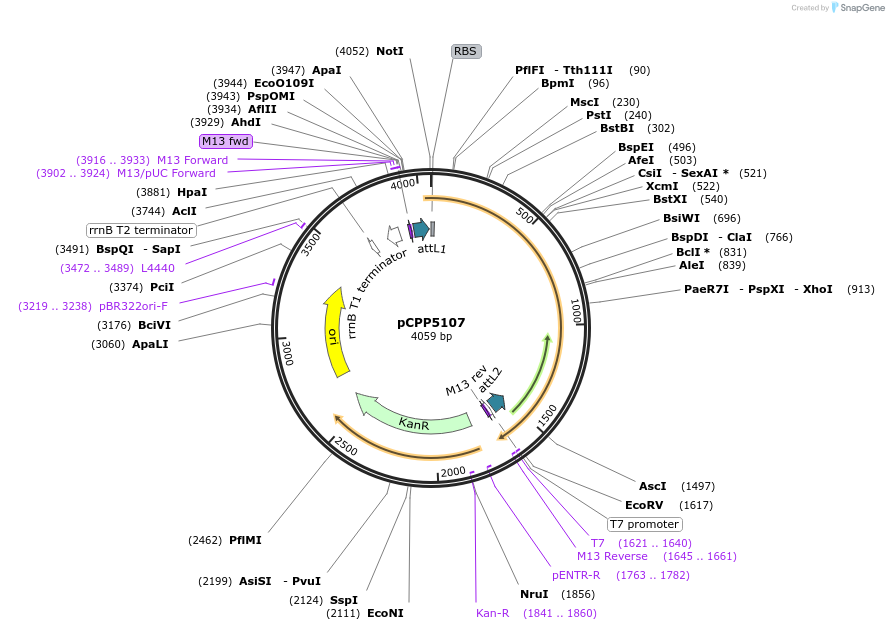
pCPP5107
Plasmid#128725PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopAA1-1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
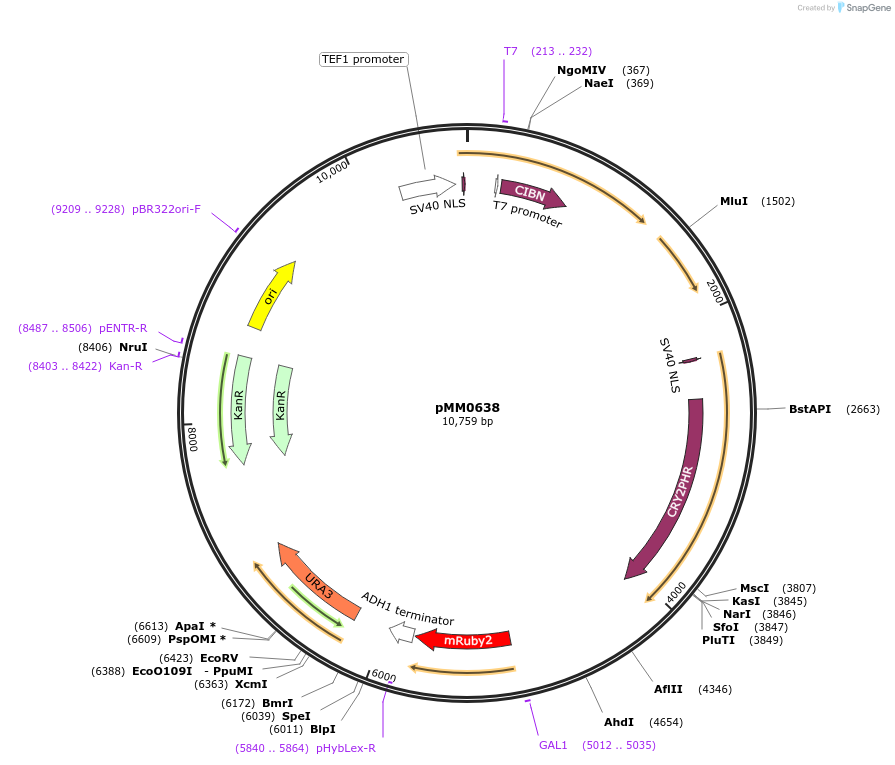
pMM0638
Plasmid#129005PurposeIntegration vector for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) and pZF-mRUBY2DepositorInsert5' URA3 -pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZiF268DBD-CRY2PHR-tSSA1-pZF(BS)-mRUBY2-tADH1-URA3 URA3 3'- KanR-ColE1
ExpressionYeastAvailable SinceOct. 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
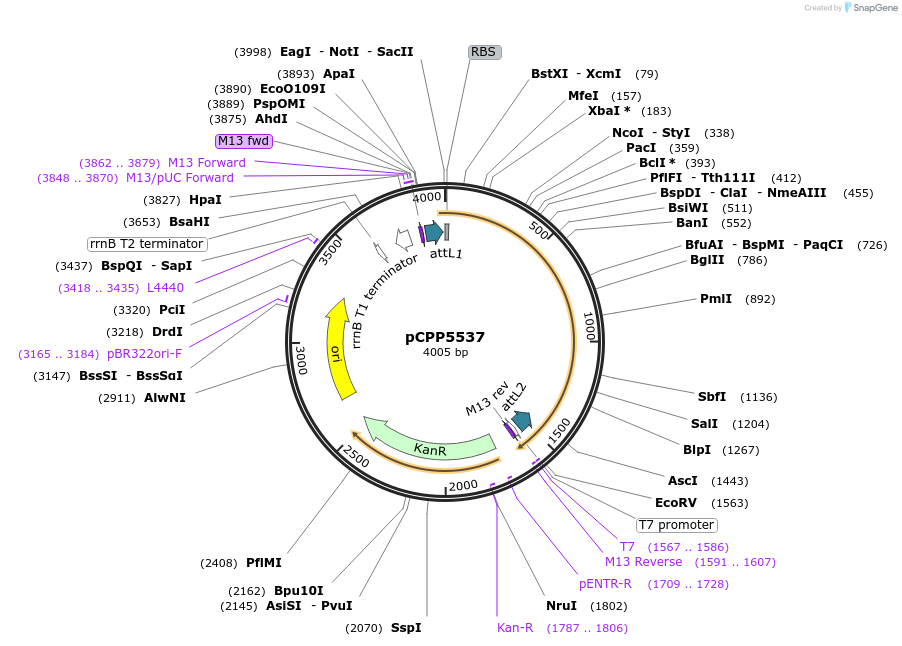
pCPP5537
Plasmid#128730PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInsertHopAO1
ExpressionBacterialAvailable SinceJan. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
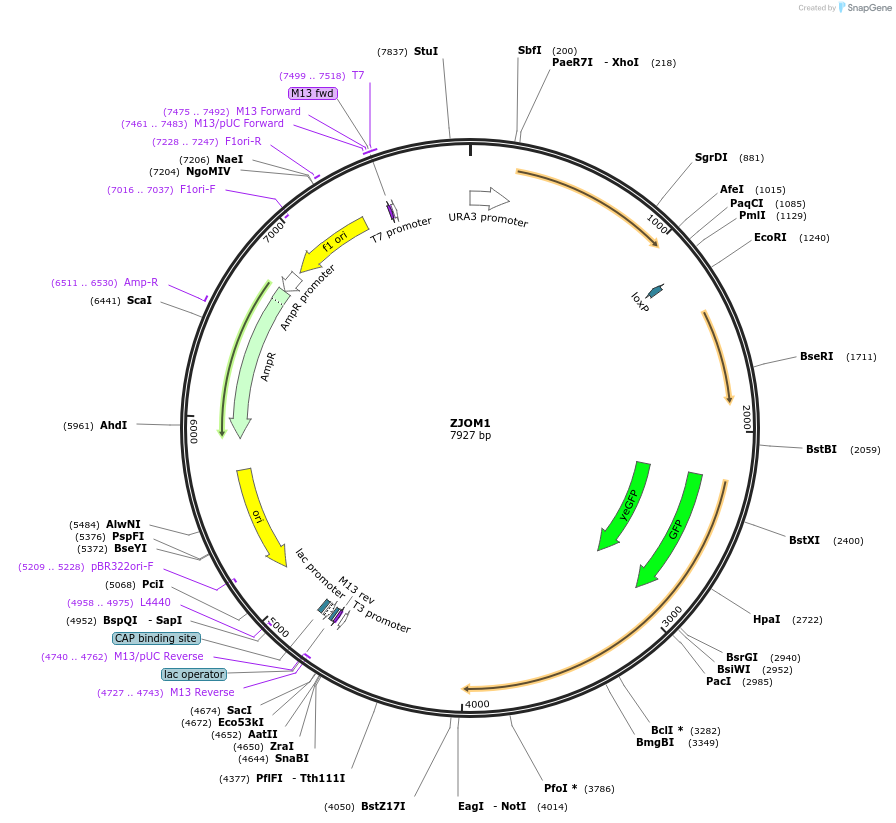
ZJOM1
Plasmid#133632PurposeIntegration of GFP-SED5 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
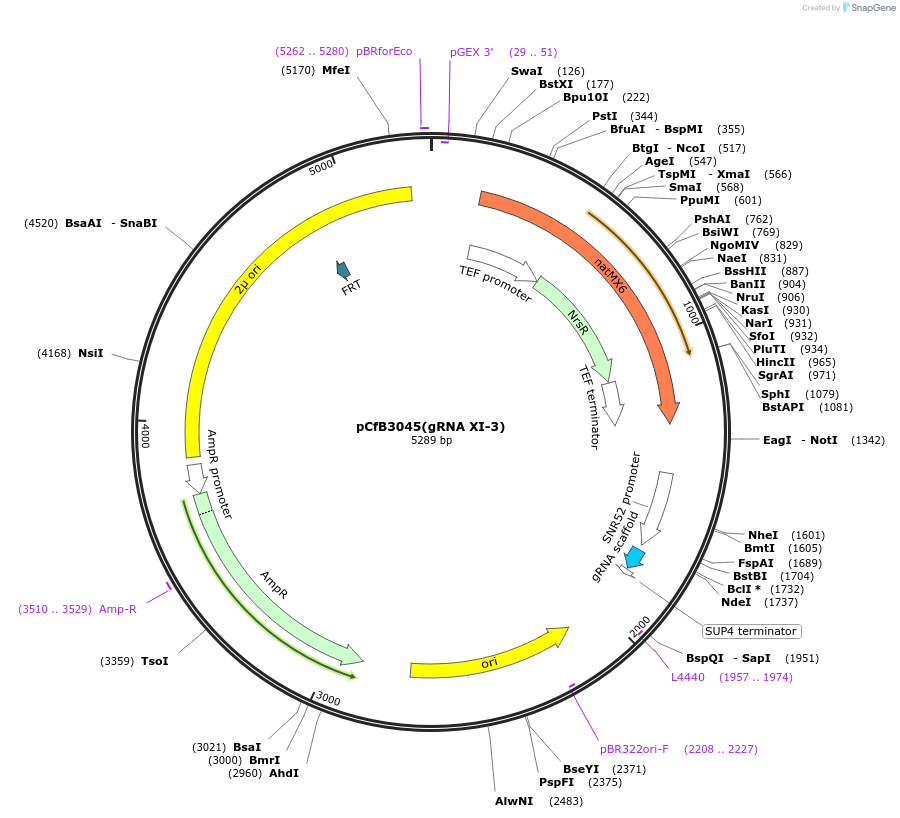
pCfB3045(gRNA XI-3)
Plasmid#73287PurposeEasyClone-MarkerFree guiding RNA vector to direct Cas9 to cut at site XI-3DepositorInsertguiding RNA
UseCRISPR; GrnaExpressionYeastAvailable SinceNov. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
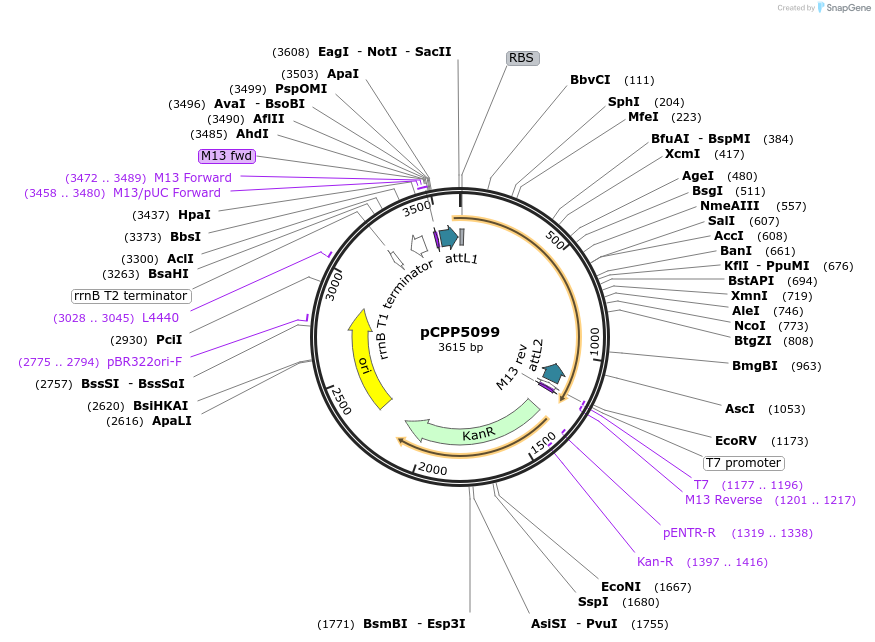
pCPP5099
Plasmid#128714PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopK1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
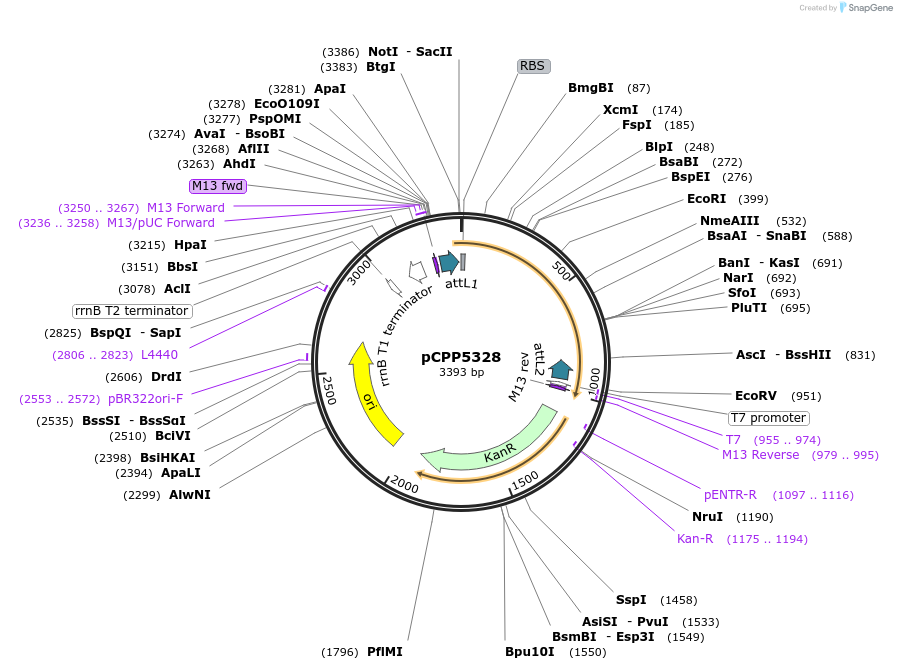
pCPP5328
Plasmid#128721PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopU1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
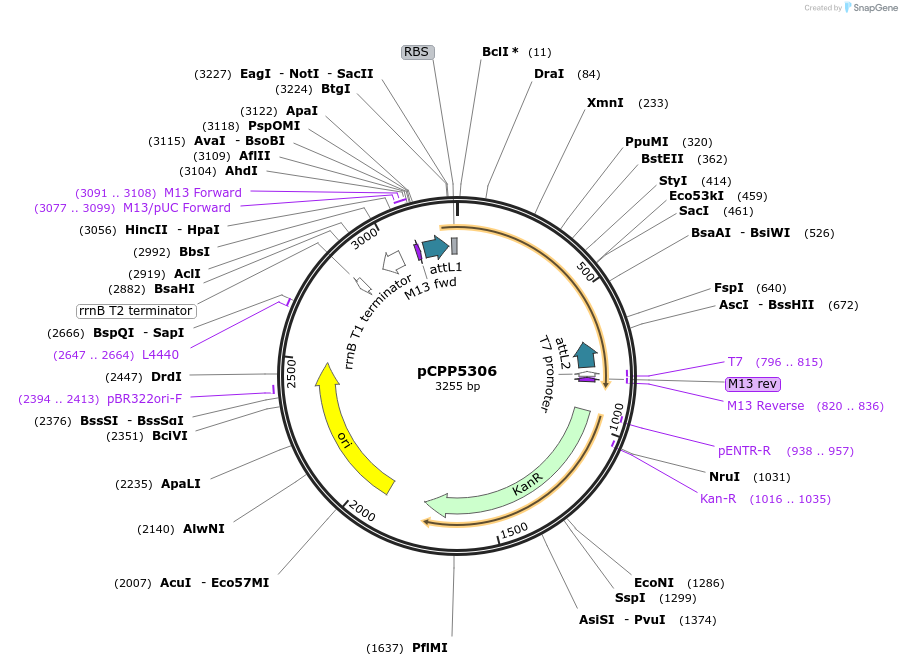
pCPP5306
Plasmid#128712PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopH1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
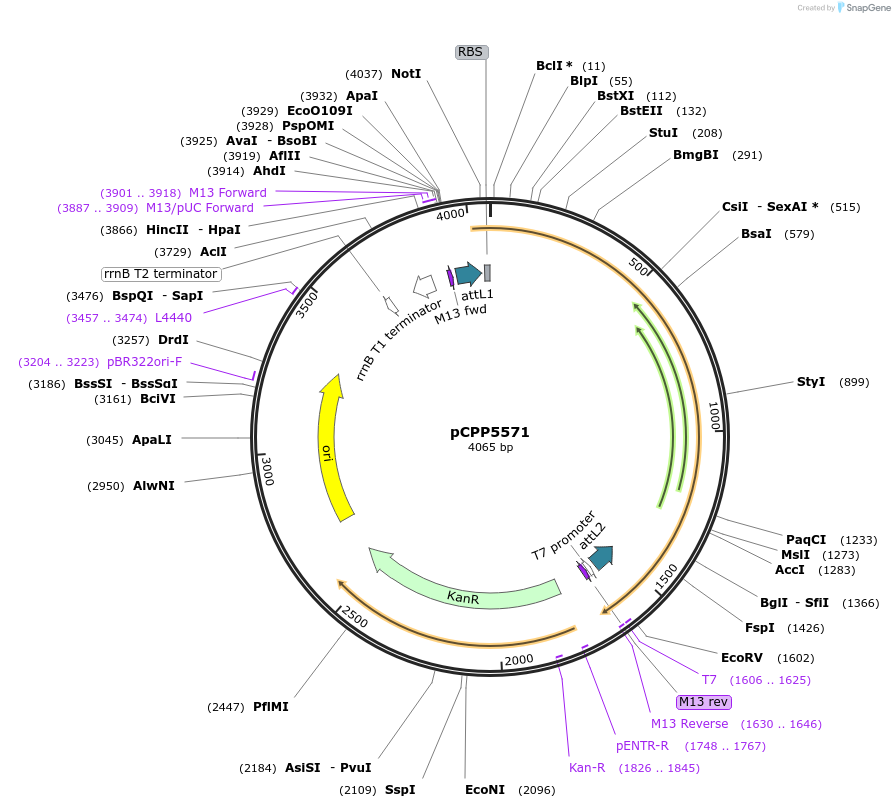
pCPP5571
Plasmid#128713PurposePhytopathogen Pseudomonas syringae pv. tomato type III secretion system effector (gene lacking stop codon) Gateway donor cloneDepositorInserthopI1
ExpressionBacterialAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only