We narrowed to 10,371 results for: otos
-
Plasmid#37305DepositorInsertSee comments
ExpressionMammalianAvailable SinceNov. 29, 2012AvailabilityAcademic Institutions and Nonprofits only -
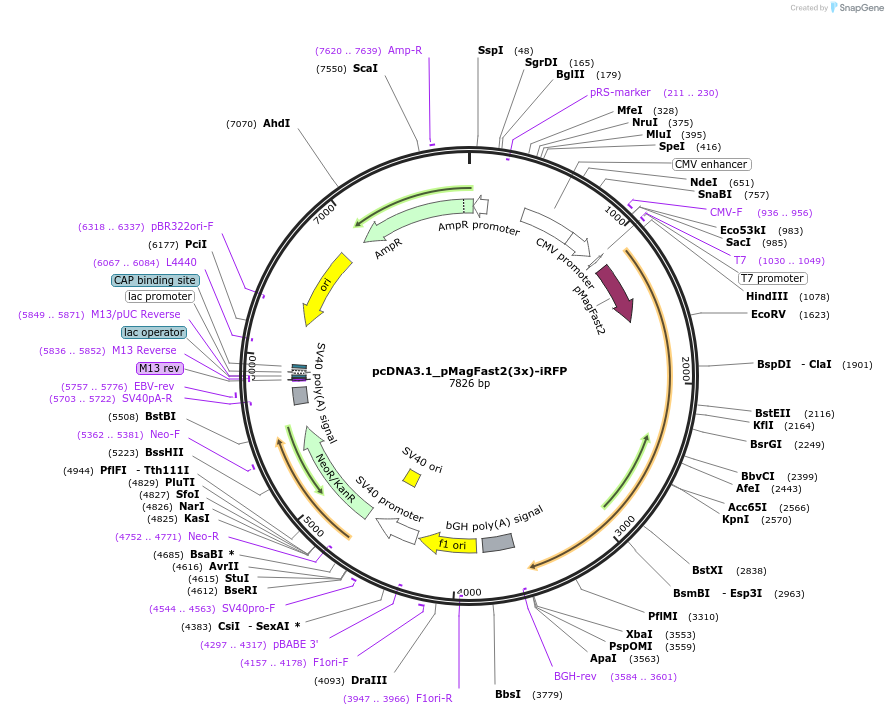
pcDNA3.1_pMagFast2(3x)-iRFP
Plasmid#67297PurposeBlue light dependent photoswitching systemDepositorInsertpMagFast2(3x)-iRFP
ExpressionMammalianPromoterCMVAvailable SinceJuly 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
mPA-GFP-CD151-7
Plasmid#57125PurposeLocalization: Tetraspanin/Membrane, Excitation: 400 / 504, Emission: 515 / 517DepositorAvailable SinceJan. 29, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
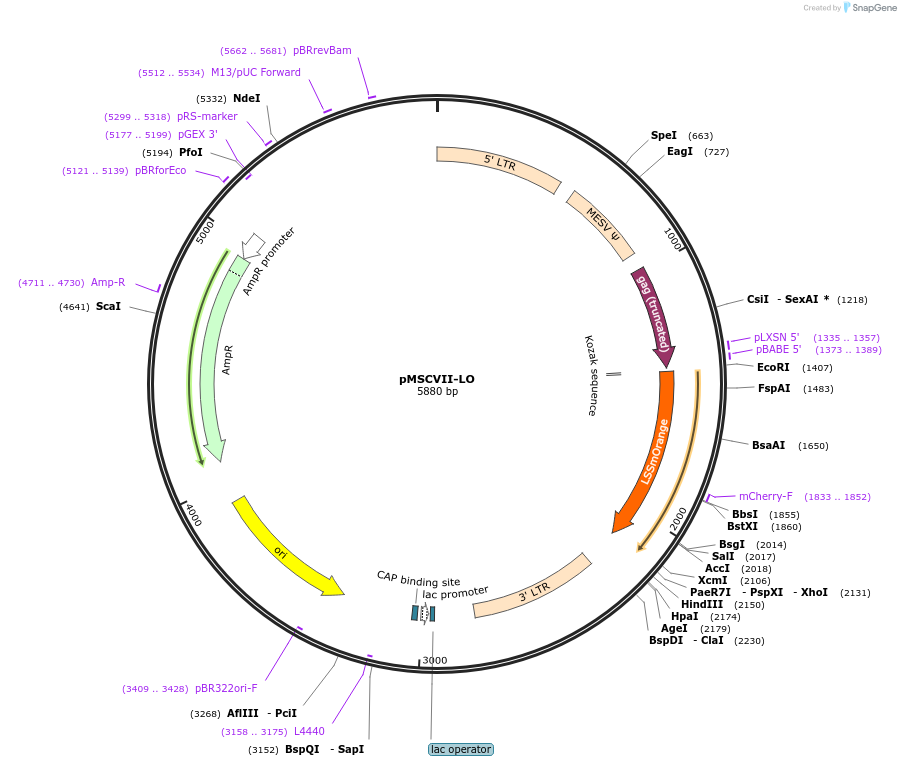
pMSCVII-LO
Plasmid#162754PurposeRetroviral (MSCV) expression of LSSmOrange fluorescent proteinDepositorInsertLSSmOrange
UseRetroviralExpressionMammalianAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
mPA-GFP-Caveolin-C-10
Plasmid#57123PurposeLocalization: Membrane, Excitation: 400 / 504, Emission: 515 / 517DepositorAvailable SinceJan. 29, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
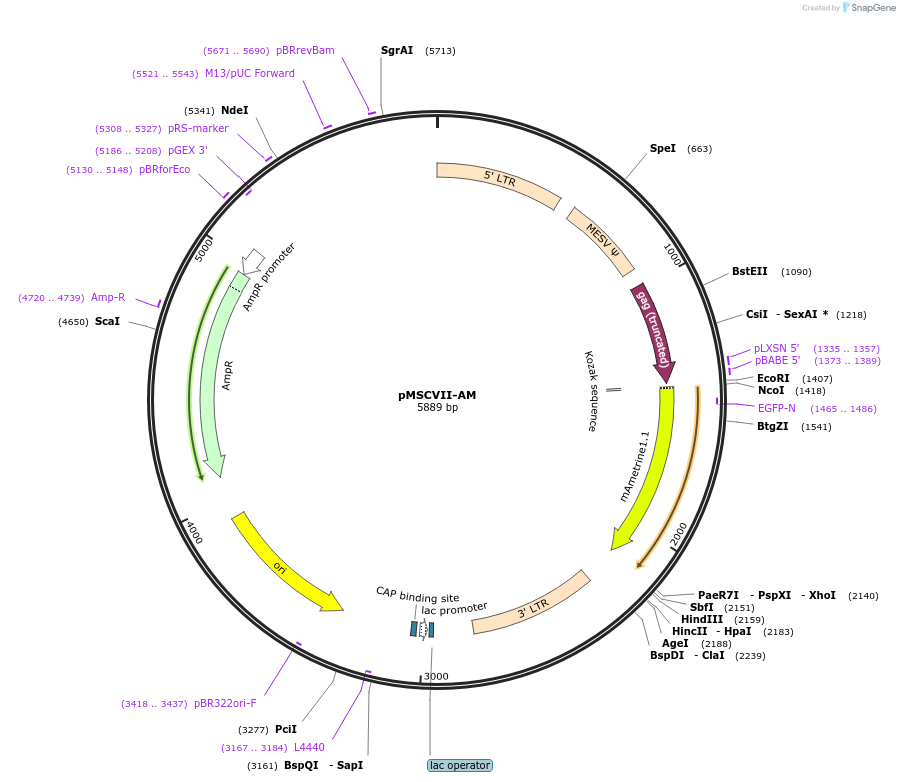
pMSCVII-AM
Plasmid#162751PurposeRetroviral (MSCV) expression of ametrine fluorescent proteinDepositorInsertametrine
UseRetroviralExpressionMammalianAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
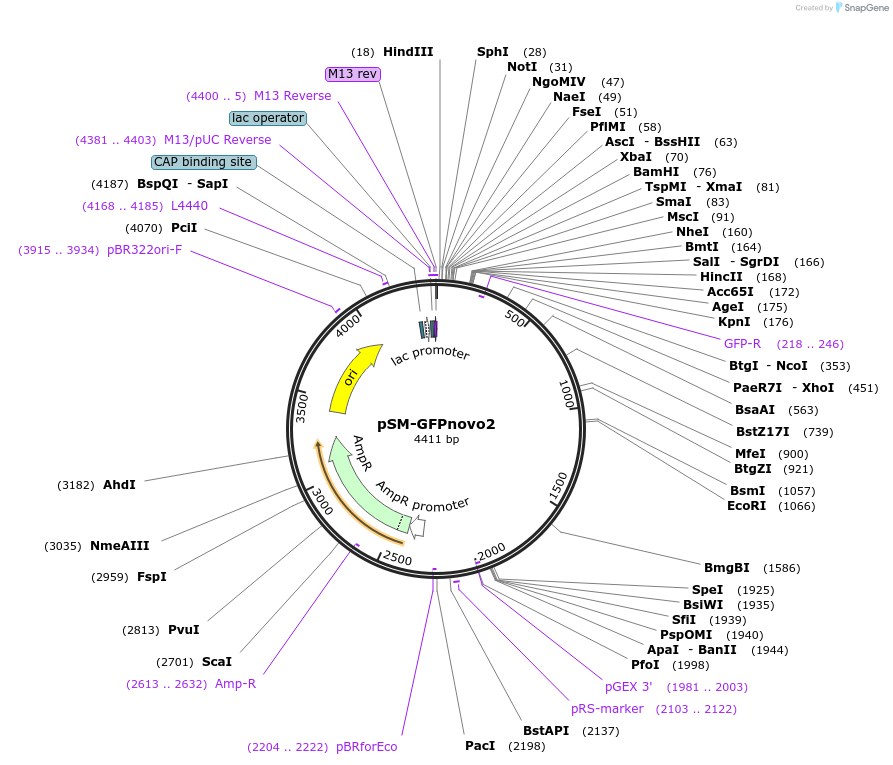
pSM-GFPnovo2
Plasmid#116943PurposeGFPnovo2_unc-54 utr vector for expression in C. elegansDepositorInsertC. elegans codon optimized GFPnovo2
TagsGFPnovo2ExpressionWormMutationY145F, V163A, S202T, L221VPromoternoneAvailable SinceOct. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
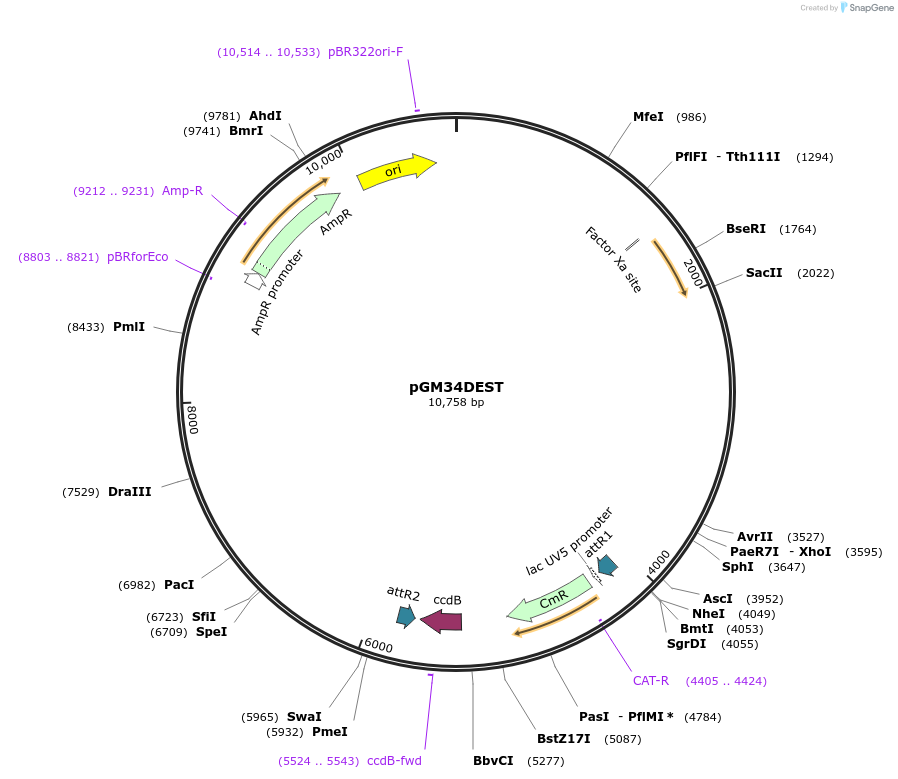
pGM34DEST
Plasmid#74756PurposeEncodes for the 5X repeat of the QUAS response element, followed by the delta pes-10 minimal promoterDepositorInsertGateway cassette
ExpressionWormAvailable SinceOct. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
mPA-GFP-Actin-C-18
Plasmid#57121PurposeLocalization: Actin, Excitation: 400 / 504, Emission: 515 / 517DepositorAvailable SinceJan. 29, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
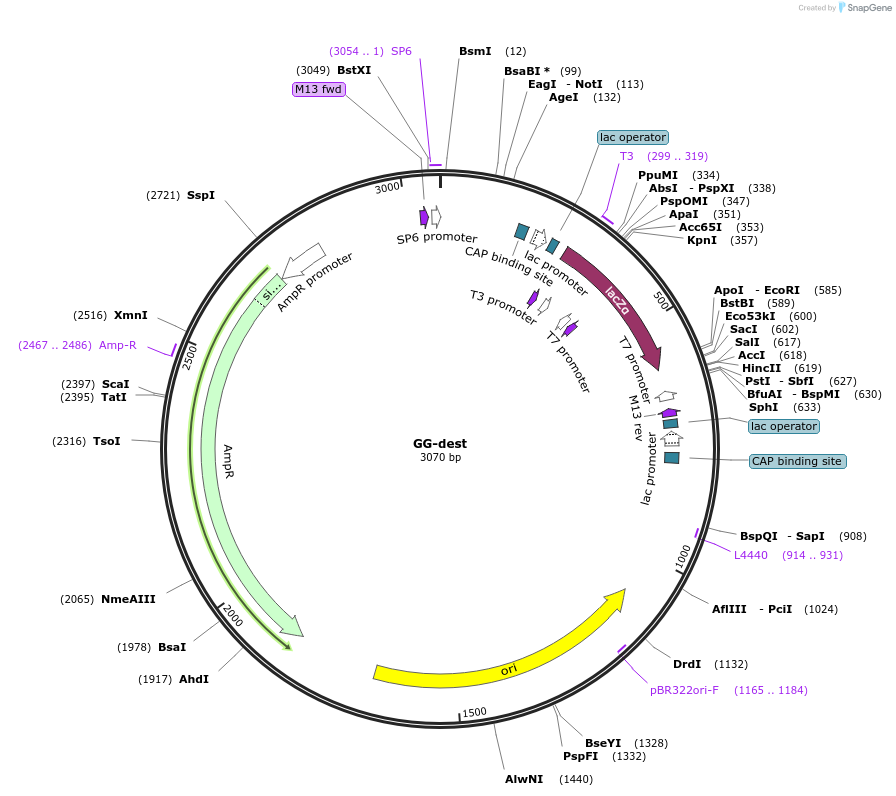
GG-dest
Plasmid#69538PurposeDestination plasmid for gRNA concatenation Golden Gate cloningDepositorTypeEmpty backboneAvailable SinceNov. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
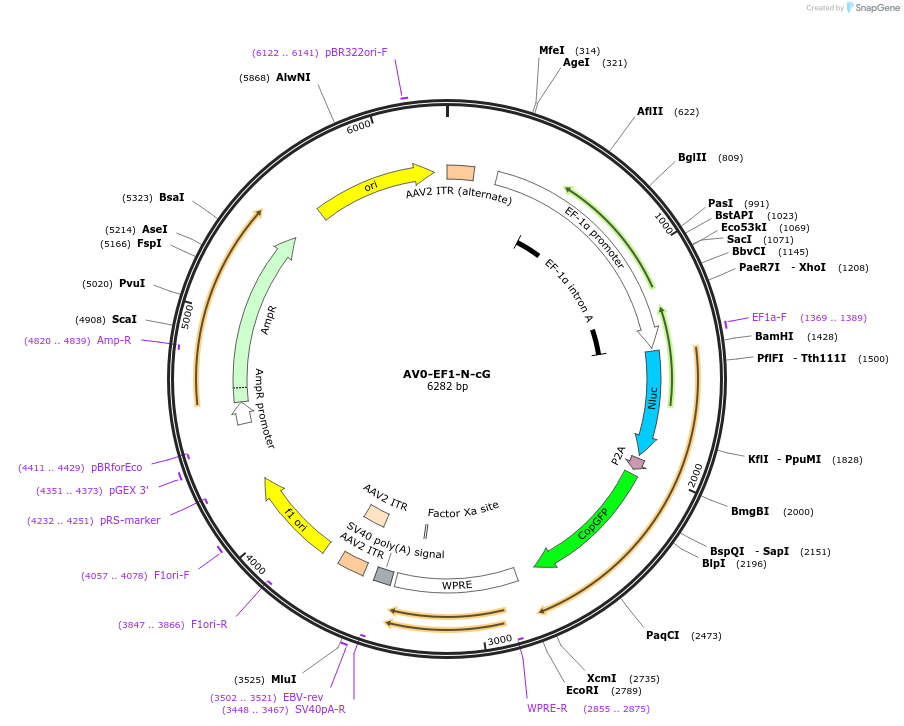
AV0-EF1-N-cG
Plasmid#192888PurposeExpression of Nanoluc-P2A-copGFPDepositorInsertNanoluc-P2A-copGFP
UseAAVAvailable SinceDec. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
tdEos-Integrin-Beta1-N-18
Plasmid#57627PurposeLocalization: Focal Adhesions, Excitation: 505 / 569, Emission: 516 / 581DepositorAvailable SinceFeb. 5, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mPA-GFP-Mito-7
Plasmid#57148PurposeLocalization: Mitochondria, Excitation: 400 / 504, Emission: 515 / 517DepositorAvailable SinceJan. 29, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
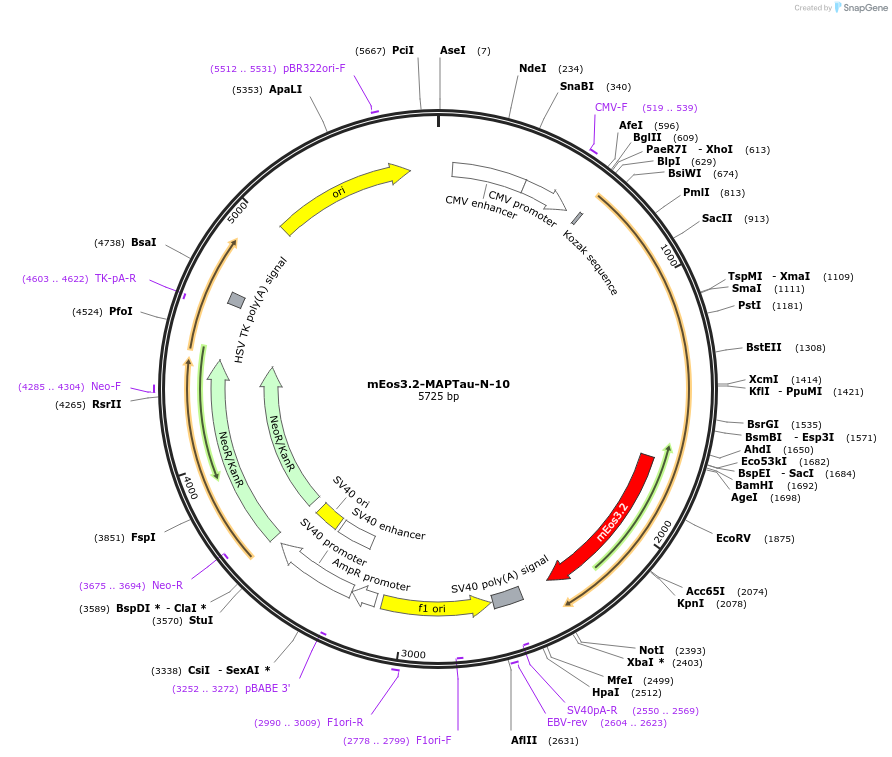
mEos3.2-MAPTau-N-10
Plasmid#57468PurposeLocalization: Microtubules, Excitation: 507 / 572, Emission: 516 / 580DepositorAvailable SinceFeb. 5, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
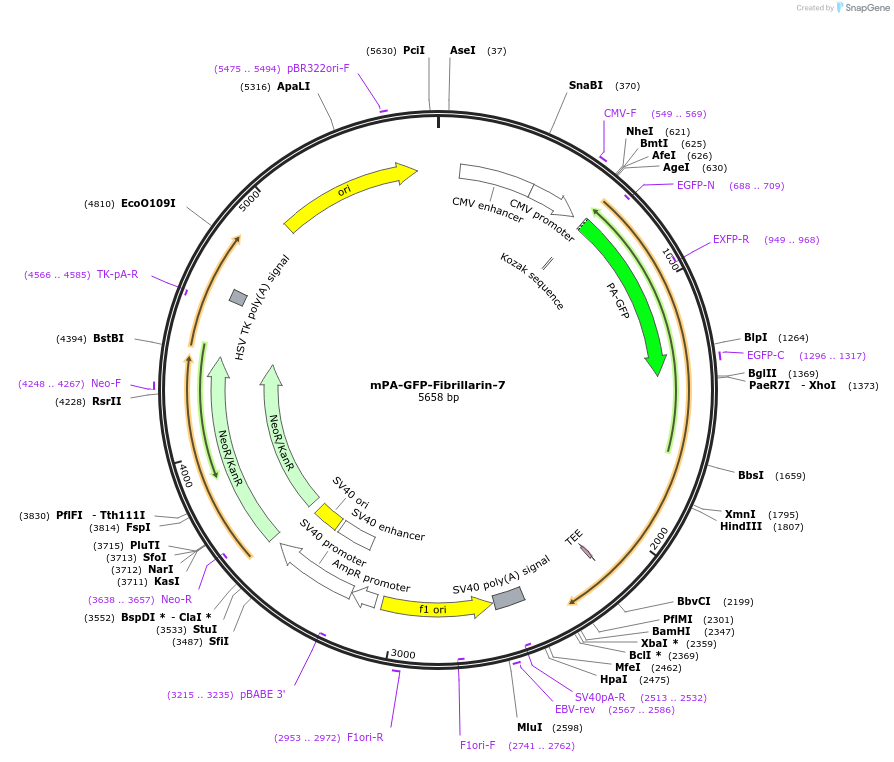
mPA-GFP-Fibrillarin-7
Plasmid#57134PurposeLocalization: Nucleoli, Excitation: 400 / 504, Emission: 515 / 517DepositorAvailable SinceFeb. 5, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
-
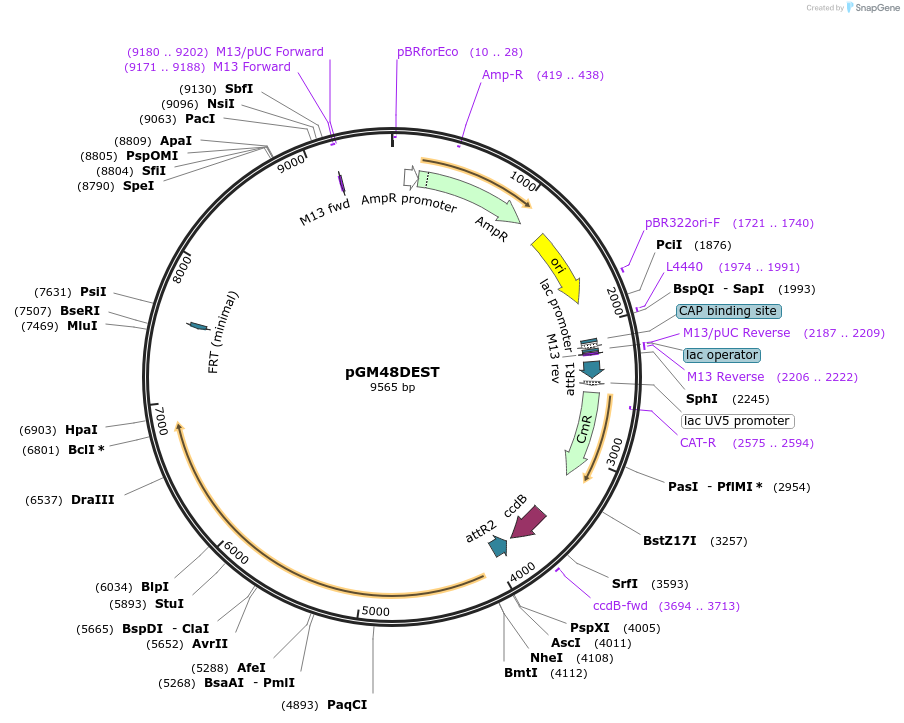
pGM48DEST
Plasmid#74757PurposeEncodes for the QS repressor gene, splice linker, and mCherry reporter geneDepositorInsertsQS
Gateway cassette
ExpressionWormAvailable SinceOct. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
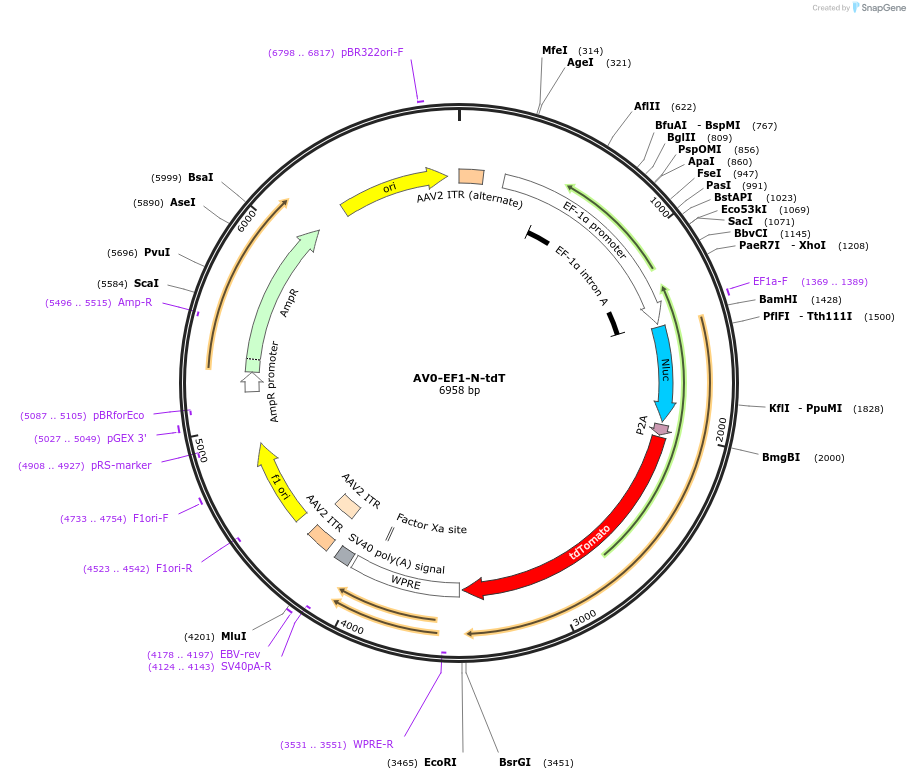
AV0-EF1-N-tdT
Plasmid#192889PurposeExpression of Nanoluc-P2A-tdTomatoDepositorInsertNanoluc-P2A-tdTomato
UseAAVAvailable SinceDec. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
mPA-GFP-EB3-7
Plasmid#57130PurposeLocalization: MT End Binding Protein, Excitation: 400 / 504, Emission: 515 / 517DepositorAvailable SinceApril 1, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -