We narrowed to 13,968 results for: SHI
-
Plasmid#184977PurposeExpress -Eco1 LYP1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 12
ExpressionYeastMutationLYP1 donor E27stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
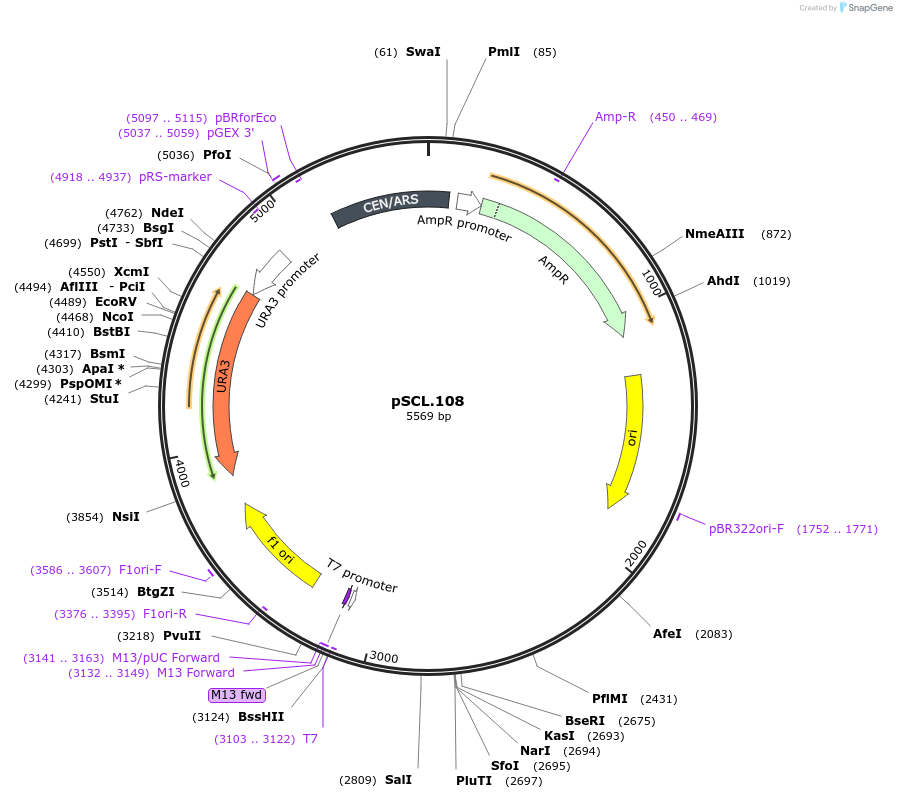
pSCL.109
Plasmid#184978PurposeTest effect of extending a1/a2 on LYP1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 27 v1
ExpressionYeastMutationLYP1 donor E27stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
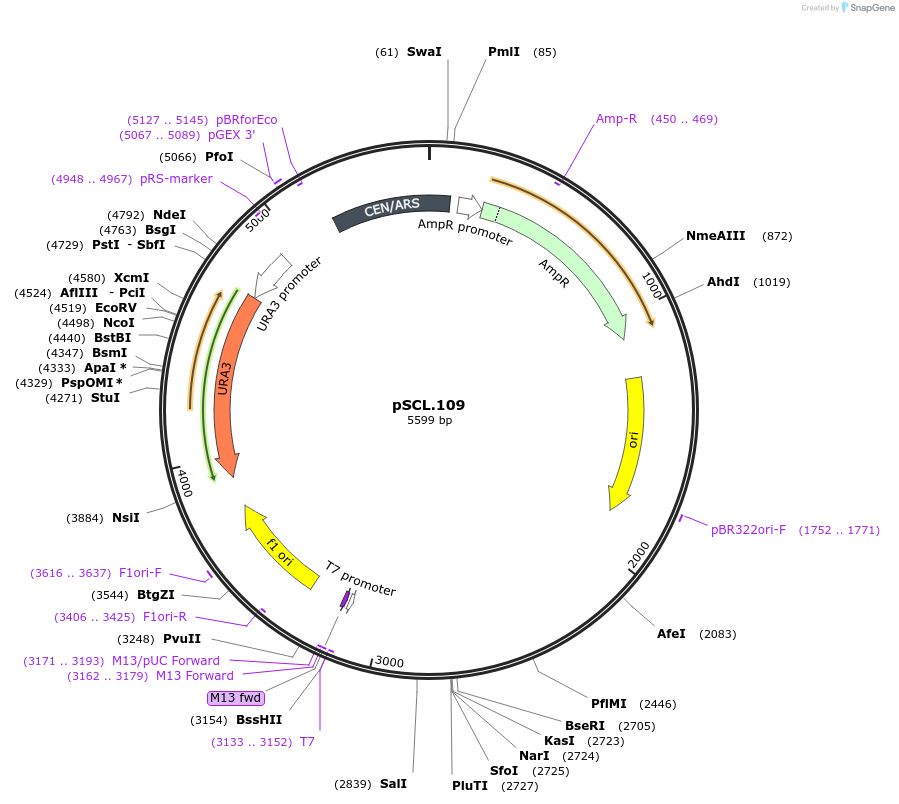
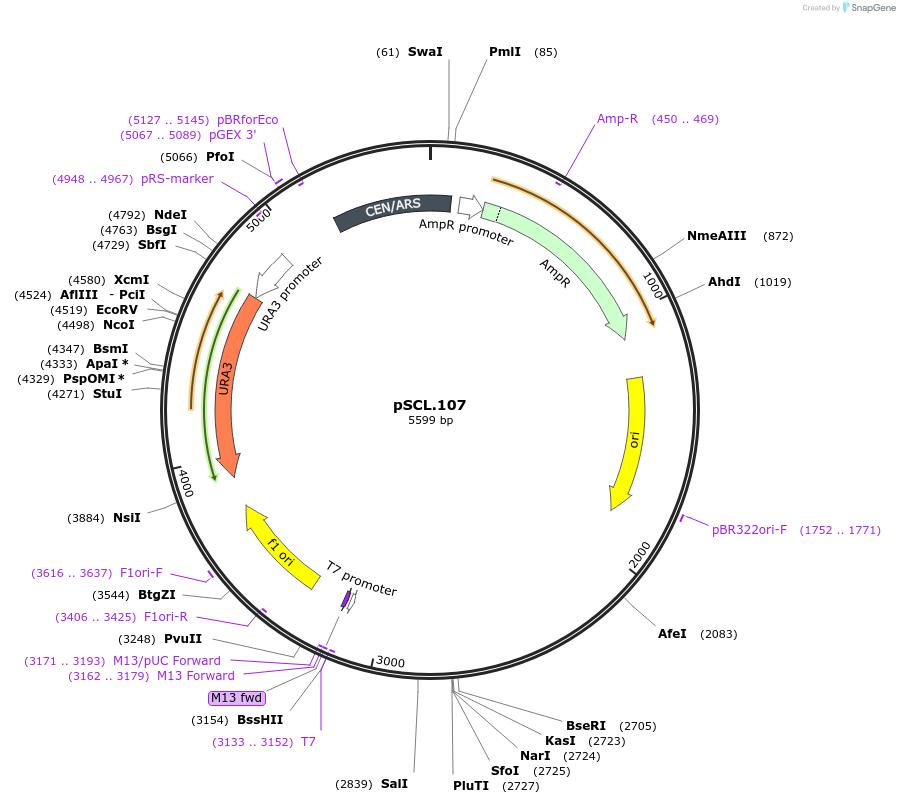
pSCL.107
Plasmid#184976PurposeTest effect of extending a1/a2 on CAN1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 27 v1
ExpressionYeastMutationCAN1 donor G444stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
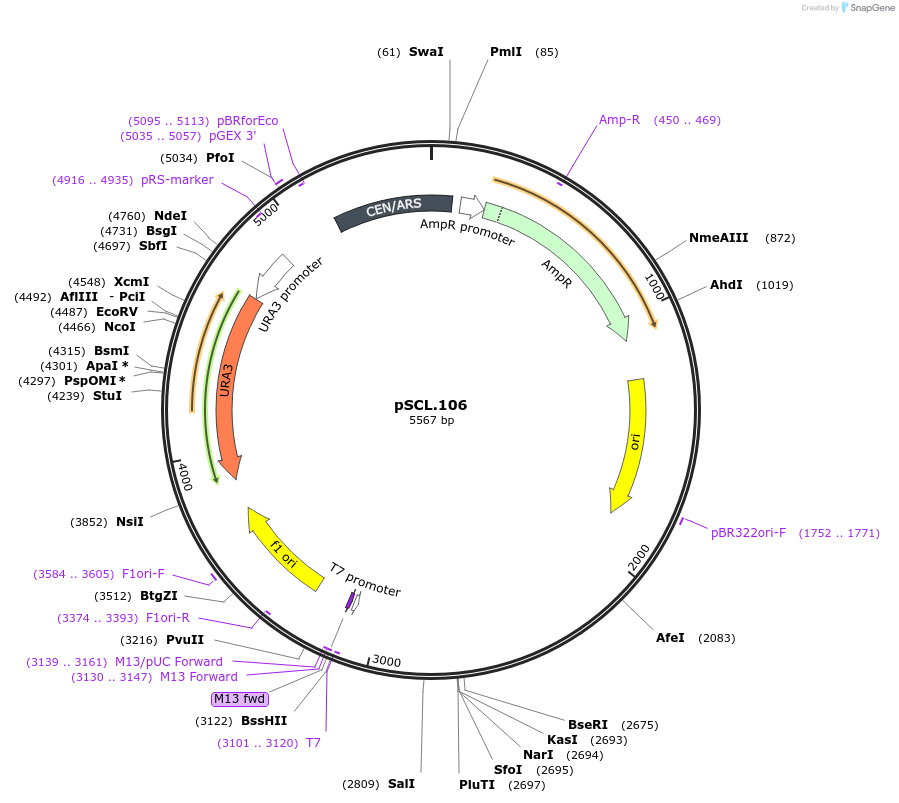
pSCL.106
Plasmid#184975PurposeExpress -Eco1 CAN1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 12
ExpressionYeastMutationCAN1 donor G444stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
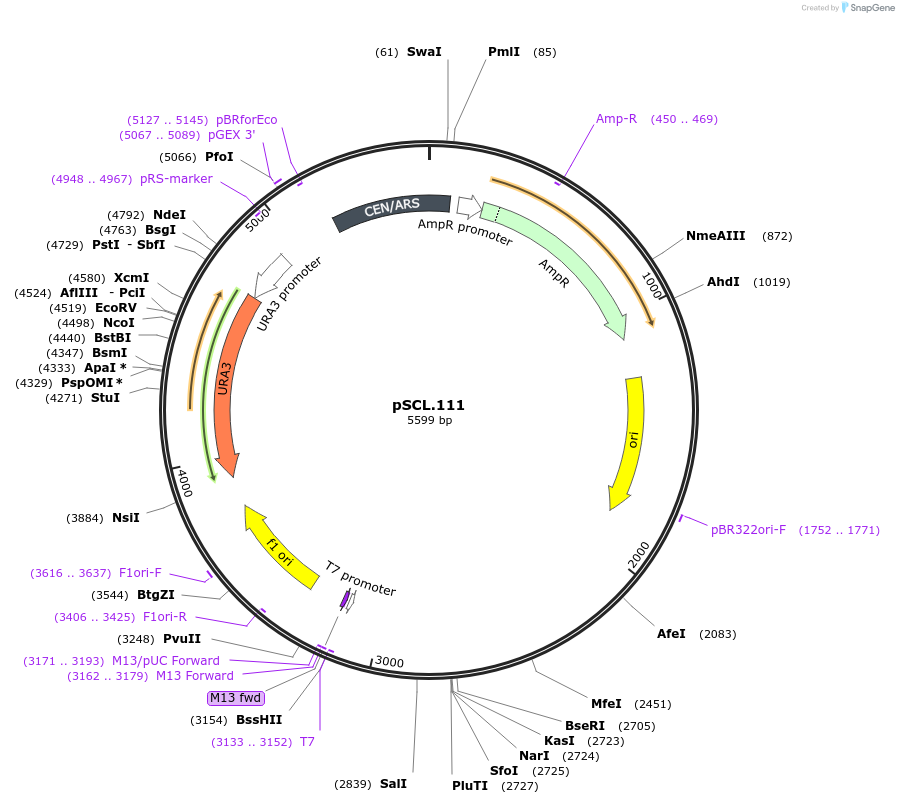
pSCL.111
Plasmid#184980PurposeTest effect of extending a1/a2 on TRP2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 27 v1
ExpressionYeastMutationTRP2 donor E64stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
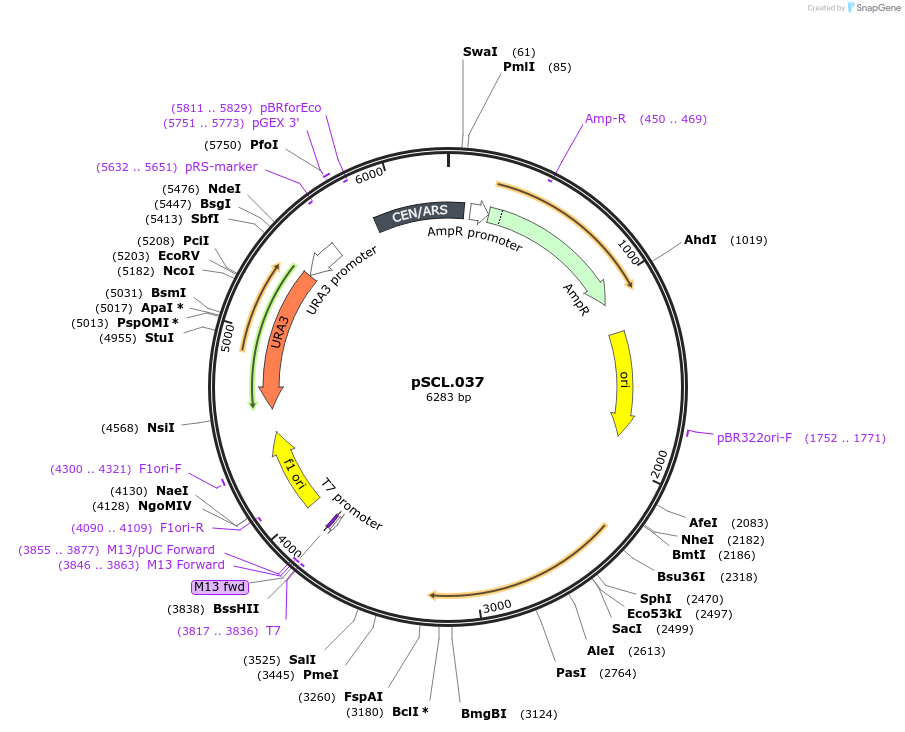
pSCL.037
Plasmid#184966PurposeExpress -Eco1 dead RT and ncRNA in yeasstDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12, dead RT
ExpressionYeastMutationHuman codon optimized RT, YXDD catalytic region m…PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
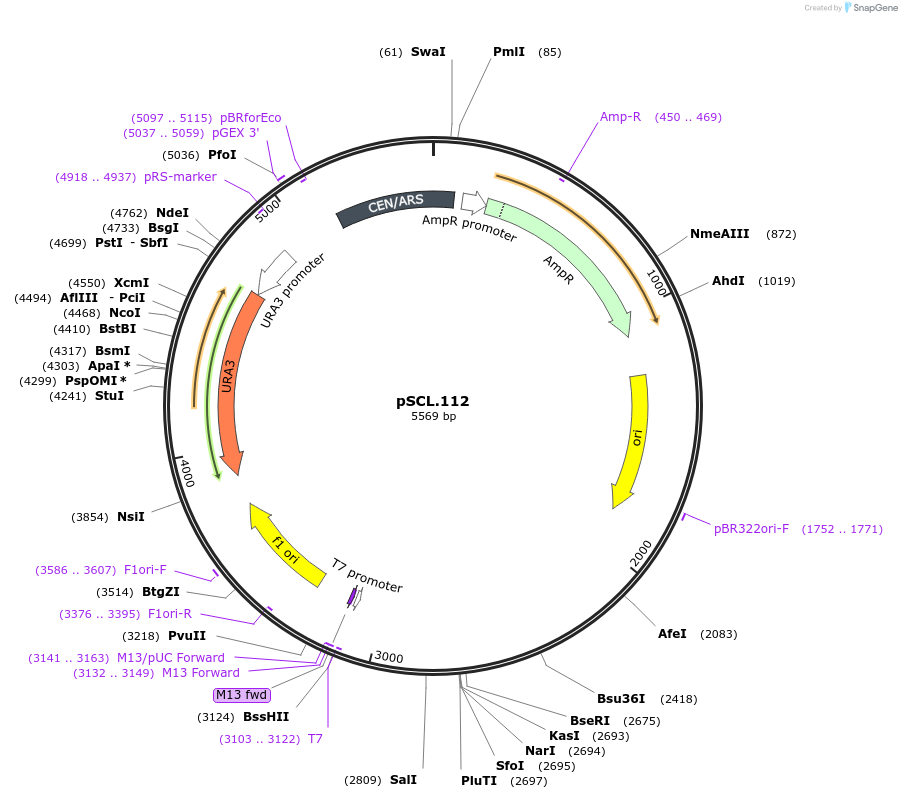
pSCL.112
Plasmid#184981PurposeExpress -Eco1 FAA1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 12
ExpressionYeastMutationFAA1 donor P233stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
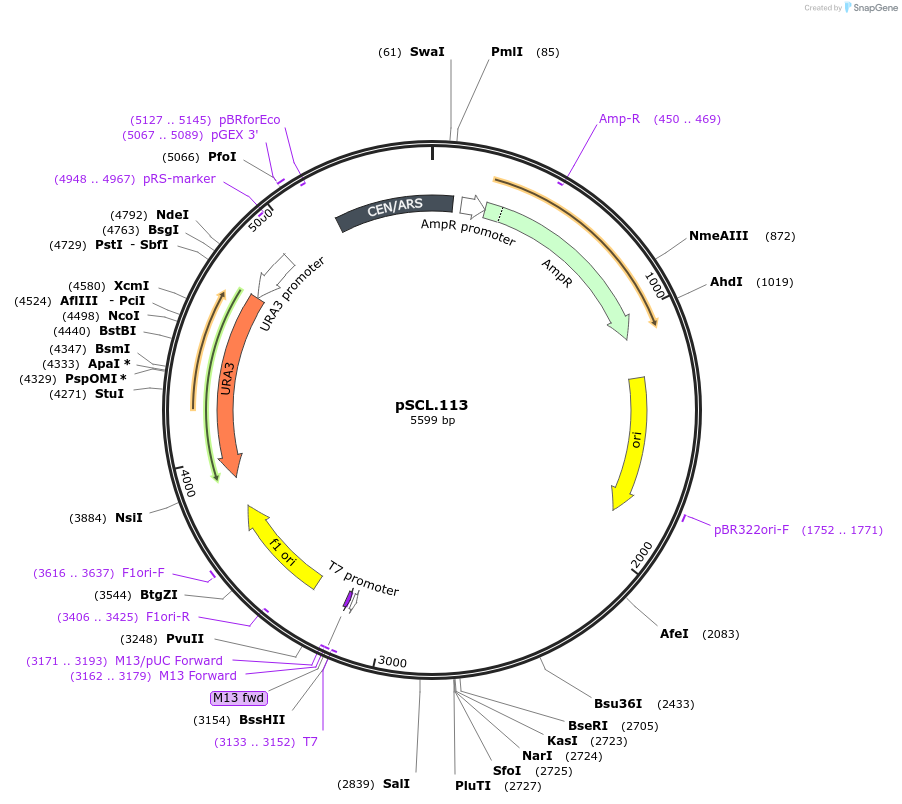
pSCL.113
Plasmid#184982PurposeTest effect of extending a1/a2 on FAA1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 27 v1
ExpressionYeastMutationFAA1 donor P233stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
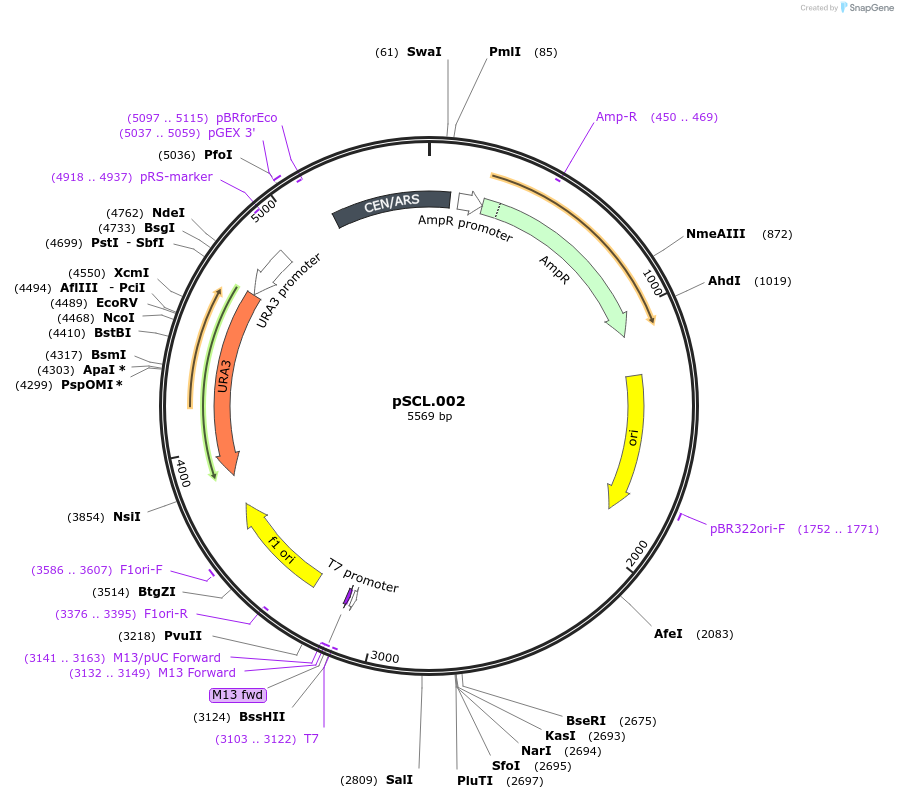
pSCL.002
Plasmid#184972PurposeExpress -Eco1 ADE2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 12
ExpressionYeastMutationADE2 donor P272stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
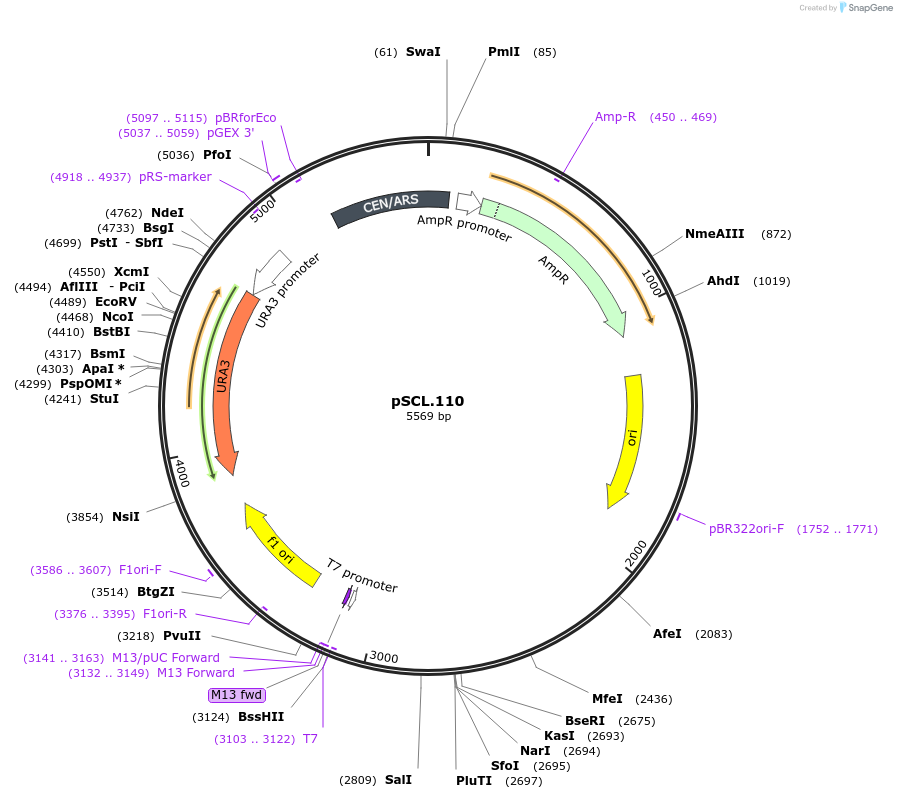
pSCL.110
Plasmid#184979PurposeExpress -Eco1 TRP2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 12
ExpressionYeastMutationTRP2 donor E64stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
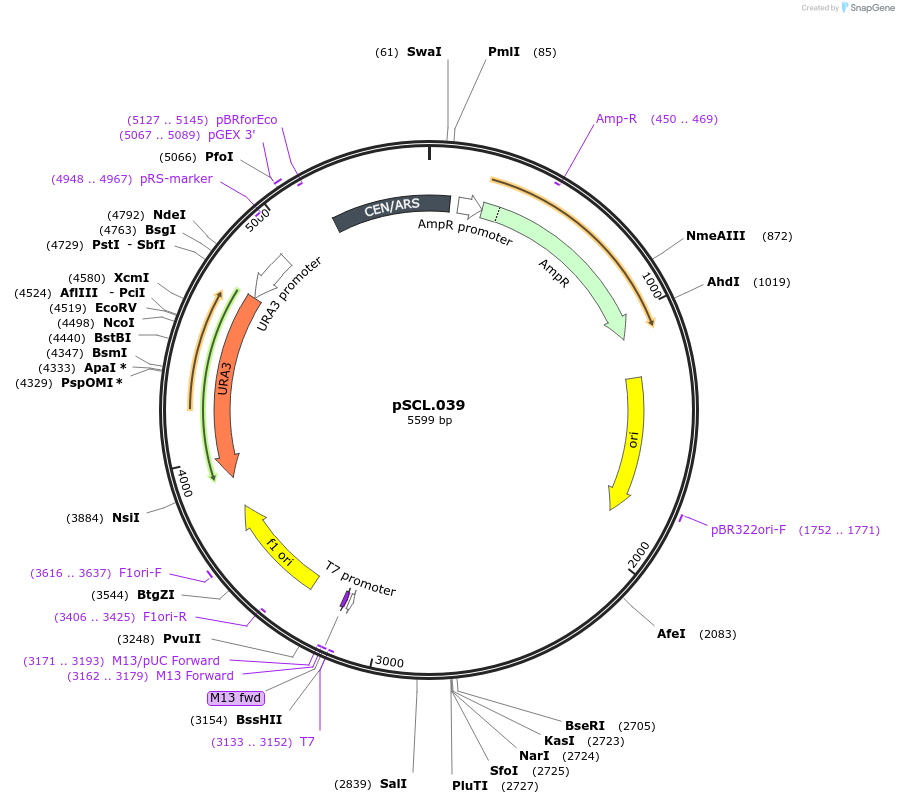
pSCL.039
Plasmid#184973PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v1
ExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
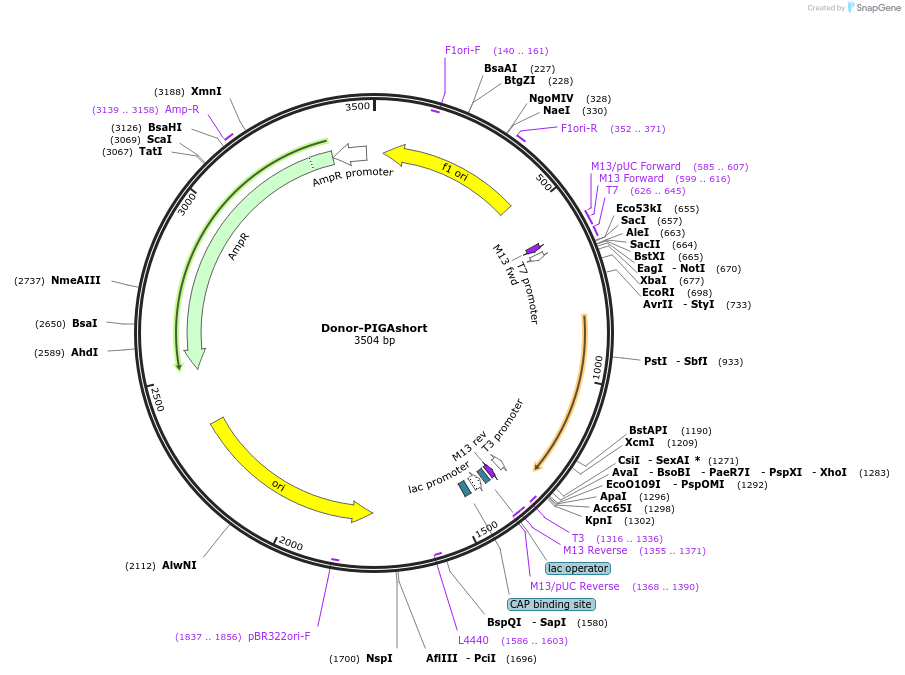
Donor-PIGAshort
Plasmid#153547PurposeUsed as a donor vector for the targeted correction of a mutation in PIGA exon 6DepositorInsertPIGA (a 603-bp fragment from intron 5 and exon 6)
UseDonor plasmidTagsN/AMutationNo mutationPromoterNo promoterAvailable SinceDec. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
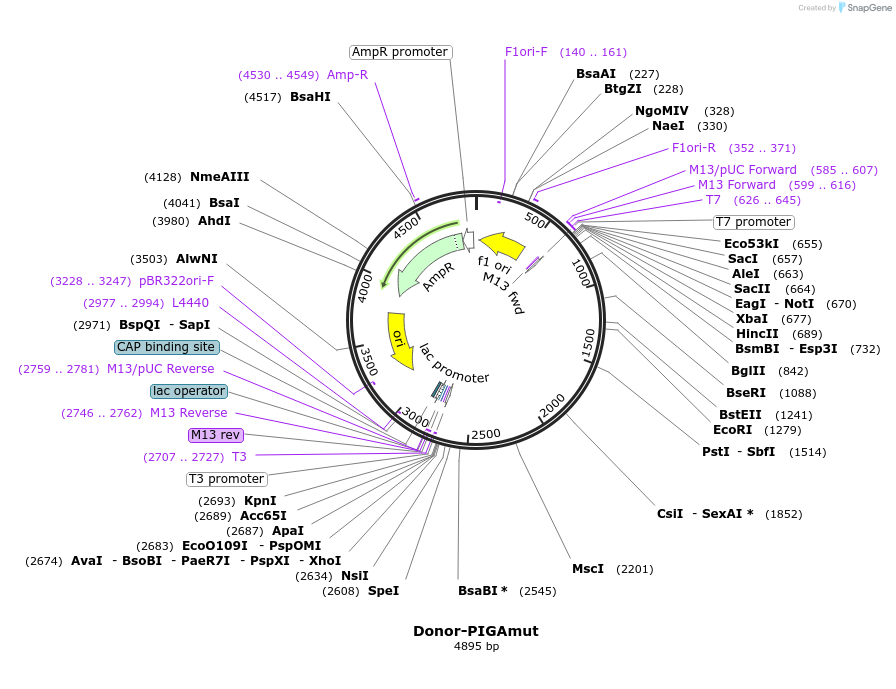
Donor-PIGAmut
Plasmid#153548PurposeUsed as a donor vector for the targeted knock-in of a mutation in PIGA exon 6DepositorInsertmutant PIGA (a 1,991-bp fragment from intron 5 and exon 6 with a 3-bp mutation in exon 6)
UseDonor plasmidTagsN/AMutationa 3-bp mutation in exon 6PromoterNo promoterAvailable SinceDec. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
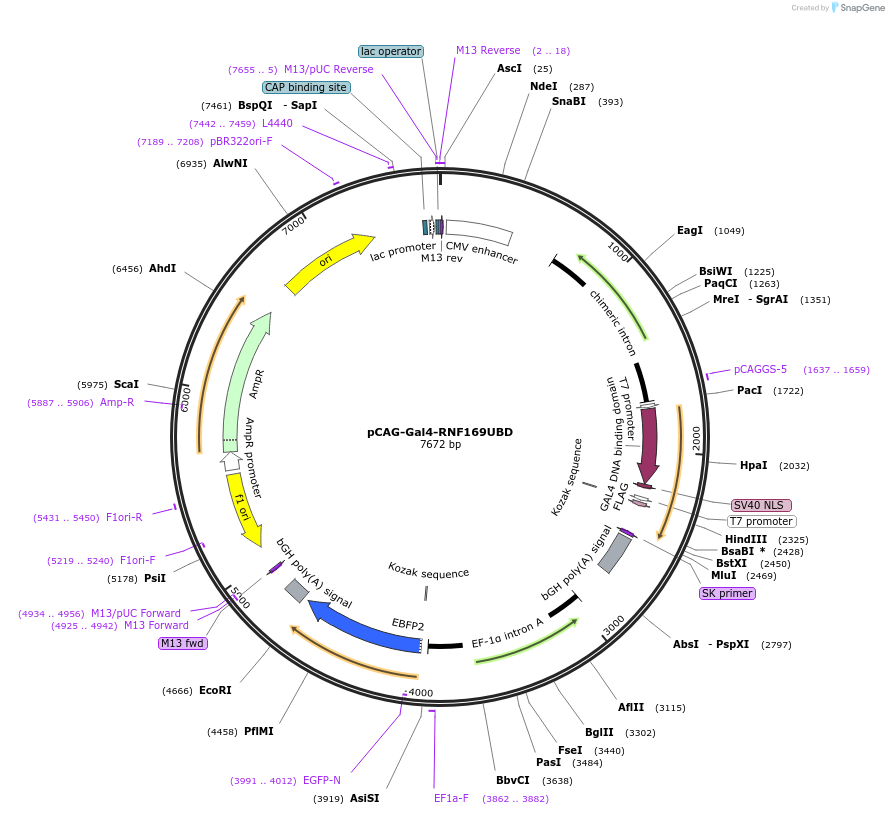
pCAG-Gal4-RNF169UBD
Plasmid#119013PurposeMammalian expression of fusion protein of yeast Gal4 with human RNF169 Ubiquitin binding domain and BFP expressionDepositorAvailable SinceOct. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
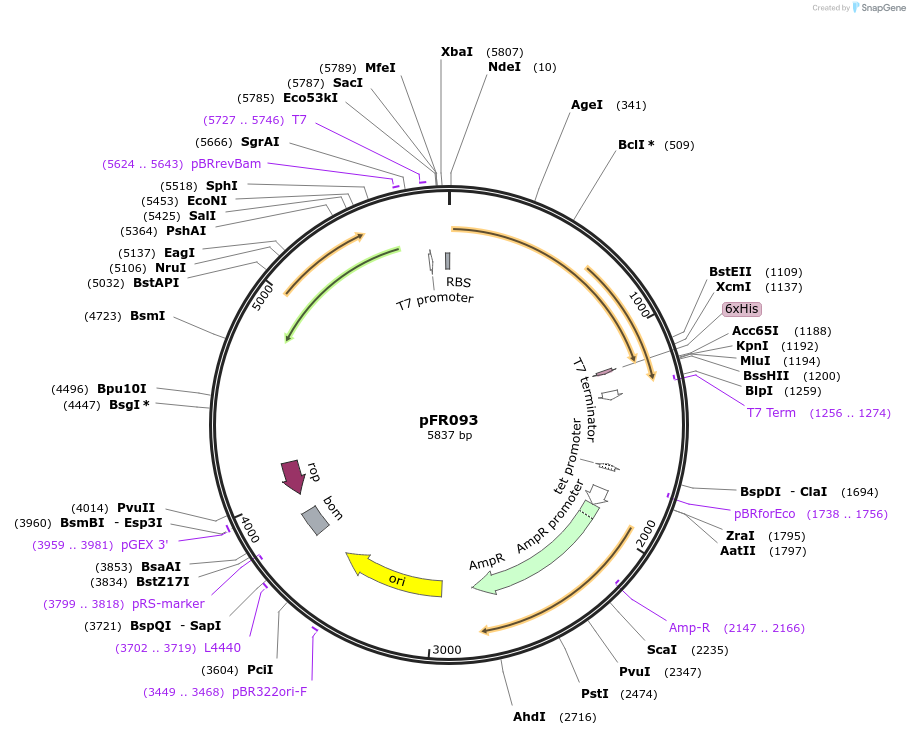
pFR093
Plasmid#134149PurposeExpression of CENP-TDepositorAvailable SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
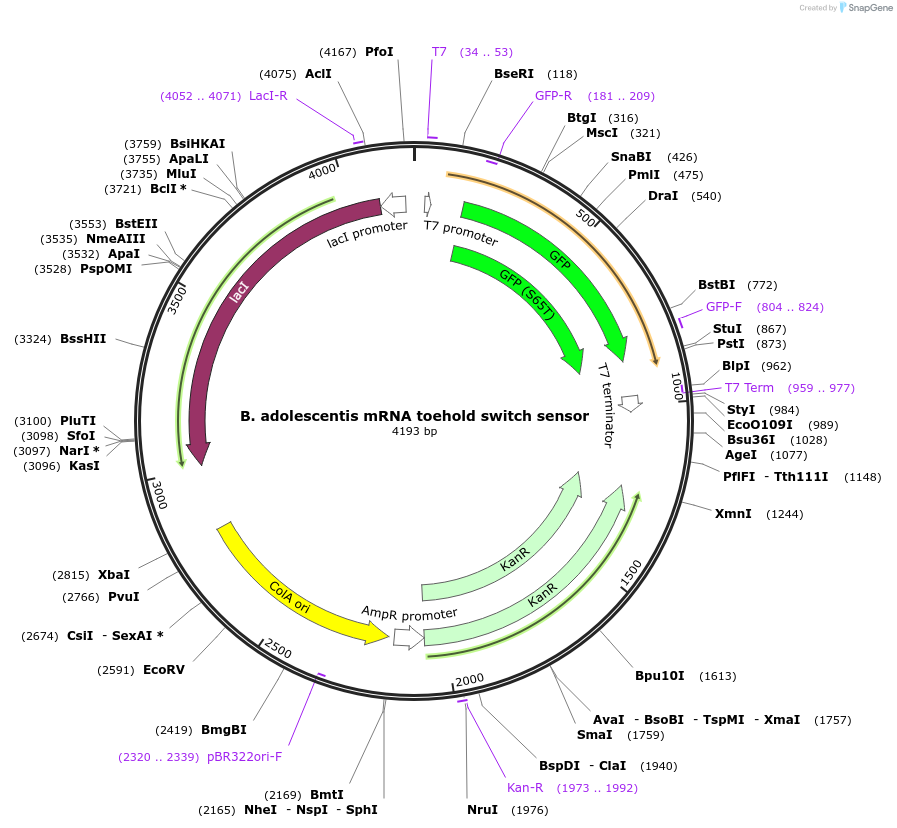
B. adolescentis mRNA toehold switch sensor
Plasmid#110710PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertB. adolescentis species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
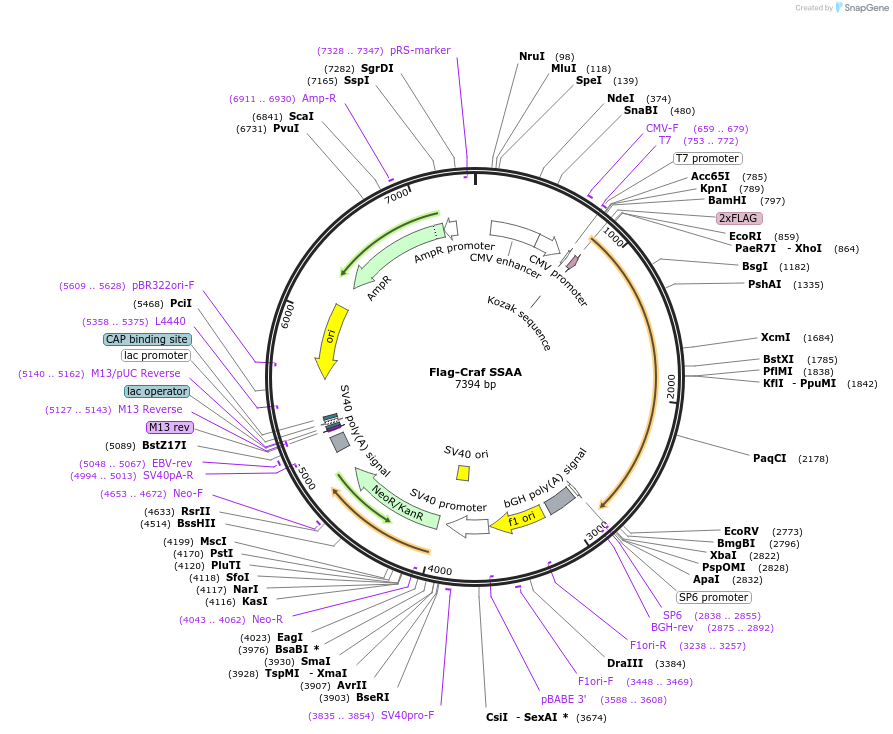
Flag-Craf SSAA
Plasmid#118336PurposeExpression of human CrafDepositorAvailable SinceNov. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
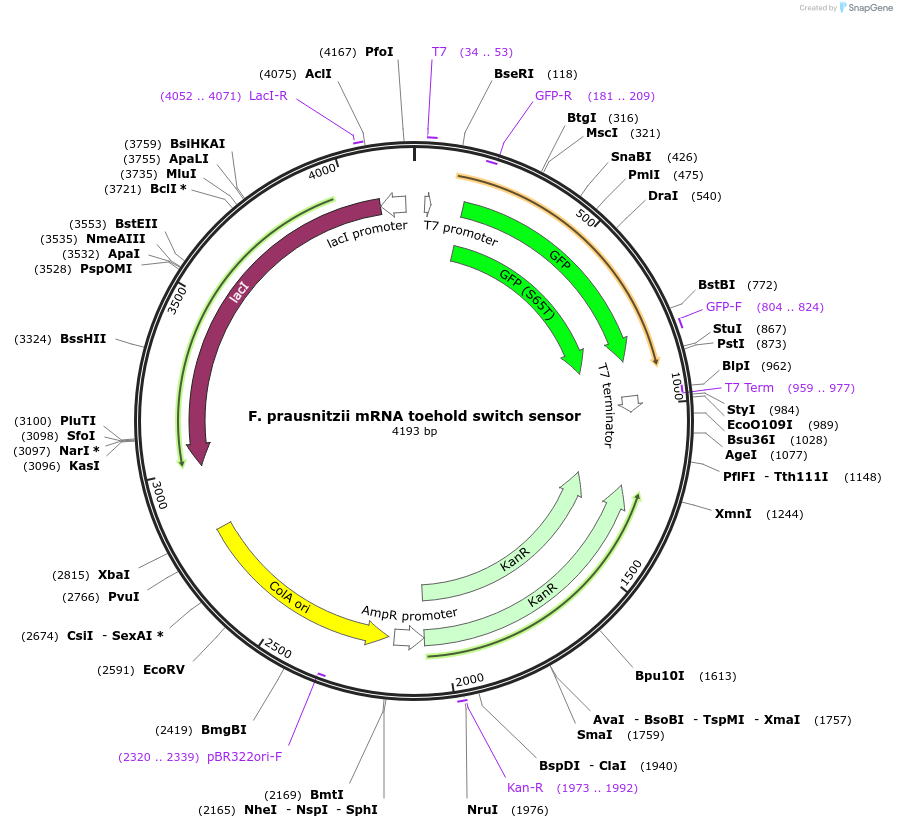
F. prausnitzii mRNA toehold switch sensor
Plasmid#110716PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertF. prausnitzii species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
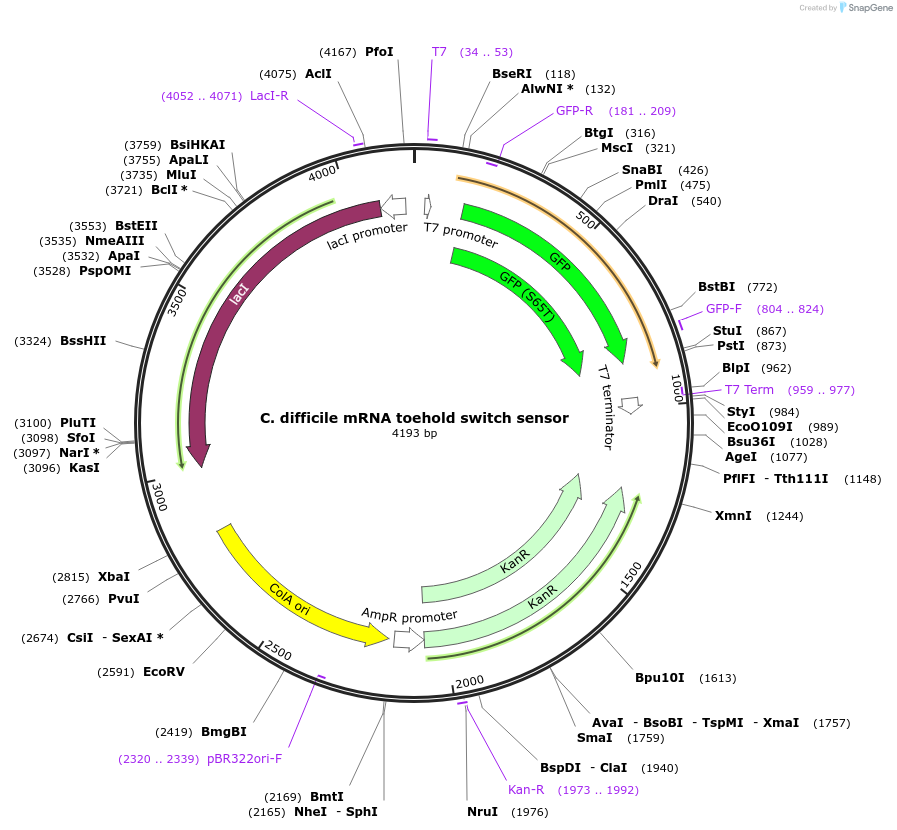
C. difficile mRNA toehold switch sensor
Plasmid#110714PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertC. difficile species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
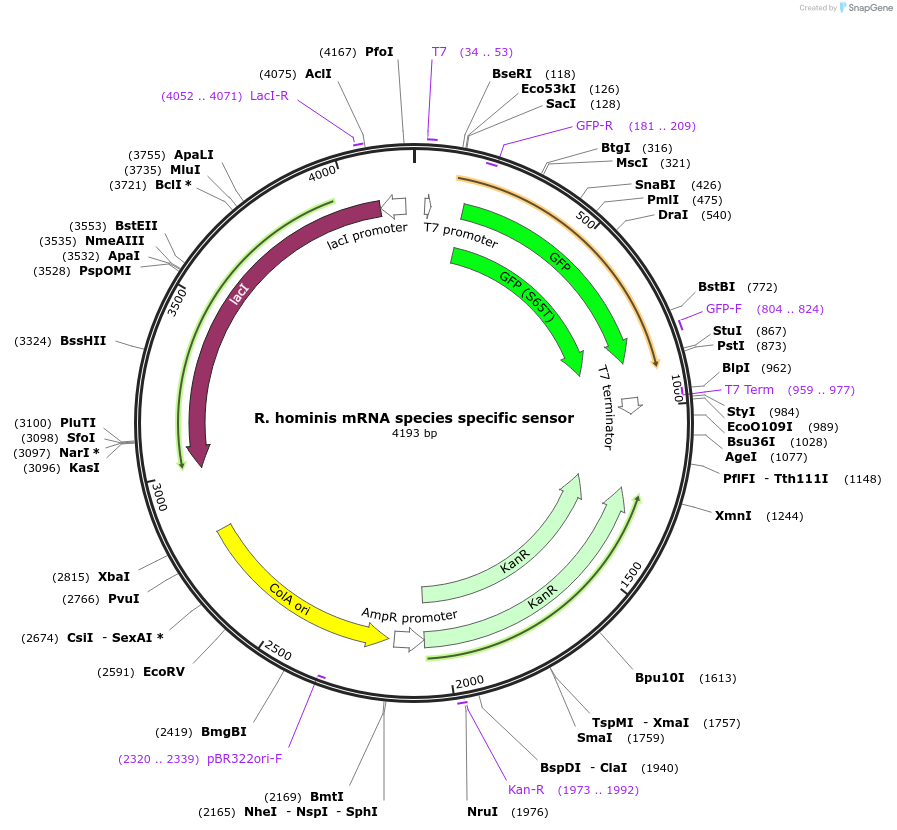
R. hominis mRNA species specific sensor
Plasmid#110713PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertR. hominis species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
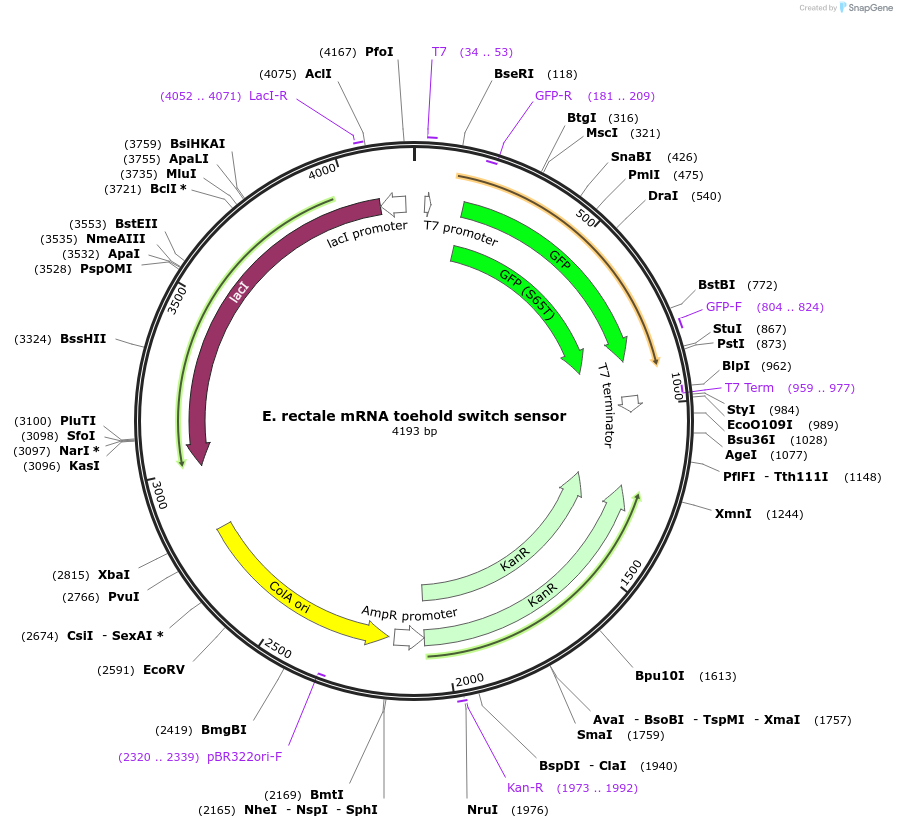
E. rectale mRNA toehold switch sensor
Plasmid#110712PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertE. rectale species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
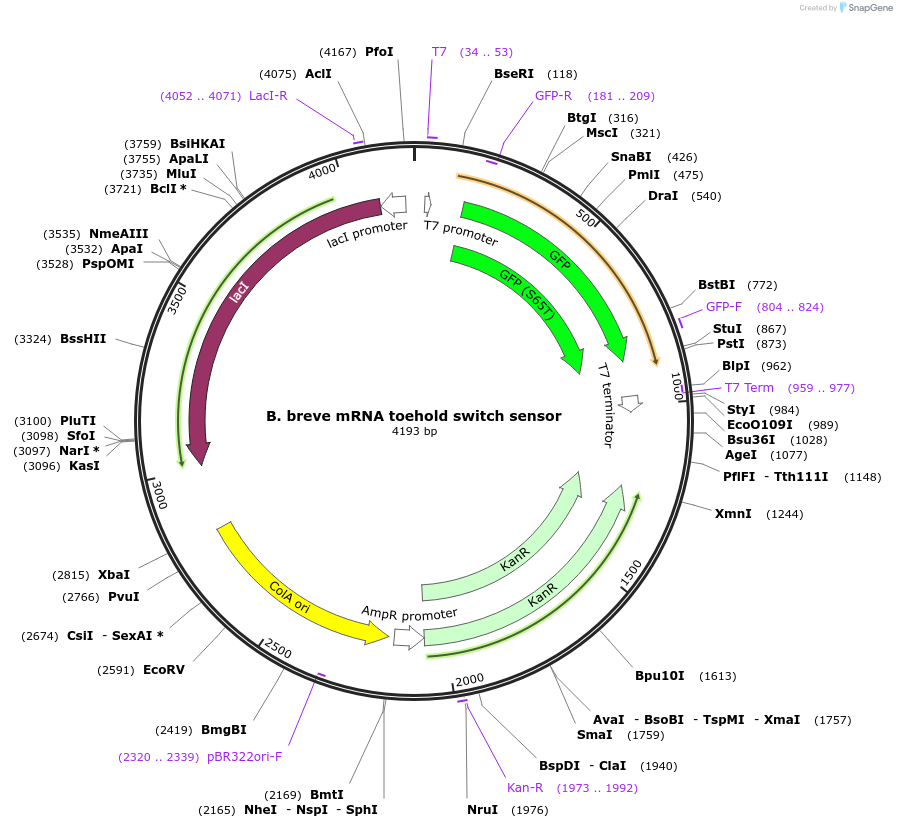
B. breve mRNA toehold switch sensor
Plasmid#110711PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertB. breve species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
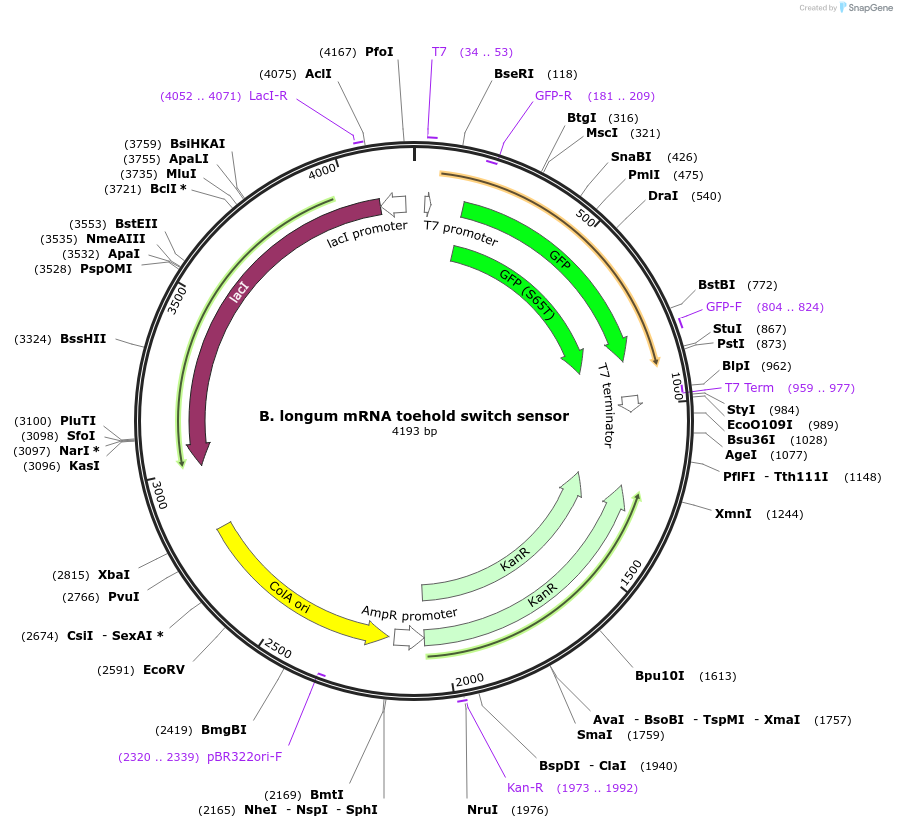
B. longum mRNA toehold switch sensor
Plasmid#110709PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertB. longum species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
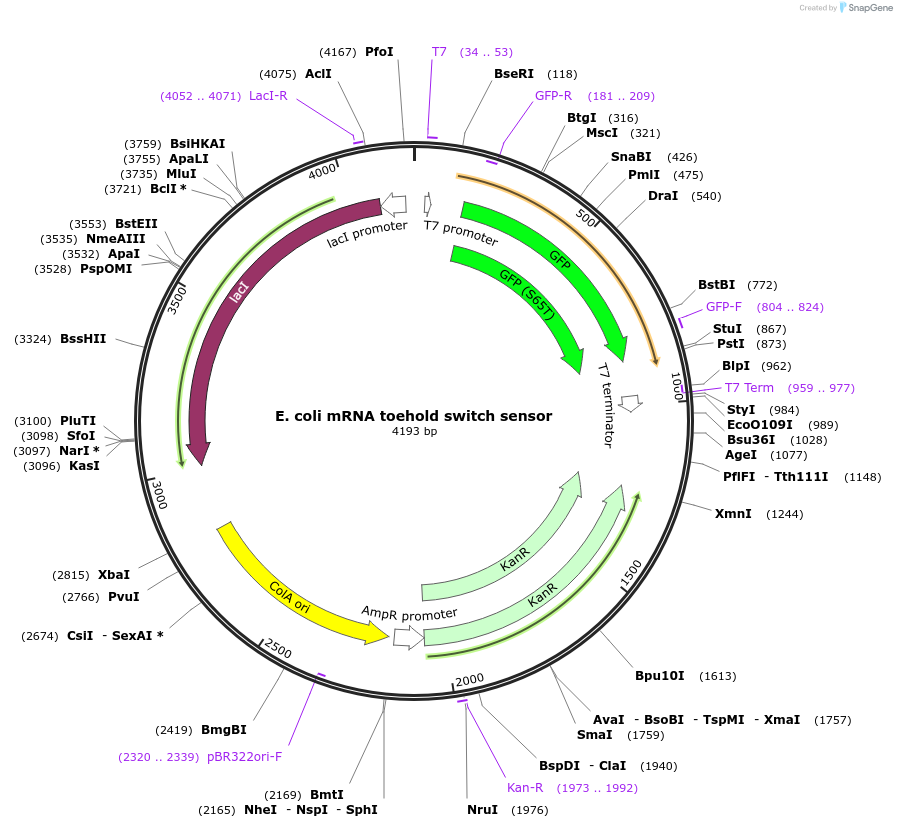
E. coli mRNA toehold switch sensor
Plasmid#110708PurposeToehold switch sensor to detect a species specific mRNA with GFP outputDepositorInsertE. coli species specific toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
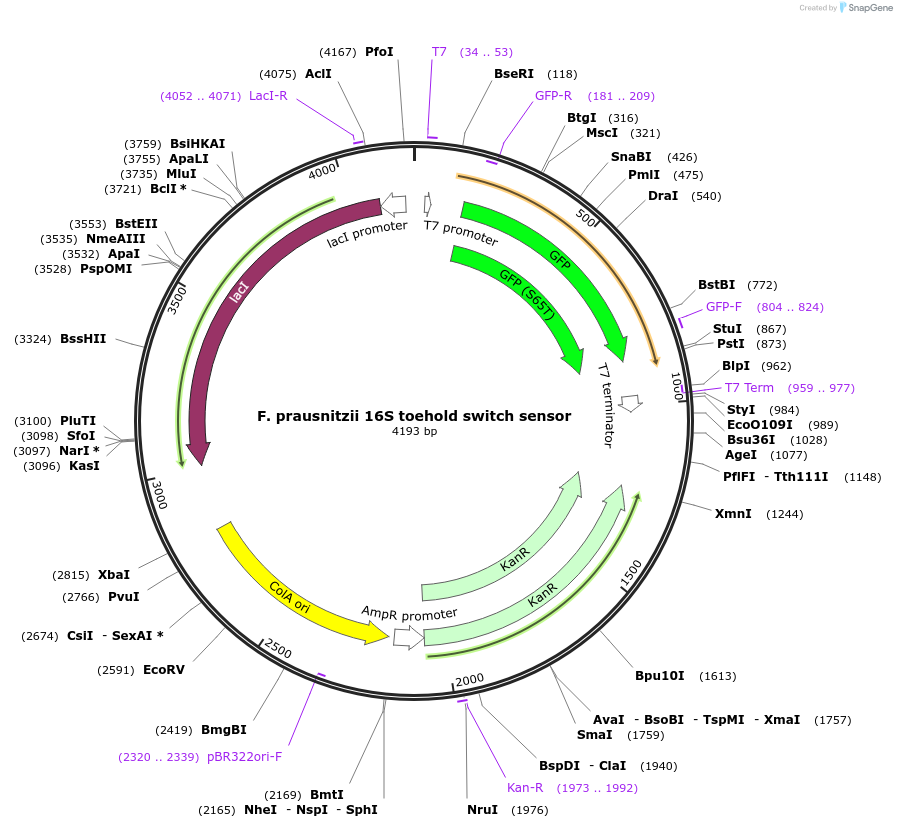
F. prausnitzii 16S toehold switch sensor
Plasmid#110705PurposeToehold switch sensor to detect the V3 hypervariable region of the 16S rRNA with GFP outputDepositorInsertF. prausnitzii 16S toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
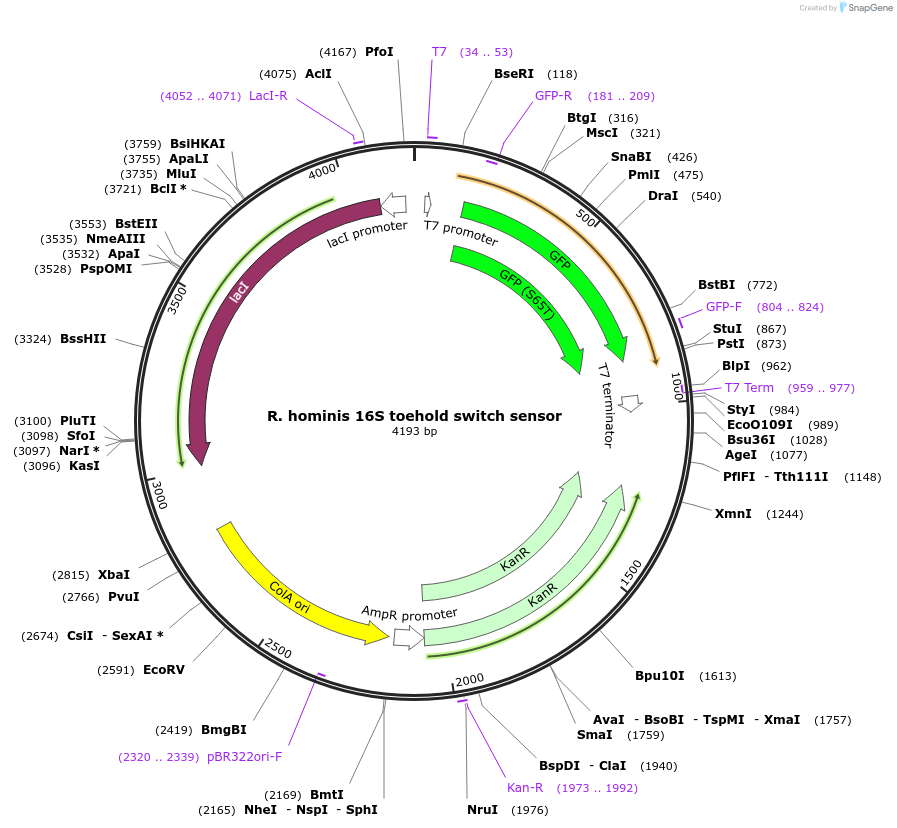
R. hominis 16S toehold switch sensor
Plasmid#110703PurposeToehold switch sensor to detect the V3 hypervariable region of the 16S rRNA with GFP outputDepositorInsertR. hominis 16S toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
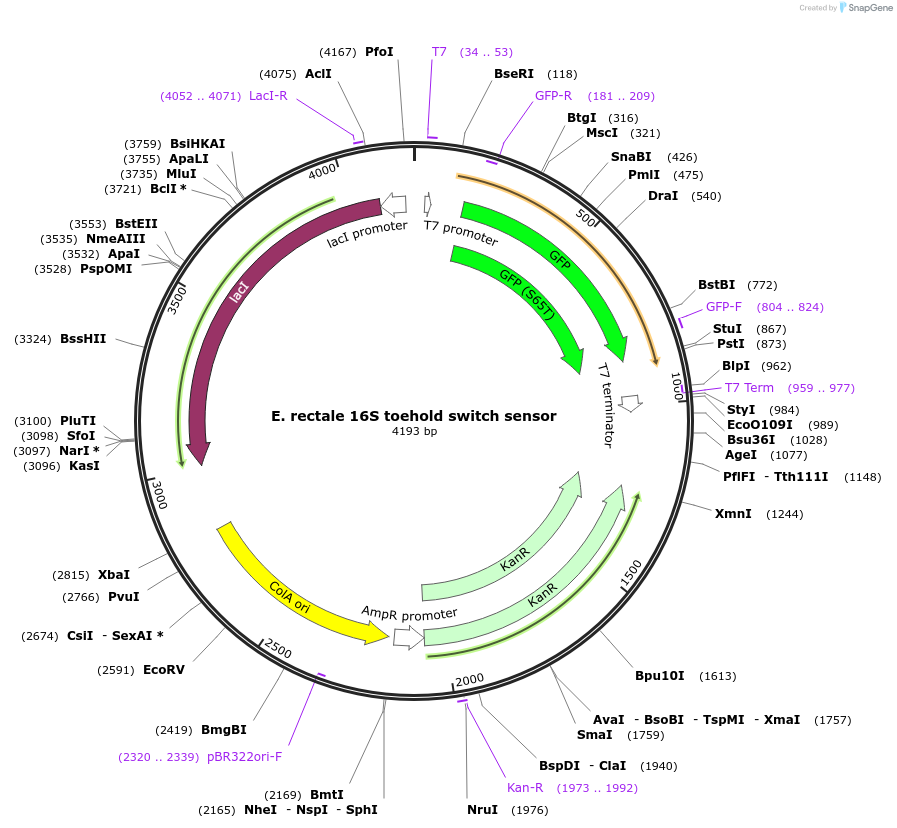
E. rectale 16S toehold switch sensor
Plasmid#110702PurposeToehold switch sensor to detect the V3 hypervariable region of the 16S rRNA with GFP outputDepositorInsertE. rectale 16S toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
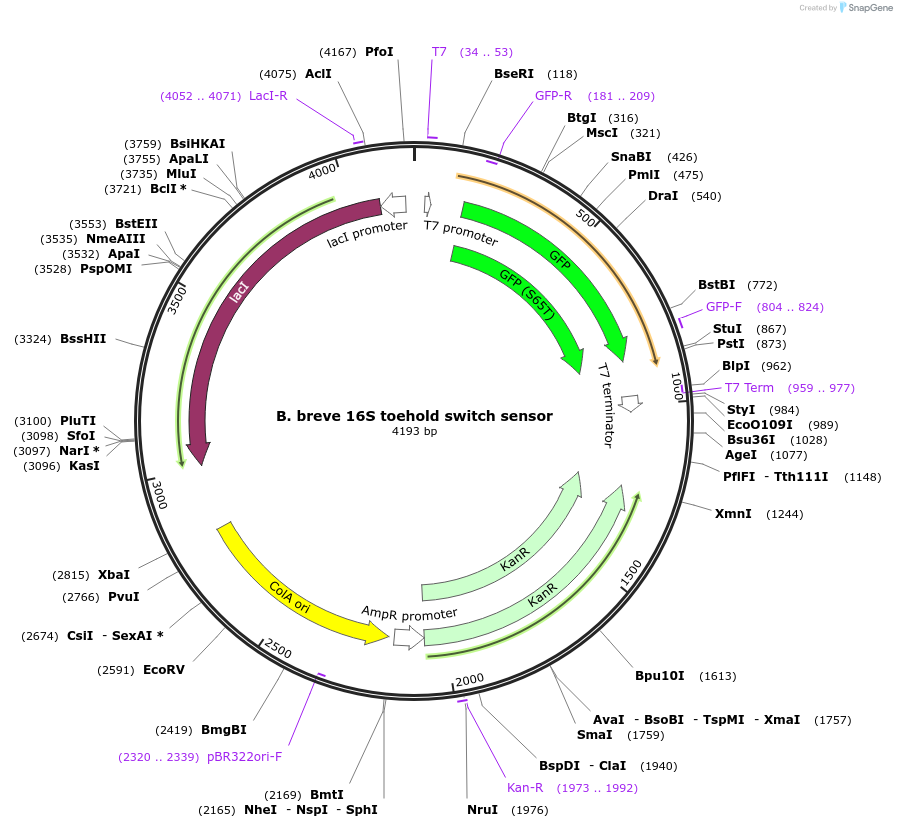
B. breve 16S toehold switch sensor
Plasmid#110701PurposeToehold switch sensor to detect the V3 hypervariable region of the 16S rRNA with GFP outputDepositorInsertB. breve 16S toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
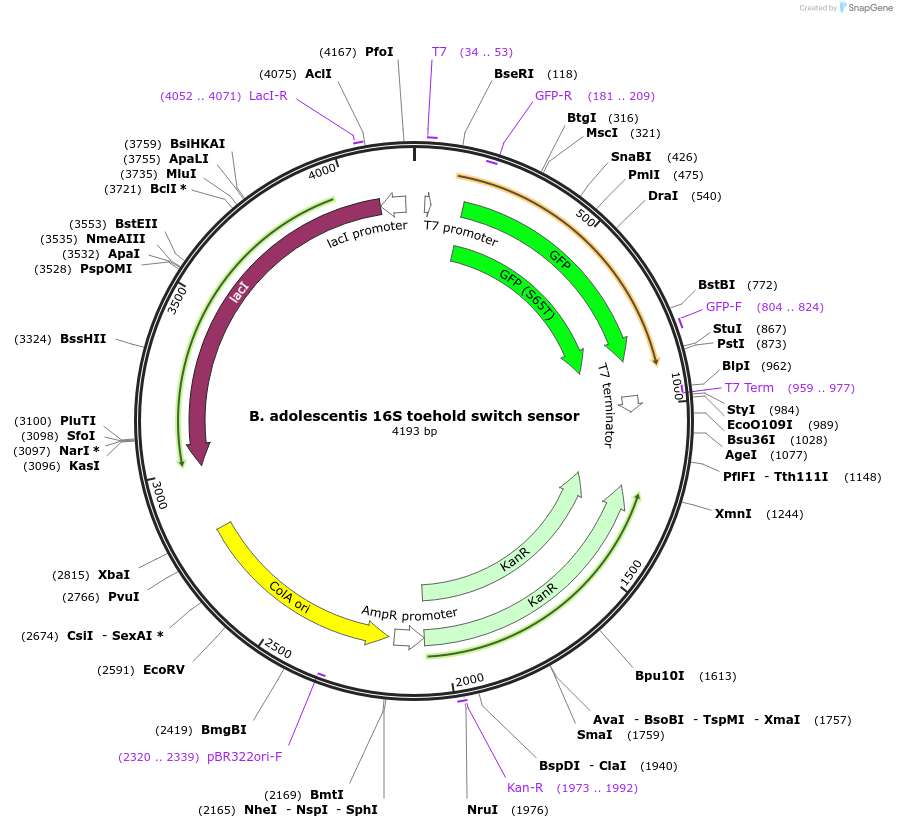
B. adolescentis 16S toehold switch sensor
Plasmid#110700PurposeToehold switch sensor to detect the V3 hypervariable region of the 16S rRNA with GFP outputDepositorInsertB. adolescentis 16S toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
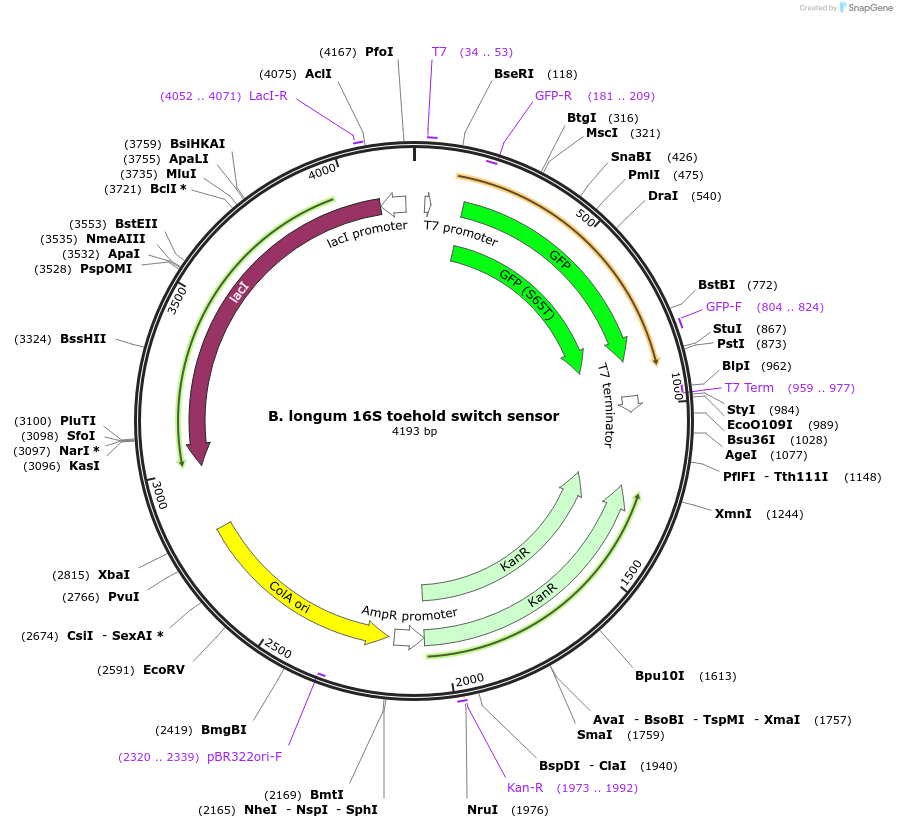
B. longum 16S toehold switch sensor
Plasmid#110699PurposeToehold switch sensor to detect the V3 hypervariable region of the 16S rRNA with GFP outputDepositorInsertB. longum 16S toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
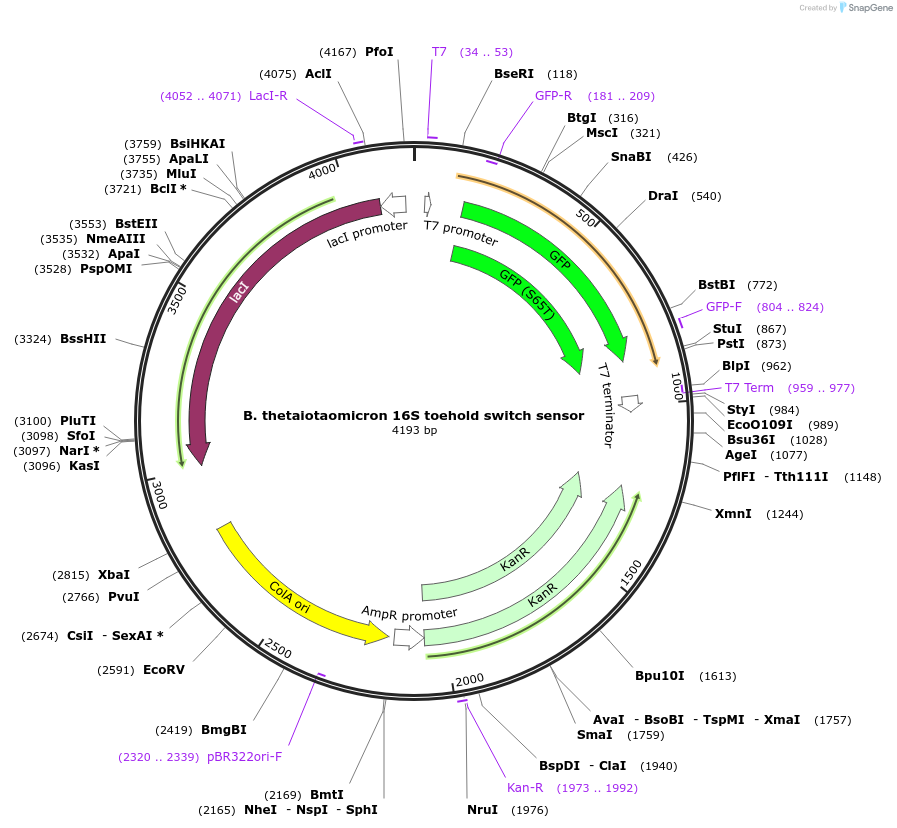
B. thetaiotaomicron 16S toehold switch sensor
Plasmid#110697PurposeToehold switch sensor to detect the V3 hypervariable region of the 16S rRNA with GFP outputDepositorInsertB. thetaiotaomicron 16S toehold switch sensor
UseSynthetic BiologyPromoterT7Available SinceSept. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
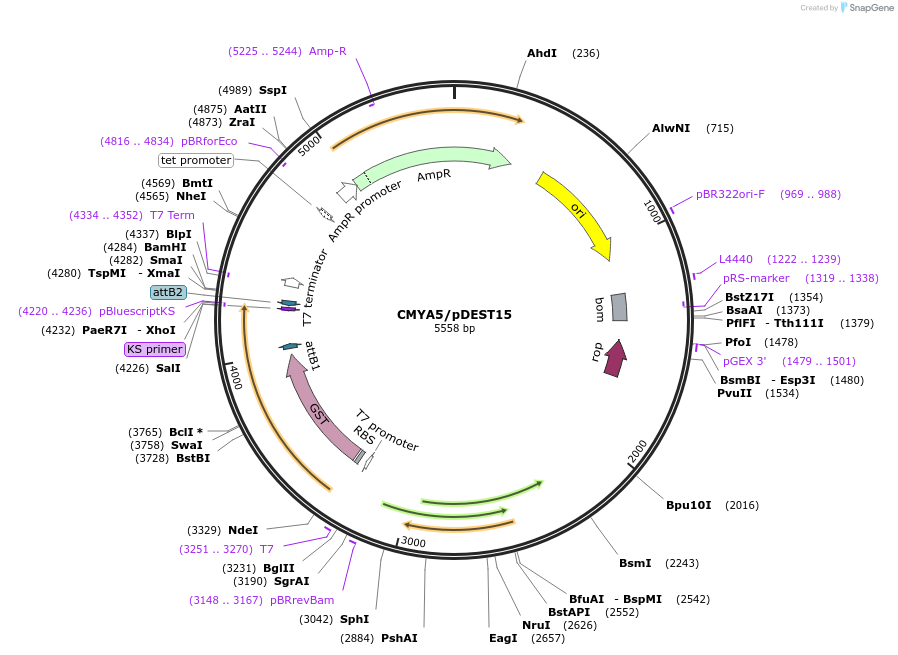
CMYA5/pDEST15
Plasmid#98106PurposeExpress CMYA5 (aa 4039 – 4069) with a N-terminal GST tag in bacteriaDepositorAvailable SinceAug. 21, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
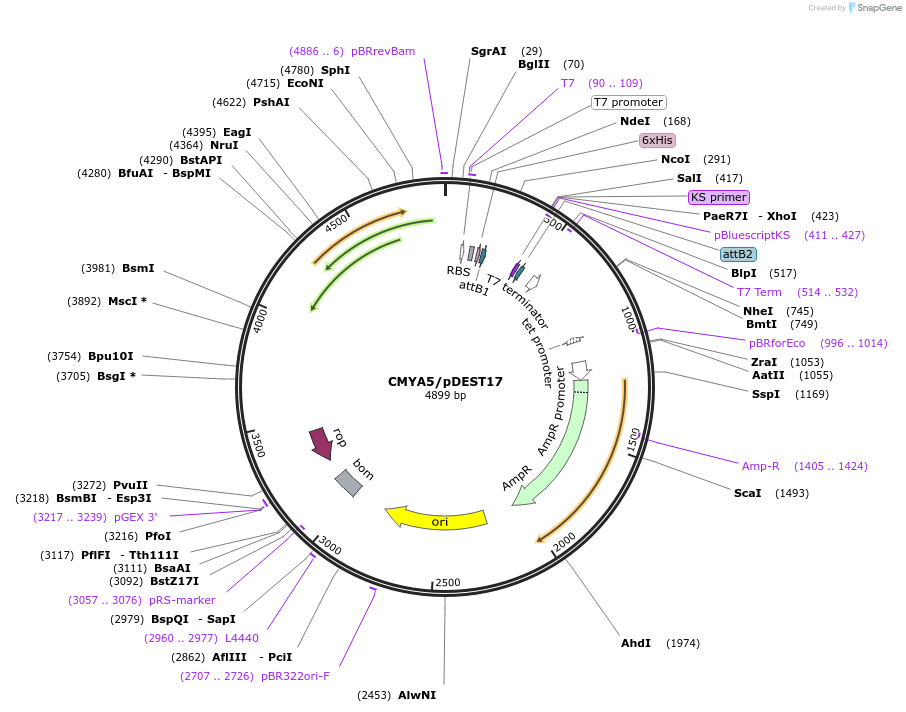
CMYA5/pDEST17
Plasmid#98115PurposeExpress CMYA5 (aa 4039 – 4069) with a N-terminal 6x-His tag in bacteriaDepositorAvailable SinceAug. 21, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
CMYA5T2/pENTR3C
Plasmid#97409PurposeCMYA5 (NM_153610, nt 12205 to 12279) T allele (rs10043986) in pENTR3C, Gateway Entry vectorDepositorAvailable SinceAug. 21, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
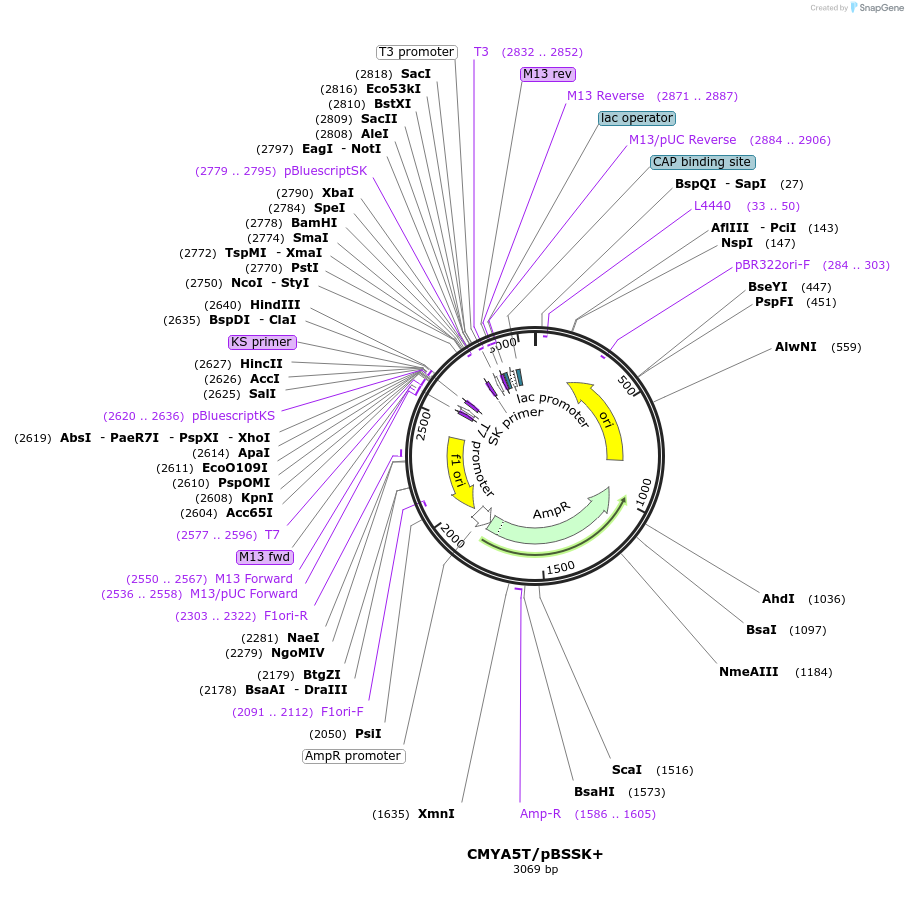
CMYA5T/pBSSK+
Plasmid#97206PurposeCMYA5 (NM_153610, nt 12205 to 12279) T allele (rs10043986) in pBSSK+DepositorAvailable SinceAug. 3, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
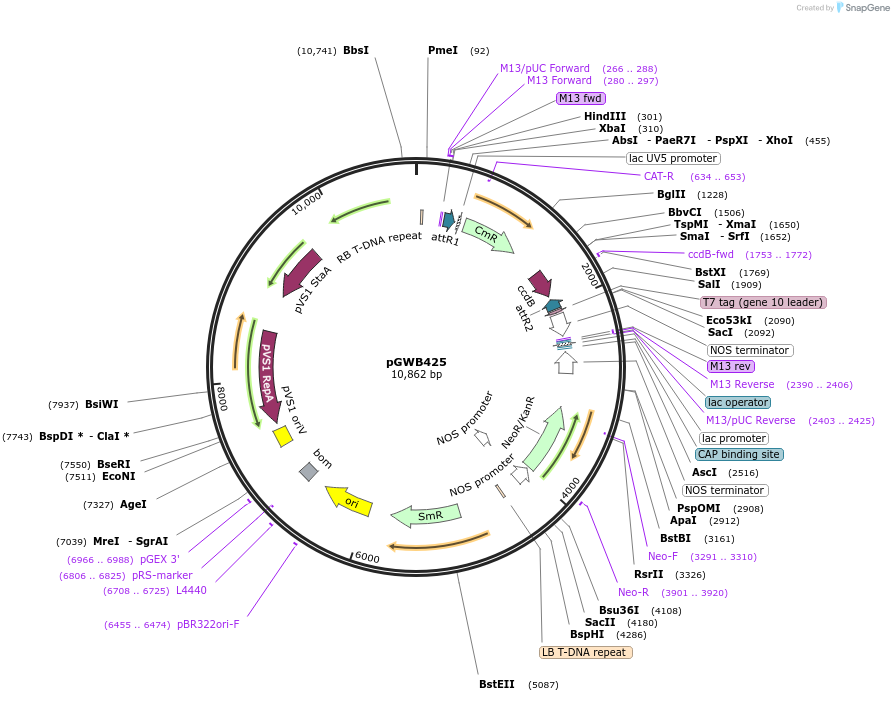
pGWB425
Plasmid#74819PurposeGateway cloning compatible binary vector for C-terminal fusion with T7 (no promoter).DepositorTypeEmpty backboneExpressionPlantAvailable SinceJuly 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
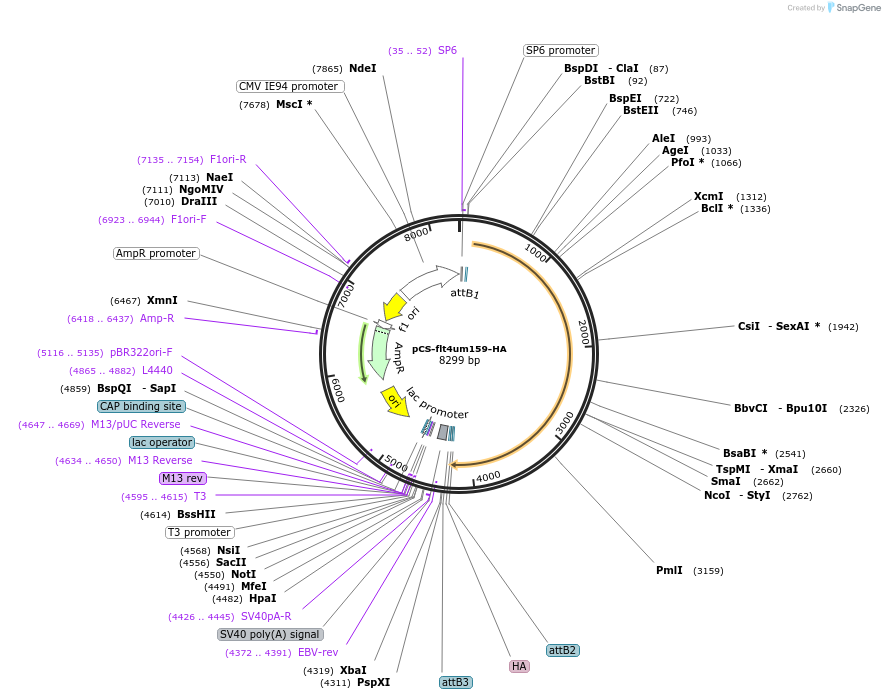
pCS-flt4um159-HA
Plasmid#67704PurposeExpression constructDepositorAvailable SinceAug. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCS-Fv2Eflt4um200-2a-tdTomato
Plasmid#67698PurposeExpression constructDepositorAvailable SinceAug. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
pGAD-hPLEKHM1
Plasmid#64145PurposeFor yeast two hybridDepositorAvailable SinceApril 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
-
-
K339CLM S330C
Plasmid#24441DepositorInsertKinesin (KIF5B Human)
Tags6XHis and GFPExpressionBacterialMutationC7S/C65A/C168A/C174S/C294A/C330S, S330CAvailable SinceMay 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
K343CLM A337C
Plasmid#24442DepositorInsertKinesin (KIF5B Human)
Tags6XHis and GFPExpressionBacterialMutationC7S/C65A/C168A/C174S/C294A/C330S, A337CAvailable SinceMay 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
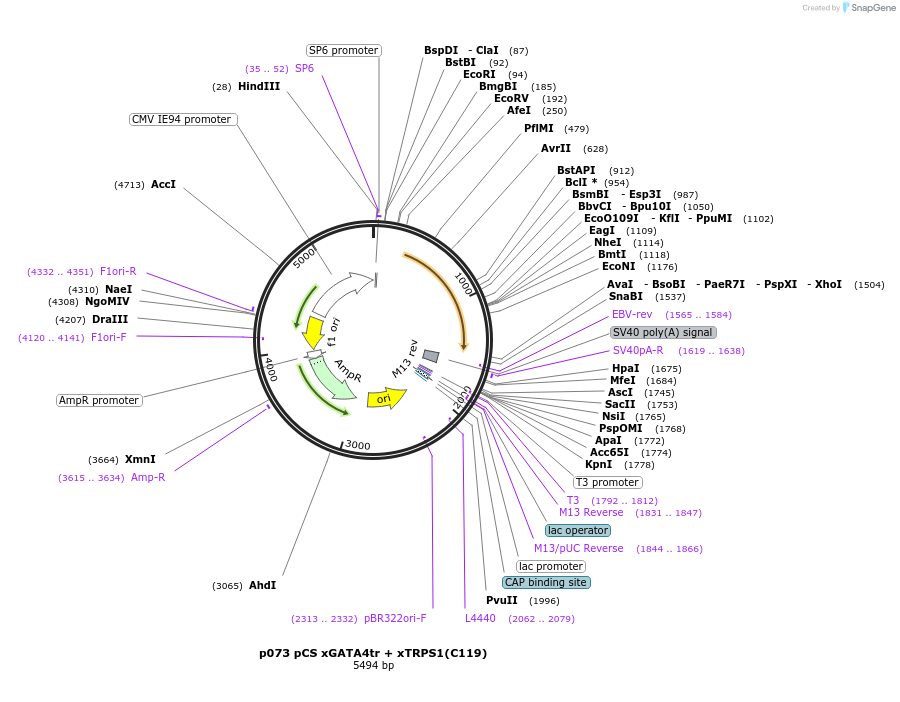
p073 pCS xGATA4tr + xTRPS1(C119)
Plasmid#11044DepositorUseXenopus, mammalian, avian, and zebrafishMutationFusion construct of XGATA4 (amino acids 1-274) wi…Available SinceDec. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
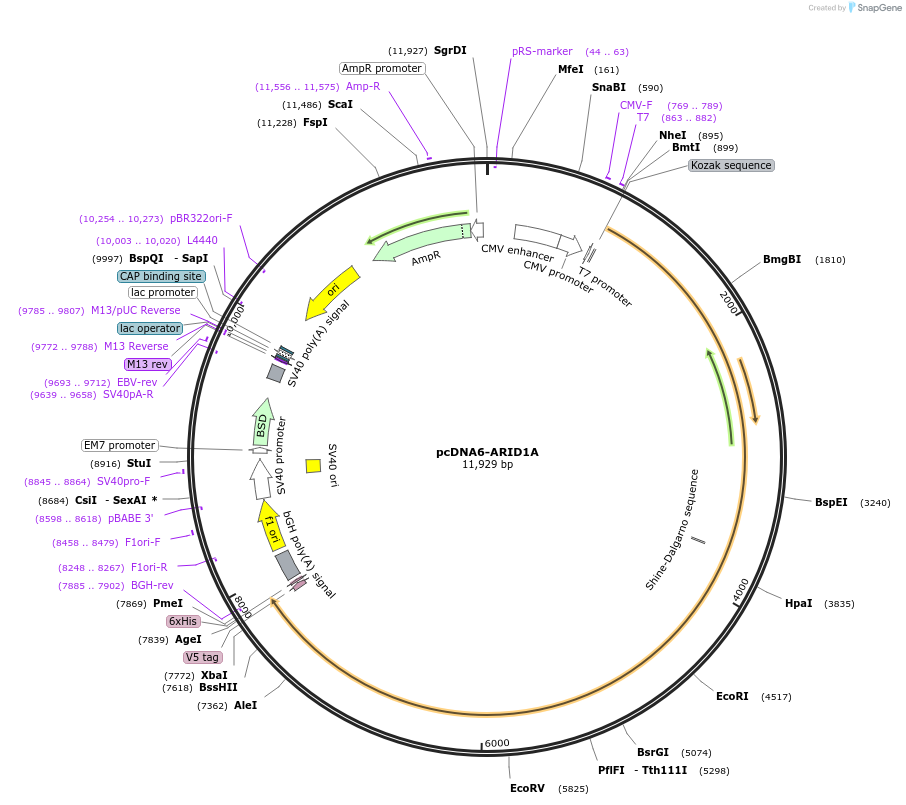
pcDNA6-ARID1A
Plasmid#39311DepositorAvailable SinceSept. 13, 2012AvailabilityAcademic Institutions and Nonprofits only -
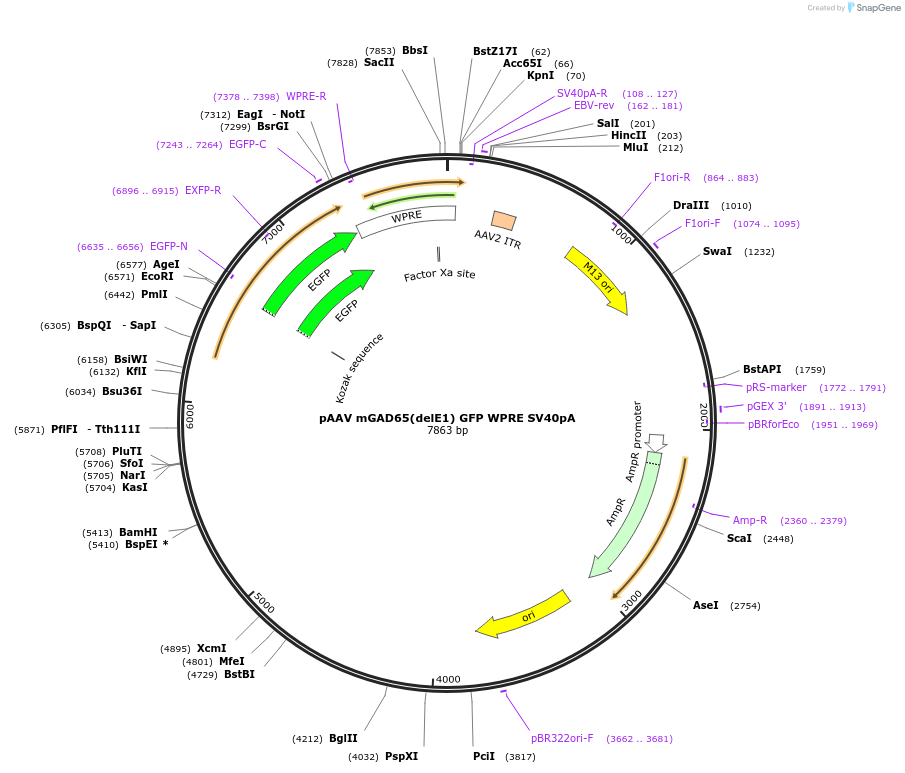
pAAV mGAD65(delE1) GFP WPRE SV40pA
Plasmid#177316PurposeTo produce the AAV vector that expresses GFP under the control of the mGAD65 promoter. The mGAD65 promoter has highly specificity to GABAergic interneurons.DepositorInsertmGAD65(delE1) promoter, GABAergic interneurons specific promoter
UseAAVAvailable SinceDec. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
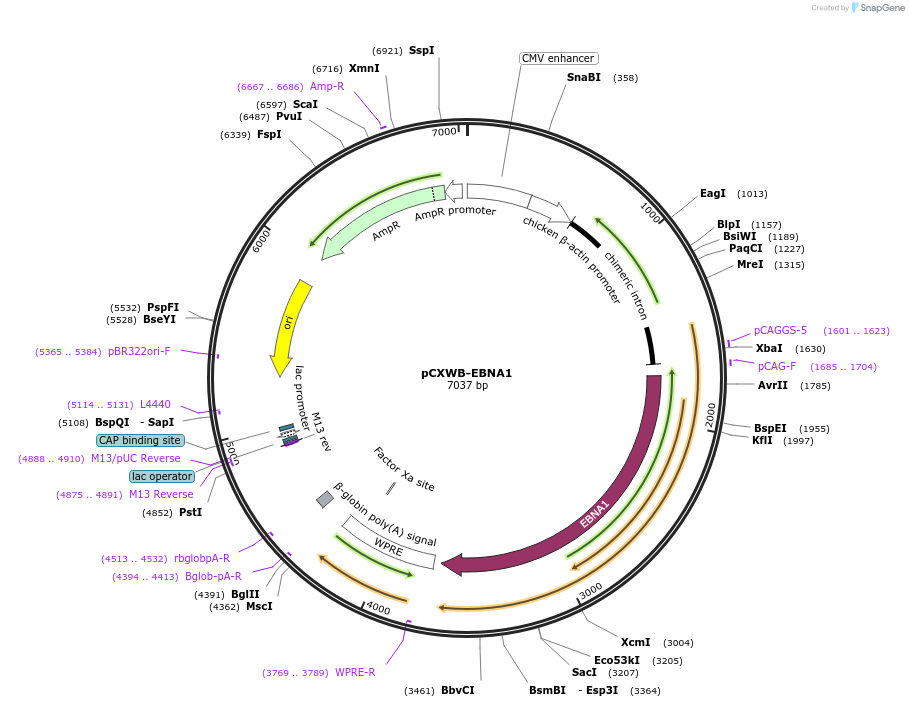
pCXWB-EBNA1
Plasmid#37624PurposeNon-replicating episomal expression of EBNA1, which enhances transfection efficiency and expression from episomal plasmidsDepositorAvailable SinceDec. 3, 2012AvailabilityAcademic Institutions and Nonprofits only