We narrowed to 17,262 results for: por
-
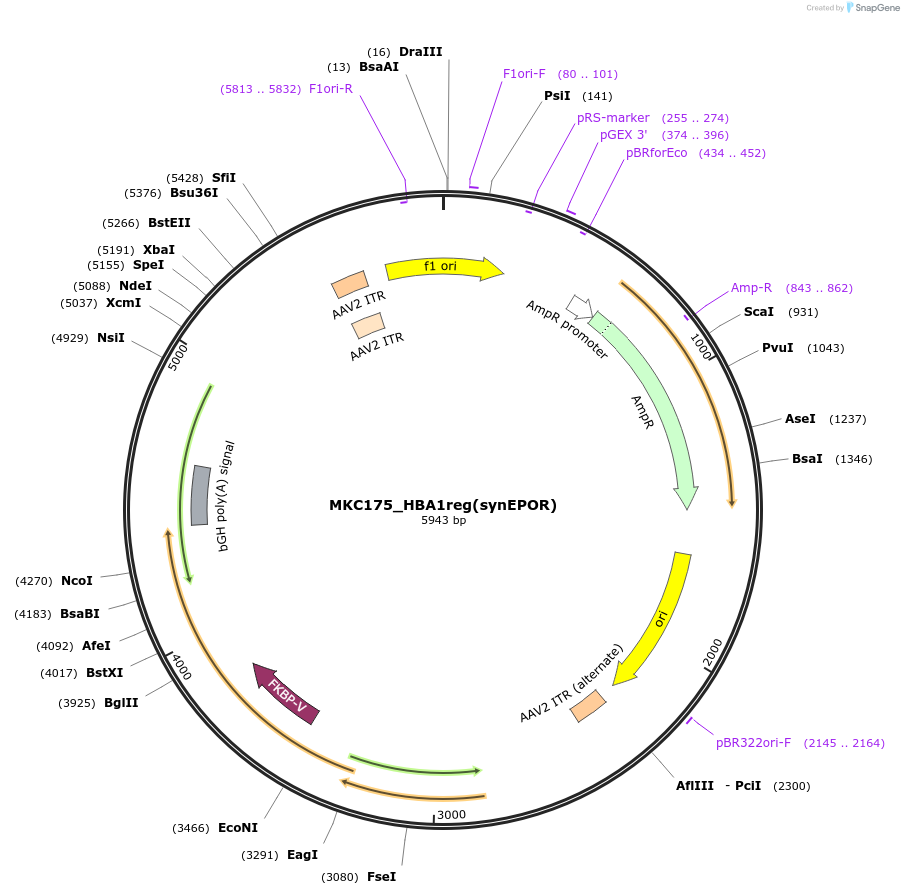
Plasmid#232413PurposeAAV production plasmid for HBA1 UTRs vector from Fig. 3 that mediates HDR at HBA1 locus using HBA1 sg5 gRNA. HBB gene includes introns; HBA1 UTRs flank H+C15BA1 cassette. HAs are ~400bp each.DepositorAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only
-
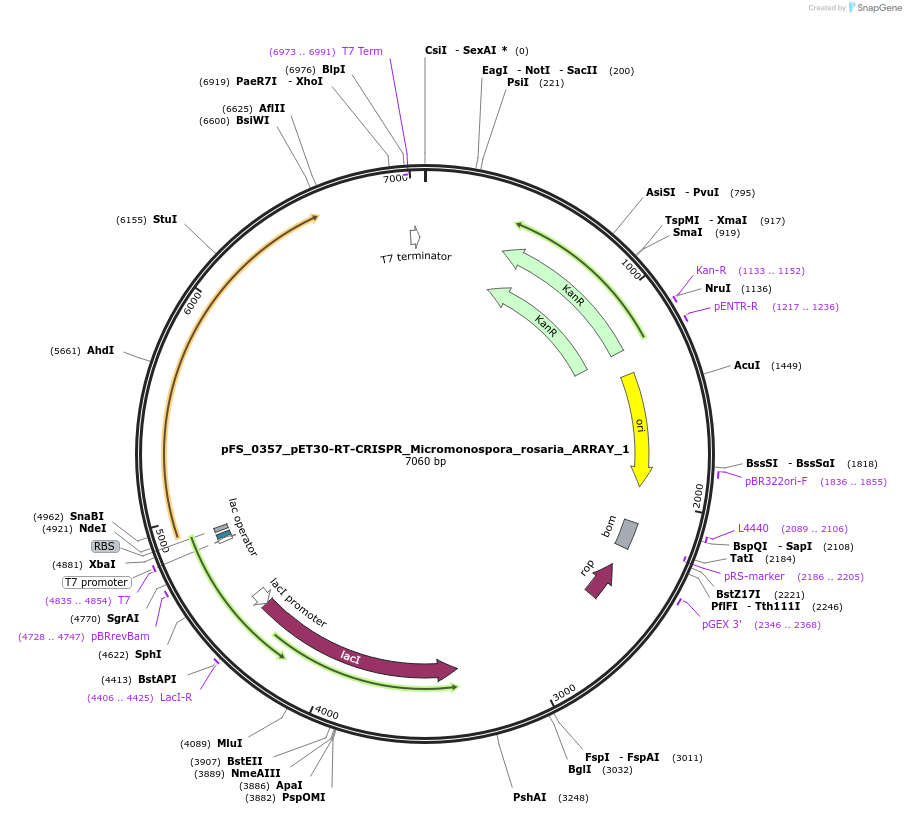
pFS_0357_pET30-RT-CRISPR_Micromonospora_rosaria_ARRAY_1
Plasmid#116969PurposeExpresses MrRT-Cas1-Cas2 under pT7lac promoter, encodes MrCRISPR array 1, compatible with SENECA acquisition readoutDepositorInsertMicromonospora rosaria RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
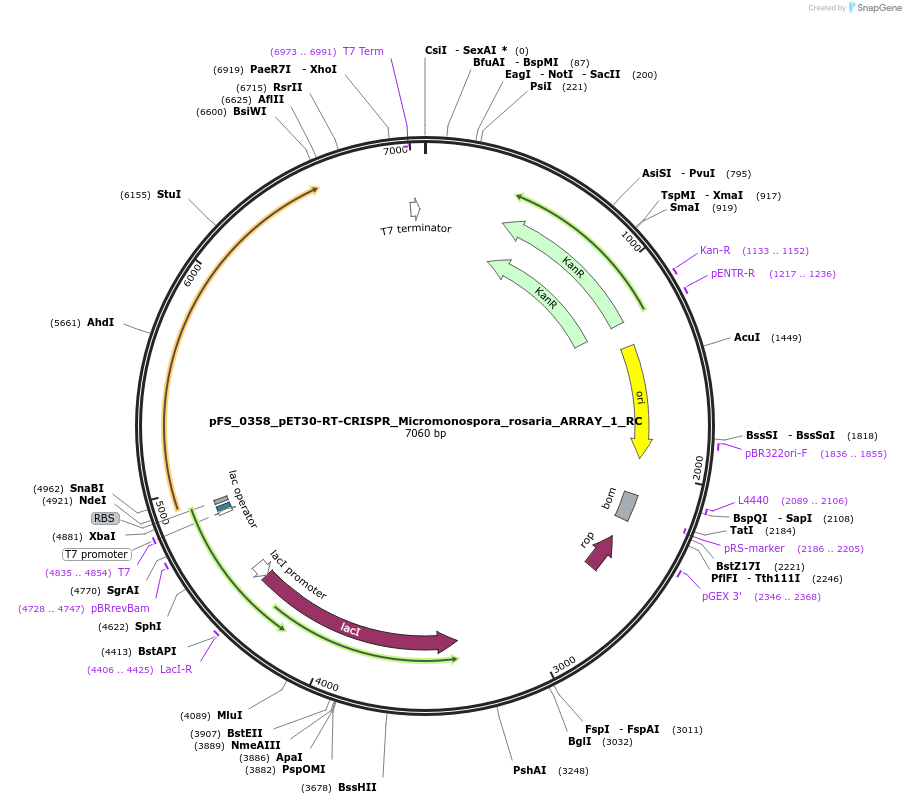
pFS_0358_pET30-RT-CRISPR_Micromonospora_rosaria_ARRAY_1_RC
Plasmid#116970PurposeExpresses MrRT-Cas1-Cas2 under pT7lac promoter, encodes MrCRISPR array 1 RC, compatible with SENECA acquisition readoutDepositorInsertMicromonospora rosaria RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
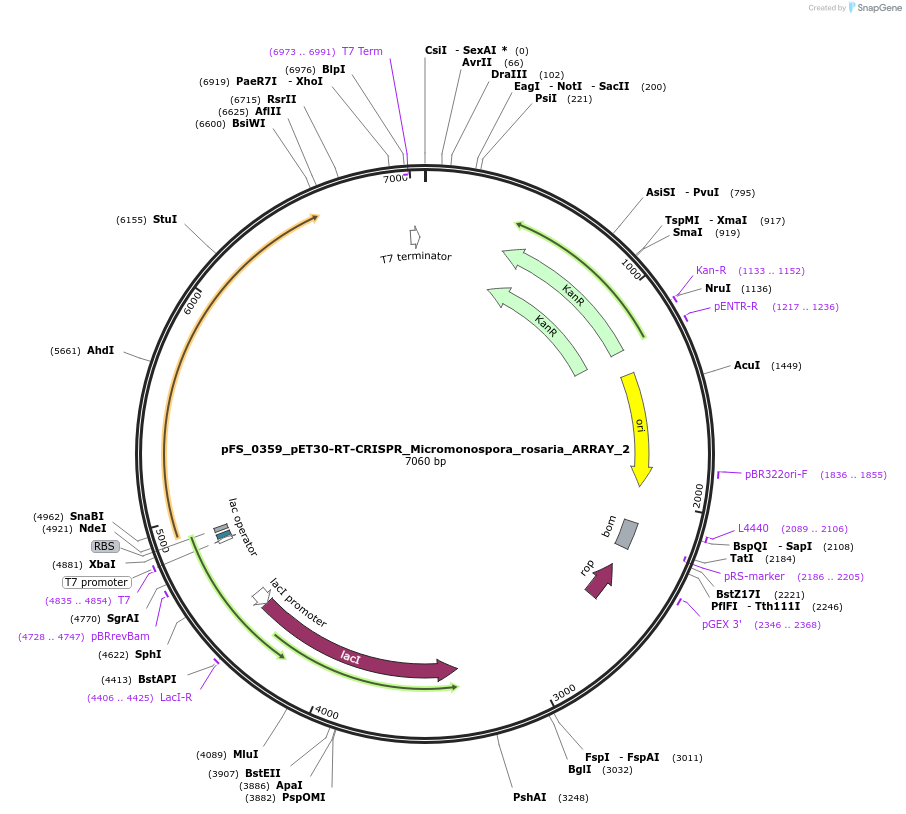
pFS_0359_pET30-RT-CRISPR_Micromonospora_rosaria_ARRAY_2
Plasmid#116971PurposeExpresses MrRT-Cas1-Cas2 under pT7lac promoter, encodes MrCRISPR array 2, compatible with SENECA acquisition readoutDepositorInsertMicromonospora rosaria RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
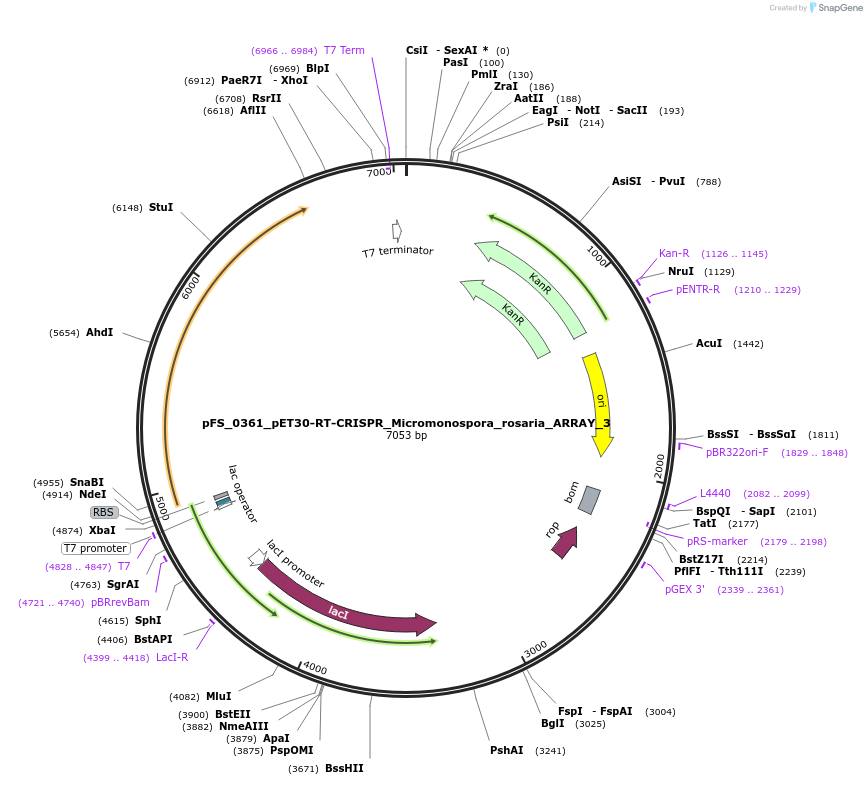
pFS_0361_pET30-RT-CRISPR_Micromonospora_rosaria_ARRAY_3
Plasmid#116973PurposeExpresses MrRT-Cas1-Cas2 under pT7lac promoter, encodes MrCRISPR array 3, compatible with SENECA acquisition readoutDepositorInsertMicromonospora rosaria RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
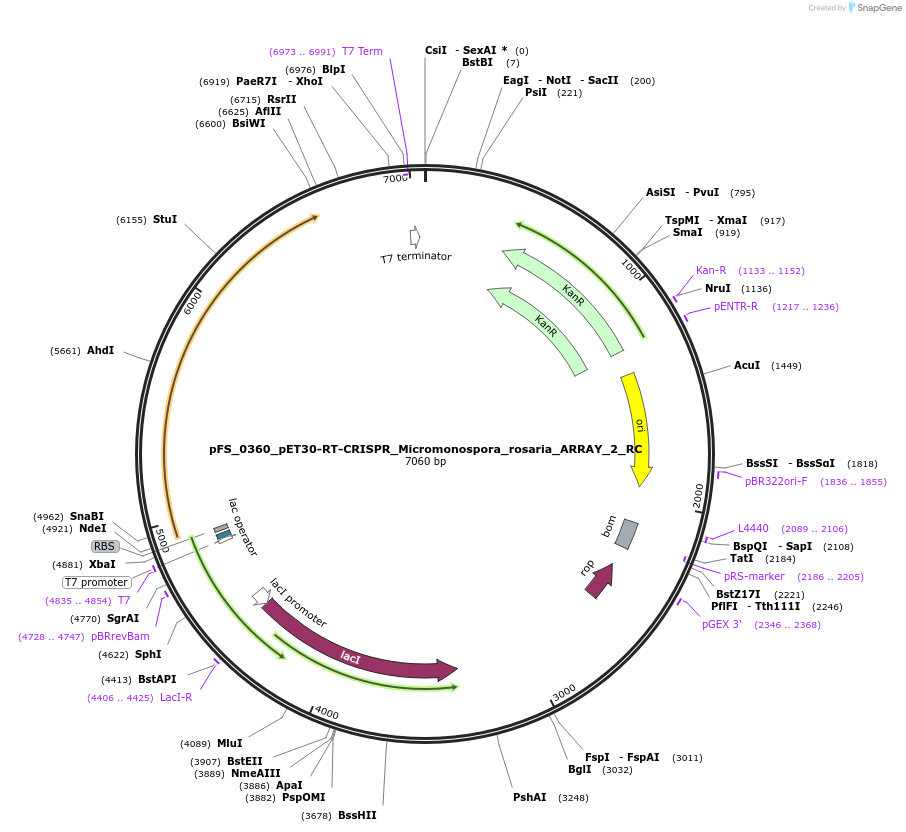
pFS_0360_pET30-RT-CRISPR_Micromonospora_rosaria_ARRAY_2_RC
Plasmid#116972PurposeExpresses MrRT-Cas1-Cas2 under pT7lac promoter, encodes MrCRISPR array 2 RC, compatible with SENECA acquisition readoutDepositorInsertMicromonospora rosaria RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
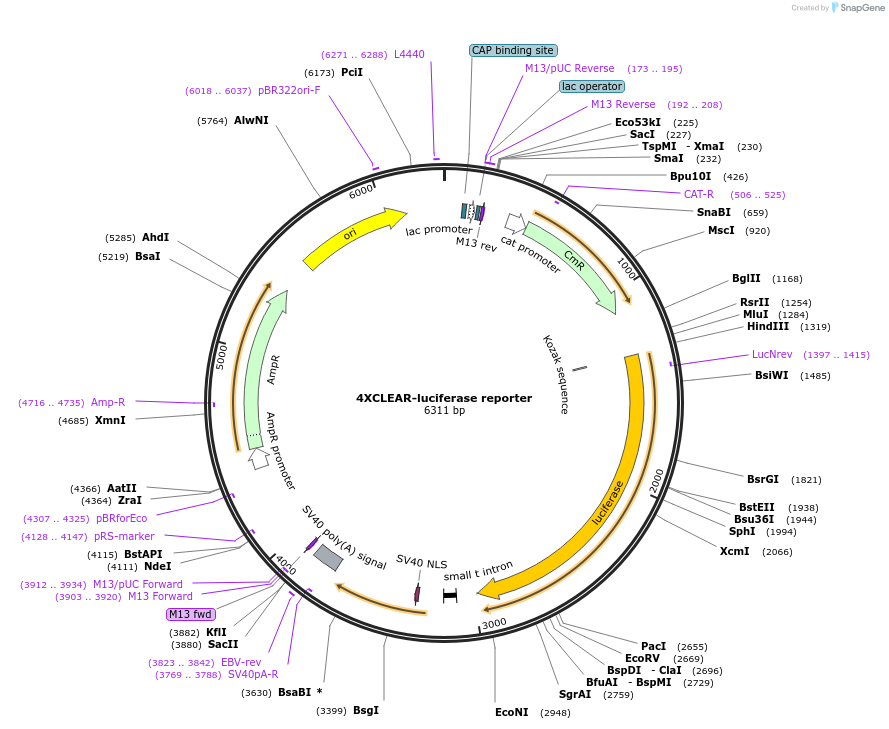
4XCLEAR-luciferase reporter
Plasmid#66800PurposeReporter for activity of the CLEAR networkDepositorInsert4XCLEAR
UseLuciferasePromoterFour CLEAR elements (GTCACGTGAC) in tandem derive…Available SinceNov. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
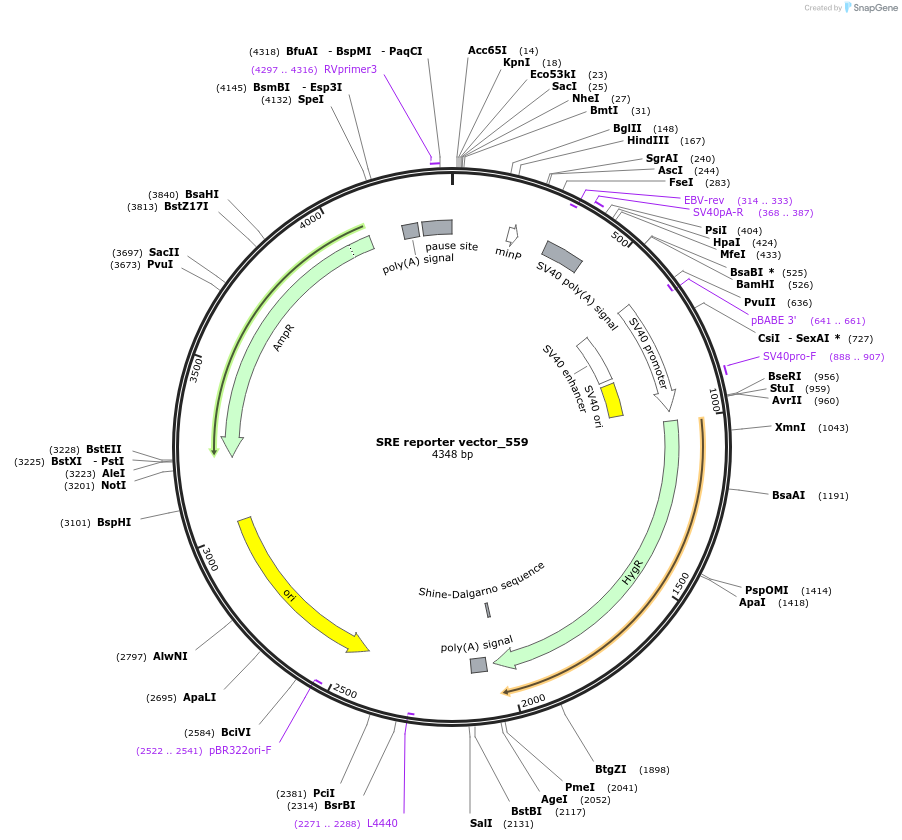
SRE reporter vector_559
Plasmid#82686PurposeCloning vector containing serum response element (SRE) followed by an AscI restriction siteDepositorTypeEmpty backboneExpressionMammalianAvailable SinceNov. 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
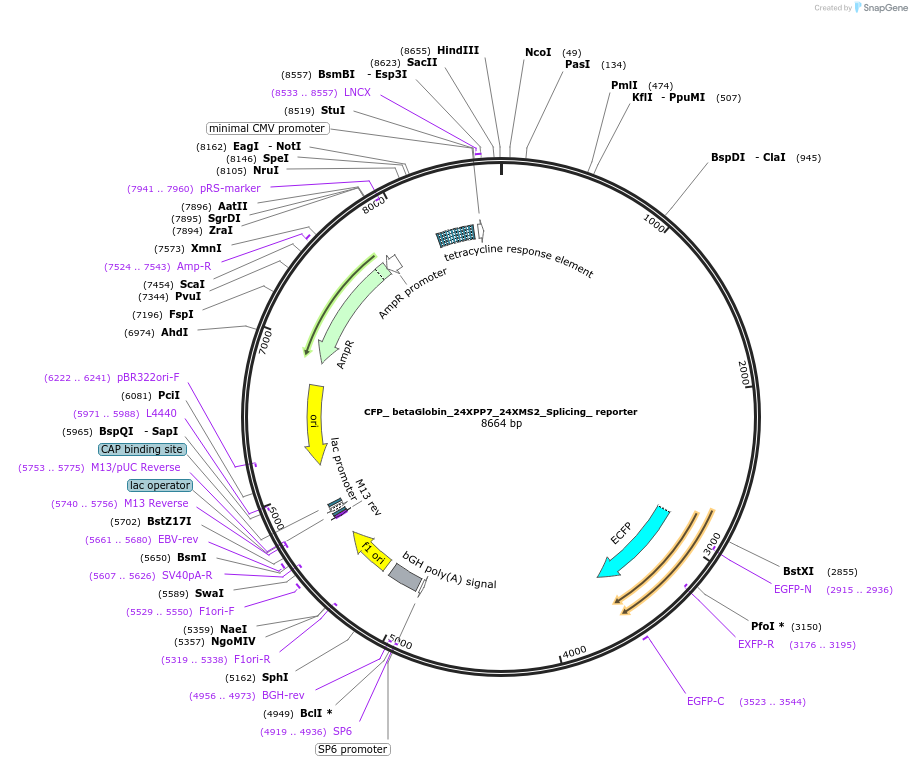
CFP_ betaGlobin_24XPP7_24XMS2_Splicing_ reporter
Plasmid#61762PurposeDual Color betaGlobin reporter to study splicing kinetics in vivoDepositorInsertb-Globin
Tags24 X MS2 hairpin cassette in 3'UTR of b-Glob…ExpressionMammalianPromoterDox inducible Tet-ON promoterAvailable SinceFeb. 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
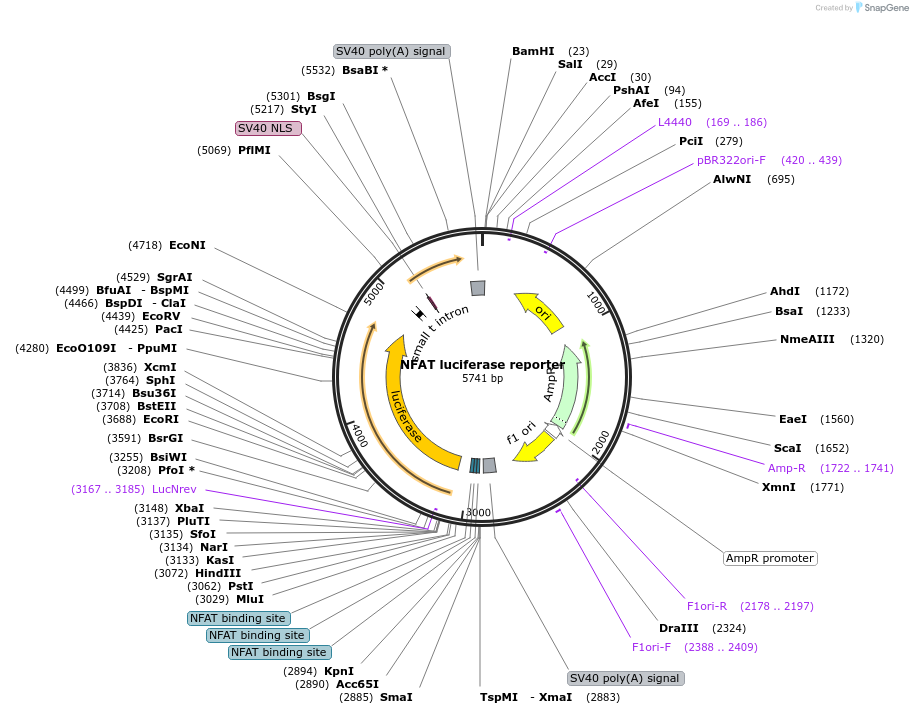
NFAT luciferase reporter
Plasmid#10959DepositorAvailable SinceJan. 6, 2006AvailabilityAcademic Institutions and Nonprofits only -
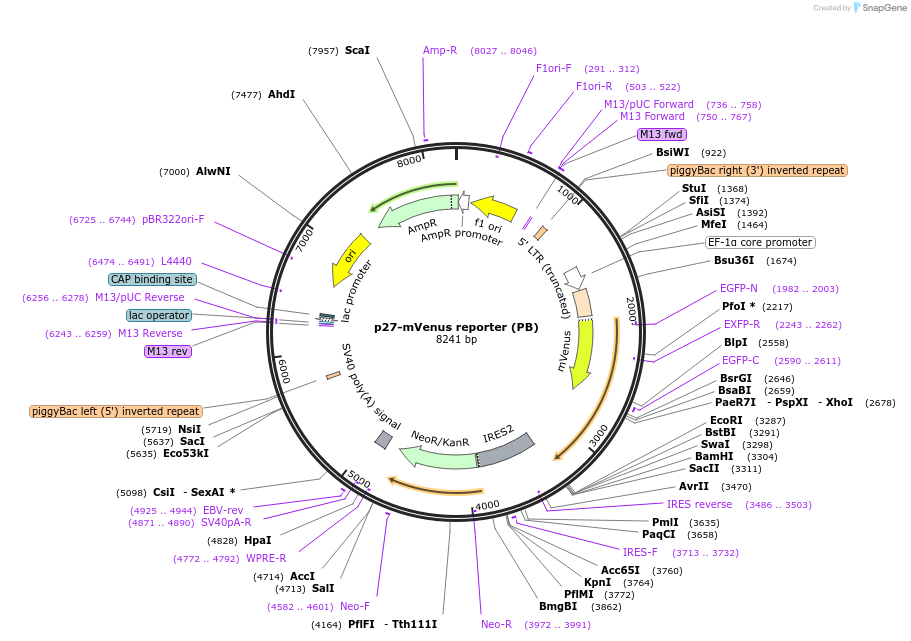
p27-mVenus reporter (PB)
Plasmid#190731PurposeThe p27 reporter vector, which expresses mVenus-fused, mutant (K–) p27 protein with a defective CDK binding.DepositorInsertsmVenus
p27
ExpressionBacterial and MammalianMutationp27-F62A-F64APromoterEF1αAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
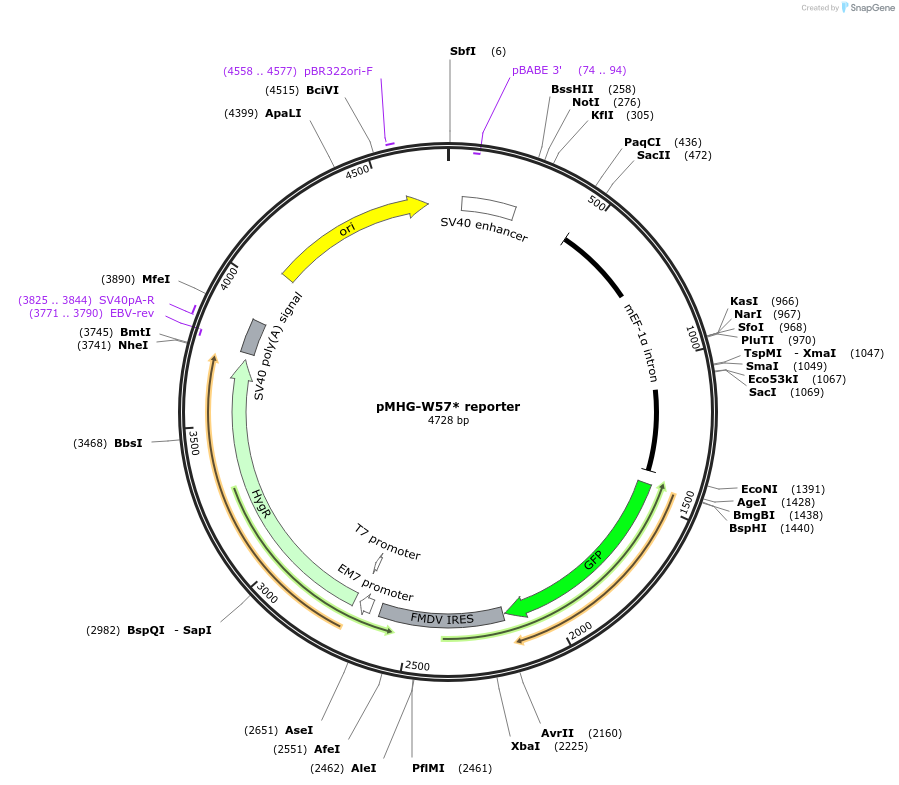
pMHG-W57* reporter
Plasmid#72850PurposeGFP-based reporter construct pMHG-W57* capable of detecting dose-dependent drug-induced PTC readthrough both by fluorescence microscopy and flow cytometry.DepositorInsertGFP
ExpressionMammalianAvailable SinceJan. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
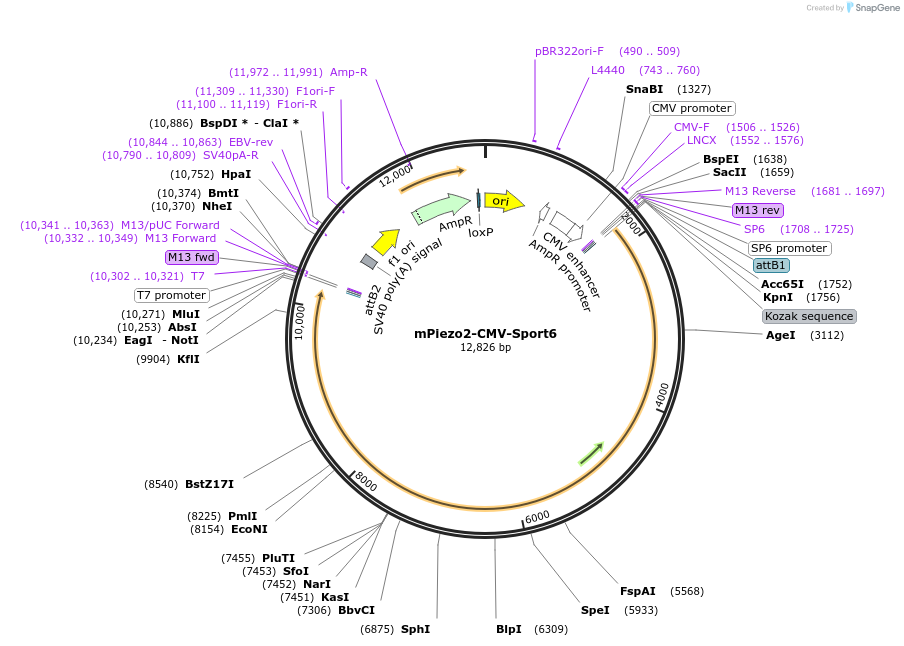
mPiezo2-CMV-Sport6
Plasmid#81073Purposemouse Piezo2DepositorInsertPiezo2 (please see depositor comments below) (Piezo2 Mouse)
ExpressionMammalianAvailable SinceNov. 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
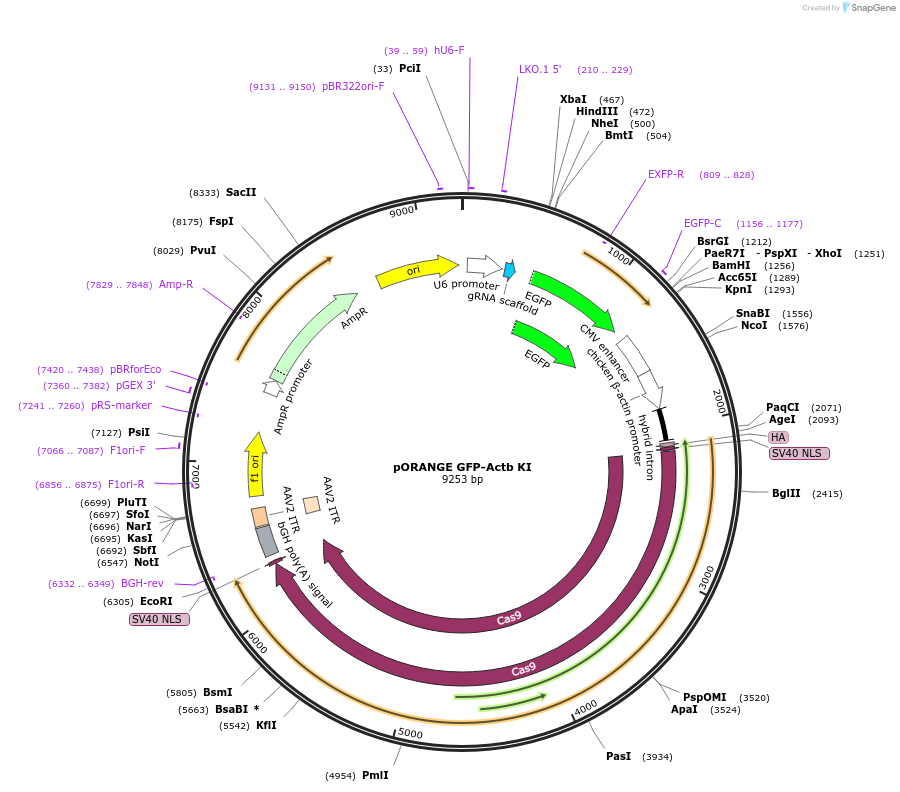
pORANGE GFP-Actb KI
Plasmid#131479PurposeEndogenous tagging of βactin: N-terminal (amino acid position: D2)DepositorAvailable SinceSept. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
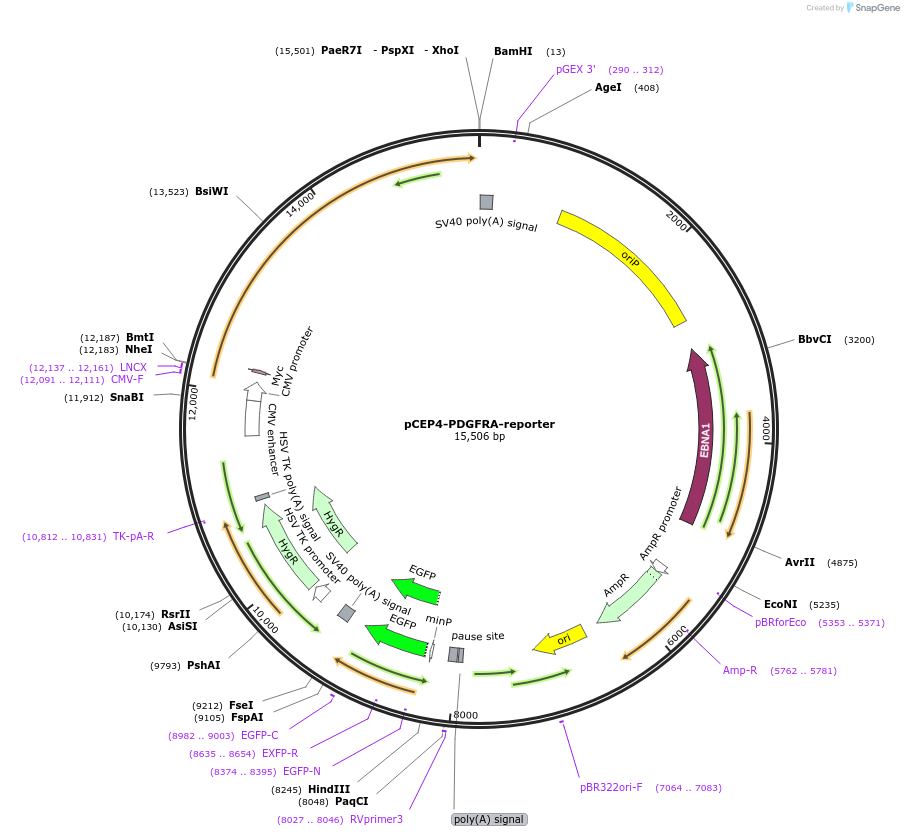
pCEP4-PDGFRA-reporter
Plasmid#136456PurposeMammalian fluorescent reporter plasmid for PDGFRA signaling.DepositorInsertsPDGFRalpha (PDGFRA Human)
Array of serum response elements (SRE) to drive destabilized GFP expression from a minimal promoter.
TagsExtracellular c-myc epitope tag, Signal peptide f…ExpressionMammalianPromoterCMV and Minimal promoter with artificial SRE enha…Available SinceJuly 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
pCMV Sport2 mPer2
Plasmid#16204DepositorInsertPer2 (Per2 Mouse)
ExpressionMammalianAvailable SinceDec. 6, 2007AvailabilityAcademic Institutions and Nonprofits only -
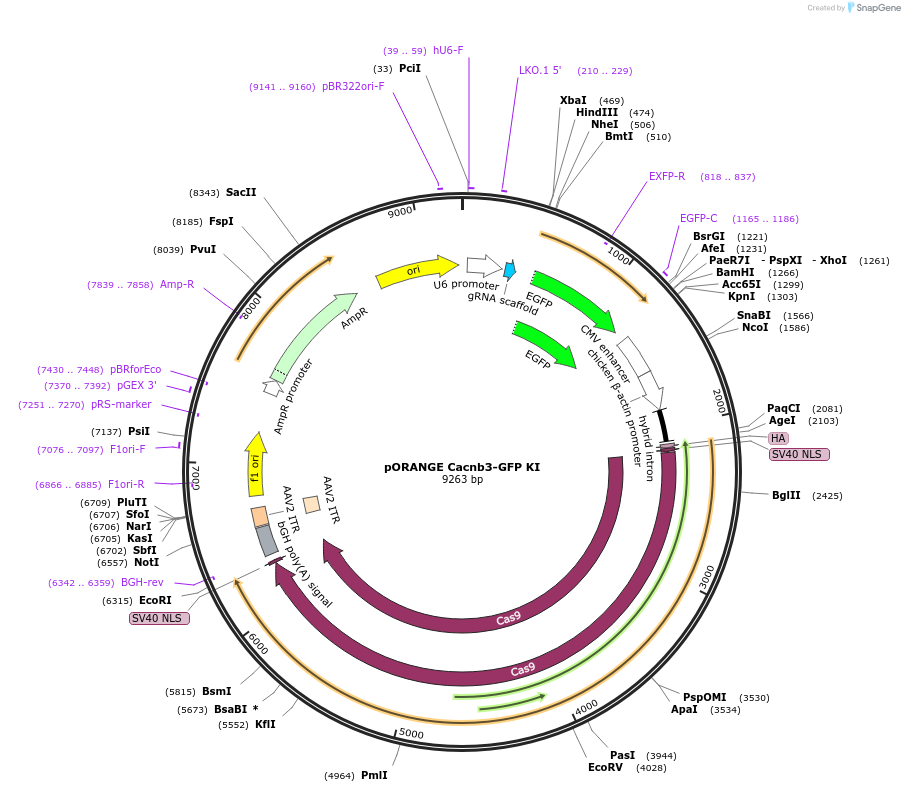
pORANGE Cacnb3-GFP KI
Plasmid#139662PurposeEndogenous tagging of CaVβ3: C-terminal (amino acid position: W479)DepositorInsertgRNA and GFP donor
ExpressionMammalianPromoterU6 and CbhAvailable SinceApril 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
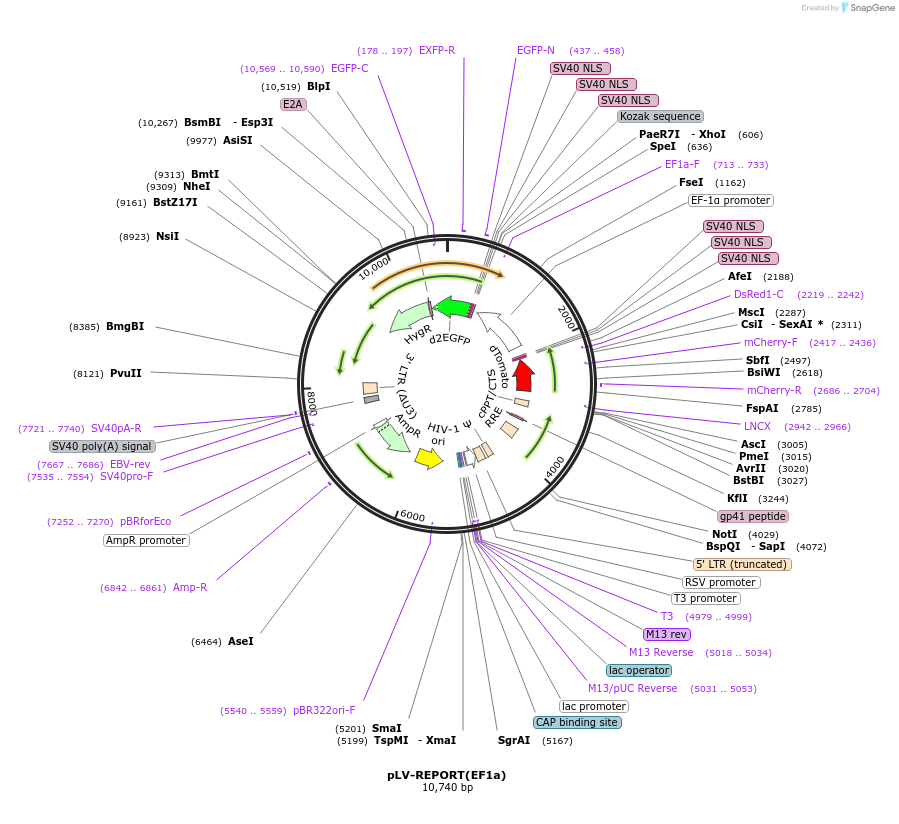
pLV-REPORT(EF1a)
Plasmid#172326PurposeLV REPORT containing minimal promoter driving monomeric nuclear Tomato followed by an EF1a promoter driving nuclear d2eGFP E2A HygromycinDepositorInsertsmonomeric nuclear Tomato
d2eGFP
HygR
UseLentiviral and Synthetic BiologyTags3x SV40 NLSExpressionMammalianPromoterEF1-alphaAvailable SinceJune 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
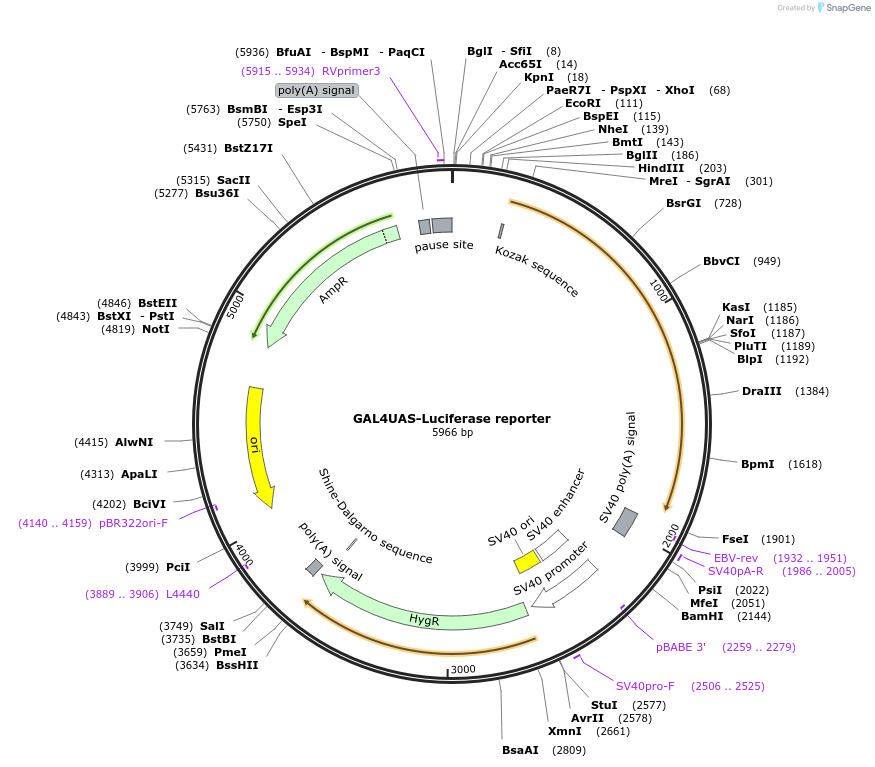
GAL4UAS-Luciferase reporter
Plasmid#64125PurposePhotoactivatable transcription system. Luciferase reporter under the control of the upstream activator sequence (UAS) of Gal4.DepositorInsertGAL4UAS
UseLuciferaseTagsluciferaseExpressionMammalianPromoterGAL4UASAvailable SinceApril 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
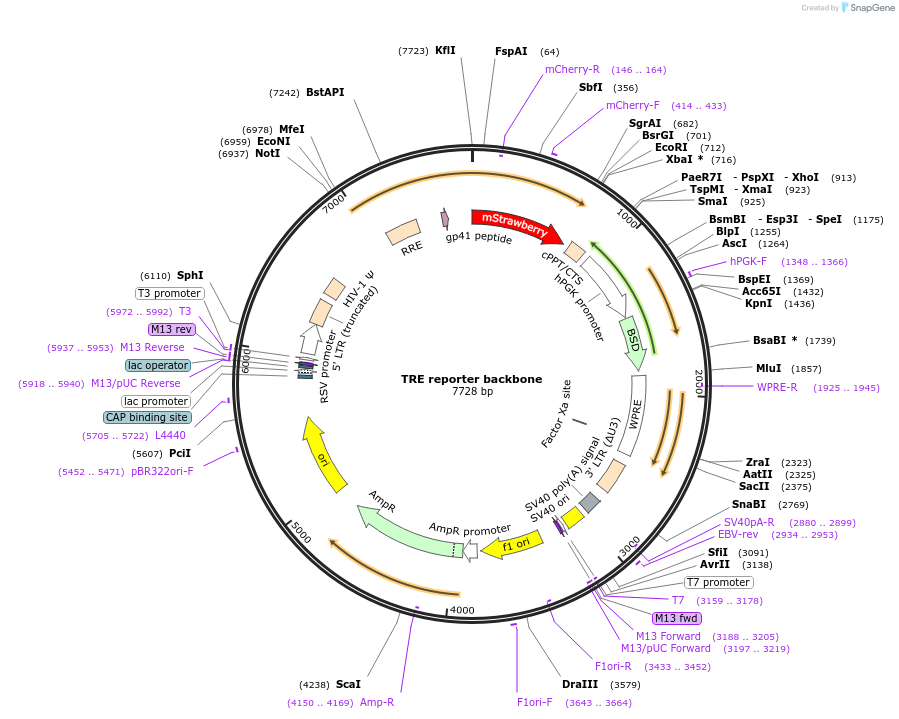
TRE reporter backbone
Plasmid#158678PurposeThis plasmid serves as the backbone for the TRE reporters described in our paper. It's a promoterless-mStrawberry-pGK-BSD backbone to which we insert a pathway specific promoter before the mStrawberryDepositorTypeEmpty backboneUseLentiviralTagsmStrawberryExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only