We narrowed to 5,944 results for: pCas
-
Plasmid#124896PurposeFor genome editing in plant by SpCas9 variant(SpCas9-NGv1)DepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceJan. 9, 2020AvailabilityAcademic Institutions and Nonprofits only
-
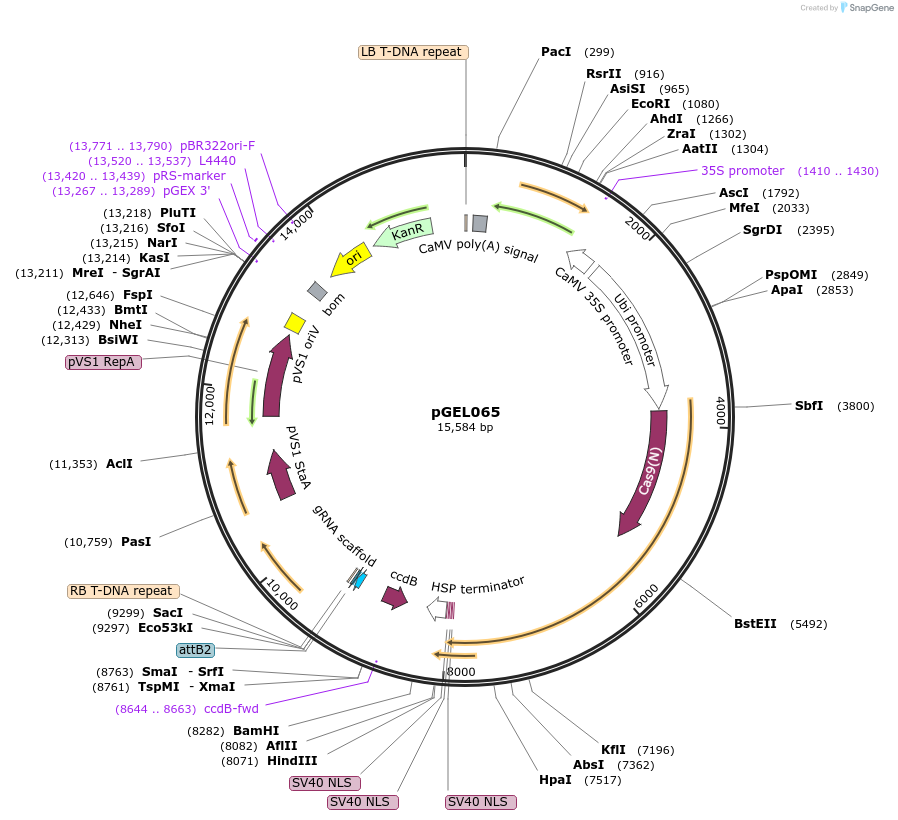
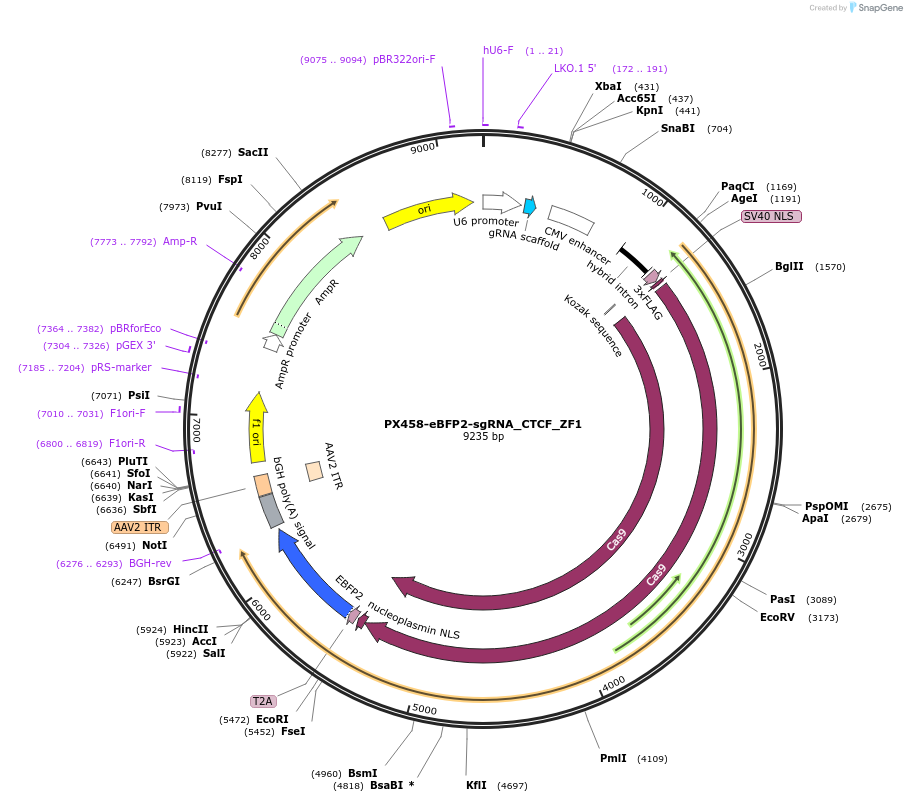
PX458-eBFP2-sgRNA_CTCF_ZF1
Plasmid#233090PurposeExpression vector for a sgRNA against the mouse CTCF ZF1 region and SpCas9-T2A-eBFP2.DepositorInsertspCas9-T2A-eBFP2 (Ctcf )
ExpressionMammalianAvailable SinceMarch 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
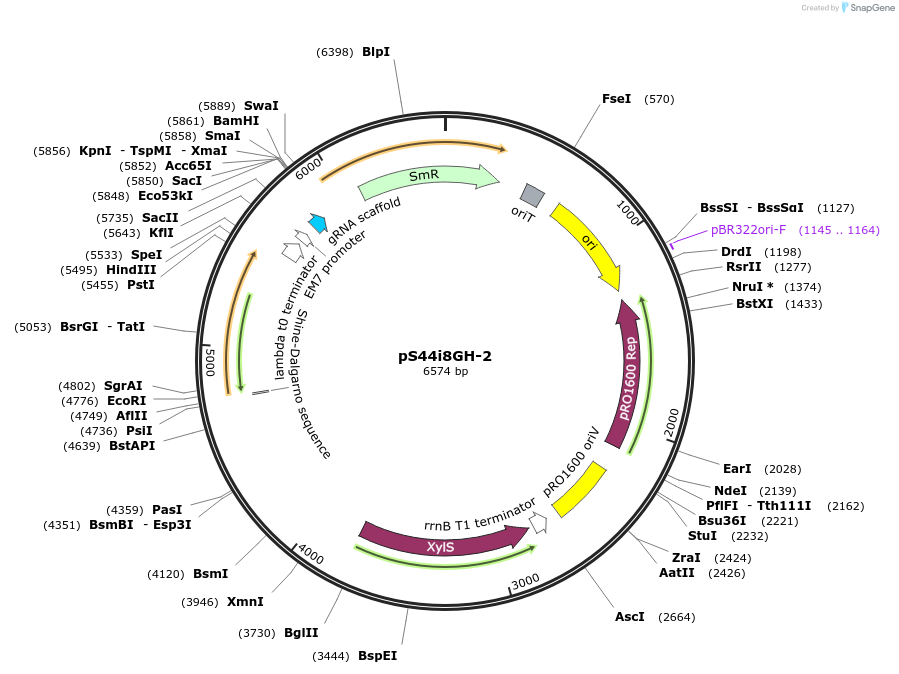
pS44i8GH-2
Plasmid#207526PurposeConditionally-replicating in Pseudomonas vector, high-copy-number; oriV(pRO1600/ColE1), xylS (cured of BsaI-sites), Pm→repA, P14b→msfGFP; SmR/SpRDepositorInsertoriV(pRO1600/ColE1), xylS (cured of BsaI-sites), Pm→repA, P14b→msfGFP
UseCRISPR; Pseudomonas vectorAvailable SinceDec. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
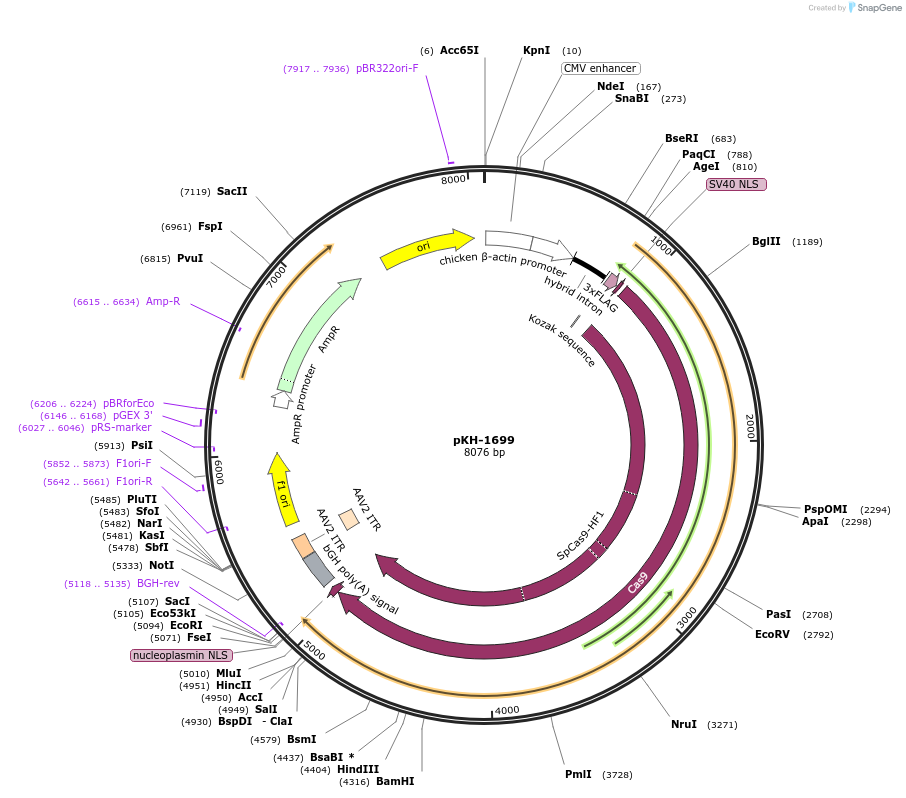
pKH-1699
Plasmid#124227PurposeMammalian SpCas9-HF1 expressionDepositorInsertSpCas9-HF1
UseCRISPRExpressionMammalianPromoterCMVAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
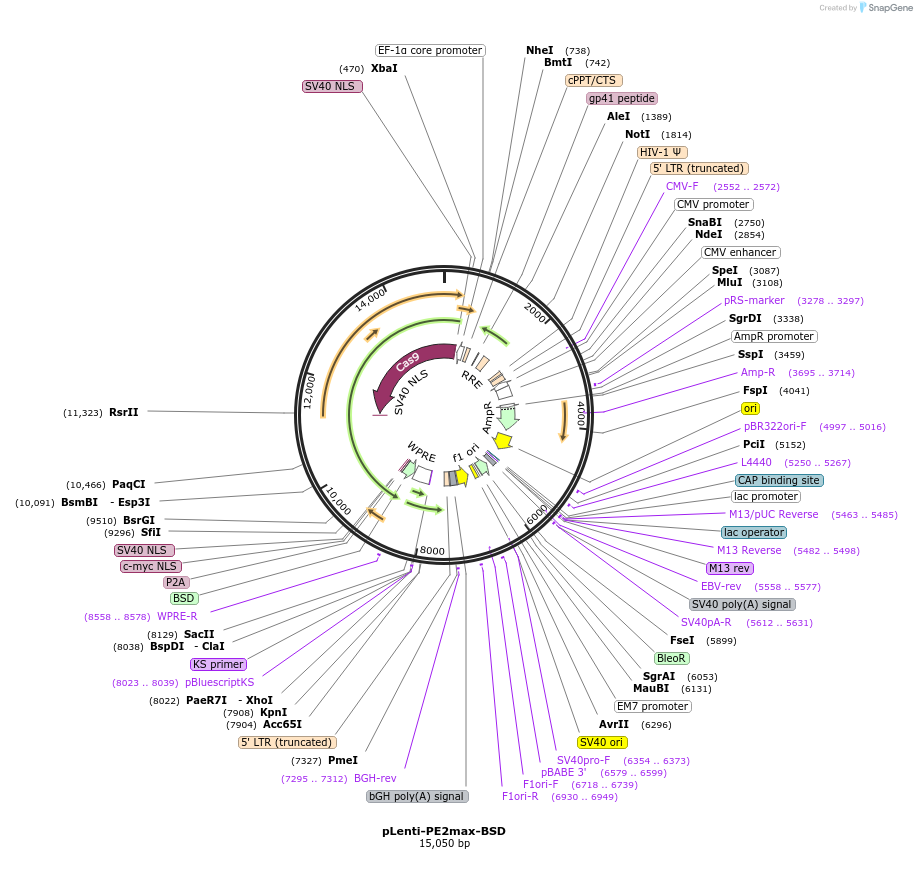
pLenti-PE2max-BSD
Plasmid#191102PurposeLentiviral expression plasmid of PEmax prime editor with P2A-BSD markerDepositorInsertPEmax-P2A-BSD
UseCRISPR and LentiviralExpressionMammalianMutationChanged R221K, N394K, H840A from SpCas9PromoterEF-1aAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
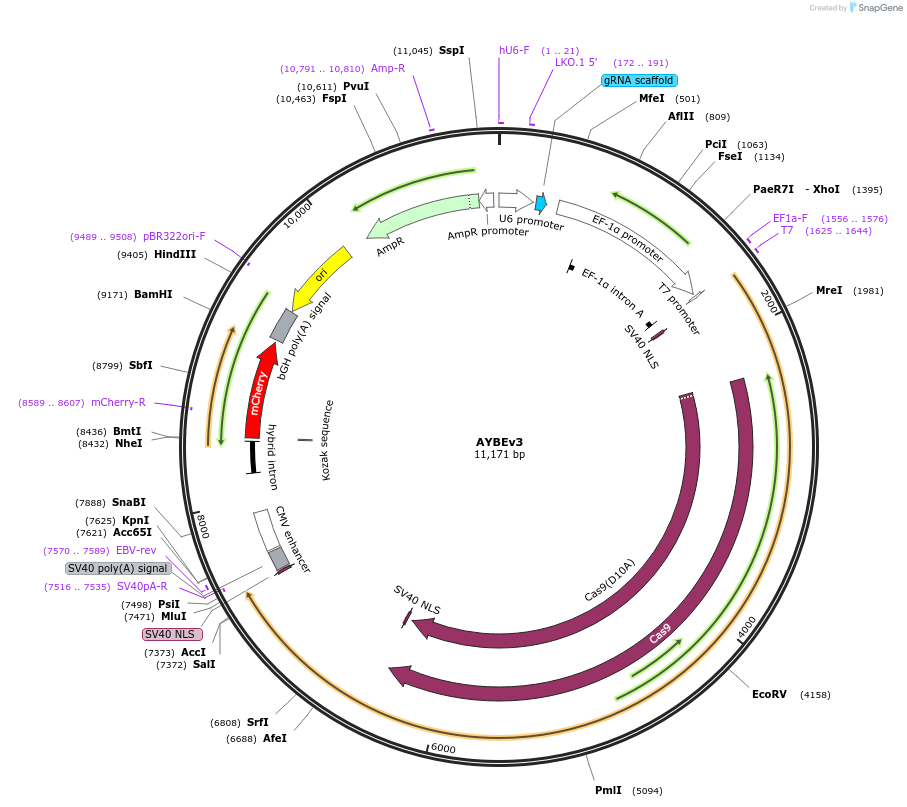
AYBEv3
Plasmid#193967PurposeAn engineered adenine transversion base editor (AYBEv3, A-to-C and/or A-to-T base editor) driven by EF1a promoter, guide RNAs compatible with spCas9 driven by hU6, mCherry driven by CBh promoterDepositorInsertecTadA8e-nSpCas9(D10A)-MPG(v3)
ExpressionMammalianMutationG163R, N169S, S198A, K202A, G203A, S206A, K210APromoterhU6, EF1a, CBhAvailable SinceJan. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
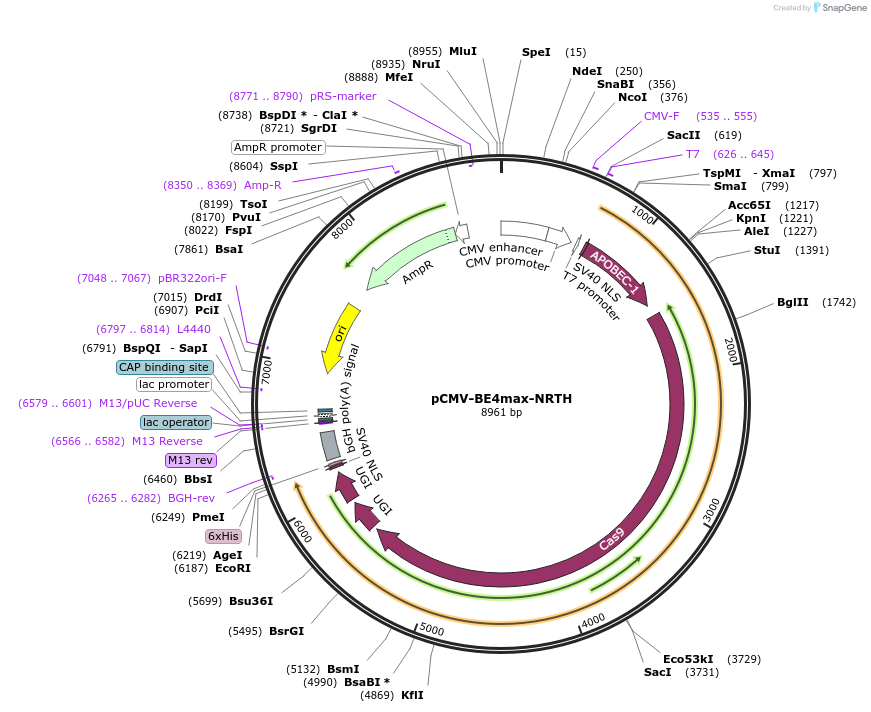
pCMV-BE4max-NRTH
Plasmid#136919PurposeExpresses BE4max-NRTH in mammalian cellsDepositorInsertBE4max-NRTH
UseCRISPRExpressionMammalianMutationsee manuscriptPromoterCMVAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
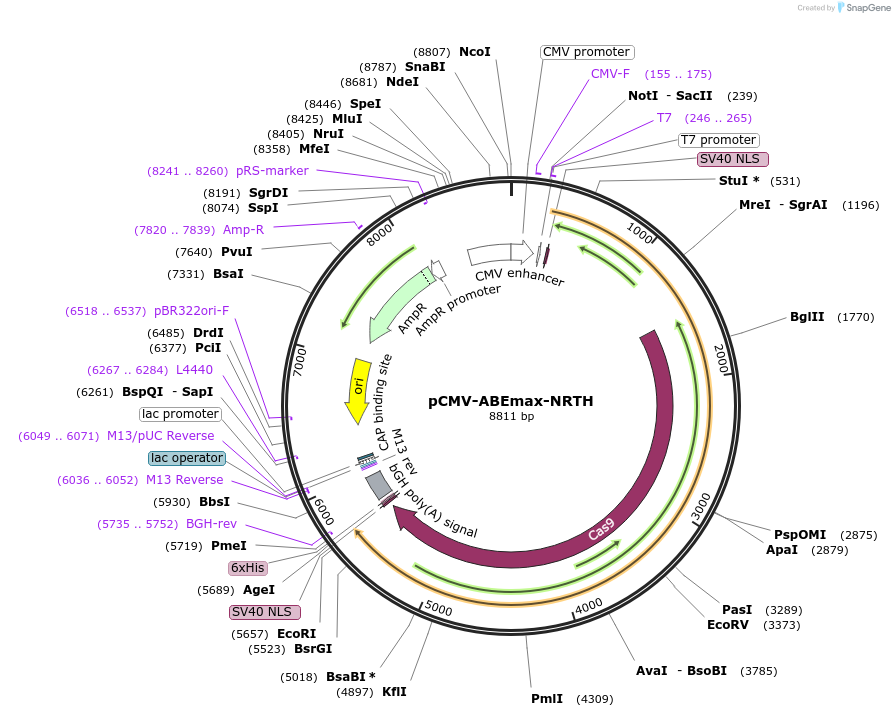
pCMV-ABEmax-NRTH
Plasmid#136922PurposeExpresses ABEmax-NRTH in mammalian cellsDepositorInsertABEmax-NRTH
UseCRISPRExpressionMammalianMutationsee manuscriptPromoterCMVAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
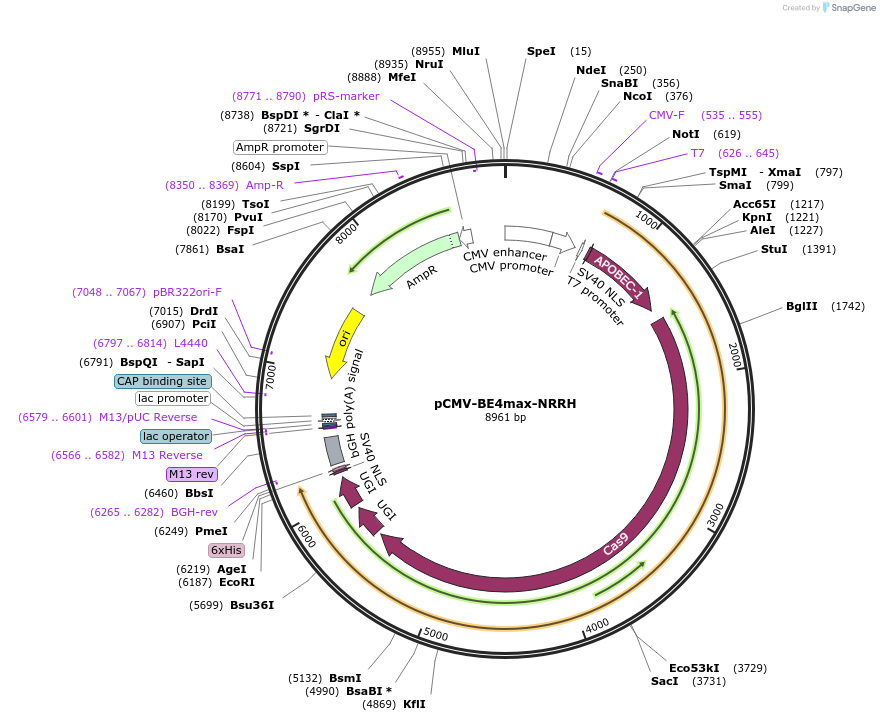
pCMV-BE4max-NRRH
Plasmid#136918PurposeExpresses BE4max-NRRH in mammalian cellsDepositorInsertBE4max-NRRH
UseCRISPRExpressionMammalianMutationsee manuscriptPromoterCMVAvailable SinceApril 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
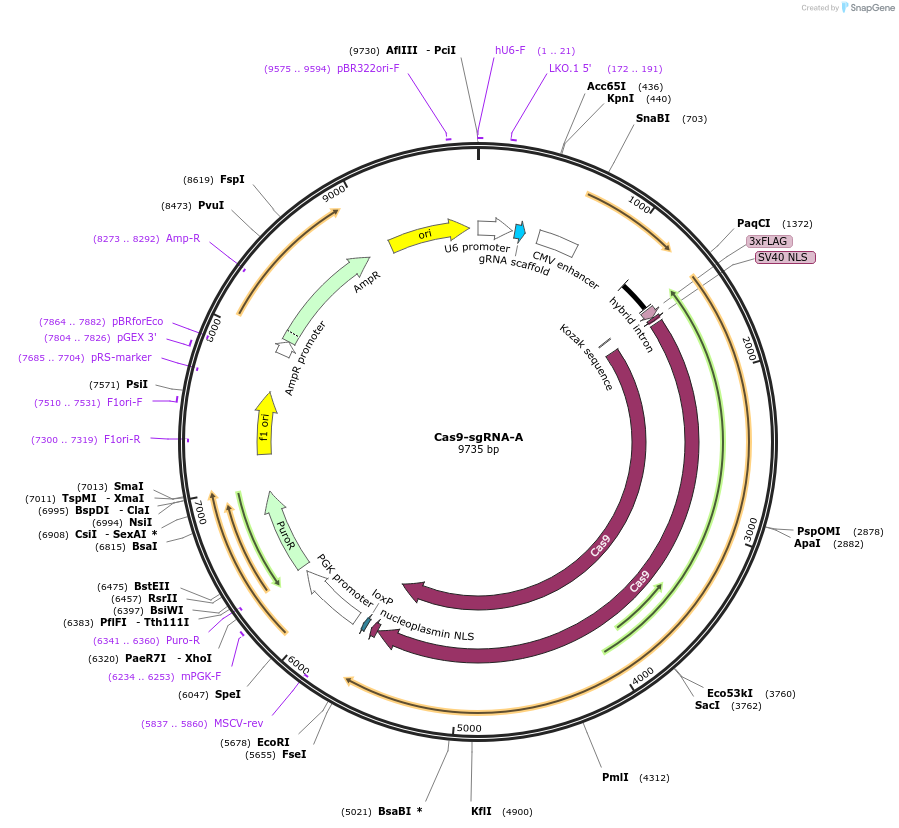
Cas9-sgRNA-A
Plasmid#149369PurposeExpresses SpCas9 and sgRNA targeting the lenti-CDDR reporterDepositorInsertsCas9
sgRNA-A
Puromycin resistance
UseCRISPRTags3xFLAG, Nucleoplasmin NLS, and SV40 NLSExpressionMammalianPromoterCBh, PGK, and U6Available SinceNov. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
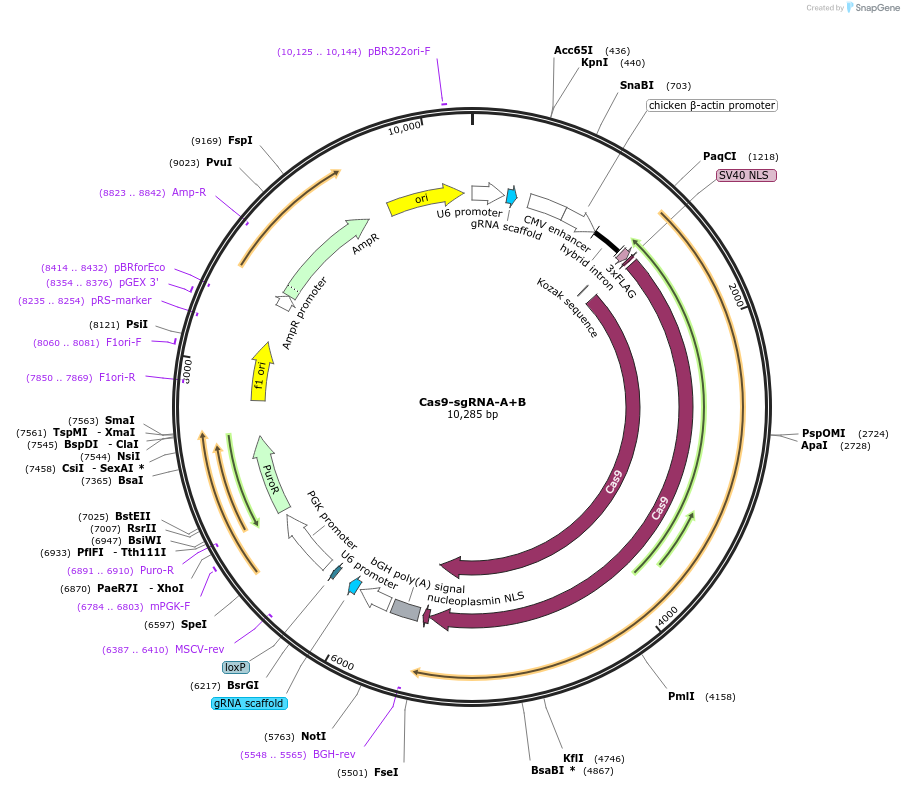
Cas9-sgRNA-A+B
Plasmid#149370PurposeExpresses SpCas9 and two sgRNA targeting the lenti-CDDR reporter at two distal sitesDepositorInsertsCas9
sgRNA-A
sgRNA-B
Puromycin resistance
UseCRISPRTags3xFLAG, Nucleoplasmin NLS, and SV40 NLSExpressionMammalianPromoterCBh, PGK, and U6Available SinceOct. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
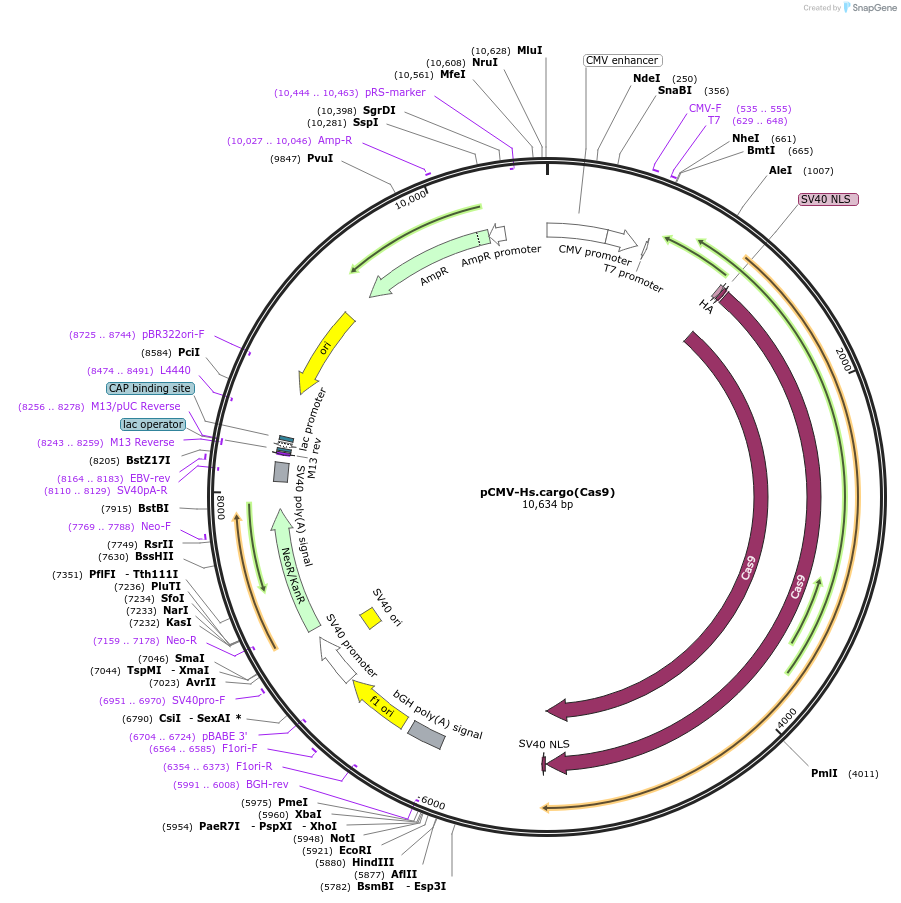
pCMV-Hs.cargo(Cas9)
Plasmid#174864PurposeExpresses human Peg10 cargoRNA encoding SpCas9DepositorInsertSpCas9 flanked by HsPeg10 UTRs
TagsHA-SV40 NLS and SV40 NLSExpressionMammalianPromoterCMVAvailable SinceSept. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
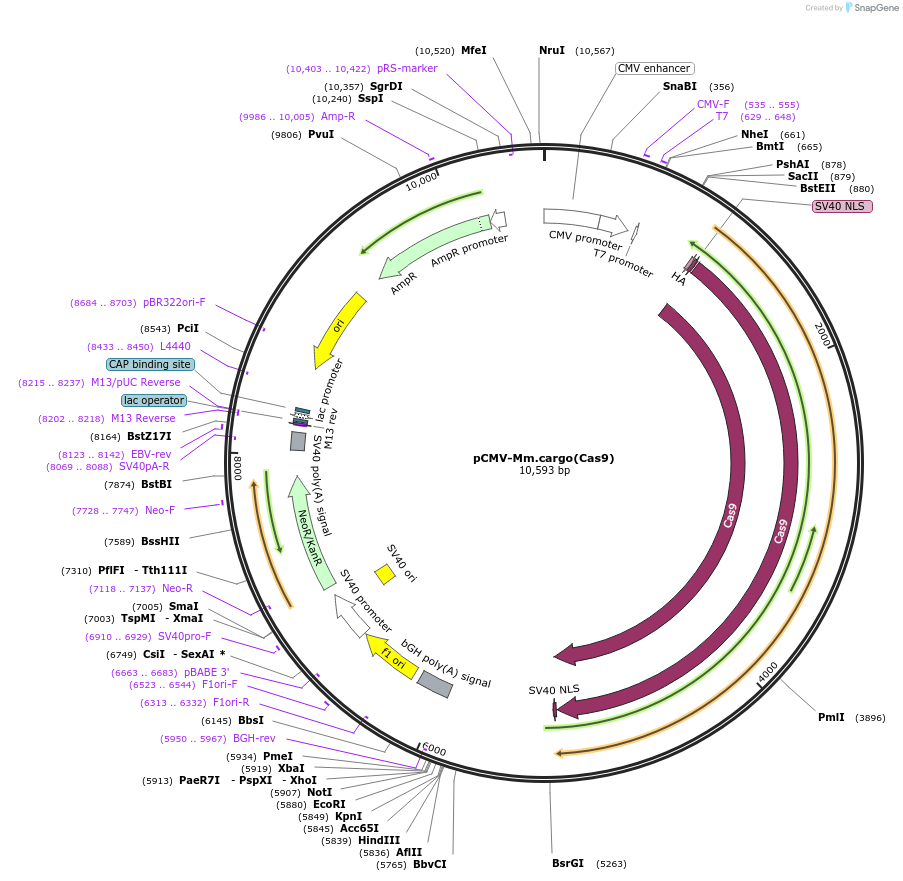
pCMV-Mm.cargo(Cas9)
Plasmid#174865PurposeExpresses mouse Peg10 cargoRNA encoding SpCas9DepositorInsertSpCas9 flanked by MmPeg10 UTRs
TagsHA-SV40 NLS and SV40 NLSExpressionMammalianPromoterCMVAvailable SinceSept. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
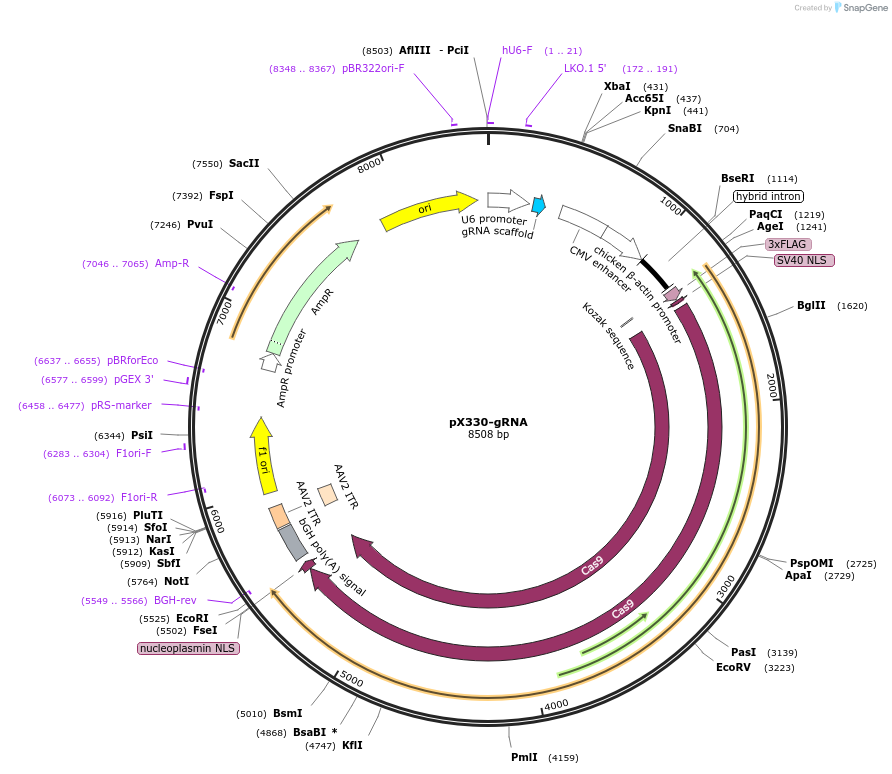
pX330-gRNA
Plasmid#158973PurposeA human codon-optimized SpCas9 and chimeric guide RNA (gRNA3) expression plasmid.DepositorInsertgRNA3
ExpressionMammalianPromoterU6Available SinceOct. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
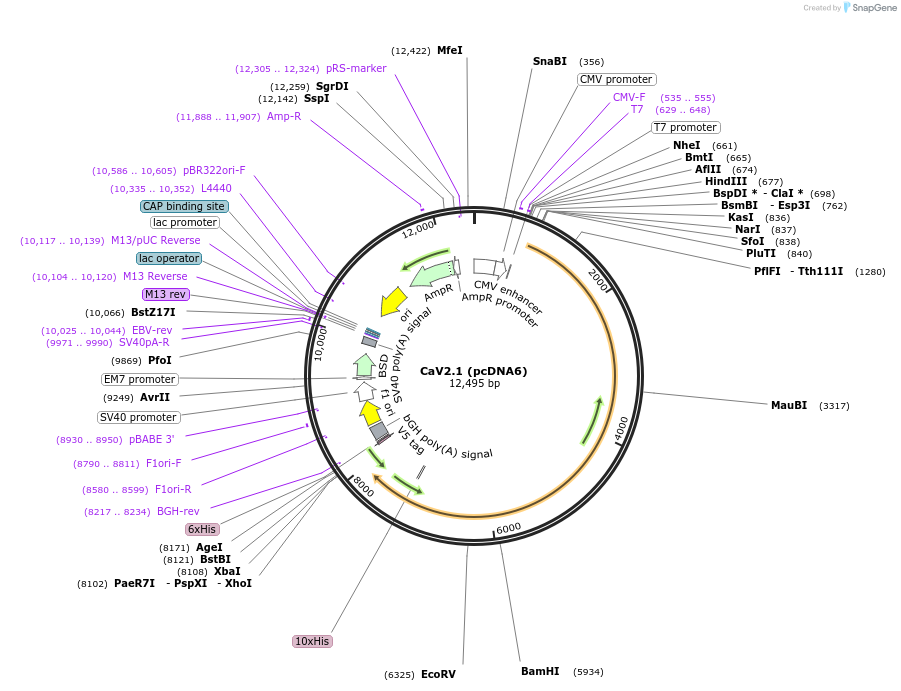
CaV2.1 (pcDNA6)
Plasmid#26578DepositorAvailable SinceNov. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
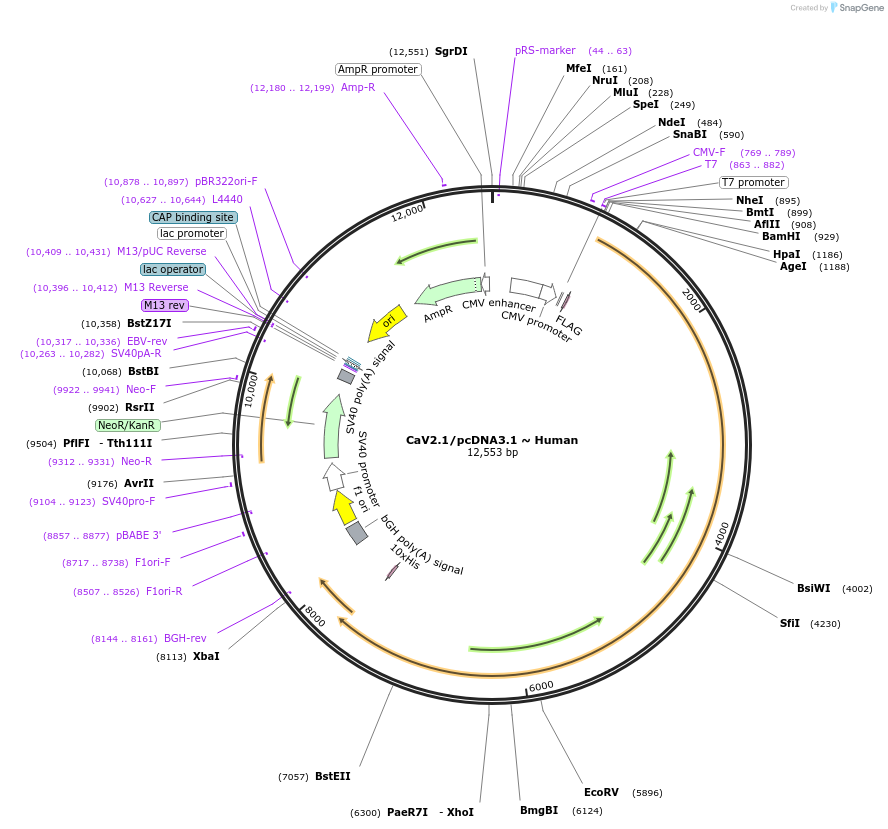
CaV2.1/pcDNA3.1 ~ Human
Plasmid#140575PurposeExpression in mammallian cells or oocytesDepositorAvailable SinceFeb. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
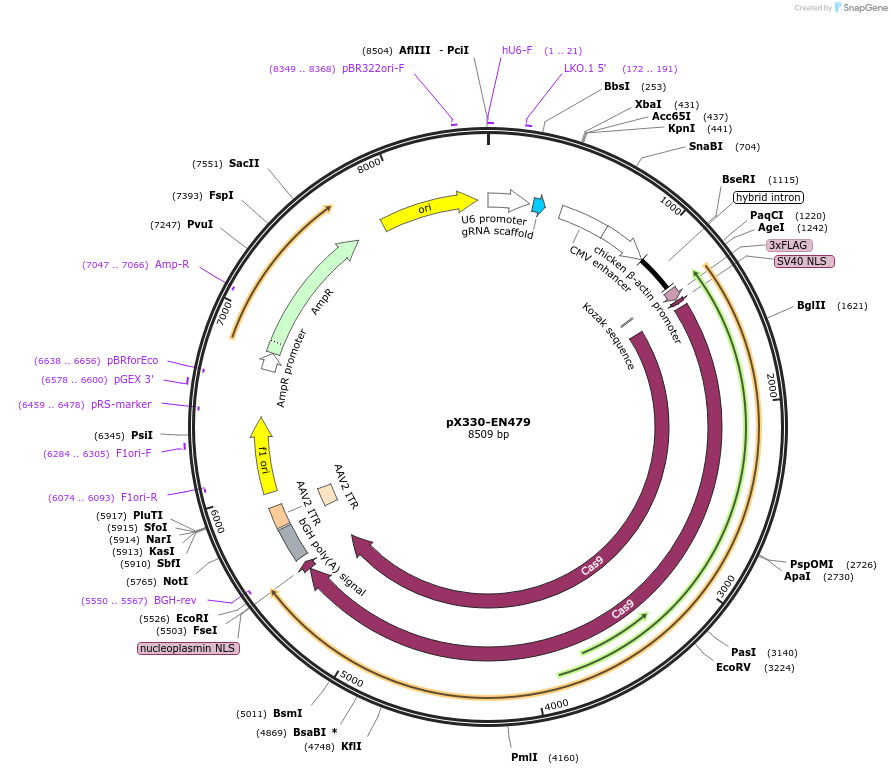
pX330-EN479
Plasmid#86234PurposeFor transient expression of spCas9-nuclease and a sgRNA targeting the mouse Rosa26 locusDepositorInsertspCas9-nickase and sgRNA against mouse rosa26 locus
UseMouse TargetingExpressionMammalianAvailable SinceMay 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
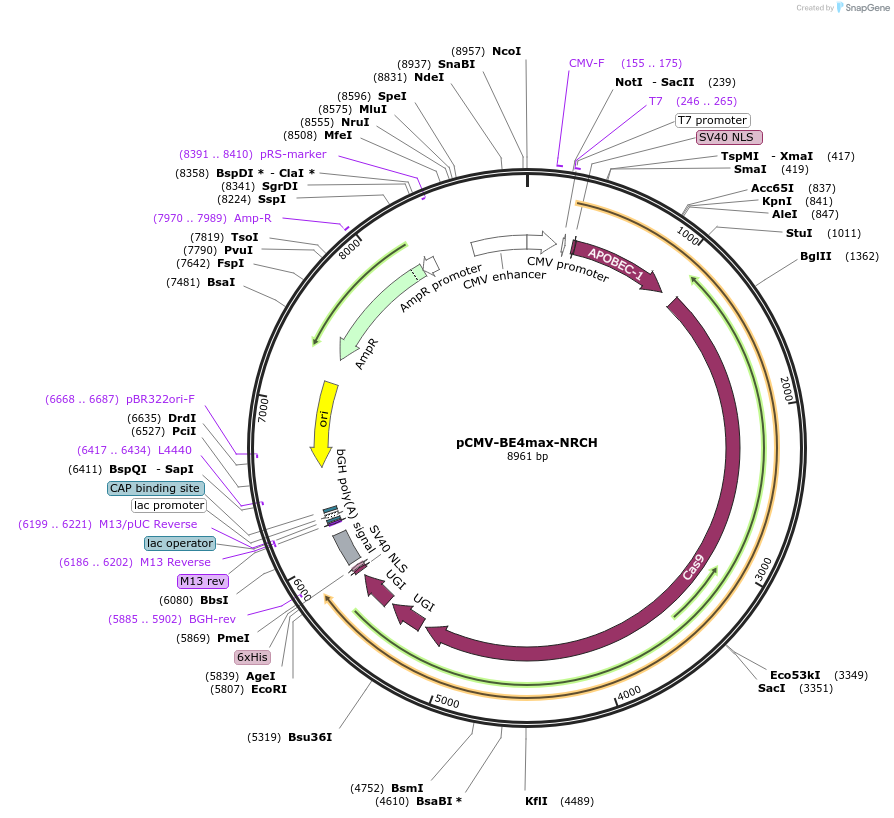
pCMV-BE4max-NRCH
Plasmid#136920PurposeExpresses BE4max-NRCH in mammalian cellsDepositorInsertBE4max-NRCH
UseCRISPRExpressionMammalianMutationsee manuscriptPromoterCMVAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
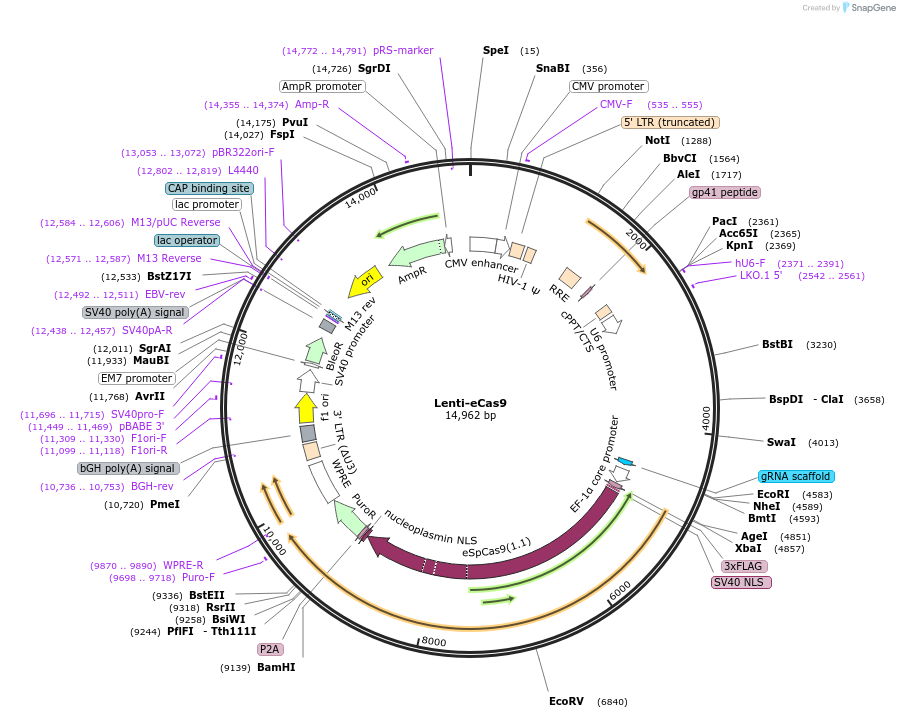
Lenti-eCas9
Plasmid#140237PurposeLentiviral vector for expression of high-specificity eSpCas9(1.1) and sgRNA scaffoldDepositorTypeEmpty backboneUseCRISPR and LentiviralTags3xFLAGExpressionMammalianPromoterEF-1a coreAvailable SinceMay 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
pePEmax-pPuro
Plasmid#215688PurposeCloning vector for Uni-Vector prime editing systemDepositorInsertPEmax
UseUnspecifiedPromoterEFS promoterAvailable SinceDec. 16, 2024AvailabilityAcademic Institutions and Nonprofits only