We narrowed to 13,664 results for: 109
-
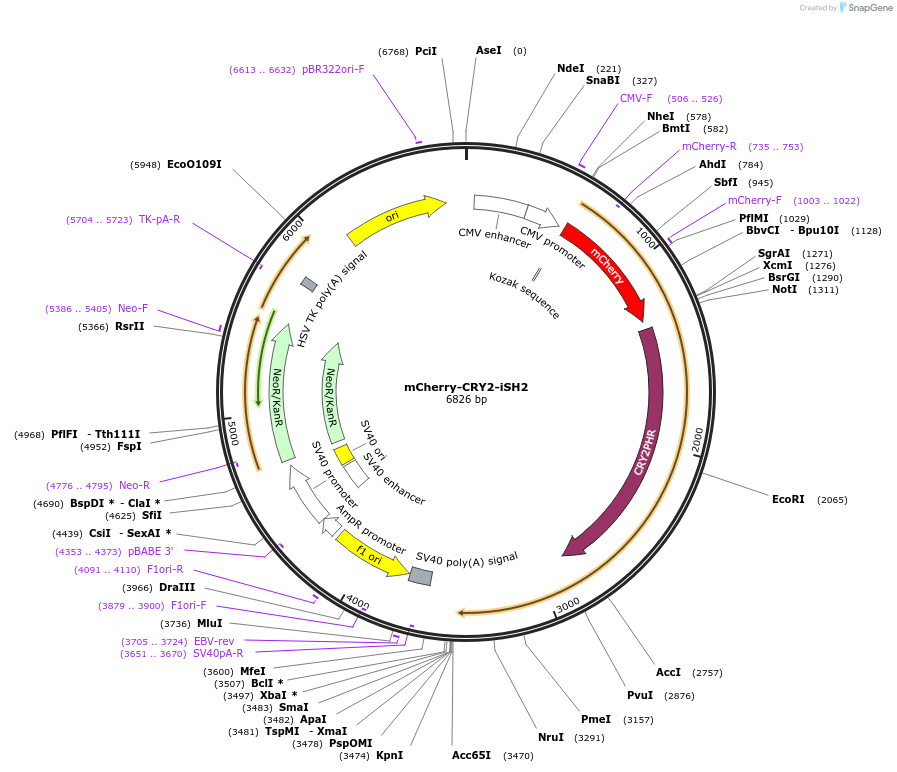
Plasmid#66839PurposeExpression of a light-inducible PI3-kinase, generated by fusing the iSH2 domain of human p85α (residues 420–615) to mCherry-CRY2. Binds endogenous PI3-kinase p110α catalytic subunits constitutively.DepositorInsertiSH2
TagsmCherry-CRY2ExpressionMammalianPromoterCMVAvailable SinceJune 22, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRK5 FLAG metap2
Plasmid#32004DepositorAvailable SinceSept. 27, 2011AvailabilityAcademic Institutions and Nonprofits only -
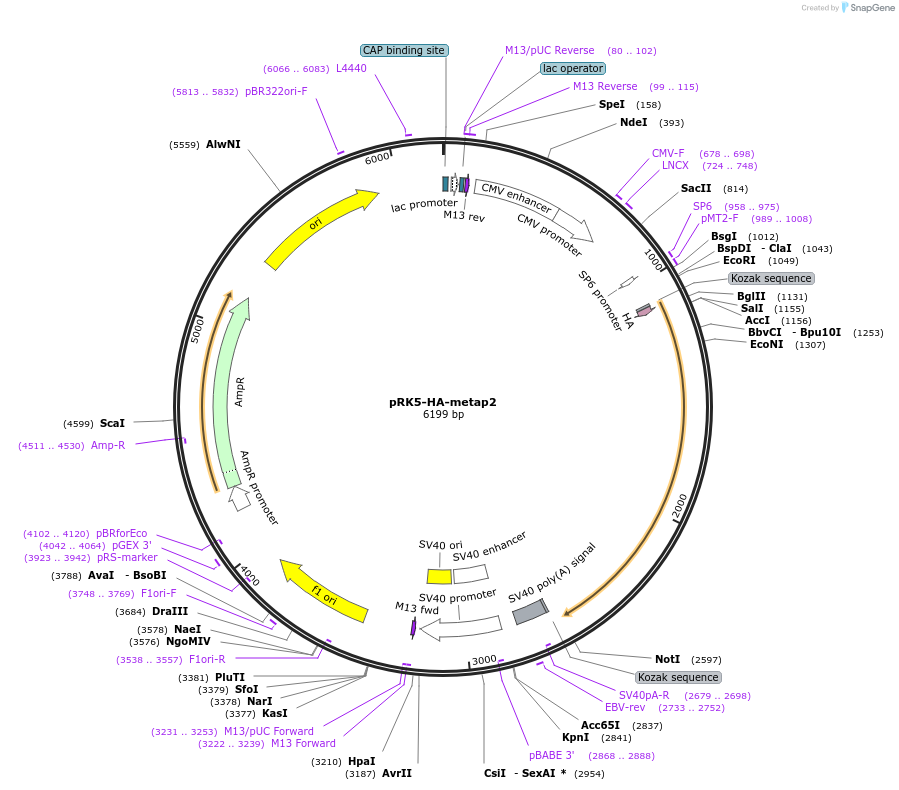
pRK5-HA-metap2
Plasmid#100512PurposeExpresses HA tagged metap2 in mammalian cellsDepositorAvailable SinceNov. 10, 2017AvailabilityAcademic Institutions and Nonprofits only -
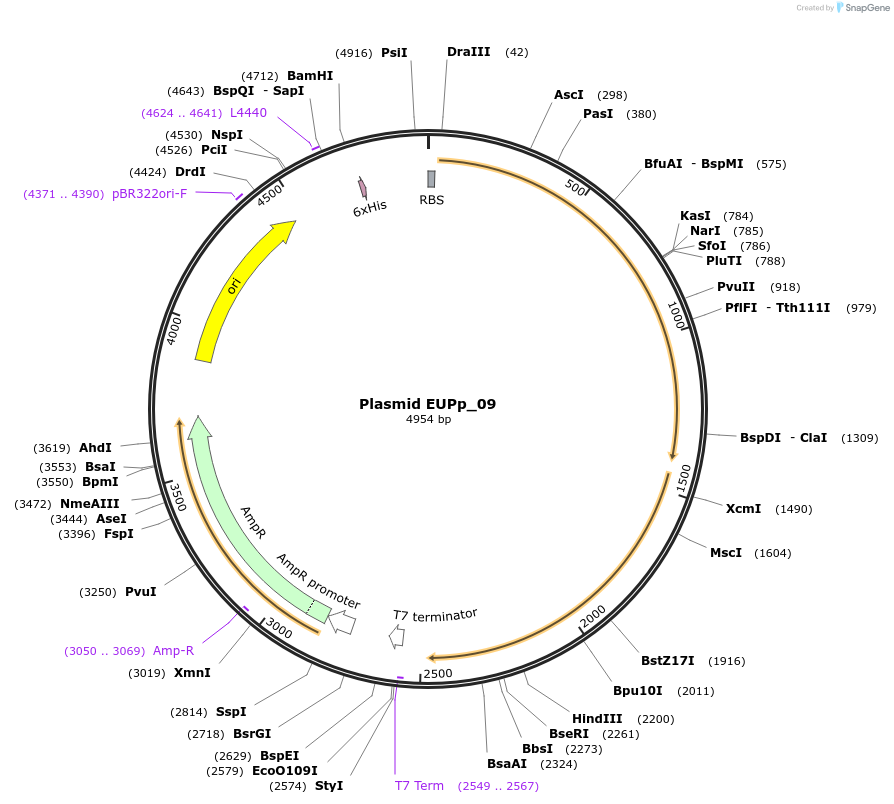
Plasmid EUPp_09
Plasmid#157745PurposePgyrA_ada _adh2 _pMB1_ampR (for expressing two enzymes (Ada and Adh2) to allow E. coli to consume ethanol)DepositorInsertsExpressionBacterialPromoterPgyrAAvailable SinceJune 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
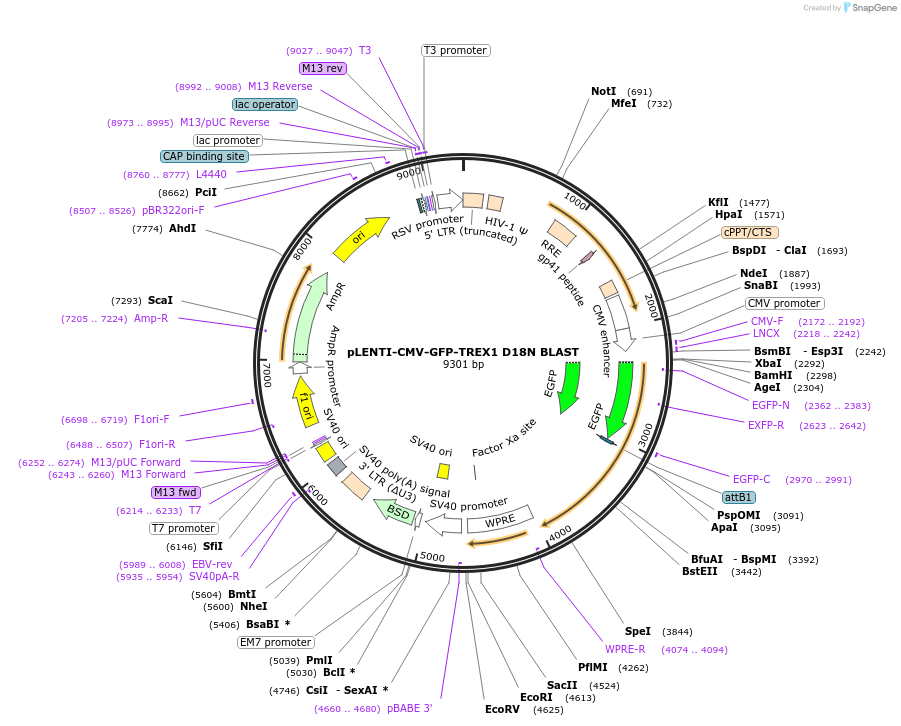
pLENTI-CMV-GFP-TREX1 D18N BLAST
Plasmid#164225Purposelentiviral plasmid for GFP-hTREX1 D18N (catalytically-deficient)DepositorAvailable SinceMarch 31, 2021AvailabilityAcademic Institutions and Nonprofits only -
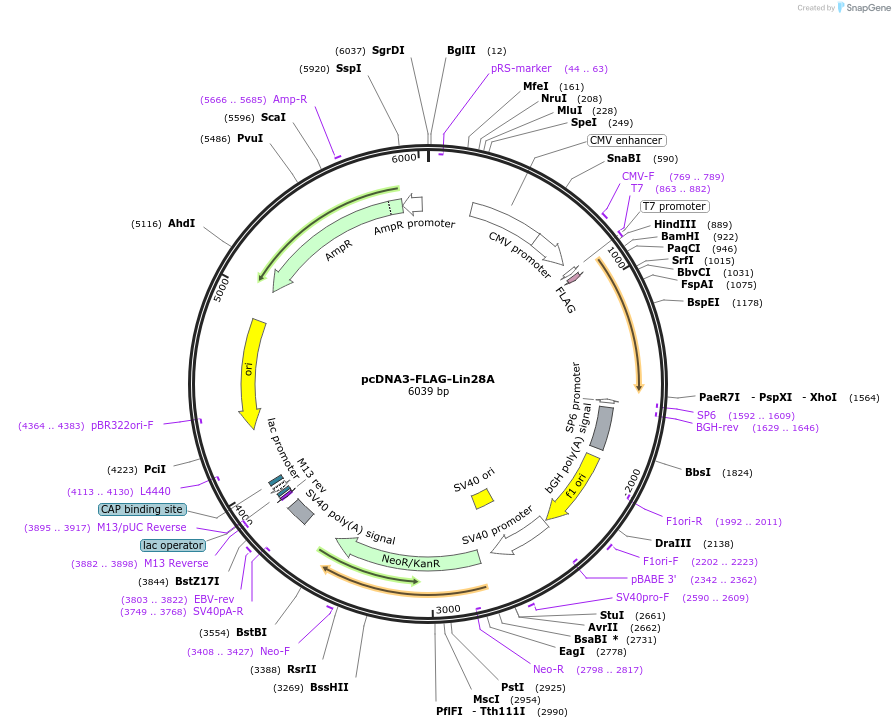
pcDNA3-FLAG-Lin28A
Plasmid#51371DepositorAvailable SinceMarch 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
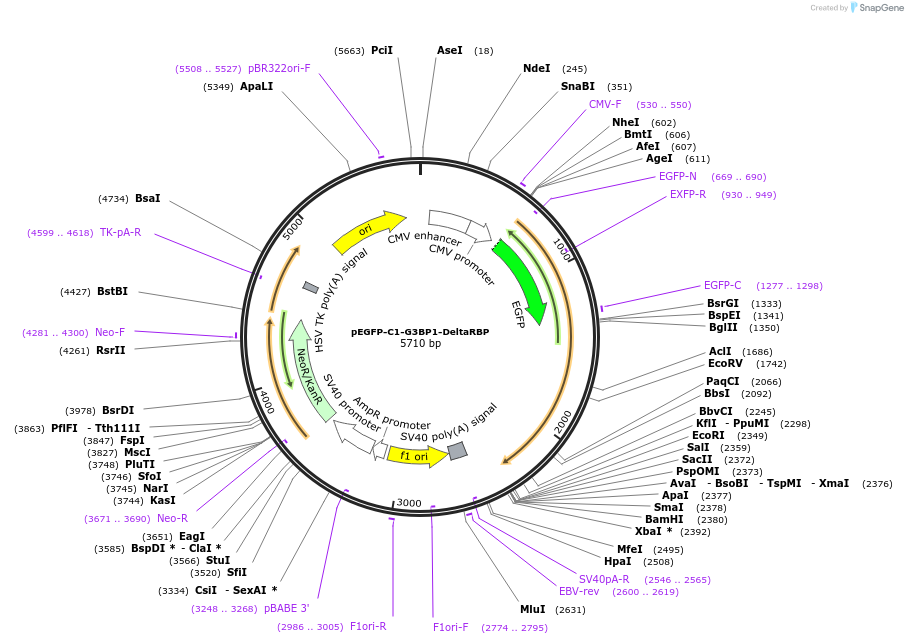
pEGFP-C1-G3BP1-DeltaRBP
Plasmid#135999PurposeG3BP1 with RNA binding domains deleted inserted with GFP tagged on the N-terminusDepositorInsertG3BP1 (G3BP1 Human)
TagsEGFPExpressionMammalianMutationC-term of G3BP1 Deleted for Delta RBPAvailable SinceFeb. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
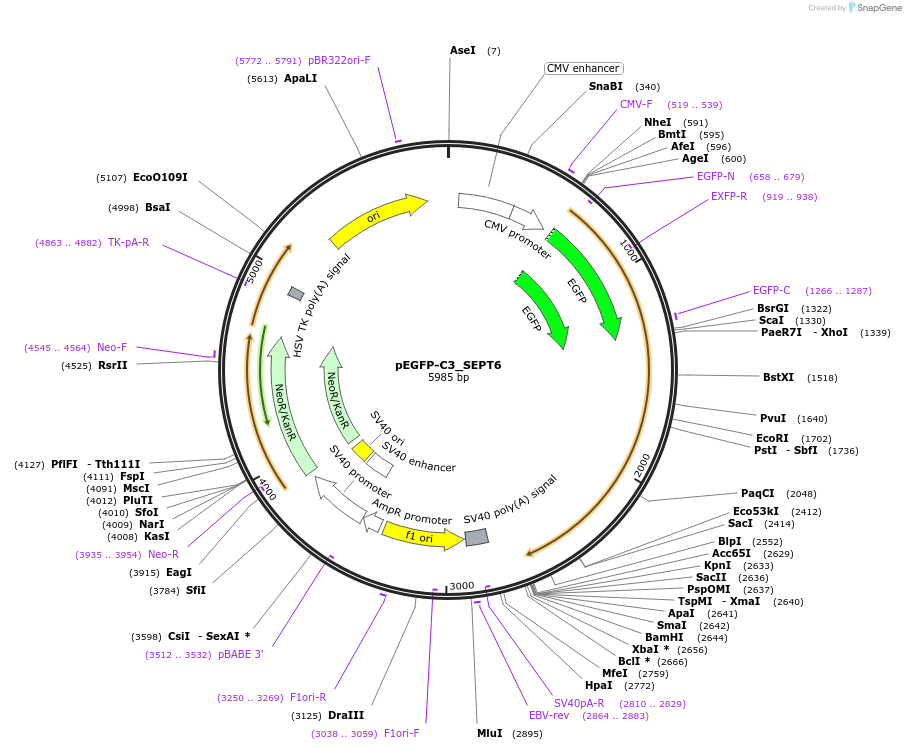
pEGFP-C3_SEPT6
Plasmid#71583Purposemammalian expression of human SEPT6 with a GFP fusionDepositorAvailable SinceDec. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
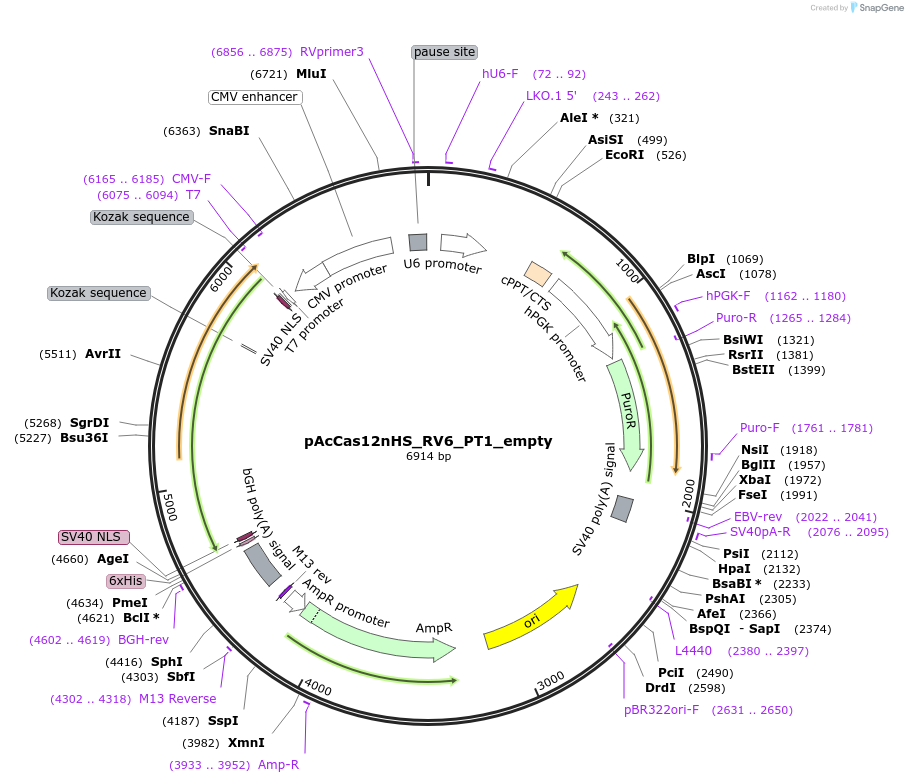
pAcCas12nHS_RV6_PT1_empty
Plasmid#203810PurposePlasmid containing a CMV-driven NLS-AcCas12n-PT1-NLS(human-codonoptimized)cassette and a U6-driven sgRNA_V6 cassette(containing BsaI sites for spacer insertion). Carbenicillin resistance.DepositorInsertAcCas12nHS_RV6_PT1
UseCRISPRPromoterCMVAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
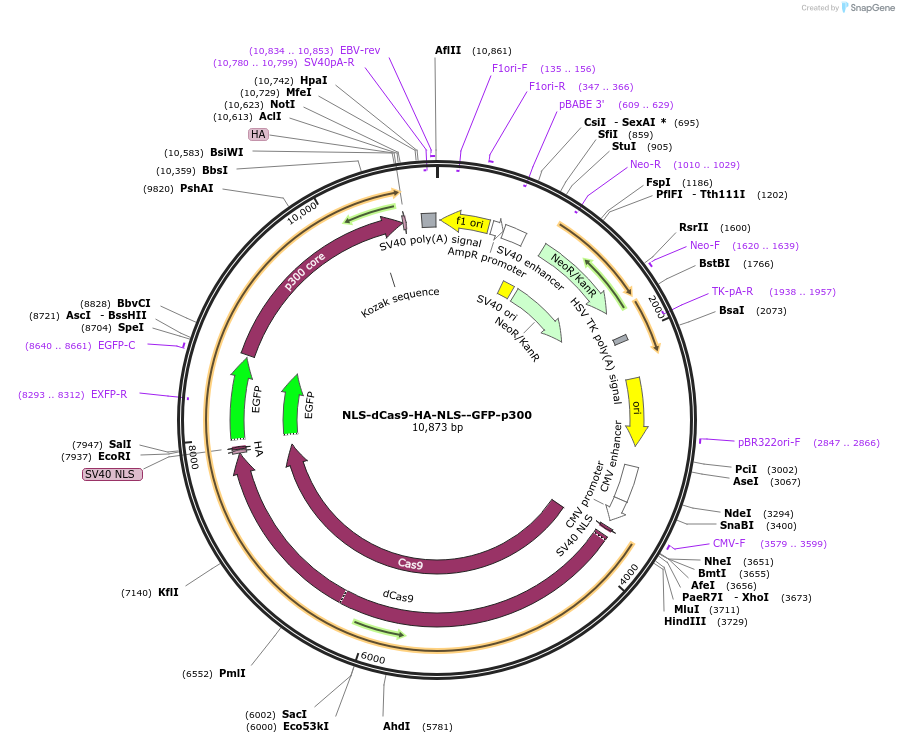
NLS-dCas9-HA-NLS--GFP-p300
Plasmid#183925Purposep300 HAT core domain coupled to catalytically dead dCas9 for localization; labeled with GFPDepositorInsertdCas9-GFP-p300
UseCRISPRTagsNLSExpressionMammalianMutationnonePromoterCMVAvailable SinceJuly 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
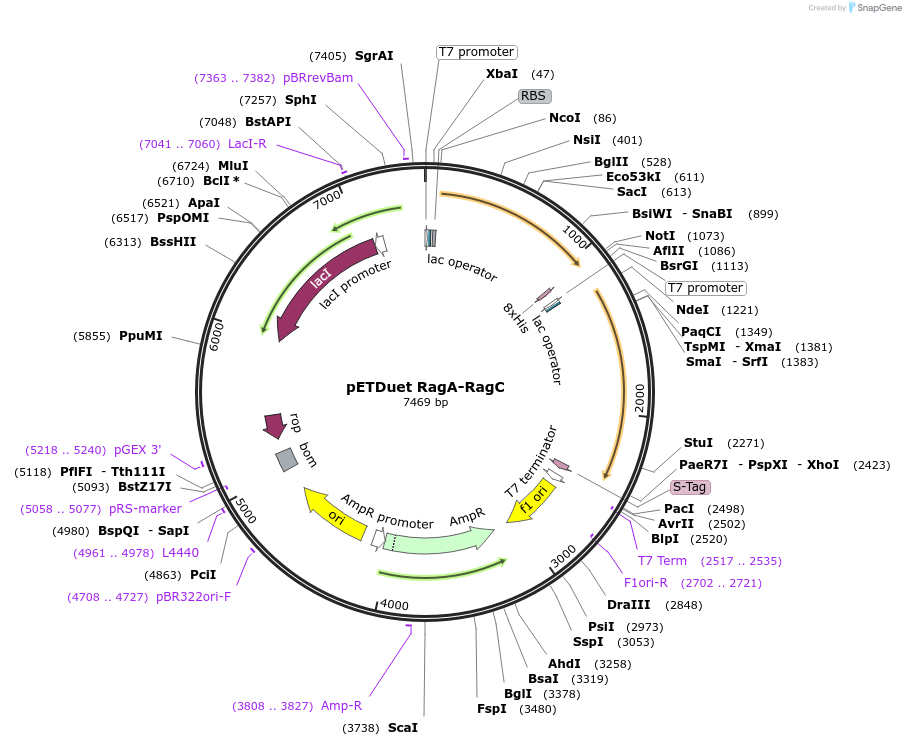
pETDuet RagA-RagC
Plasmid#99641PurposeBacterial expression of wildtype Rag GTPase dimerDepositorAvailable SinceDec. 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
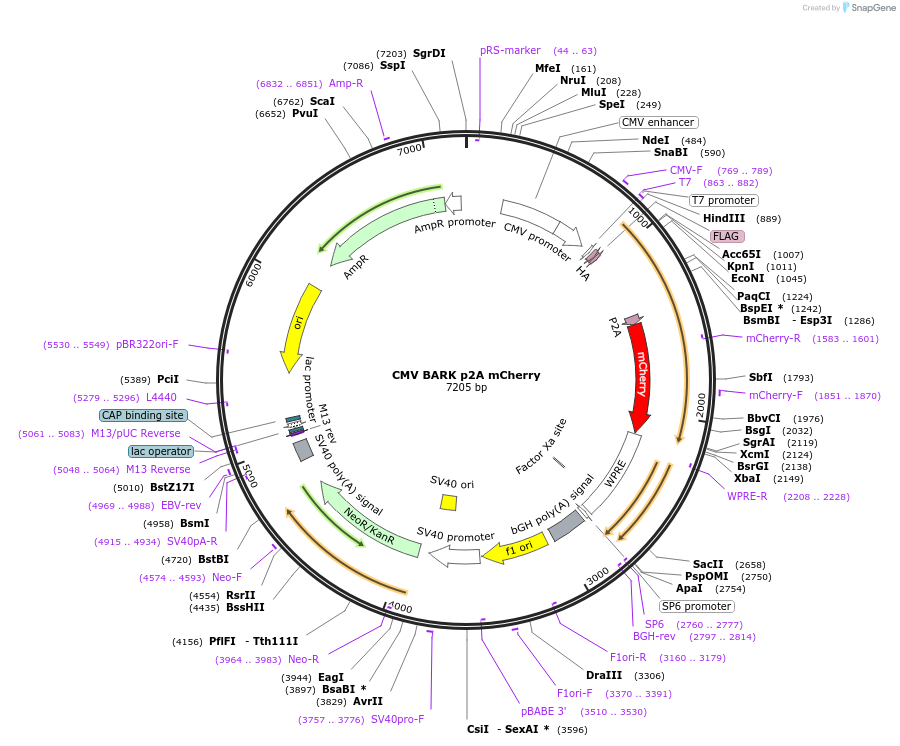
CMV BARK p2A mCherry
Plasmid#117689PurposeCMV-iBARK-p2A-mCherry: Mammalian expression vector for Gaq silencingDepositorInsertBARKrgs peptide
TagsHA / FLAGExpressionMammalianAvailable SinceMay 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
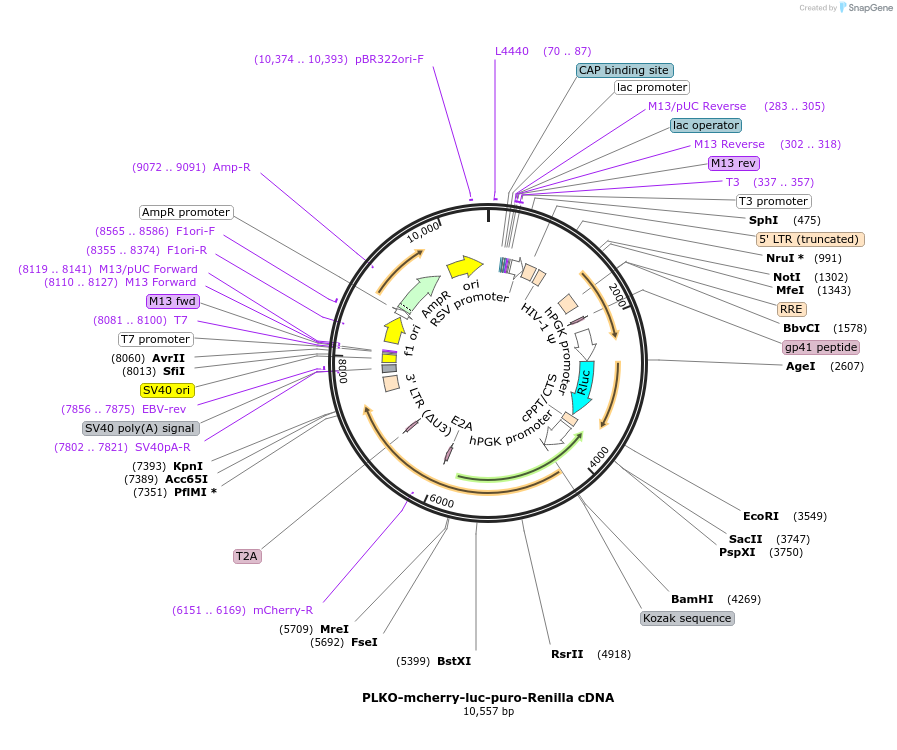
PLKO-mcherry-luc-puro-Renilla cDNA
Plasmid#29783DepositorInsertRenilla
UseLentiviralExpressionMammalianAvailable SinceJune 1, 2011AvailabilityAcademic Institutions and Nonprofits only -
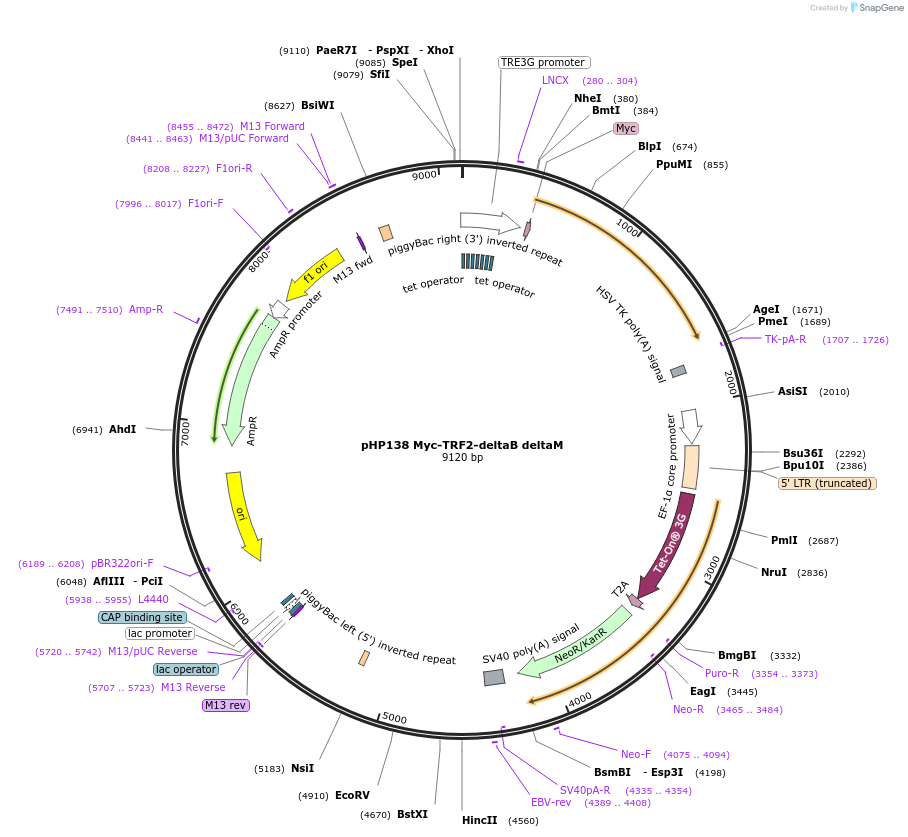
pHP138 Myc-TRF2-deltaB deltaM
Plasmid#164255Purposefor generation of mammalian cells with inducible expression of dominant negative TRF2 for induction of telomere crisis used together with any PiggyBac transposase plasmidDepositorInsertTRF2
TagsMycExpressionMammalianMutationdeletion of amino acids-86 and 496-542PromoterTetracycline inducible promotorAvailable SinceApril 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
-
pET28a-MH6-RspWYL1
Plasmid#108306PurposeExpresses E. coli codon optimized RspWYL1 in the pET28a backbone.DepositorInsertRspWYL1
UseCRISPRTagsmH6 tagExpressionBacterialPromoterlacAvailable SinceApril 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
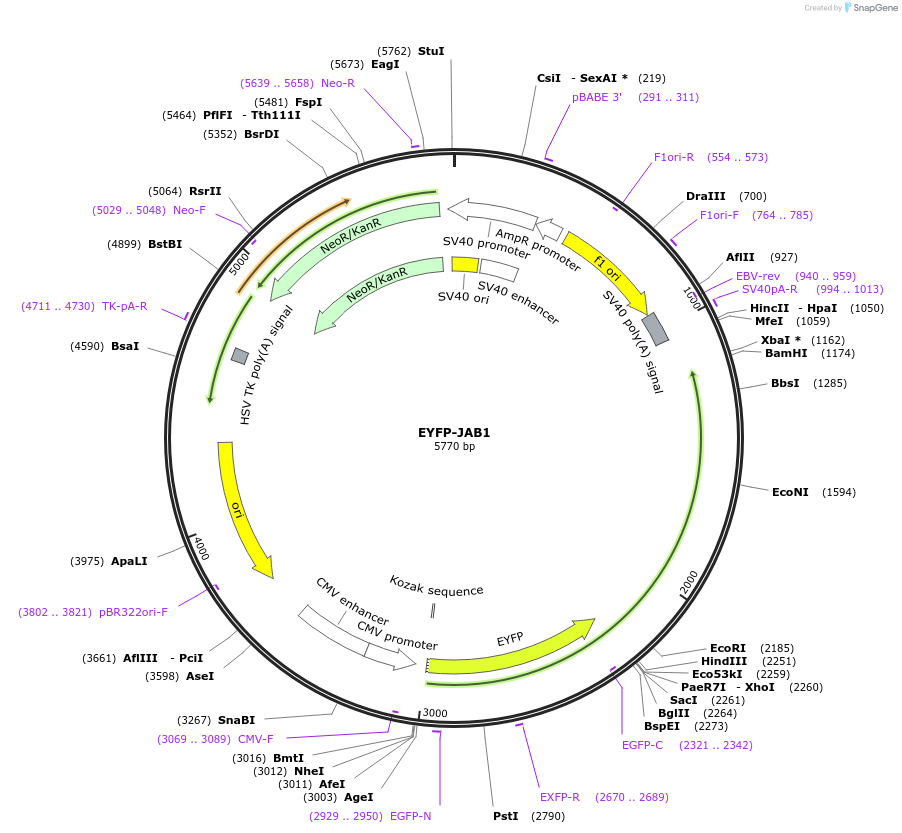
EYFP-JAB1
Plasmid#111213Purposefluorescent fusion proteinDepositorAvailable SinceSept. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
pCDNA3 MKK7b1(K149A)
Plasmid#14674DepositorAvailable SinceJuly 25, 2007AvailabilityAcademic Institutions and Nonprofits only -
GFP-Dynamin 2 K44A
Plasmid#22301DepositorAvailable SinceDec. 10, 2009AvailabilityAcademic Institutions and Nonprofits only -
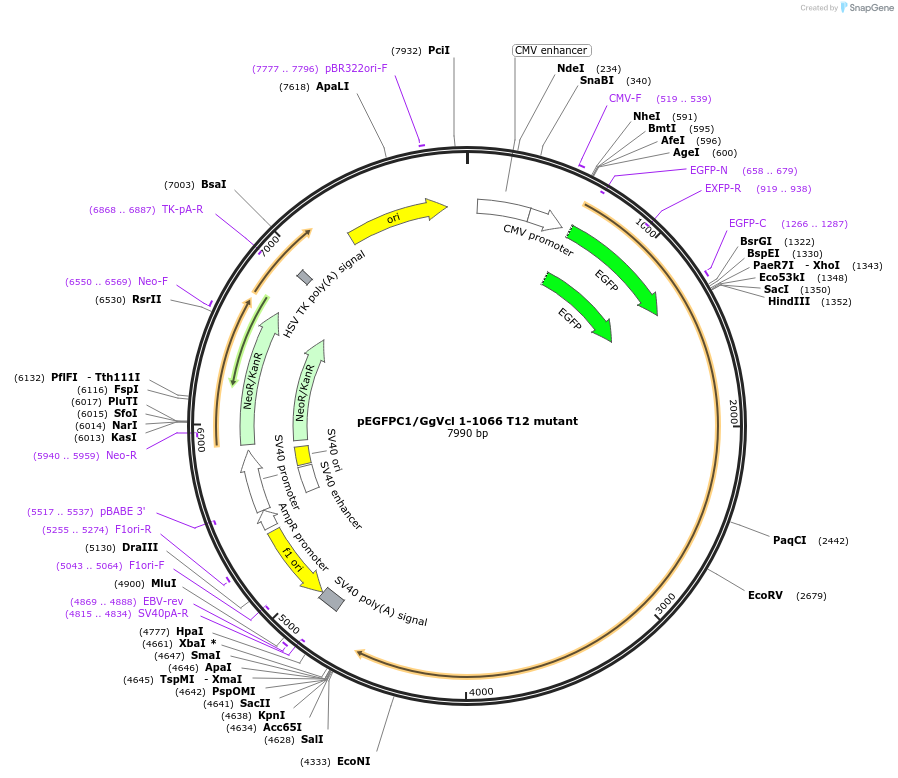
pEGFPC1/GgVcl 1-1066 T12 mutant
Plasmid#46266DepositorInsertVcl T12 mutant (VCL Chicken)
TagsEGFPExpressionMammalianMutationT12 mutant (D974;K975;R976; R978A) autoinhibition…PromoterCMVAvailable SinceJuly 11, 2013AvailabilityAcademic Institutions and Nonprofits only