We narrowed to 17,798 results for: bon
-
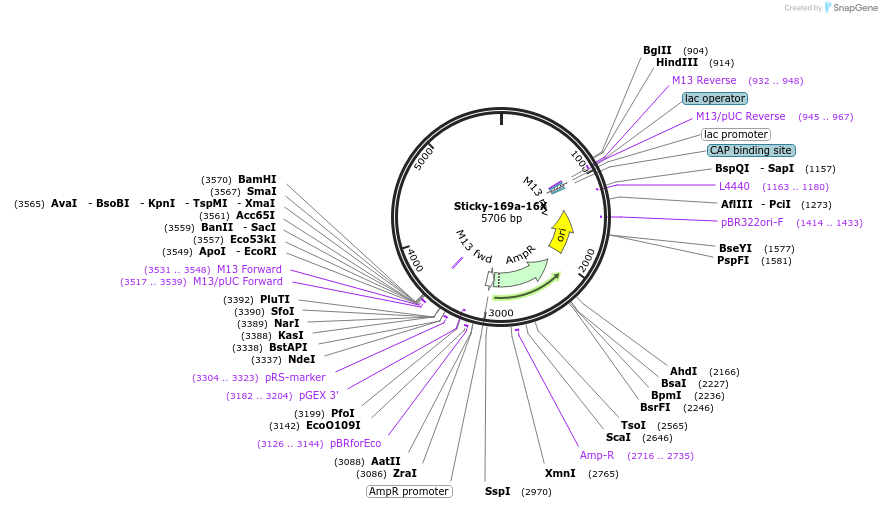
Plasmid#164495PurposeDNA fragment production for nucleosome preparationDepositorTypeEmpty backboneUseBacterial vector for dna productionAvailable SinceApril 22, 2021AvailabilityAcademic Institutions and Nonprofits only
-
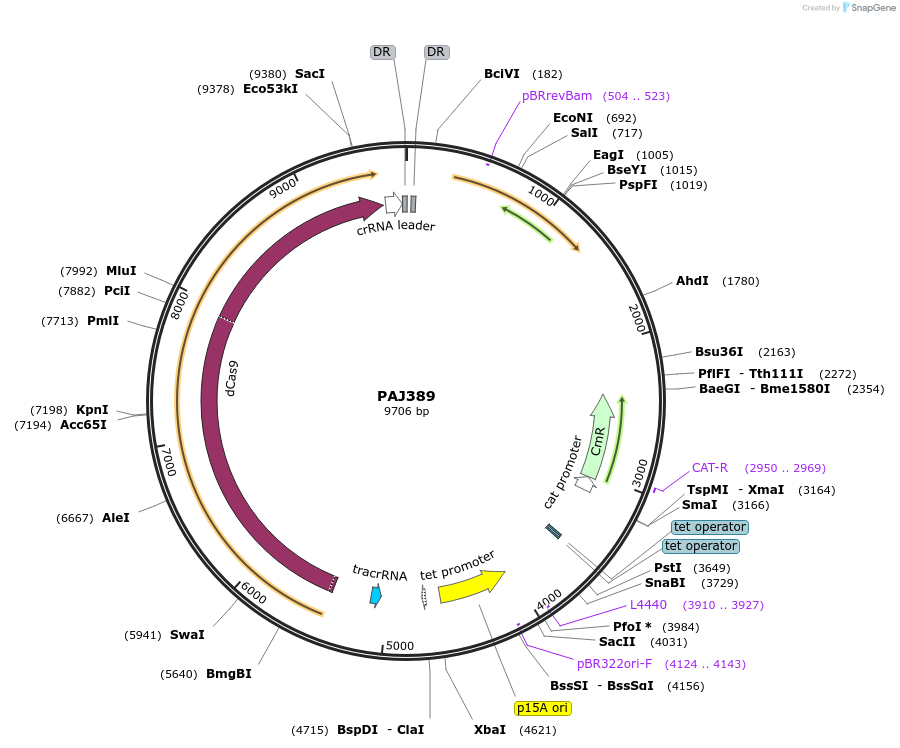
PAJ389
Plasmid#104147PurposedCas9 plasmid expressing 2A sRNA triggerDepositorTypeEmpty backboneUseCRISPRExpressionBacterialPromoterpLtetO-1Available SinceFeb. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
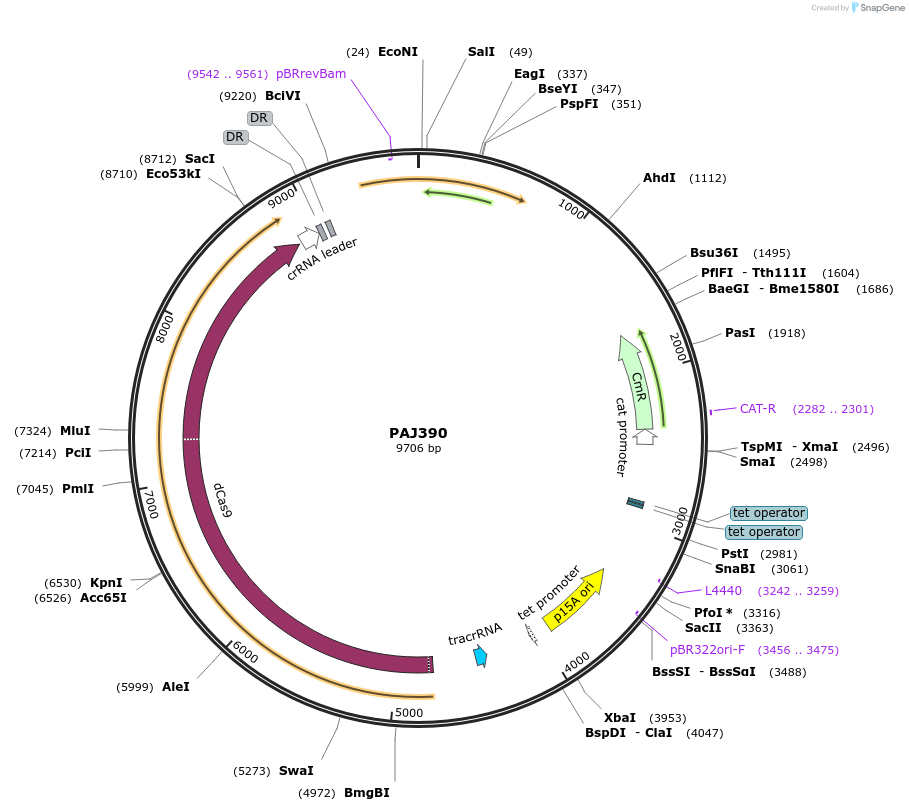
PAJ390
Plasmid#104148PurposedCas9 plasmid expressing 3A sRNA triggerDepositorTypeEmpty backboneUseCRISPRExpressionBacterialPromoterpLtetO-1Available SinceFeb. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
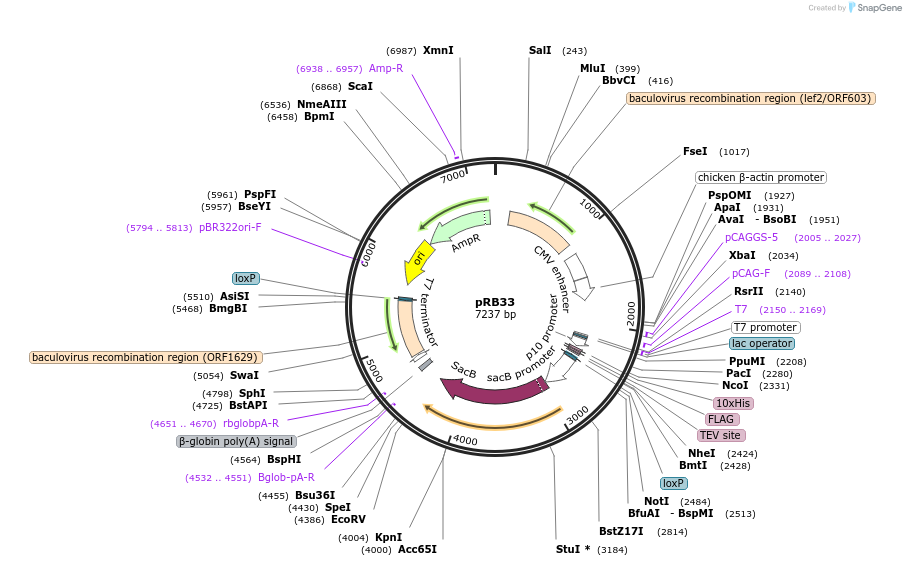
pRB33
Plasmid#165482PurposepTriEx based vector for protein expression in mammalian cells for targets fused with N-terminal Flag and HisDepositorTypeEmpty backboneTags10xHis-FLAG-TEVExpressionMammalianAvailable SinceFeb. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
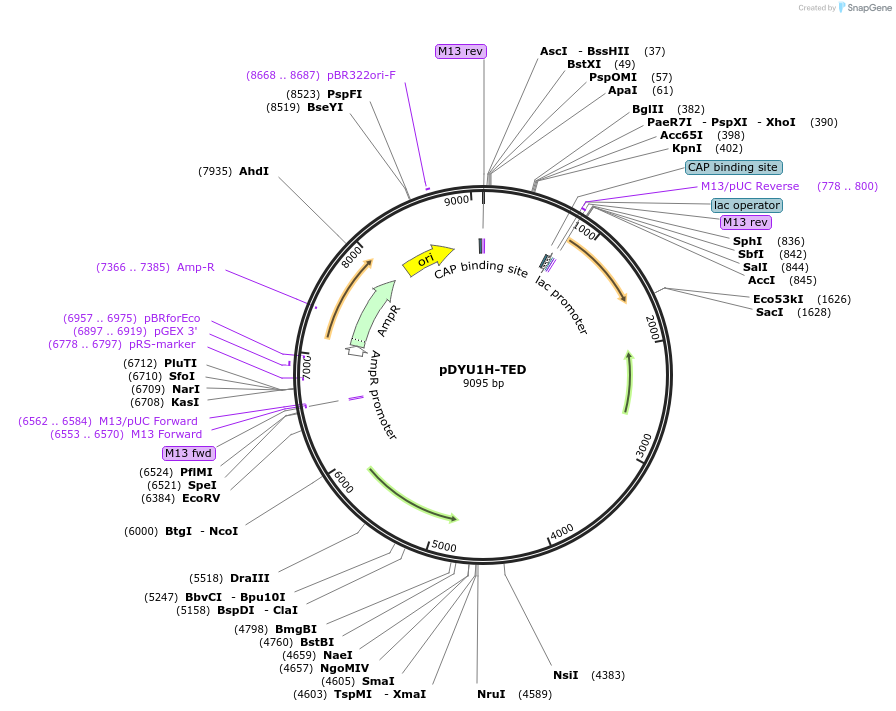
pDYU1H-TED
Plasmid#154300PurposeA expression vector in Dictyostelium discoideum containing the Hygromycin B resistant gene and the TED module for GFP-negative screeningDepositorTypeEmpty backboneUseDictyostelium expressionPromoteractin15Available SinceJan. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
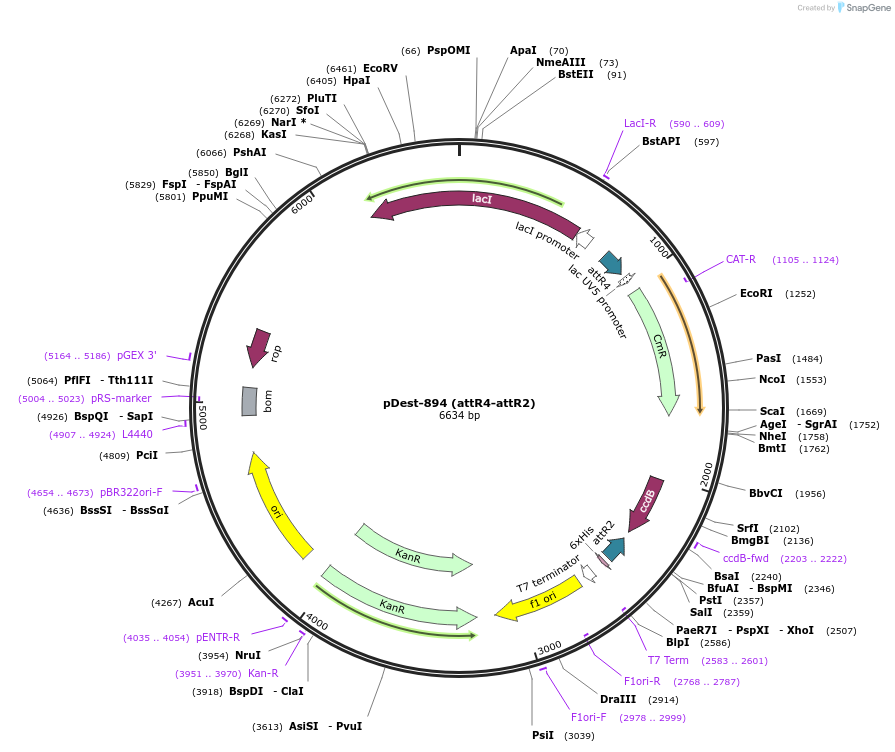
pDest-894 (attR4-attR2)
Plasmid#161893PurposeMultisite Gateway (attR4-attR2) vector for expression of multiple proteins in E. coliDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJan. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
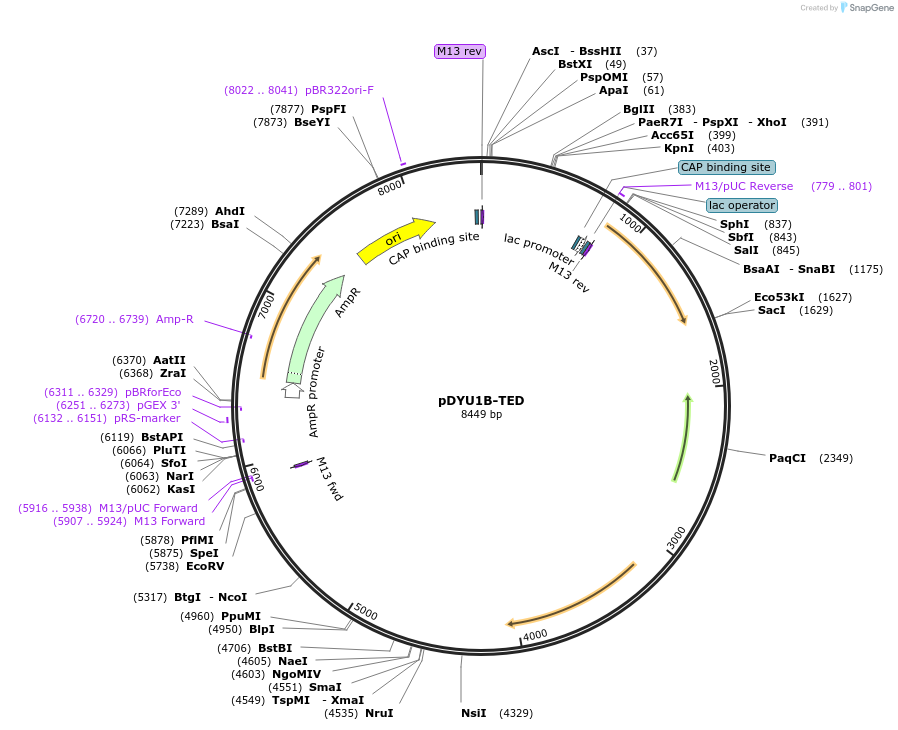
pDYU1B-TED
Plasmid#154299PurposeA expression vector in Dictyostelium discoideum containing the Blasticidin S resistant gene and the TED module for GFP-negative screeningDepositorTypeEmpty backboneUseDictyostelium expressionPromoteractin15Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
pEPMY1CB0001
Plasmid#162317PurposeAcceptor for golden gate assembly with BsaI overhangs GCTT-CCAT. Generates plasmids for T7-driven expression in E. coli. Contains mRFP1 reporter.DepositorTypeEmpty backboneExpressionBacterialAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
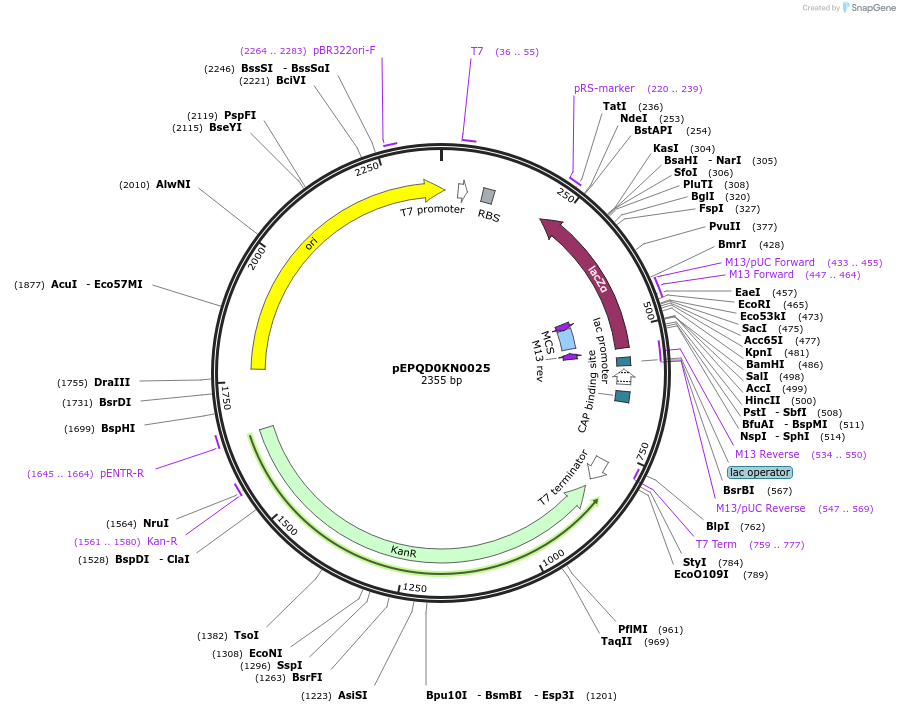
pEPQD0KN0025
Plasmid#162283PurposeAcceptor for golden gate assembly with BsaI overhangs GCTT-AATG. Generates plasmids for T7-driven E. coli cell-free protein synthesisDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
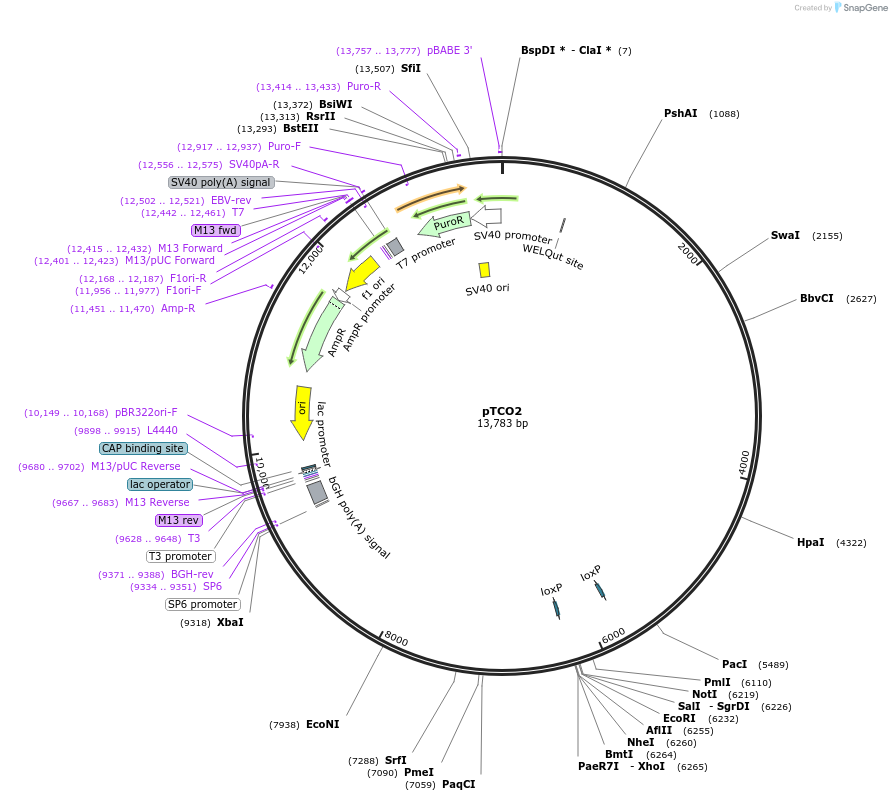
pTCO2
Plasmid#149428Purposecloning vector with regulatory regions of the murine Tdg geneDepositorTypeEmpty backboneUseCre/LoxExpressionMammalianPromotermurine Tdg promoterAvailable SinceSept. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
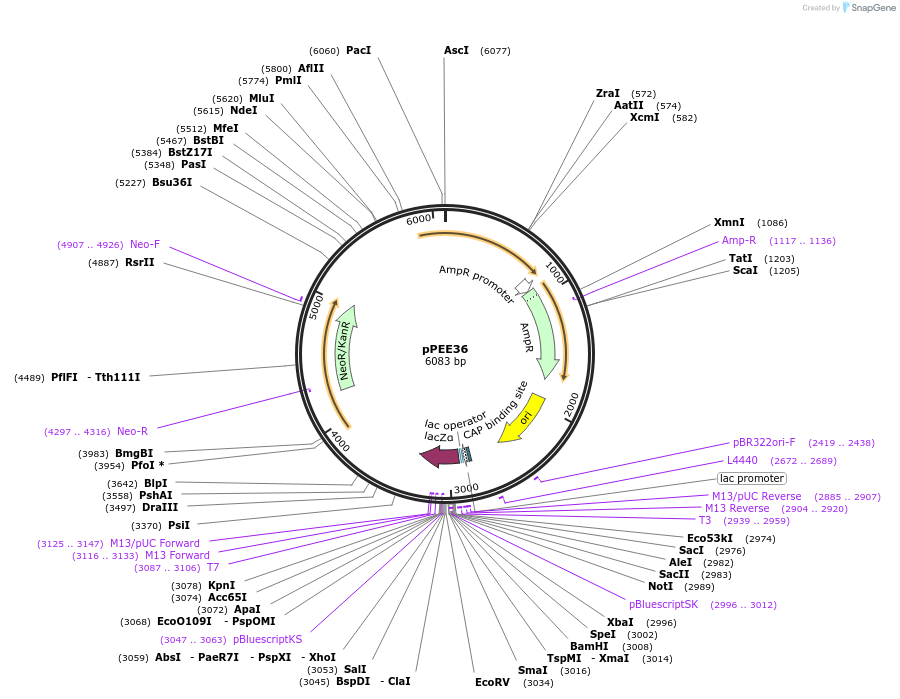
pPEE36
Plasmid#128376PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceApril 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
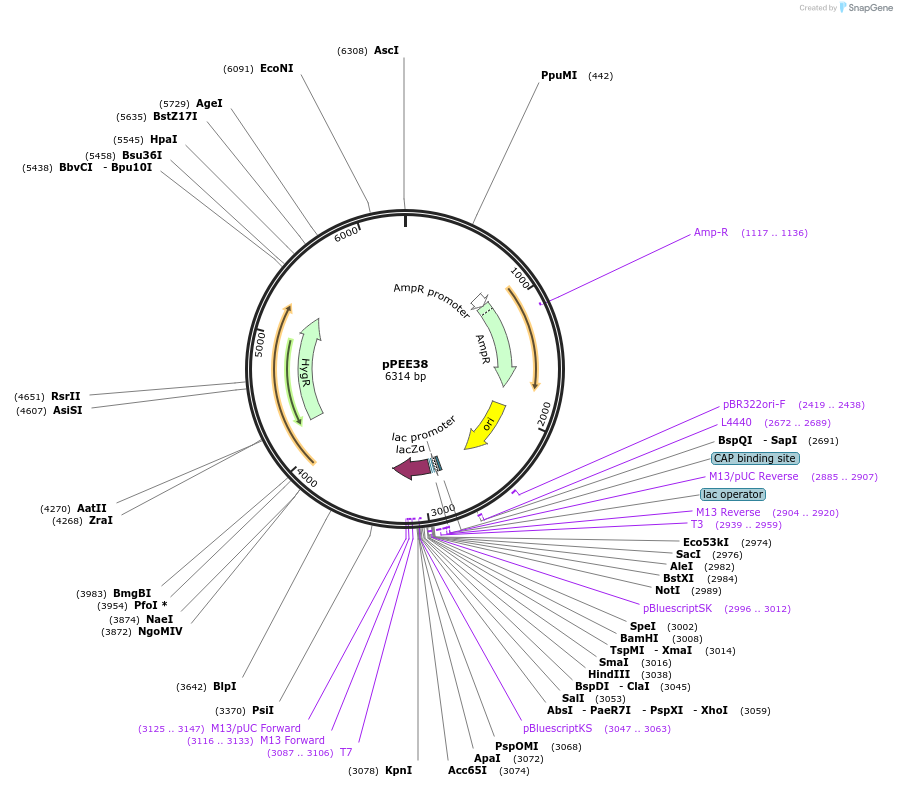
pPEE38
Plasmid#128378PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
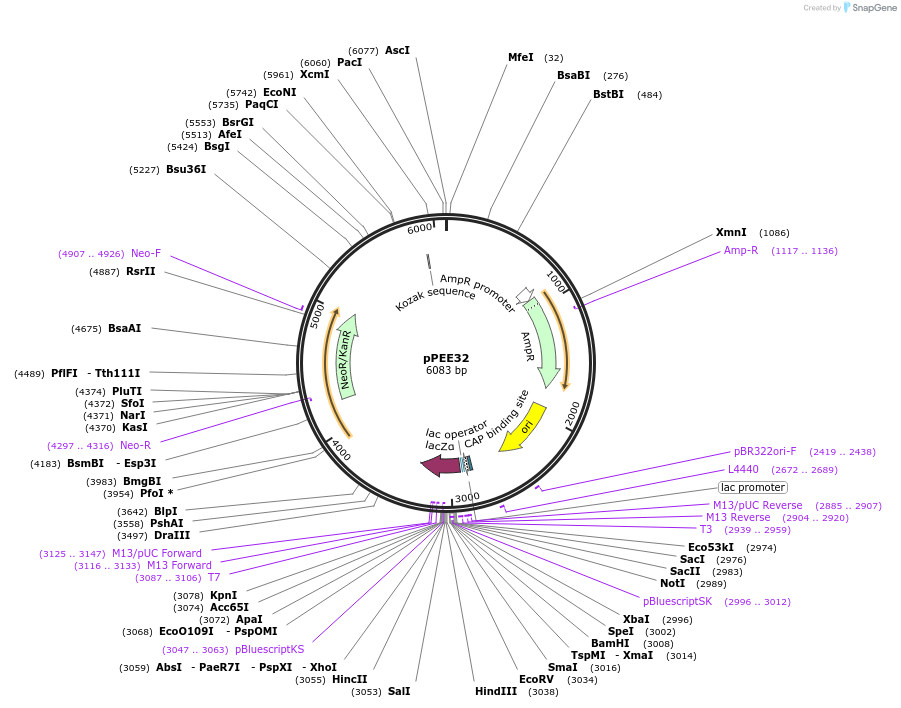
pPEE32
Plasmid#128372PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
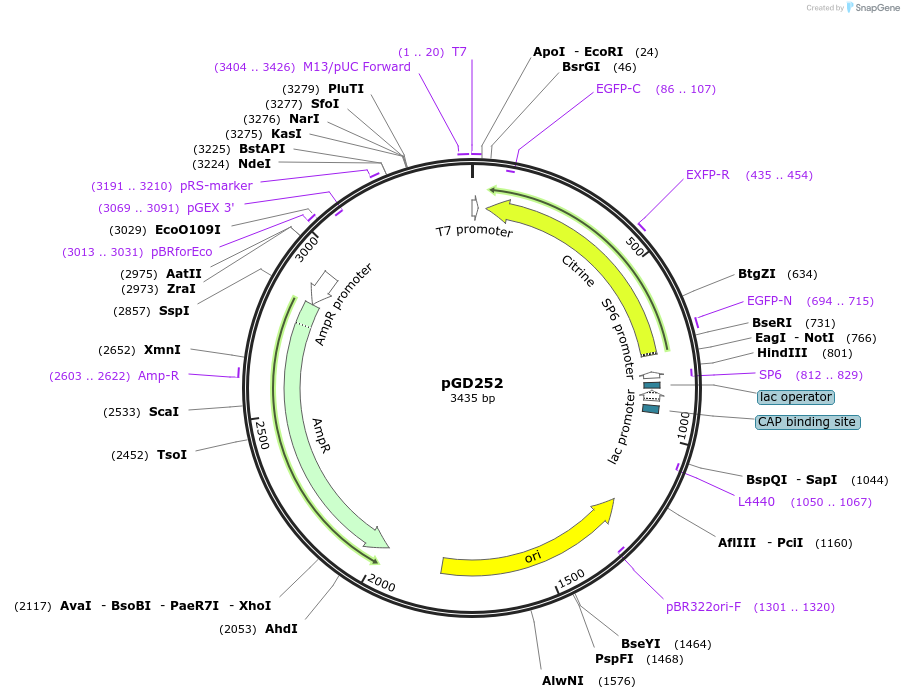
pGD252
Plasmid#132373Purposecontains the sequence of TSapphire for C-term fusions in the pGGD000 backbone (GreenGate)DepositorInsertlinker-TSapphire
UseGreengate entry plasmidAvailable SinceOct. 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
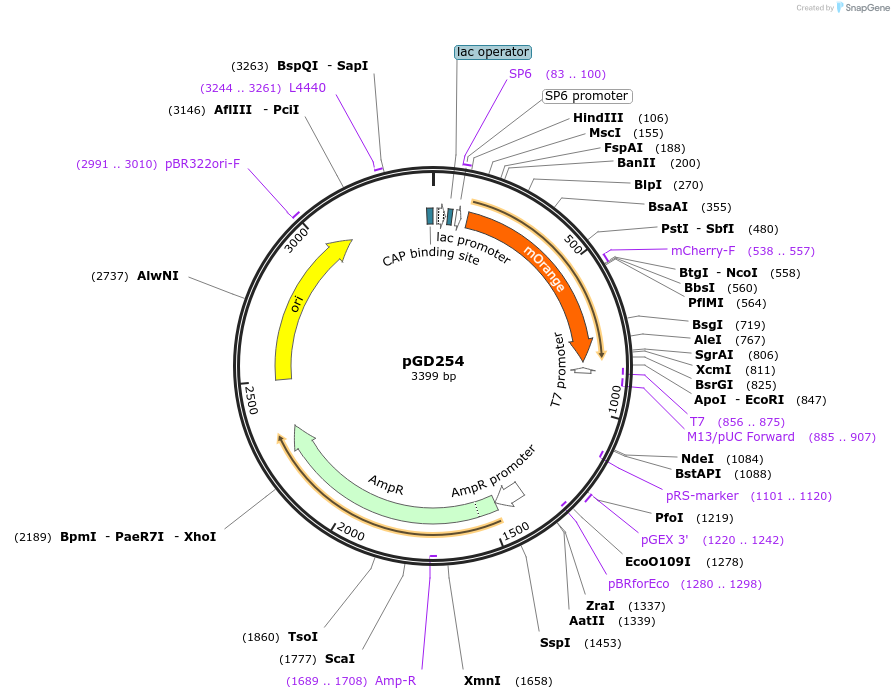
pGD254
Plasmid#132375Purposecontains the sequence of mOrange for C-term fusions in the pGGD000 backbone (GreenGate)DepositorInsertmOrange
UseGreengate entry plasmidAvailable SinceOct. 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
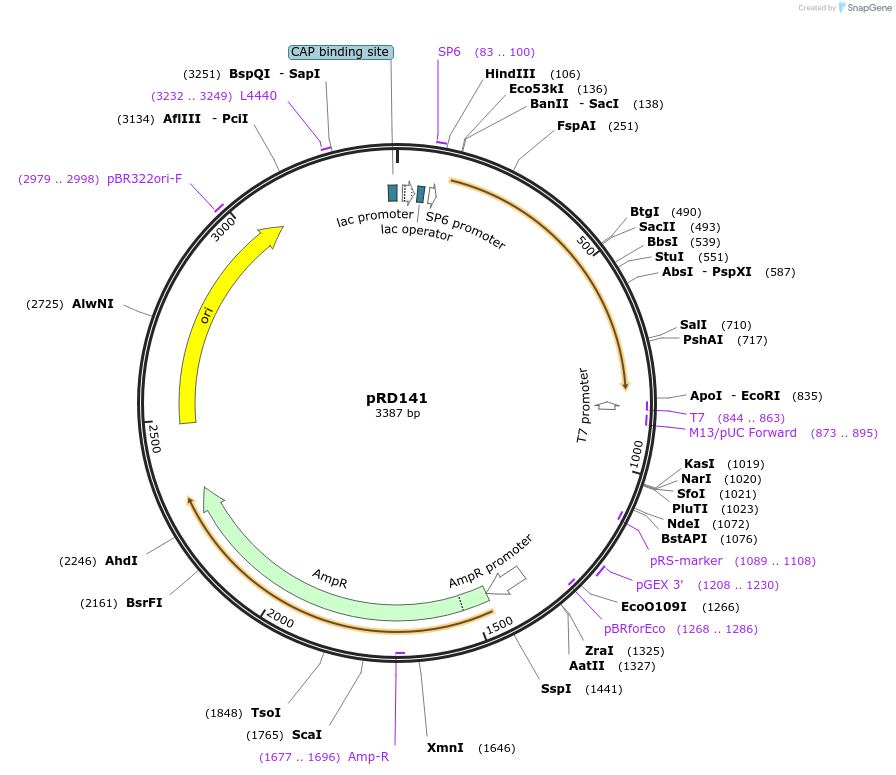
pRD141
Plasmid#132379Purposecontains the sequence of mKate2 for C-term fusions in the pGGD000 backbone (GreenGate)DepositorInsertmKate2
UseGreengate entry plasmidAvailable SinceOct. 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
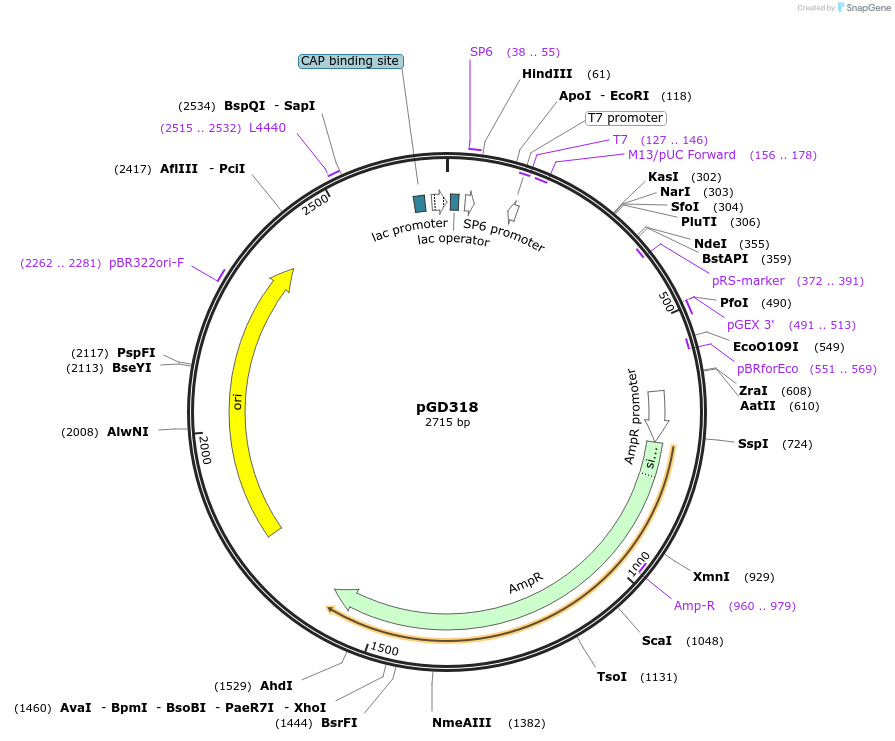
pGD318
Plasmid#129735PurposeContains a myristoylation sequence in the pGGC000 backbone (GreenGate)DepositorInsertMyristoylation sequence (MGRRKRKPK-)
UseGreengate entry plasmidAvailable SinceOct. 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
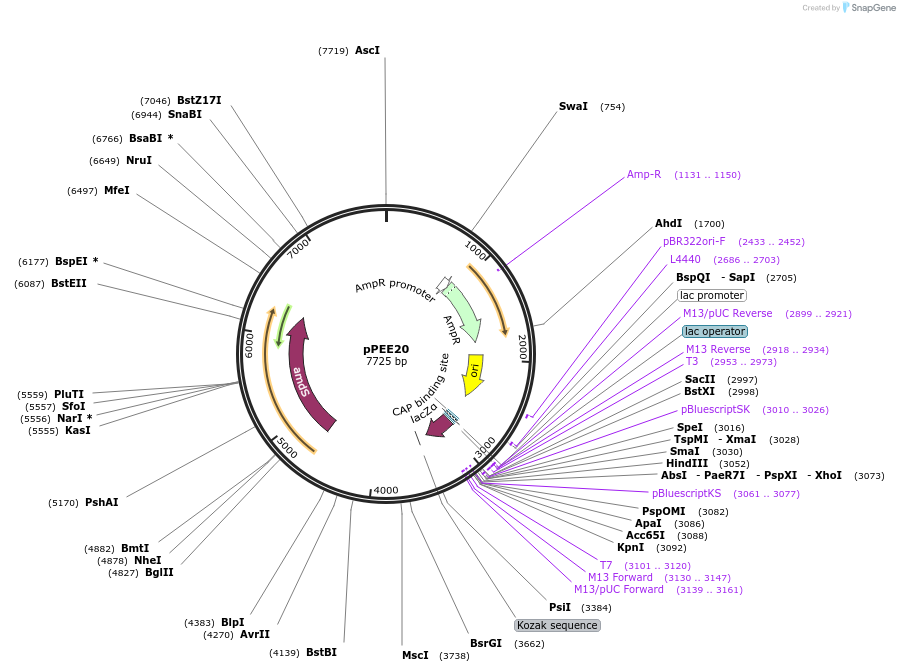
pPEE20
Plasmid#128363PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
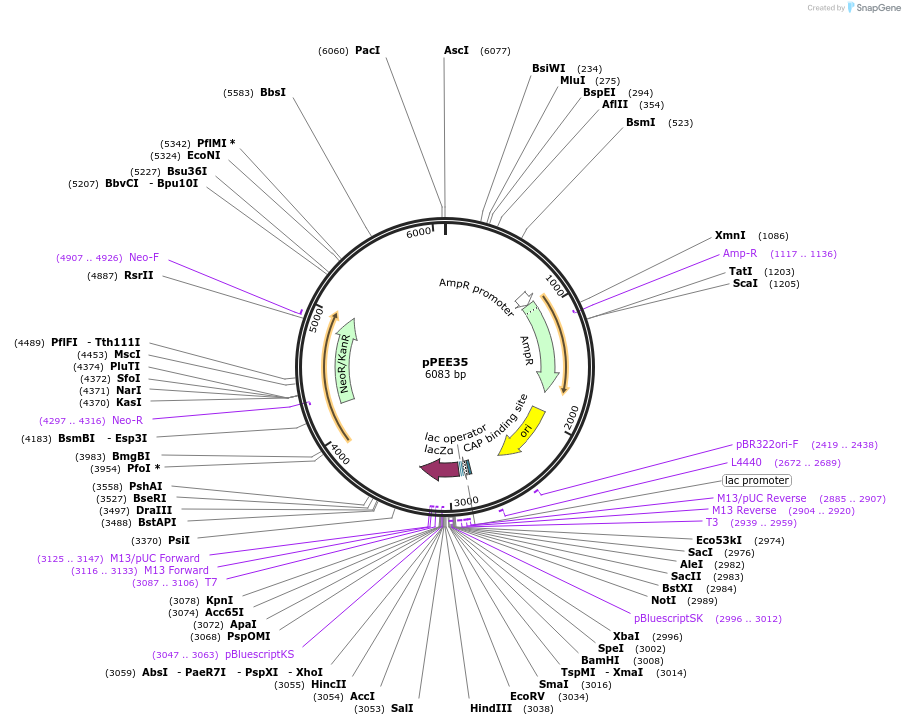
pPEE35
Plasmid#128375PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
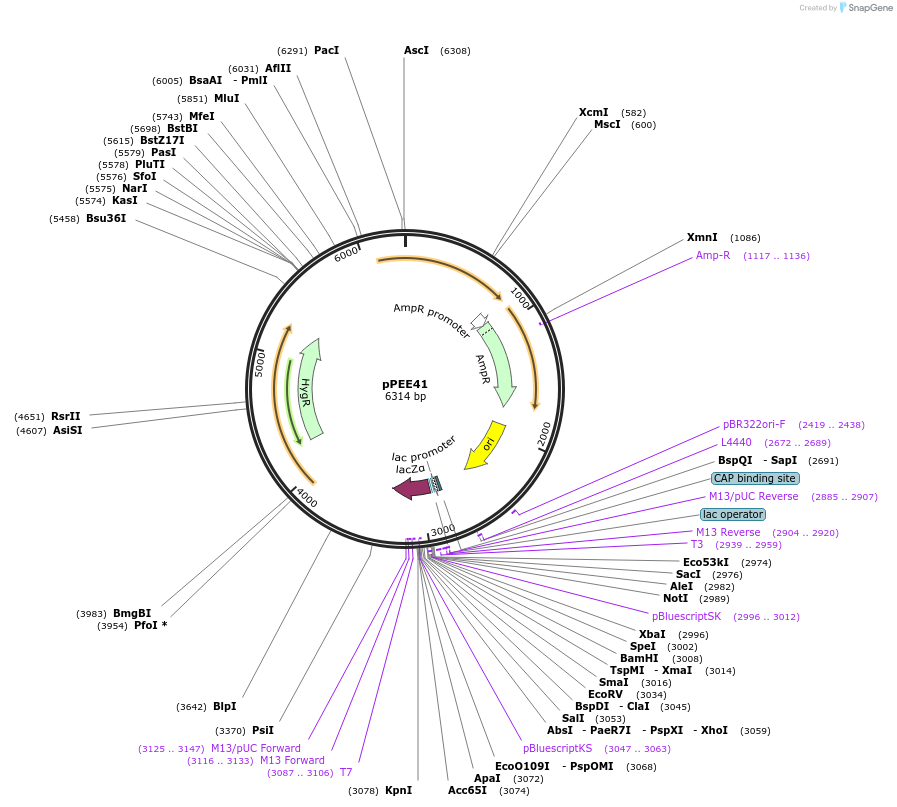
pPEE41
Plasmid#128381PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
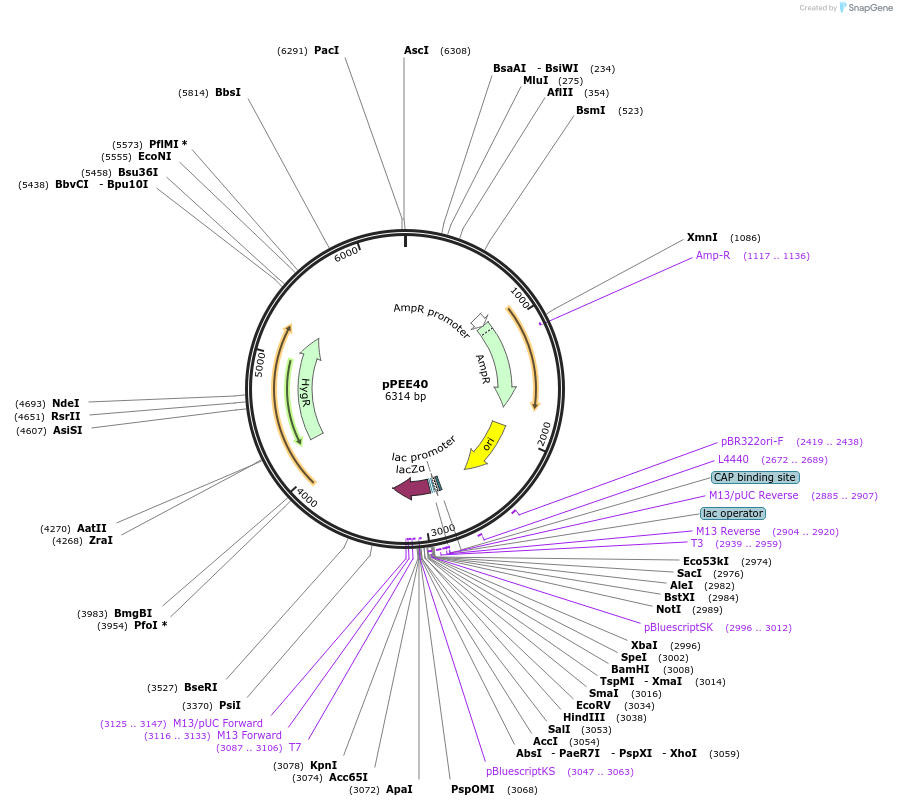
pPEE40
Plasmid#128380PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
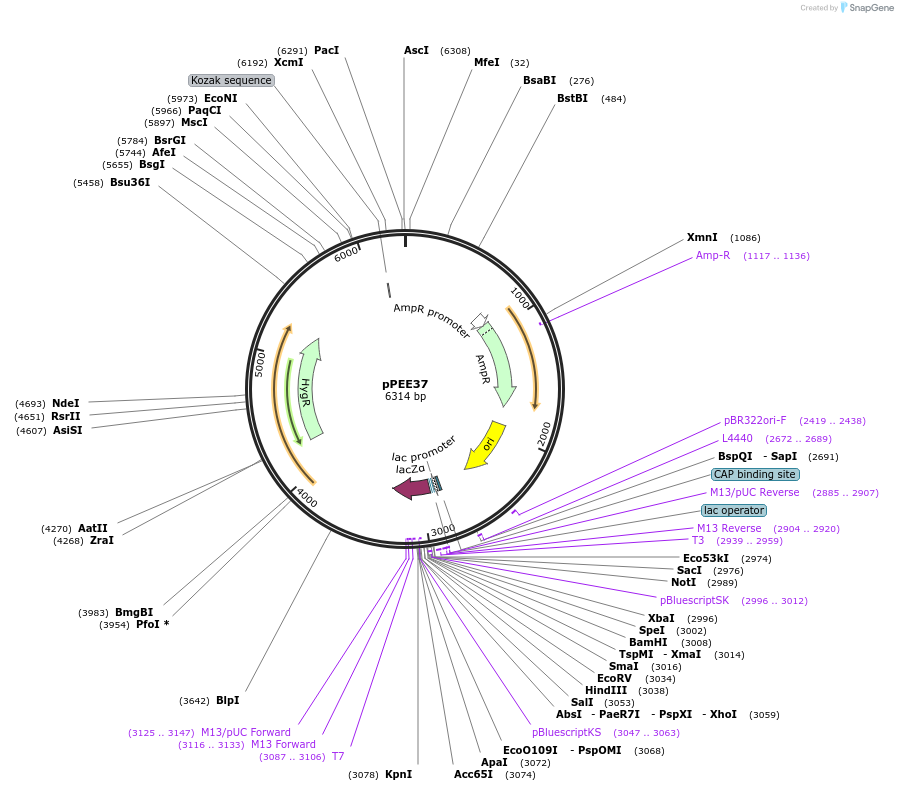
pPEE37
Plasmid#128377PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
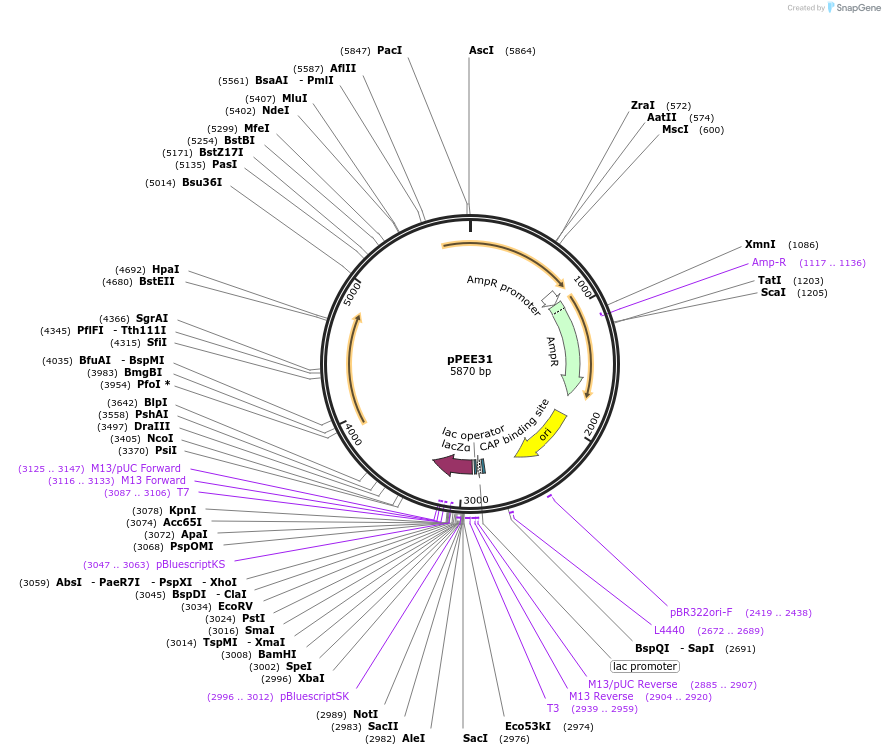
pPEE31
Plasmid#128371PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
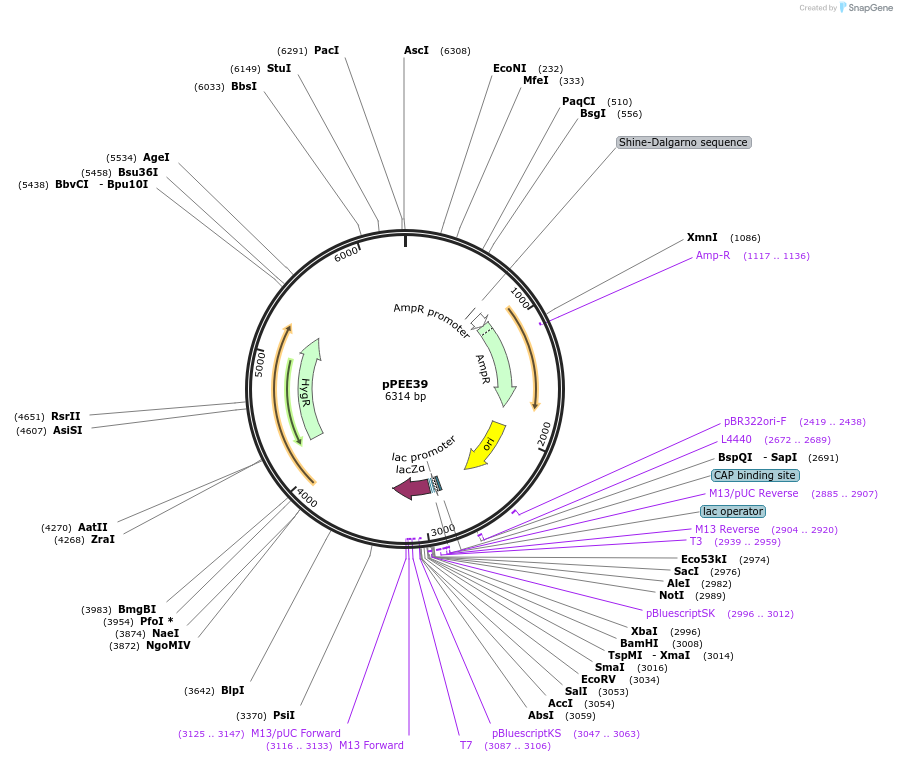
pPEE39
Plasmid#128379PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
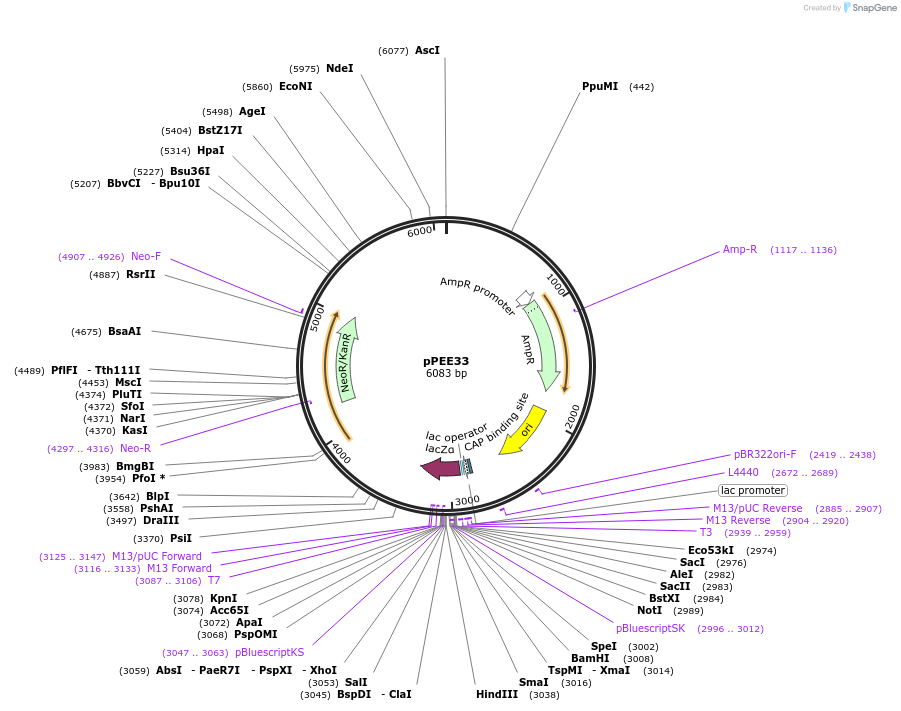
pPEE33
Plasmid#128373PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
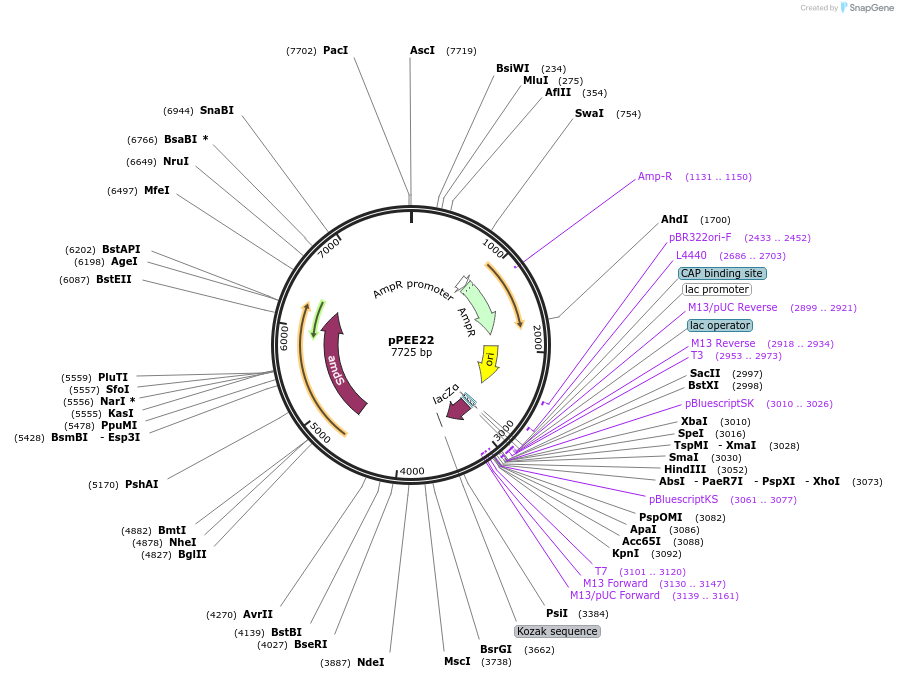
pPEE22
Plasmid#128365PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
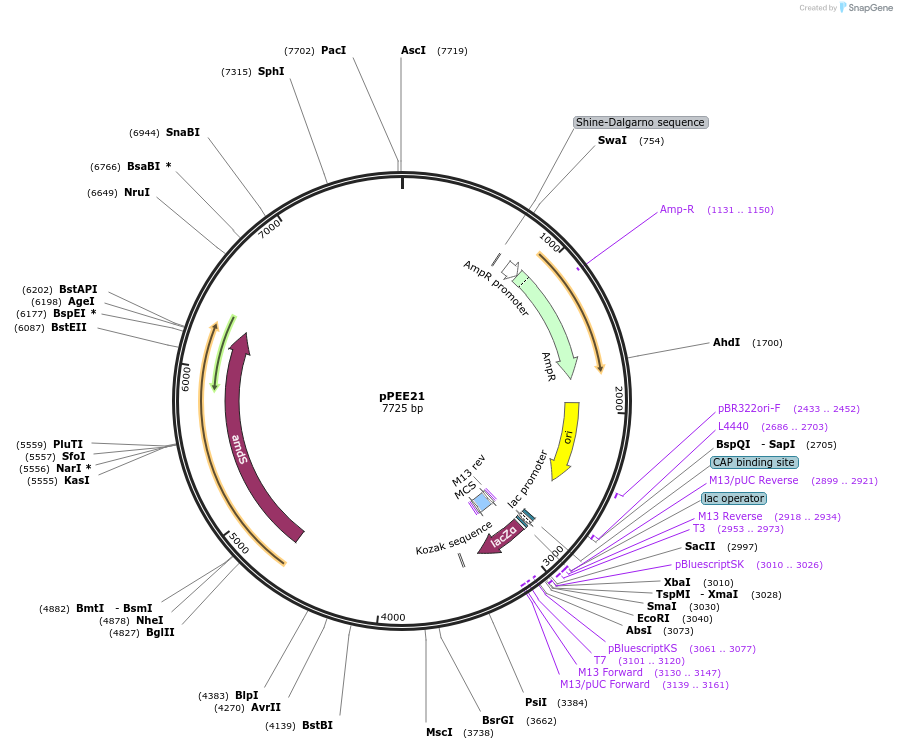
pPEE21
Plasmid#128364PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
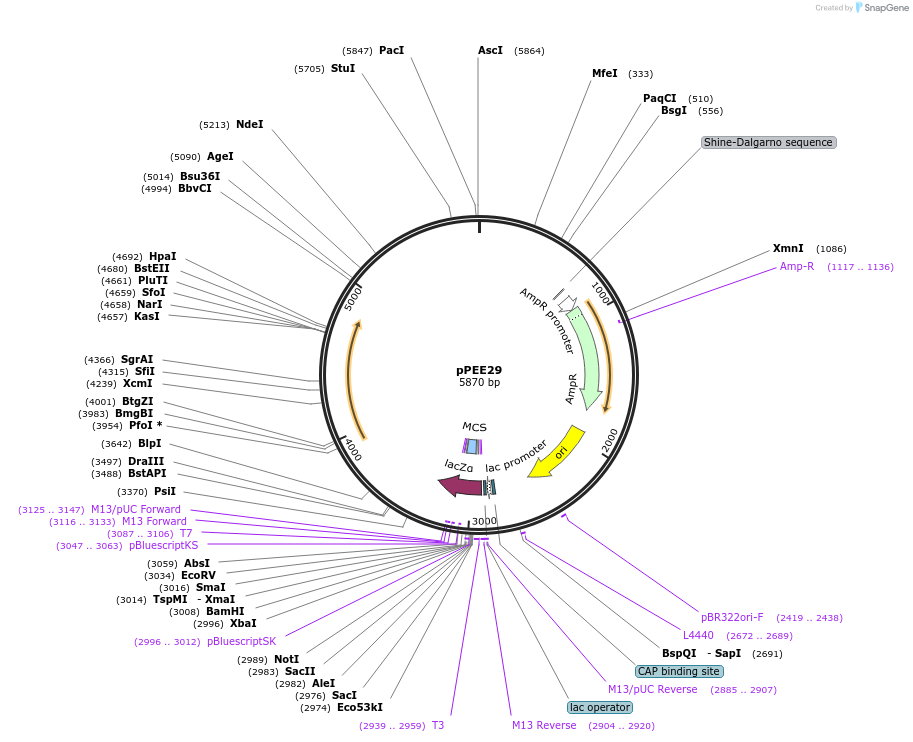
pPEE29
Plasmid#128369PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
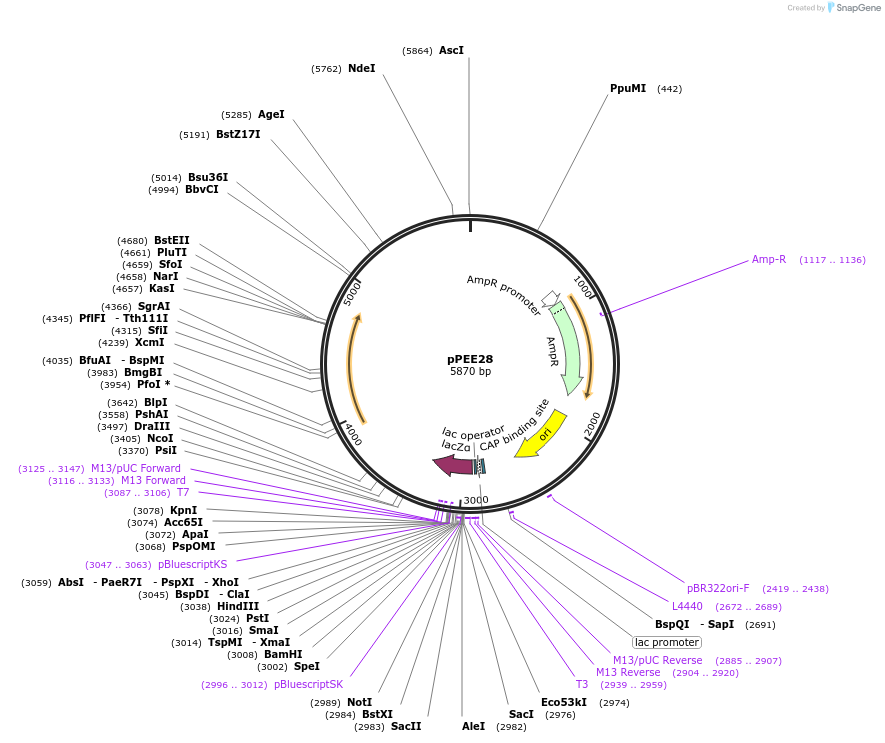
pPEE28
Plasmid#128368PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
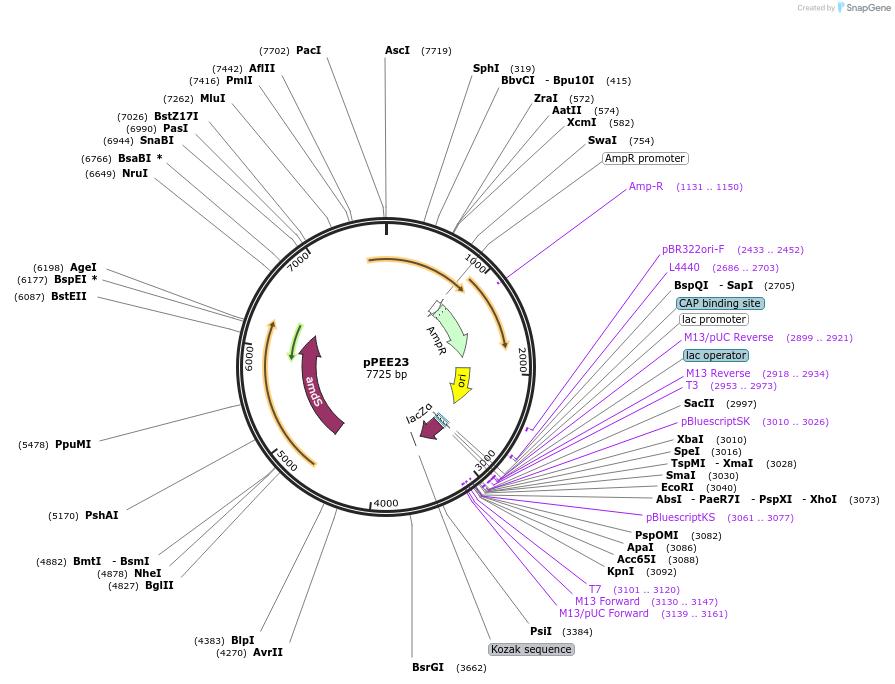
pPEE23
Plasmid#128366PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
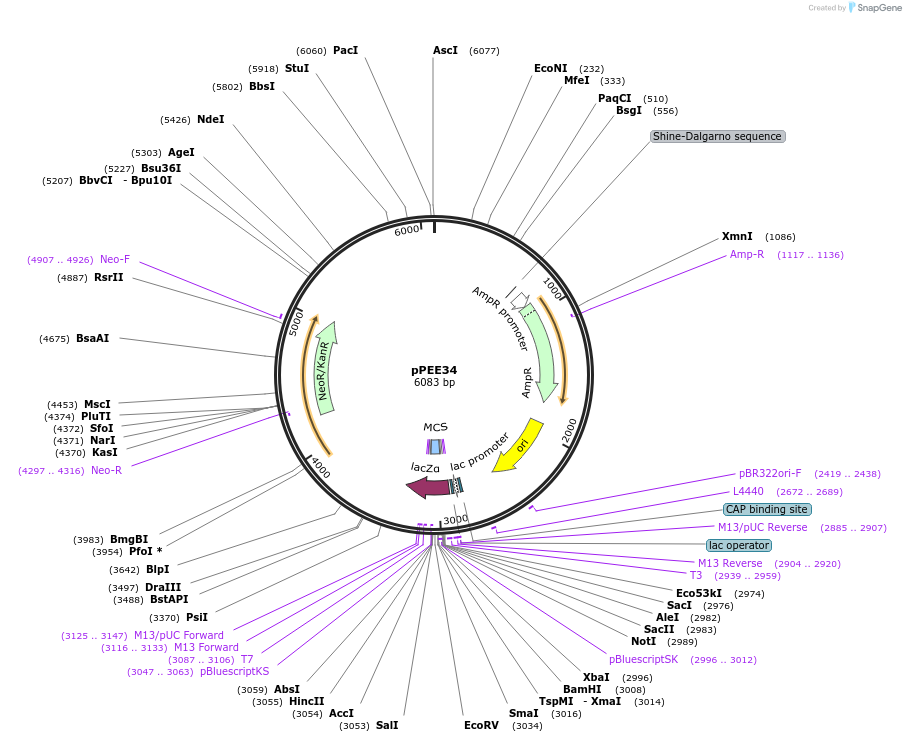
pPEE34
Plasmid#128374PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
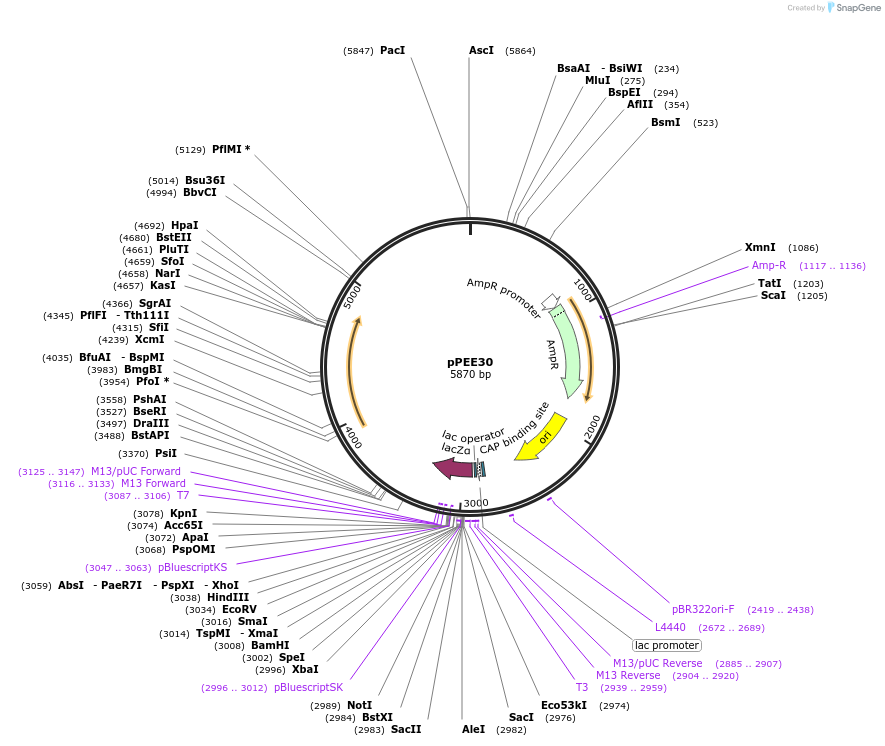
pPEE30
Plasmid#128370PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
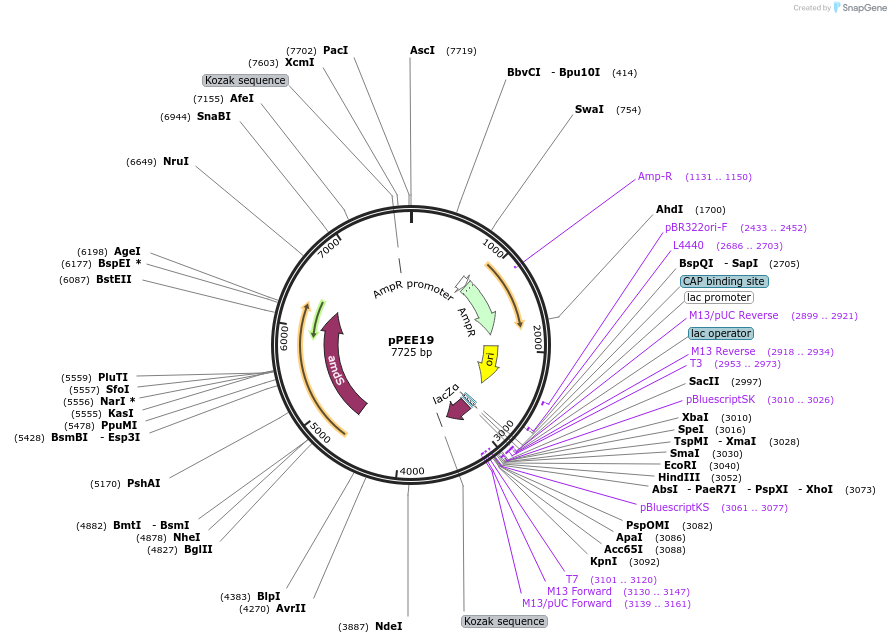
pPEE19
Plasmid#128362PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
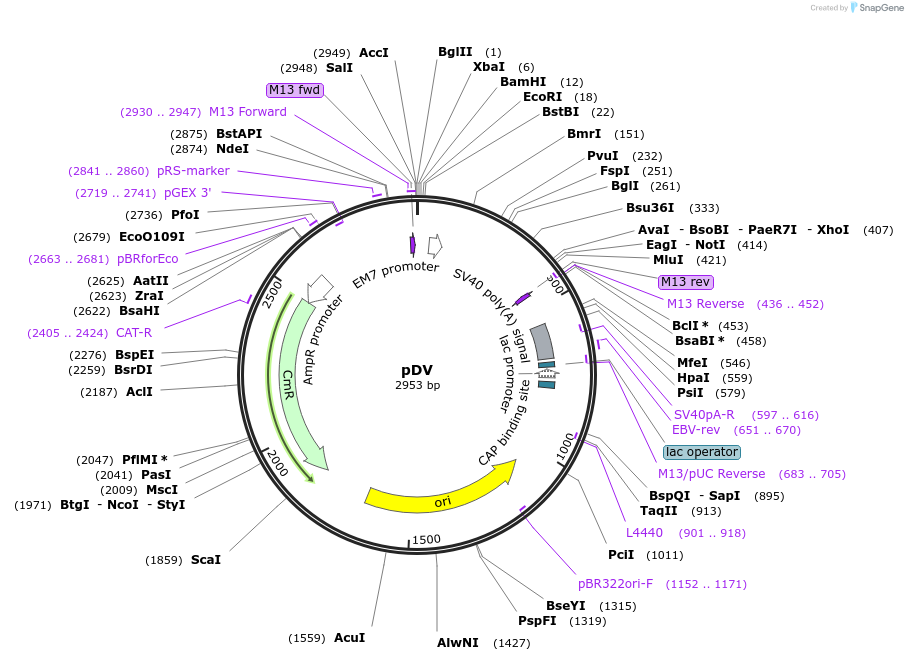
pDV
Plasmid#128860PurposeDestination vector for modules assemblyDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial, Mammalian, and Y…Available SinceAug. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
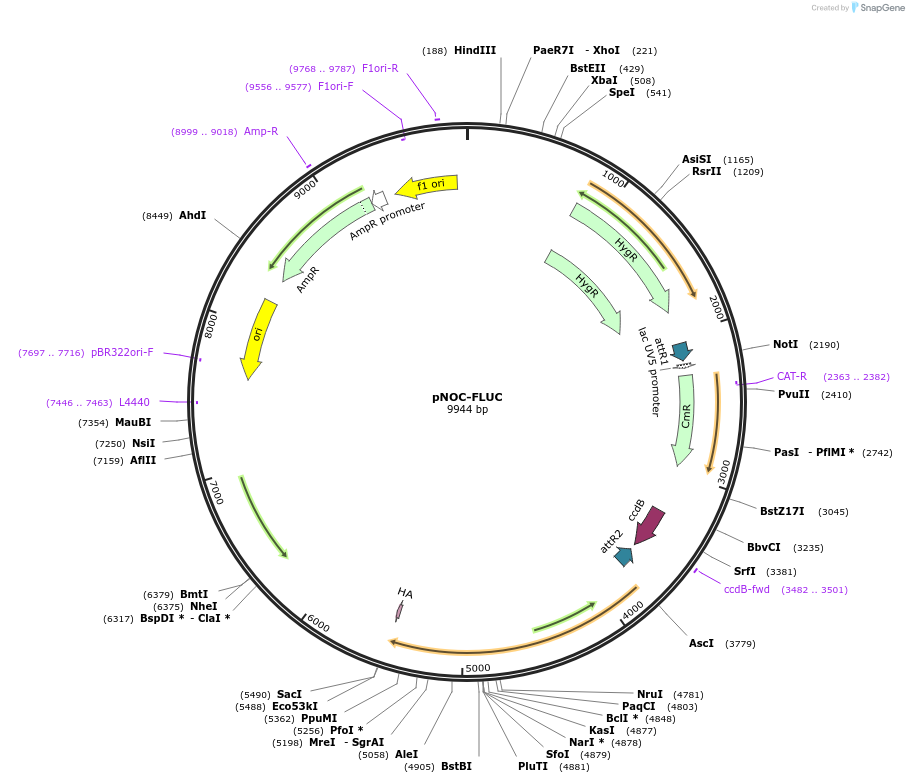
pNOC-FLUC
Plasmid#98140PurposeGateway compatible construct for N' terminal fusion or promoter driven firefly luciferase reporter, hygromycin resistance cassetteDepositorTypeEmpty backboneUseAlgae, nannochloropsisTagsHAAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
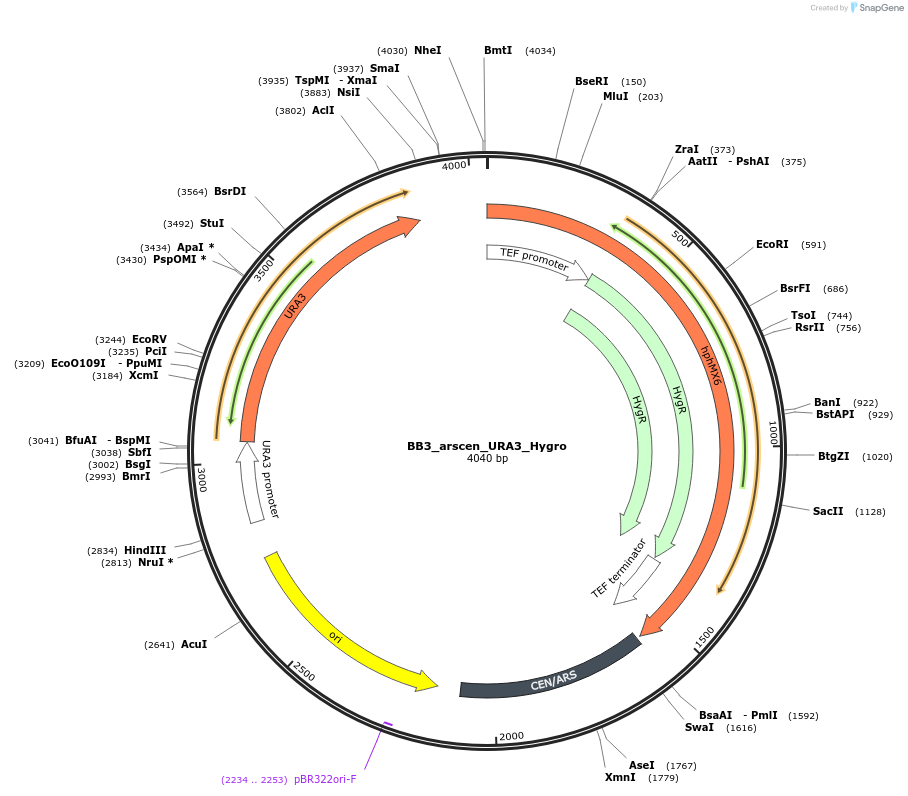
BB3_arscen_URA3_Hygro
Plasmid#118077Purposelow copy plasmid, neg. controlDepositorTypeEmpty backboneExpressionYeastAvailable SinceFeb. 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
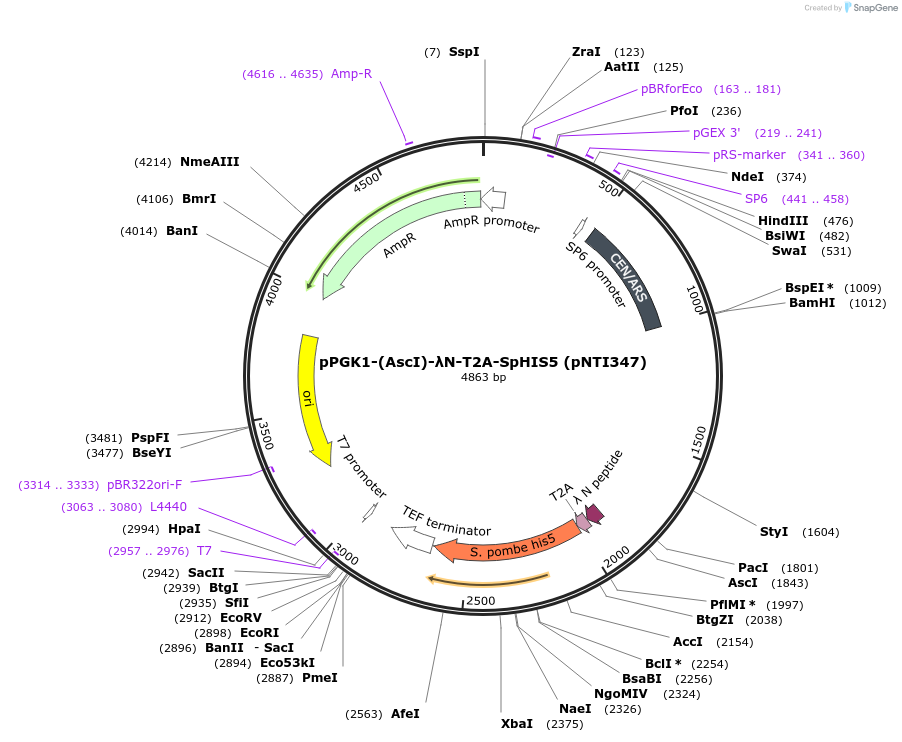
pPGK1-(AscI)-λN-T2A-SpHIS5 (pNTI347)
Plasmid#115429PurposeSelection for in-frame fusion expression constructsDepositorTypeEmpty backboneTagsT2A-SpHIS5 and λNExpressionYeastPromoterPGK1Available SinceOct. 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
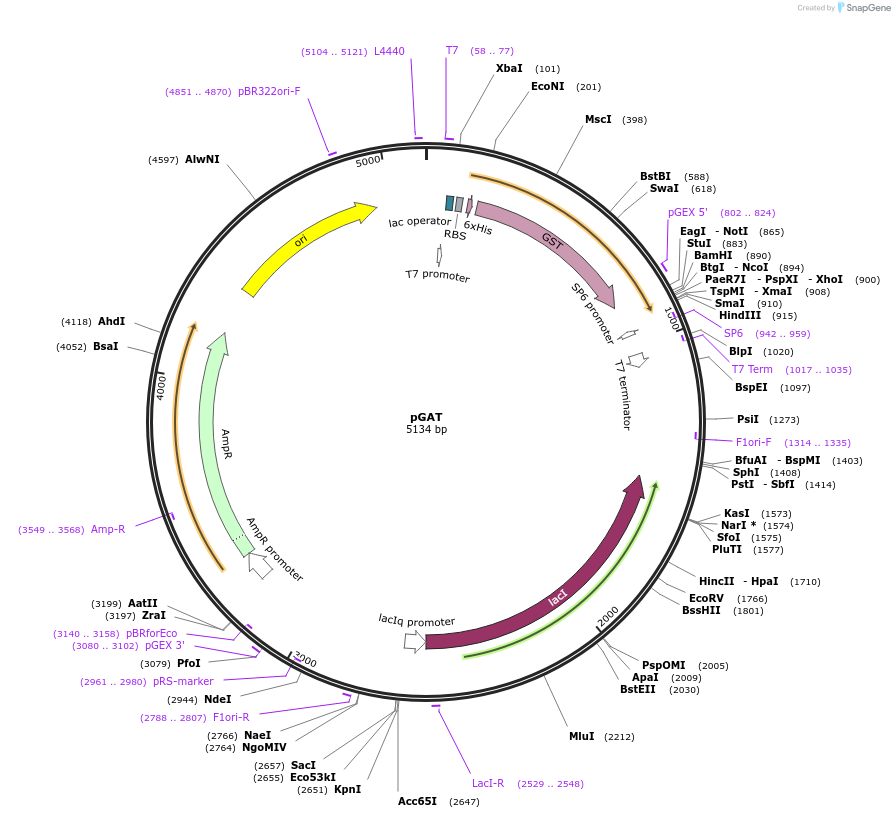
pGAT
Plasmid#112587PurposeUsed for expression of protein in E. coli as Thrombin cleavable 6His-tag-GST fusions with a blunt end restriction site immediately after thrombin recognition sequenceDepositorTypeEmpty backboneTags6His-GSTExpressionBacterialAvailable SinceSept. 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
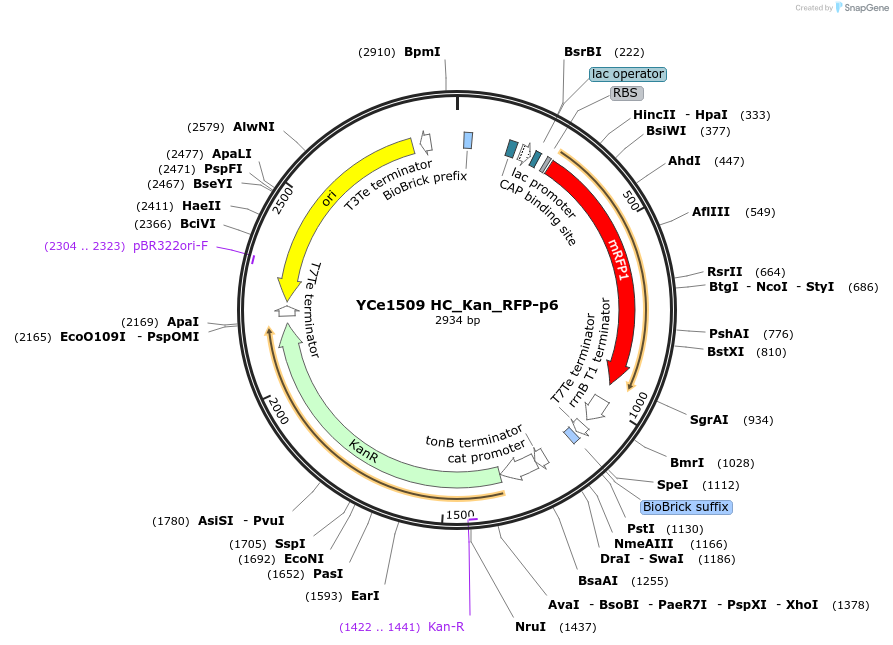
YCe1509 HC_Kan_RFP-p6
Plasmid#100614PurposeEMMA toolkit part-entry vector position 6DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
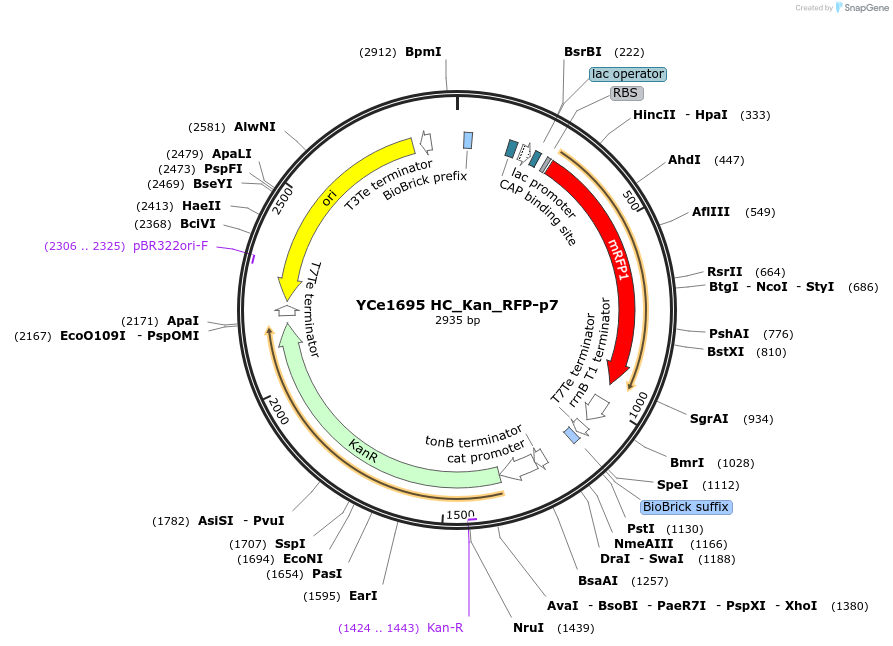
YCe1695 HC_Kan_RFP-p7
Plasmid#100615PurposeEMMA toolkit part-entry vector position 7DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
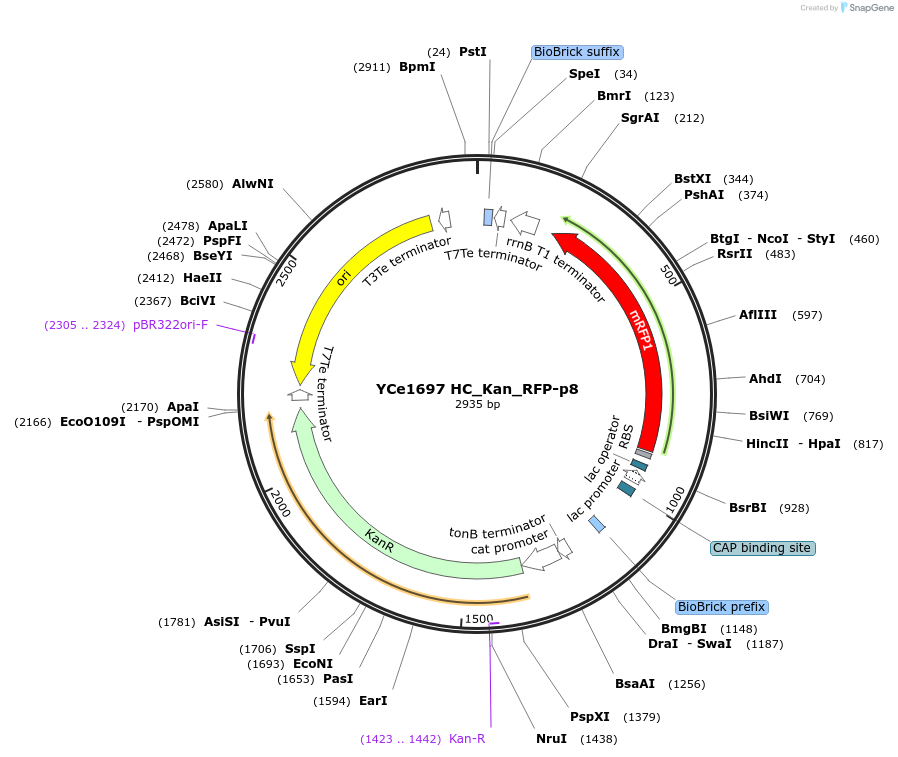
YCe1697 HC_Kan_RFP-p8
Plasmid#100616PurposeEMMA toolkit part-entry vector position 8DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
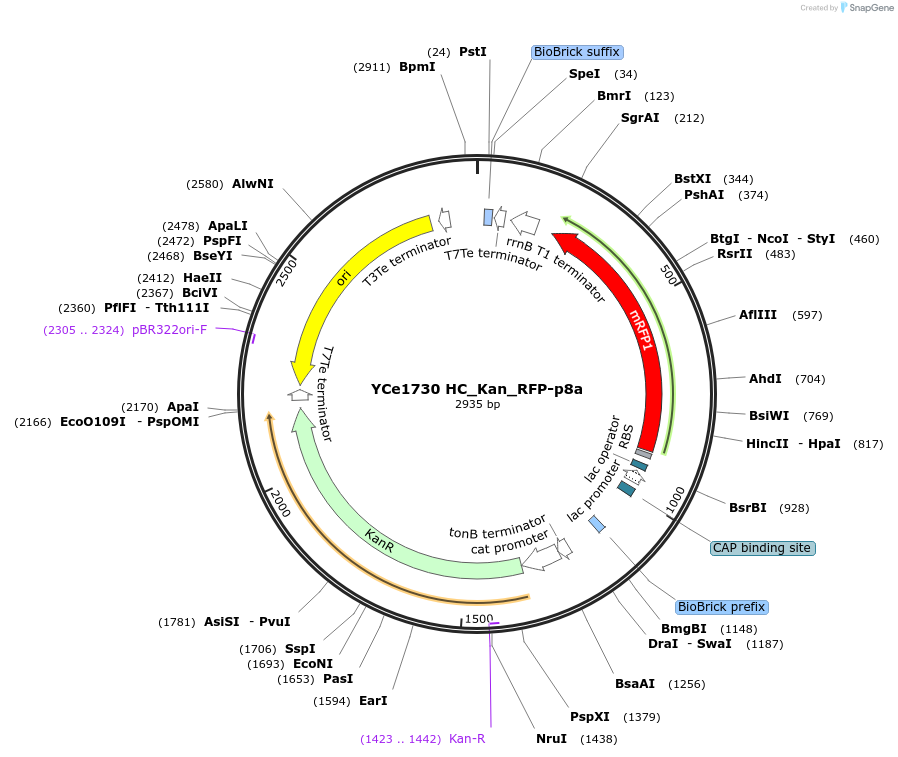
YCe1730 HC_Kan_RFP-p8a
Plasmid#100617PurposeEMMA toolkit part-entry vector position 8aDepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
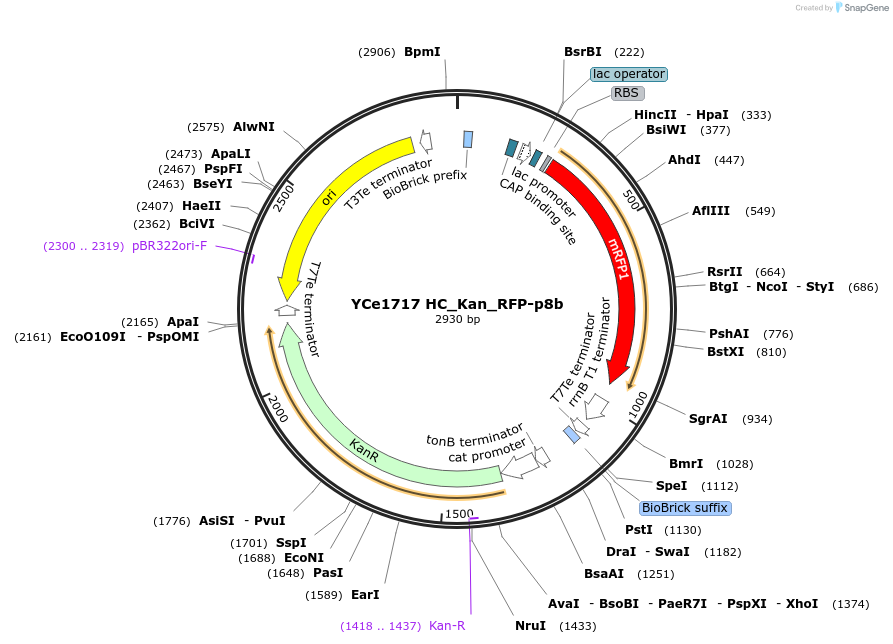
YCe1717 HC_Kan_RFP-p8b
Plasmid#100618PurposeEMMA toolkit part-entry vector position 8bDepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
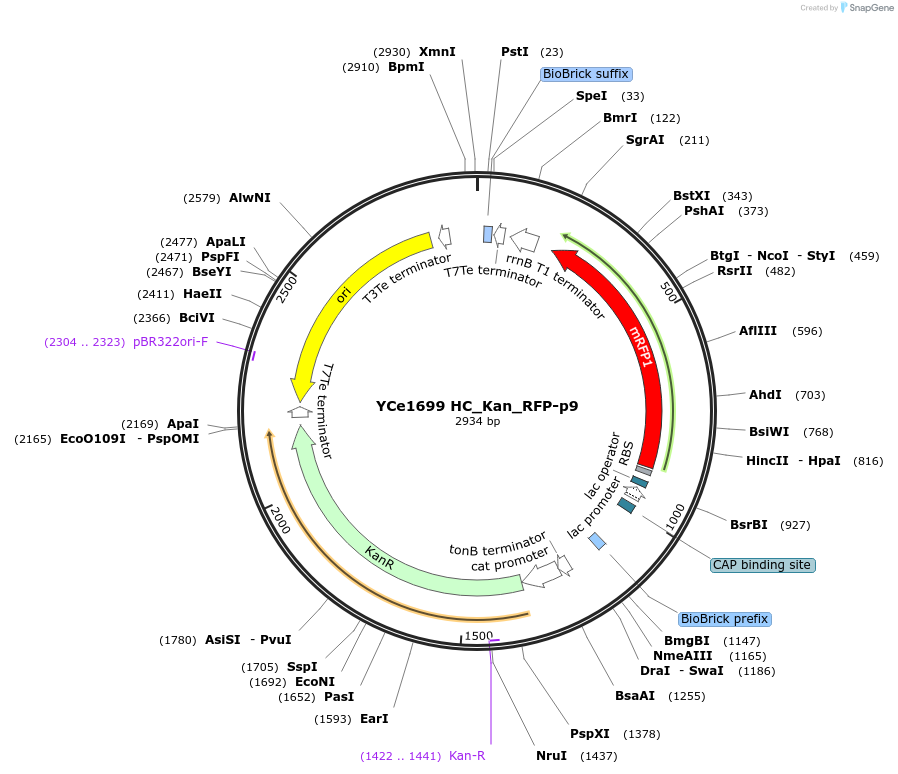
YCe1699 HC_Kan_RFP-p9
Plasmid#100619PurposeEMMA toolkit part-entry vector position 9DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
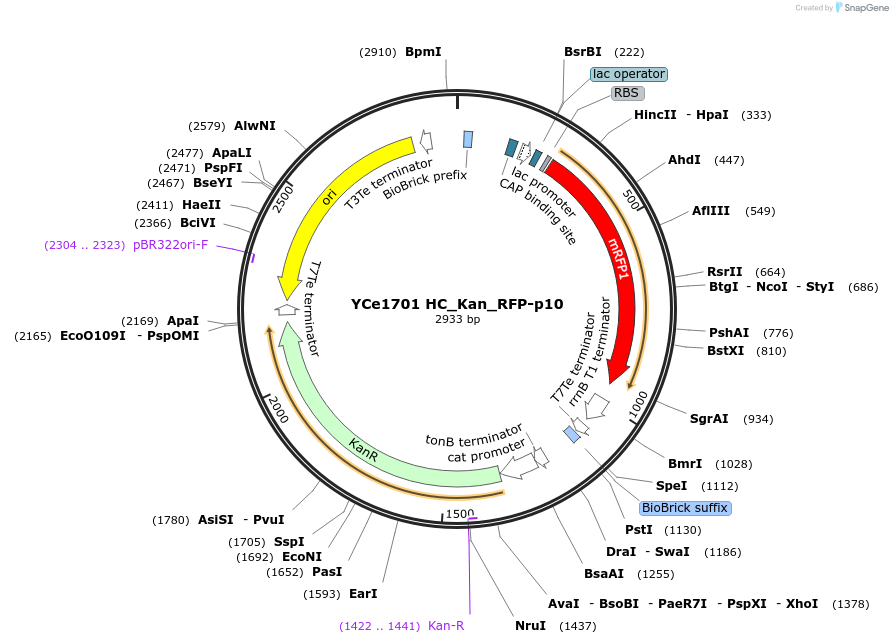
YCe1701 HC_Kan_RFP-p10
Plasmid#100620PurposeEMMA toolkit part-entry vector position 10DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
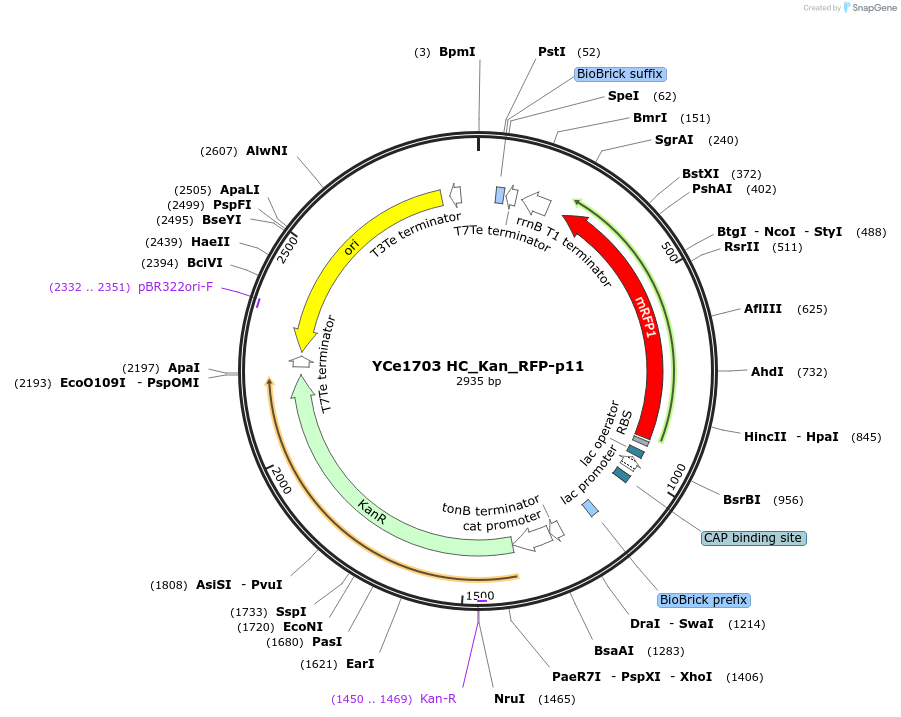
YCe1703 HC_Kan_RFP-p11
Plasmid#100621PurposeEMMA toolkit part-entry vector position 11DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
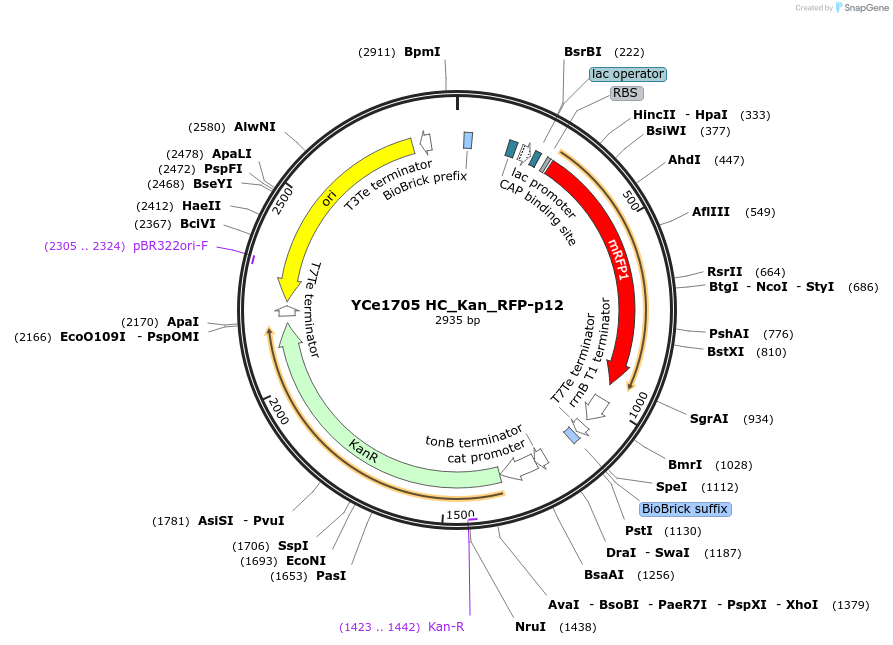
YCe1705 HC_Kan_RFP-p12
Plasmid#100622PurposeEMMA toolkit part-entry vector position 12DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
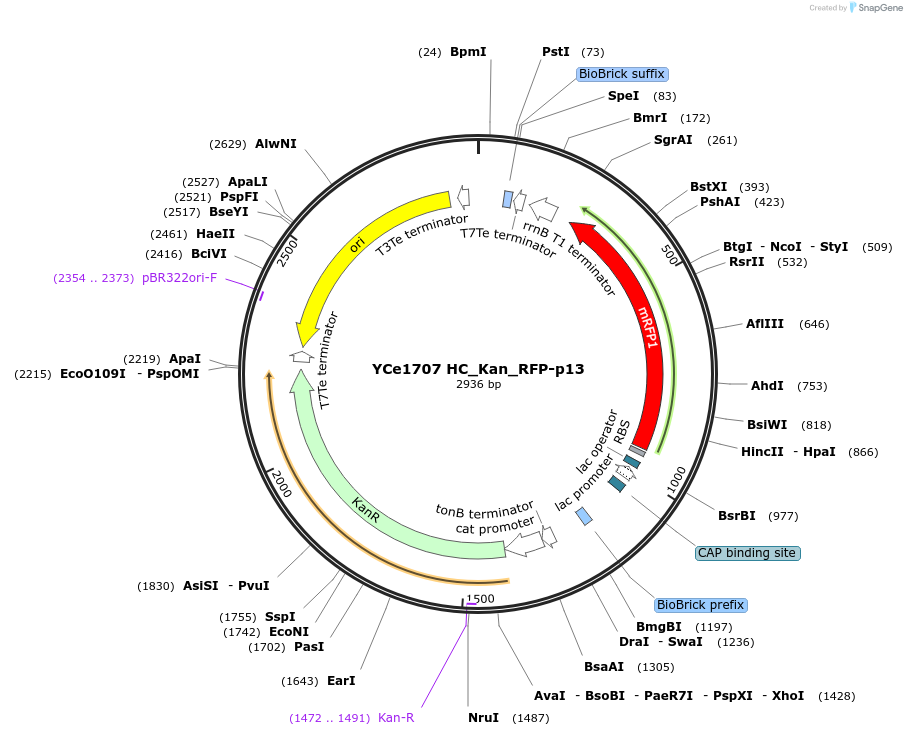
YCe1707 HC_Kan_RFP-p13
Plasmid#100623PurposeEMMA toolkit part-entry vector position 13DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
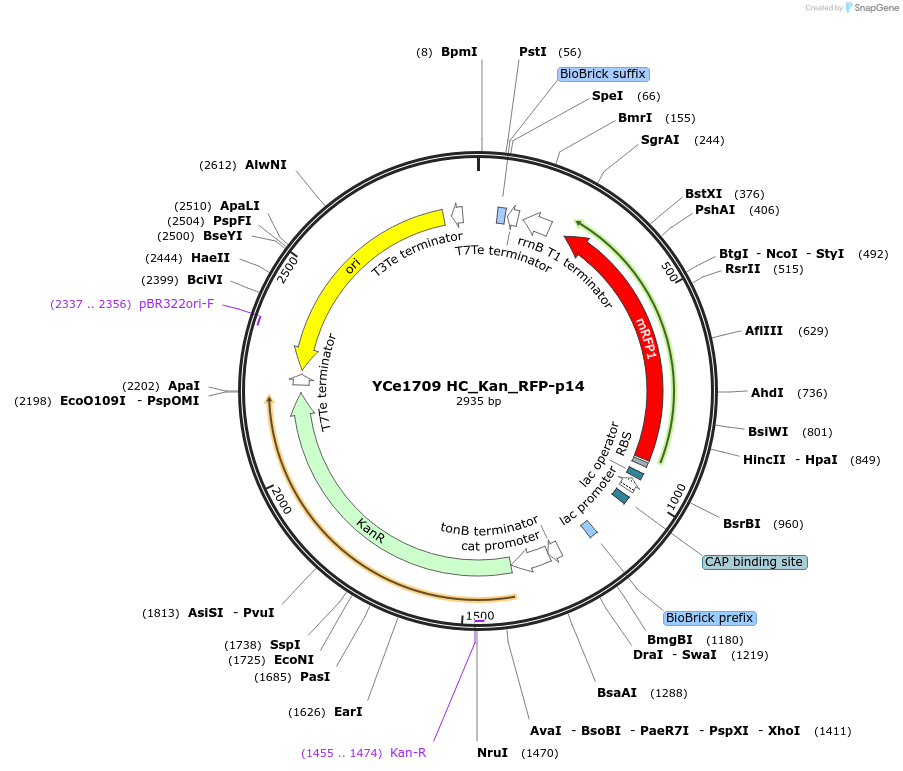
YCe1709 HC_Kan_RFP-p14
Plasmid#100624PurposeEMMA toolkit part-entry vector position 14DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
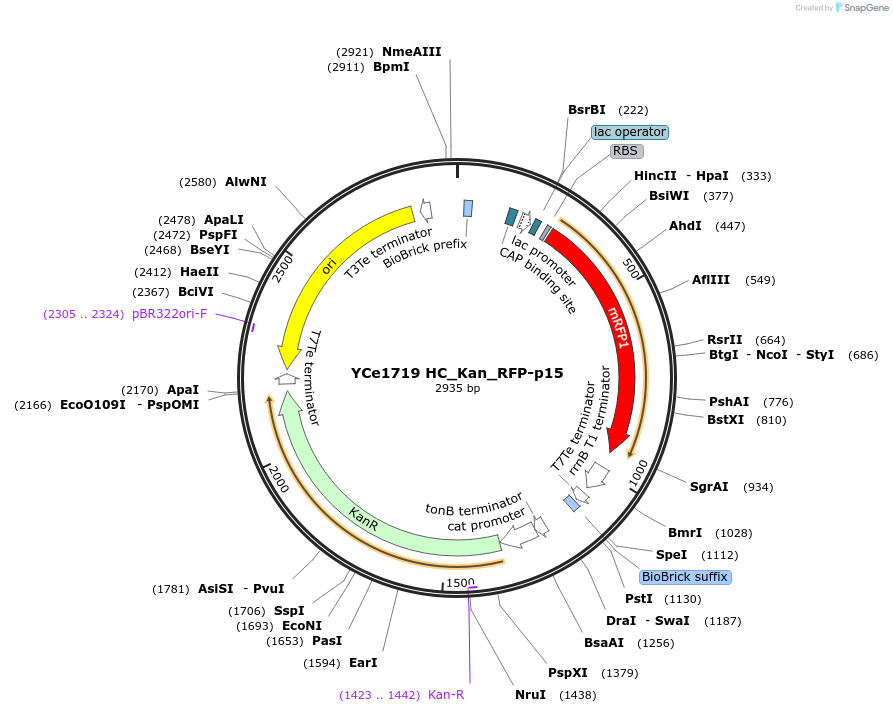
YCe1719 HC_Kan_RFP-p15
Plasmid#100625PurposeEMMA toolkit part-entry vector position 15DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only