We narrowed to 81,061 results for: myc
-
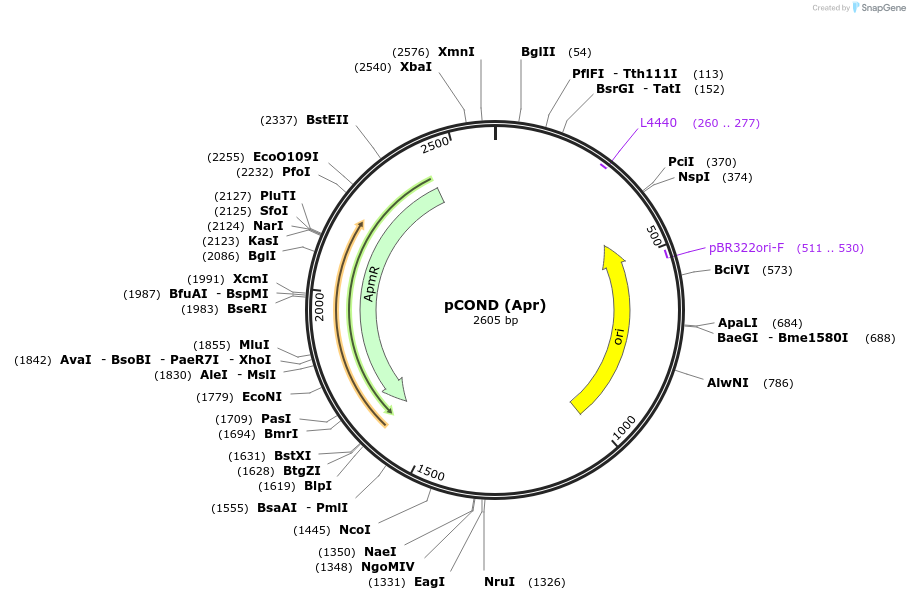
Plasmid#110094PurposeSuicide vector based on the TetR/Pip OFF system.DepositorTypeEmpty backboneUseBacterial knockout generationAvailable SinceMay 22, 2018AvailabilityAcademic Institutions and Nonprofits only
-
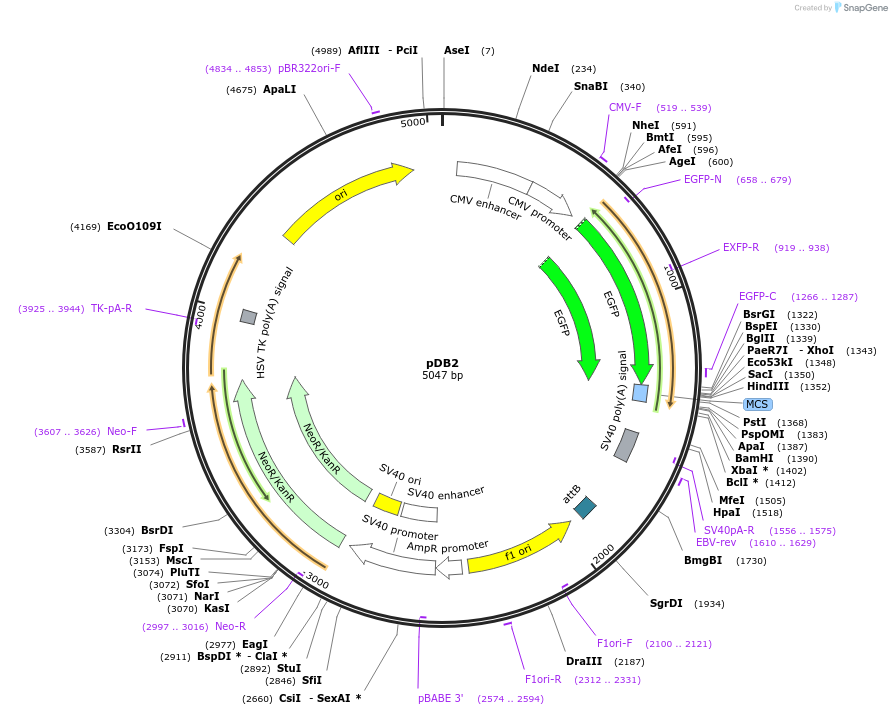
pDB2
Plasmid#18954DepositorInsertphiC31 attB
ExpressionMammalianAvailable SinceOct. 14, 2008AvailabilityAcademic Institutions and Nonprofits only -
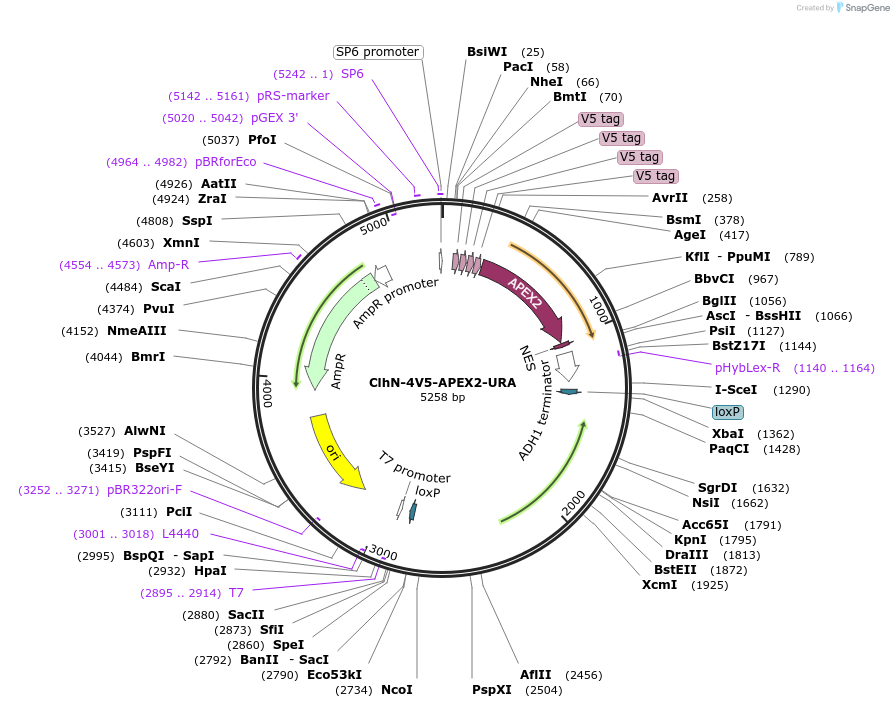
ClhN-4V5-APEX2-URA
Plasmid#207056PurposeFor ORF tagging with 4V5-APEX2DepositorInsert4xV5-APEX2
Tags4xV5-APEX2ExpressionYeastAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
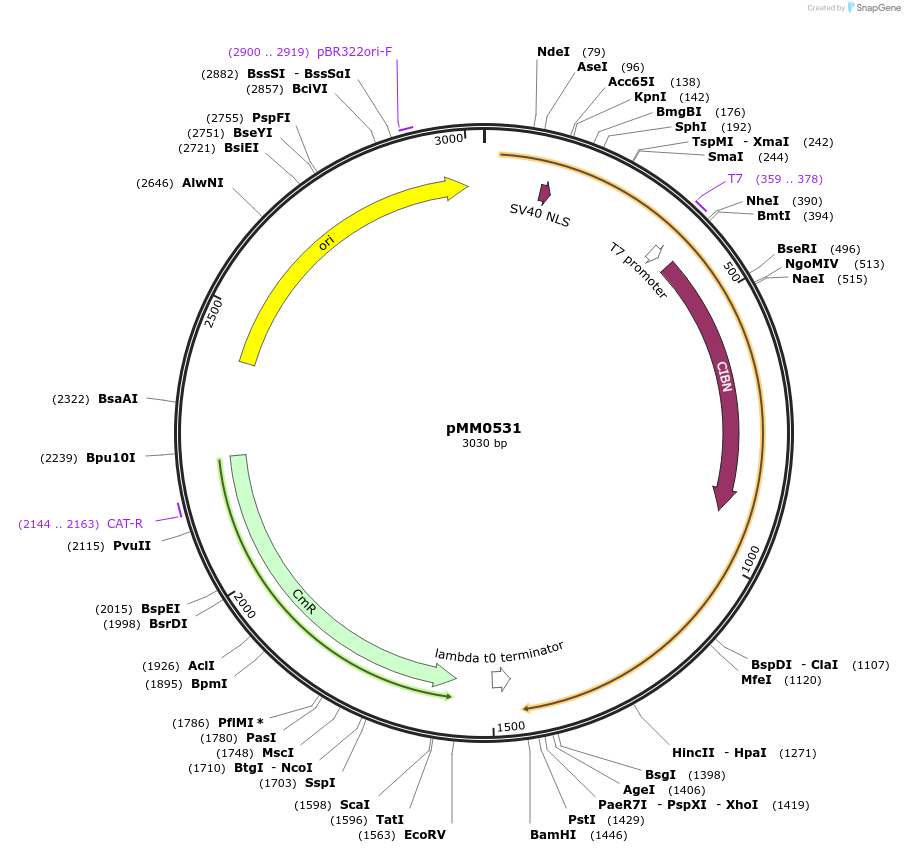
pMM0531
Plasmid#128988PurposePart plasmid for VP16-CIB1, Type 3 (coding sequence) partDepositorInsertSV40NLS-VP16-CIB1
UseSynthetic BiologyAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
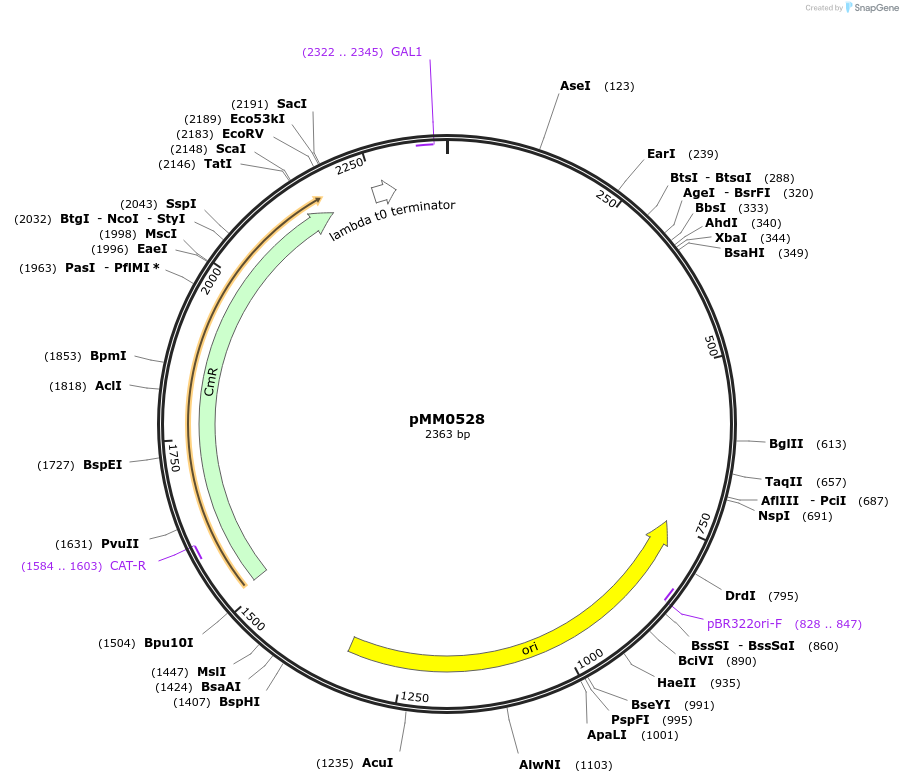
pMM0528
Plasmid#128985PurposePart plasmid for pZF(3BS), Type 2 (promoter) partDepositorInsertpZF(3BS)
UseSynthetic BiologyAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
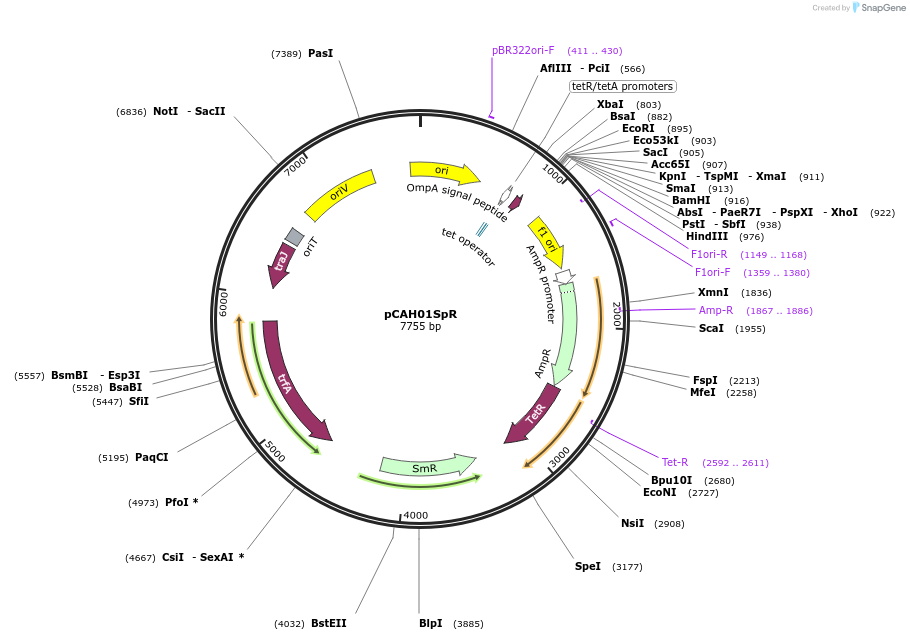
pCAH01SpR
Plasmid#128159PurposepCAH01 derivative (Ptet inducible promoter) with ahp (kanamycin resistance) CDS replaced with the aadA (streptomycin/spectinomycin resistance) CDSDepositorTypeEmpty backboneExpressionBacterialPromoterPtetAvailable SinceJuly 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
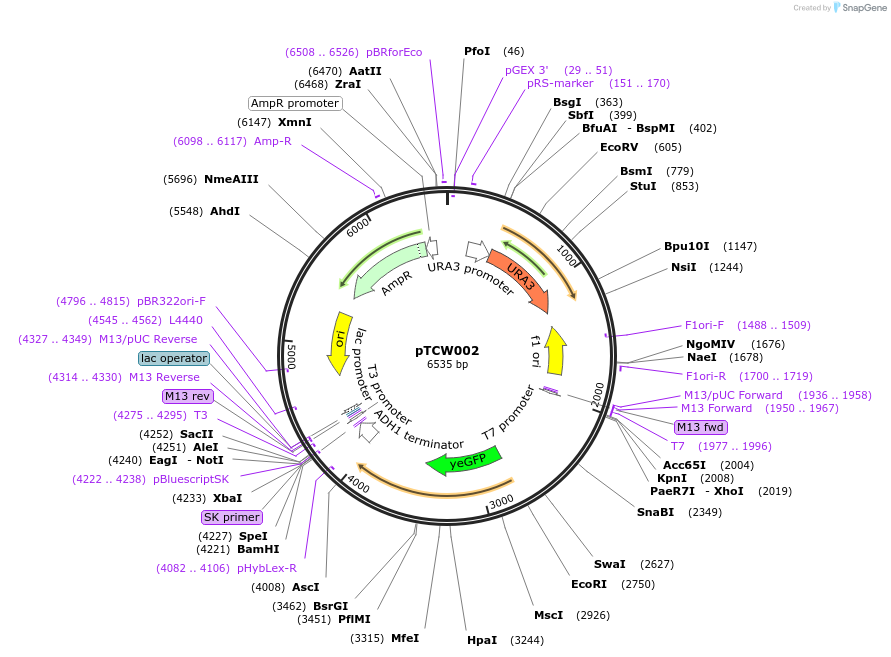
pTCW002
Plasmid#83522Purposepheromone mediated destablised-GFP expression in yeastDepositorInsertpFUS1-yEGFPCLN2PEST
ExpressionYeastPromoterpFUS1Available SinceDec. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
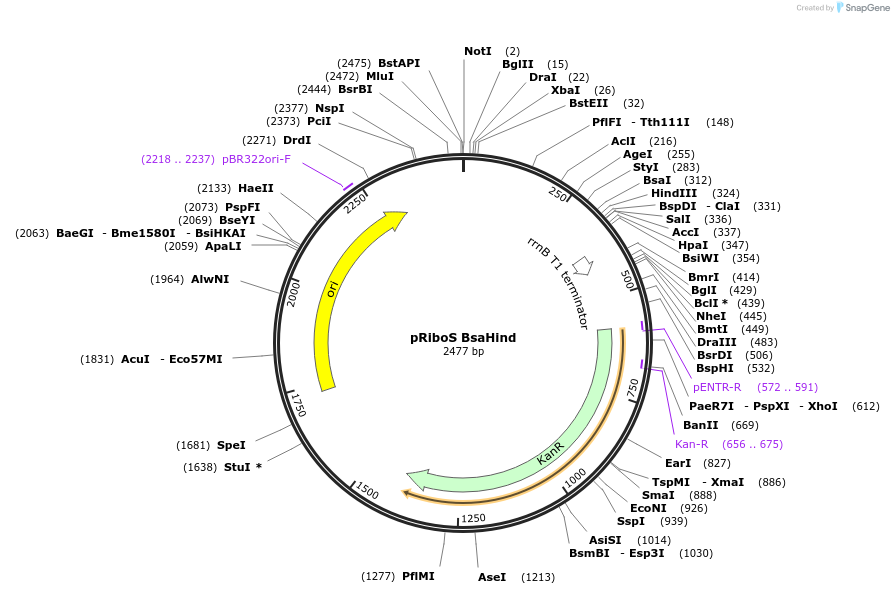
pRiboS BsaHind
Plasmid#36253DepositorTypeEmpty backboneUseSynthetic Biology; Mycobacteria replicating vectorExpressionBacterialPromoterModified Hsp60Available SinceJune 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
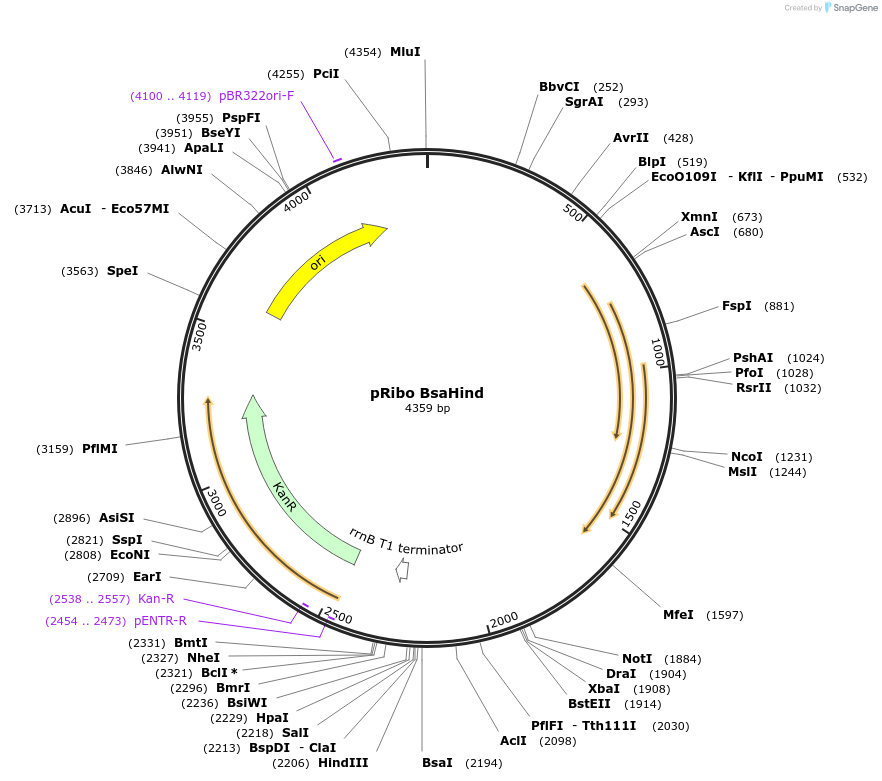
pRibo BsaHind
Plasmid#36251DepositorTypeEmpty backboneUseSynthetic Biology; Mycobacteria replicating vectorExpressionBacterialPromoterModified Hsp60Available SinceMay 7, 2012AvailabilityAcademic Institutions and Nonprofits only -
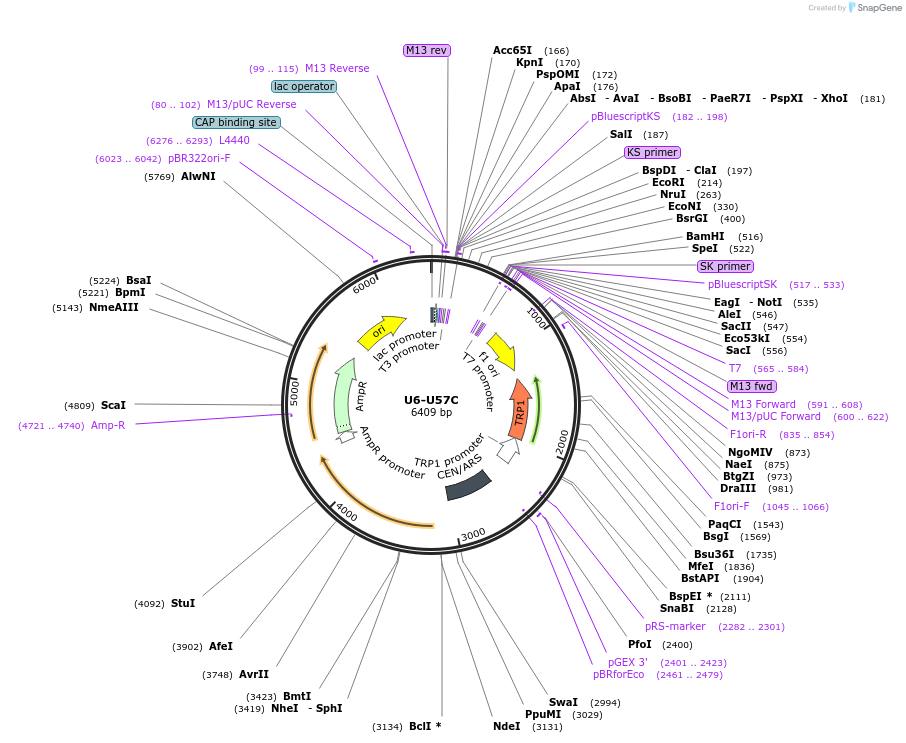
U6-U57C
Plasmid#169928PurposeWT U6 with a U57C mutationDepositorInsertU6 U57C
ExpressionBacterial and YeastAvailable SinceAug. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
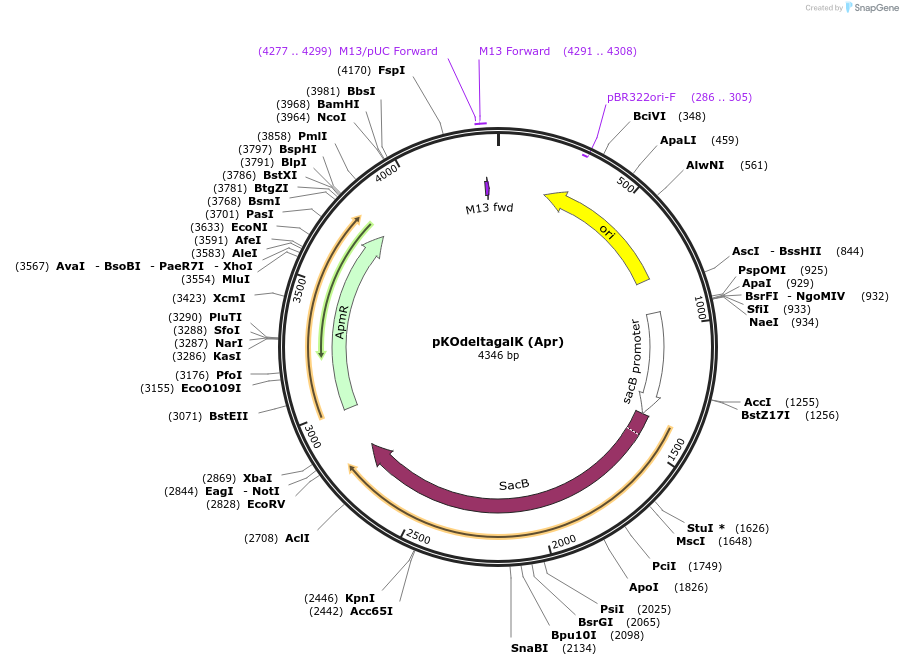
pKOdeltagalK (Apr)
Plasmid#110091PurposeAnalogous to pKO but without the negative selection gene galK.DepositorTypeEmpty backboneUseBacterial knockout generationAvailable SinceMay 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
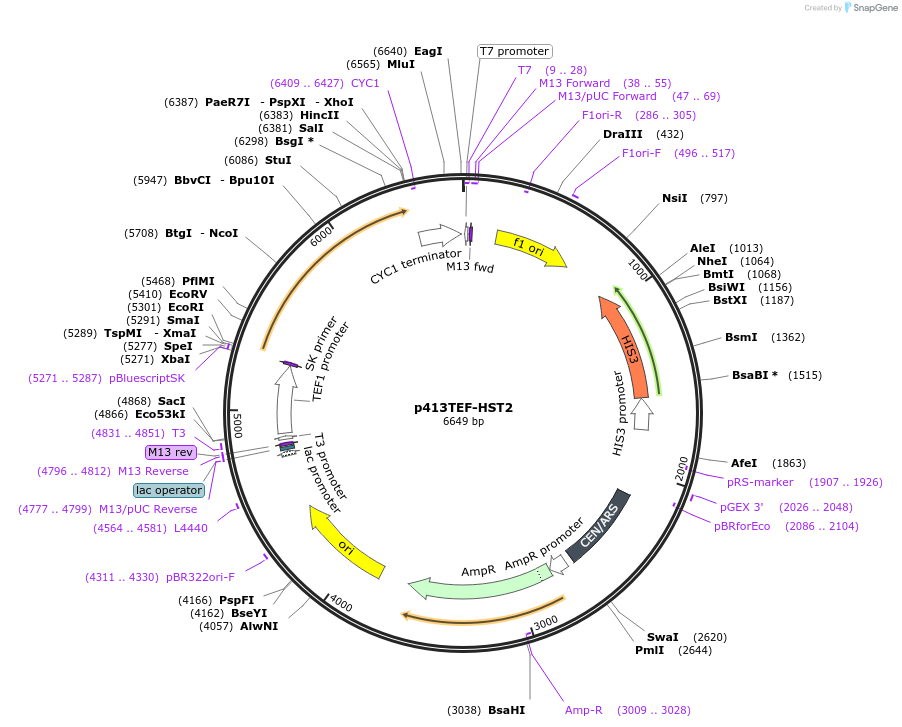
p413TEF-HST2
Plasmid#51745Purposeoverexpression of HST2 in S. cerevisiae, HIS3 markerDepositorAvailable SinceMarch 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
-
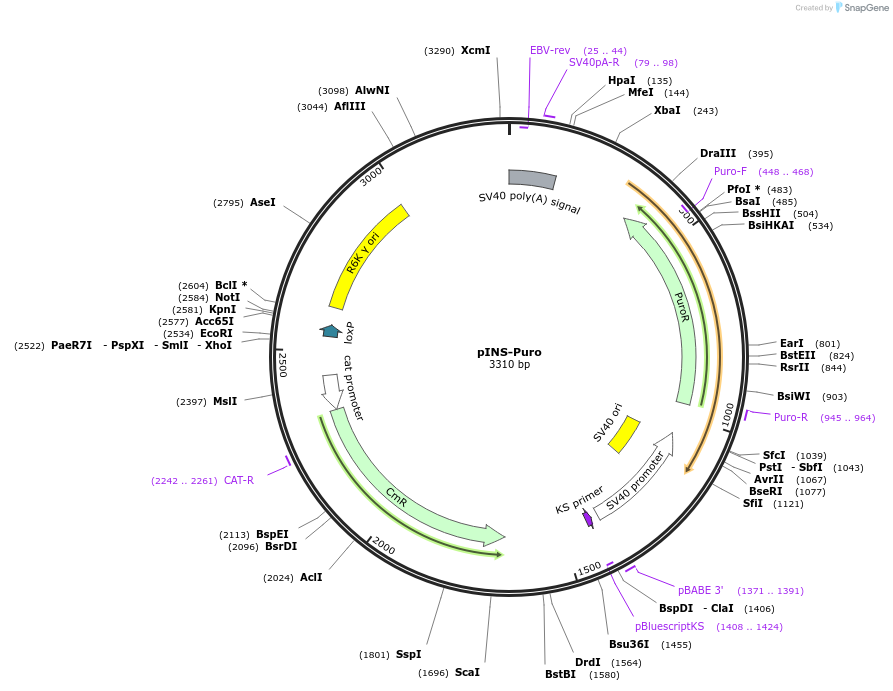
pINS-Puro
Plasmid#31389DepositorInsertpac
UseCre/LoxAvailable SinceSept. 13, 2012AvailabilityAcademic Institutions and Nonprofits only -
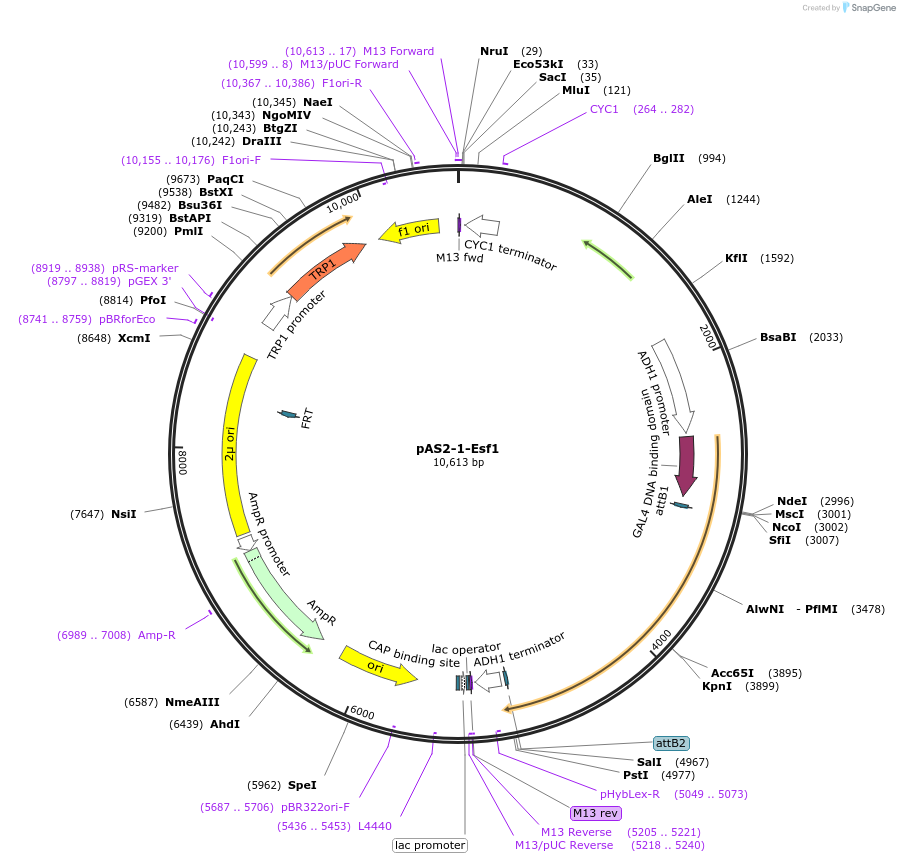
pAS2-1-Esf1
Plasmid#99987PurposeExpress yeast Esf1 with Y2H binding domain fusionDepositorAvailable SinceNov. 20, 2017AvailabilityAcademic Institutions and Nonprofits only -
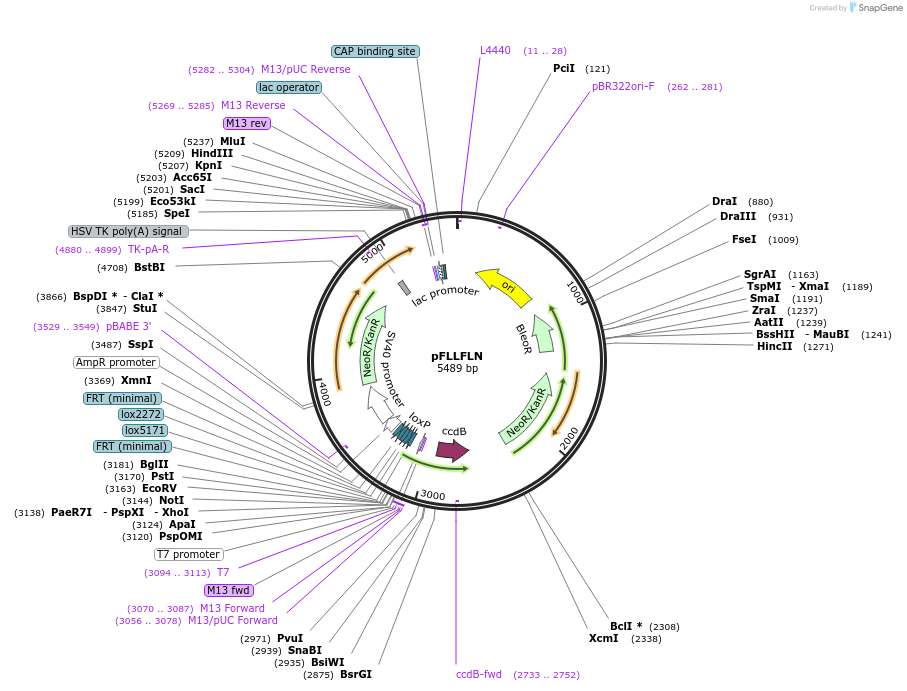
pFLLFLN
Plasmid#40030DepositorInsertsFRT-Lox5171-Lox2272-FRT-loxP
Neo
ExpressionBacterial and MammalianPromoterNone and SV40Available SinceOct. 15, 2012AvailabilityAcademic Institutions and Nonprofits only -
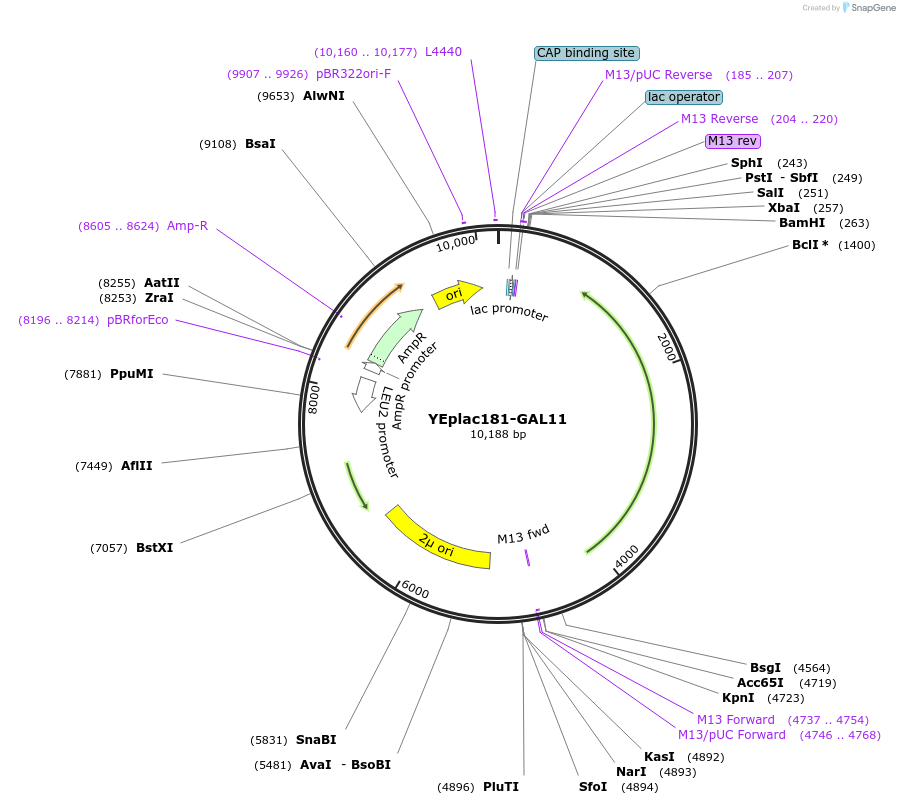
YEplac181-GAL11
Plasmid#8628DepositorAvailable SinceNov. 15, 2005AvailabilityAcademic Institutions and Nonprofits only -
-
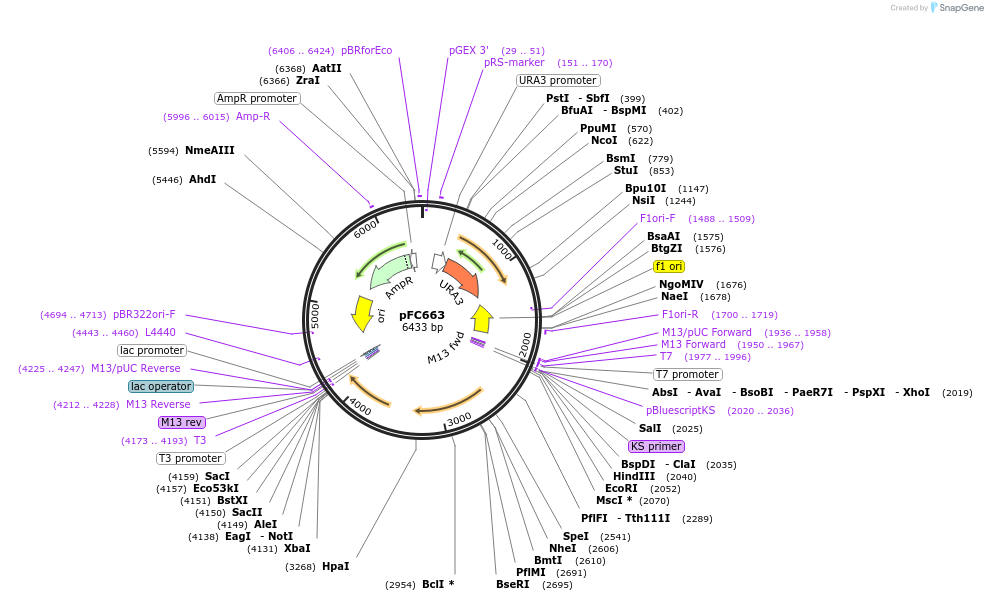
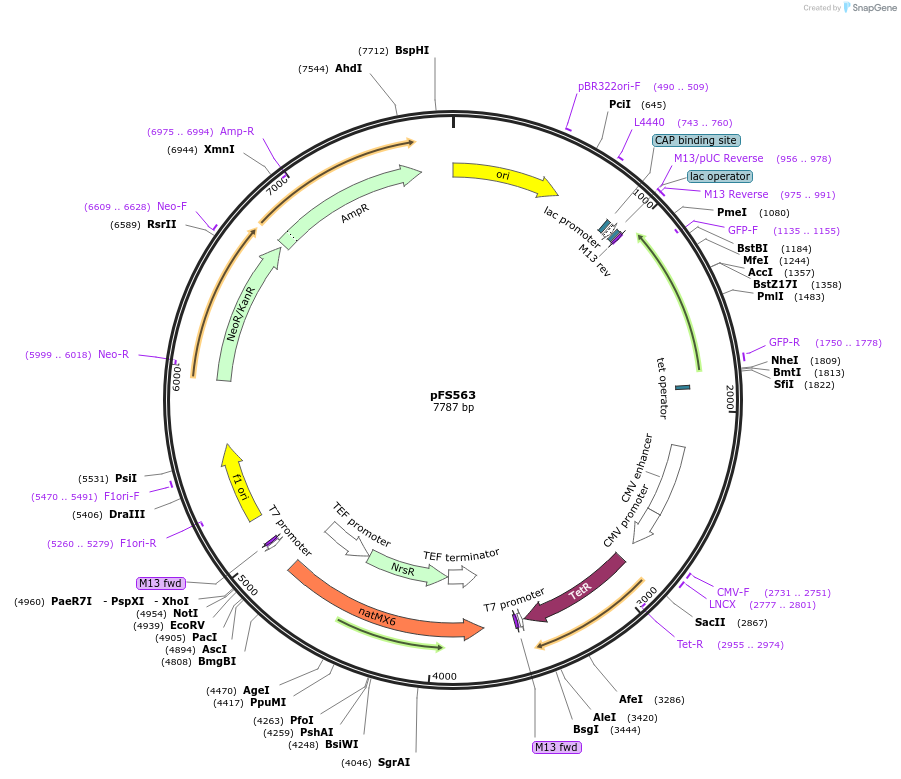
pFS563
Plasmid#242863PurposeThis plasmid will be used to replace the promoter of a gene with the PenotetSW2 promoter at its endogenous locusDepositorInsertsTetR
mECitrine
ExpressionYeastPromoterCMV promoter and enotetSW2 promoterAvailable SinceOct. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
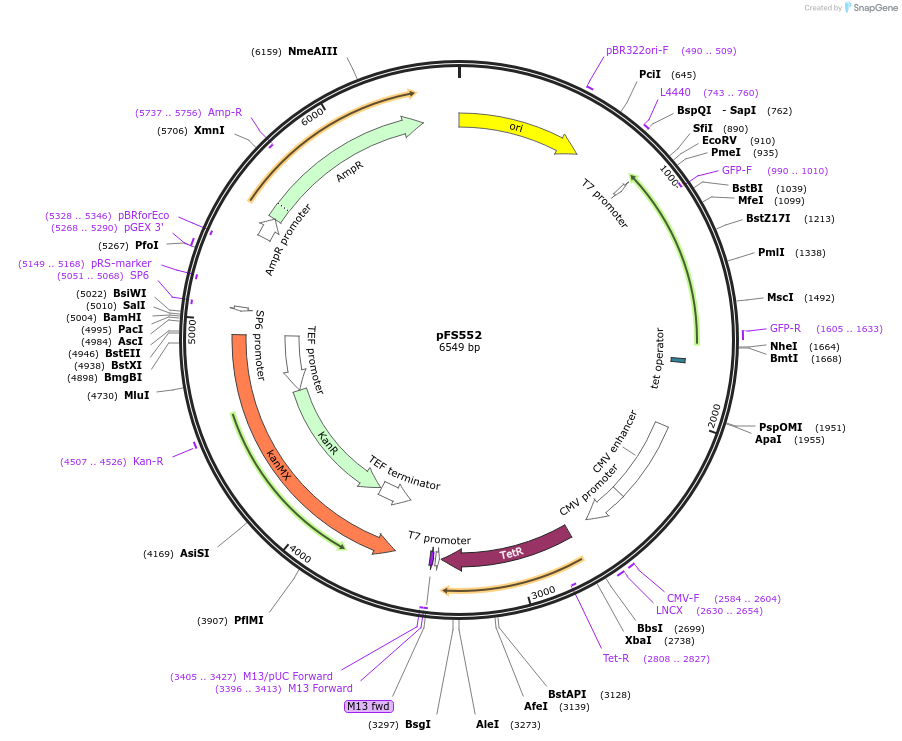
pFS552
Plasmid#242852PurposeThis plasmid will be used to replace the promoter of a gene with the PenotetSW1 promoter at its endogenous locusDepositorInsertsTetR
mECitrine
ExpressionYeastPromoterCMV promoter and enotetSW1 promoterAvailable SinceSept. 9, 2025AvailabilityAcademic Institutions and Nonprofits only