We narrowed to 30,195 results for: REP
-
Plasmid#51191DepositorInsertOrf2
UseSynthetic BiologyTags6x-hisExpressionBacterialPromoterT7Available SinceAug. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
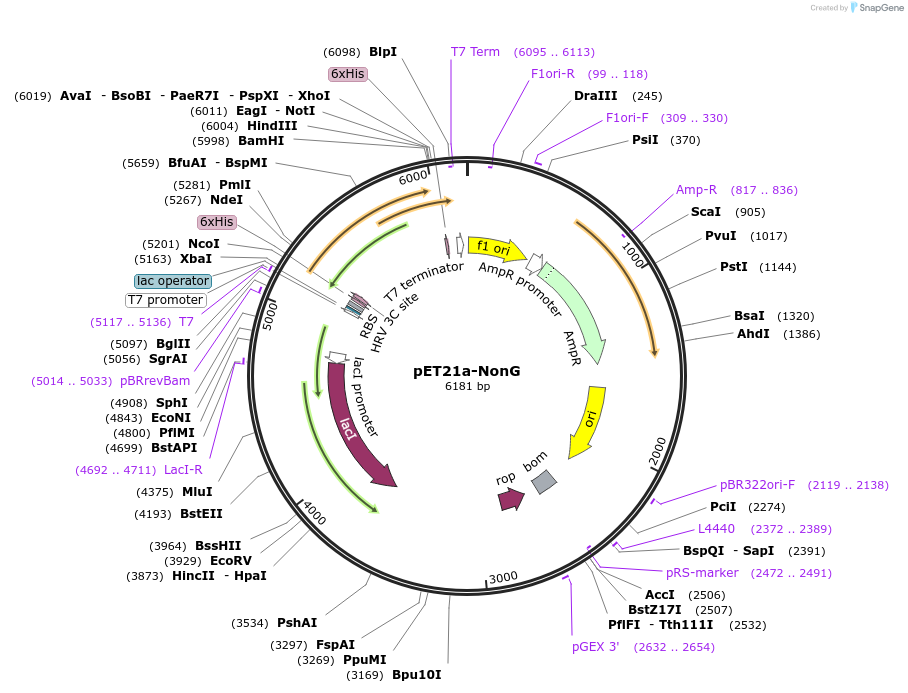
pET21a-NonG
Plasmid#51189DepositorInsertNonG
UseSynthetic BiologyTags6x-hisExpressionBacterialPromoterT7Available SinceAug. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
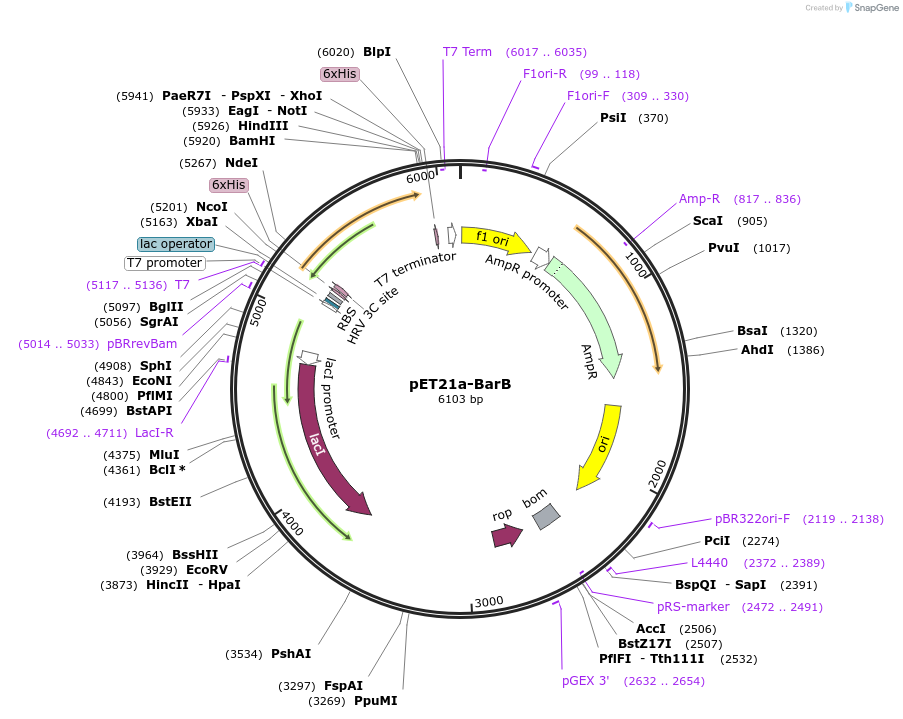
pET21a-BarB
Plasmid#51155DepositorInsertBarB
UseSynthetic BiologyTags6x-hisExpressionBacterialPromoterT7Available SinceAug. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
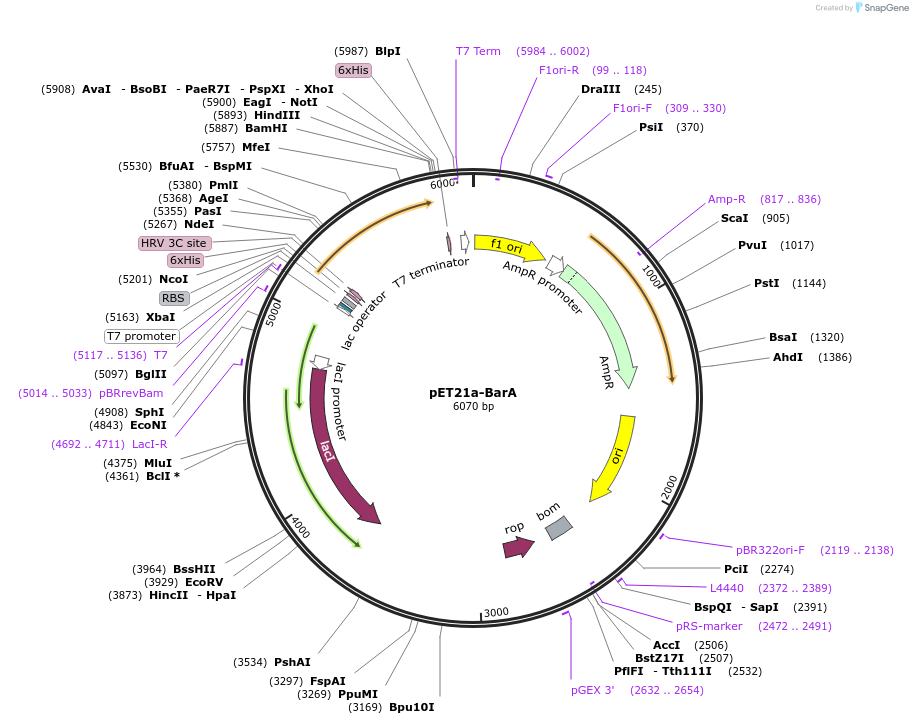
pET21a-BarA
Plasmid#51154DepositorInsertBarA
UseSynthetic BiologyTags6x-hisExpressionBacterialPromoterT7Available SinceAug. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
pSaI14
Plasmid#8614DepositorInsertRibonuclease inhibitor SaI14
ExpressionBacterialMutationN terminal threonine changed to alanineAvailable SinceJan. 17, 2007AvailabilityAcademic Institutions and Nonprofits only -
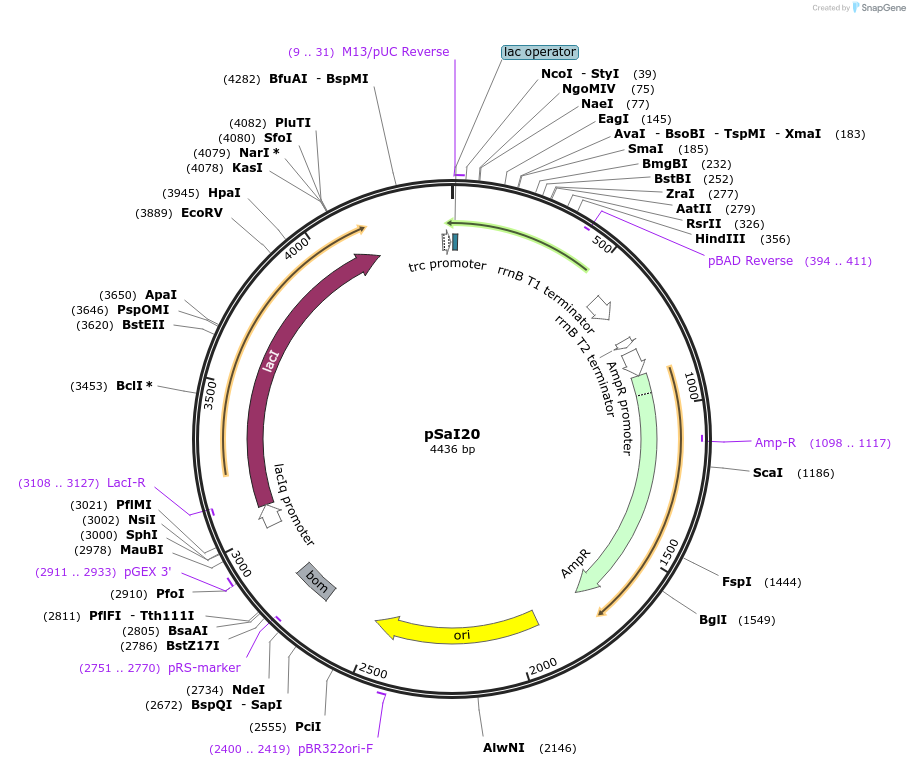
pSaI20
Plasmid#8615DepositorInsertRibonuclease inhibitor SaI20
ExpressionBacterialMutationN terminal threnine changed to alanineAvailable SinceJan. 17, 2007AvailabilityAcademic Institutions and Nonprofits only -
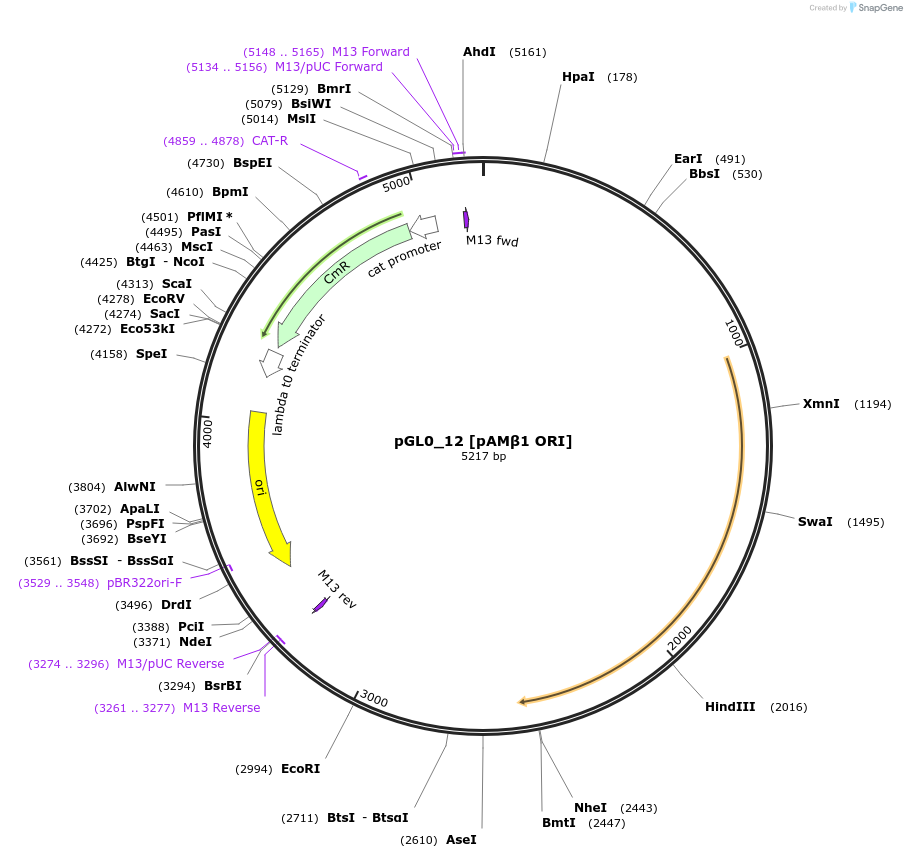
pGL0_12 [pAMβ1 ORI]
Plasmid#198926PurposeLevel 0 partDepositorInsertpAMβ1 ORI
UseSynthetic BiologyAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
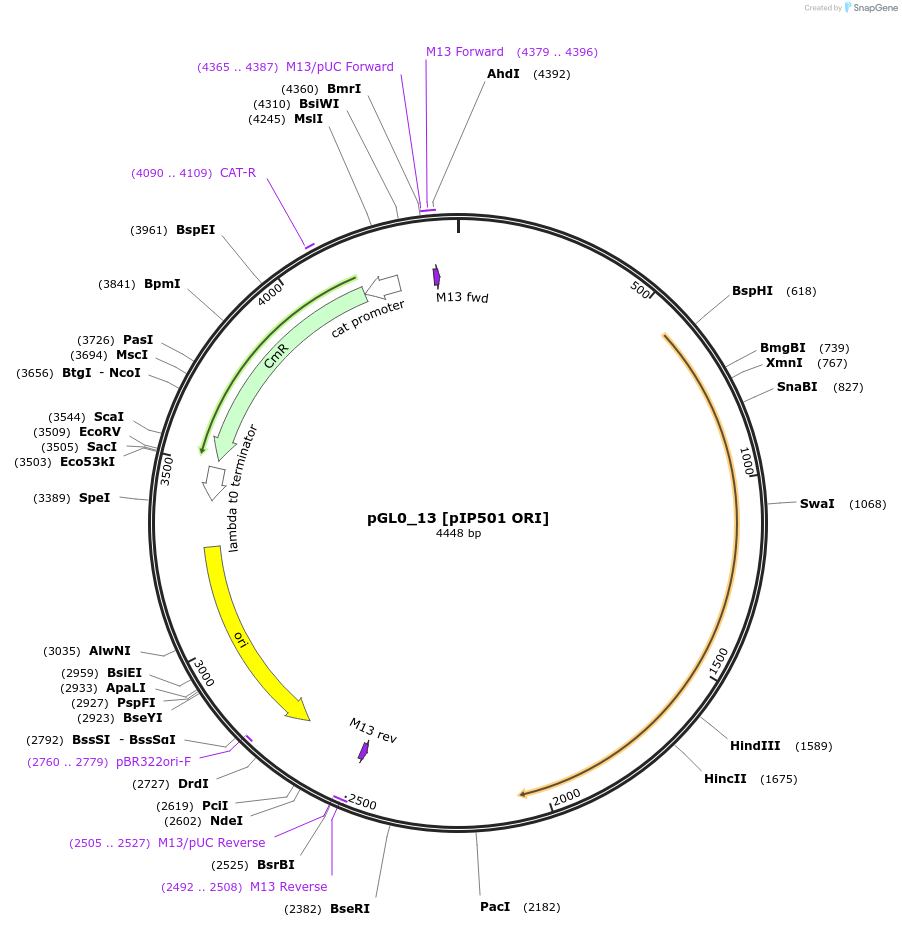
pGL0_13 [pIP501 ORI]
Plasmid#198927PurposeLevel 0 partDepositorInsertpIP501 ORI
UseSynthetic BiologyAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
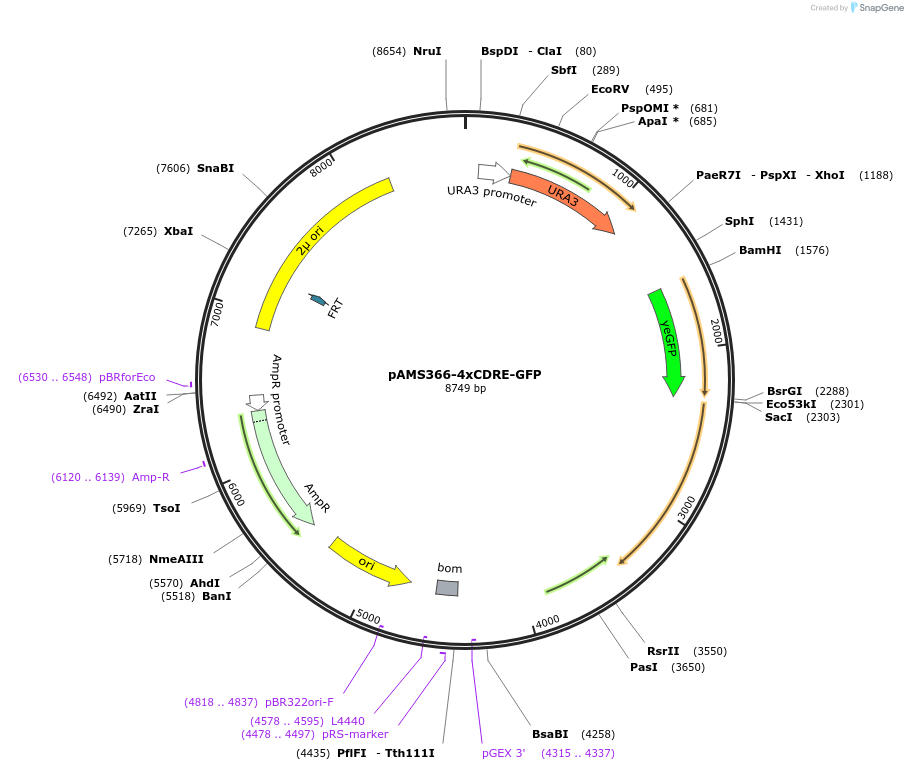
pAMS366-4xCDRE-GFP
Plasmid#138657Purposestable GFP reporter to monitor calcineurin activity via calcineurin-dependent response element (CDRE) in S. cerevisiaeDepositorInsert4xCDRE-CYC1(PM)-yeGFP
UseYeast reporter plasmidPromoterCYC1Available SinceMarch 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
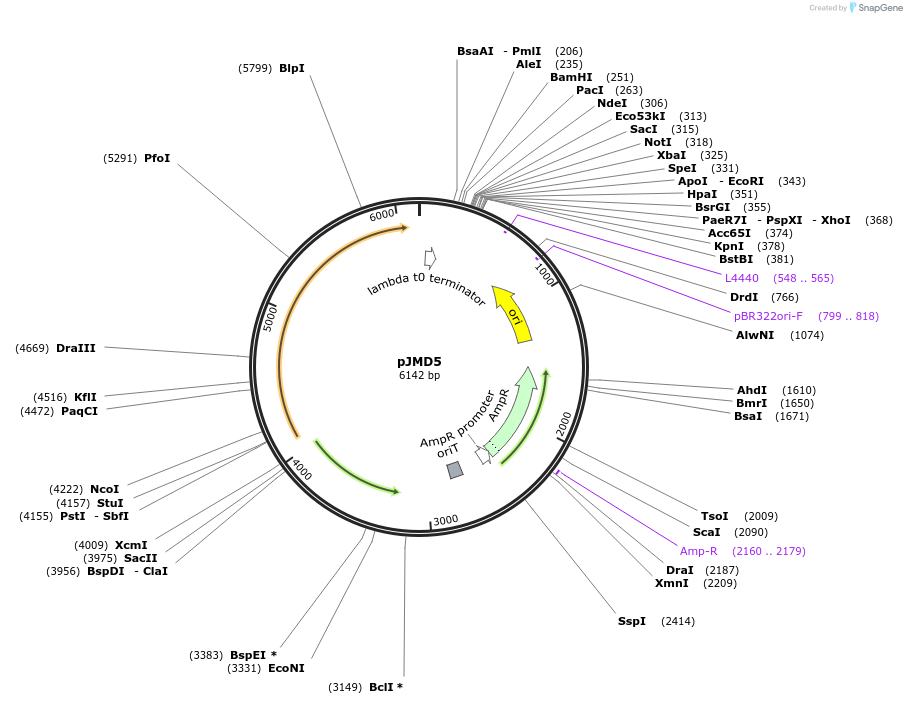
pJMD5
Plasmid#138652PurposephiBT1 integrating Streptomyces expression vectorDepositorTypeEmpty backboneExpressionBacterialPromoterPermE* variant sequence from pDA1652Available SinceApril 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
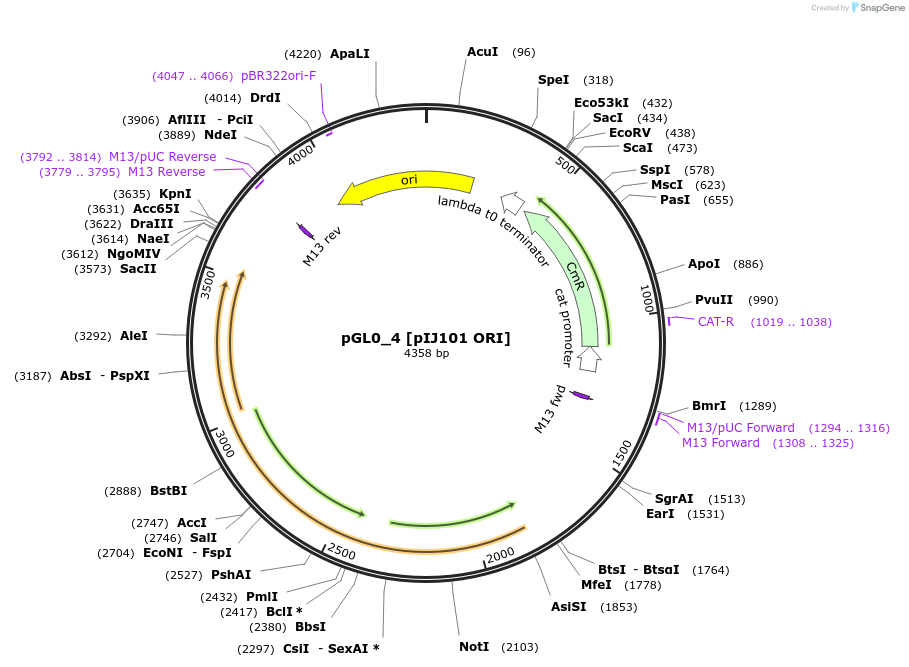
pGL0_4 [pIJ101 ORI]
Plasmid#198921PurposeLevel 0 partDepositorInsertpIJ101 ORI
UseSynthetic BiologyAvailable SinceSept. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
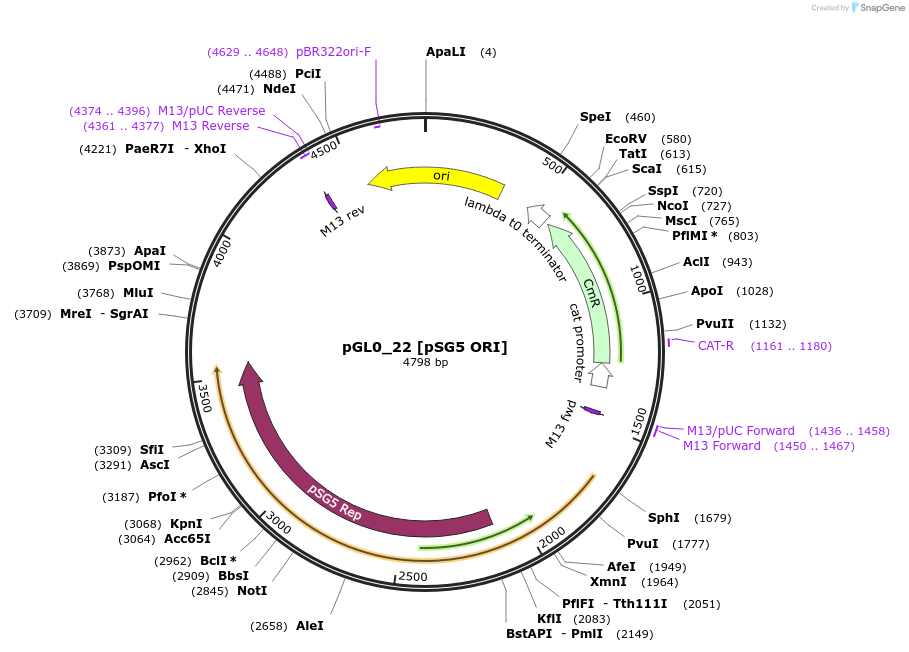
pGL0_22 [pSG5 ORI]
Plasmid#198935PurposeLevel 0 partDepositorInsertpSG5 ORI
UseSynthetic BiologyAvailable SinceJuly 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
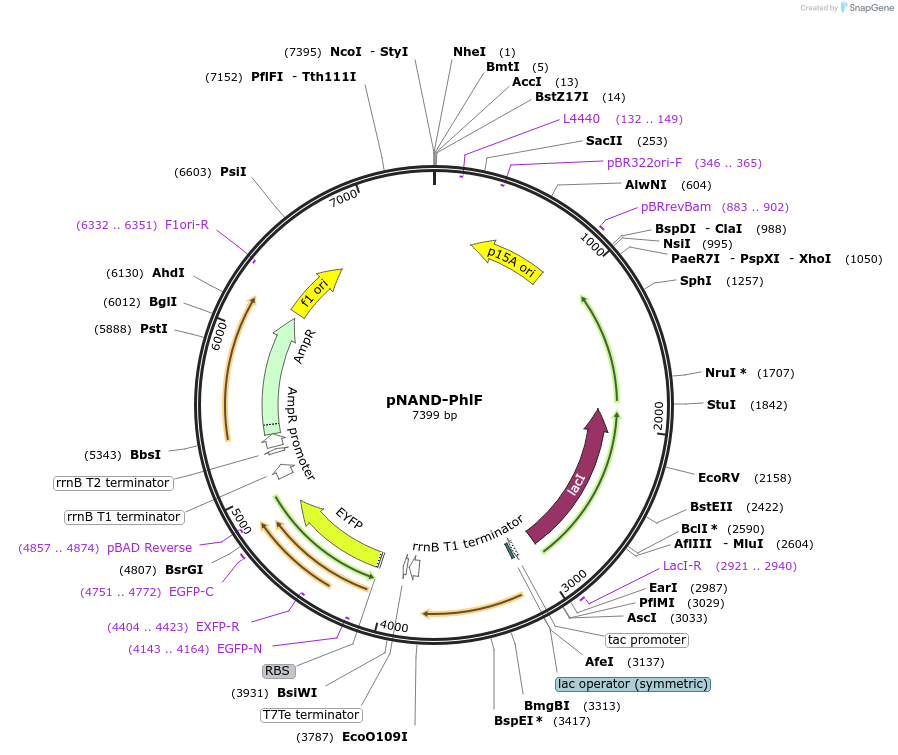
pNAND-PhlF
Plasmid#49376DepositorInsertPhlF repressor, pPhlF-pLmrA reporter
UseSynthetic BiologyExpressionBacterialAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
pFastBac K-Flag Ring1A
Plasmid#1995DepositorAvailable SinceMarch 23, 2006AvailabilityAcademic Institutions and Nonprofits only -
pFastBac mph1 flag
Plasmid#1985DepositorAvailable SinceMarch 23, 2006AvailabilityAcademic Institutions and Nonprofits only -
pFastBac flag Mel18
Plasmid#1968DepositorAvailable SinceMay 24, 2006AvailabilityAcademic Institutions and Nonprofits only -
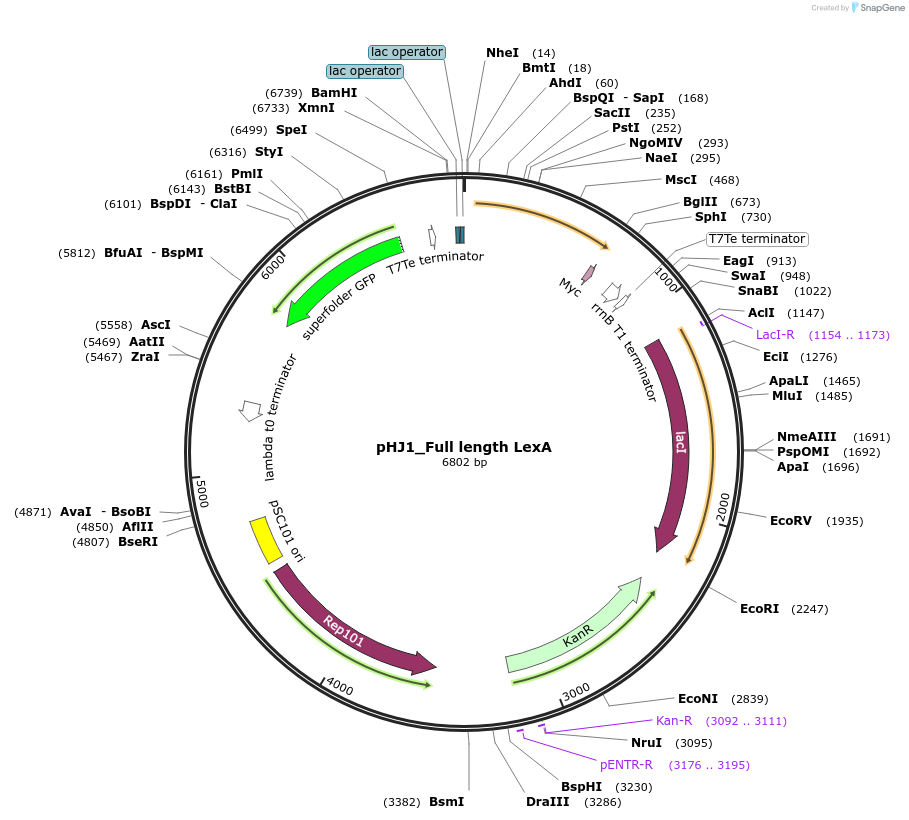
pHJ1_Full length LexA
Plasmid#107237PurposesfGFP expression repressed by transcriptional factor LexADepositorInsertFull length LexA repressor
UseSynthetic BiologyPromoterpLacO1Available SinceMay 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
pFastBac Mel18 flag
Plasmid#1969DepositorAvailable SinceMarch 23, 2006AvailabilityAcademic Institutions and Nonprofits only -
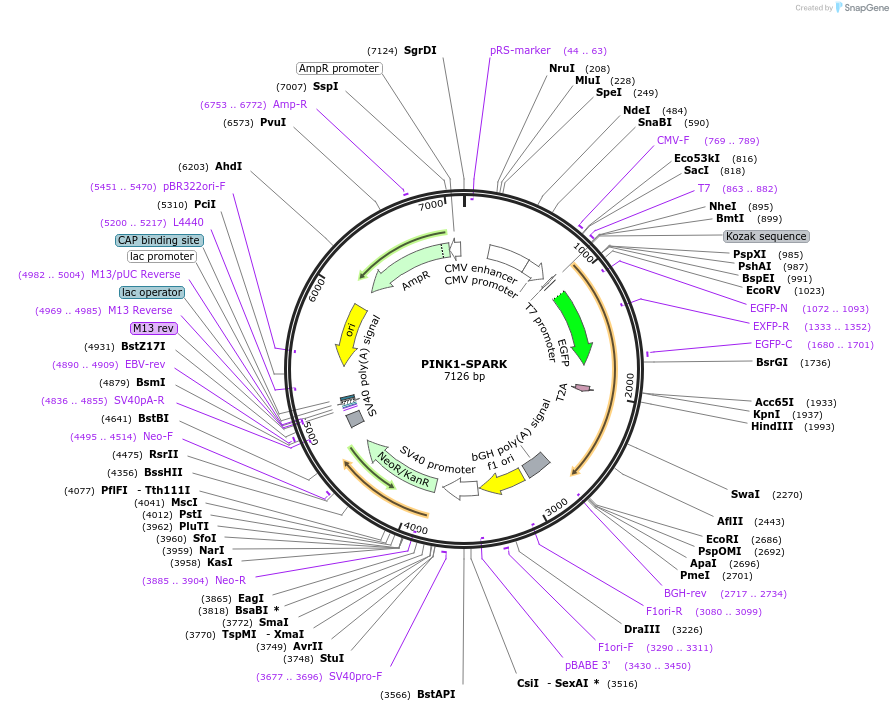
PINK1-SPARK
Plasmid#248086PurposePINK1 phase separation-based biosensorDepositorInsertPINK1-SPARK
ExpressionMammalianPromoterCMVAvailable SinceNov. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
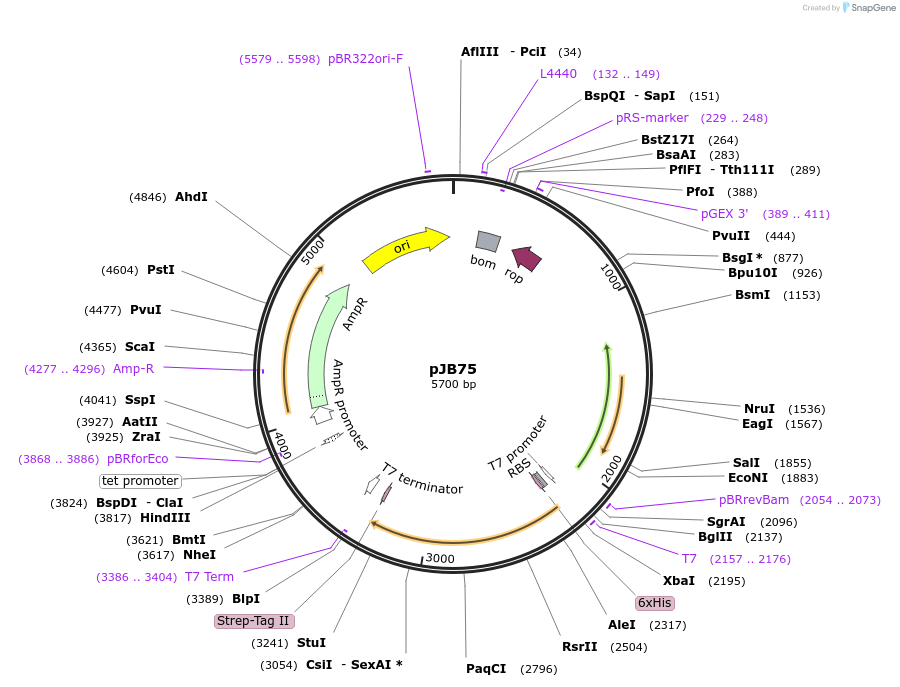
pJB75
Plasmid#226320PurposeE. coli protein expression for protein purificationDepositorInsert6xHis-(Y to A MR1-6)-strep
Tags6xHis and StrepExpressionBacterialMutationN-terminal domain deleted, C-terminal domain dele…PromoterT7Available SinceOct. 22, 2024AvailabilityAcademic Institutions and Nonprofits only