We narrowed to 20,523 results for: OMP;
-
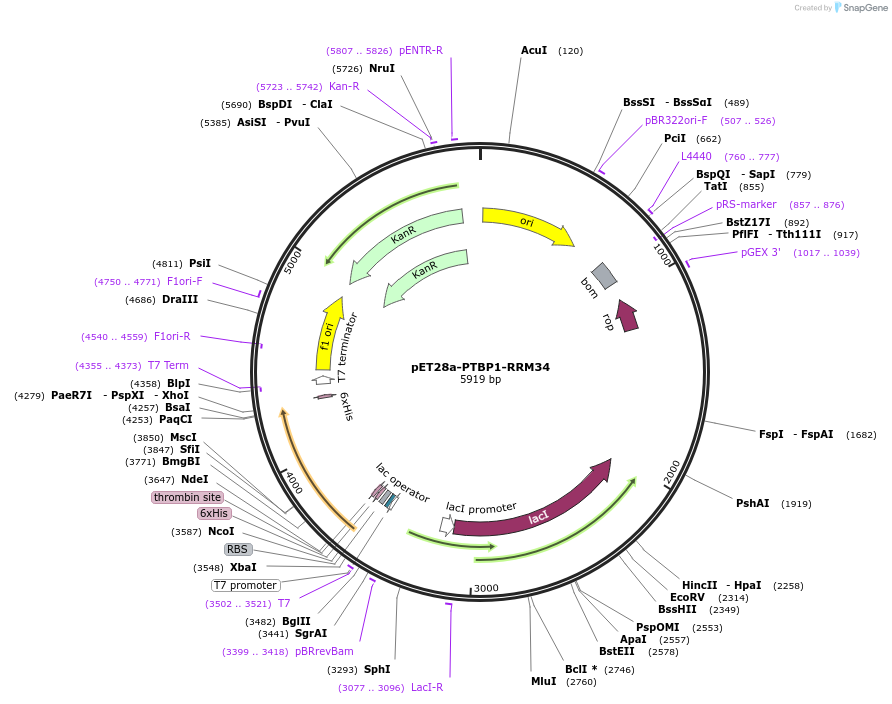
Plasmid#89156PurposeRecombinant expression of PTBP1-RRM34 in E.coliDepositorInsertPolypyrimidine Tract Binding Protein 1 RNA Recognition Motif 3+4 (PTBP1 Human)
Tagshexa HisExpressionBacterialMutationcontains RNA Recognition Motif 3+4PromoterT7Available SinceApril 12, 2017AvailabilityAcademic Institutions and Nonprofits only -
hRAD18 dSAP-EGFP
Plasmid#68825PurposeExpresses human RAD18 (deleting SAP domain) tagged with EGFPDepositorInserthuman RAD18 (deleting SAP domain) tagged with EGFP (RAD18 Human)
ExpressionMammalianAvailable SinceOct. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
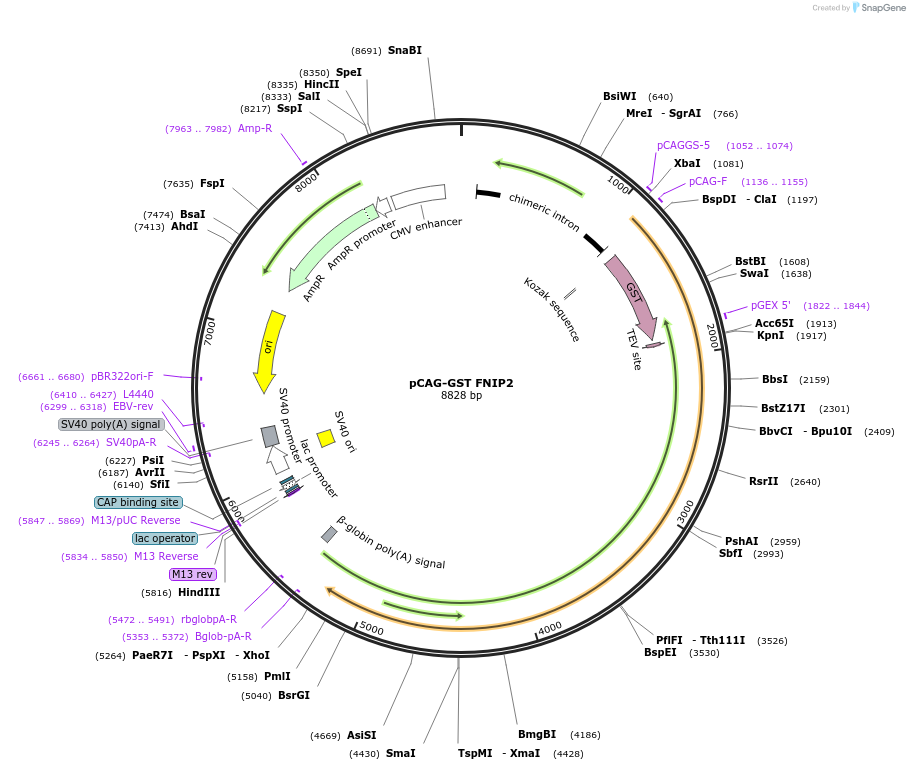
pCAG-GST FNIP2
Plasmid#164982PurposeFNIP2 expressing vector for mammalian cellsDepositorAvailable SinceMay 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLKO.1 MP1 shRNA
Plasmid#26632DepositorAvailable SinceNov. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
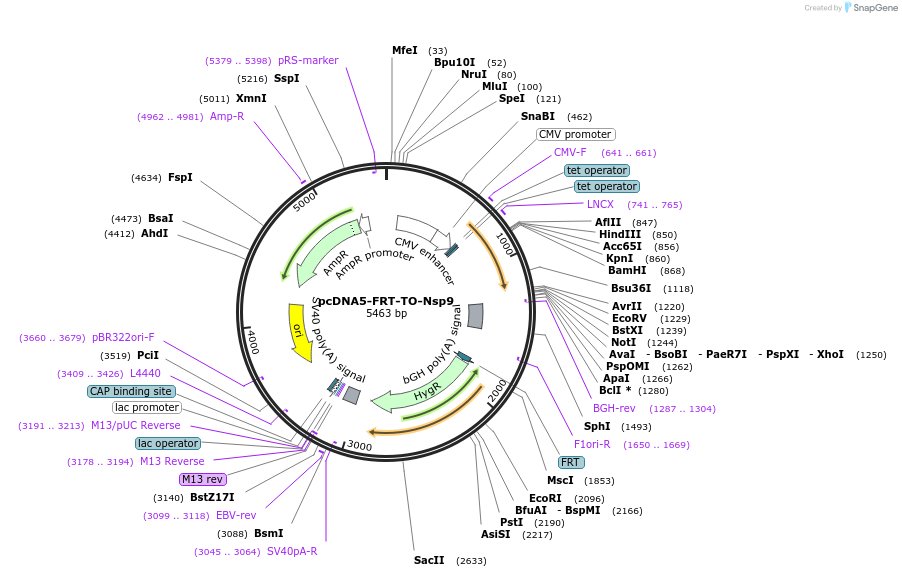
pcDNA5-FRT-TO-Nsp9
Plasmid#157703Purposemammalian expression of untagged SARS-CoV-2 Nsp9 under control of a tetracycline-inducible promoterDepositorAvailable SinceAug. 5, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGEM-human TCR zeta/2470
Plasmid#11507DepositorAvailable SinceMarch 8, 2006AvailabilityAcademic Institutions and Nonprofits only -
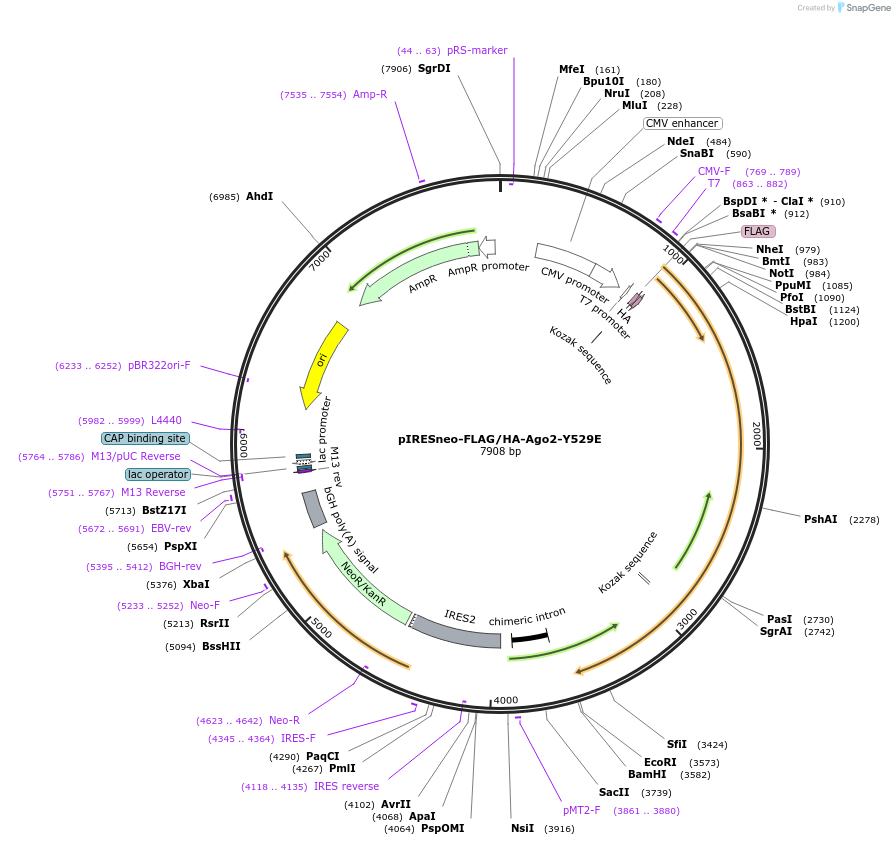
pIRESneo-FLAG/HA-Ago2-Y529E
Plasmid#215842PurposeExpression of AGO2-Y529E mutantDepositorAvailable SinceApril 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
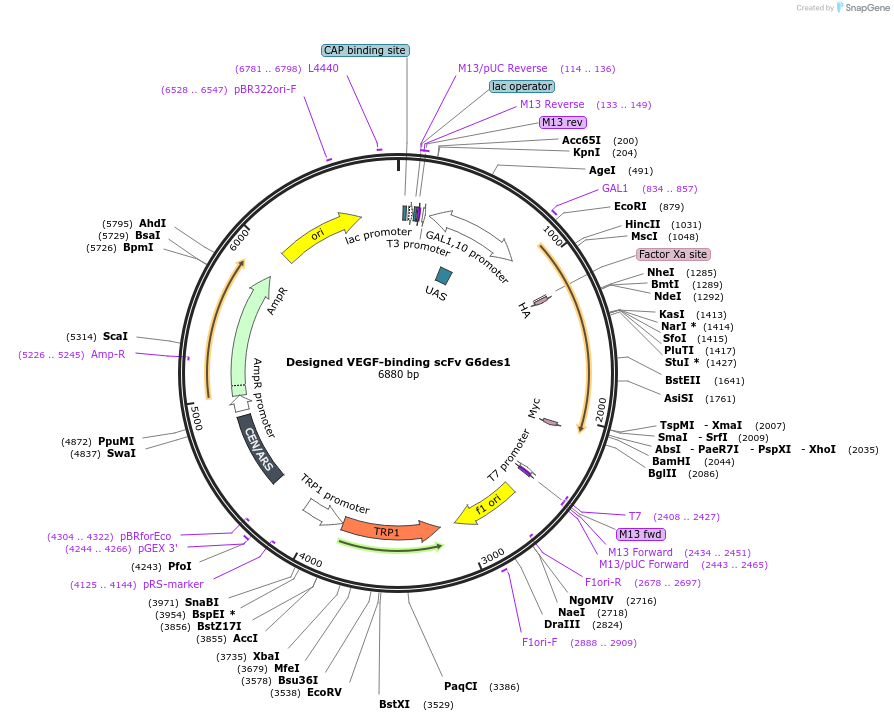
Designed VEGF-binding scFv G6des1
Plasmid#110212PurposeYeast Surface Display of G6des1DepositorInsertG6des1
Tagscmyc tagExpressionYeastMutationL A43P, L Y49F, L Y55F ,L Q89L, L T94V, L P95T, L…Available SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
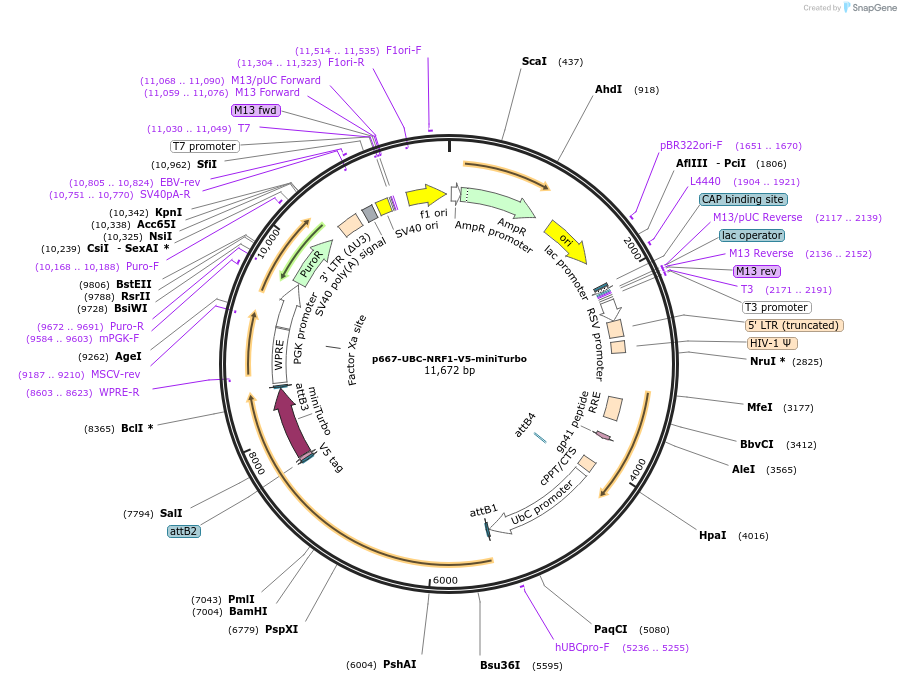
p667-UBC-NRF1-V5-miniTurbo
Plasmid#207810PurposeC-terminal tagged NRF1 for miniTurbo proximity experimentsDepositorAvailable SinceJan. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
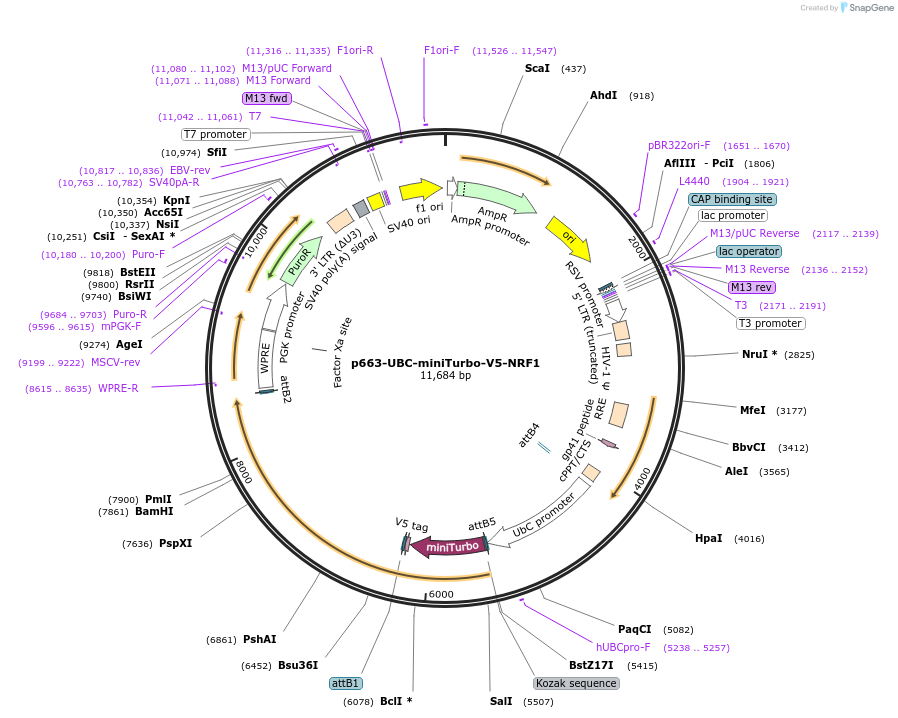
p663-UBC-miniTurbo-V5-NRF1
Plasmid#207809PurposeN-terminal tagged NRF1 for miniTurbo proximity experimentsDepositorAvailable SinceJan. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
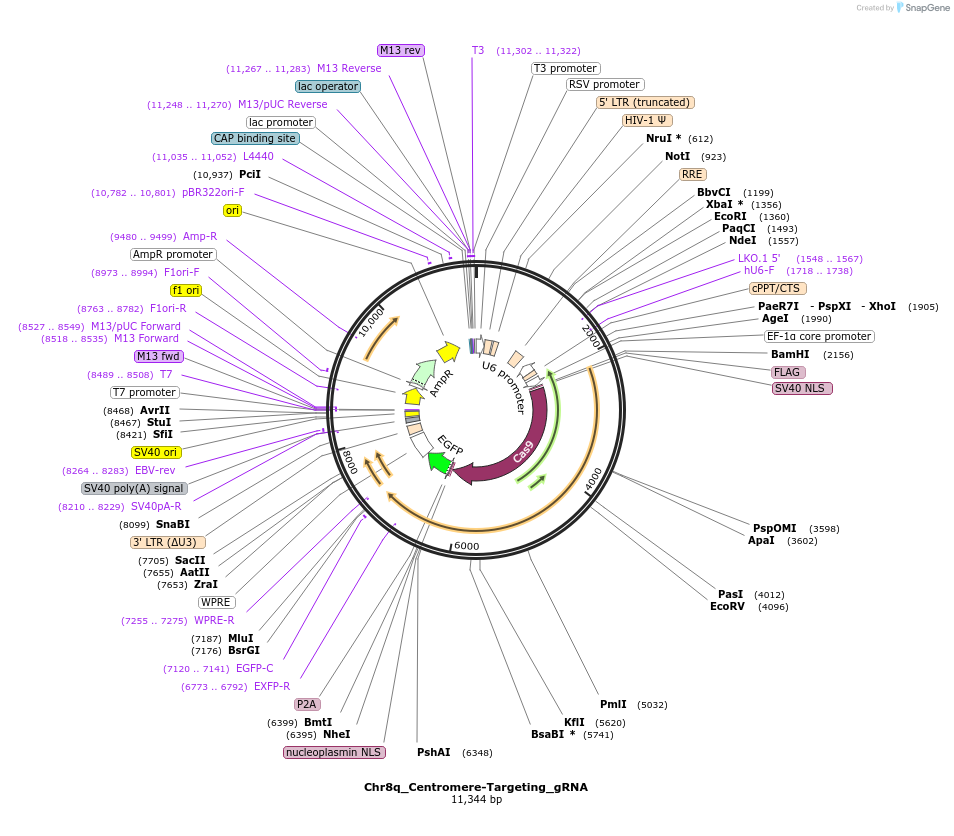
Chr8q_Centromere-Targeting_gRNA
Plasmid#195128PurposegRNA targeting centromere-proximal location on Chromosome 8q in a third generation Cas9 backbone with GFPDepositorInsertChr8q gRNA
ExpressionMammalianAvailable SinceFeb. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAd-Track Flag GCN5 Y621A/P622A
Plasmid#14425DepositorInsertGCN5 Y621A/P622A (KAT2A Human)
UseAdenoviralTagsflagExpressionMammalianMutationY621A, P622A. Catalytically inactive acetyltransf…Available SinceFeb. 23, 2007AvailabilityAcademic Institutions and Nonprofits only -
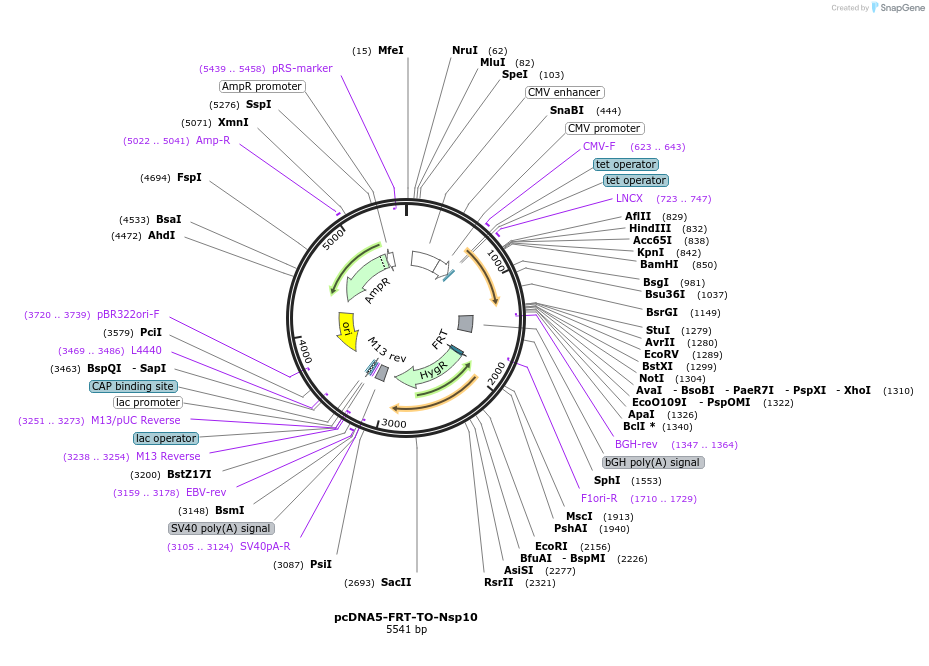
pcDNA5-FRT-TO-Nsp10
Plasmid#157706Purposemammalian expression of untagged SARS-CoV-2 Nsp10 under control of a tetracycline-inducible promoterDepositorAvailable SinceAug. 5, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
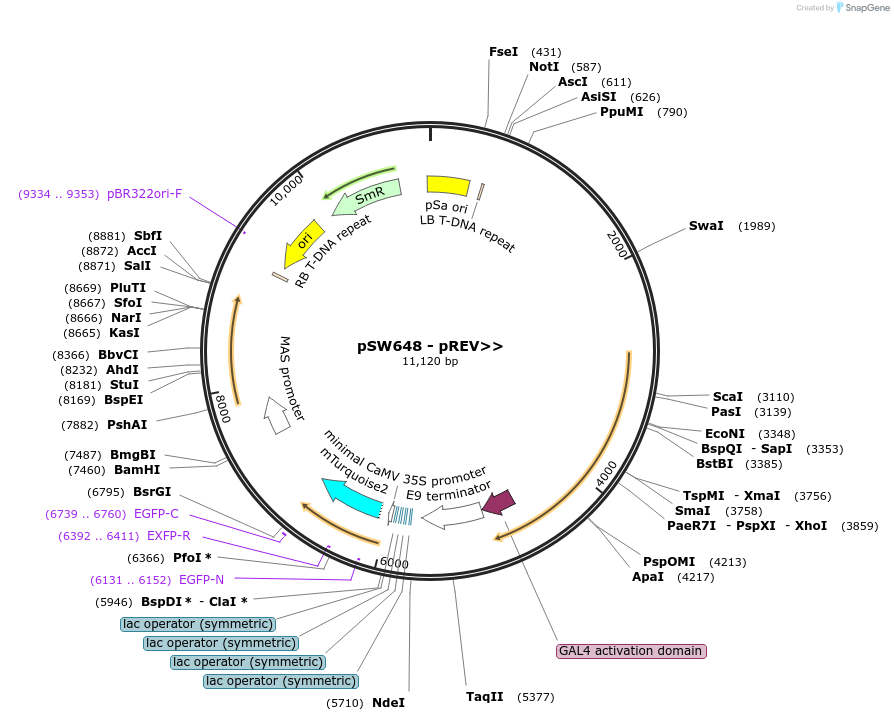
pSW648 - pREV>>
Plasmid#115996Purposedestination vector for GreenGate cloning method, contains 2x a set of modules from A-F, "Driver line", tissues-specific promoter, Dex-inducible mTurquoise2 expressionDepositorInsertpREV:GR-LhG4:tRBCS:F-H adapter-H-A adapter::pOp4:SP(ER)-mTurquoise2-HDEL:tUBQ10::SulfR
TagsHDEL and SP(ER)ExpressionPlantAvailable SinceApril 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
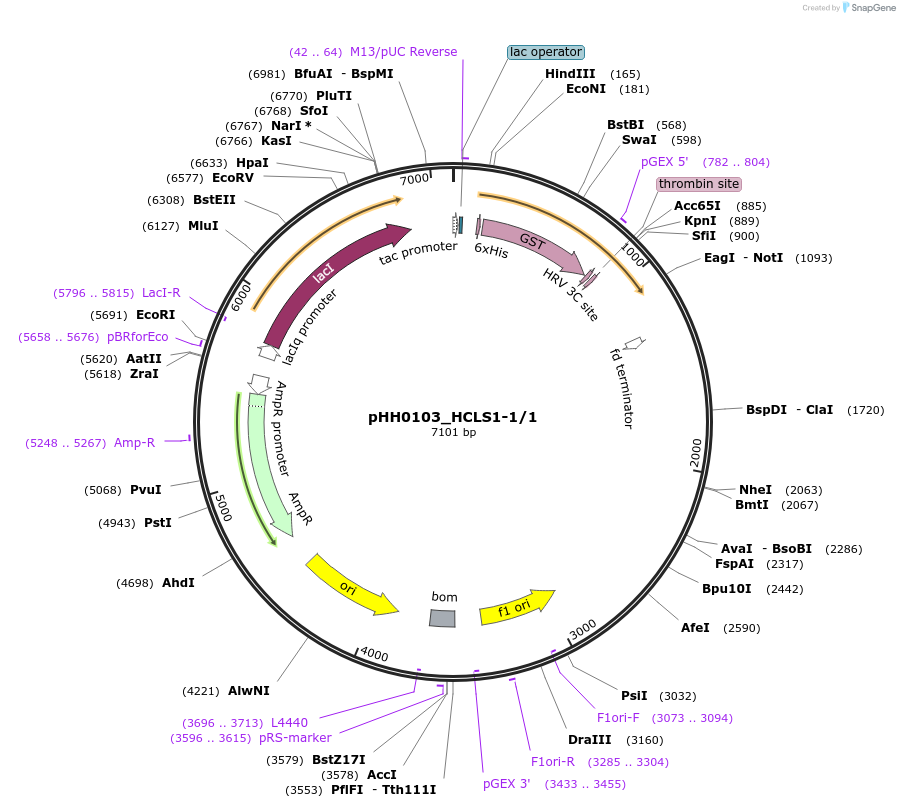
pHH0103_HCLS1-1/1
Plasmid#91437PurposeProtein expression and purification of human SH3 domain construct HCLS1-1/1DepositorInsertHCLS1-1/1 (HCLS1 Human)
ExpressionBacterialAvailable SinceMarch 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
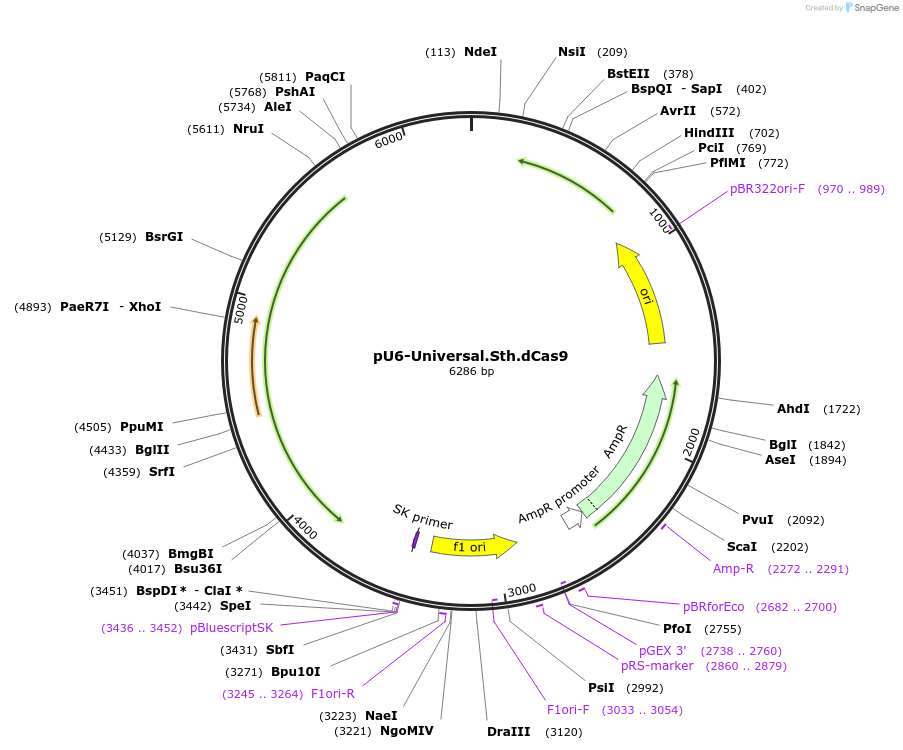
pU6-Universal.Sth.dCas9
Plasmid#171088PurposeCRISPR interference. Recipient construct for cloning 20-nt spacers into the sgRNA scaffold compatible with Streptococcus thermophilus dCas9 (CRISPR1 system) via BsaI restriction sites.DepositorInsertsToxoplasma U6 upstream region – spacer cloning site - sgRNA scaffold (compatible with S. thermophilus Cas9 CRISPR1)
DHFR-TSc3
UseToxoplasma gondii expressionMutationNote: A S245F mutation is present but did not app…PromoterDHFR-TS (Toxoplasma gondii) and U6 (Toxoplasma go…Available SinceOct. 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
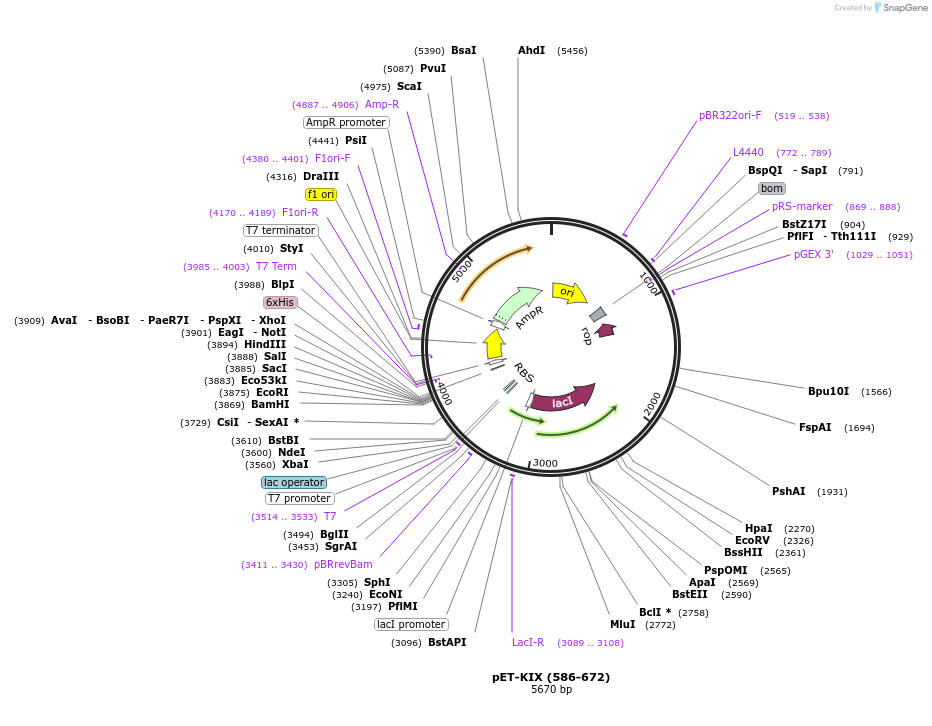
pET-KIX (586-672)
Plasmid#99336PurposeBacterial expression of mouse CBP KIX domainDepositorAvailable SinceSept. 6, 2017AvailabilityAcademic Institutions and Nonprofits only -
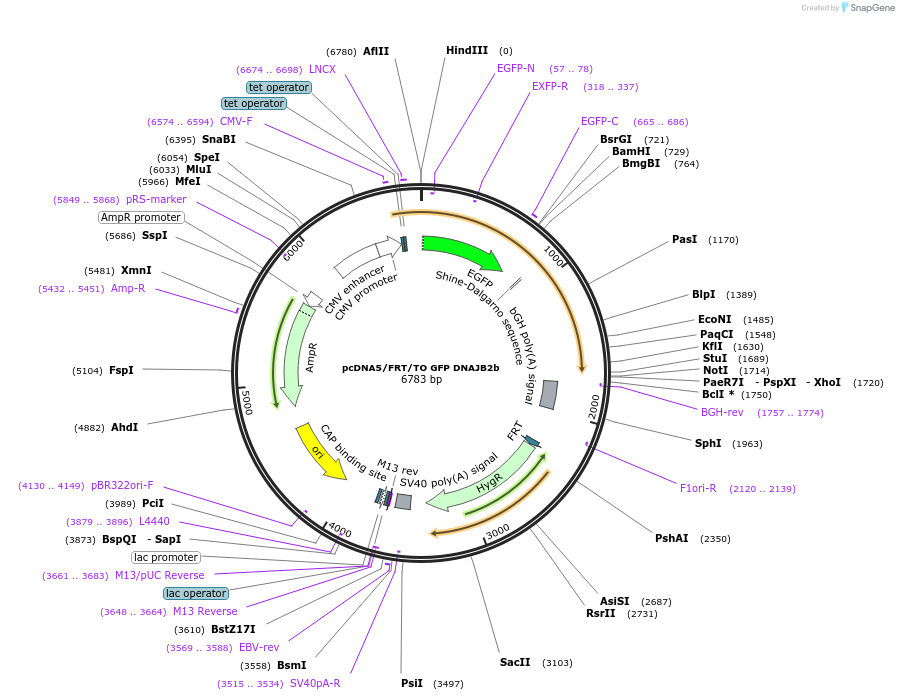
pcDNA5/FRT/TO GFP DNAJB2b
Plasmid#19496DepositorAvailable SinceOct. 6, 2008AvailabilityAcademic Institutions and Nonprofits only -
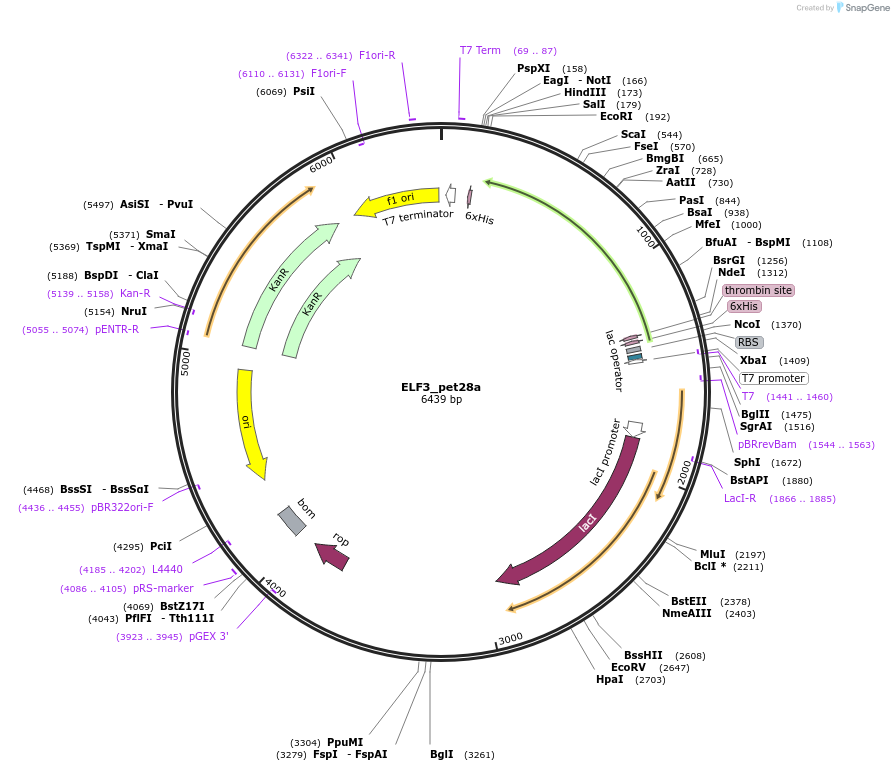
ELF3_pet28a
Plasmid#131658PurposeBacterial expression of ETS gene ELF3DepositorAvailable SinceOct. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
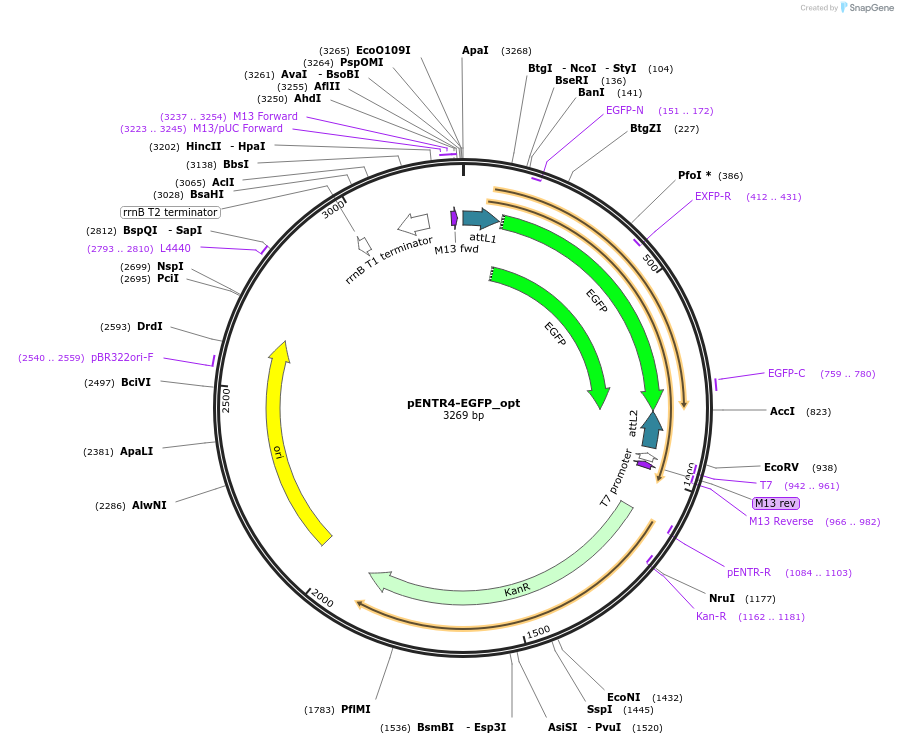
pENTR4-EGFP_opt
Plasmid#106334PurposeGateway Entry vector for EGFP (codon optimized for efficient gene synthesis)DepositorInsertEGFP
UseGateway entry vectorExpressionBacterial and MammalianMutationcodon optimized for gene synthesis without affect…Available SinceMarch 26, 2018AvailabilityAcademic Institutions and Nonprofits only