We narrowed to 38,370 results for: NAM
-
Plasmid#169815PurposeExpresses codon optimised full length His tagged PARP1 in bacterial cellsDepositorInsertPoly[ADP-ribose] polymerase 1 (PARP1 Synthetic, Codon optimised, Human)
TagsMKHHHHHHMKQExpressionBacterialMutationValine 762 mutated to an alaninePromoterT7 promoterAvailable SinceAug. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
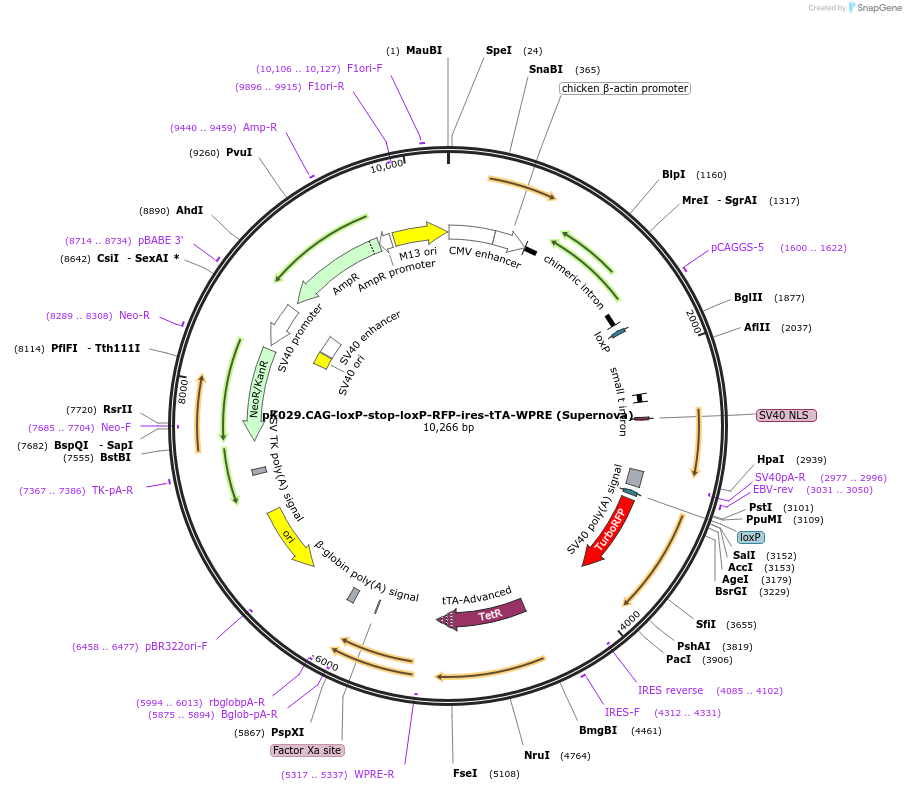
pK029.CAG-loxP-stop-loxP-RFP-ires-tTA-WPRE (Supernova)
Plasmid#69138PurposeFor the Supernova, a series of vector systems for single-cell labeling and gene function sudies in vivoDepositorInsertCAG-loxP-stop-loxP-RFP-ires-tTA-WPRE
TagsnoneExpressionMammalianAvailable SinceNov. 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
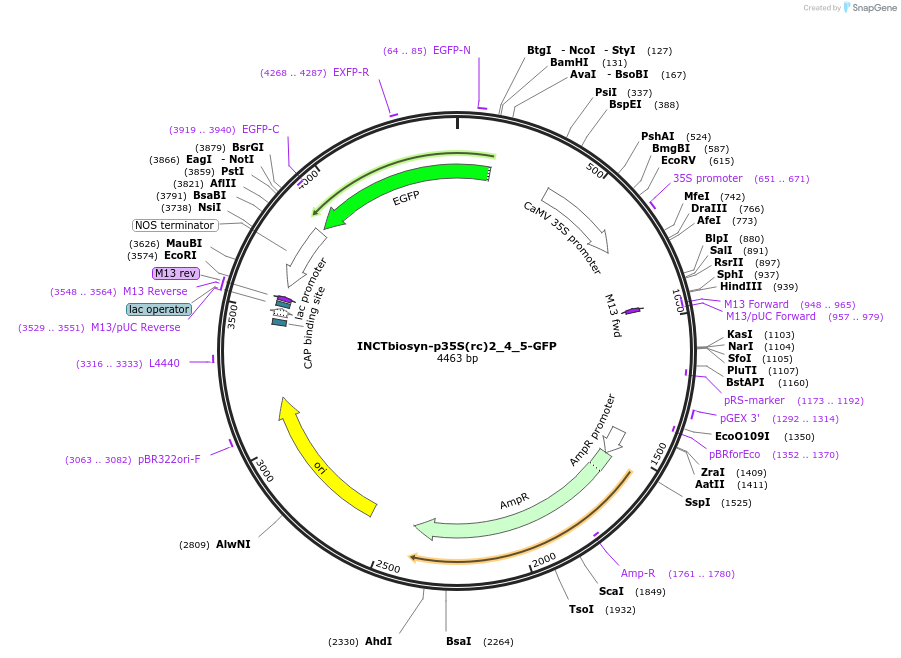
INCTbiosyn-p35S(rc)2_4_5-GFP
Plasmid#127520PurposePlasmid has an inverted CaMV 35S promoter sequence (reverse complement) flanked by attB and attP attachment sites of integrases 2, 4, and 5. EGFP coding sequence is in the forward orientation.DepositorInsertCaMV 35S reverse complement promoter sequence flanked by attB/attP Integrase 2, 4 and 5 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
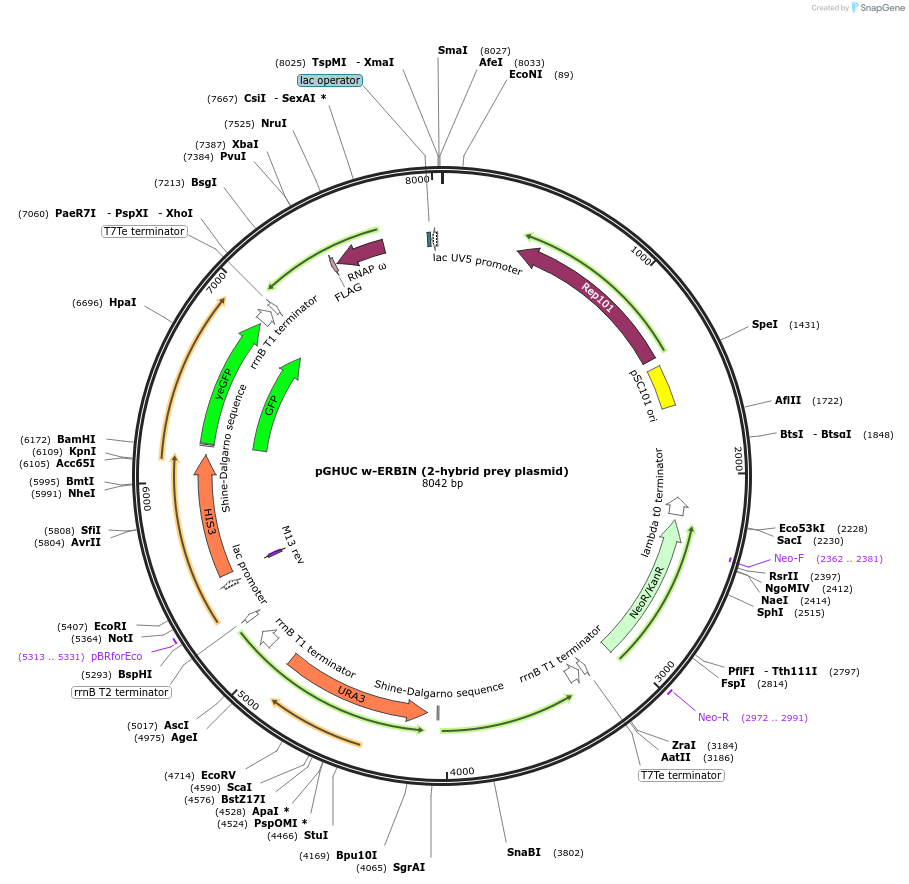
pGHUC w-ERBIN (2-hybrid prey plasmid)
Plasmid#128163PurposeUV5 promoter drives expression of omega-erbin fusion. Pair with pB1H2 UV5 Zif268-cons as a positive control (turns on HIS3/GFP).DepositorInsertERBIN PDZ domain (ERBIN Synthetic)
UseSynthetic BiologyTagsFLAG and Omega subunit of RNA polymeraseExpressionBacterialPromoterUV5Available SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
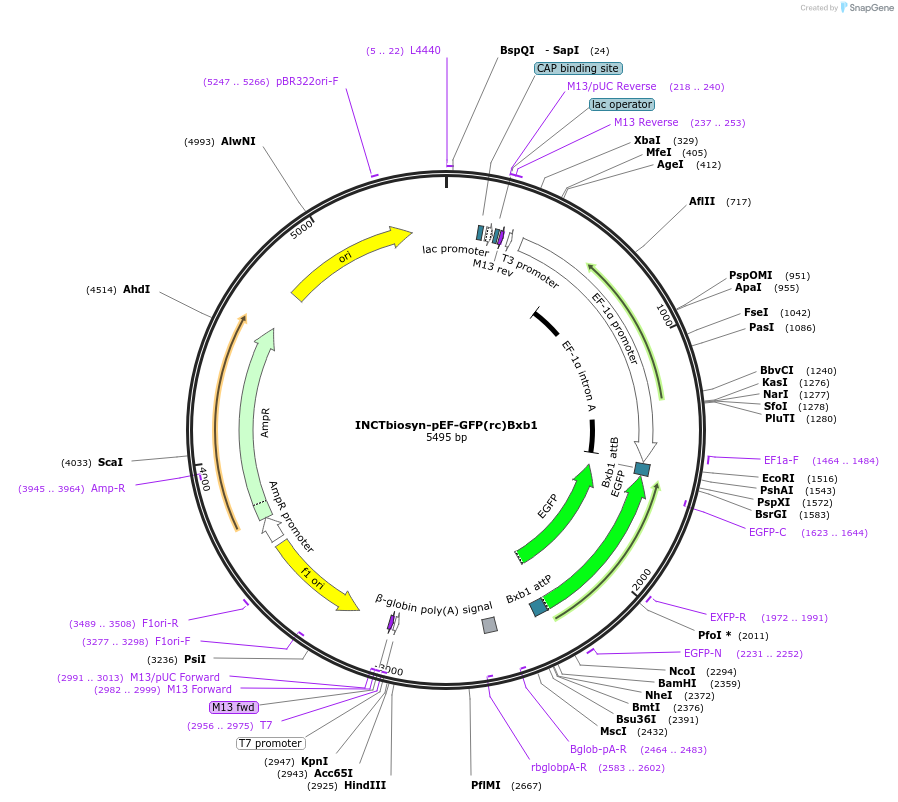
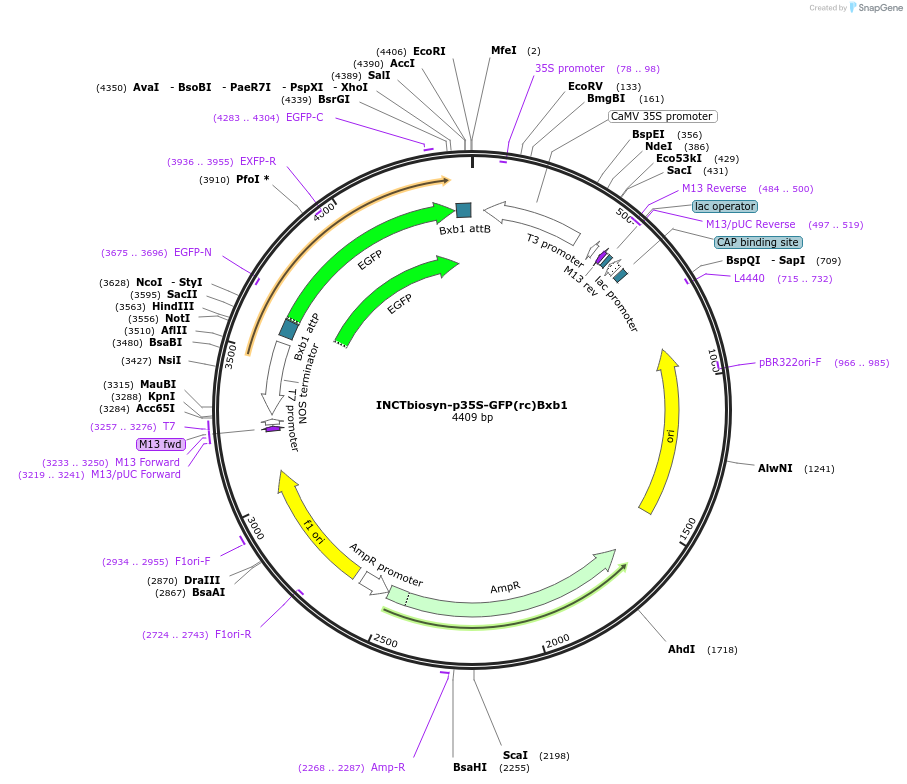
INCTbiosyn-pEF-GFP(rc)Bxb1
Plasmid#127511PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase Bxb1 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase Bxb1 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
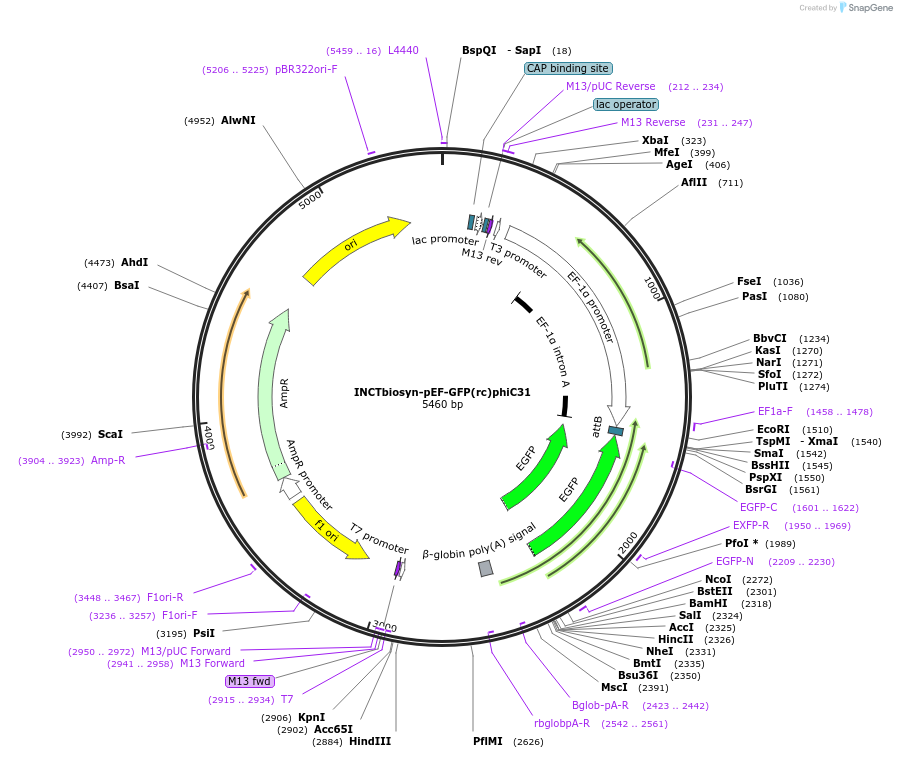
INCTbiosyn-pEF-GFP(rc)phiC31
Plasmid#127510PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase phiC31 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase phiC31 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
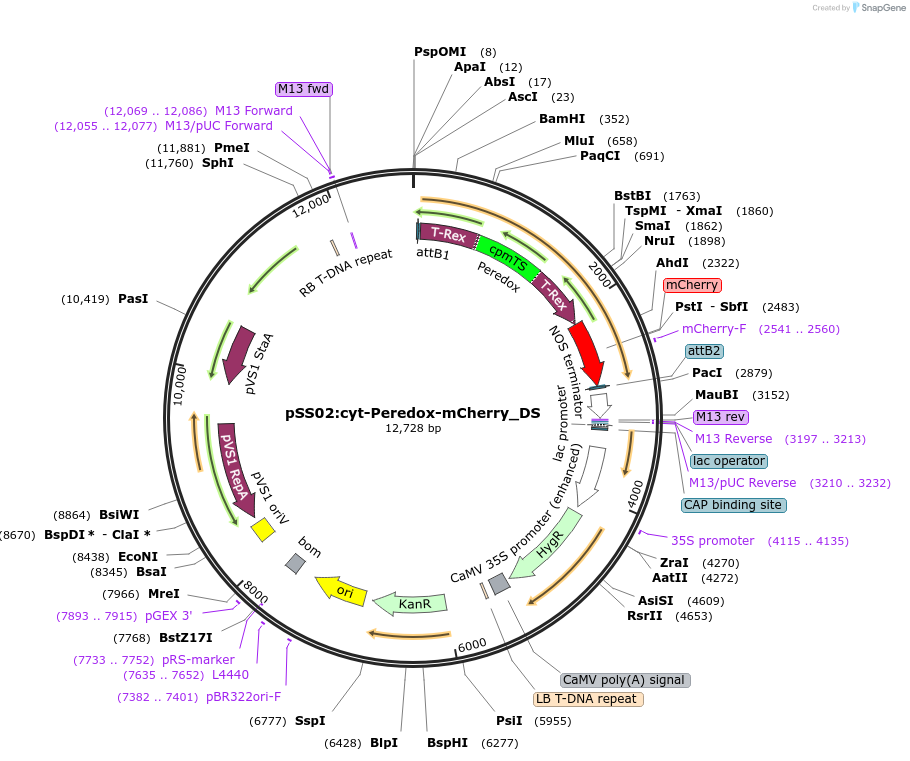
pSS02:cyt-Peredox-mCherry_DS
Plasmid#161747Purposecytosolic expression of fluorescent NADH/NAD+ biosensor Peredox-mCherry in plantsDepositorInsertcyt-Peredox-mCherry_DS
ExpressionPlantMutationamino acid substitution D115S in first T-Rex doma…PromoterUbiquitin 10Available SinceNov. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
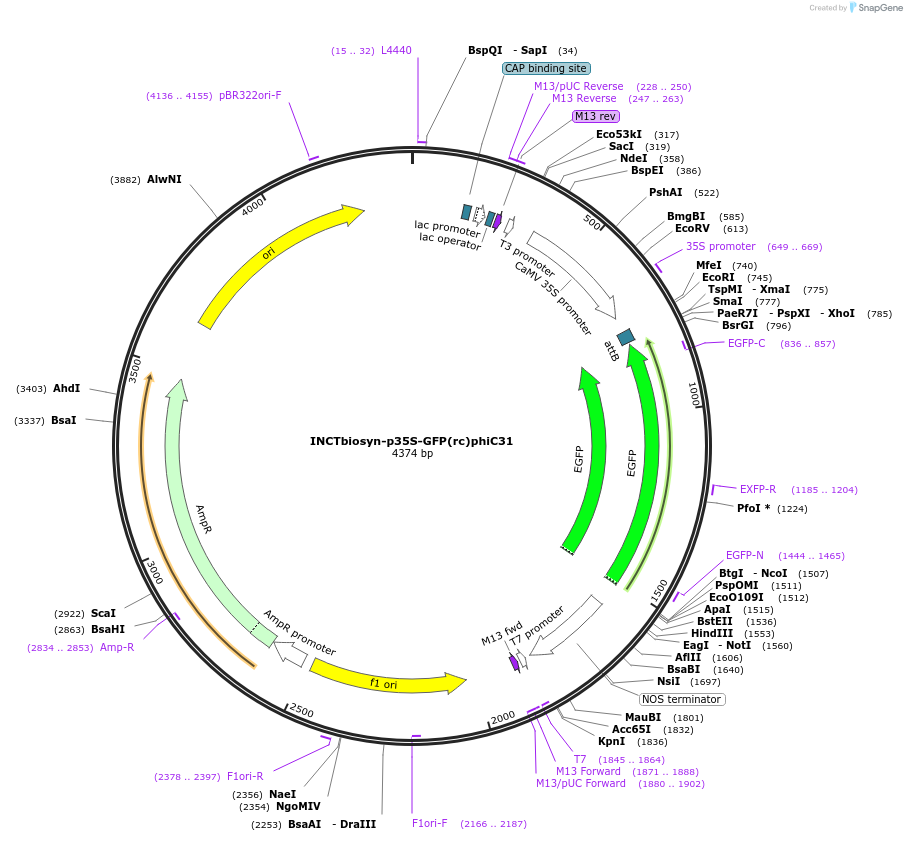
INCTbiosyn-p35S-GFP(rc)phiC31
Plasmid#127527PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase phiC31 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase phiC31 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
INCTbiosyn-p35S-GFP(rc)Bxb1
Plasmid#127528PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase Bxb1 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase Bxb1 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
pEGFP C1-Eps8 ∆cap/∆bund
Plasmid#74891Purposemammalian expression of the capping and bundling activity mutant Eps8 fused to GFPDepositorAvailable SinceMay 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
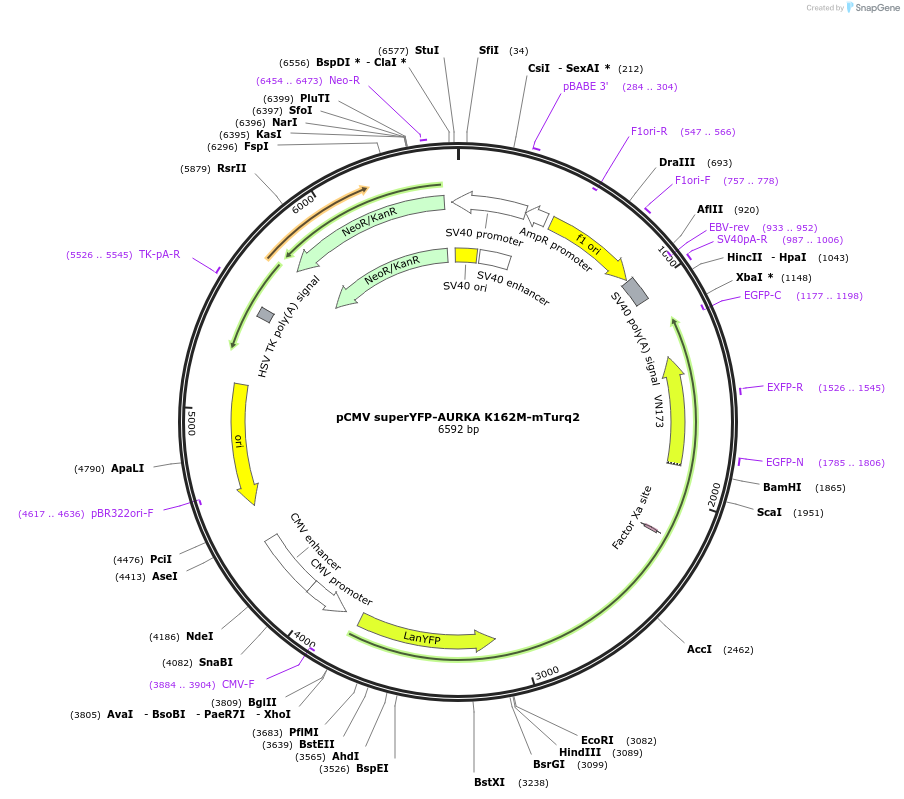
pCMV superYFP-AURKA K162M-mTurq2
Plasmid#157773PurposeExpression of AuroraA kinase-dead biosensor under CMV promoterDepositorInsertAURKA (AURKA Human)
TagsmTurquoise2 and superYFPExpressionMammalianMutationAURKA K162M is a kinase-dead version of AURKAPromoterCMVAvailable SinceFeb. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
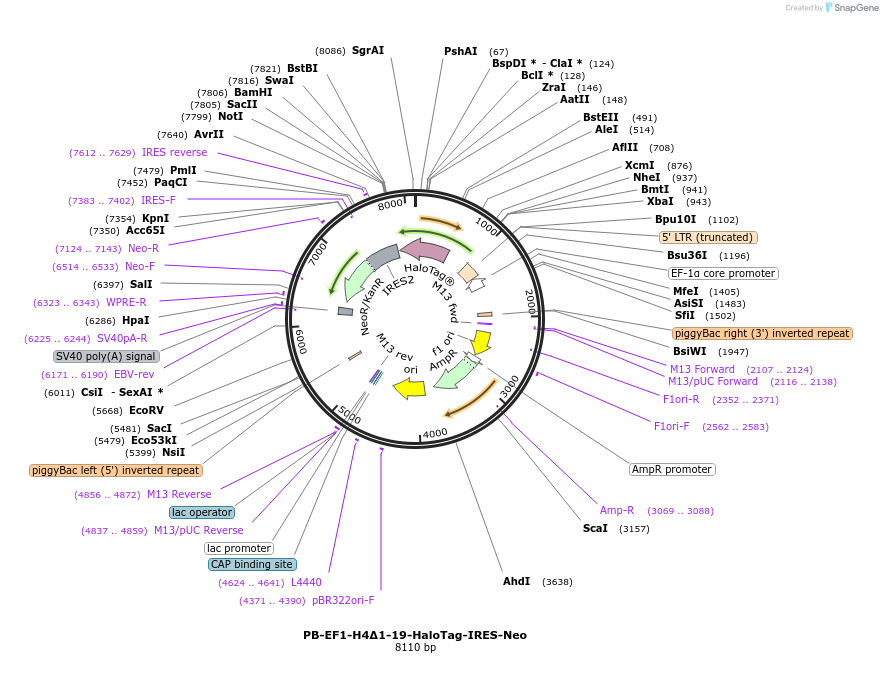
PB-EF1-H4Δ1-19-HaloTag-IRES-Neo
Plasmid#247451PurposeExpresses H4Δ1-19-Halo under EF1 promoter and can be randomly integrated by PiggyBac transposaseDepositorInsertH4 (H4C1) (H4C1 Human)
TagsHaloTagExpressionMammalianMutationH4Δ1-19 lacks the initial 19 residues at the N-te…PromoterEF1αAvailable SinceNov. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
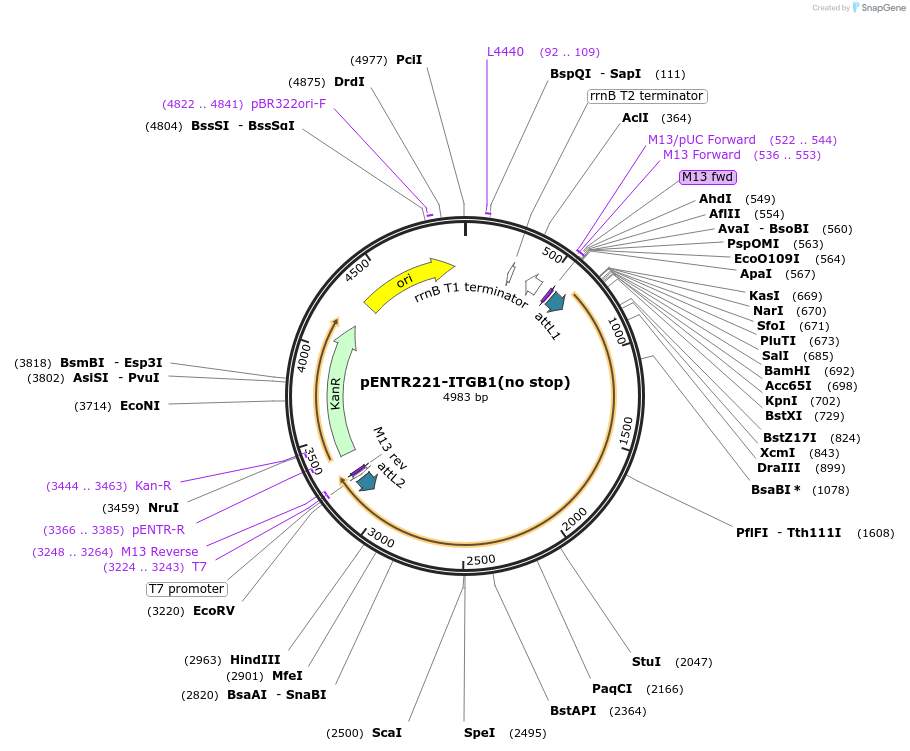
pENTR221-ITGB1(no stop)
Plasmid#215453PurposeGateway entry vector with integrin beta1 wild-type (WT) without a stop codonDepositorInsertITGB1 (ITGB1 Human)
UseGateway entry vectorMutationSilent mutations added to disrupt shRNA binding a…PromoternoneAvailable SinceSept. 4, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
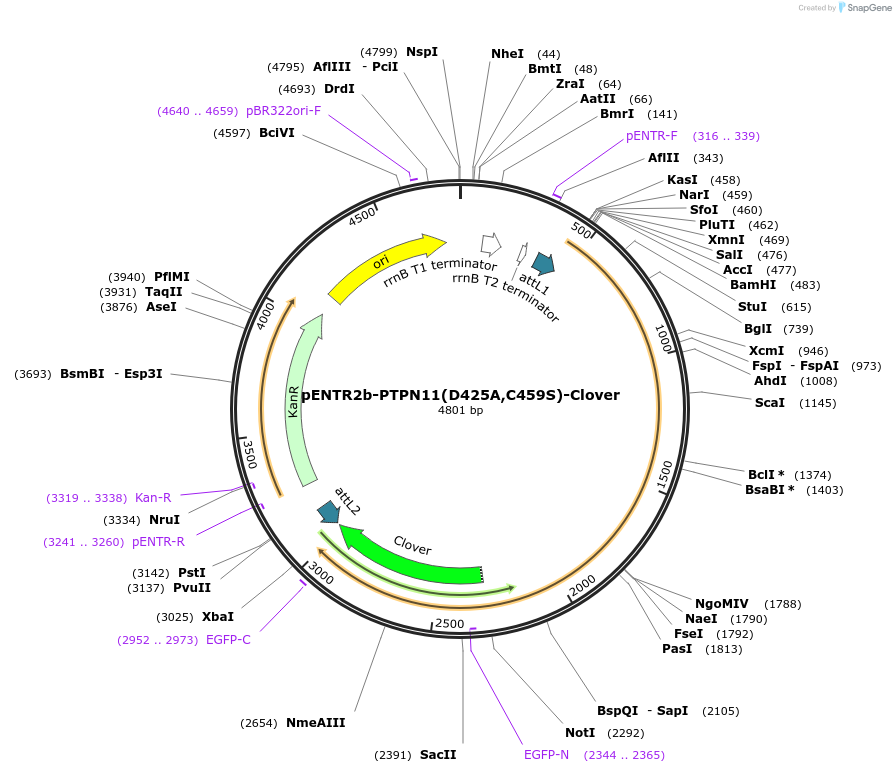
pENTR2b-PTPN11(D425A,C459S)-Clover
Plasmid#215520PurposeGateway entry vector with a clover-tagged phosphatase-inactive Shp2 mutant (D425A,C459S)DepositorInsertPTPN11 (PTPN11 Human)
UseGateway entry vectorTagsCloverMutationD425A, C459S (Phosphatase inactive)PromoternoneAvailable SinceSept. 3, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
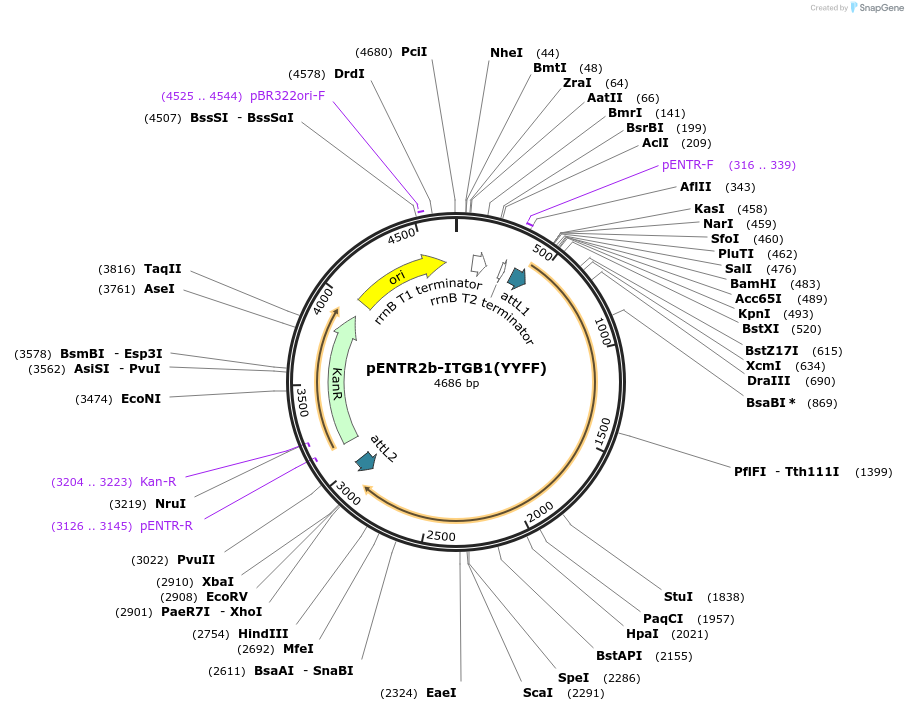
pENTR2b-ITGB1(YYFF)
Plasmid#215451PurposeGateway entry vector with integrin beta1 NPxY mutant (YYFF)DepositorInsertITGB1 (ITGB1 Human)
UseGateway entry vectorMutationMutations in the NPxY sites Y783F, Y795F; Silent …PromoternoneAvailable SinceMay 29, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
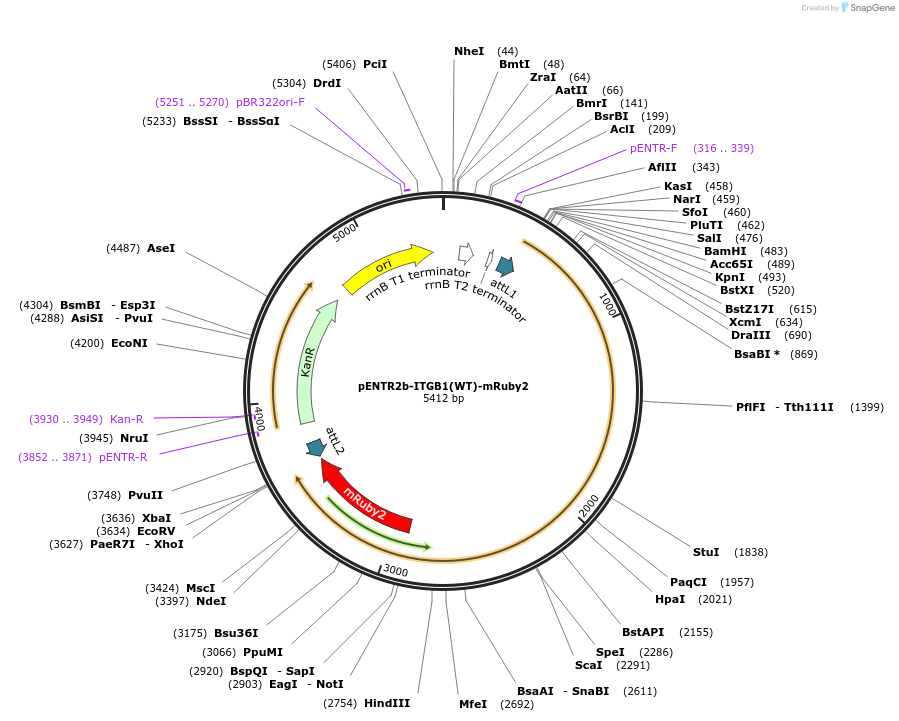
pENTR2b-ITGB1(WT)-mRuby2
Plasmid#215446PurposeGateway entry vector with integrin beta1 tagged with mRuby2DepositorInsertITGB1 (ITGB1 Human)
UseGateway entry vectorTagsmRuby2MutationSilent mutations added to disrupt shRNA binding a…PromoternoneAvailable SinceMay 29, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
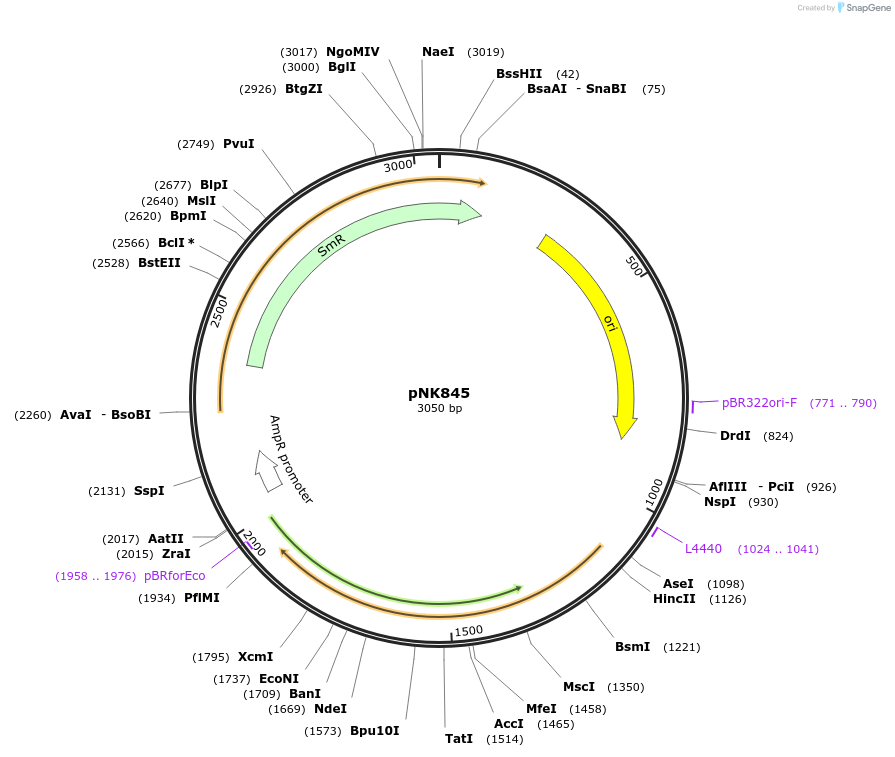
pNK845
Plasmid#219746PurposeMoClo-compatible Level 0 promoterless vector encoding mutant of Neonothopanus nambi luciferase nnLuz_v4 codon-optimised for expression in Pichia pastoris, Homo sapiensDepositorInsertmutant of fungal luciferase
UseLuciferase and Synthetic BiologyMutationI3S, N4T, F11L, I63T, T99P, T192S, A199PAvailable SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
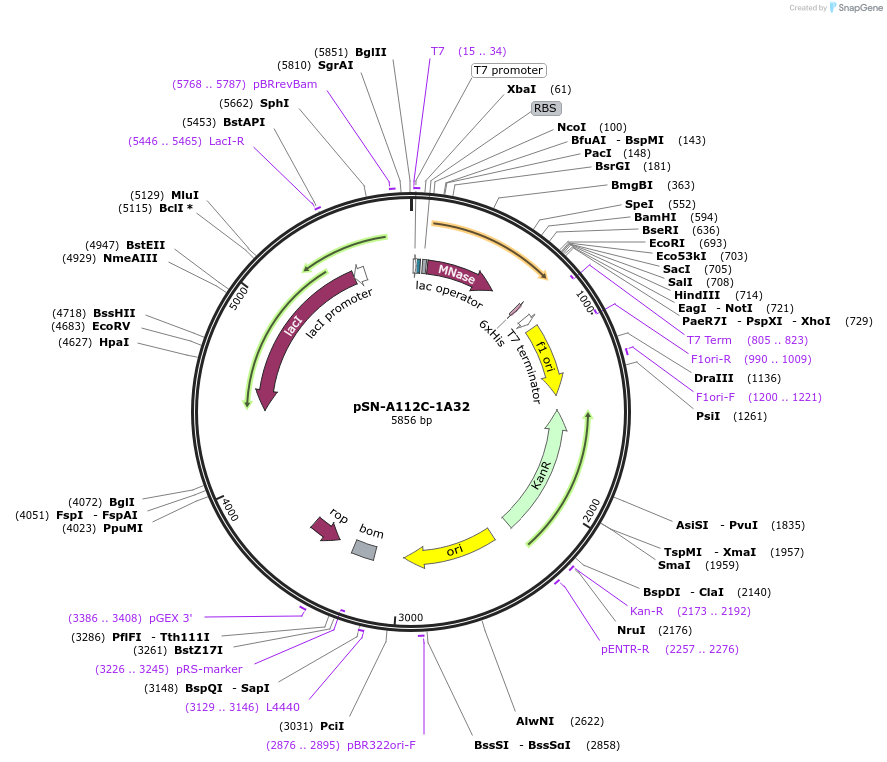
pSN-A112C-1A32
Plasmid#191241PurposeExpresses nuclease A-H124L from Staphylococcus aureus, with an A112C mutation, fused through a flexible linker to the transmembrane domain of poly-Leu-1A32 and a His-tag, for fluorescent studies.DepositorInsertnuc (nuclease A), HLA-A (HLAA) (predicted interfacial residues of the transmembrane domain in a poly-leu background)
Tagsnuclease A from S. aureus fused to the poly-Leu T…ExpressionBacterialMutationChanged Alanine 112 to Cysteine in SN, and predic…PromoterT7 promoterAvailable SinceFeb. 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
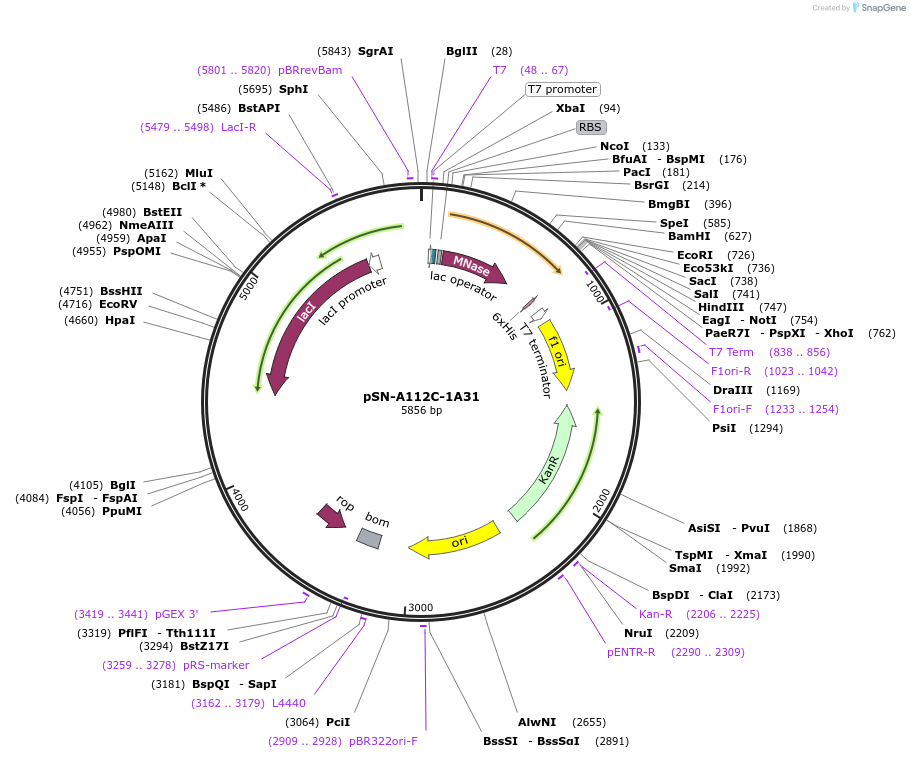
pSN-A112C-1A31
Plasmid#191242PurposeExpresses nuclease A-H124L from Staphylococcus aureus, with an A112C mutation, fused through a flexible linker to the transmembrane domain of poly-Leu-1A31 and a His-tag, for fluorescent studies.DepositorInsertnuc (nuclease A), HLA-A (HLAA) (predicted interfacial residues of the transmembrane domain in a poly-leu background)
Tagsnuclease A from S. aureus fused to the poly-Leu T…ExpressionBacterialMutationChanged Alanine 112 to Cysteine in SN, and predic…PromoterT7 promoterAvailable SinceJan. 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
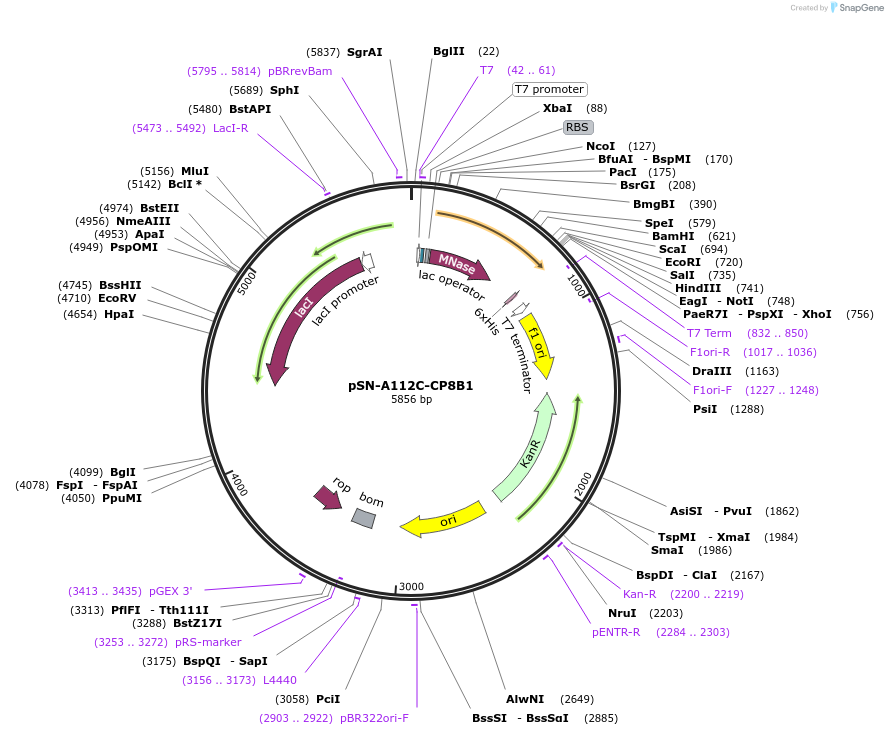
pSN-A112C-CP8B1
Plasmid#191240PurposeExpresses nuclease A-H124L from Staphylococcus aureus, with an A112C mutation, fused through a flexible linker to the transmembrane domain of poly-Leu-CP8B1 and a His-tag, for fluorescent studies.DepositorInsertnuc (nuclease A), CYP8B1 (CYP12) (predicted interfacial residues of the transmembrane domain in a poly-leu background)
Tagsnuclease A from S. aureus fused to the poly-Leu T…ExpressionBacterialMutationChanged Alanine 112 to Cysteine in SN, and predic…PromoterT7 promoterAvailable SinceDec. 21, 2022AvailabilityAcademic Institutions and Nonprofits only