We narrowed to 13,777 results for: ckb
-
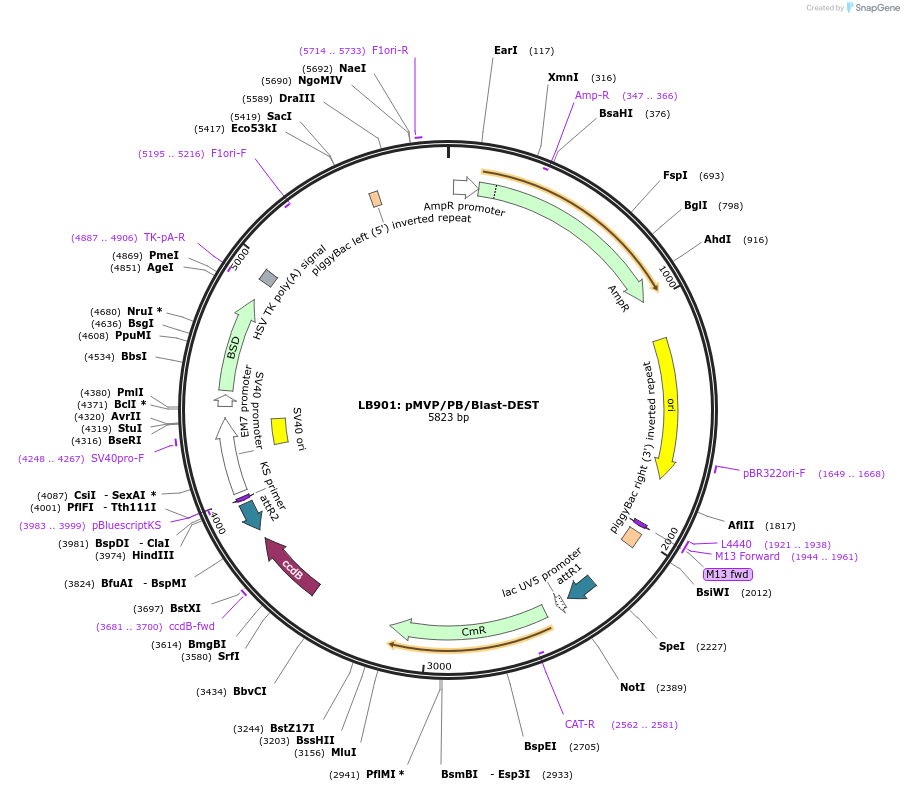
Plasmid#121873PurposepMVP destination vector, empty PiggyBac transposon vector backbone w/ Blast selection cassetteDepositorTypeEmpty backboneUsePiggybac transposon, pmvp gateway destination vec…ExpressionMammalianAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only
-
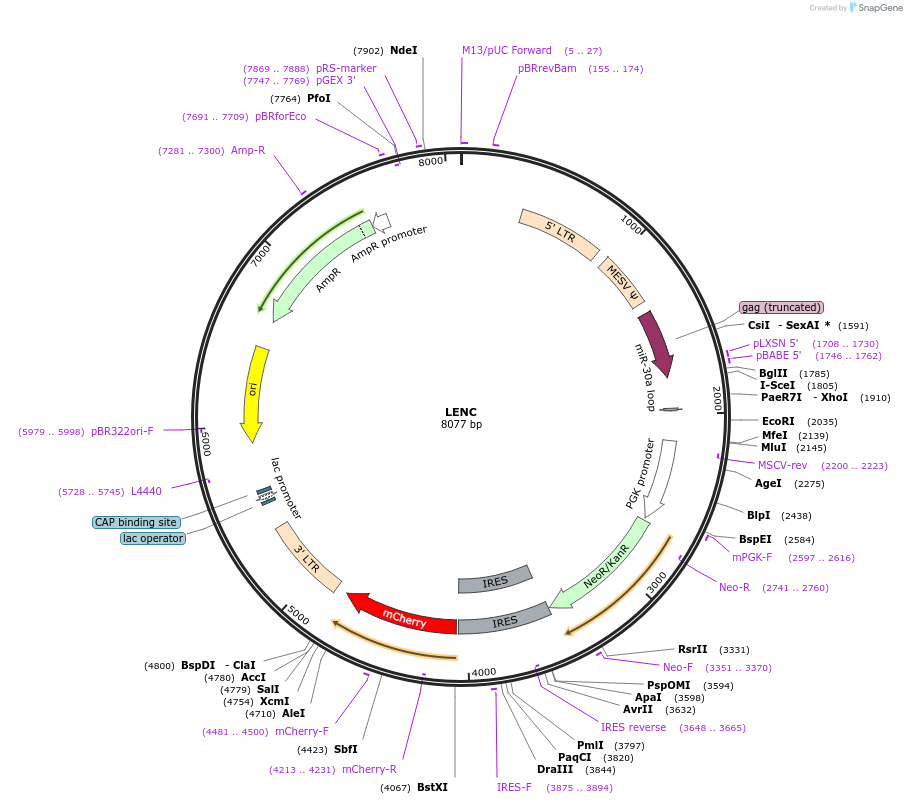
LENC
Plasmid#111163PurposemiR-E (miR-30 variant)-based RNAi with mCherry reporterDepositorInsertmiR-E (miR-30 variant)
UseRetroviralMutationWTAvailable SinceMay 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
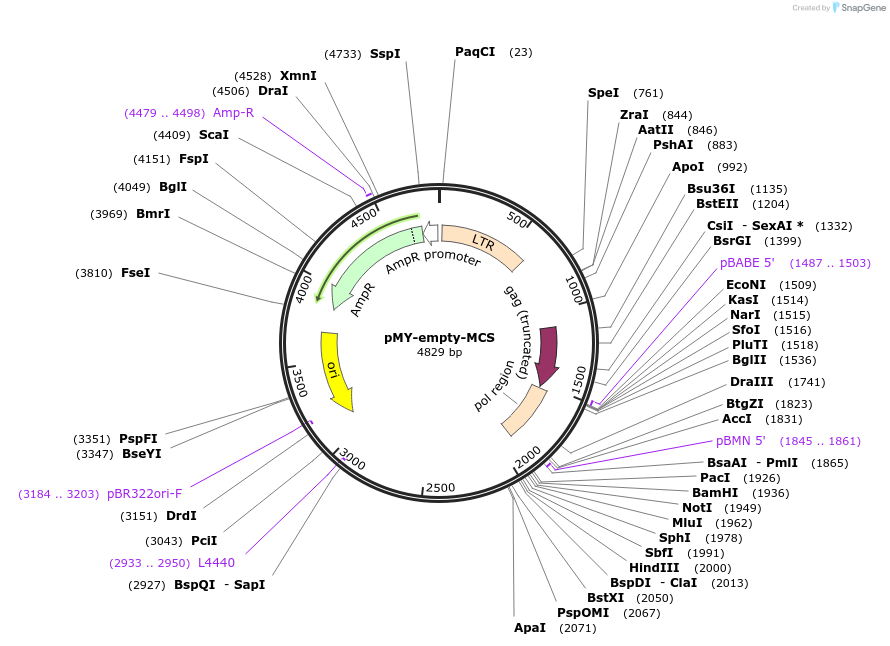
pMY-empty-MCS
Plasmid#163351PurposeThe empty pMY backbone with multiple cloning siteDepositorTypeEmpty backboneUseRetroviralExpressionMammalianAvailable SinceJan. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
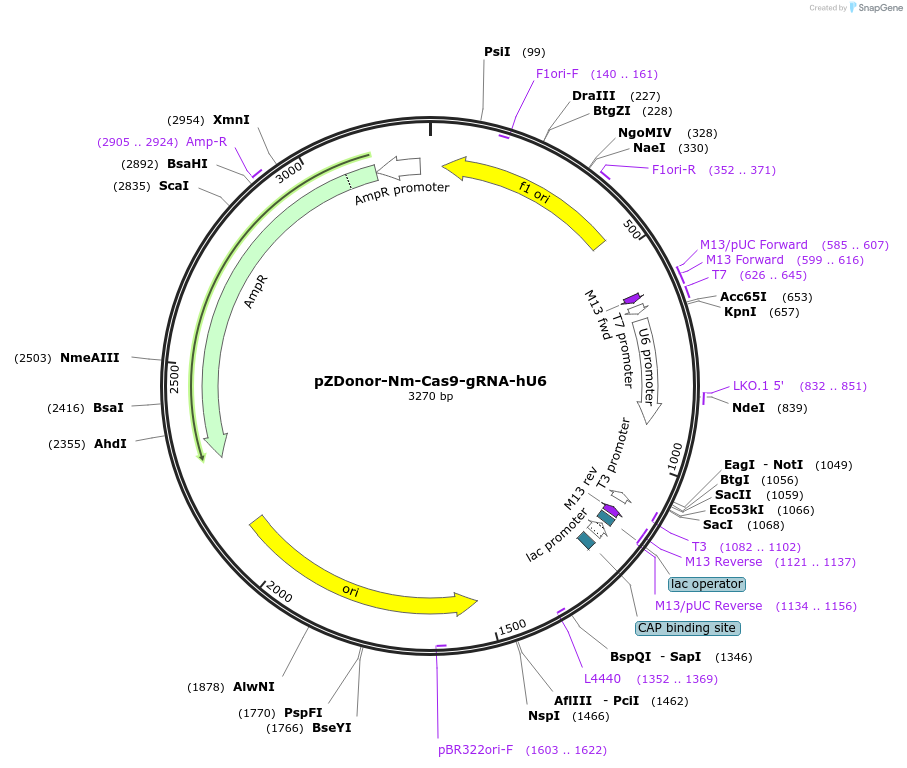
pZDonor-Nm-Cas9-gRNA-hU6
Plasmid#61366Purposeempty backbone for cloning N. meningiditis protospacers. Encodes constant region of N. meningiditis Cas9 synthetic gRNADepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionMammalianPromoterhuman U6Available SinceApril 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
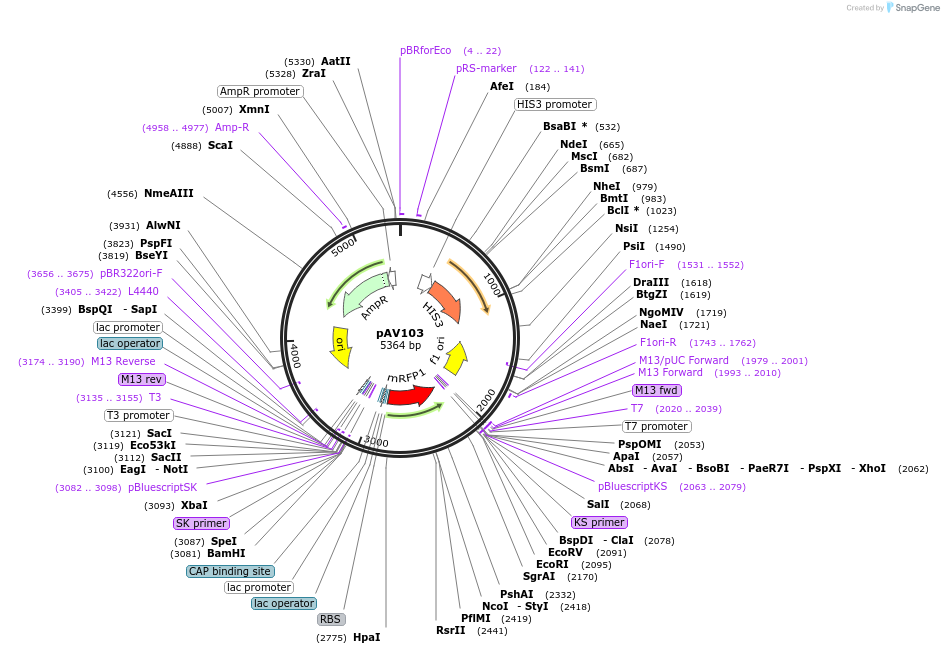
pAV103
Plasmid#63193PurposepRS403 backbone, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for transcription unit (TU) assembly using yGG.DepositorTypeEmpty backboneUseSynthetic BiologyExpressionWorm and YeastAvailable SinceJan. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
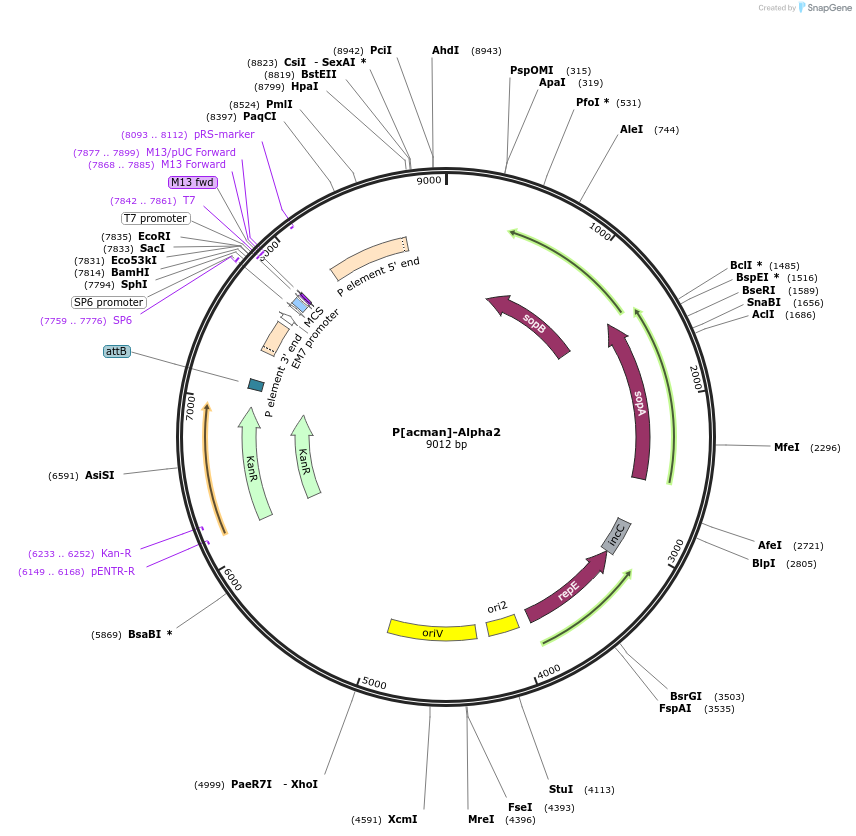
P[acman]-Alpha2
Plasmid#165859PurposeAlpha2 Basic cloning vector GB2.0 compatible P[acman]-derived backbone. Allows for copy induction from low to high using AutoFOS or similar. Requires EPI300 bacterial strain (DH10B Derived) or similar. uses blue-white screeningDepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
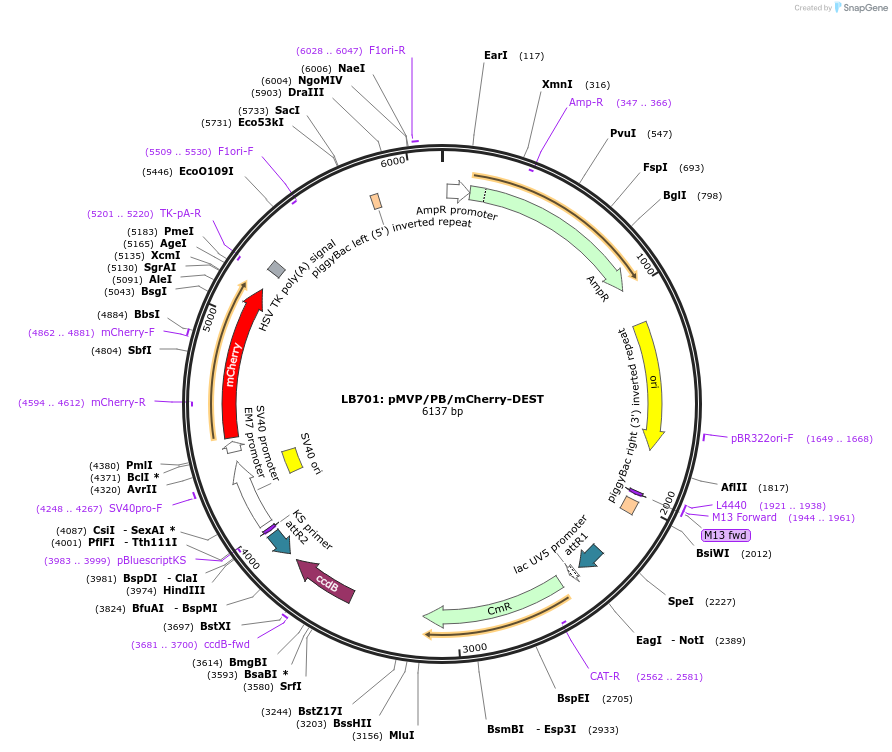
LB701: pMVP/PB/mCherry-DEST
Plasmid#121871PurposepMVP destination vector, empty PiggyBac transposon vector backbone w/ mCherry selection cassetteDepositorTypeEmpty backboneUsePiggybac transposon, pmvp gateway destination vec…ExpressionMammalianAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
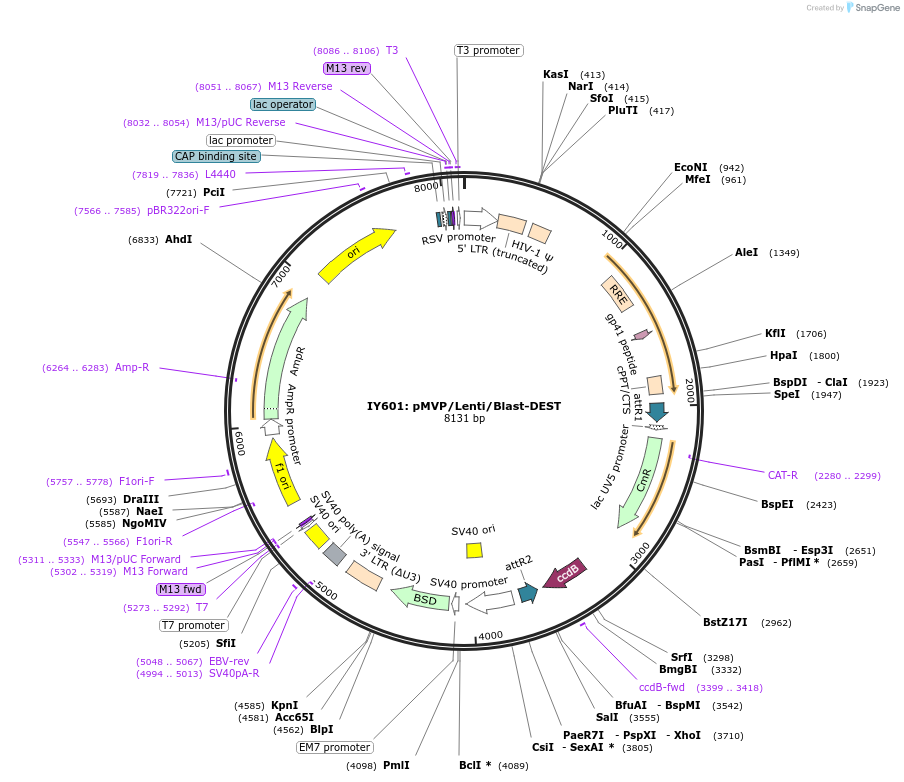
IY601: pMVP/Lenti/Blast-DEST
Plasmid#121849PurposepMVP destination vector, empty Lentivirus vector backbone w/ Blast selection cassetteDepositorTypeEmpty backboneUseLentiviral; Pmvp gateway destination vectorAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
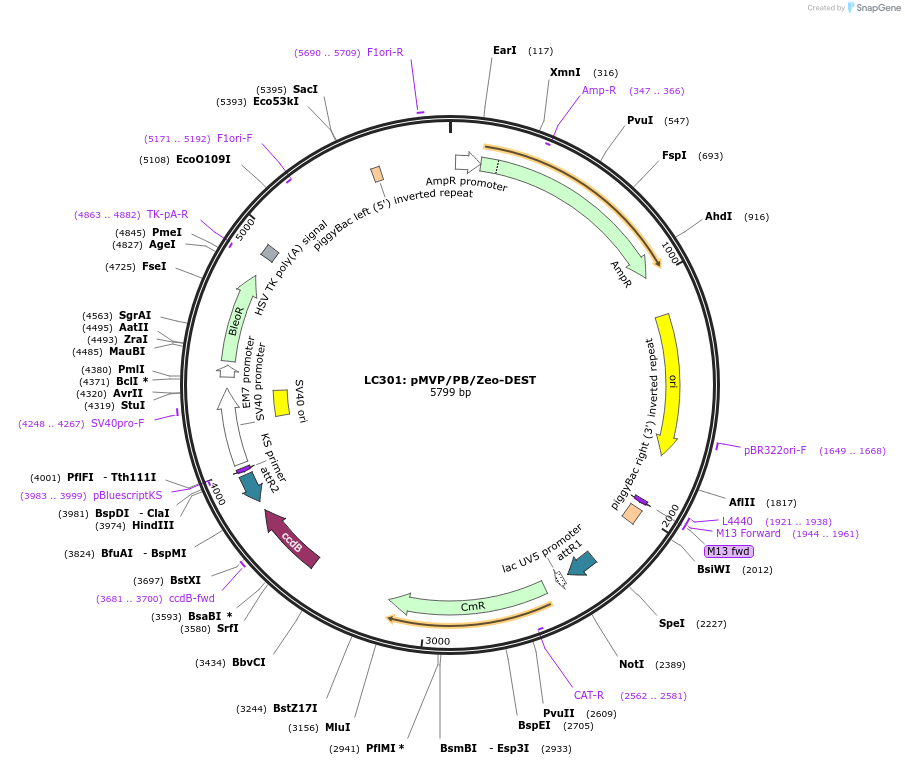
LC301: pMVP/PB/Zeo-DEST
Plasmid#121877PurposepMVP destination vector, empty PiggyBac transposon vector backbone w/ Zeo selection cassetteDepositorTypeEmpty backboneUsePiggybac transposon, pmvp gateway destination vec…ExpressionMammalianAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
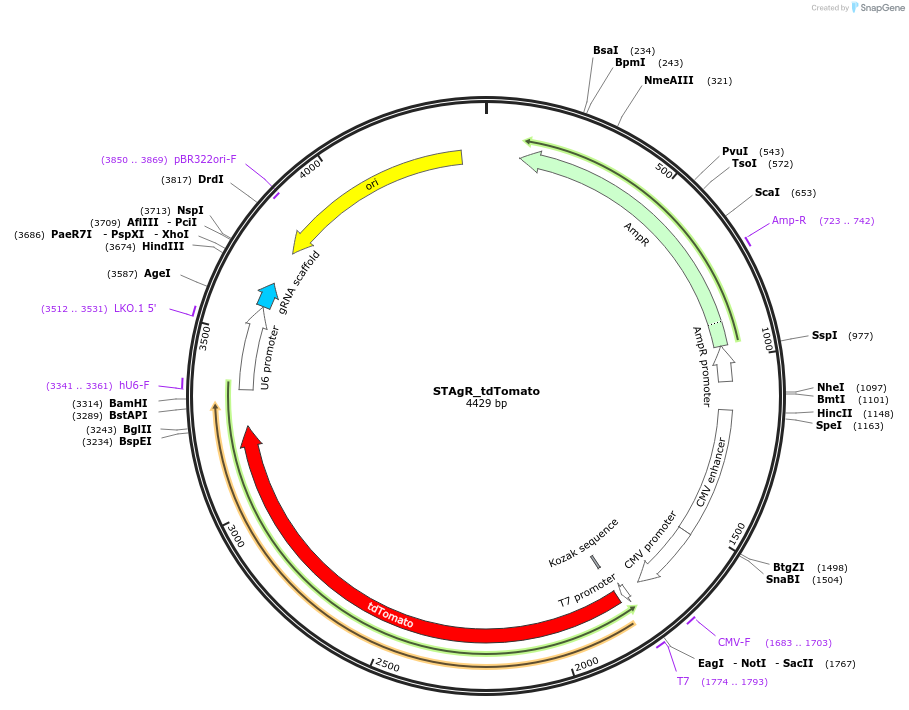
STAgR_tdTomato
Plasmid#102993PurposeCan be used as backbone for a STAgR reactionDepositorTypeEmpty backboneUseCRISPR; Pcr template for stagr vectorsPromoterU6Available SinceMarch 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
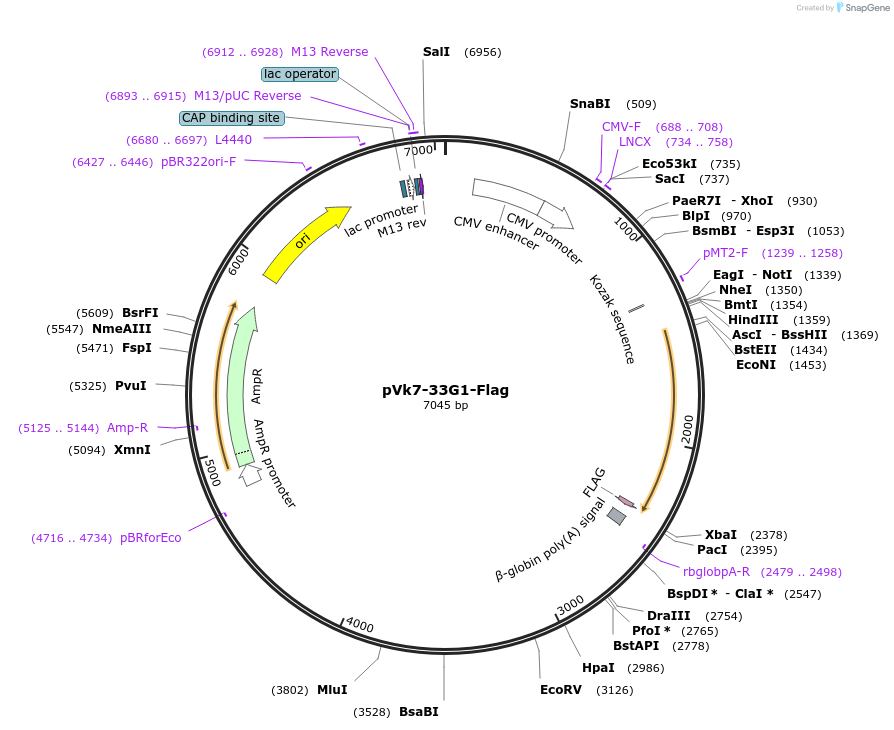
pVk7-33G1-Flag
Plasmid#58950Purposebackbone for mouse recombinant antibody with added C-terminal FLAG tagDepositorTypeEmpty backboneTagsFLAG tag (DYKDDDDK)ExpressionMammalianPromoterCMVAvailable SinceJune 11, 2015AvailabilityAcademic Institutions and Nonprofits only -
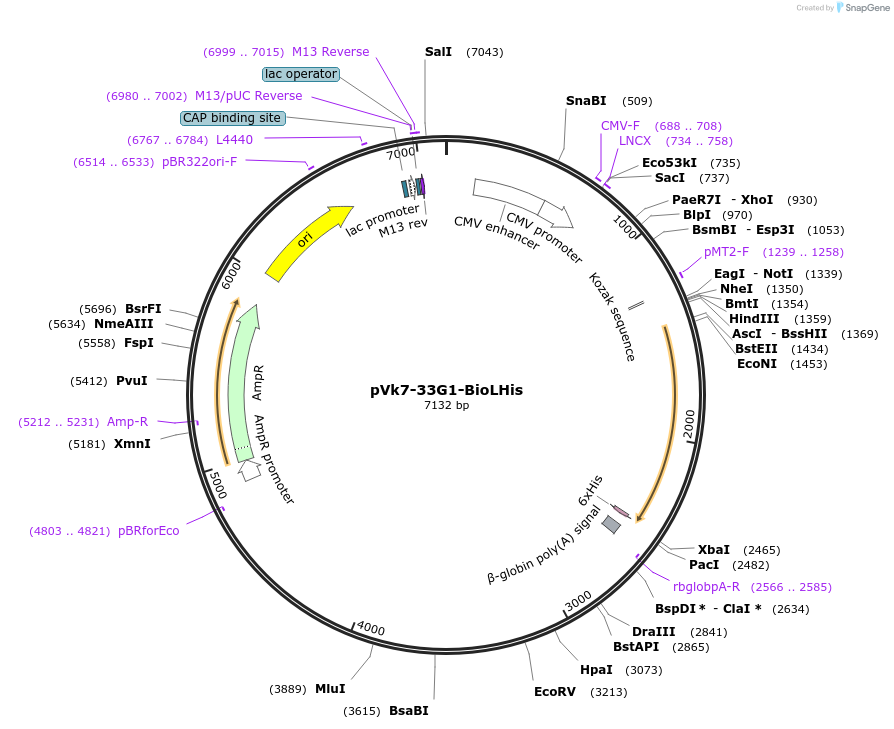
pVk7-33G1-BioLHis
Plasmid#58949Purposebackbone for mouse recombinant antibody with added C-terminal biotinylation site, linker, 6His tagDepositorTypeEmpty backboneTags6His tag and biotinylation peptideExpressionMammalianPromoterCMVAvailable SinceJune 11, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAAV-CMV-SauriCas9-Puro
Plasmid#135965PurposeExpresses SauriCas9 and puromycin resistance genes, and cloning backbone for sgRNADepositorTypeEmpty backboneUseAAVExpressionMammalianAvailable SinceApril 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
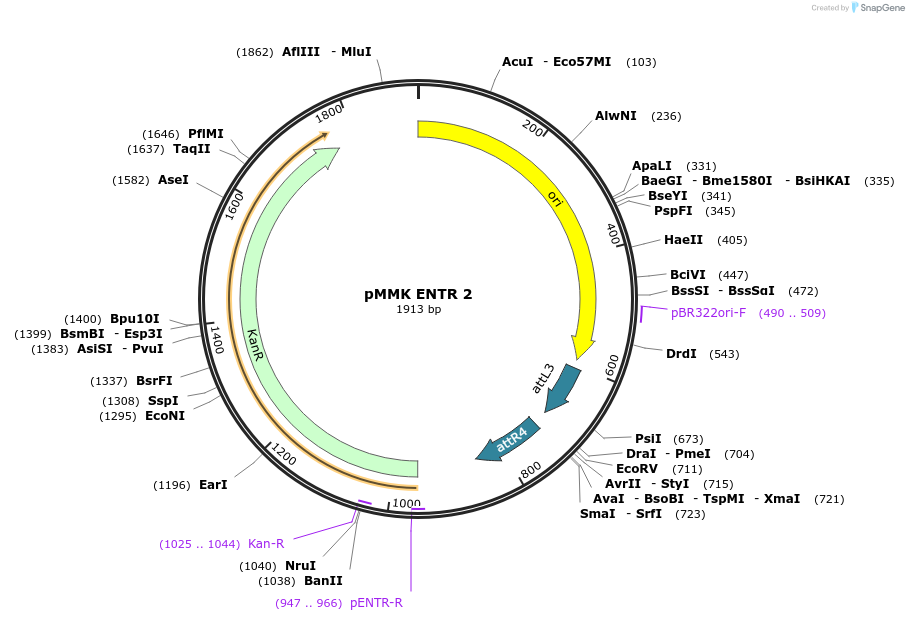
pMMK ENTR 2
Plasmid#206261PurposeENTR Vector 1 for MultiSite Gateway assembly and production of recombinant baculovirus using MultiMate assembly. Empty backbone.DepositorTypeEmpty backboneUseMultimate/gateway entr 2Available SinceSept. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
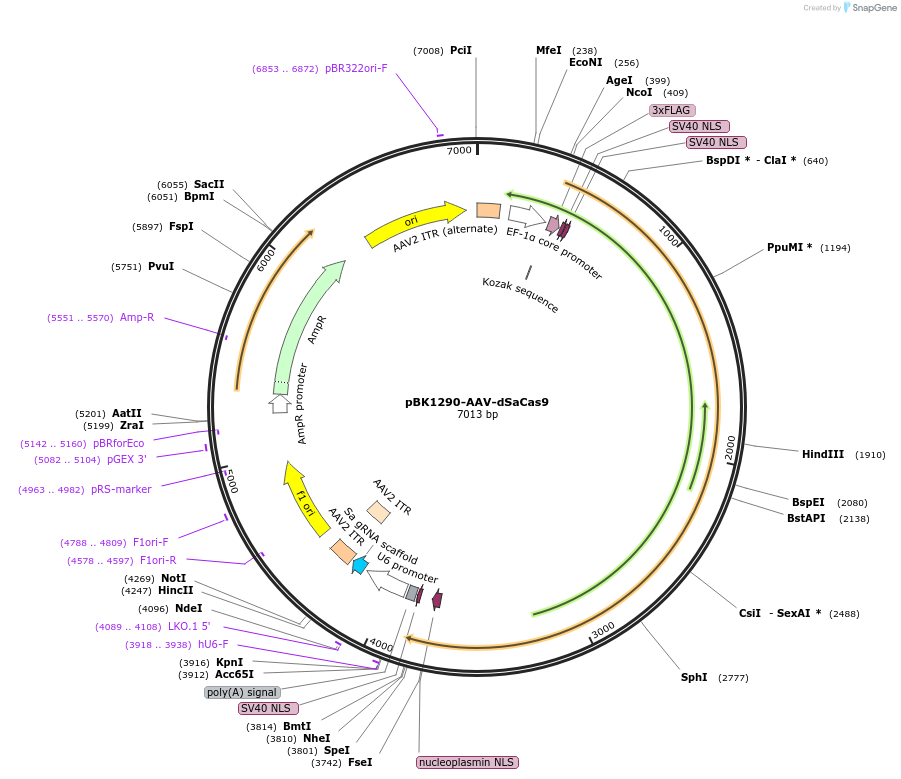
pBK1290-AAV-dSaCas9
Plasmid#223139PurposeAAV backbone for dSaCas9 (endonuclease dead Cas9 from Staphylococcus aureus) and Sa-gRNA ScaffoldDepositorTypeEmpty backboneUseAAV and CRISPRTags3xFLAGExpressionMammalianAvailable SinceAug. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
Clover-pBAD
Plasmid#54575PurposeLocalization: Protein Expression Vector , Excitation: 505, Emission: 515. This plasmid encodes Clover H231L based on reference from Bajar et. al. Sci Rep. 2016.DepositorTypeEmpty backboneTags6xHis and CloverExpressionBacterialAvailable SinceJuly 25, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
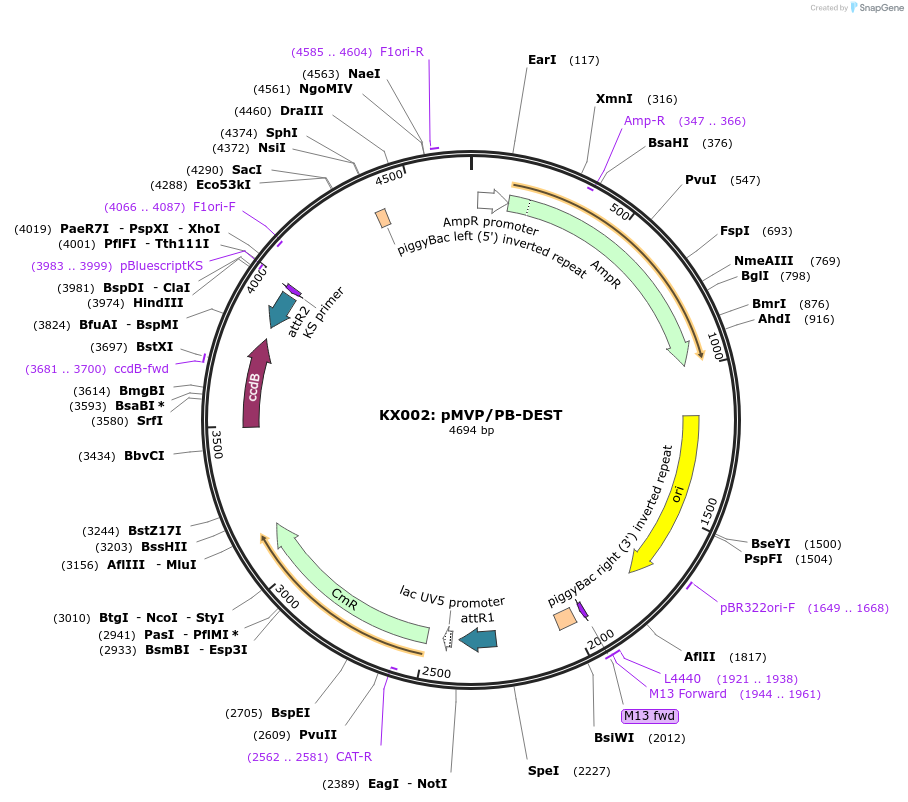
KX002: pMVP/PB-DEST
Plasmid#121869PurposepMVP destination vector, empty PiggyBac transposon vector backboneDepositorTypeEmpty backboneUsePiggybac transposon, pmvp gateway destination vec…ExpressionMammalianAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
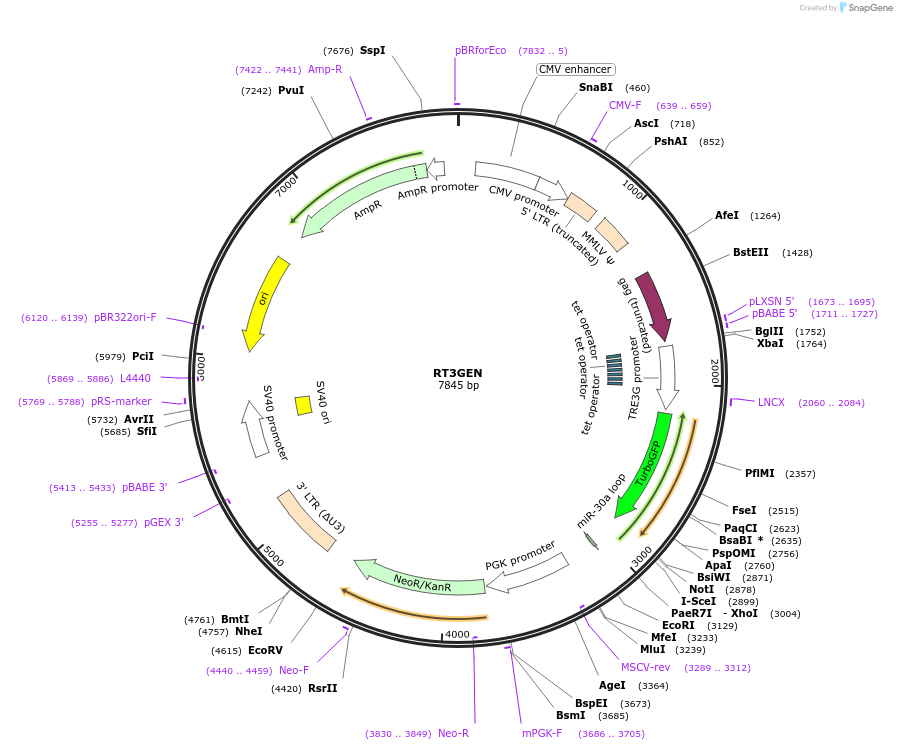
RT3GEN
Plasmid#111165PurposeTet-regulated miR-E (miR-30 variant)-based RNAi with tGFP reporterDepositorInsertmiR-E (miR-30 variant)
UseRetroviralMutationWTPromoterT3GAvailable SinceMay 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
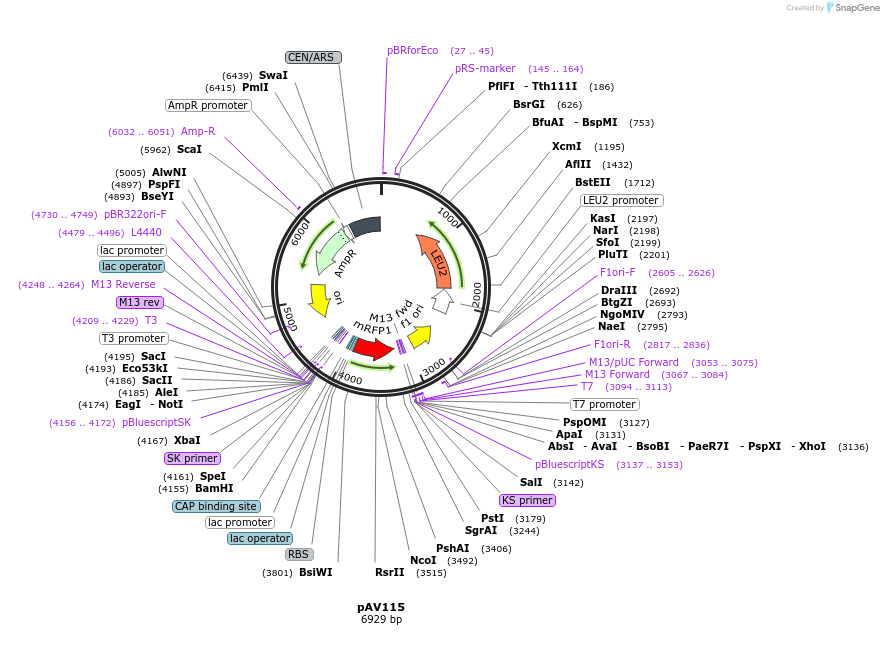
pAV115
Plasmid#63182PurposepRS415 backbone, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for transcription unit (TU) assembly using yGG.DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailable SinceSept. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
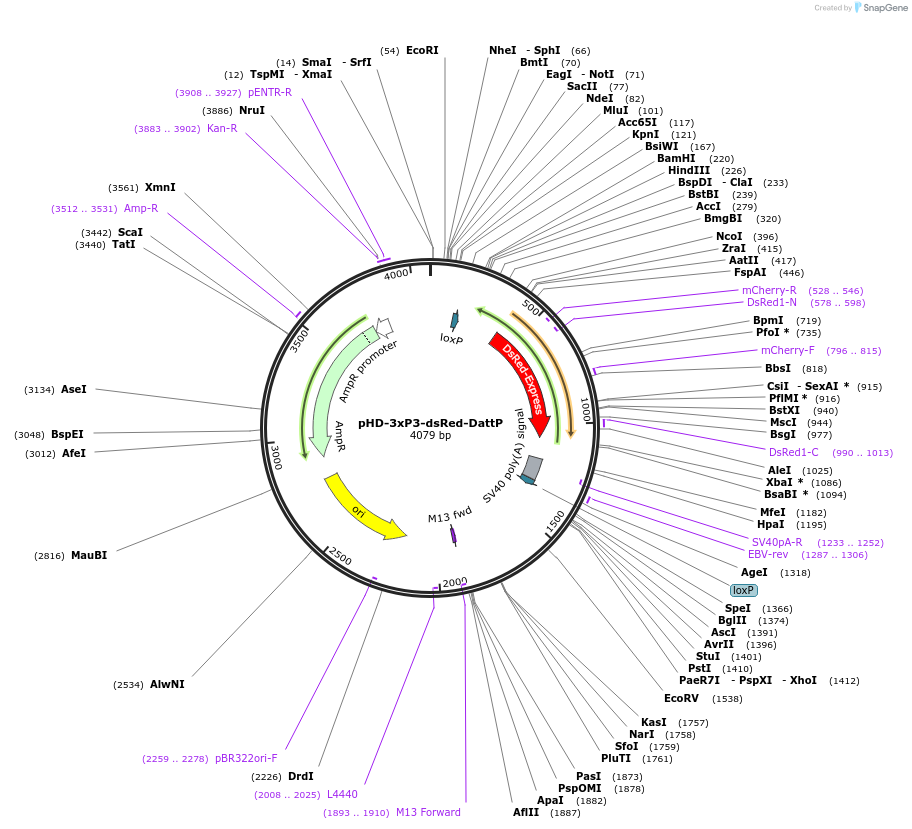
pHD-3xP3-dsRed-DattP
Plasmid#133559PurposeBackbone vector for CRISPR-HDR insertion of SPARC-Variant constructsDepositorTypeEmpty backboneUseCRISPRExpressionInsectAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only