We narrowed to 5,933 results for: chia
-
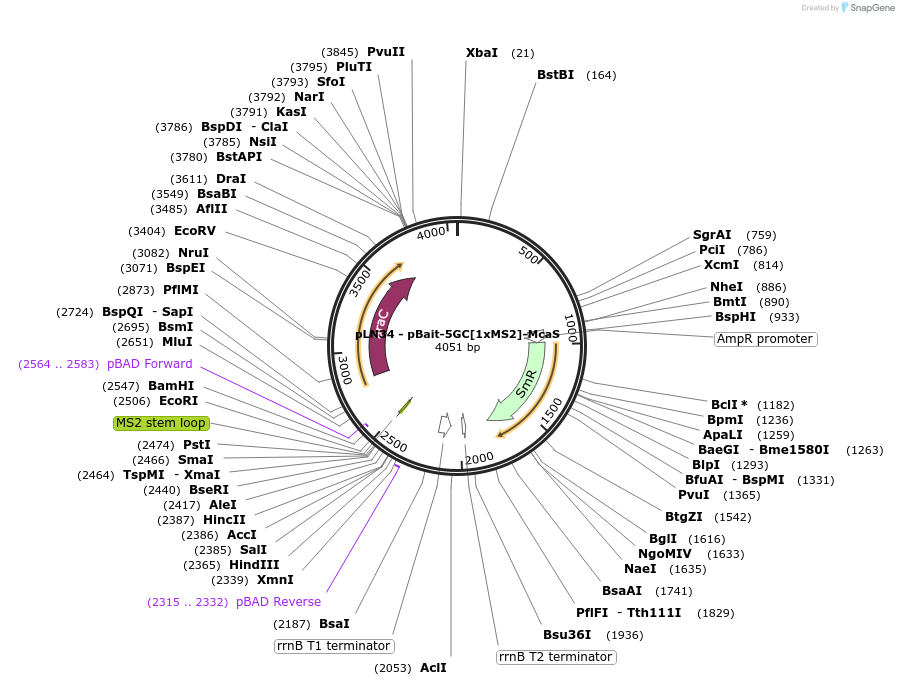
Plasmid#222403Purposearabinose-induced synthesis of a hybrid RNA with an MS2 hairpin surrounded by a 5bp GC clamp with E coli McaS inserted between XmaI and HindIII sites. Insert can be replaced with RNA bait of interest.DepositorAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only
-
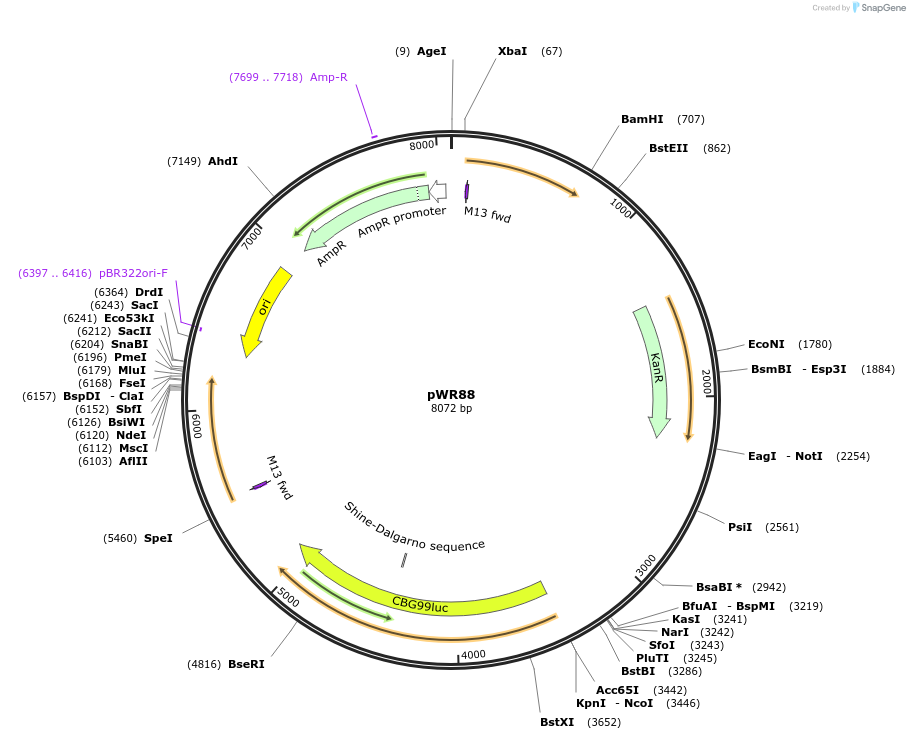
pWR88
Plasmid#203422Purposeempty BLINCAR plasmid for payload delivery, no payload, no genomic targetDepositorInsertsCAR copy 1
P(TEF1)/coKanMX/T(ACT1) + P(TDH3)/coCBG/T(ADH2)
CAR copy 2
ExpressionYeastAvailable SinceJune 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
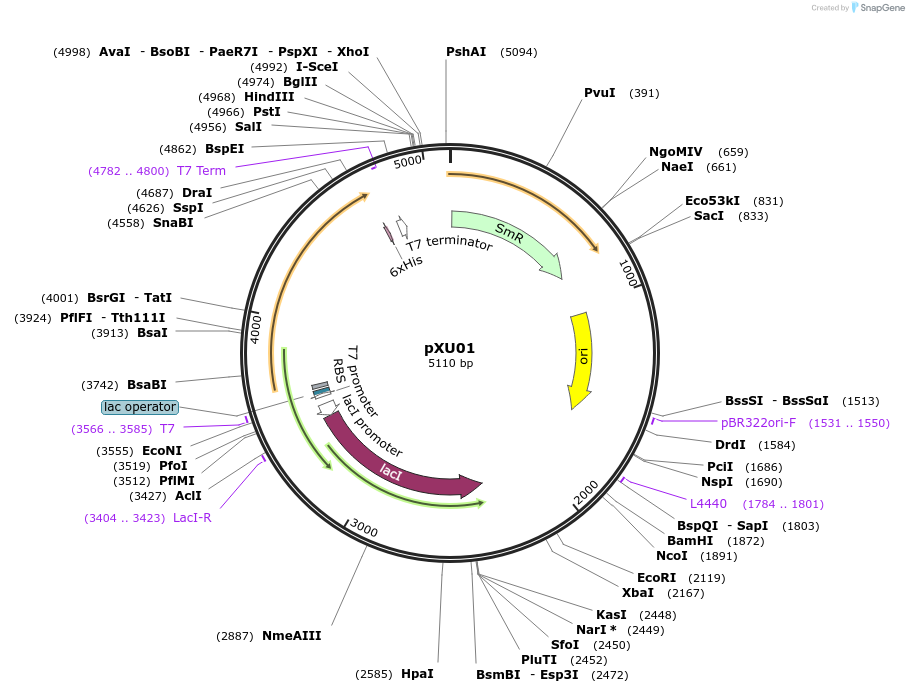
pXU01
Plasmid#212151PurposePlasmid expressing xylitol dehydrogenase under inducible promoter PT7 control. Plasmid confers the ability to use xylitol as major carbon source to E. coli.DepositorInsertxylitol dehydrogenase
Tags6x HisExpressionBacterialPromoterPT7Available SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
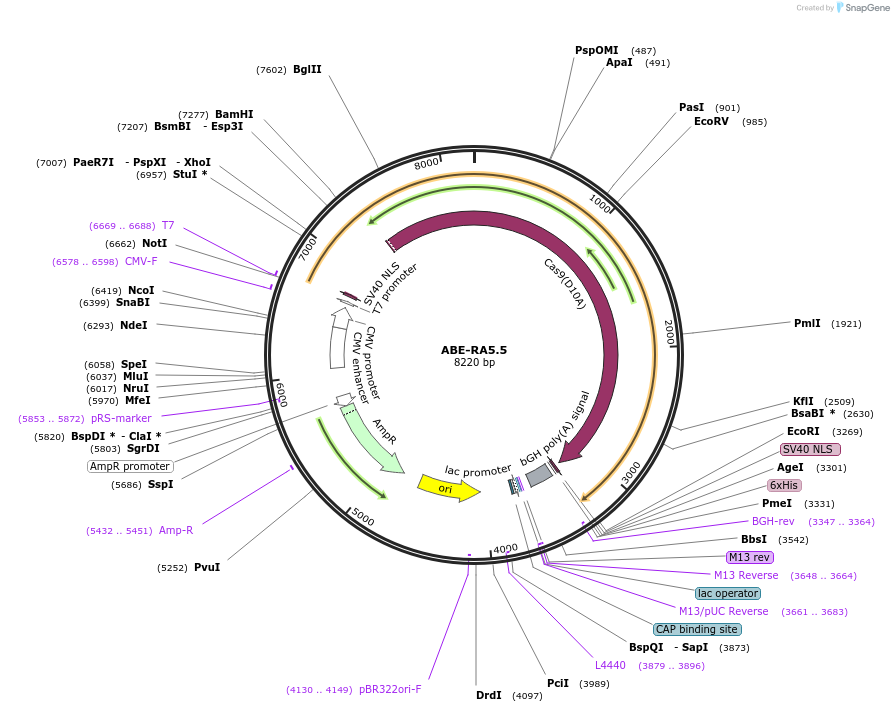
ABE-RA5.5
Plasmid#208316PurposeExpresses ABE-RA5.5 in mammalian cellsDepositorInsertTadA-RA5.5-SpCas9 D10A
ExpressionMammalianMutationW23R, H36L, R47K, P48A, R51L, I76Y, V82S, A106V, …Available SinceJan. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHAGE-PGUS
Plasmid#207827PurposeFLAG-tagged PGUS driven by CMV promoterDepositorInsertPGUS
UseLentiviralTagsFLAGPromoterCMVAvailable SinceDec. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
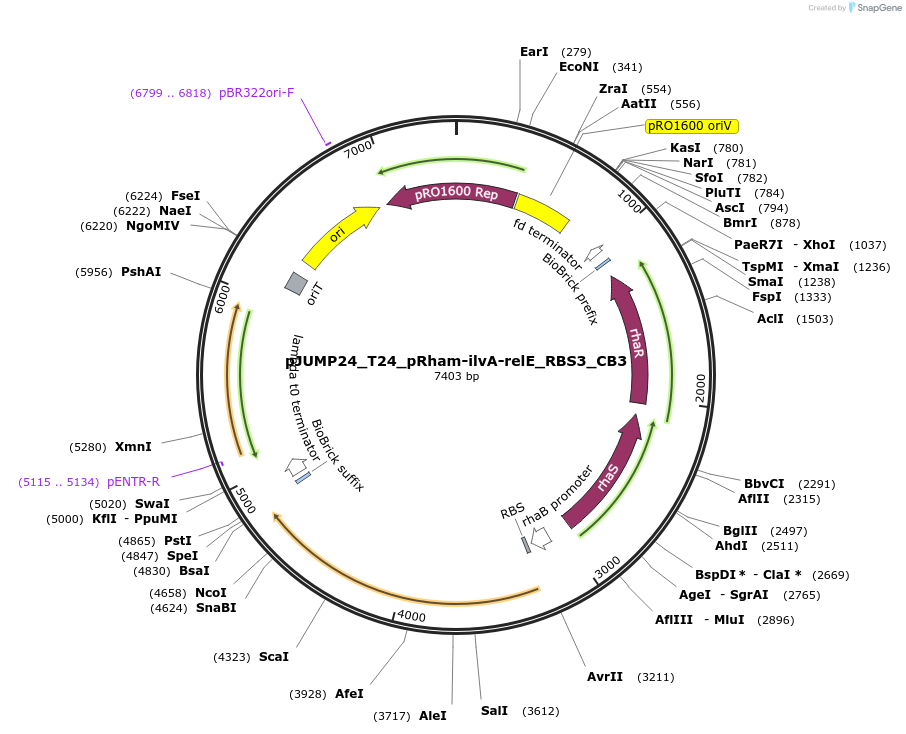
pJUMP24_T24_pRham-ilvA-relE_RBS3_CB3
Plasmid#201534PurposeExpressing synthetic overlapping gene, ilvA-relE with internal RBS modification. Evolved strain with lower ilvA-relE expression. Origin pRO1600/ColE1 (E.coli - Pseudomonas shuttle)DepositorInsertilvA-relE_RBS3_CB3
UseSynthetic BiologyMutationrhaS W190GPromoterPrhaBAD (rhamnose)Available SinceSept. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
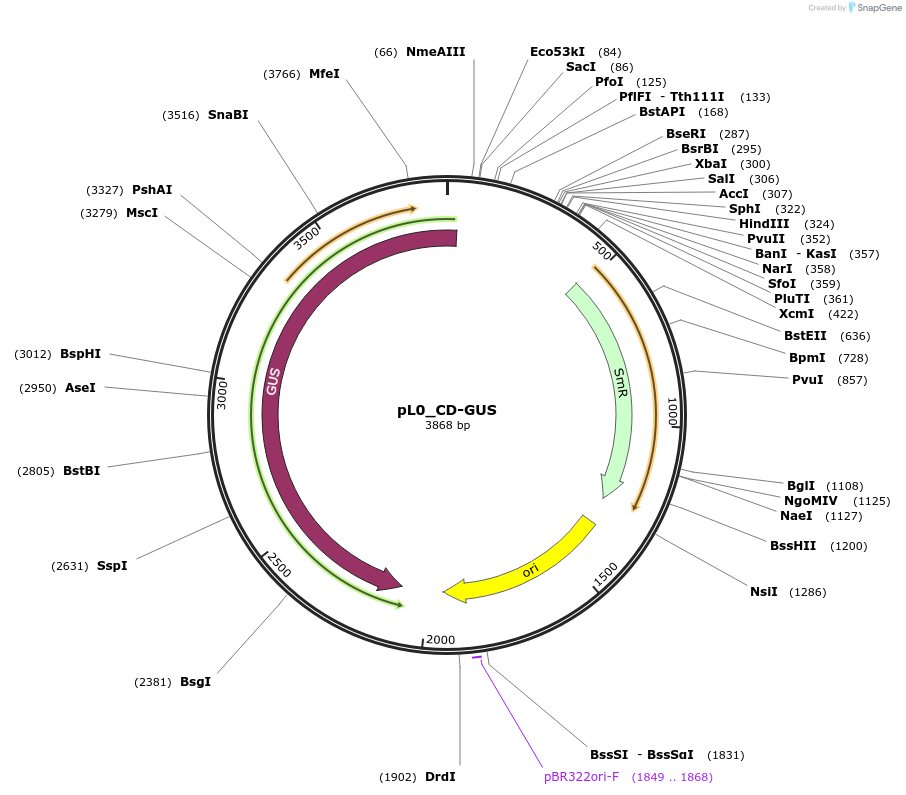
pL0_CD-GUS
Plasmid#202137PurposeLevel 0 plasmid containing a beta glucuronidase reporter gene with CD overhangs used to build a level 1 construct.DepositorInsertBeta glucuronidase gene
UseSynthetic BiologyMutationNoneAvailable SinceAug. 25, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
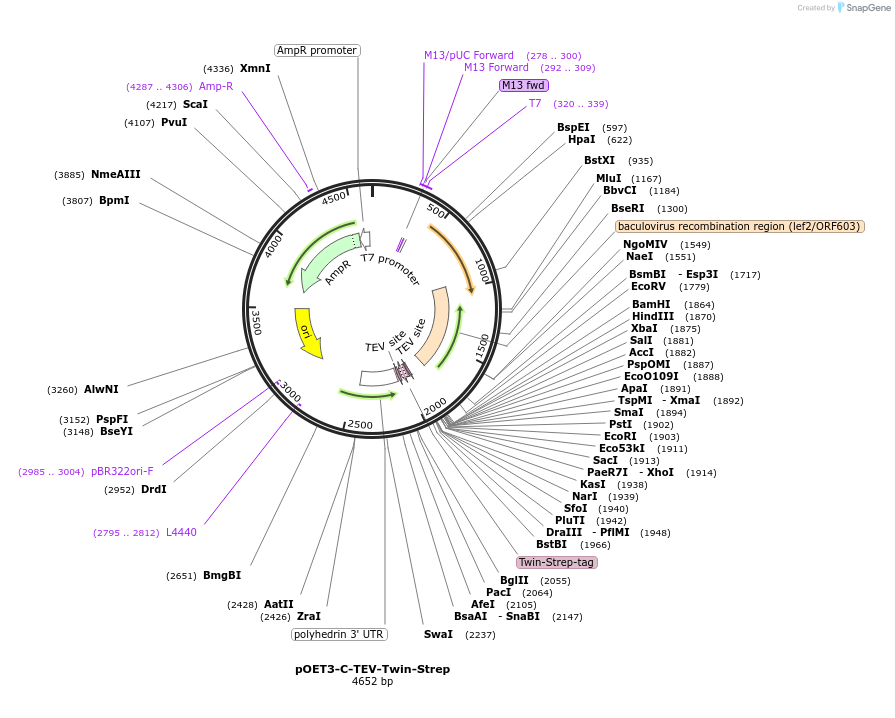
pOET3-C-TEV-Twin-Strep
Plasmid#203324PurposeGenerate the baculovirus to express N-terminal Tobacco Etch Virus (TEV) protease cleavable Twin-Strep-tagged protein (ENLYFQGS-WSHPQFEK-(GGGS)2-GGSA-WSHPQFEK) in insect cellsDepositorTypeEmpty backboneTagsTEV cleavable site (ENLYFQG) and Twin-strep-tagExpressionInsectAvailable SinceJuly 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
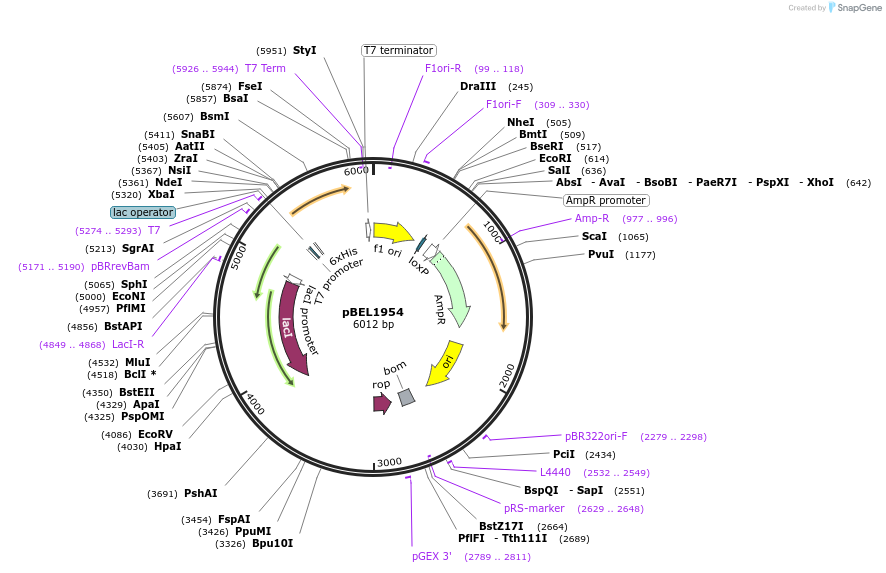
pBEL1954
Plasmid#195689PurposeExpression/purification of 6xHis-TEV-MlaD(F118A/L123A)DepositorInsertMlaD
ExpressionBacterialMutation6xHis-TEV-MlaD(F118A/L123A)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL1936
Plasmid#195692PurposeComplementation of MlaF-MlaE-MlaDdelta153-183-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaDdelta153-183-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
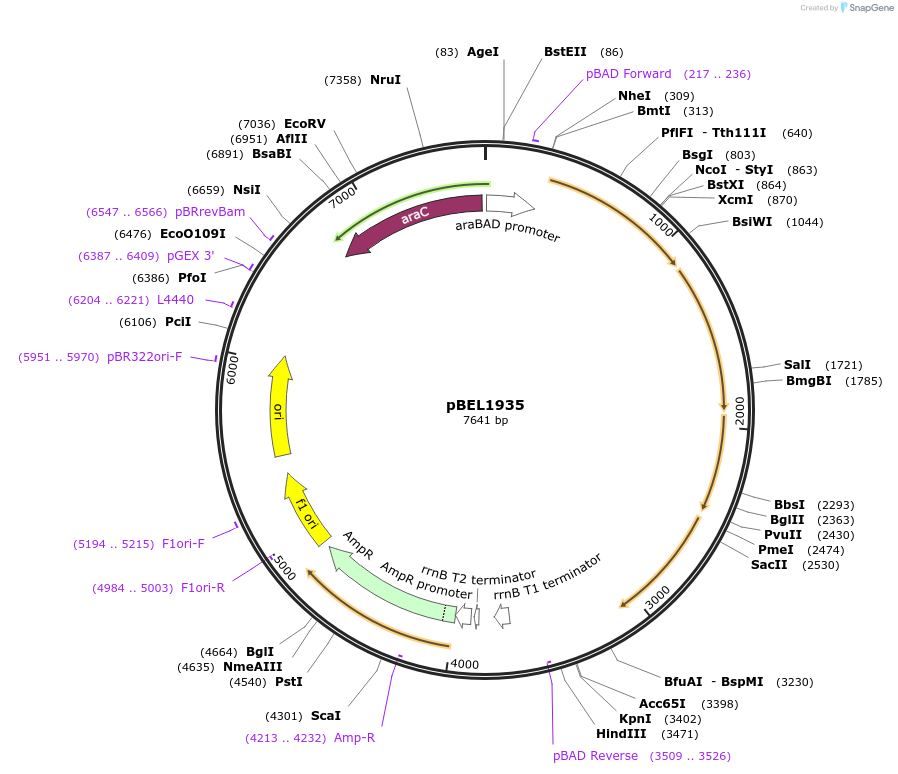
pBEL1935
Plasmid#195693PurposeComplementation of MlaF-MlaE-MlaDdelta173-183-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaDdelta173-183-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
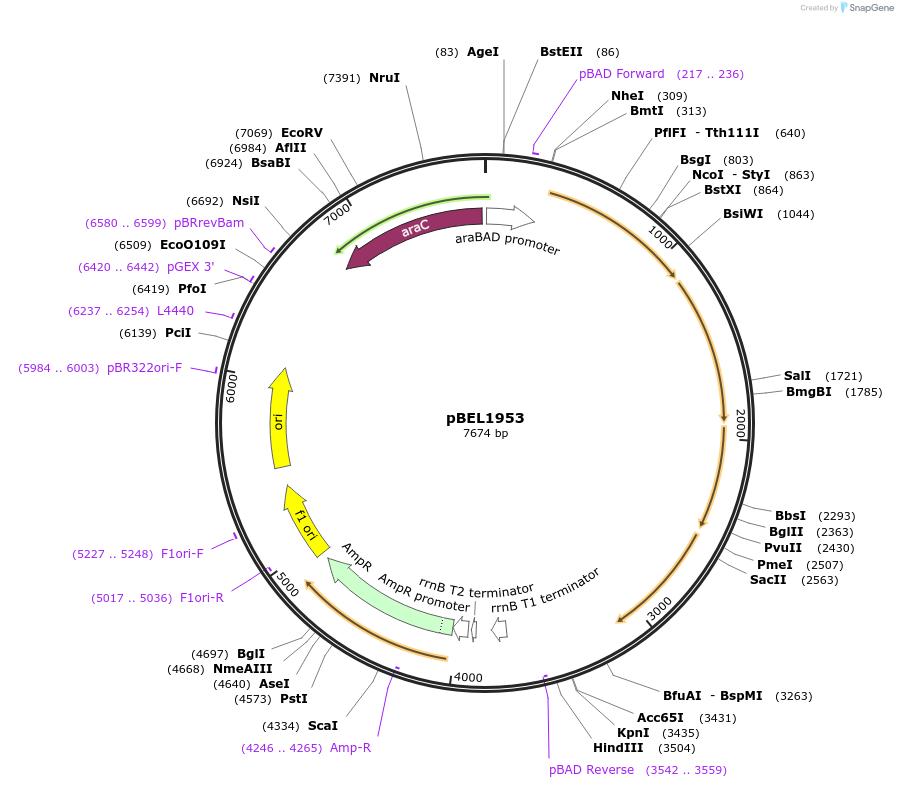
pBEL1953
Plasmid#195694PurposeComplementation of MlaF-MlaE-MlaD(F118A/L123A)-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaD(F118A/L123A)-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
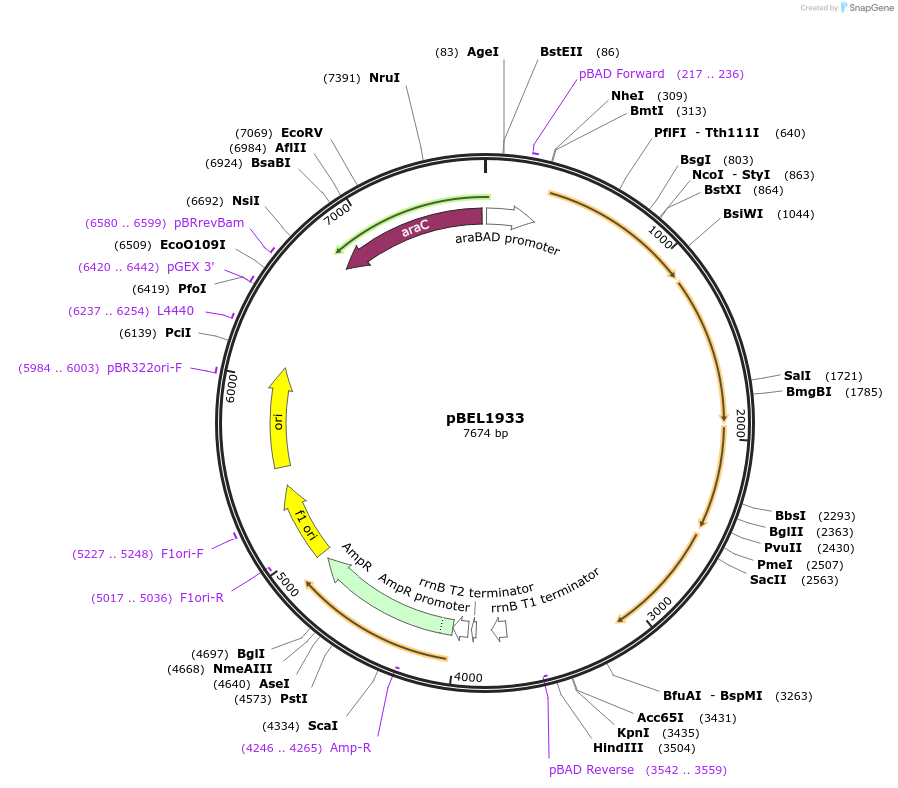
pBEL1933
Plasmid#195695PurposeComplementation of MlaF-MlaE-MlaD(F118A)-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaD(F118A)-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
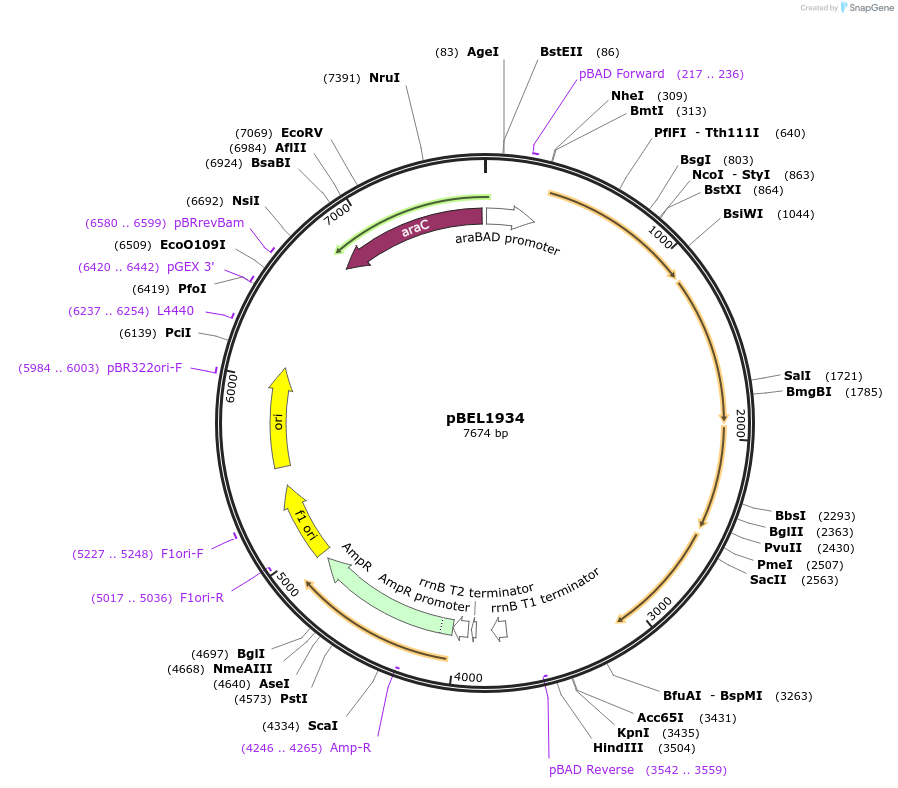
pBEL1934
Plasmid#195696PurposeComplementation of MlaF-MlaE-MlaD(L123A)-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaD(L123A)-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
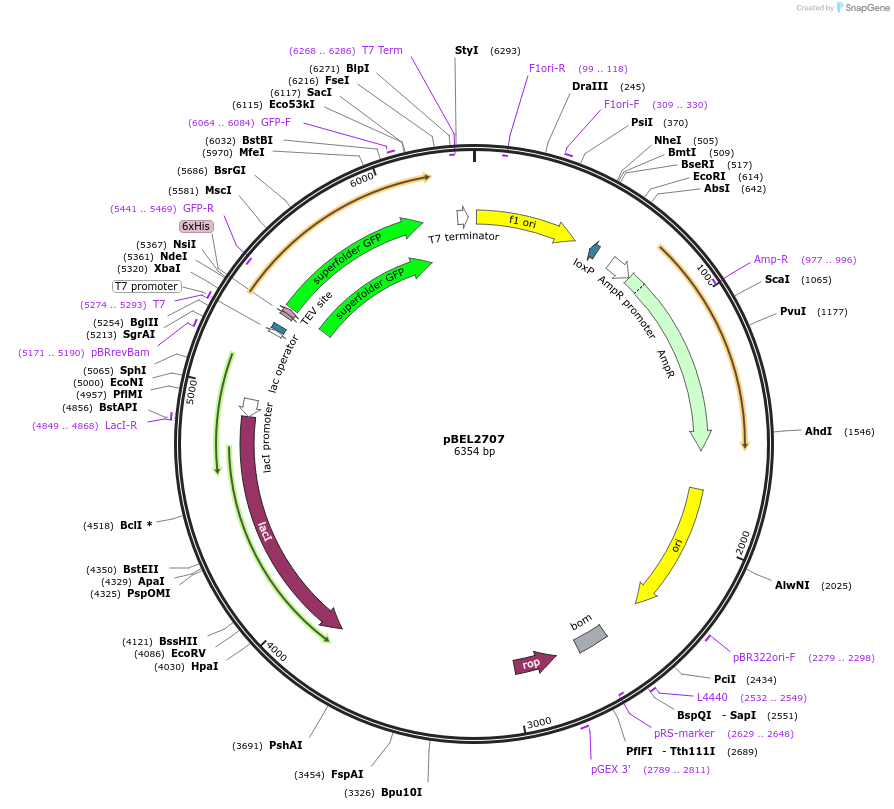
pBEL2707
Plasmid#195636PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
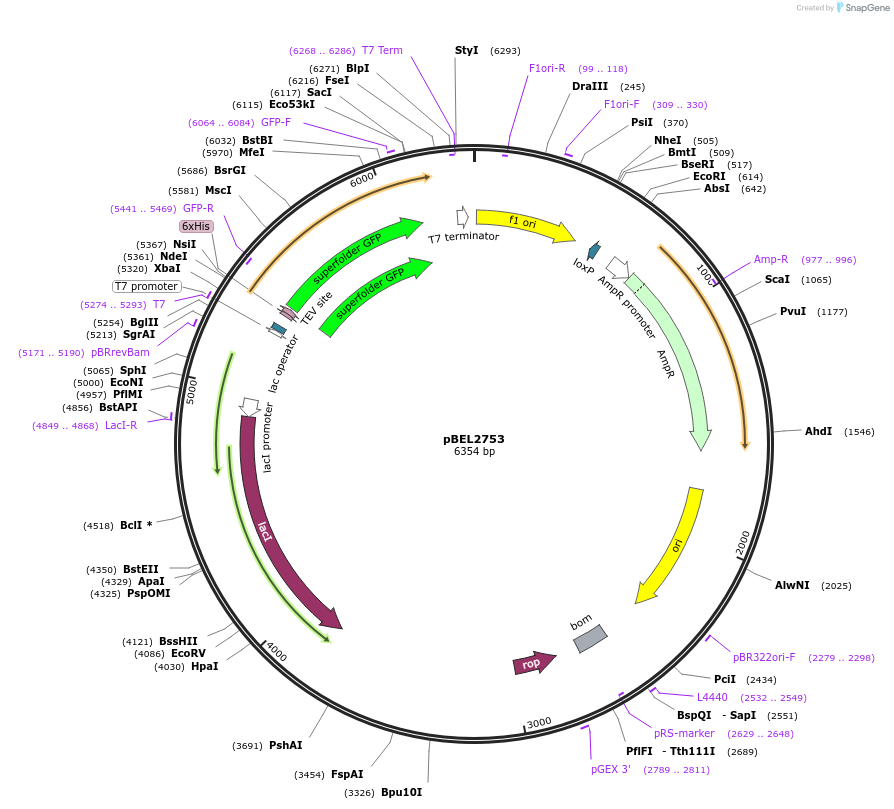
pBEL2753
Plasmid#195637PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251; I241N)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251; I241N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
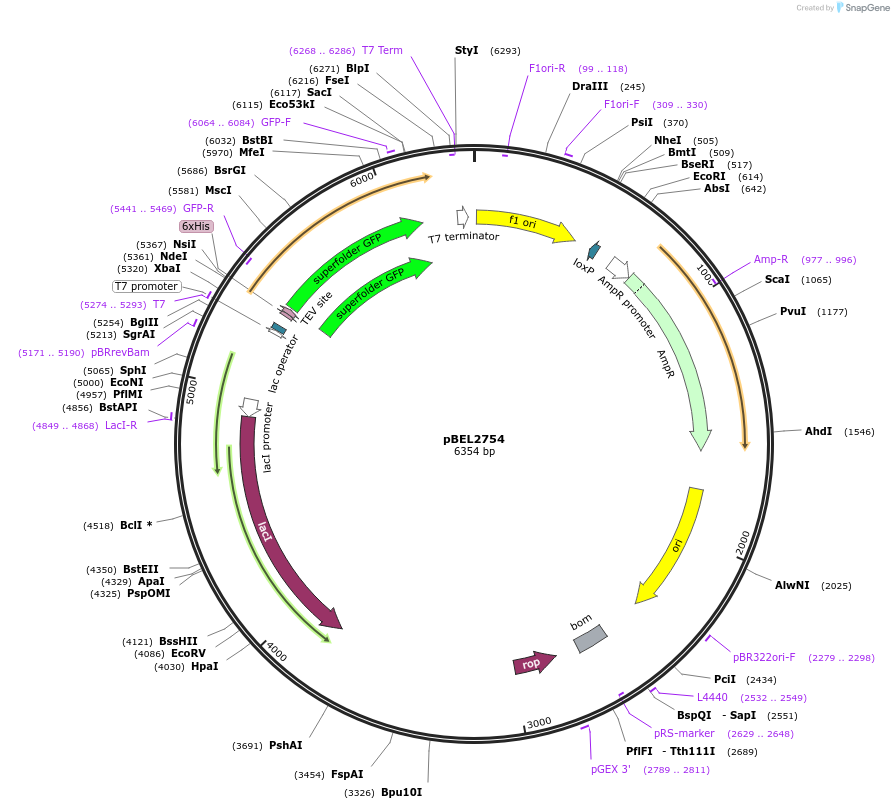
pBEL2754
Plasmid#195638PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251; L245N)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251; L245N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
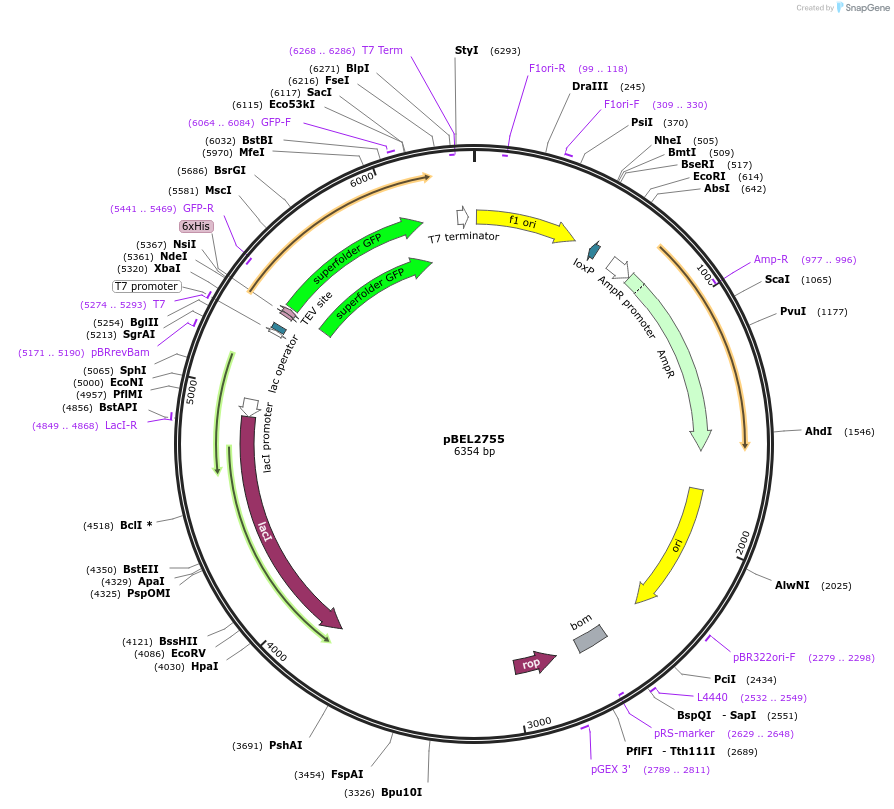
pBEL2755
Plasmid#195639PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251; I248N)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251; I248N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
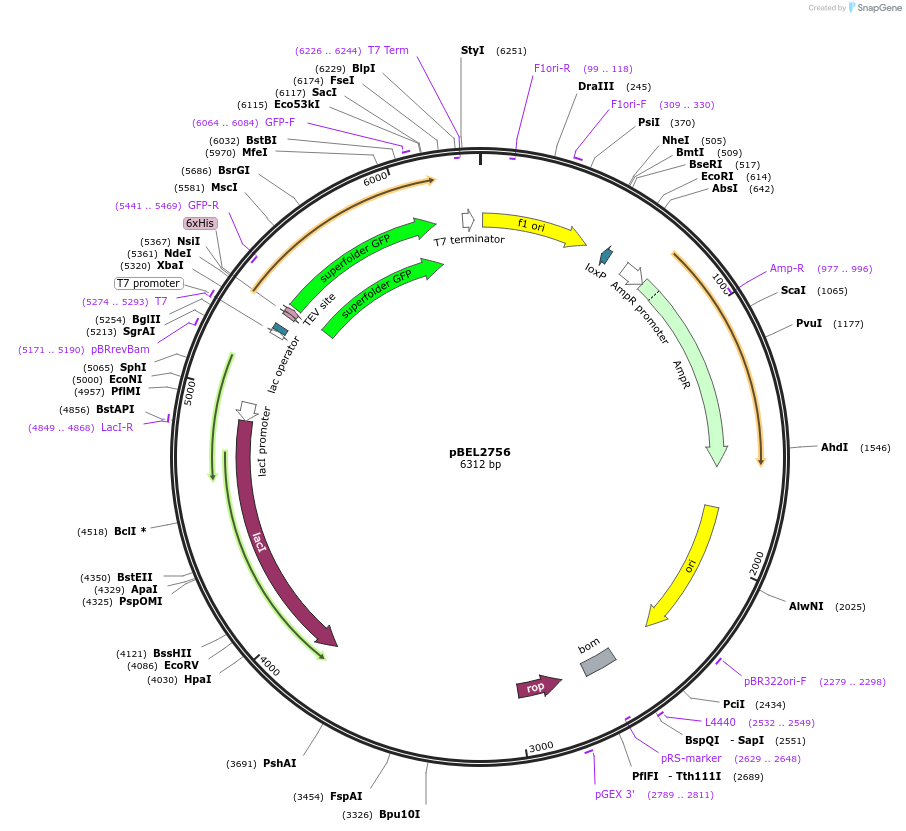
pBEL2756
Plasmid#195640PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA(227-251; delta238-251)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA(227-251; delta238-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
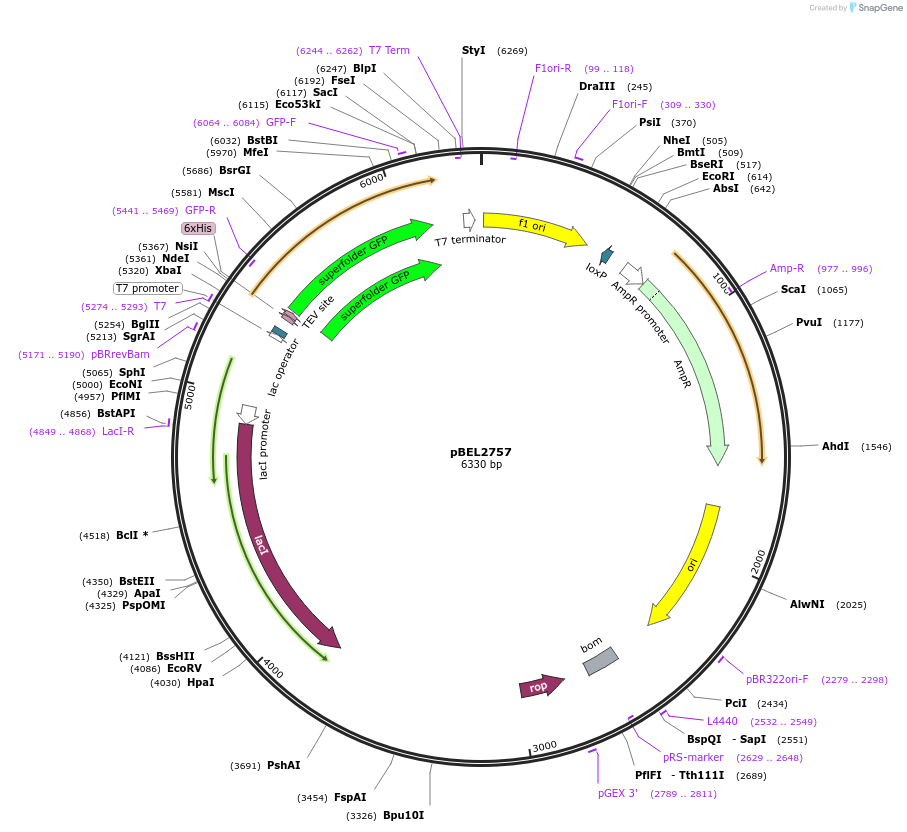
pBEL2757
Plasmid#195641PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA(227-251; delta244-251)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA(227-251; delta244-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
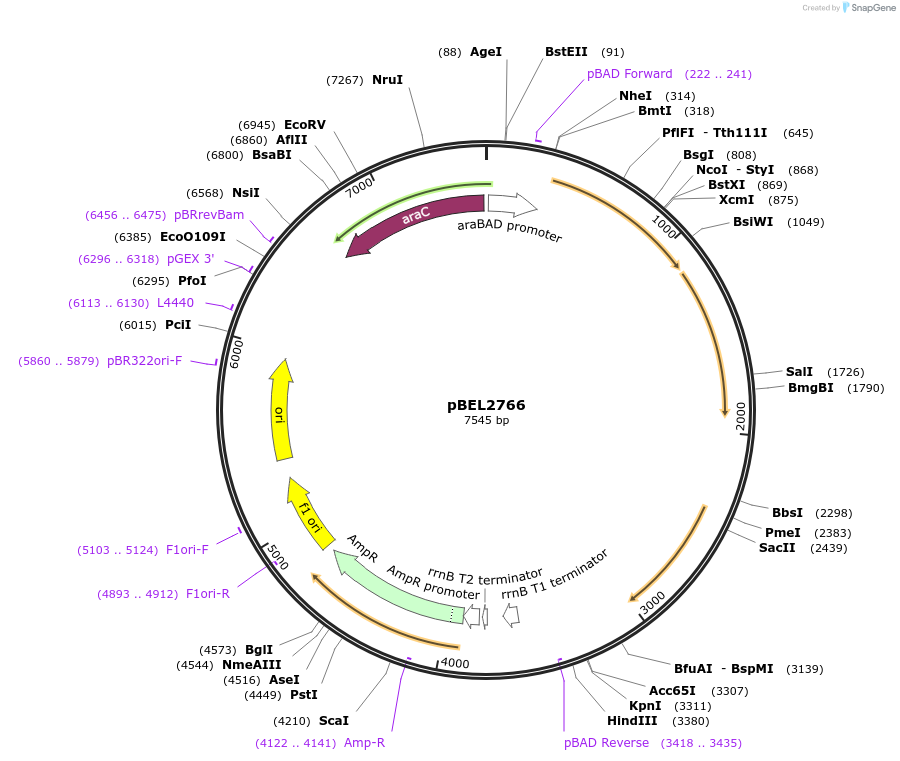
pBEL2766
Plasmid#196028PurposeComplementation of MlaF-MlaE-MlaDdelta141-183-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaDdelta141-183-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
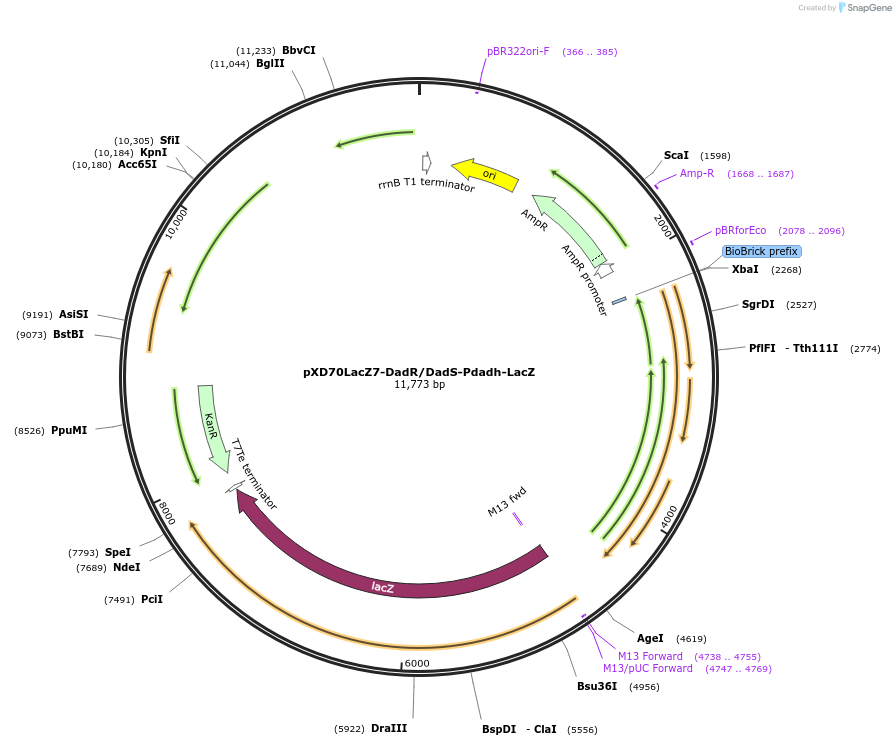
pXD70LacZ7-DadR/DadS-Pdadh-LacZ
Plasmid#191622PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Pdadh dopamine dehydroxylase promoter-lacZ with transcriptional regulators DadR/DadSDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
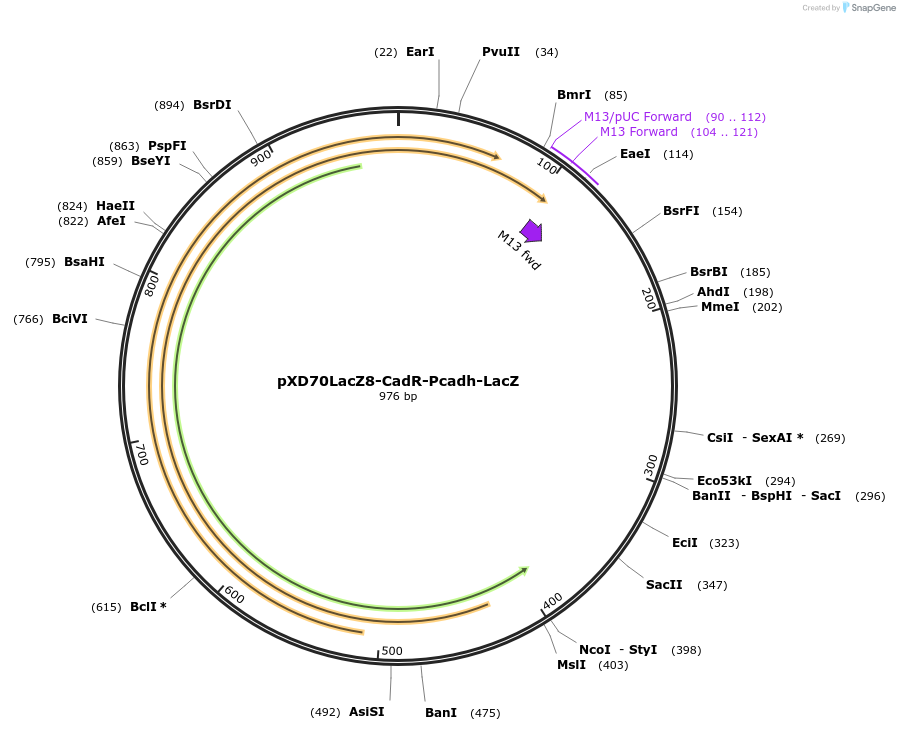
pXD70LacZ8-CadR-Pcadh-LacZ
Plasmid#191623PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Pcadh catechin dehydroxylase promoter-lacZ with transcriptional regulator CadRDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
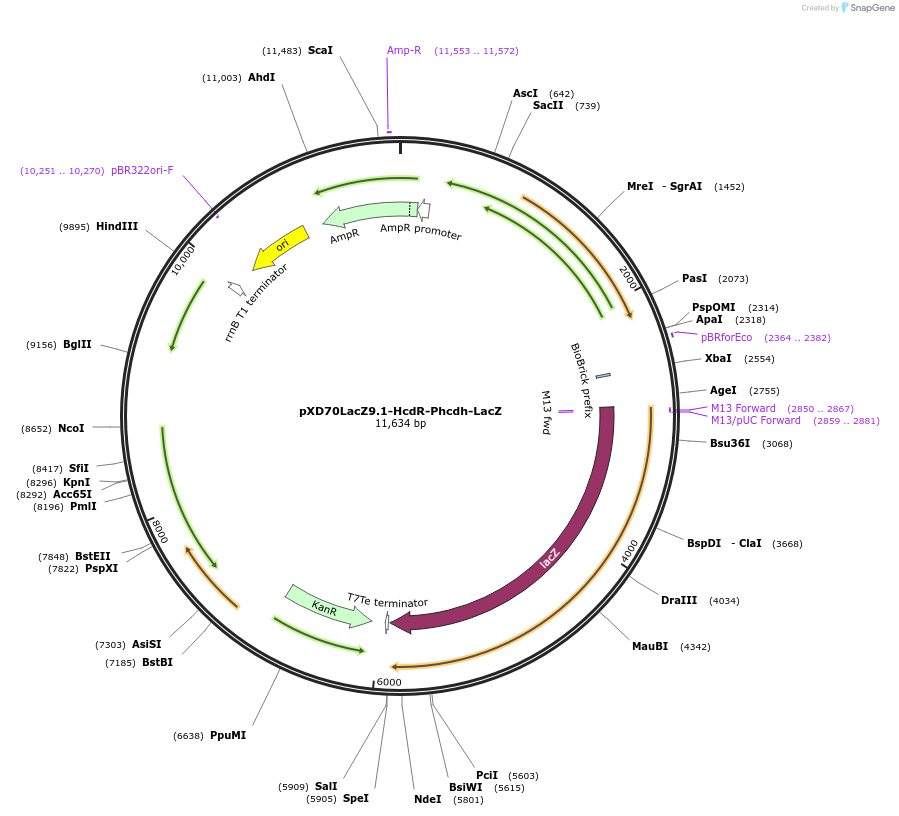
pXD70LacZ9.1-HcdR-Phcdh-LacZ
Plasmid#191629PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Phcdh hydrocaffeic acid dehydroxylase promoter-lacZ with transcriptional regulator HcdRDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
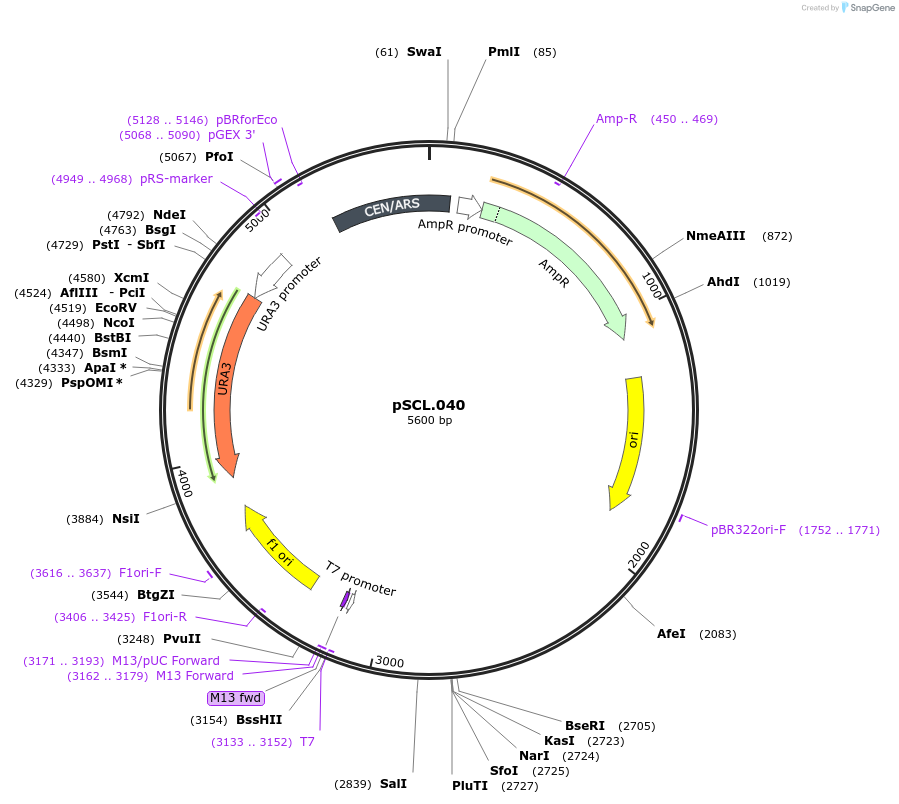
pSCL.040
Plasmid#184974PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v2
ExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
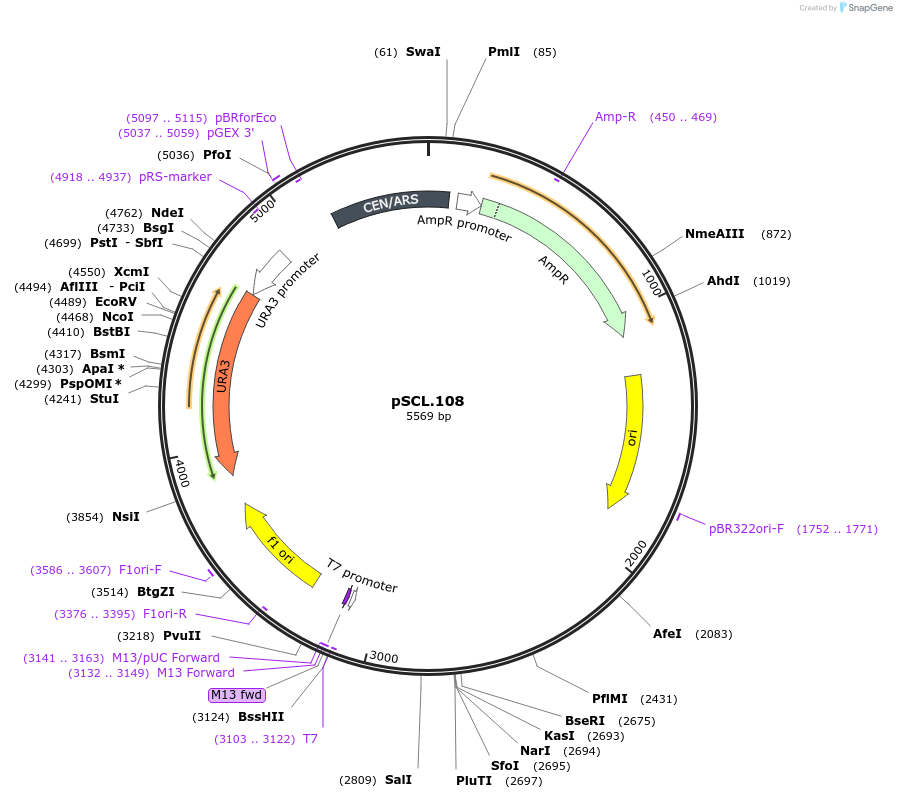
pSCL.108
Plasmid#184977PurposeExpress -Eco1 LYP1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 12
ExpressionYeastMutationLYP1 donor E27stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
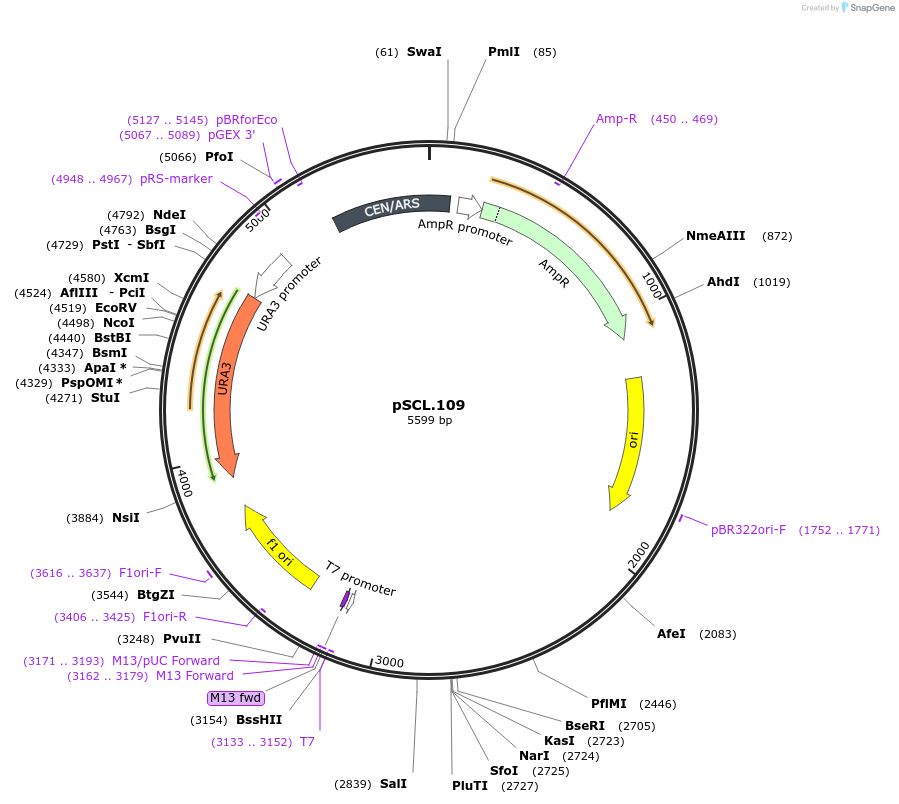
pSCL.109
Plasmid#184978PurposeTest effect of extending a1/a2 on LYP1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 27 v1
ExpressionYeastMutationLYP1 donor E27stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
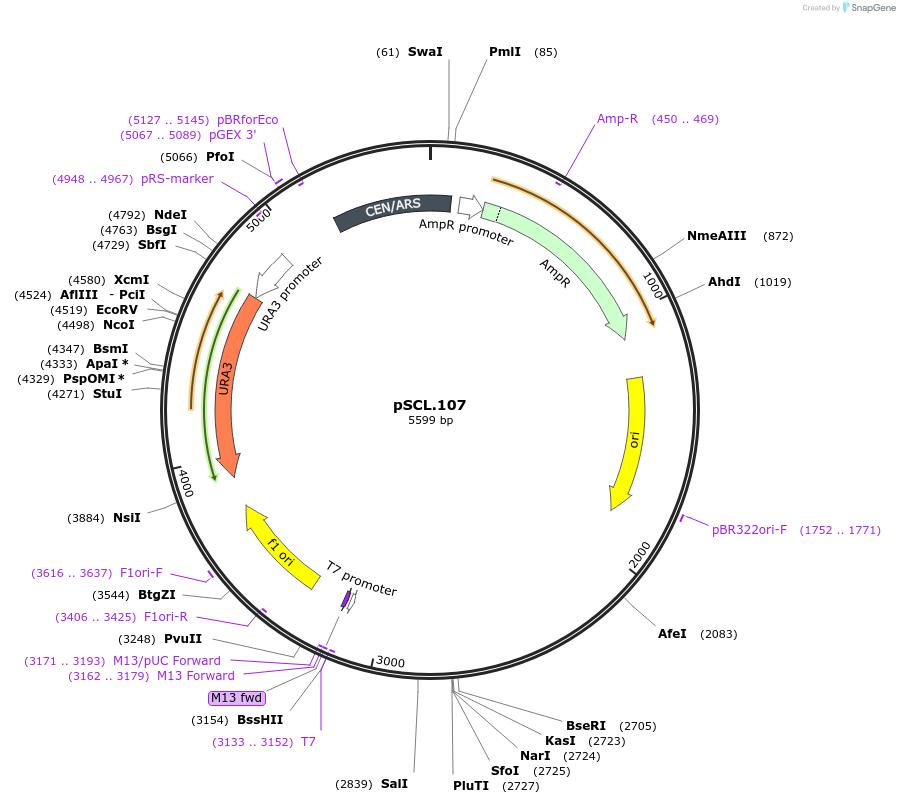
pSCL.107
Plasmid#184976PurposeTest effect of extending a1/a2 on CAN1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 27 v1
ExpressionYeastMutationCAN1 donor G444stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
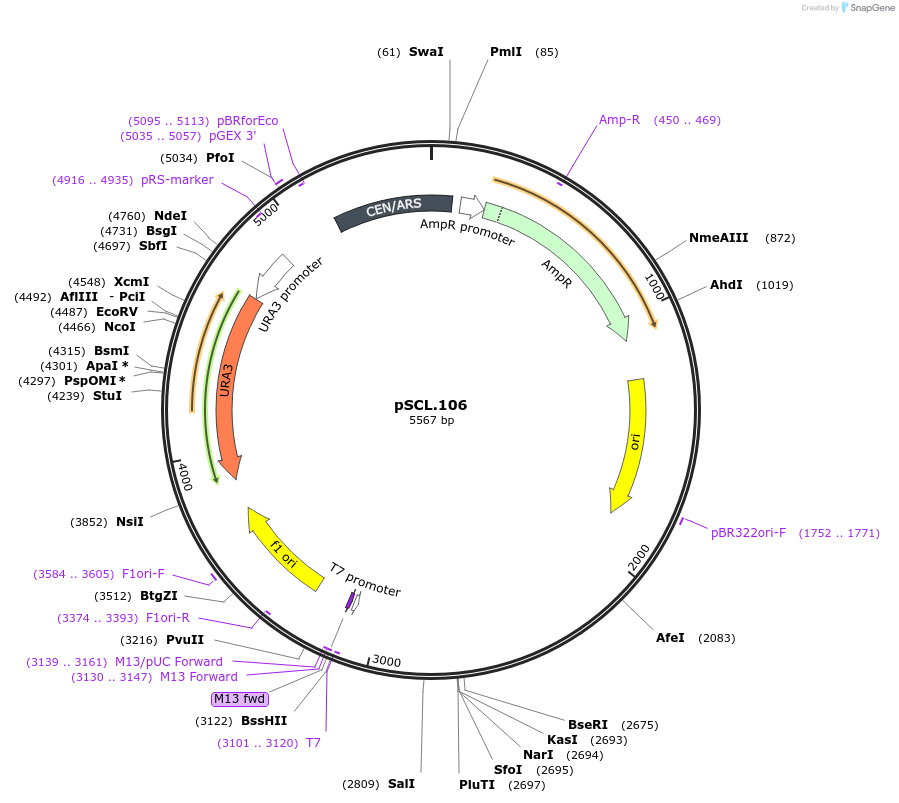
pSCL.106
Plasmid#184975PurposeExpress -Eco1 CAN1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 12
ExpressionYeastMutationCAN1 donor G444stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
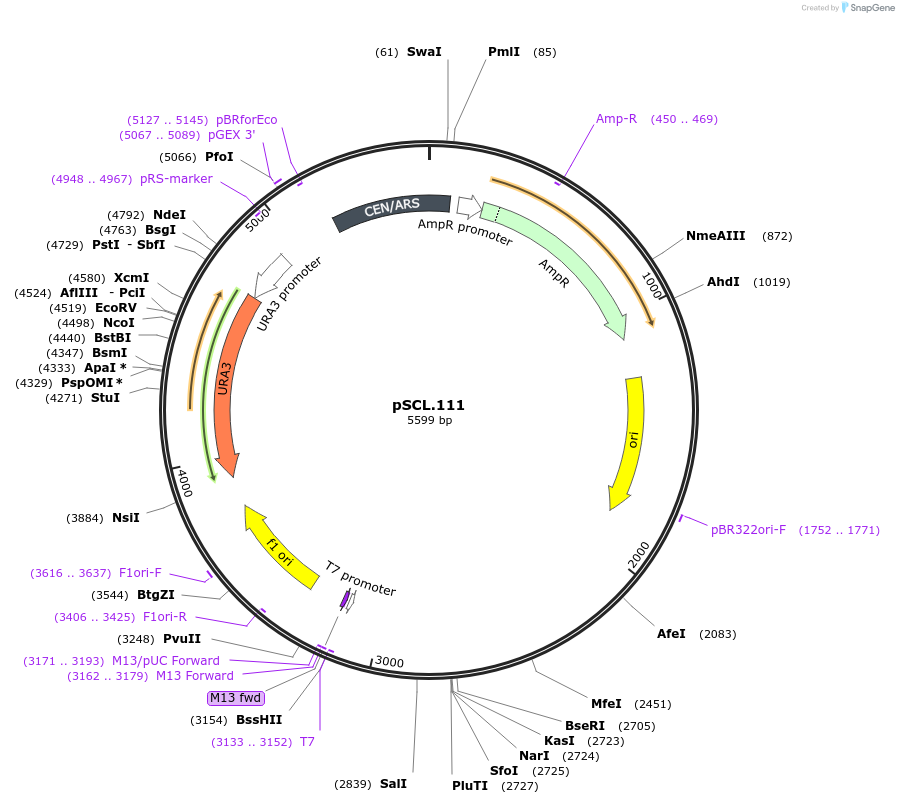
pSCL.111
Plasmid#184980PurposeTest effect of extending a1/a2 on TRP2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 27 v1
ExpressionYeastMutationTRP2 donor E64stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
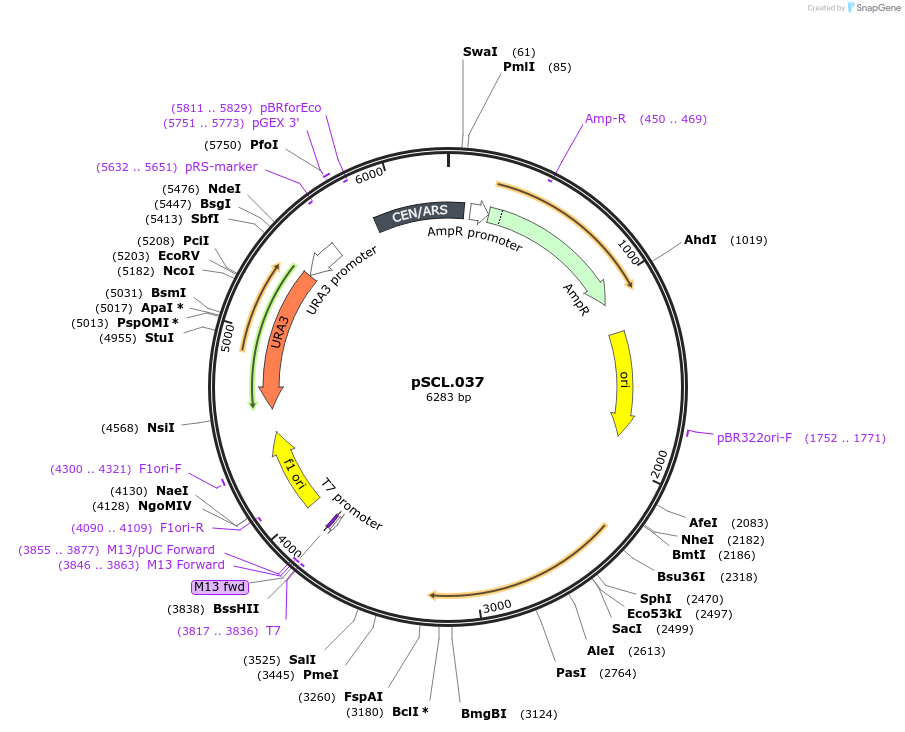
pSCL.037
Plasmid#184966PurposeExpress -Eco1 dead RT and ncRNA in yeasstDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12, dead RT
ExpressionYeastMutationHuman codon optimized RT, YXDD catalytic region m…PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
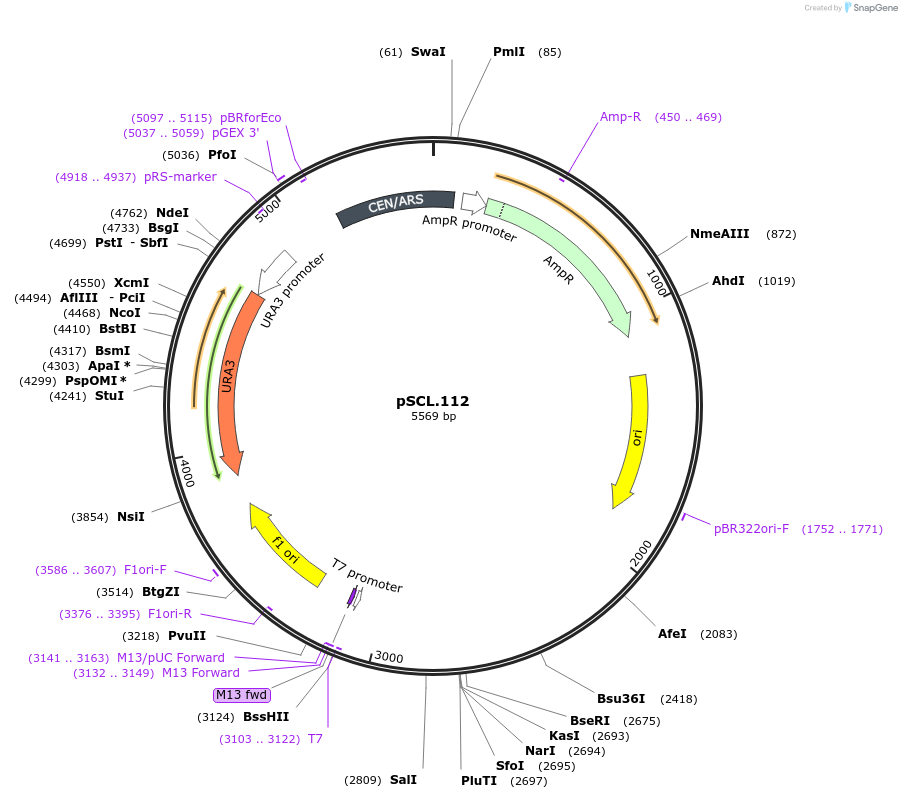
pSCL.112
Plasmid#184981PurposeExpress -Eco1 FAA1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 12
ExpressionYeastMutationFAA1 donor P233stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
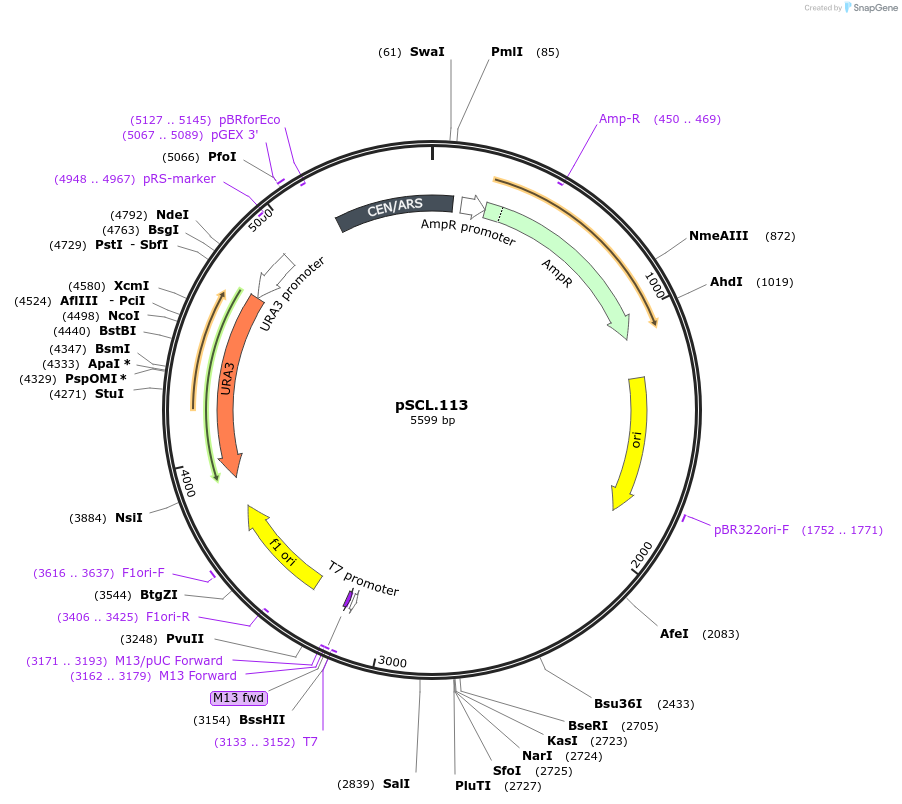
pSCL.113
Plasmid#184982PurposeTest effect of extending a1/a2 on FAA1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 27 v1
ExpressionYeastMutationFAA1 donor P233stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
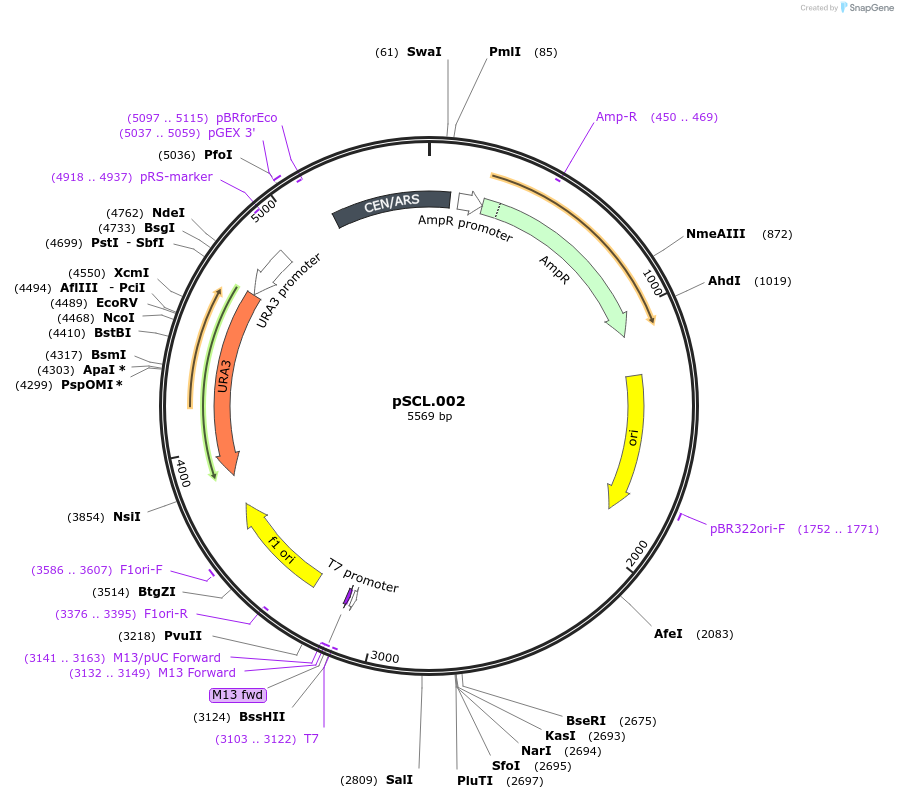
pSCL.002
Plasmid#184972PurposeExpress -Eco1 ADE2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 12
ExpressionYeastMutationADE2 donor P272stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
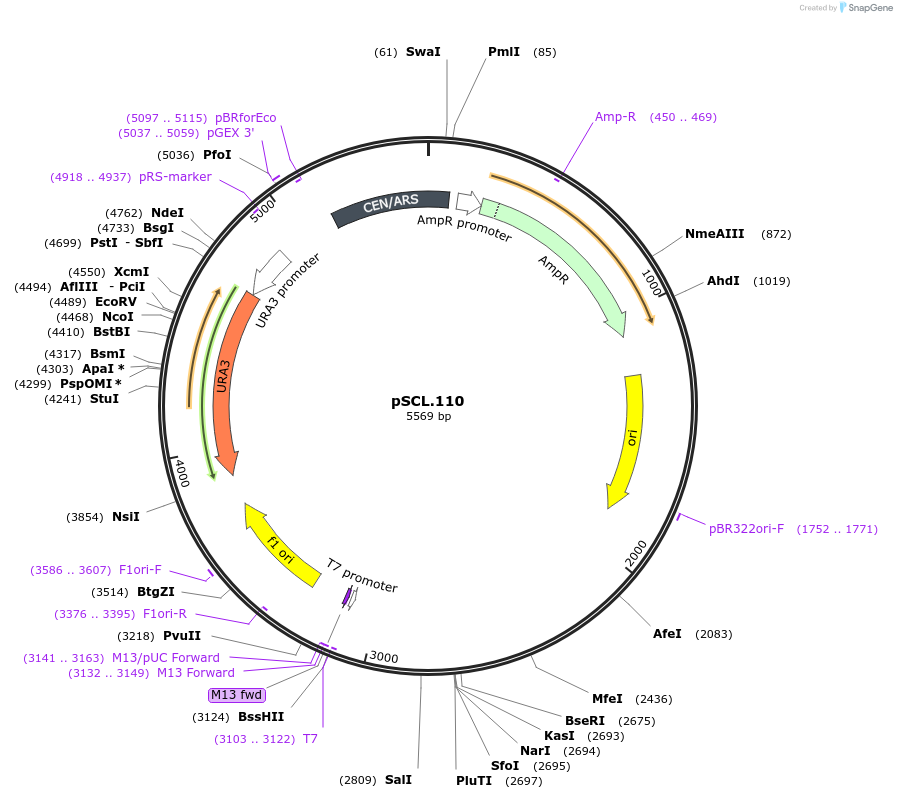
pSCL.110
Plasmid#184979PurposeExpress -Eco1 TRP2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 12
ExpressionYeastMutationTRP2 donor E64stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
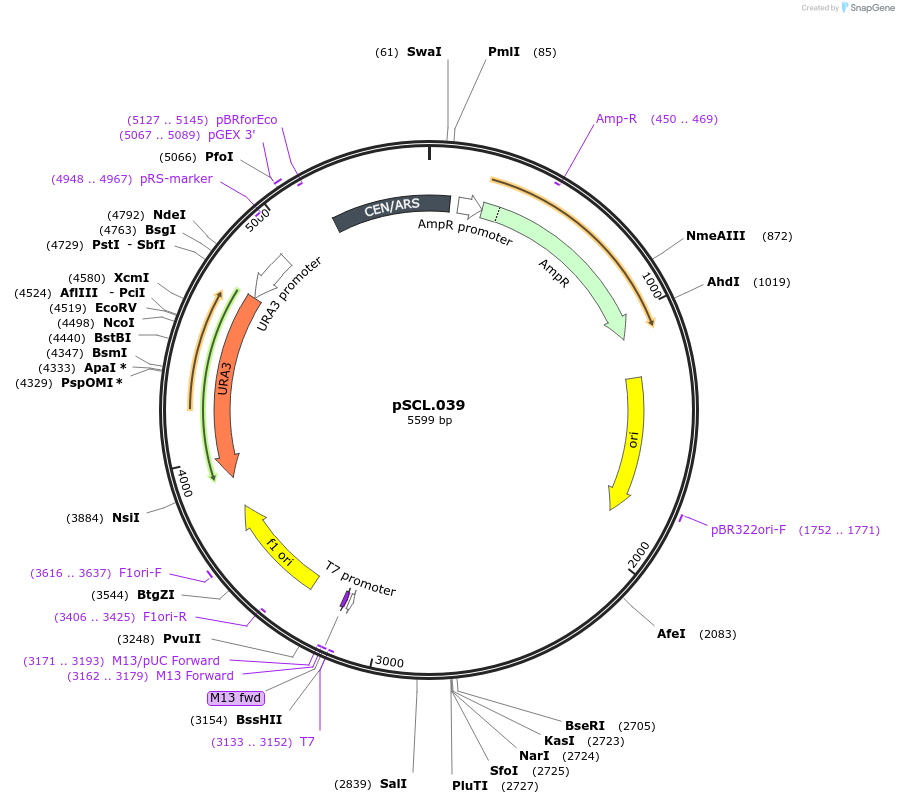
pSCL.039
Plasmid#184973PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v1
ExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
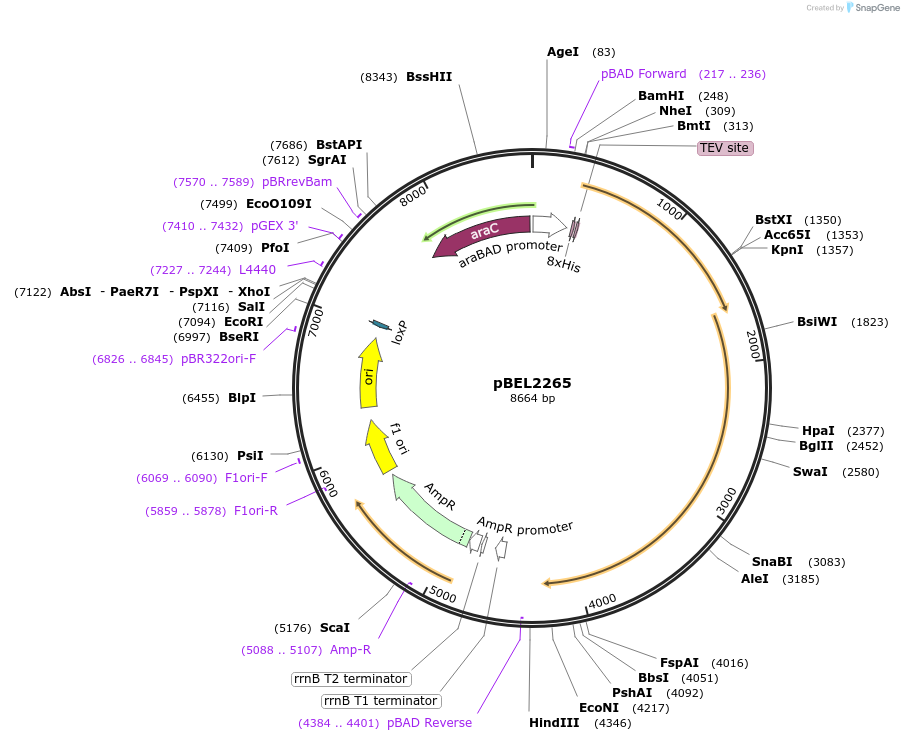
pBEL2265
Plasmid#175962PurposeFor expresson of LetA-[LetB PLL3->6] operon from E. coli. N-terminal his tag on LetA, araBAD promoterDepositorInsertsLetA
LetB
Tags8xHis 2xQH and TEV cleavage site on LetAExpressionBacterialMutationΔ347-361, replace with 702-716PromoteraraBADAvailable SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
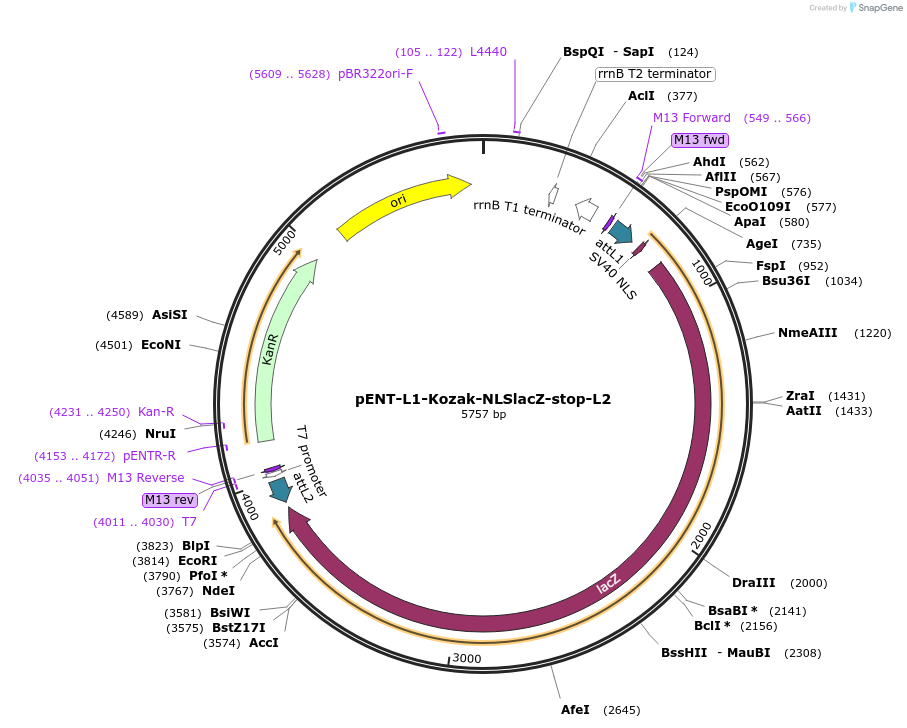
pENT-L1-Kozak-NLSlacZ-stop-L2
Plasmid#186358PurposeEntry clone with ORF encoding NLS lacZ flanked by Gateway recombination sequences.DepositorInsertlacZ (lacZ Synthetic, E. coli)
UseExpression of nuclear lacz reporterTagsNuclear localization signalAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
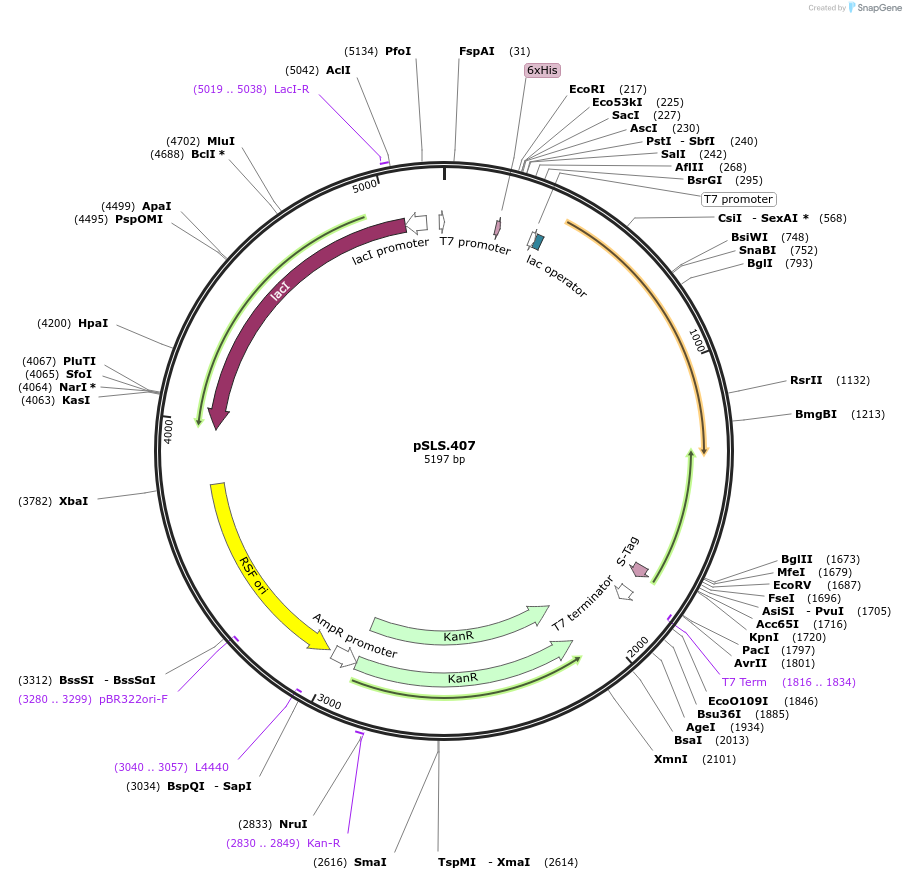
pSLS.407
Plasmid#187204PurposeExpresses retron Eco1 ncRNA v32, and Cas1 + Cas2DepositorInsertsRetron Eco1 ncRNA v32
Cas1 + Cas2
UseSynthetic BiologyExpressionBacterialPromoterT7/LacAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
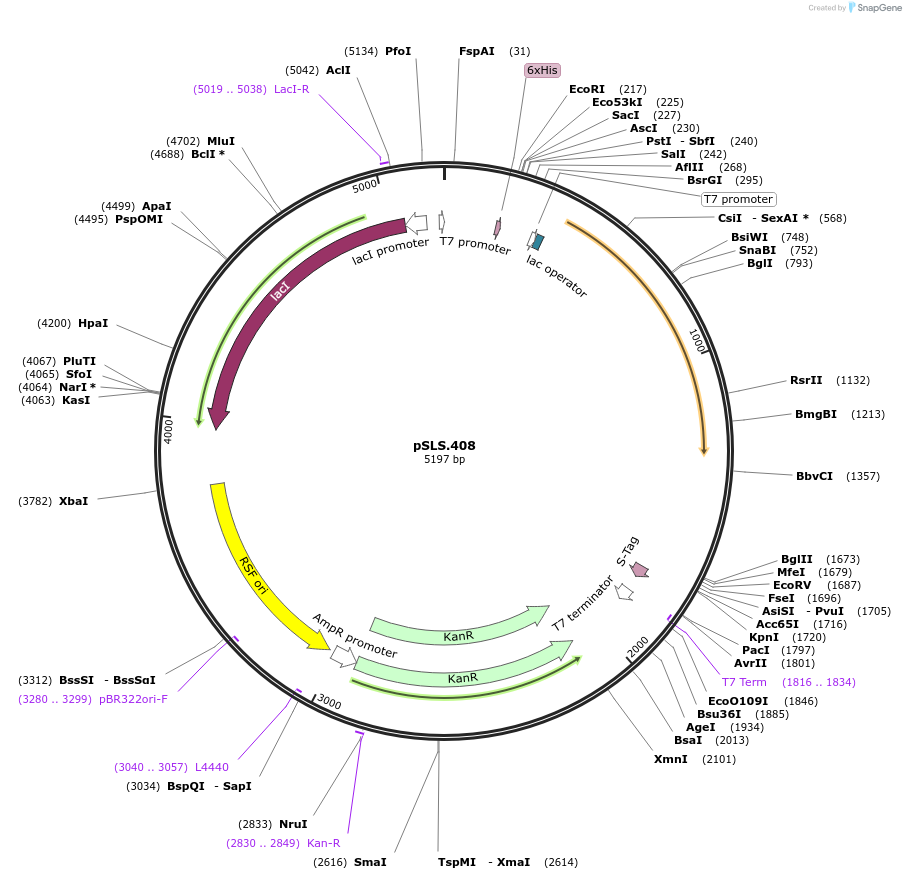
pSLS.408
Plasmid#187205PurposeExpresses retron Eco1 ncRNA v35, and Cas1 + Cas2DepositorInsertsRetron Eco1 ncRNA v35
Cas1 + Cas2
UseSynthetic BiologyExpressionBacterialPromoterT7/LacAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
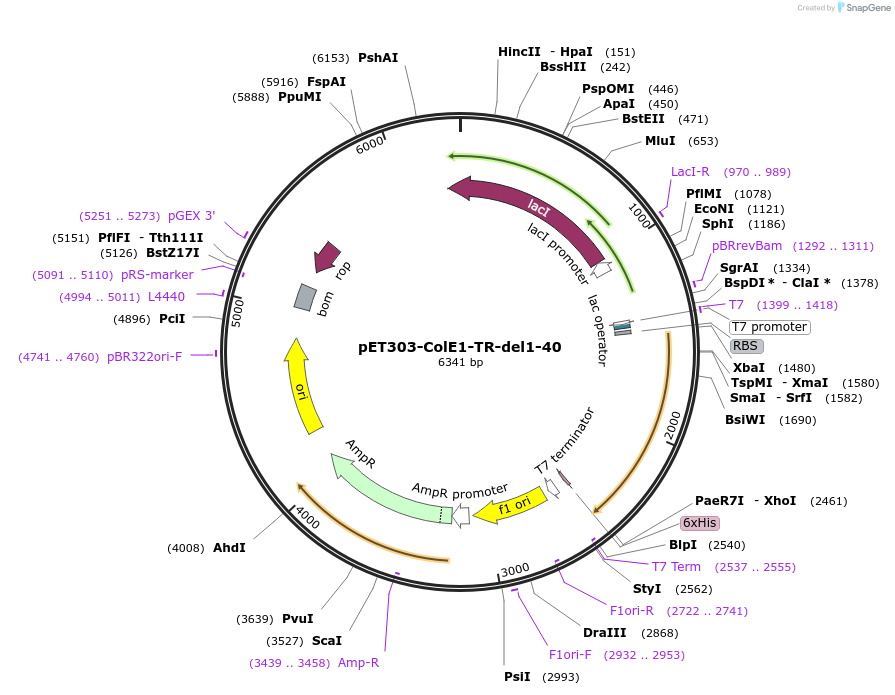
pET303-ColE1-TR-del1-40
Plasmid#180236PurposeExpresses T and R domains of Colicin E1 with N-terminal 40 residue deletion (residues 41-364)DepositorInsertColicin E1
Tags6x His-tagExpressionBacterialMutationTruncation of ColE1 containing T and R domains (r…PromoterT7Available SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
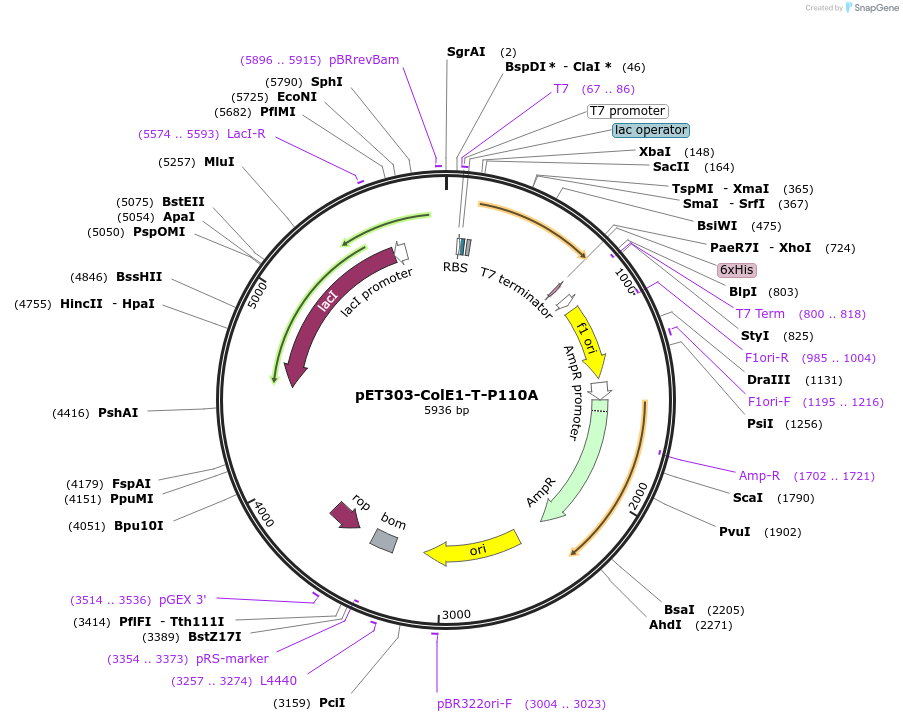
pET303-ColE1-T-P110A
Plasmid#180237PurposeExpresses T domain of Colicin E1 (residues 1-190) with proline to alanine mutation at residue 110DepositorInsertColicin E1
Tags6x His-tagExpressionBacterialMutationTruncation of ColE1 containing T domain of ColE1 …PromoterT7Available SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
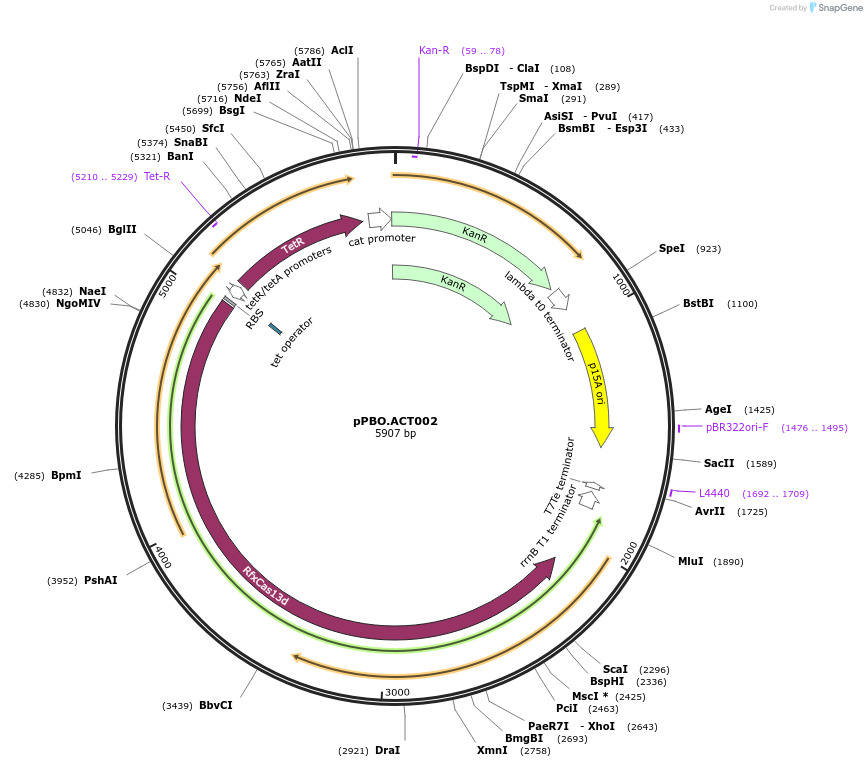
pPBO.ACT002
Plasmid#182712PurposeaTc inducible dCasRx-IF1DepositorInsertdCasRx
UseCRISPRTags3x(GGGS)-Escherichia Coli Initiation Factor 1ExpressionBacterialMutationCatalytically deactivated R295A, H300A, R849A, H8…PromoterpTetAvailable SinceAug. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
pTS2343_Tier3(SB)-YPet_Blast
Plasmid#169641PurposeTier-3 vector for stable stable integration of up to three cassettes via sleeping beauty transposase including a YPet marker and BlastR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-YPet-p2A-BlastR-pA-3'ITR)DepositorInsertPRPBSA-driven YPet and BlastR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
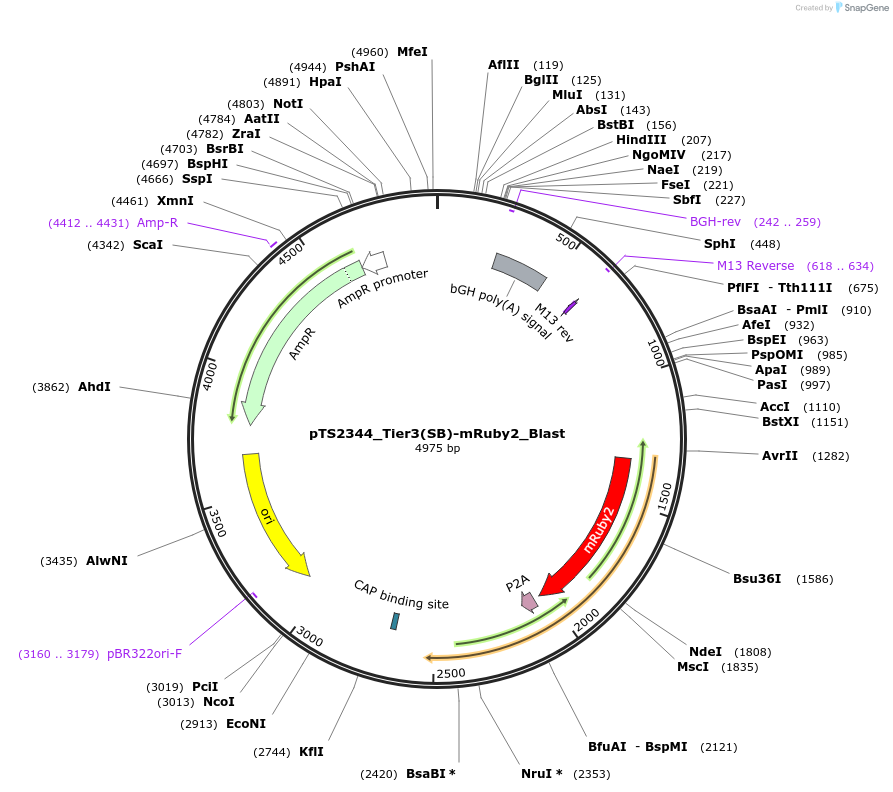
pTS2344_Tier3(SB)-mRuby2_Blast
Plasmid#169642PurposeTier-3 vector for stable stable integration of up to three cassettes via sleeping beauty transposase including a mRuby marker and BlastR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-mRuby2-p2A-BlastR-pA-3'ITR)DepositorInsertPRPBSA-driven mRuby2 and BlastR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
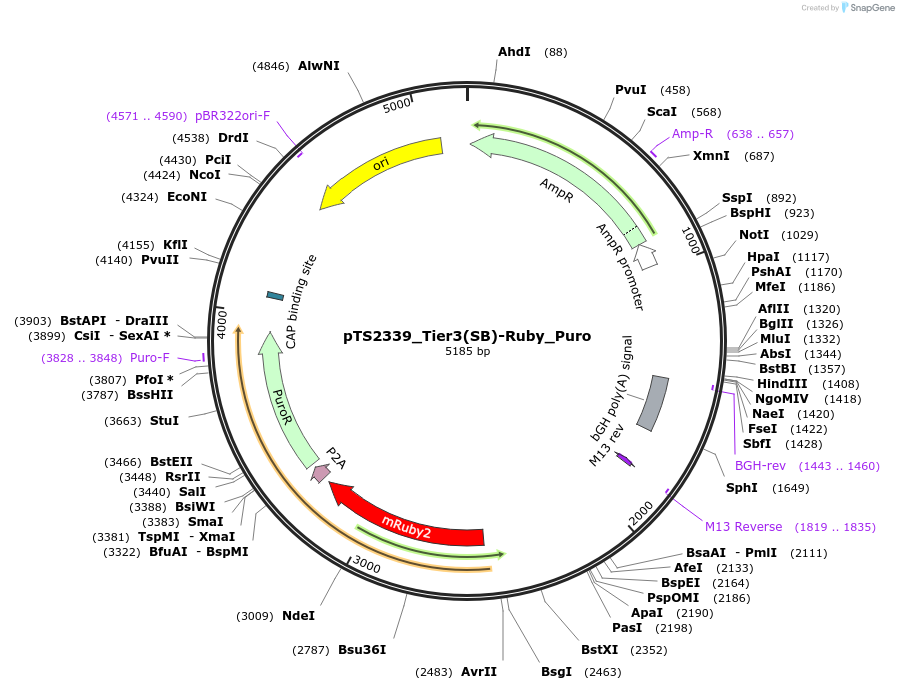
pTS2339_Tier3(SB)-Ruby_Puro
Plasmid#169637PurposeTier-3 vector for stable stable integration of up to three cassettes via sleeping beauty transposase including a mRuby2 marker and PuroR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-mRuby2-p2A-PuroR-pA-3'ITR)DepositorInsertPRPBSA-driven mRuby2 and PuroR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
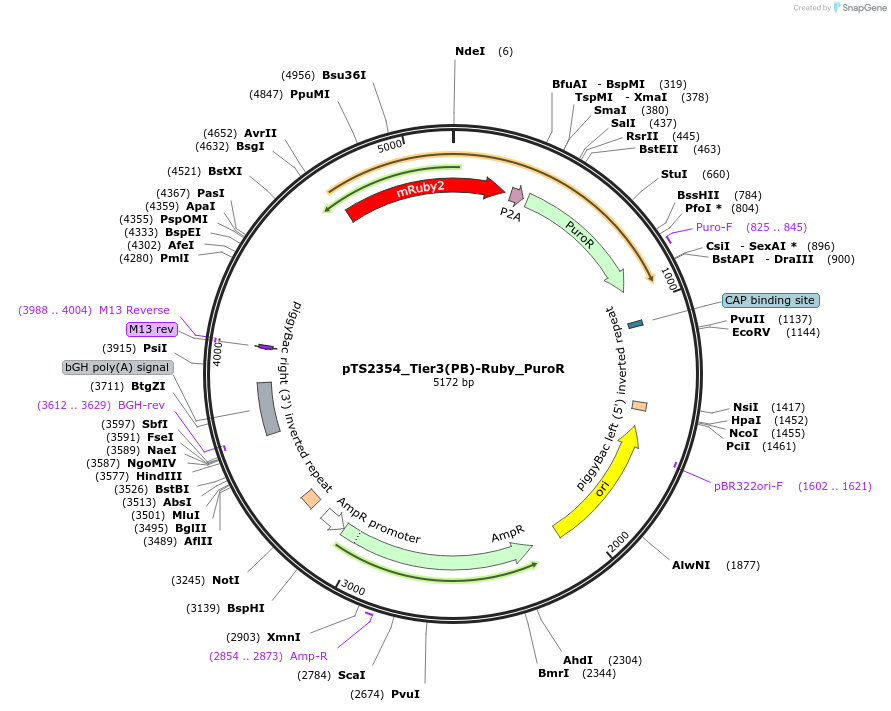
pTS2354_Tier3(PB)-Ruby_PuroR
Plasmid#169653PurposeTier-3 vector for stable stable integration of up to three cassettes via Piggy Bac transposase including a mRuby2 marker and PuroR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-mRuby2-p2A-PuroR-pA-3'ITR)DepositorInsertPRPBSA-driven mRuby2 and PuroR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
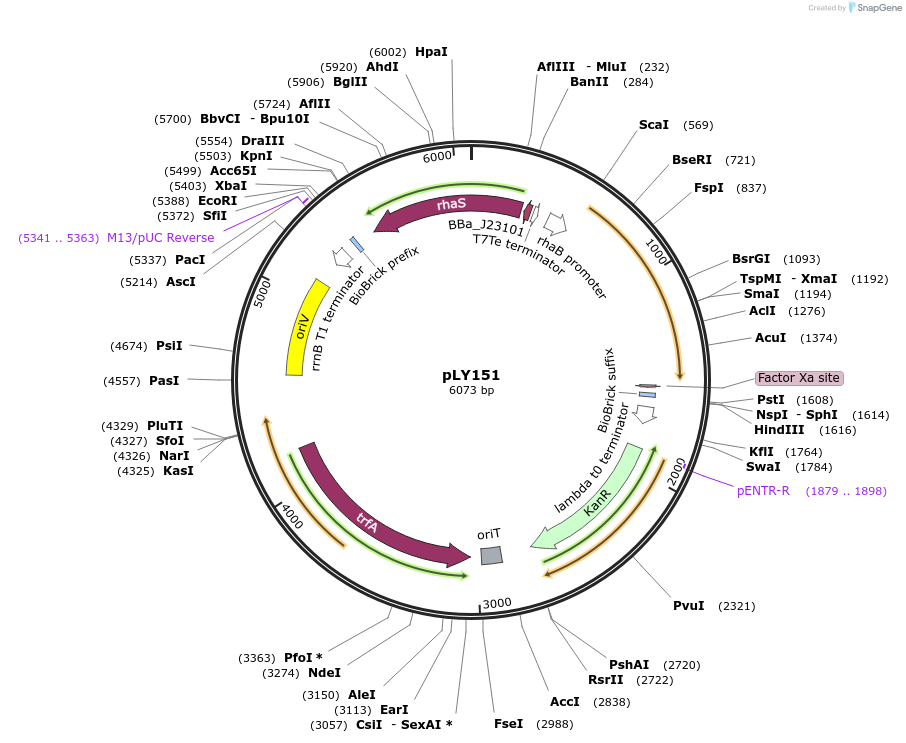
pLY151
Plasmid#130947PurposepspFΔHTH::λN22plus gene controlled by promoter PrhaBDepositorInsertsrhaS
pspFΔHTH::λN22plus
UseSynthetic BiologyTagspspFΔHTH::λN22plusExpressionBacterialMutationThe HTH domain of the pspF gene was deleted (297-…PromoterPcon and PrhaBAvailable SinceOct. 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
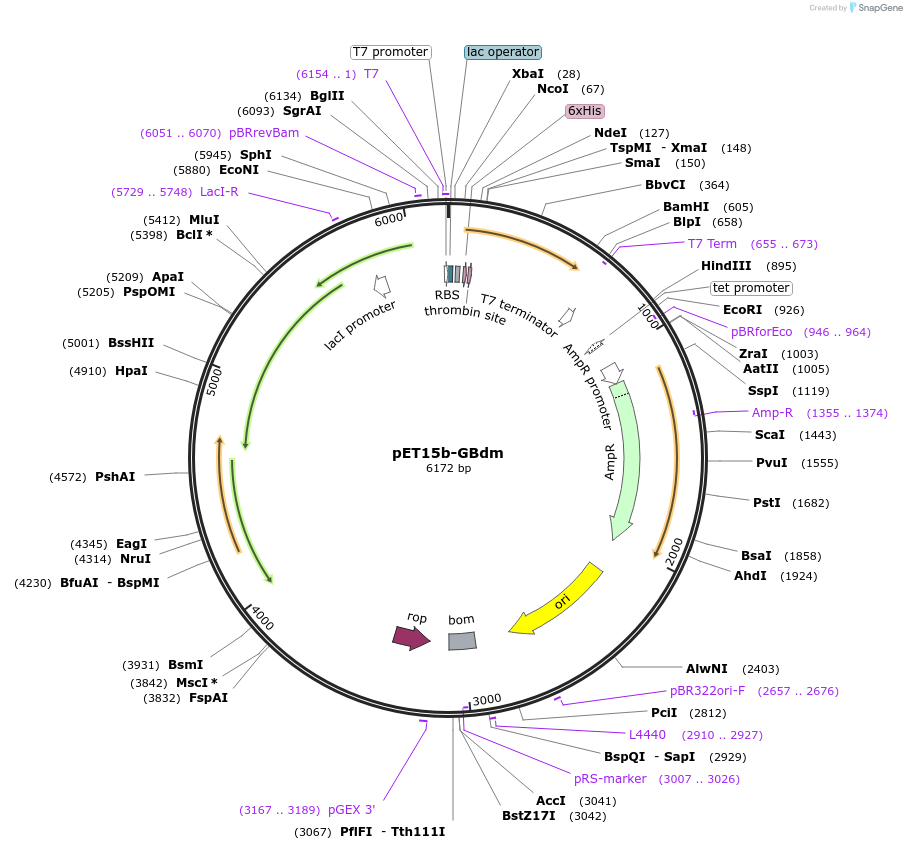
pET15b-GBdm
Plasmid#129690Purposeexpress E. coli GreB E82C/C68SDepositorAvailable SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
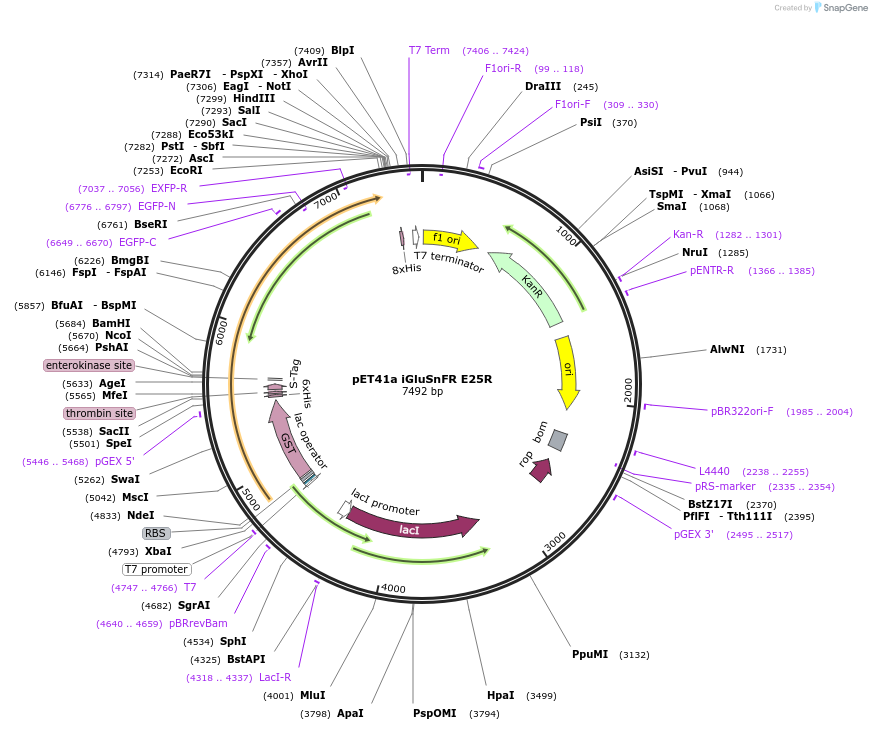
pET41a iGluSnFR E25R
Plasmid#119831PurposeProduction of recombinant iGluSnFR E25R variant, a single-wavelength glutamate indicator, in E. coliDepositorInsertiGluSnFR E25R
TagsGSTExpressionBacterialMutationE25RPromoterT7Available SinceMarch 12, 2019AvailabilityAcademic Institutions and Nonprofits only