We narrowed to 23,294 results for: SCR
-
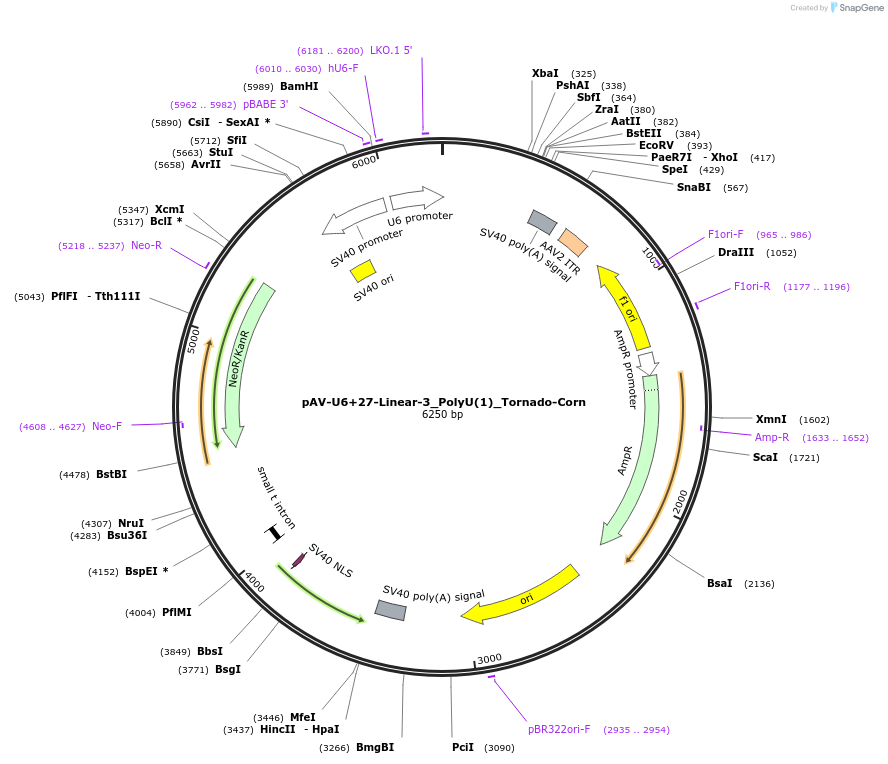
Plasmid#159503PurposeTests for the impact of 1 uracil in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-3_PolyU(1)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
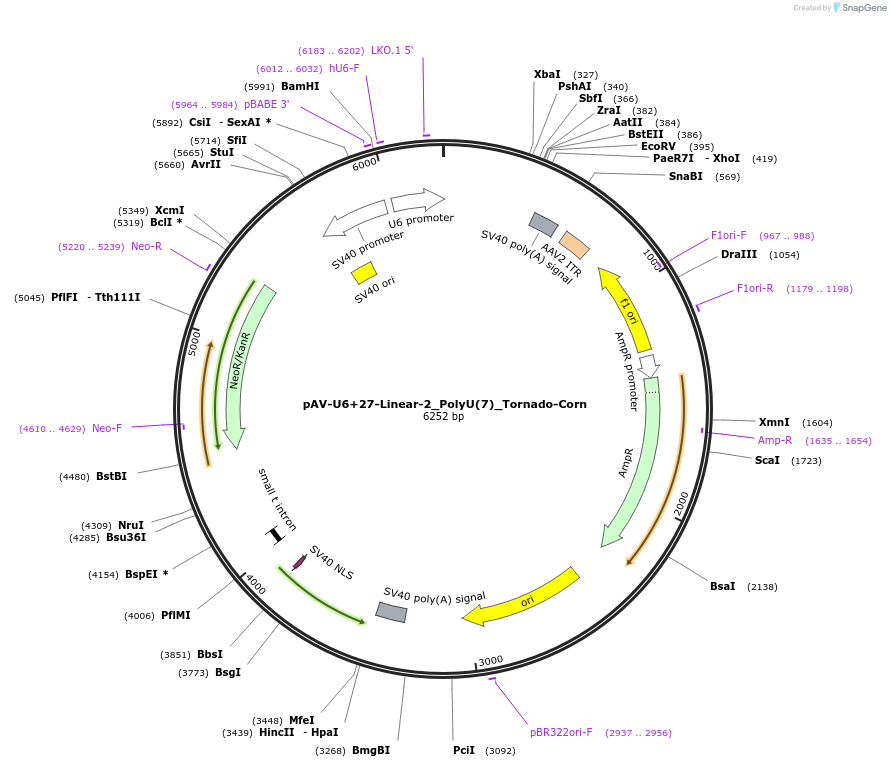
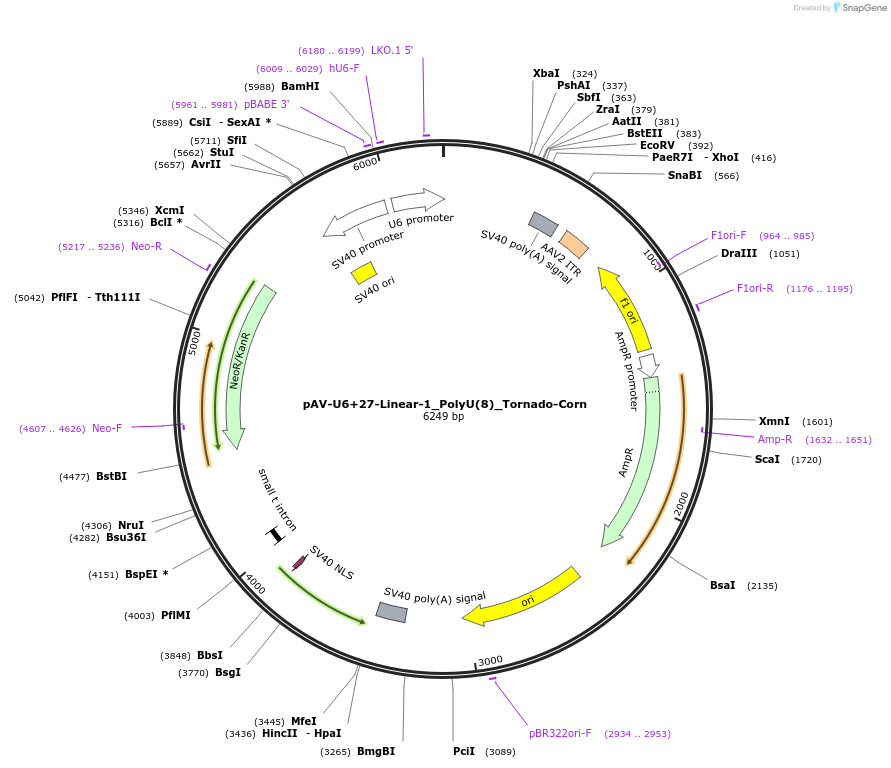
pAV-U6+27-Linear-2_PolyU(7)_Tornado-Corn
Plasmid#159493PurposeTests for the impact of 7 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-2_PolyU(7)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
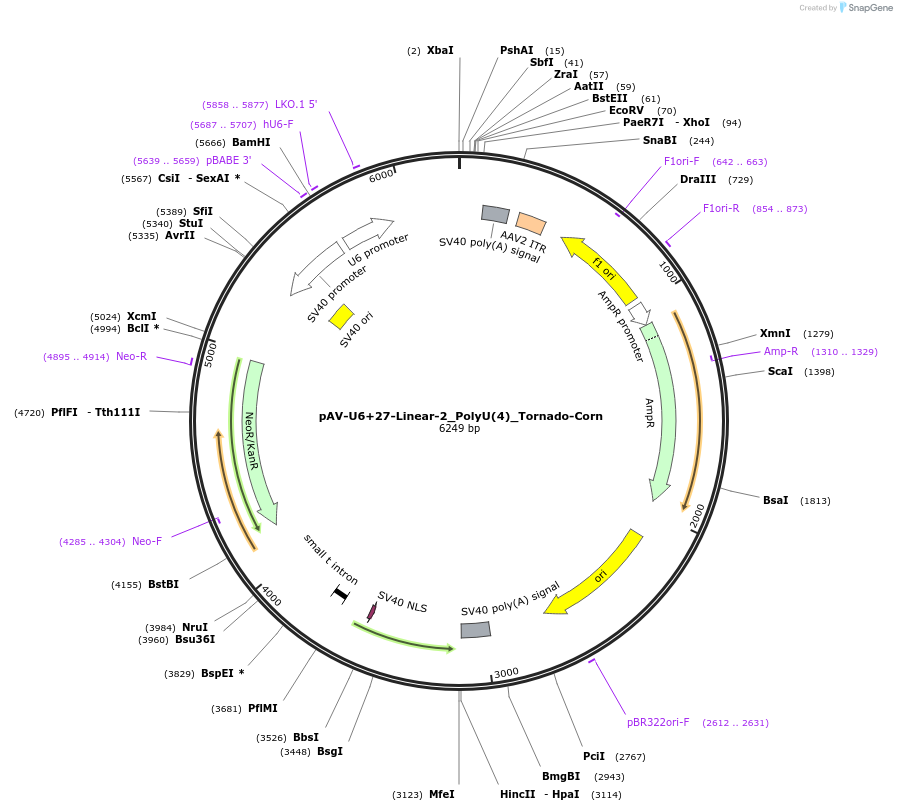
pAV-U6+27-Linear-2_PolyU(4)_Tornado-Corn
Plasmid#159490PurposeTests for the impact of 4 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-2_PolyU(4)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
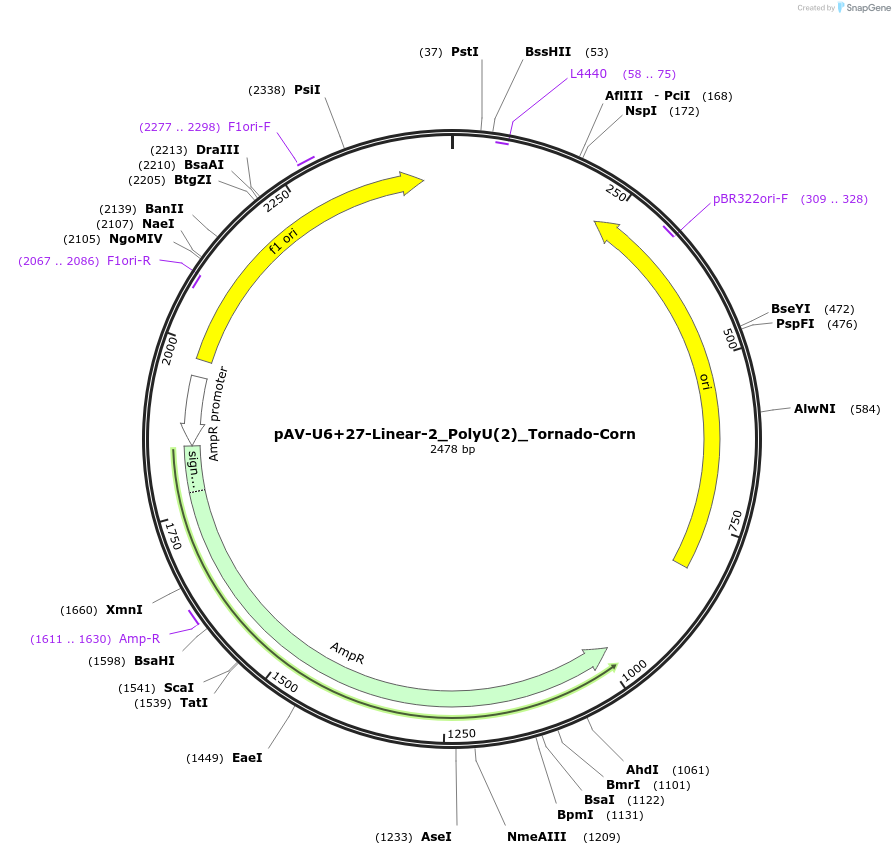
pAV-U6+27-Linear-2_PolyU(2)_Tornado-Corn
Plasmid#159488PurposeTests for the impact of 2 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-2_PolyU(2)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
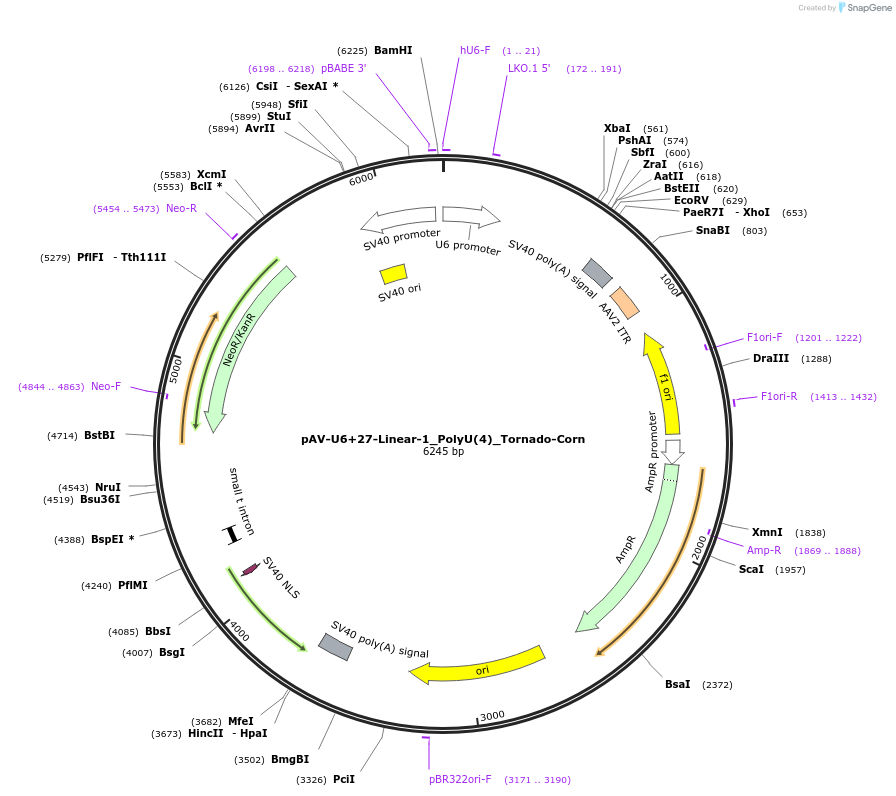
pAV-U6+27-Linear-1_PolyU(4)_Tornado-Corn
Plasmid#159482PurposeTests for the impact of 4 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-1_PolyU(4)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
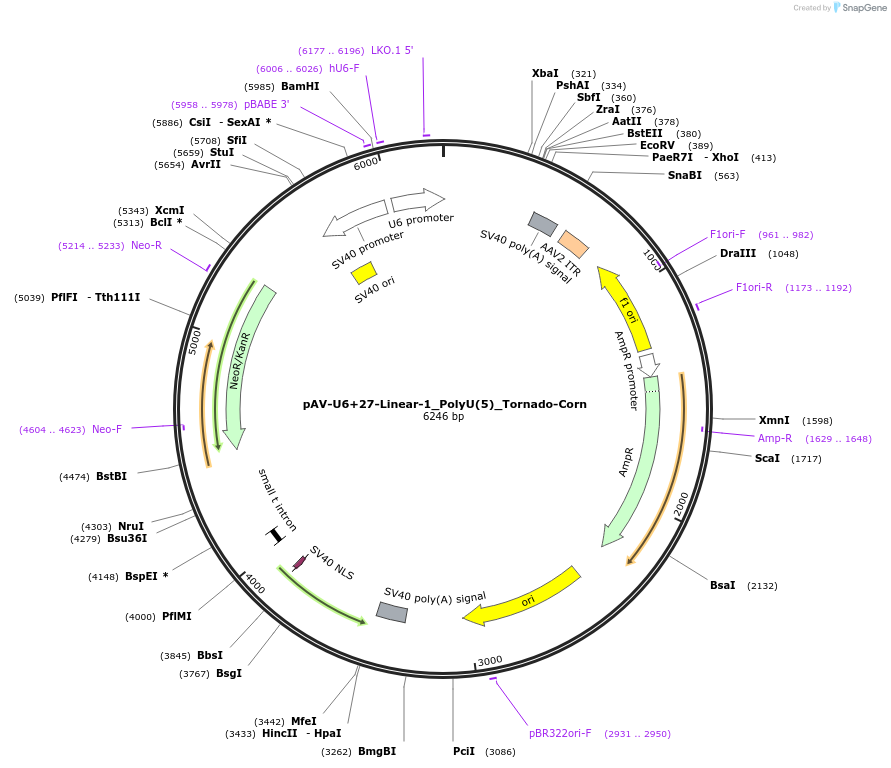
pAV-U6+27-Linear-1_PolyU(5)_Tornado-Corn
Plasmid#159483PurposeTests for the impact of 5 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-1_PolyU(5)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
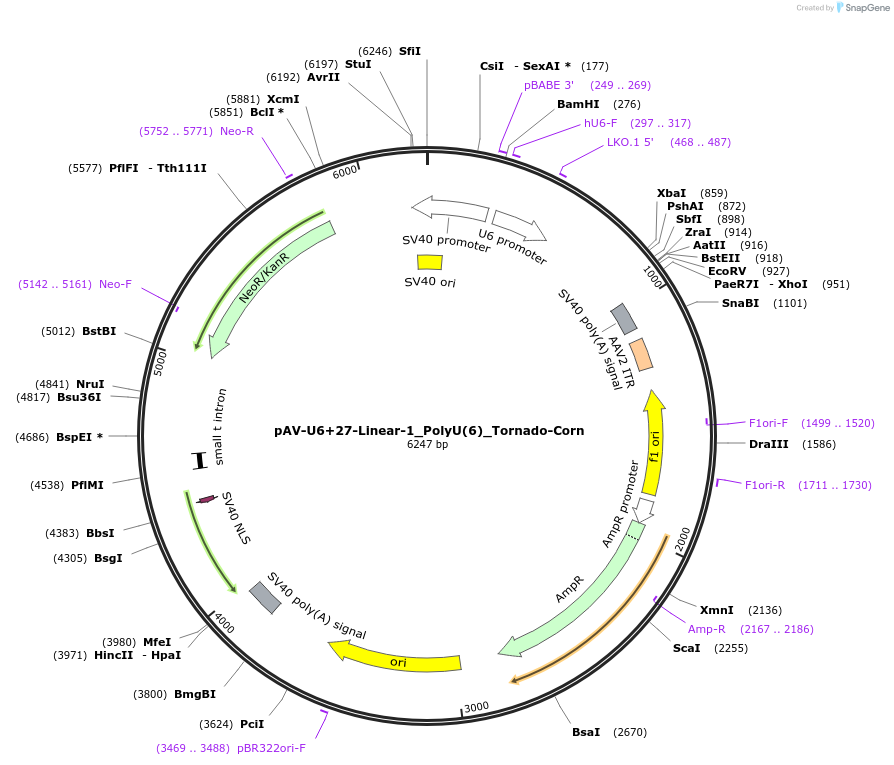
pAV-U6+27-Linear-1_PolyU(6)_Tornado-Corn
Plasmid#159484PurposeTests for the impact of 6 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-1_PolyU(6)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
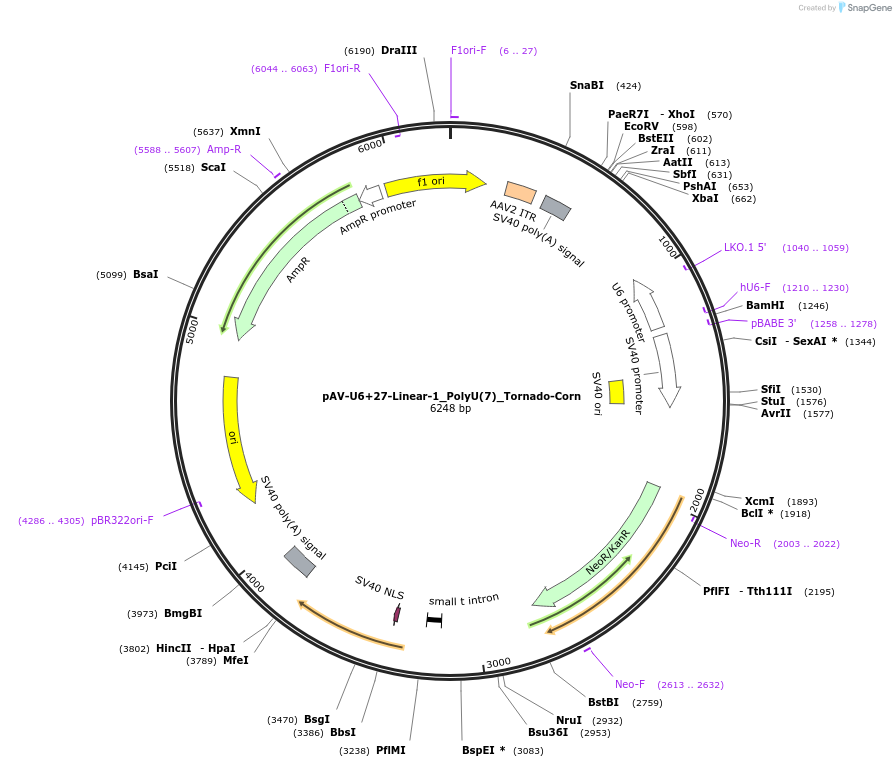
pAV-U6+27-Linear-1_PolyU(7)_Tornado-Corn
Plasmid#159485PurposeTests for the impact of 7 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-1_PolyU(7)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
pAV-U6+27-Linear-1_PolyU(8)_Tornado-Corn
Plasmid#159486PurposeTests for the impact of 8 uracils in a poly-uracil tract on human polymerase III transcription from a U6 promoter.DepositorInsertTermination Module:Linear-1_PolyU(8)
ExpressionMammalianPromoterU6-27Available SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
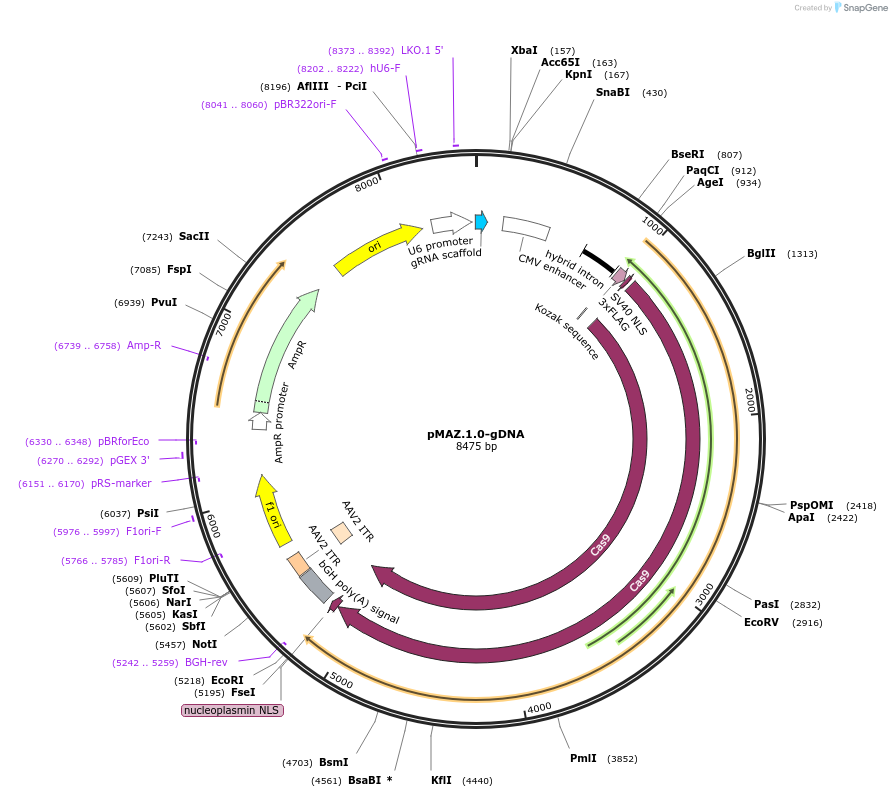
pMAZ.1.0-gDNA
Plasmid#112455PurposeCRISPR plasmid for expression of Cas9 and gRNA targeting human transcription factor MAZDepositorAvailable SinceDec. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
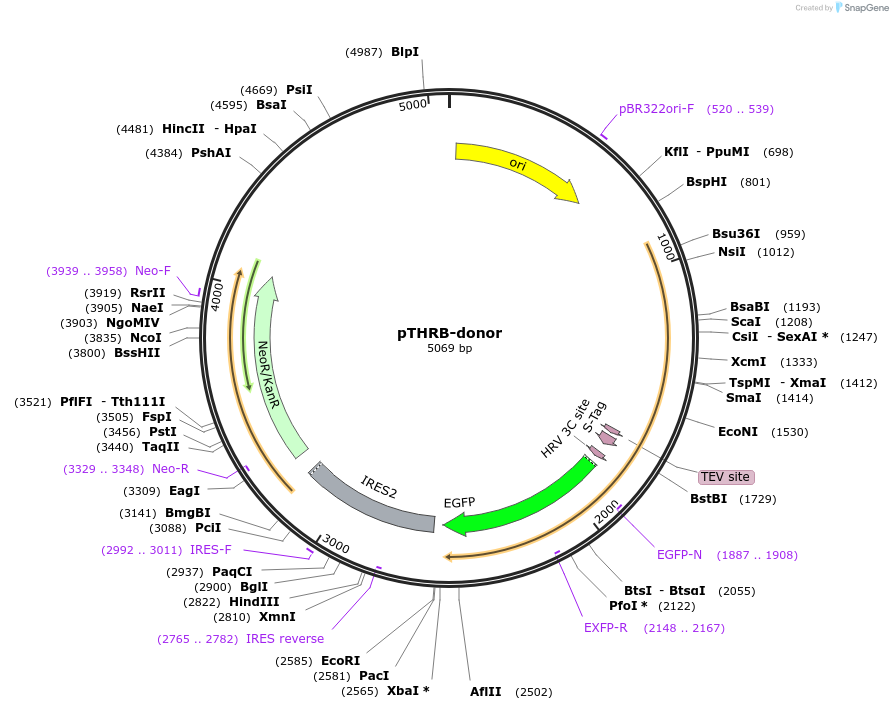
pTHRB-donor
Plasmid#112333PurposeCRISPR donor plasmid to tag human transcription factor THRB with GFPDepositorInsertTHRB homology arms flanking EGFP-IRES-Neo cassette (THRB Human)
UseCRISPRAvailable SinceNov. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
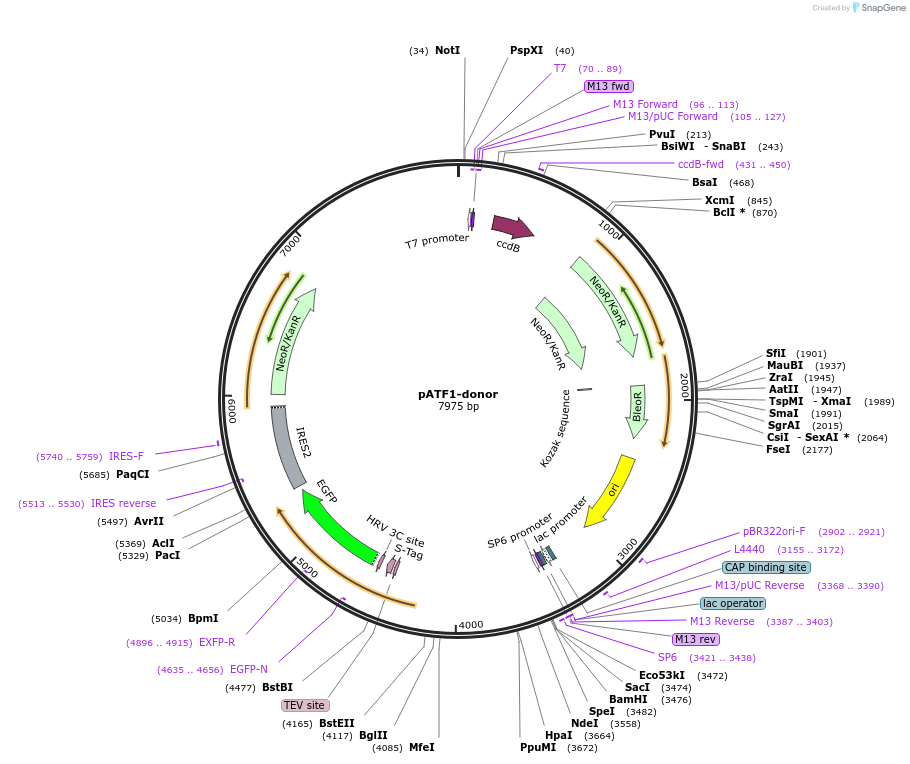
pATF1-donor
Plasmid#112328PurposeCRISPR donor plasmid to tag human transcription factor ATF1 with GFPDepositorInsertATF1 homology arms flanking EGFP-IRES-Neo cassette (ATF1 Human)
UseCRISPRMutationhg38:12:50819876:T>A, hg38:12:50819888:T>C,…Available SinceNov. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
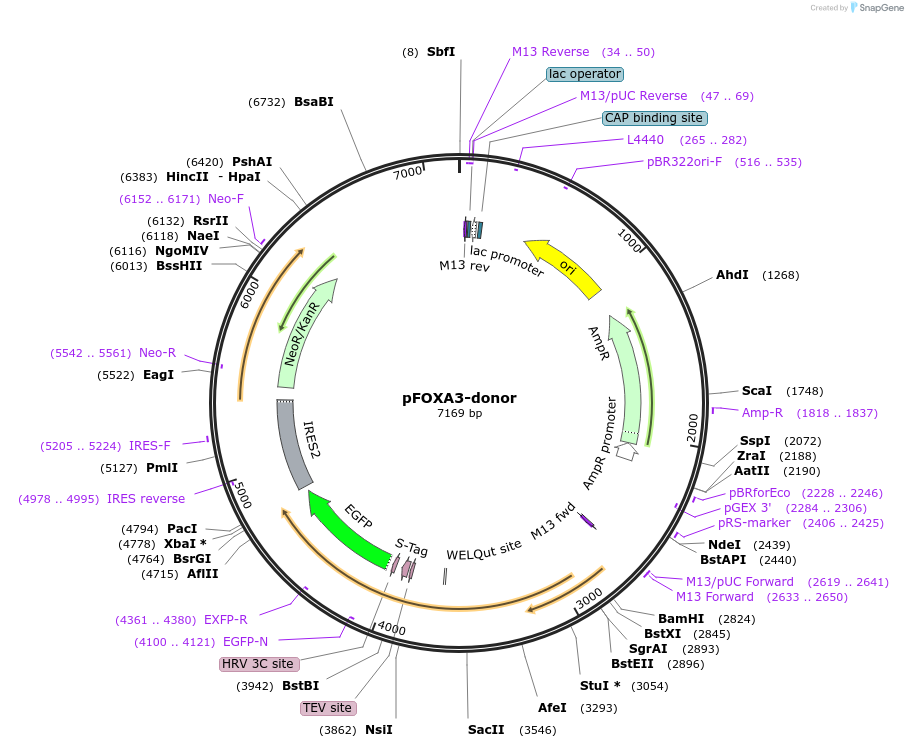
pFOXA3-donor
Plasmid#112321PurposeCRISPR donor plasmid to tag human transcription factor FOXA3 with GFPDepositorInsertFOXA3 homology arms flanking EGFP-IRES-Neo cassette (FOXA3 Human)
UseCRISPRMutationhg38:19:45872401:G>T, hg38:19:45872421:C>T,…Available SinceNov. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
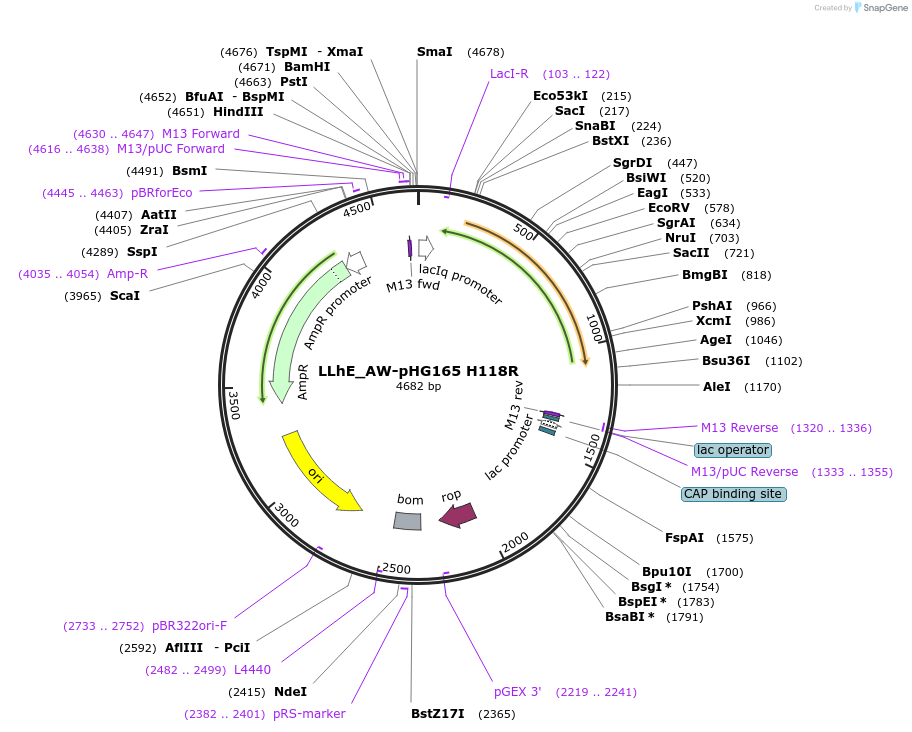
LLhE_AW-pHG165 H118R
Plasmid#90063PurposeConstitutive expression of a chimeric LacI:CelR transcription repressorDepositorInsertLLhE_AW/H118R
ExpressionBacterialMutationI48V/Q55A/Q60R/H118RPromoterIqAvailable SinceJune 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
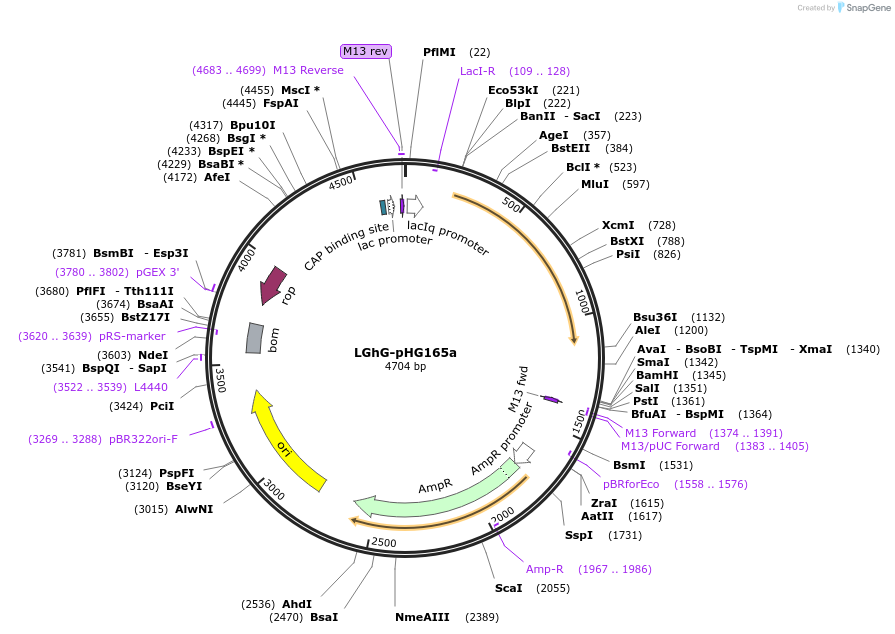
LGhG-pHG165a
Plasmid#90048PurposeConstitutive expression of a chimeric LacI:GalR transcription repressorDepositorInsertLGhG
ExpressionBacterialPromoterIqAvailable SinceJune 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
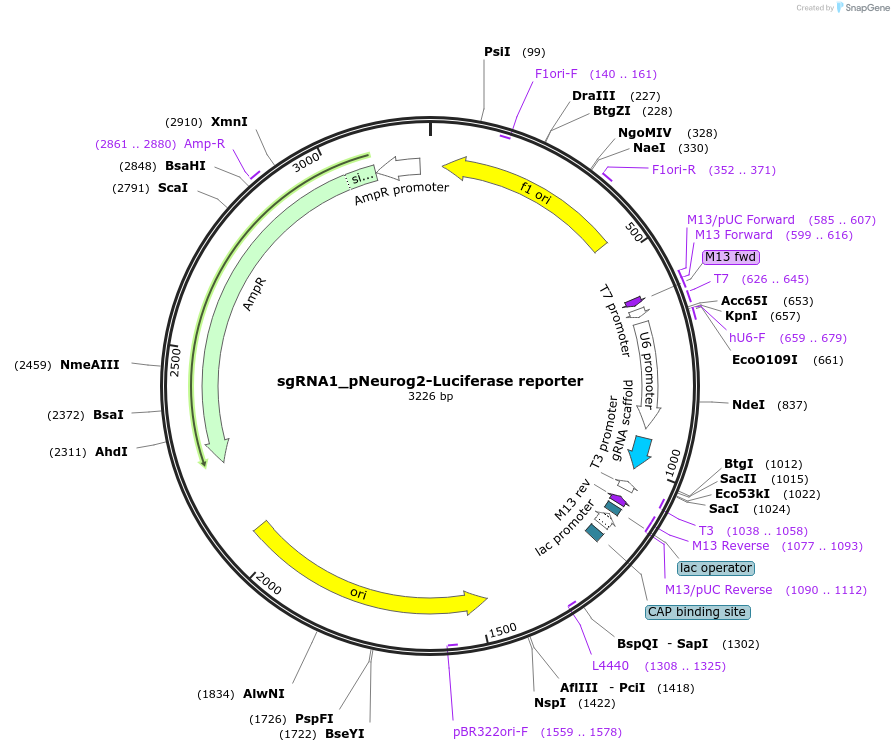
sgRNA1_pNeurog2-Luciferase reporter
Plasmid#64160PurposePhotoactivatable transcription system. Mammalian expression of sgRNA1 to target pNeurog2-Luciferase reporter.DepositorInsertsgRNA1 for pNeurog2-Luciferase reporter
UseCRISPRExpressionMammalianPromoterhuman U6Available SinceJune 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
pCMV Ub Lex VP16 (F442A) HA (P#1711)
Plasmid#14596DepositorInsertLexA
TagsHA, Ubiquitin, and VP16 (F442A)ExpressionMammalianMutationThe VP16 sequence contains an F442A mutation. The…Available SinceJune 30, 2008AvailabilityAcademic Institutions and Nonprofits only -
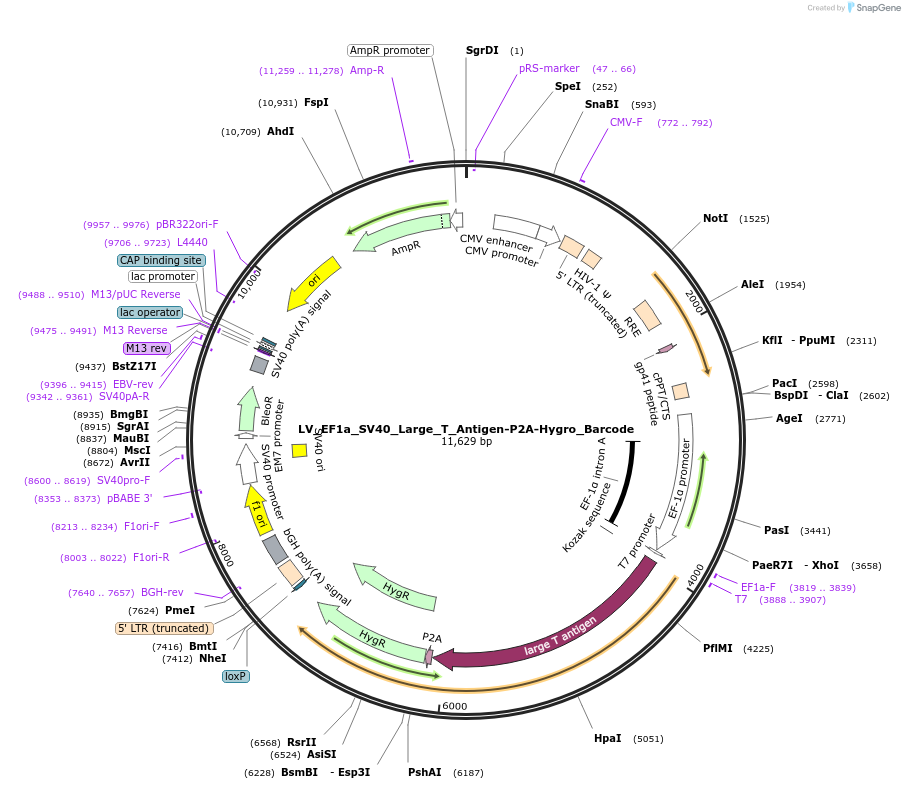
LV_EF1a_SV40_Large_T_Antigen-P2A-Hygro_Barcode
Plasmid#170255PurposeBarcoded lentiviral vector to express SV40 Large T Antigen in mammalian cells under control of EF1a promoter. Barcoded for transcript detection in single cell RNA-seqDepositorInsertSV40 Large T Antigen
UseLentiviralPromoterEF1aAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
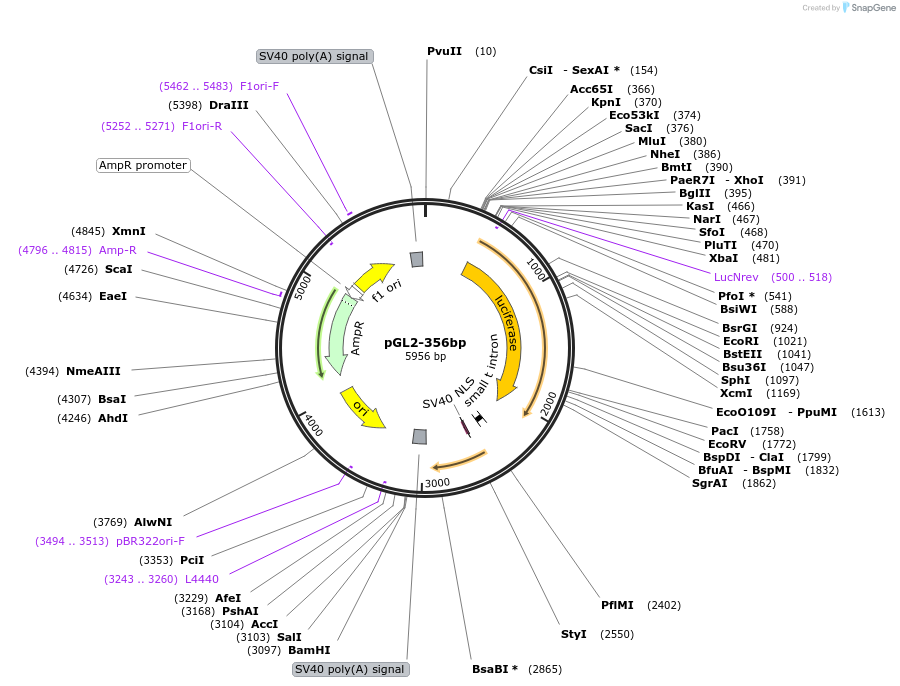
pGL2-356bp
Plasmid#16292DepositorInsertp53 promoter fragment (TP53 Human)
UseLuciferaseExpressionMammalianMutationfragment -344 to +12 relative to first transcript…Available SinceDec. 14, 2007AvailabilityAcademic Institutions and Nonprofits only -
pGL2-200bp
Plasmid#16293DepositorInsertp53 promoter fragment (TP53 Human)
UseLuciferaseExpressionMammalianMutationfragment -188 to +12 relative to first transcript…Available SinceDec. 14, 2007AvailabilityAcademic Institutions and Nonprofits only -
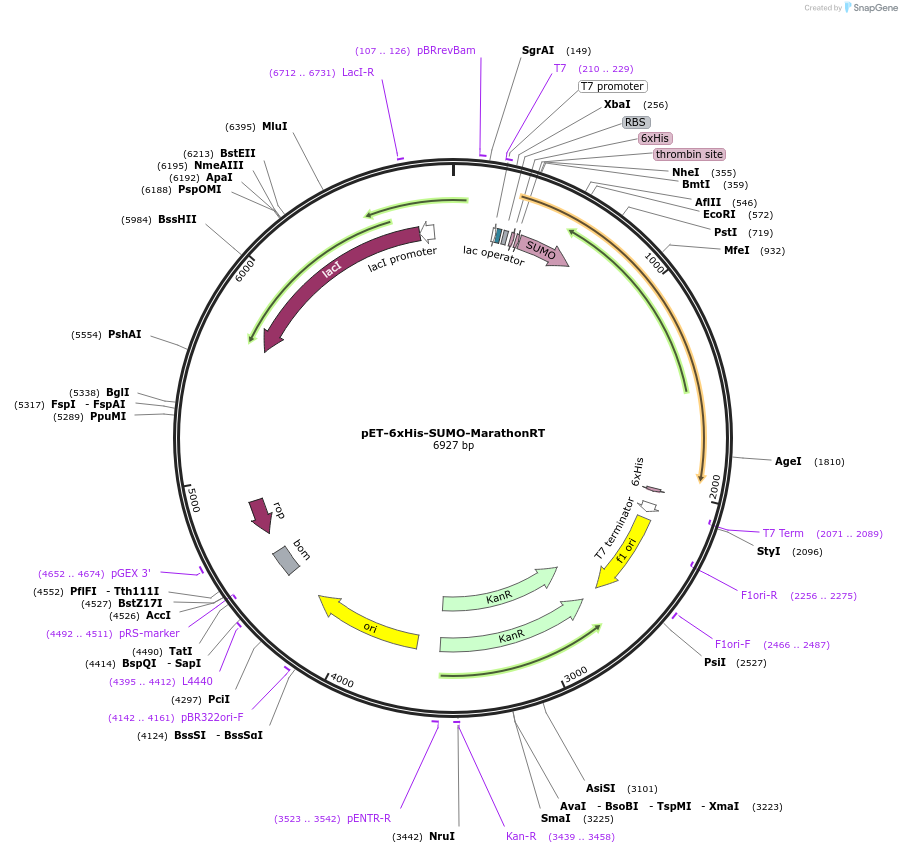
pET-6xHis-SUMO-MarathonRT
Plasmid#109029PurposeGroup II intron maturase from Eubacterium rectale (wild-type sequence). This is a highly processive reverse transcriptase that is called “MarathonRT”DepositorInsertERE_01490
Tags6xHis-SUMO tagExpressionBacterialPromoterT7Available SinceApril 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
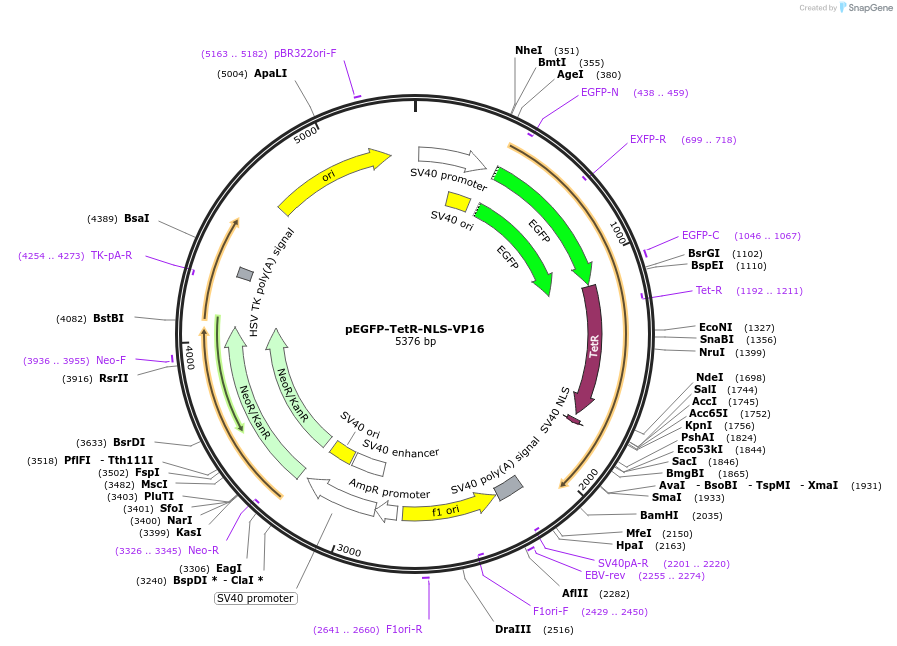
pEGFP-TetR-NLS-VP16
Plasmid#103834PurposeTranscription activator (transactivation domain of viral VP16 protein) that is constitutively recruited tetO arraysDepositorAvailable SinceDec. 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
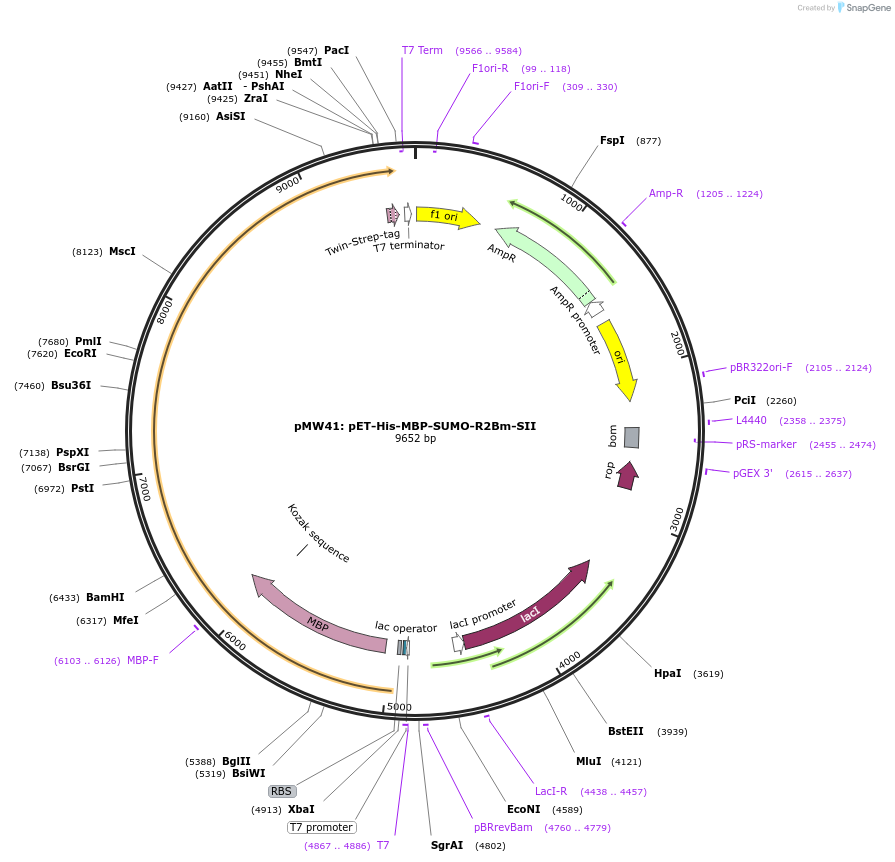
pMW41: pET-His-MBP-SUMO-R2Bm-SII
Plasmid#209290PurposeExpresses the R2Bm ORF in E. coliDepositorInsertR2
TagsHis, MBP, StrepII, and bdSUMOExpressionBacterialPromoterT7Available SinceNov. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
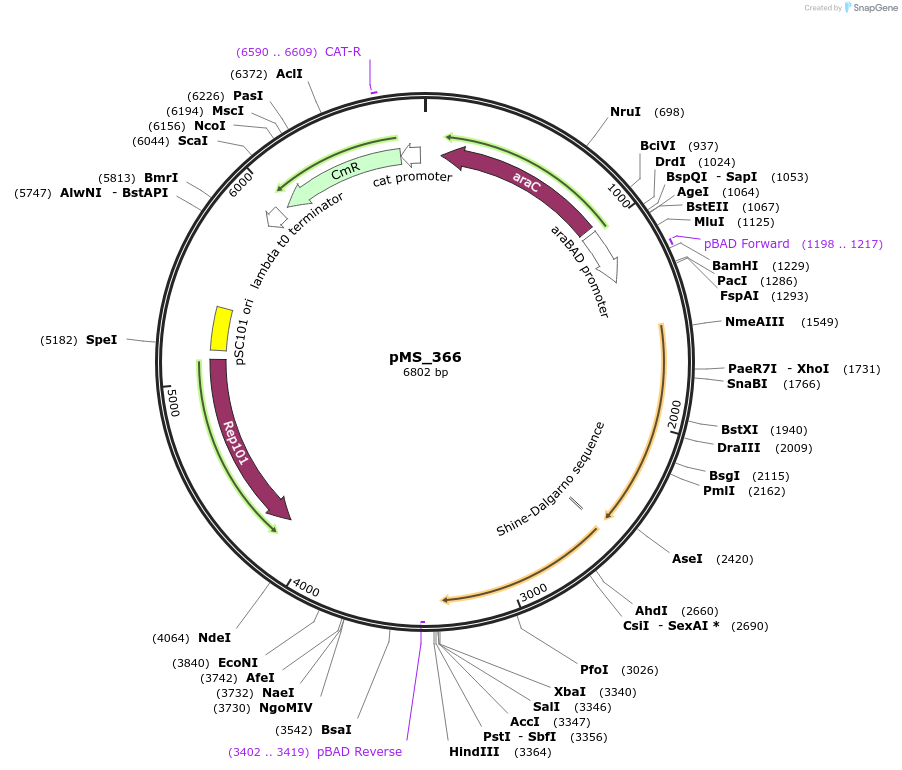
pMS_366
Plasmid#182131Purposeexpresses retron RNA, ec.86 Reverse Transcriptase, and CspRecT SSAP, to create a specific rifampicin resistance mutationDepositorInsertsretron RNA
ec.86 RT
CspRecT
Mutationincorporated rifampicin-resistance-conferring don…Available SinceSept. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
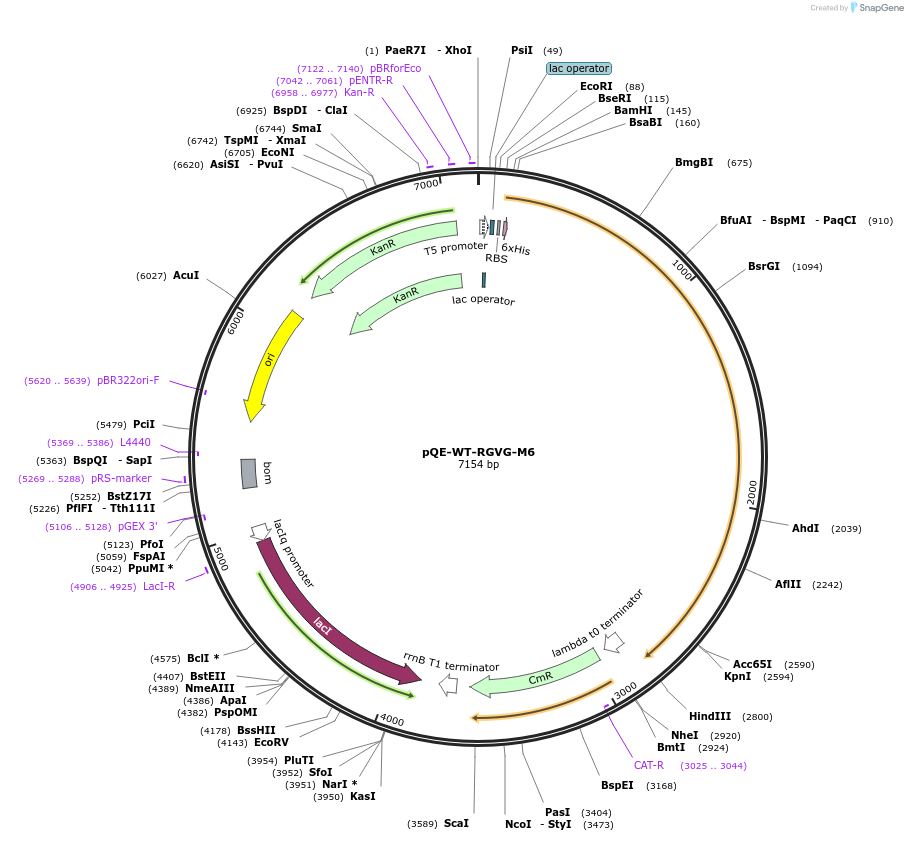
pQE-WT-RGVG-M6
Plasmid#70172PurposeExpresses RGVG-M6 T7 RNA polymerase variant with enhanced transcription of 2' modified RNADepositorInsertRGVG-M6 T7 RNA polymerase
TagsHisExpressionBacterialPromoterT5/LacAvailable SinceDec. 16, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
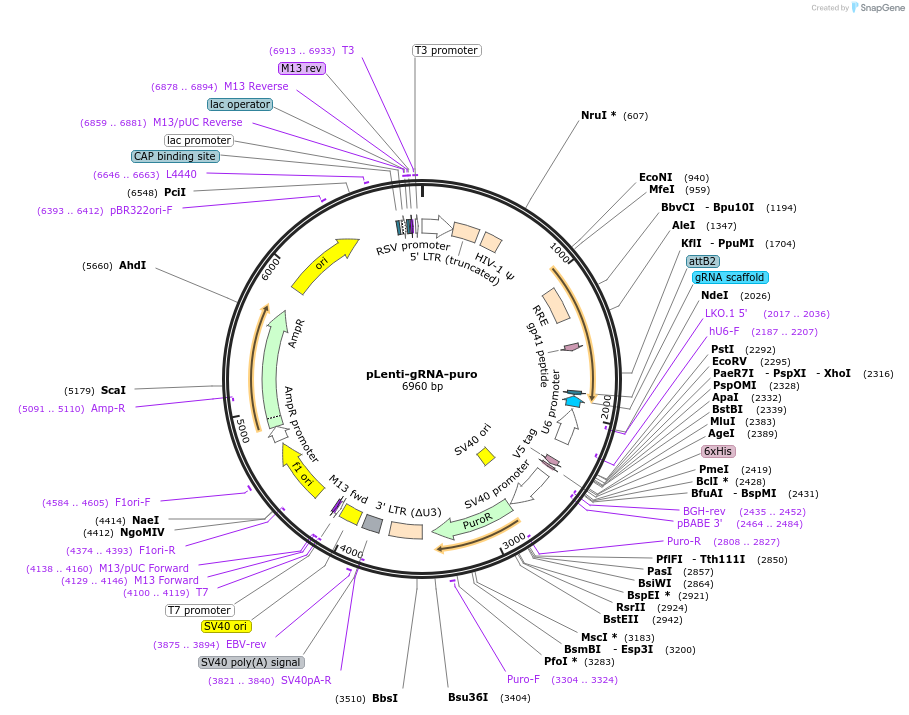
pLenti-gRNA-puro
Plasmid#180426PurposeExpresses puromycin resistance gene and scrambled gRNA for stable integration in mammalian cells; useful as control and template for new gRNAs by mutagenesis.DepositorInsertgRNAscr
UseCRISPR and LentiviralExpressionMammalianPromoterhuman U6Available SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
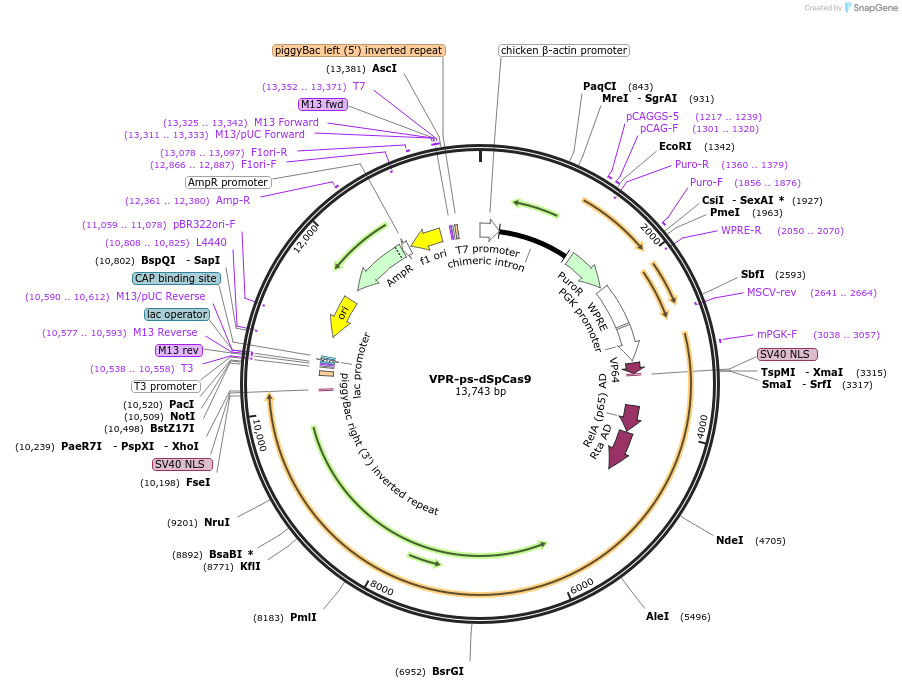
VPR-ps-dSpCas9
Plasmid#102855PurposeA single-chain light-controllable dSpCas9 with pdDronpa1.2 domainsDepositorInsertdSpCas9, pdDronpa1.2
UseCRISPR and Synthetic BiologyTagsNLS and VP64-p64-Rta transcription activatorExpressionMammalianPromoterPGKAvailable SinceNov. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
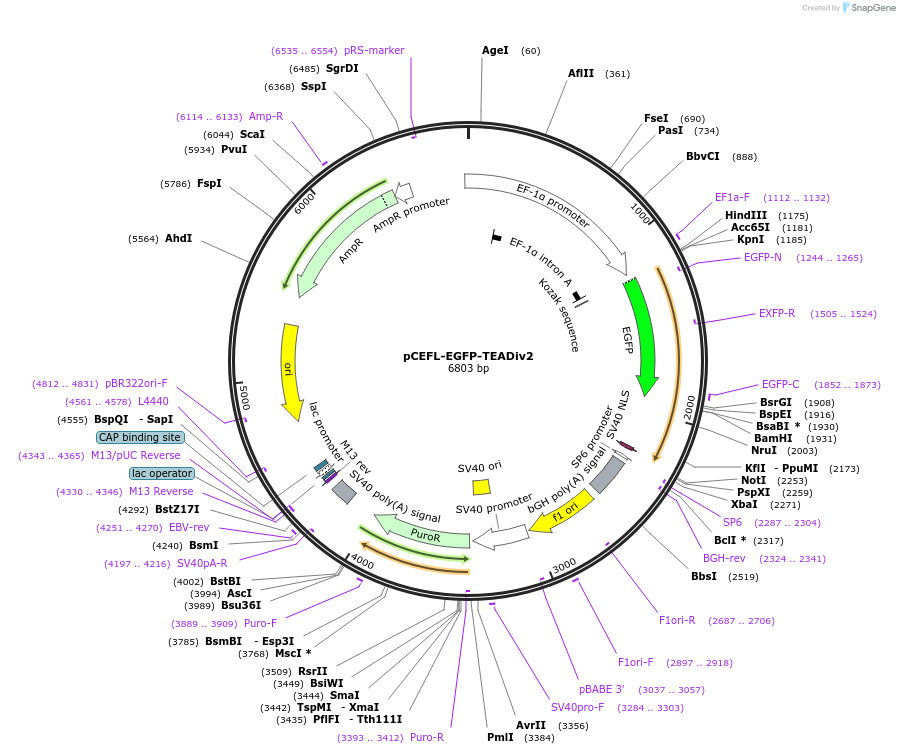
pCEFL-EGFP-TEADiv2
Plasmid#228872PurposeExpression of optimized TEAD inhibitor (TEADiv2): GFP-tagged inhibitor of the interaction of YAP1 and TAZ with TEAD transcription factors with nuclear localization signalDepositorInsertTEADiv2
TagsGFPExpressionMammalianAvailable SinceJan. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
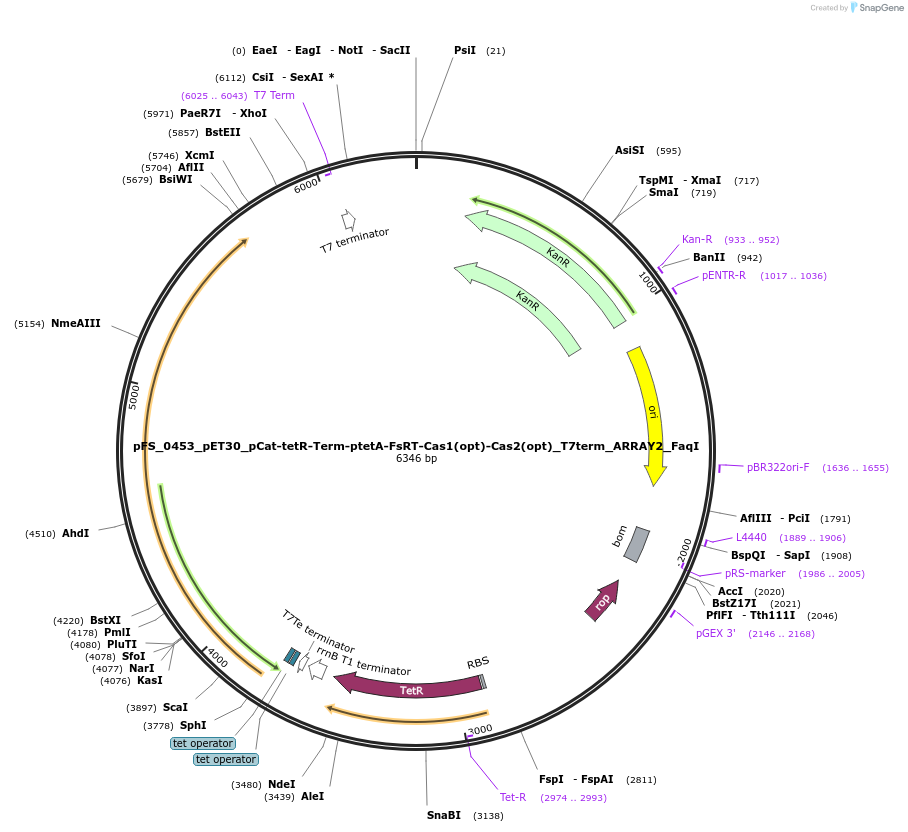
pFS_0453_pET30_pCat-tetR-Term-ptetA-FsRT-Cas1(opt)-Cas2(opt)_T7term_ARRAY2_FaqI
Plasmid#117006PurposeExpresses E. coli codon-optimized FsRT-Cas1-Cas2 under ptetA promoter, encodes FsCRISPR array 2, compatible with SENECA acquisition readoutDepositorInsertFusicatenibacter saccharivorans RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
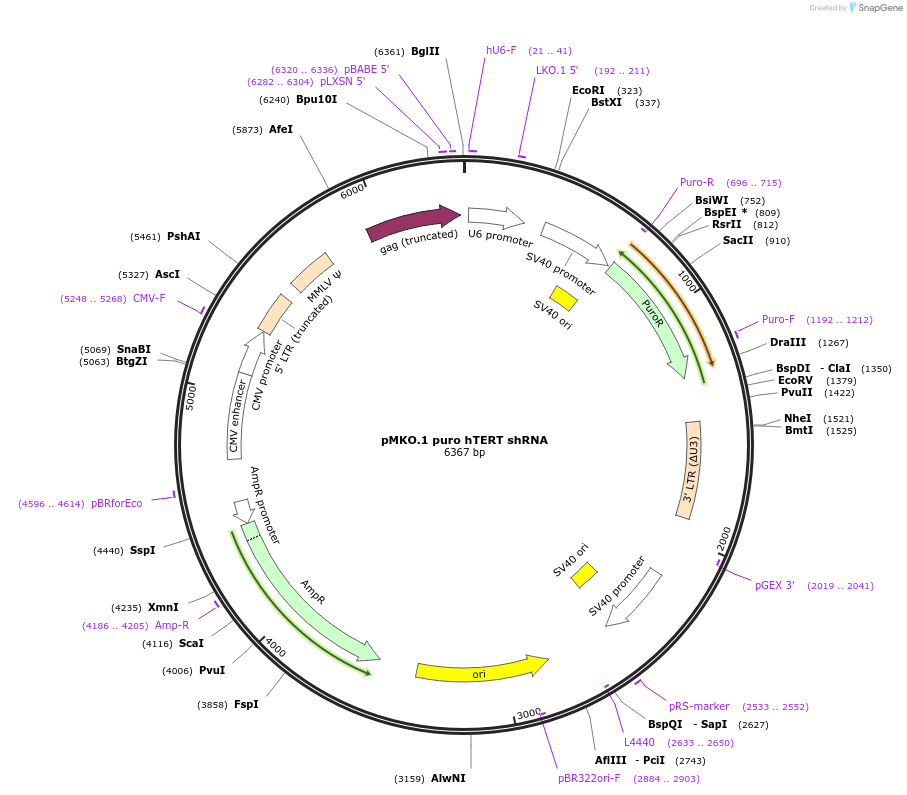
pMKO.1 puro hTERT shRNA
Plasmid#10688DepositorAvailable SinceJan. 6, 2006AvailabilityAcademic Institutions and Nonprofits only -
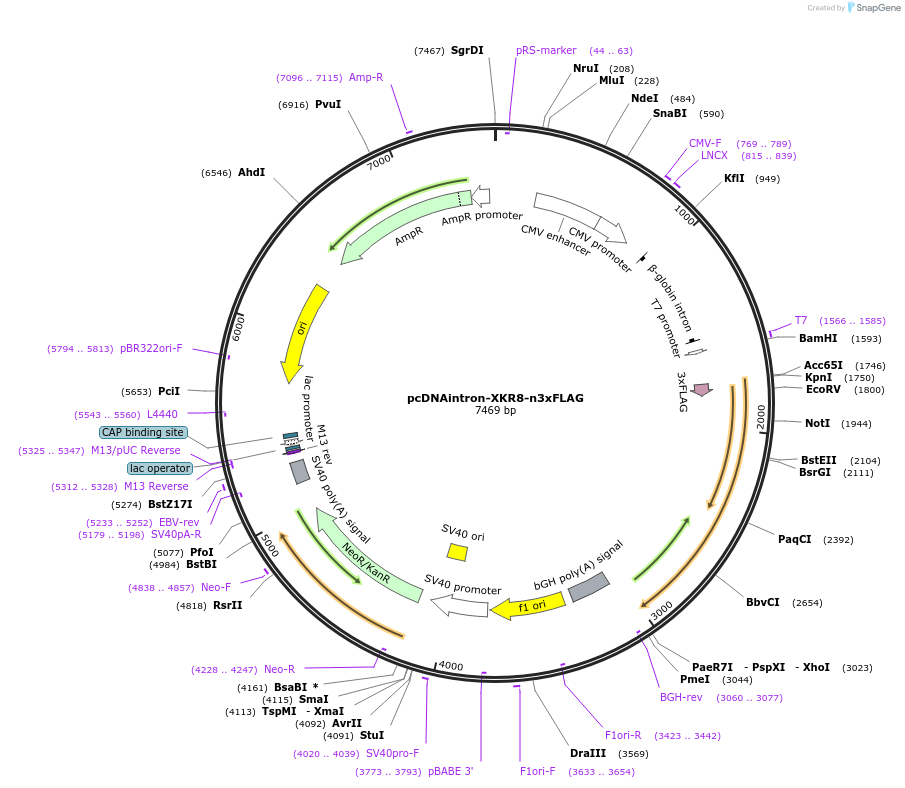
pcDNAintron-XKR8-n3xFLAG
Plasmid#215693PurposeProduces the XKR8 lipid scramblase with an n-terminal 3xFLAG tagDepositorAvailable SinceMarch 25, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
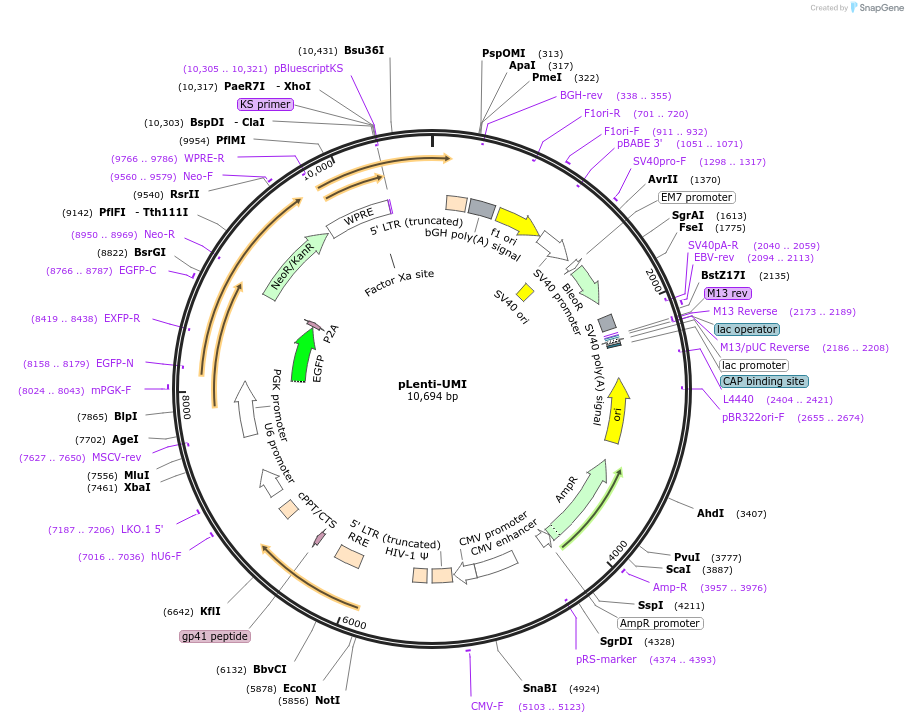
pLenti-UMI
Plasmid#222694PurposeCRISPR-screeningDepositorTypeEmpty backboneUseCRISPR and LentiviralTagsGFPExpressionMammalianMutationPGK without BsmBI cutsiteAvailable SinceAug. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
pGL2-Skp2 promoter-luciferase
Plasmid#81119PurposeReporter activity of SKP2 gene promoterDepositorInsertSKP2 (Skp2 Mouse)
UseLuciferaseTagsluciferaseMutationcontains the region from 1_kb upstream of transcr…PromoterSKP2Available SinceSept. 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
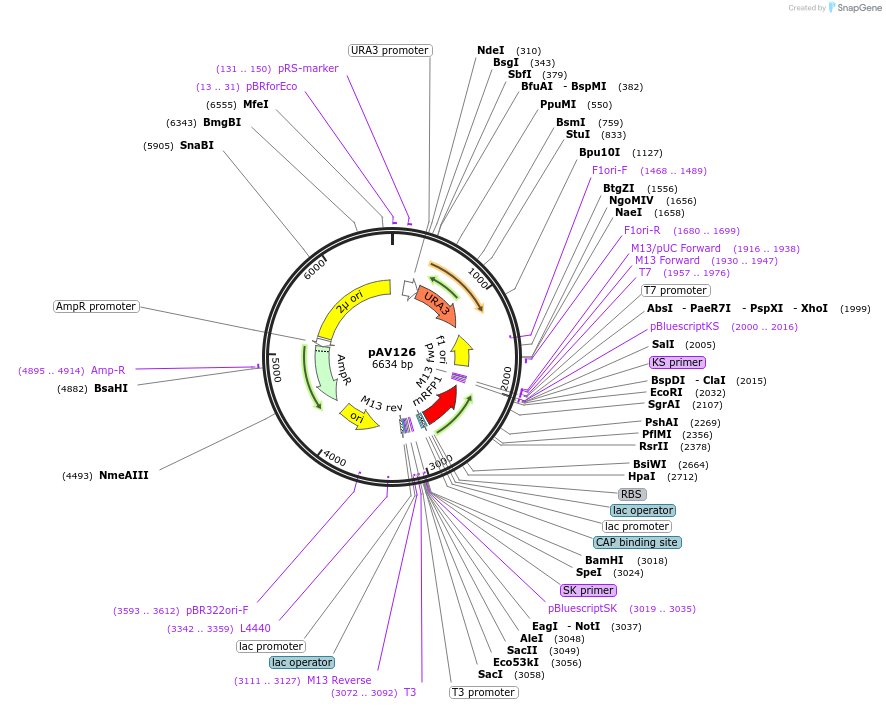
pAV126
Plasmid#63192PurposepRS426 backbone, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for transcription unit (TU) assembly using yGG.DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailable SinceAug. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
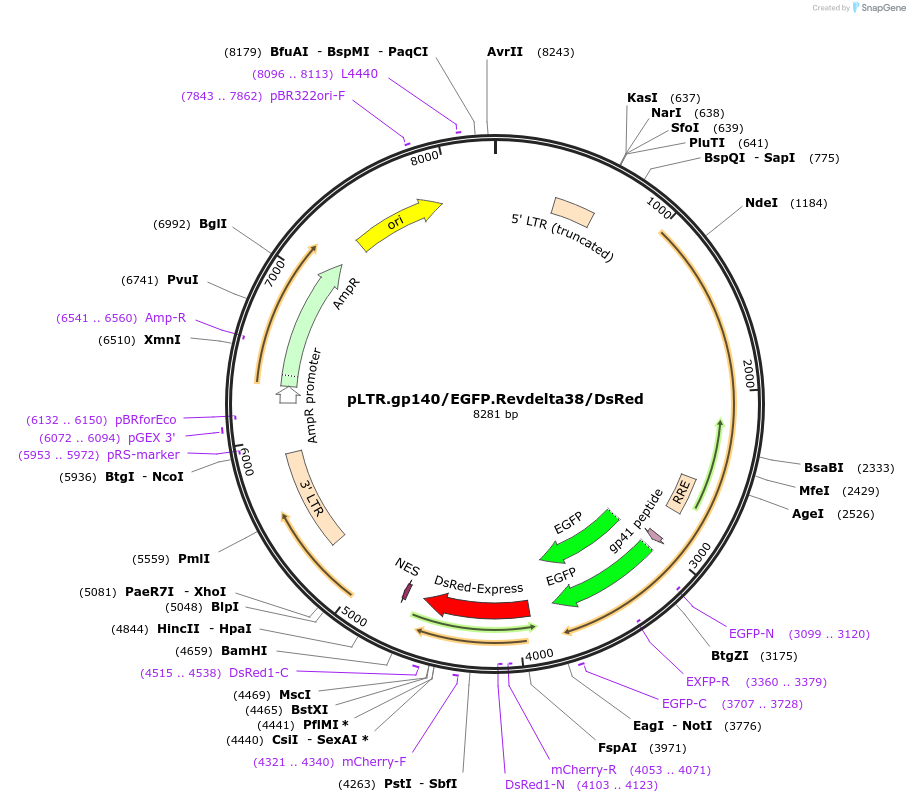
pLTR.gp140/EGFP.Revdelta38/DsRed
Plasmid#115775PurposeHIV splicing reporter allows the assessment of the effect of LRAs on HIV-1 transcription and splicing.DepositorInsertEnvelope fused to EGFP (gp140-EGFP) and non-functional Rev protein fused to DsRed fluorescent protein (Rev-delta38-DsRed).
TagsEGFP; dsReDExpressionMammalianMutationgp140 uncleaved (1190-91aa, KR > TG; truncated…PromoterHIV-1 (LTR)Available SinceOct. 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
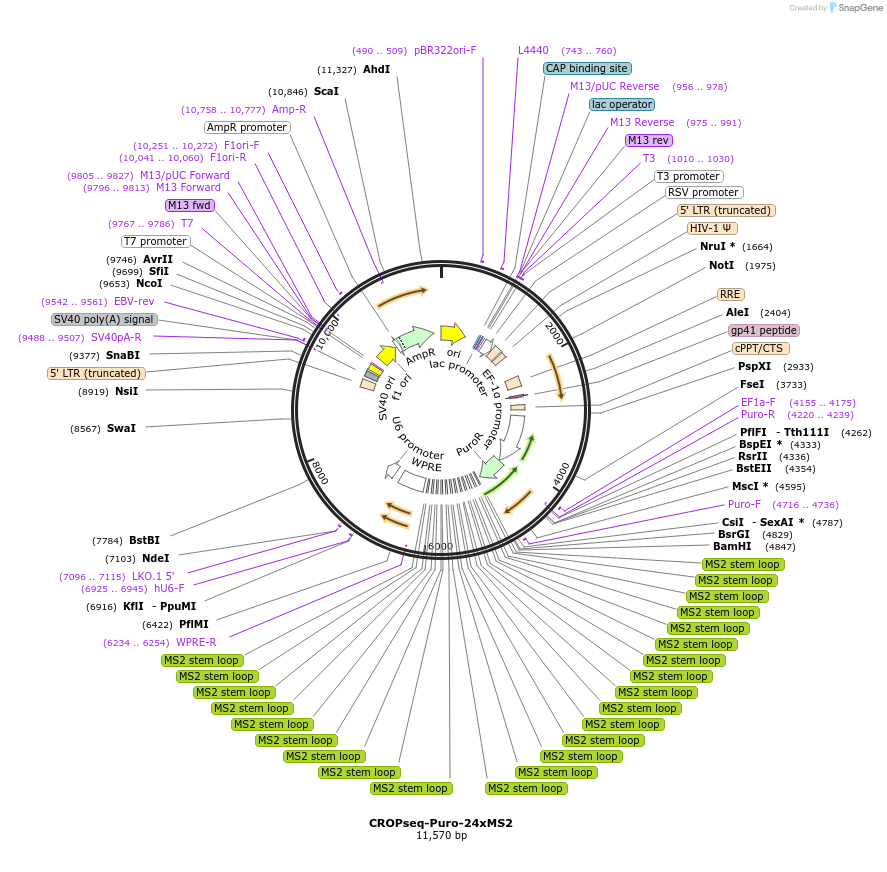
CROPseq-Puro-24xMS2
Plasmid#212629PurposeMS2 hairpin-containing expression vector and sgRNA entry plasmid for the directed export of barcode transcripts in Gag-MCP virus-like particlesDepositorInsertEF1a-PuroR-24xMS2-WPRE-hU6-sgRNA
UseCRISPR and LentiviralExpressionMammalianPromoterEF1aAvailable SinceNov. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
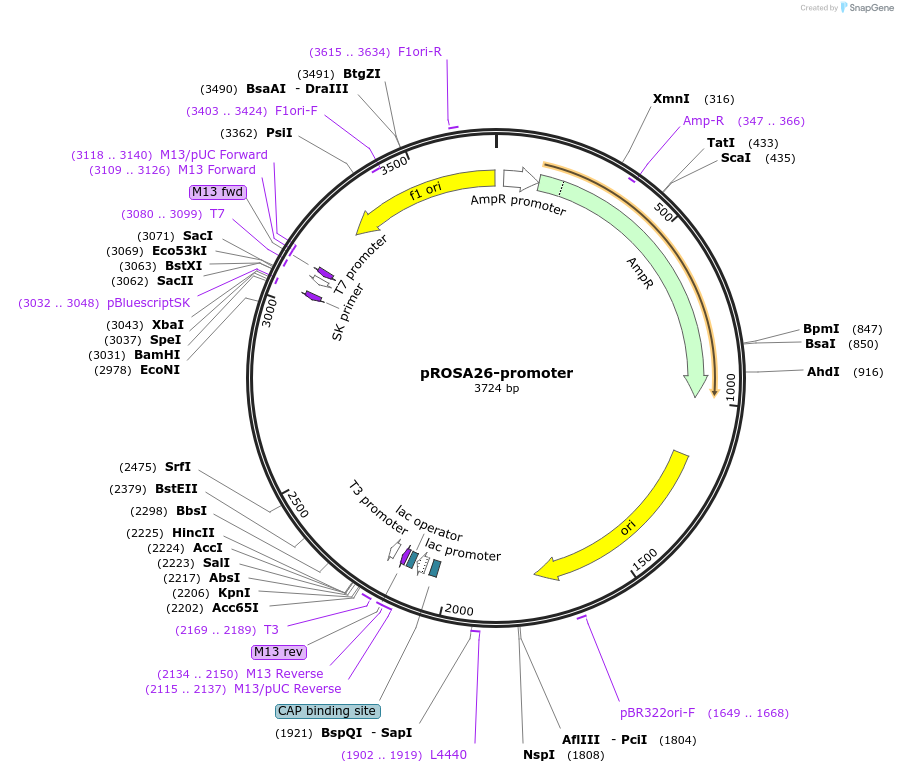
pROSA26-promoter
Plasmid#21710DepositorInsertROSA26 promoter (Gt(ROSA)26Sor Mouse)
ExpressionMammalianAvailable SinceJuly 28, 2009AvailabilityAcademic Institutions and Nonprofits only -
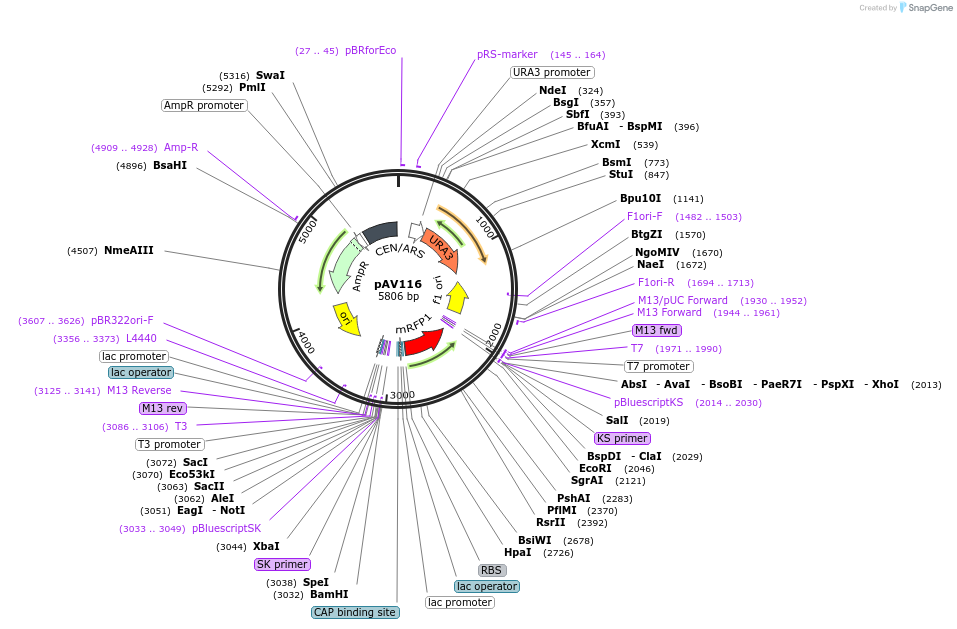
pAV116
Plasmid#63183PurposepRS416 backbone, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for transcription unit (TU) assembly using yGG.DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailable SinceSept. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
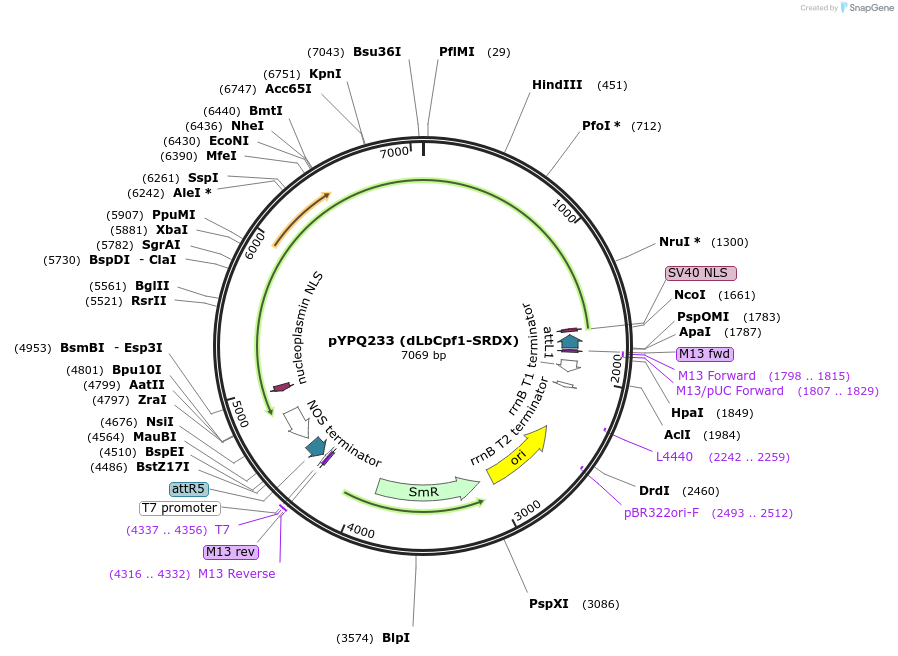
pYPQ233 (dLbCpf1-SRDX)
Plasmid#86211PurposeDeactivated LbCpf1-SRDX repressor fusion; transcriptional repressor dCpf1 Gateway entry plasmidDepositorInsertLbCpf1
UseCRISPRTags5' and 3' NLS and SRDX repressorExpressionPlantAvailable SinceApril 10, 2017AvailabilityAcademic Institutions and Nonprofits only