We narrowed to 3,439 results for: aaas
-
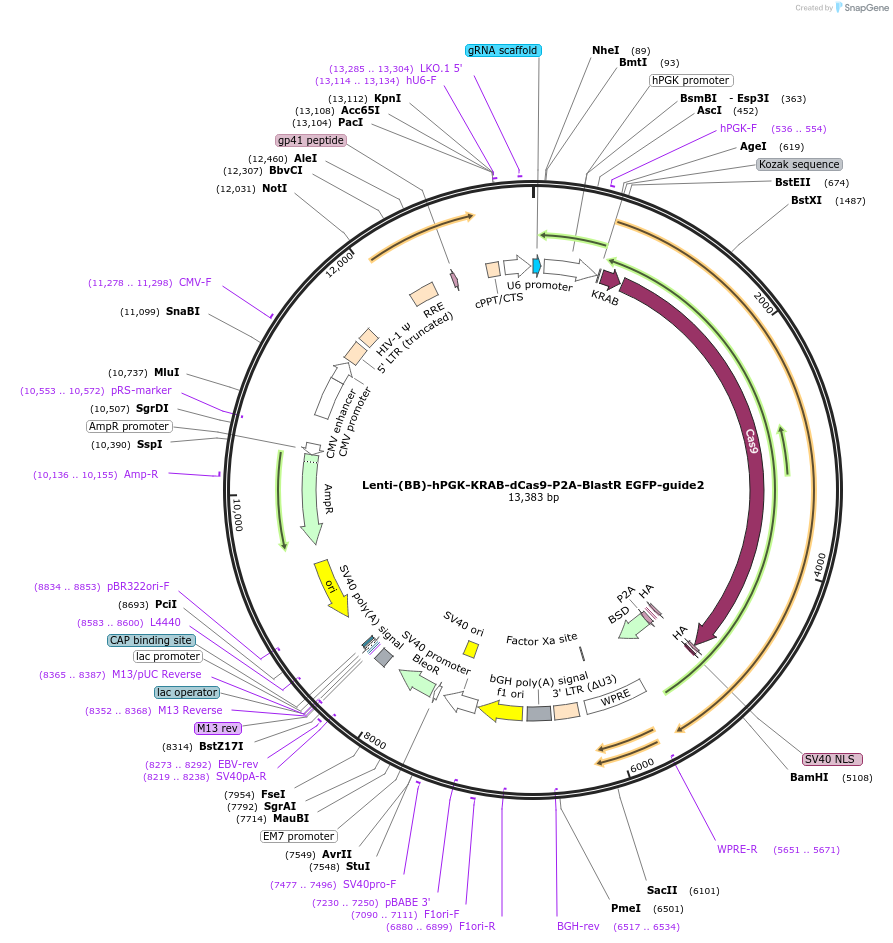
Plasmid#118163PurposeCRISPRi negative control. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter, and sgRNA2 against the EGFP CDSDepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
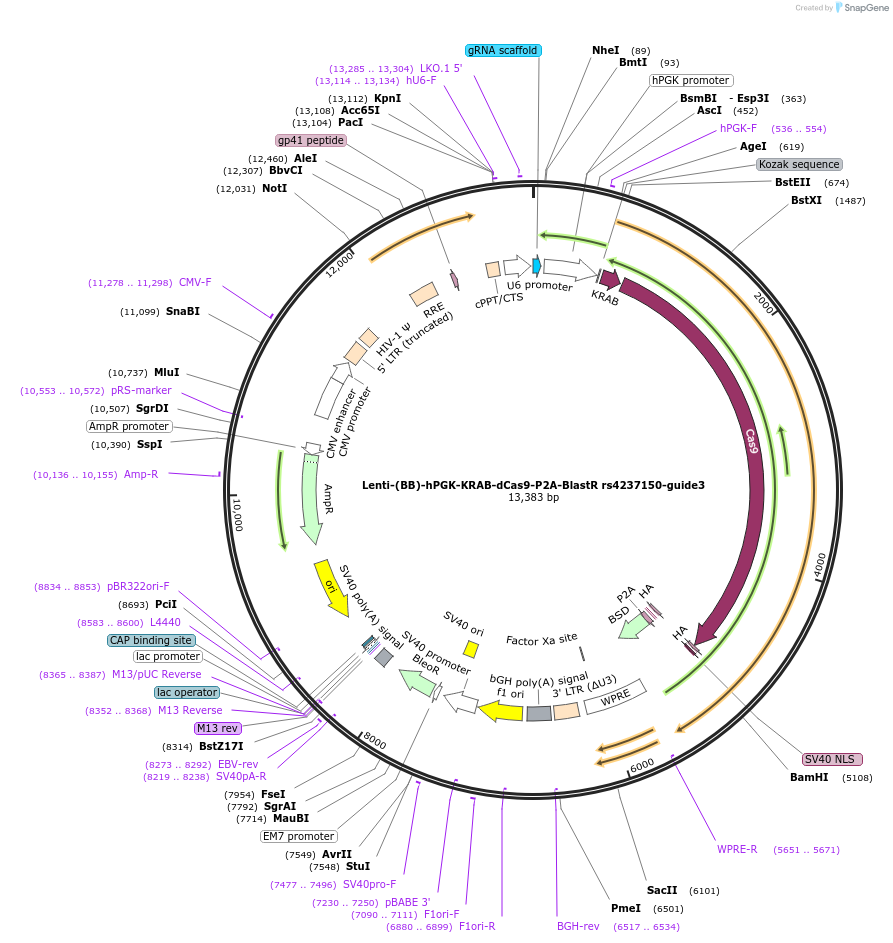
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR rs4237150-guide3
Plasmid#125398PurposeCRISPR-mediated repression of human islet enhancer containing T1D/T2D-associated variant rs4237150 (GLIS3 GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
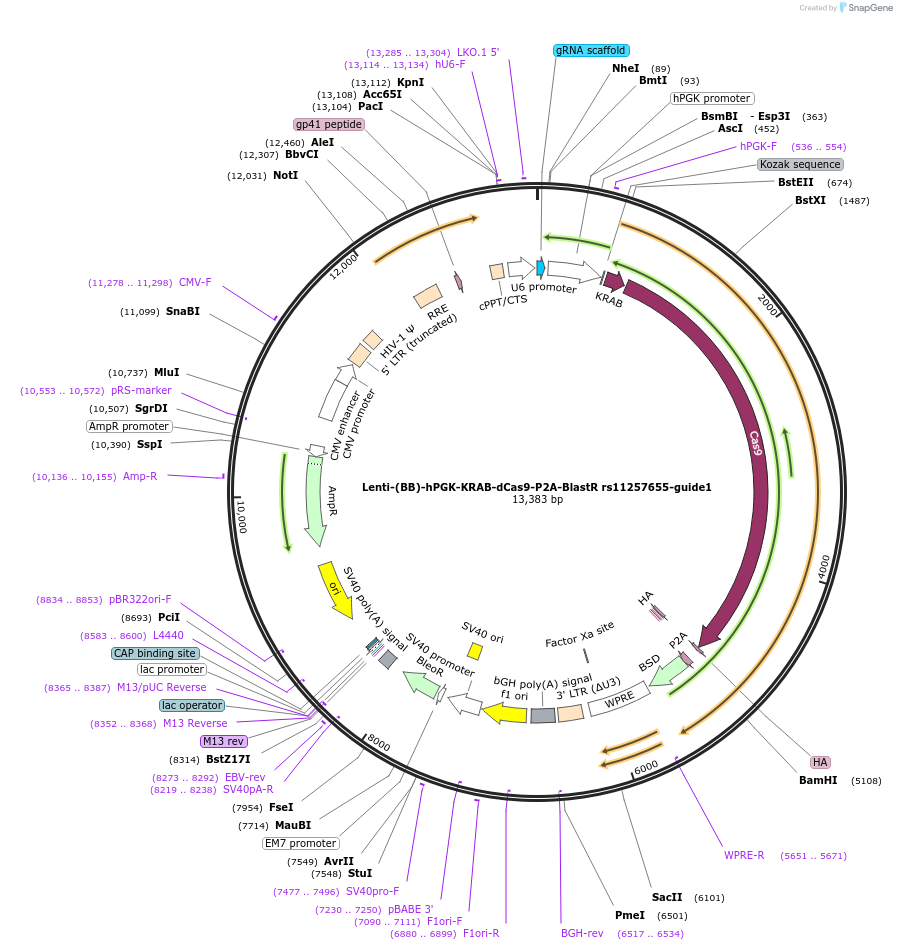
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR rs11257655-guide1
Plasmid#125393PurposeCRISPR-mediated repression of human islet enhancer containing T2D-associated variant rs11257655 (CDC123/CAMK1D GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
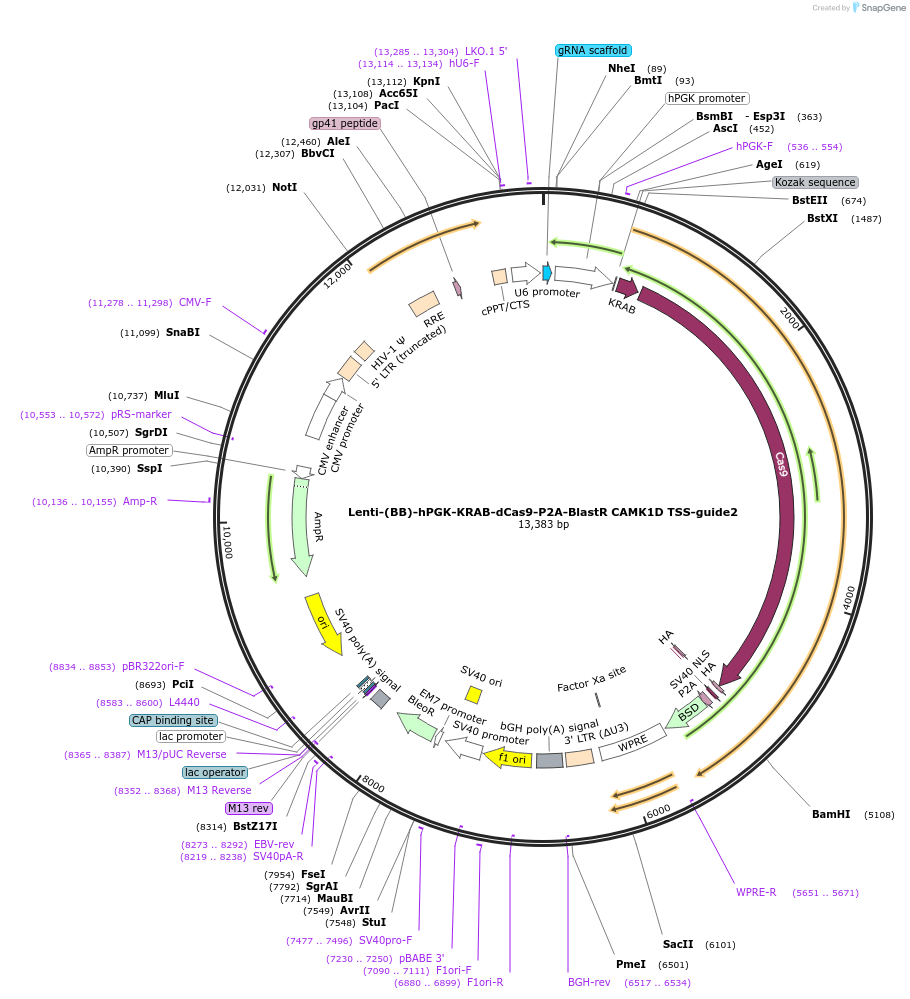
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR CAMK1D TSS-guide2
Plasmid#118178PurposeCRISPR-mediated repression of CAMK1D. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJune 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
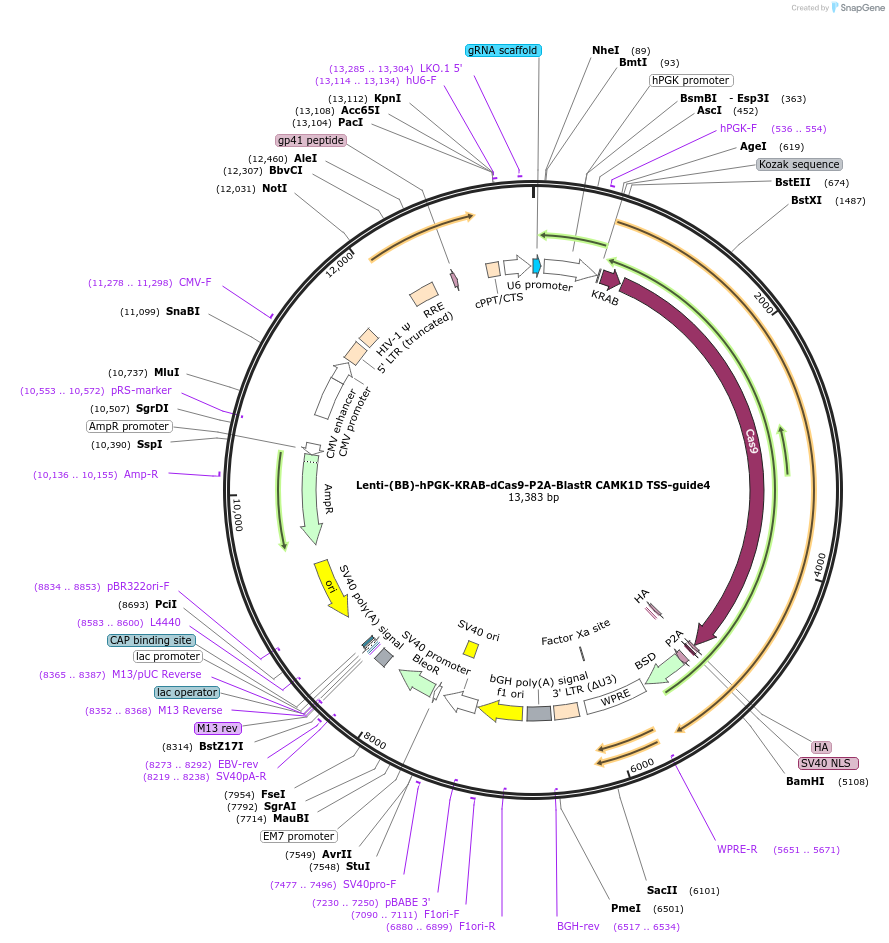
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR CAMK1D TSS-guide4
Plasmid#118180PurposeCRISPR-mediated repression of CAMK1D. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
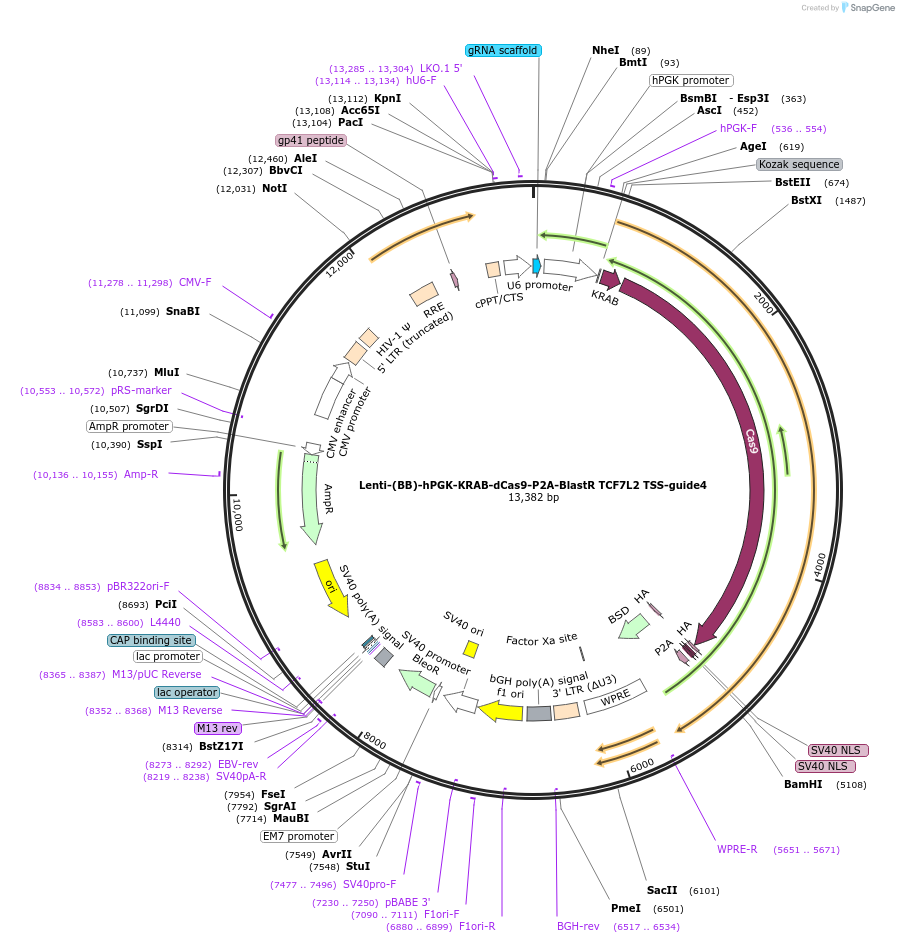
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR TCF7L2 TSS-guide4
Plasmid#118172PurposeCRISPR-mediated repression of TCF7L2. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
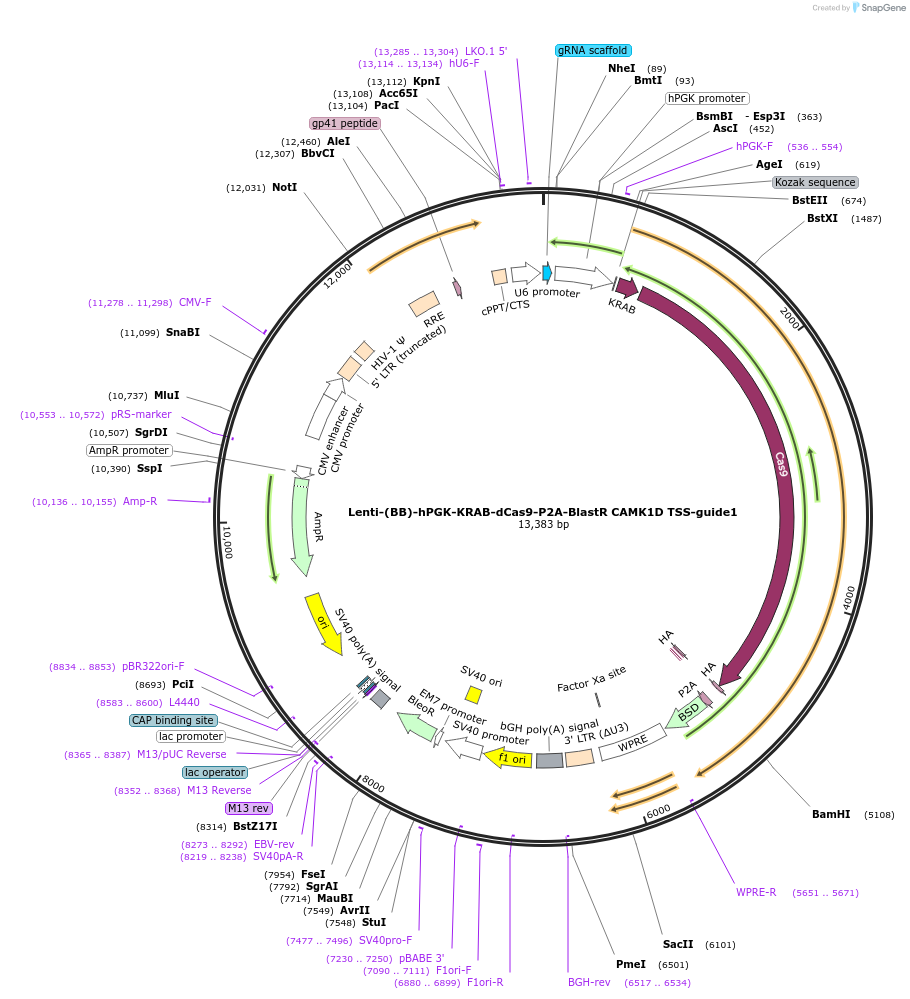
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR CAMK1D TSS-guide1
Plasmid#118177PurposeCRISPR-mediated repression of CAMK1D. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
Syn61Δ3(ev5)
Bacterial Strain#174514PurposeRecoded E. coli strain without TCG, TCA, or TAG codons and deleted serT, serU and prfA genes. Full sequence - Genbank: CP071799.1DepositorBacterial ResistanceStreptomycinAvailable SinceNov. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
eScaf
Bacterial Strain#220921PurposecssDNA production strainDepositorBacterial ResistanceNoneSpeciesEscherichia coliAvailable SinceAug. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
S4246
Bacterial Strain#156372PurposeErythromycin-resistant S2060 derivativeDepositorBacterial ResistanceStreptomycin, Tetracycline, ErythromycinAvailable SinceJan. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
Syn61
Bacterial Strain#174513PurposeRecoded E. coli strain without TCG, TCA, or TAG codons in all open reading frames. Full sequence - GenBank: CP040347.1DepositorBacterial ResistanceStreptomycinAvailable SinceNov. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
JEC027
Bacterial Strain#180310PurposeE. coli MG1655 strain with knockout of endogenous Type I-E CRISPR Cas systemDepositorBacterial ResistanceNoneAvailable SinceMay 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
TK310
Bacterial Strain#196340PurposeBacterial strain lacking adenlyate cyclase (cyaA, needed for cAMP synthesis) and phosphodiesterase (cpdA, which degrades cAMP); unable to control its intracellular cAMP levelsDepositorBacterial ResistanceNoneAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
SB39
Bacterial Strain#235121PurposeE. coli strain suitable for cloning, expression, library construction (good electroporation yield) and quantitative bacterial two-hybrid (qB2H).DepositorBacterial ResistanceNoneAvailable SinceOct. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
bSLS.114
Bacterial Strain#191530PurposeContains a knockout gene in the E. coli strain BL21-AIDepositorBacterial ResistanceNoneSpeciesEscherichia coliAvailable SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
BL21 Rosetta2 GamS
Bacterial Strain#176584PurposeE. coli BL21 strain for making cell-free extracts for Linear DNA expression. Contains pRARE2 + pBADmod1-linker2-gamS plasmids.DepositorBacterial ResistanceChloramphenicol and AmpicillinAvailable SinceDec. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
BL21 Rosetta2 ΔrecB GamS
Bacterial Strain#176585PurposeE. coli BL21 strain (with recB genomic locus deleted) for making cell-free extracts for Linear DNA expression. Contains pRARE2 + pBADmod1-linker2-gamS plasmids.DepositorBacterial ResistanceChloramphenicol and AmpicillinAvailable SinceDec. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
E. coli ER1821ΔlacI
Bacterial Strain#141407PurposeLacI-deficient E. coli host bacterium that produces β-galactosidase constitutivelyDepositorBacterial ResistanceNoneAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
CY008
Bacterial Strain#72401PurposeBacterial strain: E. coli BW25113 ΔlacI ΔaraC ΔsdiADepositorBacterial ResistanceNoneAvailable SinceMarch 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
CY007
Bacterial Strain#72337PurposeBacterial strain: E. coli BW25113 ΔlacI ΔaraC ΔsdiADepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 10, 2016AvailabilityAcademic Institutions and Nonprofits only