We narrowed to 7,924 results for: ARA-2
-
Plasmid#186360PurposeEntry clone with ORF encoding Venus fluorescent protein flanked by Gateway recombination sequencesDepositorInsertVenus yellow fluorescent protein
UseExpression of the venus fluorescent proteinAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-lambdaN-HA-DmHPat_57-499_B
Plasmid#145979PurposeInsect Expression of DmHPat_57-499DepositorInsertDmHPat_57-499 (Patr-1 Fly)
ExpressionInsectMutationtwo silent mutations compared to the sequence giv…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-EGFP-DmHPat_1-225_A
Plasmid#145870PurposeInsect Expression of DmHPat_1-225DepositorInsertDmHPat_1-225 (Patr-1 Fly)
ExpressionInsectMutationone non silent mutation N218K compared to the seq…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-lambdaN-HA-DmHPat_1-225_A
Plasmid#145875PurposeInsect Expression of DmHPat_1-225DepositorInsertDmHPat_1-225 (Patr-1 Fly)
ExpressionInsectMutationone non silent mutation N218K compared to the seq…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-lambdaN-HA-DmHPat_1-499_A
Plasmid#145876PurposeInsect Expression of DmHPat_1-499DepositorInsertDmHPat_1-499 (Patr-1 Fly)
ExpressionInsectMutationtwo silent mutations compared to the sequence giv…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-lambdaN-HA-DmXRN1_A
Plasmid#145879PurposeInsect Expression of DmXRN1DepositorInsertDmXRN1 (pcm Fly)
ExpressionInsectMutationone silent mutation compared to the sequence give…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pJJF496_dpy-10_CDS_sg6
Plasmid#164267PurposesgRNA targeting dpy-10 (CDS) in C. elegans. Backbone: pJJR50.DepositorInsertsgRNA targeting the coding sequence of dpy-10
UseCRISPRExpressionWormPromoterR07E5.16 (U6)Available SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJJF495_dpy-10_CDS_sg6
Plasmid#164268PurposesgRNA targeting dpy-10 (CDS) in C. elegans. Backbone: pJJF439.DepositorInsertsgRNA targeting the coding sequence of dpy-10
UseCRISPRExpressionWormPromoterU6 promoter from W05B2.8Available SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJJF449_dpy-10_CDS_sg1
Plasmid#163866PurposesgRNA targeting dpy-10 (CDS) in C. elegans. Backbone: pJJR50.DepositorInsertsgRNA targeting the coding sequence of dpy-10
UseCRISPRExpressionWormPromoterR07E5.16 (U6)Available SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJJF328_sqt-3_3′UTR_sg2
Plasmid#163867PurposesgRNA targeting sqt-3 (3′UTR) in C. elegans. Backbone: pJJR50.DepositorInsertsgRNA targeting the 3'UTR of sqt-3
UseCRISPRExpressionWormPromoterR07E5.16 (U6)Available SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
YSBE420:pNEO_CAC1_Vn_HOG1 terminator
Plasmid#122701PurposePlasmid for tagging proteins in Cryptococcus neoformans; N-terminal fragment of Venus proteinDepositorInsertN-terminal fragment of Venus protein
UseTagging proteins in cryptococcus neoformansAvailable SinceMay 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
YSBE418:pNAT_ACA1_Vc_HOG1 terminator
Plasmid#122700PurposePlasmid for tagging proteins in Cryptococcus neoformans; C-terminal fragment of Venus proteinDepositorInsertC-terminal fragment of Venus protein
UseTagging proteins in cryptococcus neoformansAvailable SinceApril 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
KA2896_pPB-TREtight-Tkmin-cHA-IRESH2BmKO2
Plasmid#124175PurposepiggyBac-based Dox-inducible expression (very weak)DepositorTypeEmpty backboneExpressionMammalianAvailable SinceMarch 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM535
Plasmid#84227PurposeDisturb the expression of miR535 in riceDepositorInsertmiR535
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM820
Plasmid#84228PurposeDisturb the expression of miR820 in riceDepositorInsertmiR820
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM827
Plasmid#84229PurposeDisturb the expression of miR827 in riceDepositorInsertmiR827
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM1425
Plasmid#84230PurposeDisturb the expression of miR1425 in riceDepositorInsertmiR1425
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM1430
Plasmid#84231PurposeDisturb the expression of miR1430 in riceDepositorInsertmiR1430
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM1432
Plasmid#84233PurposeDisturb the expression of miR1432 in riceDepositorInsertmiR1432
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM1861
Plasmid#84234PurposeDisturb the expression of miR1861 in riceDepositorInsertmiR1861
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM5144
Plasmid#84235PurposeDisturb the expression of miR5144 in riceDepositorInsertmiR5144
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM5338
Plasmid#84236PurposeDisturb the expression of miR5338 in riceDepositorInsertmiR5338
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM5794
Plasmid#84237PurposeDisturb the expression of miR5794 in riceDepositorInsertmiR5794
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM164
Plasmid#84196PurposeDisturb the expression of miR164 in riceDepositorInsertmiR164
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM167
Plasmid#84198PurposeDisturb the expression of miR167 in riceDepositorInsertmiR167
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM169
Plasmid#84199PurposeDisturb the expression of miR169 in riceDepositorInsertmiR169
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM171
Plasmid#84200PurposeDisturb the expression of miR171 in riceDepositorInsertmiR171
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
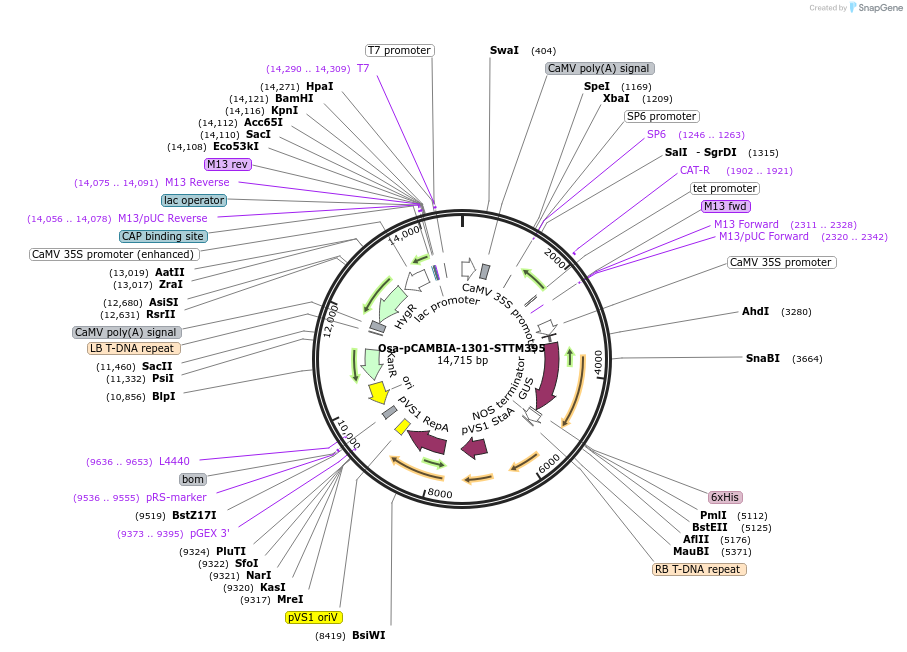
Osa-pCAMBIA-1301-STTM395
Plasmid#84205PurposeDisturb the expression of miR395 in riceDepositorInsertmiR395
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM398
Plasmid#84208PurposeDisturb the expression of miR398 in riceDepositorInsertmiR398
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
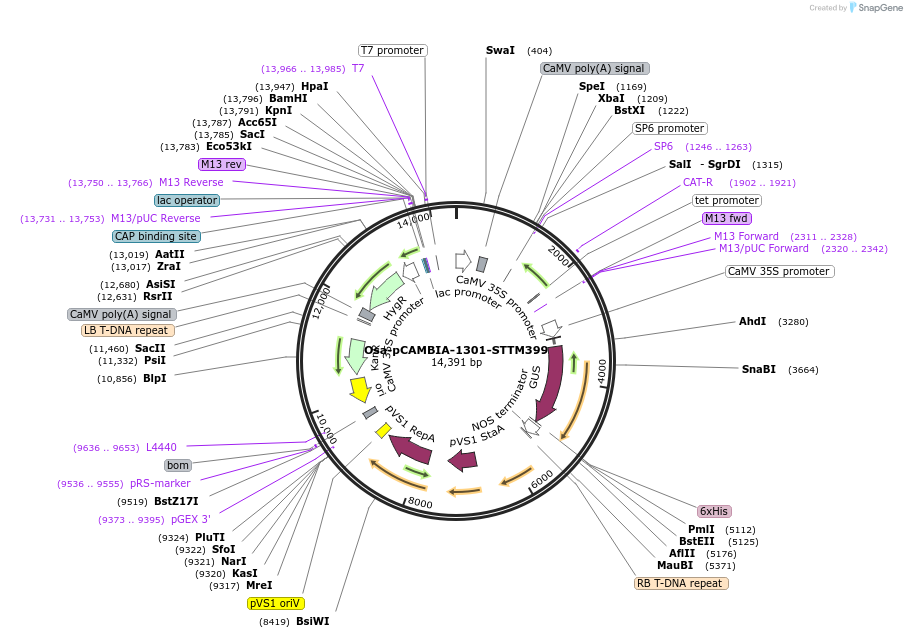
Osa-pCAMBIA-1301-STTM399
Plasmid#84216PurposeDisturb the expression of miR399 in riceDepositorInsertmiR399
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
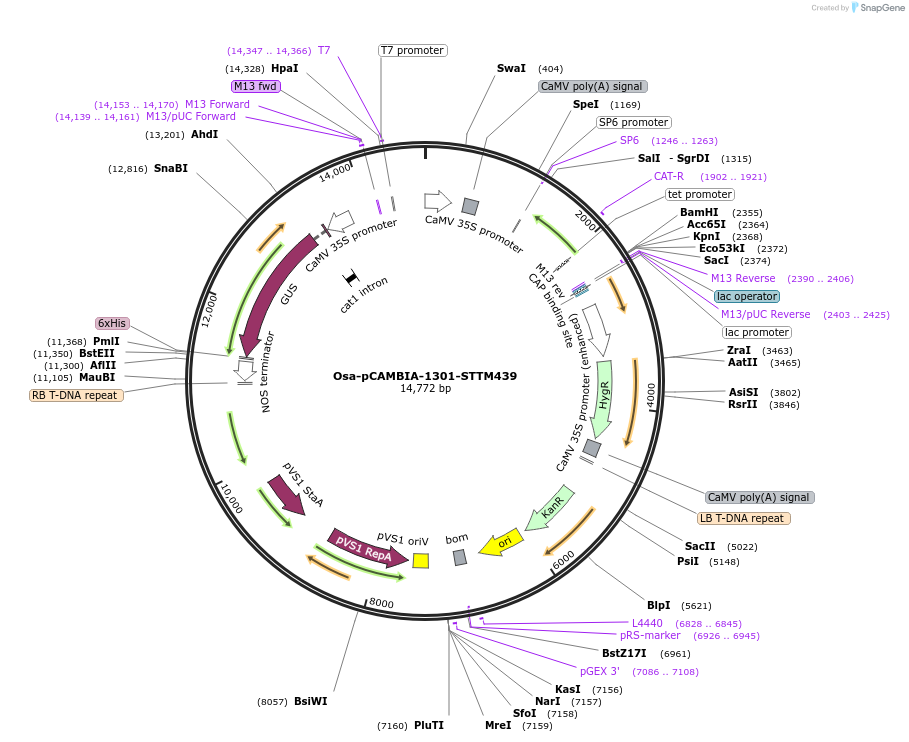
Osa-pCAMBIA-1301-STTM439
Plasmid#84222PurposeDisturb the expression of miR439 in riceDepositorInsertmiR439
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM444
Plasmid#84223PurposeDisturb the expression of miR444 in riceDepositorInsertmiR444
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
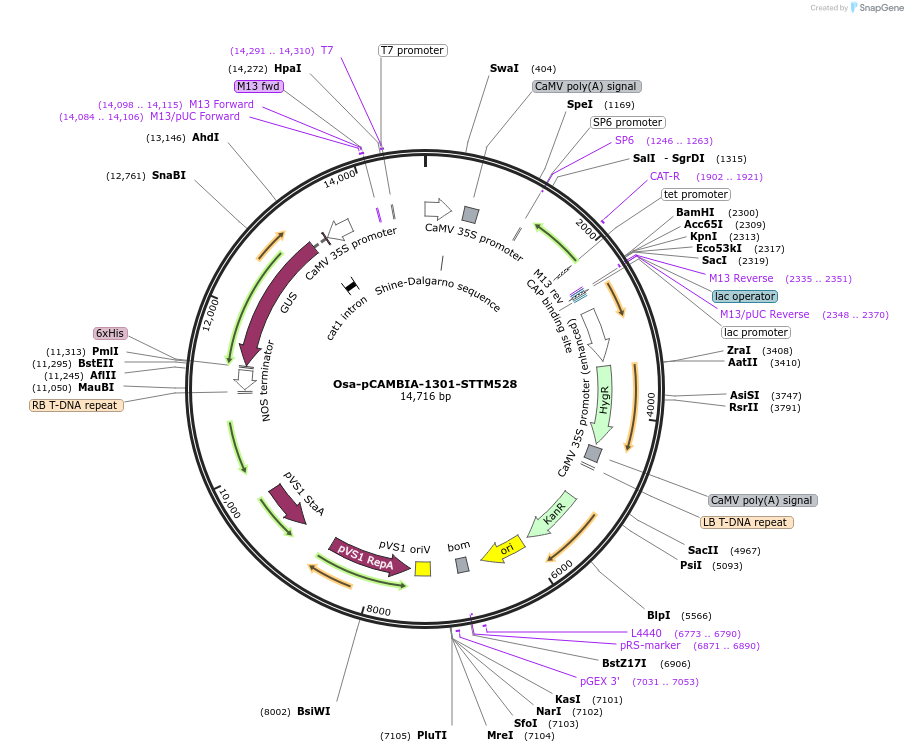
Osa-pCAMBIA-1301-STTM528
Plasmid#84224PurposeDisturb the expression of miR528 in riceDepositorInsertmiR528
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM529
Plasmid#84225PurposeDisturb the expression of miR529 in riceDepositorInsertmiR529
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Gma-pCAMBIA-1301-STTM396
Plasmid#84081PurposeDisturb the expression of miR396 in soybeanDepositorInsertmiR396
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Gma-pCAMBIA-1301-STTM408
Plasmid#84082PurposeDisturb the expression of miR408 in soybeanDepositorInsertmiR408
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
Osa-pCAMBIA-1301-STTM397
Plasmid#84207PurposeDisturb the expression of miR397 in riceDepositorInsertmiR397
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
pVH004
Plasmid#80401PurposeContains pBAD-CusR and pCusR-antiscaffold with no fluorescent protein. Used as a control for autofluorescence.DepositorUseSynthetic BiologyTagsC-terminal LZx domainExpressionBacterialAvailable SinceOct. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
COVID-SARS2 NSP14/10
Plasmid#159613PurposeBacterial co-expression vector fo Covid-SARS2 NSP14 and NSP10DepositorInsertsTagsHis6, TEV cleavage site and noneExpressionBacterialMutationCodon-optimized for E. coli expressionPromoter(Bicistronic) and T7 - LacOAvailable SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
pGL4.10/RSV_SF3B3-miniXon_Luciferase
Plasmid#174660PurposePlasmid containing the SF3B3-miniXon/Firefly luciferase cassette. Translation of Firefly luciferase is controlled by splicing modulation of the SF3B3-miniXon by LMI070.DepositorInsertminiXon cassette
UseLuciferaseExpressionMammalianMutationKozak ATG initiation sequence in middle exonPromoterRSVAvailable SinceJan. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
pGL4.10/RSV_SF3B3-Xon_Luciferase
Plasmid#174659PurposePlasmid containing the SF3B3-Xon/Firefly luciferase cassette. Translation of Firefly luciferase is controlled by splicing modulation of the SF3B3-xon by LMI070.DepositorInsertXon cassette
UseLuciferaseExpressionMammalianMutationKozak ATG initiation sequence in middle exonPromoterRSVAvailable SinceJan. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
pETduet+p300HAT
Plasmid#157793PurposeExpression of p300 HAT domainDepositorAvailable SinceAug. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
hBACCS2-IRES-mCherry
Plasmid#72892PurposeMammalian expression of hBACCS2, a human blue light-activated Ca2+ channel switch, and mCherryDepositorInserthBACCS2-IRES-mCherry
TagsIRES-mCherryExpressionMammalianPromoterCMVAvailable SinceJan. 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
hBACCS2-IRES-GFP
Plasmid#72891PurposeMammalian expression of hBACCS2, a human blue light-activated Ca2+ channel switch, and GFPDepositorInserthBACCS2-IRES-GFP
TagsIRES-GFPExpressionMammalianPromoterCMVAvailable SinceJan. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-GNPTAB
Plasmid#78108PurposeMammalian expression of human GNPTAB with C terminal myc tagDepositorAvailable SinceMay 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pUTKan-3xHA-GFP-TOM5
Plasmid#130674PurposeUbiquitously expresses 3xHA-GFP-TOM5 in the outer mitochondrial membrane of plantaDepositorInsertTranslocase of outer mitochondrial membrane 5 (TOM5 Mustard Weed)
Tags3xHA and sGFPExpressionPlantPromoterUbiquitin10Available SinceSept. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-CAG-sbGLuc-hGtARC2-EYFP
Plasmid#114111Purposefusion protein of Gaussia luciferase variant slow burn, Guillardia theta anion channelrhodopsin-2, and enhanced yellow fluorescent protein for bioluminescent optogeneticsDepositorInsertGaussia luciferase, slow burn; Guillardia theta anion channelrhodopsin-2, human codon optimized
ExpressionMammalianPromoterCAGAvailable SinceAug. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPHR-iRFP713
Plasmid#103818PurposeSoluble BLInCR mock effector that is recruited to 'localizer' sites upon blue light illuminationDepositorAvailable SinceMay 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
dmBACCS2VL-IRES-dOrai
Plasmid#67629PurposeMammalian expression of dmBACCS2VL, a modified Drosophila melanogaster blue light-activated Ca2+ channel switch, and dOraiDepositorInsertdmBACCS2VL-IRES-dOrai
ExpressionMammalianPromoterCMVAvailable SinceAug. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
pET30a-SRAP-delCys2
Plasmid#84568PurposeEncodes murine SRAP domain (250aa) of Hmces/Srap1 with deletion of catalytic Cys2DepositorInsertSrap1 (Hmces Mouse)
Tags6xHis and S-TagExpressionBacterialMutationdeletion of Cys2PromoterT7Available SinceNov. 18, 2016AvailabilityAcademic Institutions and Nonprofits only