We narrowed to 13,064 results for: LIC
-
Plasmid#14065DepositorInsertRat Fibronectin 7iBi89 (Fn1 Rat)
ExpressionMammalianMutationInsert was cut out of pGEM3 with SalI and SacI an…Available SinceMarch 2, 2007AvailabilityAcademic Institutions and Nonprofits only -
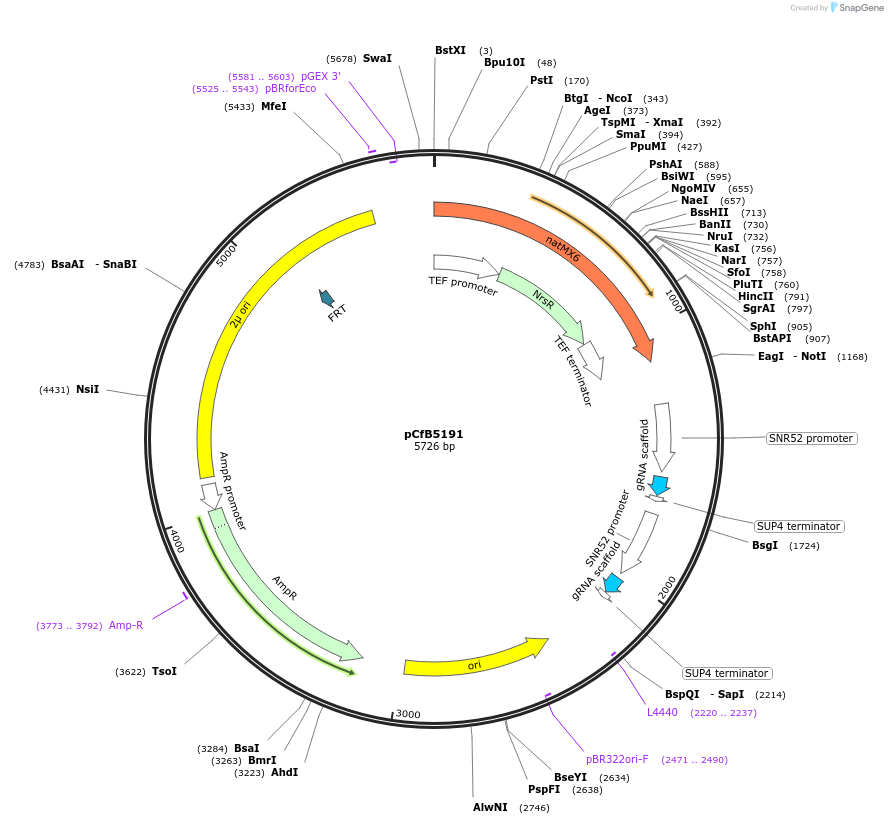
pCfB5191
Plasmid#126911PurposePlasmid containing gRNA expression cassettes targeting the X-4 and XII-5 integration sites in Saccharomyces cerevisiaeDepositorInsertX-4 and XII-5 gRNA
UseCRISPRAvailable SinceDec. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
Cav1-mRed
Plasmid#12681DepositorAvailable SinceOct. 16, 2006AvailabilityAcademic Institutions and Nonprofits only -
pTrc99-Rat FABP2
Plasmid#13578DepositorInsertintestinal fatty acid binding protein (Fabp2 Rat)
ExpressionBacterialAvailable SinceMarch 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
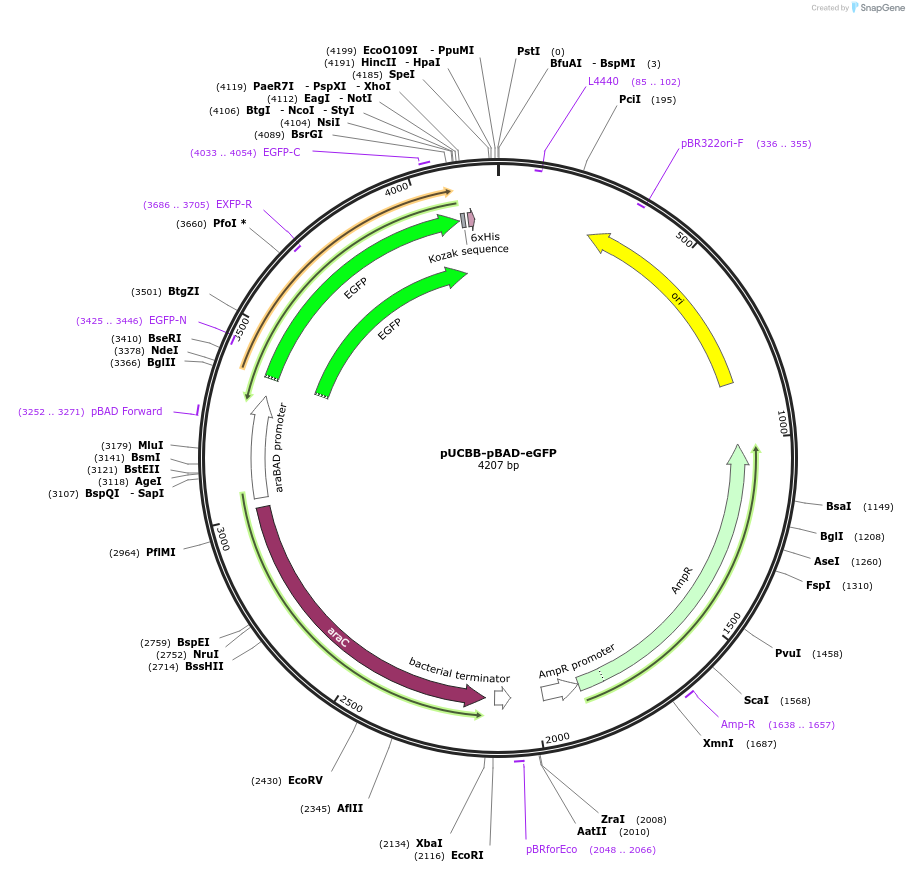
pUCBB-pBAD-eGFP
Plasmid#32553DepositorAvailable SinceOct. 18, 2011AvailabilityAcademic Institutions and Nonprofits only -
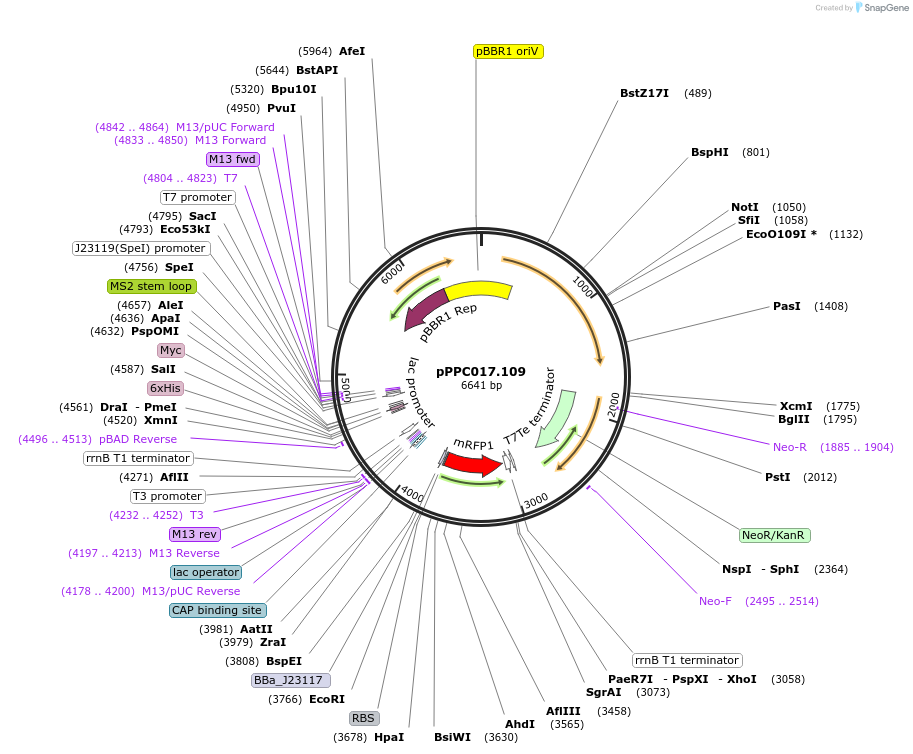
pPPC017.109
Plasmid#171145PurposeExpression of J1-BBa_J23117-mRFP and J109 scRNA on pBBR1-KmR plasmidDepositorInsertsJ109 scRNA
J1-BBa_J23117-mRFP
UseCRISPRExpressionBacterialPromoterBBa_J23119 and J1-BBa_J23117Available SinceOct. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
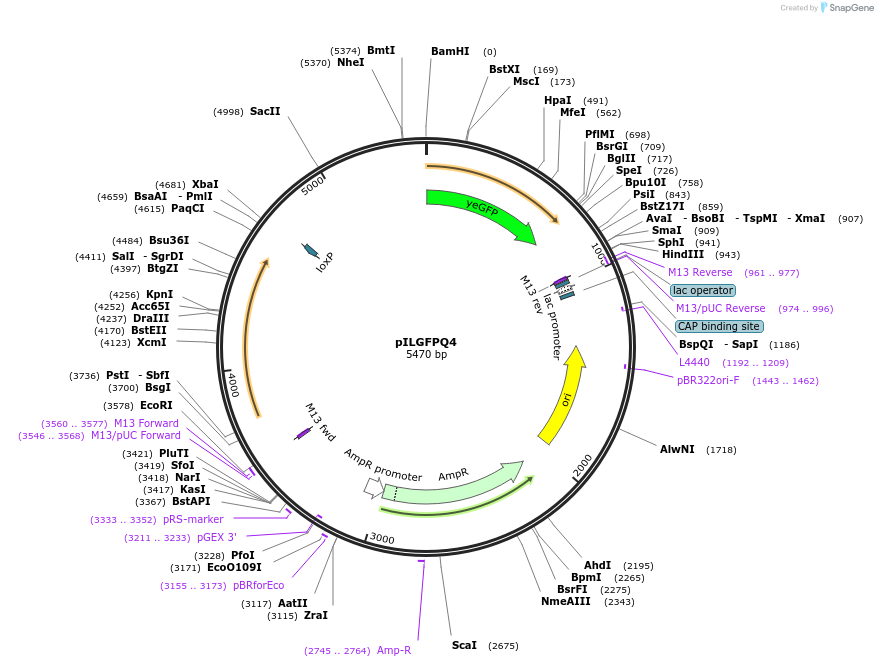
pILGFPQ4
Plasmid#83559PurposeUsed to evaluate the expression output of GAL7 promoter with yEGFP used as the reporter gene.DepositorInsertGAL7 promoter-yEGFP
ExpressionYeastPromoterGAL7Available SinceDec. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
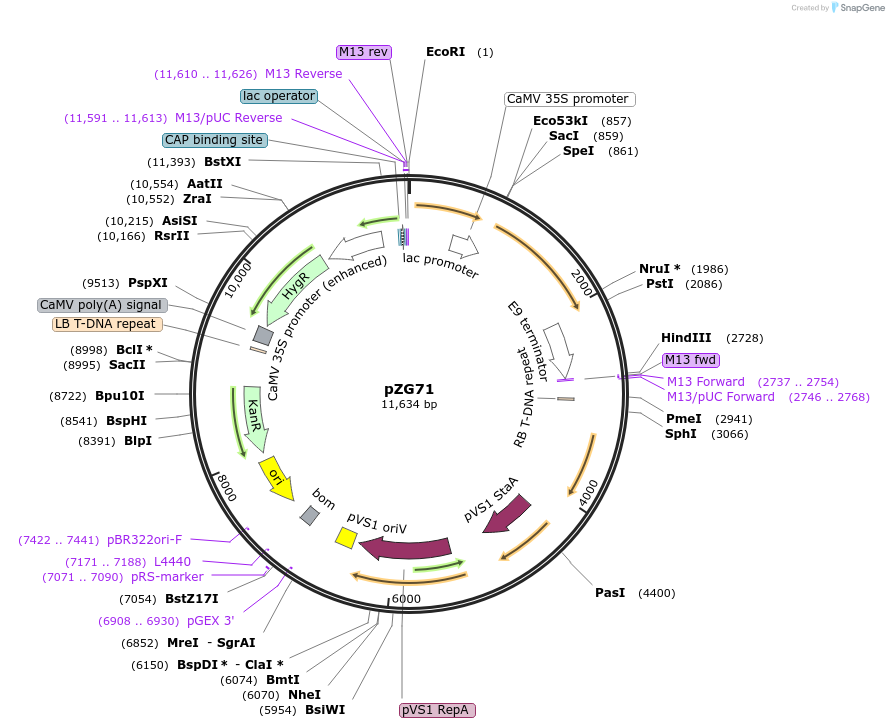
pZG71
Plasmid#202577PurposeRNA editing in plants. Plasmid contains 35S-dADARcd-LIC1-ccdB-LIC2-dADARcd-35S-hptIIDepositorInsertHyperADARcd
ExpressionPlantMutationE488QAvailable SinceDec. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
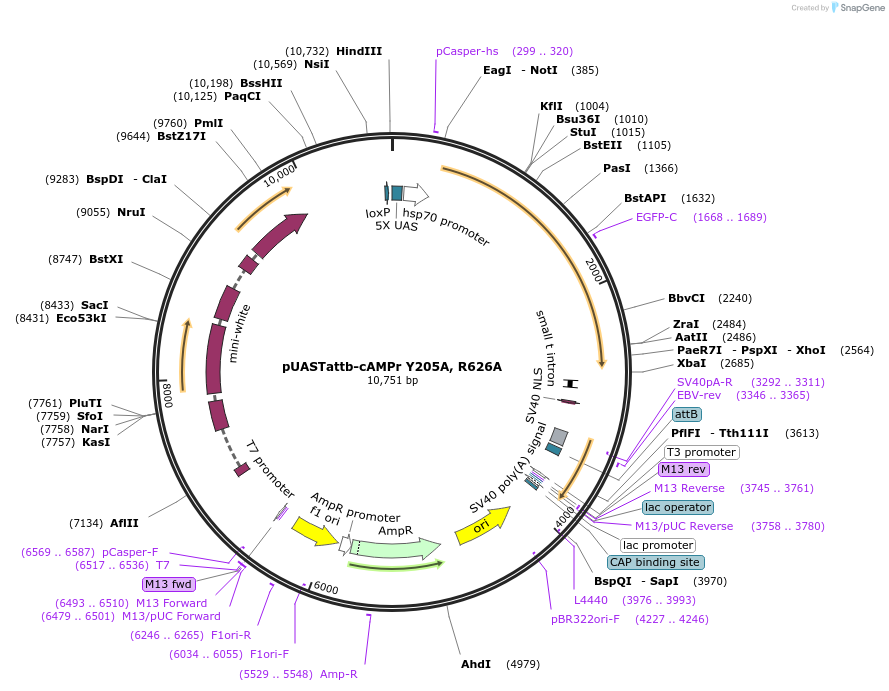
pUASTattb-cAMPr Y205A, R626A
Plasmid#84932PurposecAMPr under 5x UAS regulation. pUASTTB for phiC site mediated insertion. Y205A mutation inhibits PKA-C kinase activity. R626A should decrease affinity for cAMP without affecting specifity.DepositorInsertcAMPr Y205A R626A
ExpressionInsectMutationPKA-C Y205A, PKA-R R626APromoter5X UASAvailable SinceMarch 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
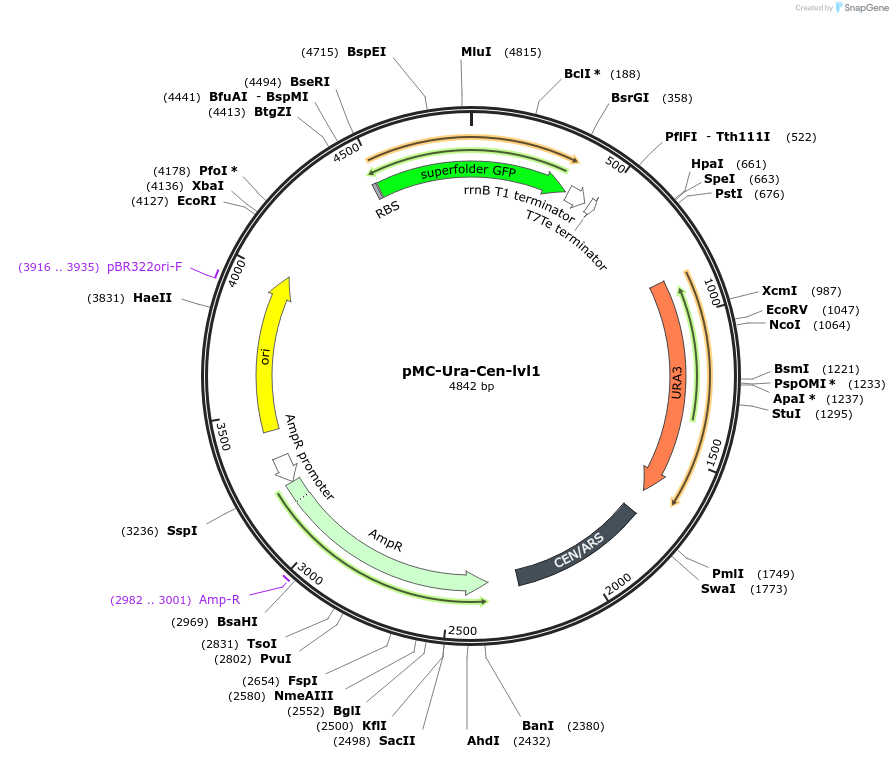
pMC-Ura-Cen-lvl1
Plasmid#176182Purposepreassembled yeast MoClo level-1 backbone with GFP dropout and URA3 markerDepositorTypeEmpty backboneExpressionYeastAvailable SinceFeb. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
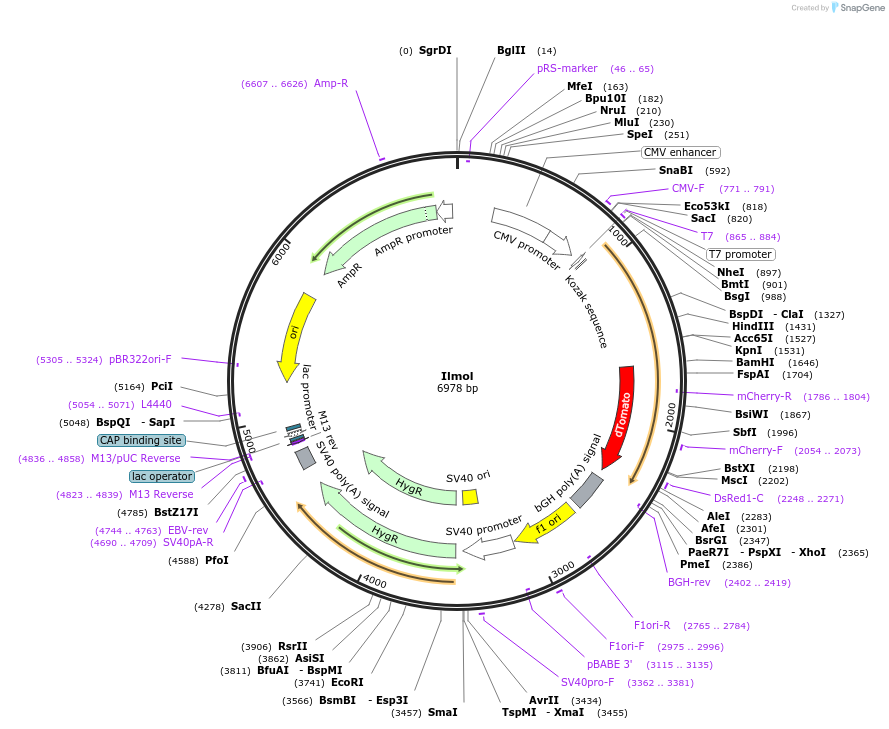
Ilmol
Plasmid#120310PurposeA red-shifted GEVI consists of triple mutant ciona based voltage sensing domain with optimized linker and dTomato K159R.DepositorInsertIlmol
ExpressionMammalianPromoterCMVAvailable SinceMay 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
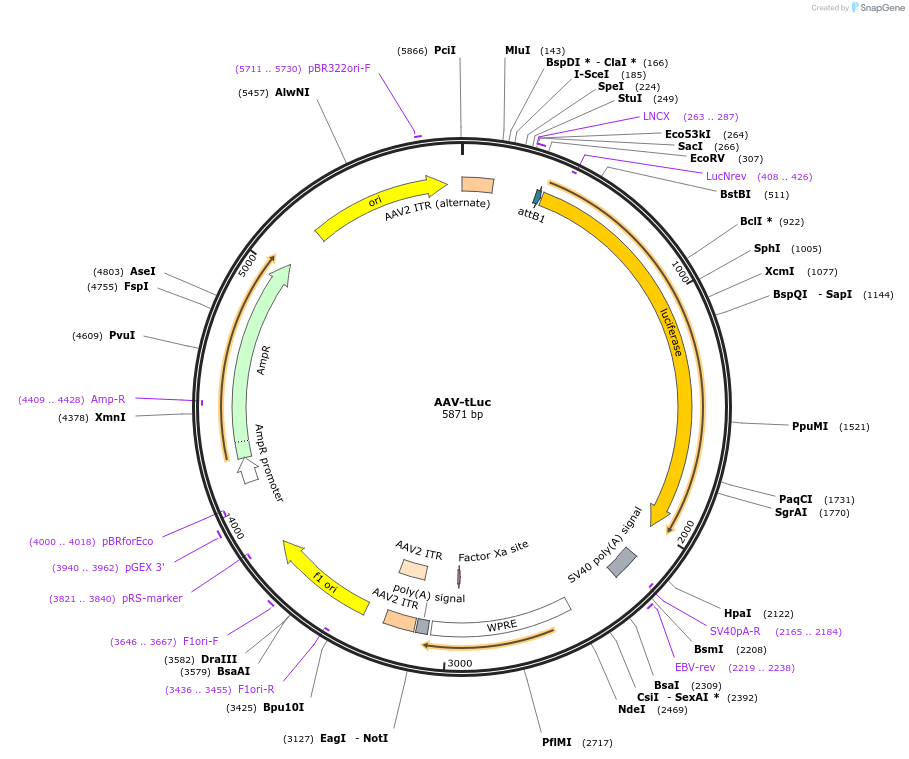
AAV-tLuc
Plasmid#107552PurposetLuc in AAV backboneDepositorInsertLuciferase
UseAAVAvailable SinceJuly 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
mTFP1-VSVG-16
Plasmid#55517PurposeLocalization: Golgi/ER Transport, Excitation: 462, Emission: 492DepositorInsertVSVG
TagsmTFP1ExpressionMammalianMutationdeltaK VSVG ts045PromoterCMVAvailable SinceOct. 2, 2014AvailabilityAcademic Institutions and Nonprofits only -
pcDNA-NES-ZapCmR1.1
Plasmid#59014PurposeCytosolic zinc-binding FRET biosensorDepositorInsertNES-ZapCmR1.1 genetically encoded cytosolic Zn(II) sensor
TagsNuclear Exclusion SignalExpressionMammalianMutationSee commentPromoterCMVAvailable SinceAug. 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
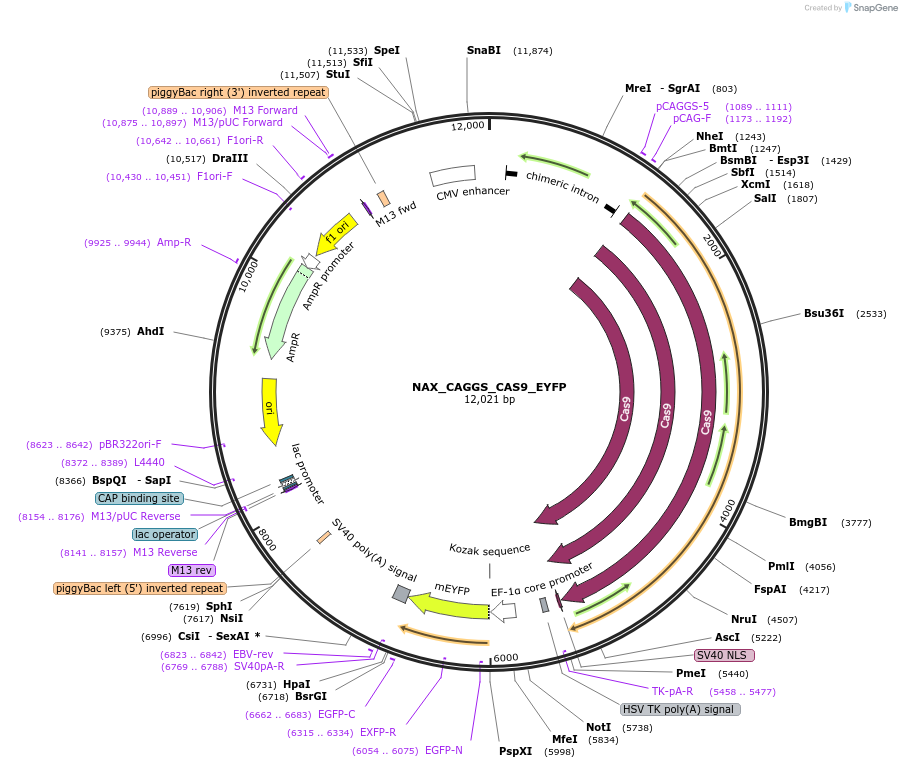
NAX_CAGGS_CAS9_EYFP
Plasmid#167858PurposePiggyBac compatible plasmid expressing spCas9DepositorInsertCas9
UseCRISPRAvailable SinceFeb. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
HGXPRT
Plasmid#64201PurposeExpression of Plasmodium falciparum HGXPRTDepositorInsertHypoxanthine guanine xanthine phosphoribosyltransferase
TagsHistidine tagExpressionBacterialAvailable SinceApril 29, 2015AvailabilityAcademic Institutions and Nonprofits only -
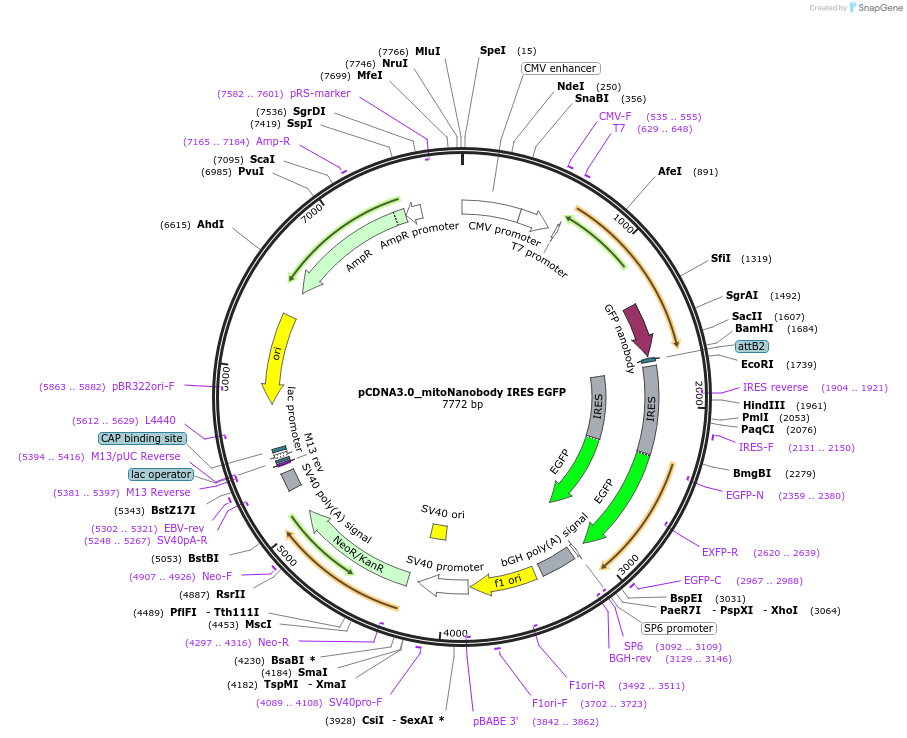
pCDNA3.0_mitoNanobody IRES EGFP
Plasmid#136621PurposePlasmid encoding outer mitochondrial localization of the Enhancer nanobody for GFP as a SNAP-Tag-fusion and EGFPDepositorInsertNTOM20-SNAP-Tag-EnhancerNb (IRES EGFP)
ExpressionMammalianPromoterCMVAvailable SinceFeb. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
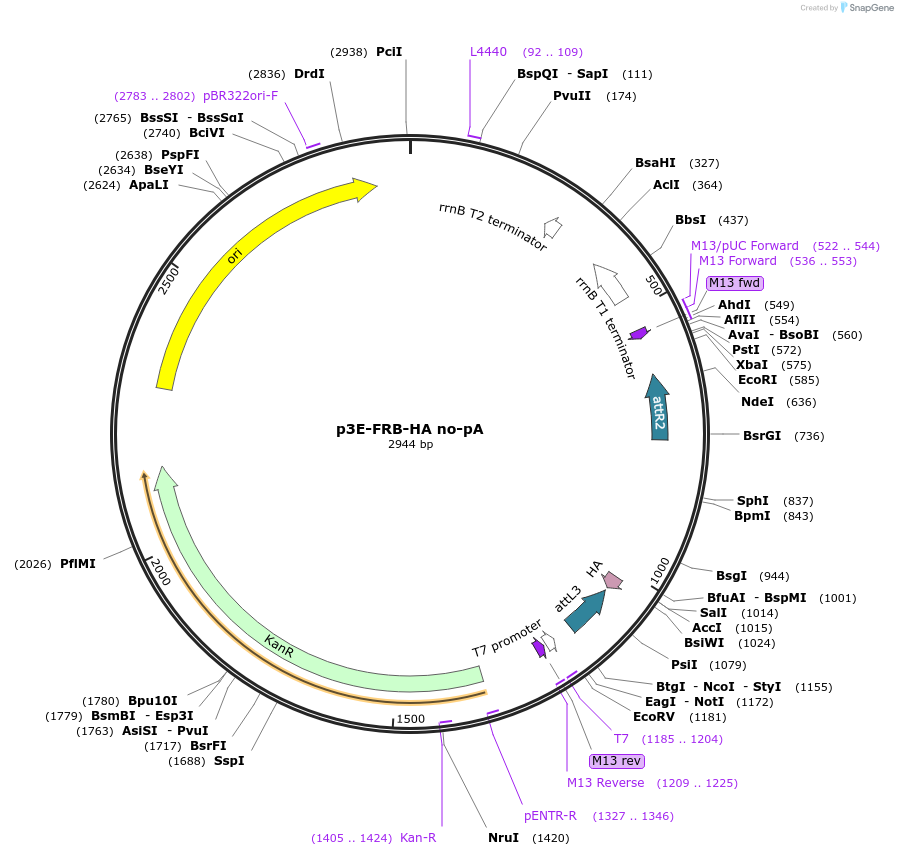
p3E-FRB-HA no-pA
Plasmid#82594Purpose3' entry vector containing HA epitope fused to C-terminus of TOR FKBP and rapamycin binding (FRB) domain without pA for rapamycin/rapalog induced dimerizationDepositorInsertFRB-HA (no pA)
PromoterNoneAvailable SinceDec. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
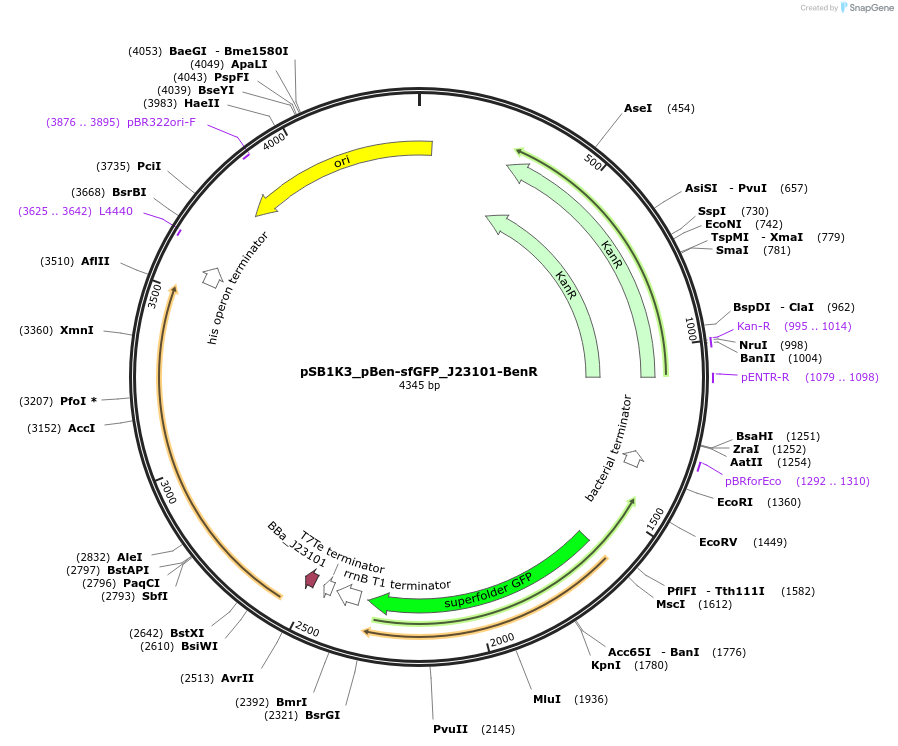
pSB1K3_pBen-sfGFP_J23101-BenR
Plasmid#128125PurposeExpressing sfGFP under control of benzoate responsive promoter (pBen) and expressing BenR transcription factor gene under constitutive promoter J23101, and RBS B0032DepositorInsertsfGFP and BenR
UseSynthetic BiologyExpressionBacterialPromoterpBen and J23101Available SinceAug. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
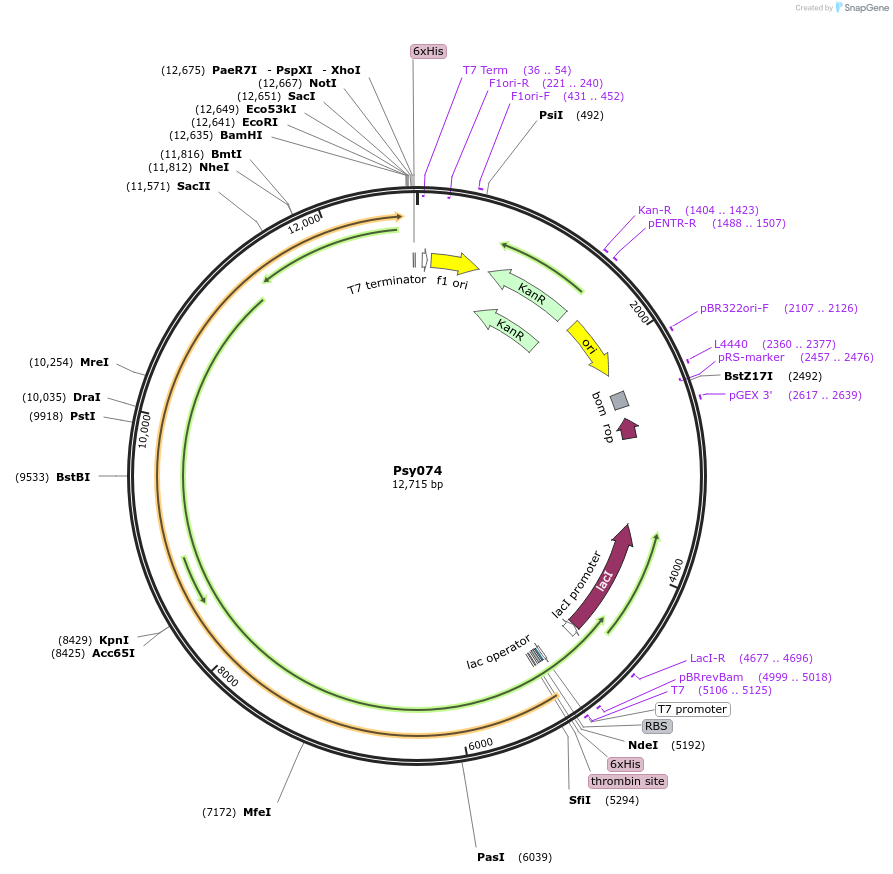
Psy074
Plasmid#99504PurposeBacterial expression of LIPS M1+TEDepositorInsertLIPS M1+TE
ExpressionBacterialAvailable SinceOct. 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
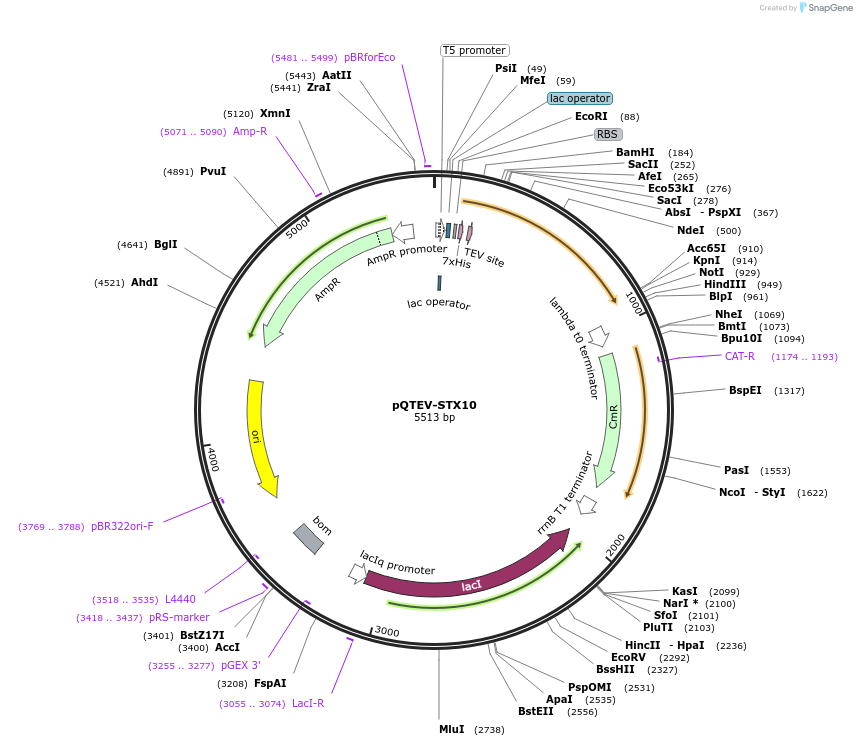
pQTEV-STX10
Plasmid#31572DepositorAvailable SinceMarch 25, 2013AvailabilityAcademic Institutions and Nonprofits only -
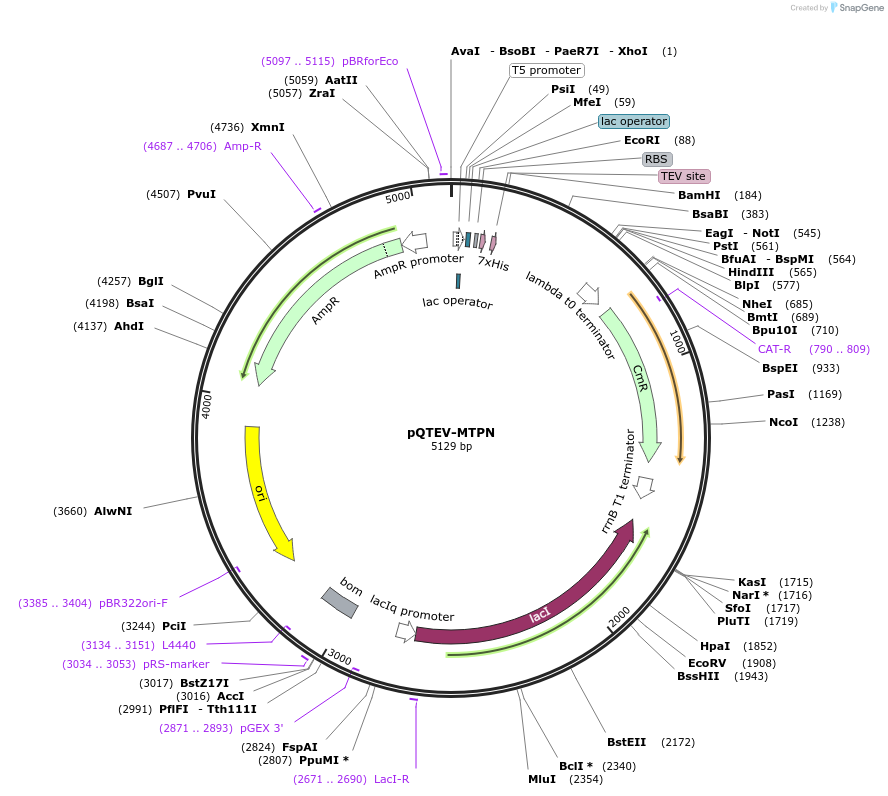
pQTEV-MTPN
Plasmid#34829DepositorAvailable SinceMarch 22, 2013AvailabilityAcademic Institutions and Nonprofits only -
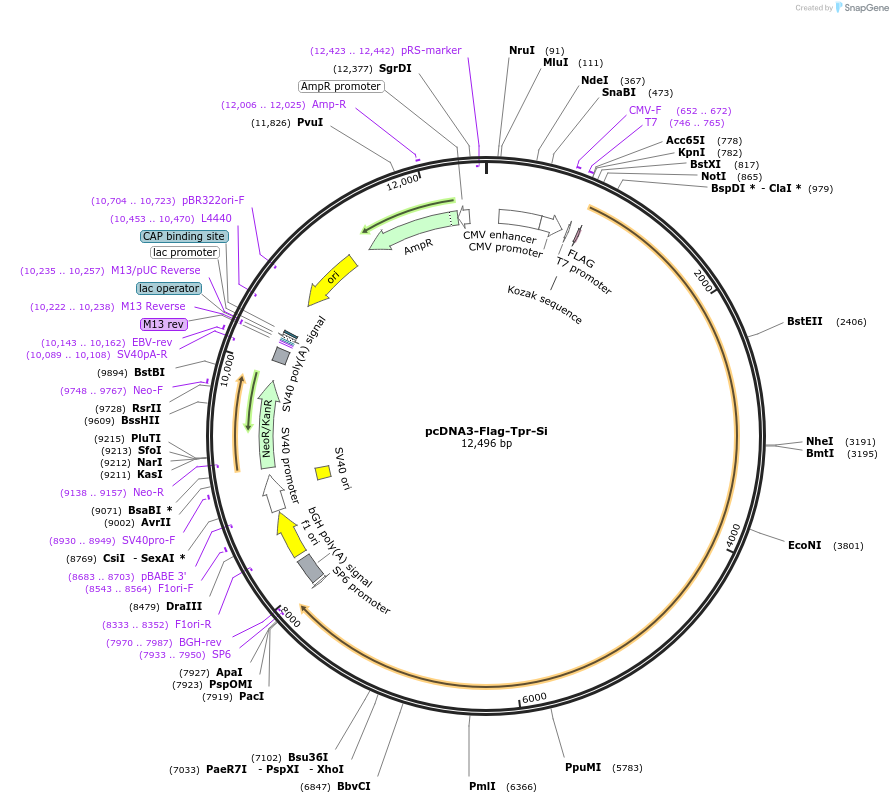
pcDNA3-Flag-Tpr-Si
Plasmid#60918PurposeExpresses Flag tagged nucleoporin Tpr resistant to siRNA GCACAACAGGATAAGGTTADepositorInsertTPR (TPR Human)
ExpressionMammalianMutationSilent point mutations in siRNA target region; E1…PromoterCMVAvailable SinceJan. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
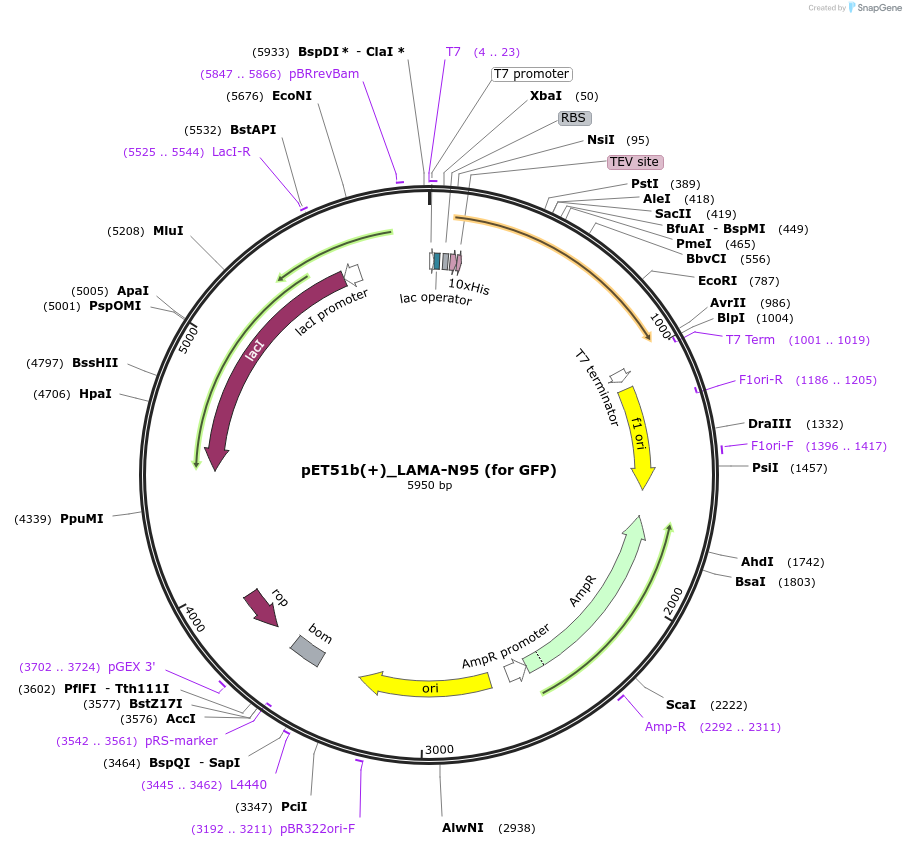
pET51b(+)_LAMA-N95 (for GFP)
Plasmid#130716PurposePlasmid encoding the bacterial expression of LAMA-N95 for GFPDepositorInsertLAMA-N95
TagsHisx10ExpressionBacterialMutationDeleted first Met, Ala and Last Ser from the Enha…Available SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
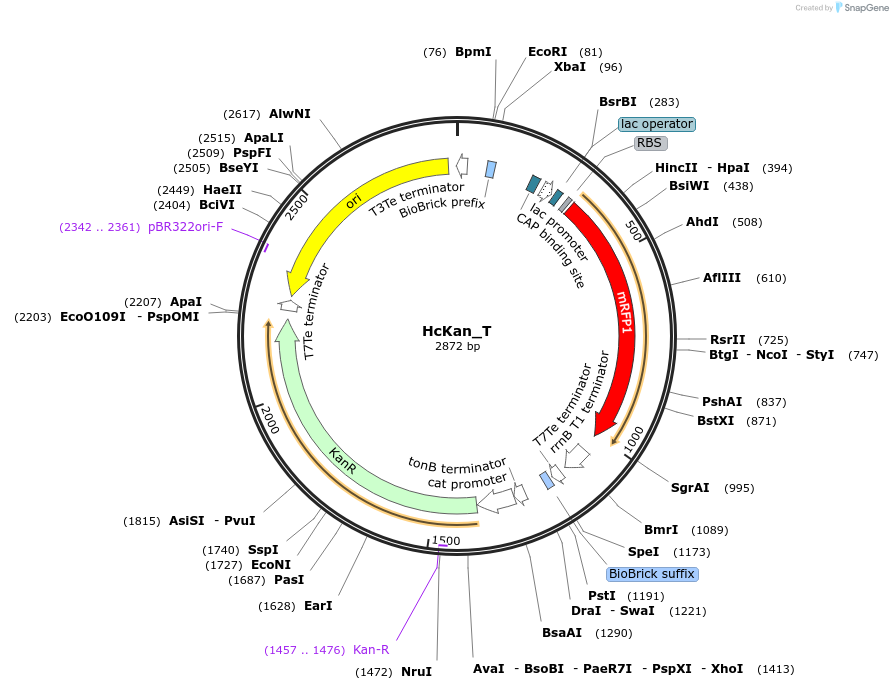
HcKan_T
Plasmid#65316Purposereceiving vector of terminators for the construction of standard terminator parts in YeastFab workDepositorInsertRFP
UseSynthetic BiologyAvailable SinceFeb. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
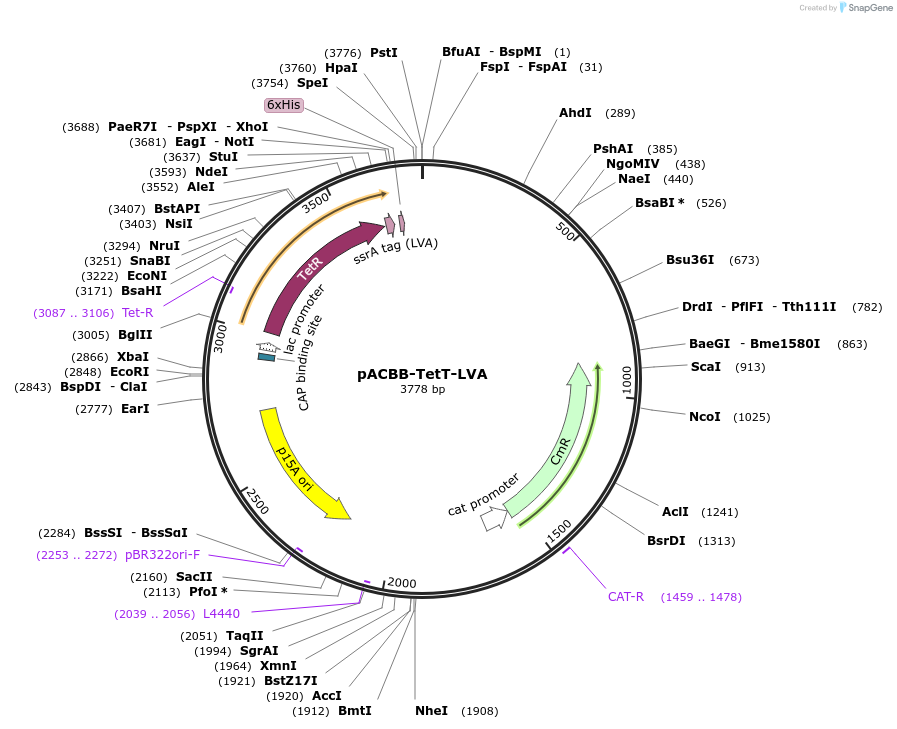
pACBB-TetT-LVA
Plasmid#32555DepositorInsertTetR
UseSynthetic BiologyTagsLVA tagExpressionBacterialPromoterConsitutive lacAvailable SinceNov. 8, 2011AvailabilityAcademic Institutions and Nonprofits only -
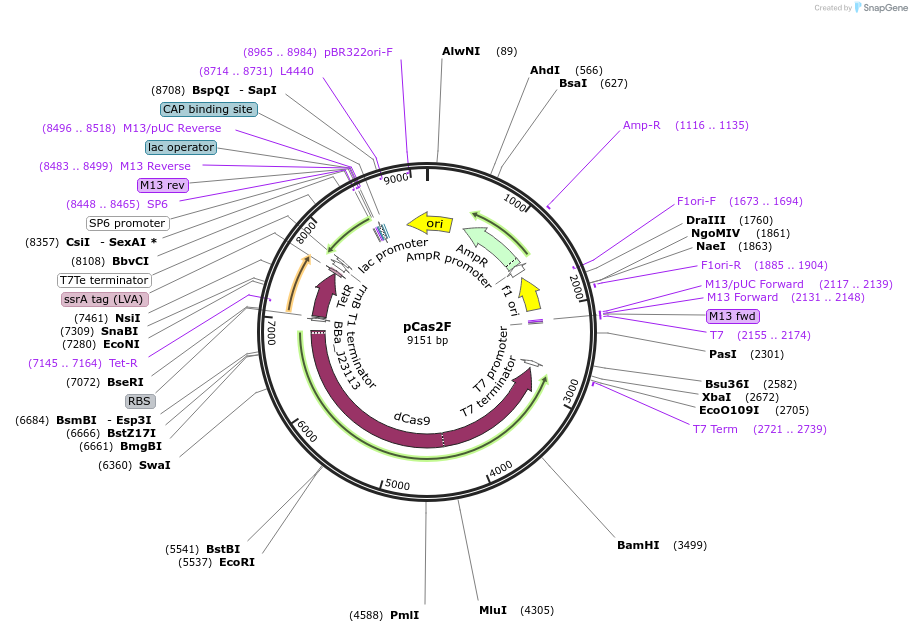
pCas2F
Plasmid#82377PurposedCas9 expressed from aTc-inducible promoter in acsA, F RBS: TGGGCGDepositorInsertdCas9
PromoterPEZ3Available SinceSept. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
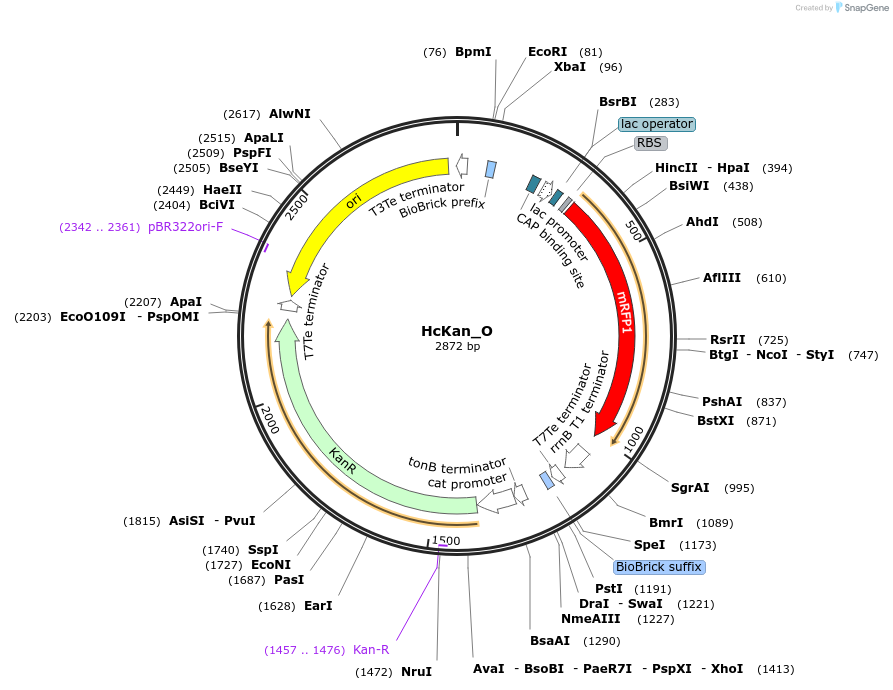
HcKan_O
Plasmid#65315Purposereceiving vector of ORFs for the construction of standard ORF parts in YeastFab workDepositorInsertRFP
UseSynthetic BiologyAvailable SinceSept. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
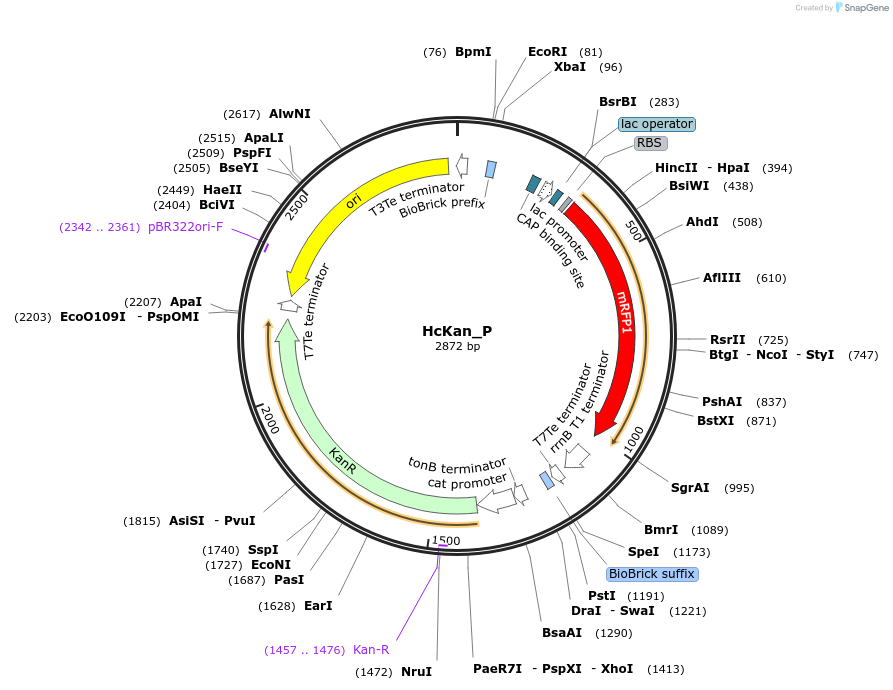
HcKan_P
Plasmid#65314Purposereceiving vector of promoters for the construction of standard promoter parts in YeastFab workDepositorInsertRFP
UseSynthetic BiologyAvailable SinceSept. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
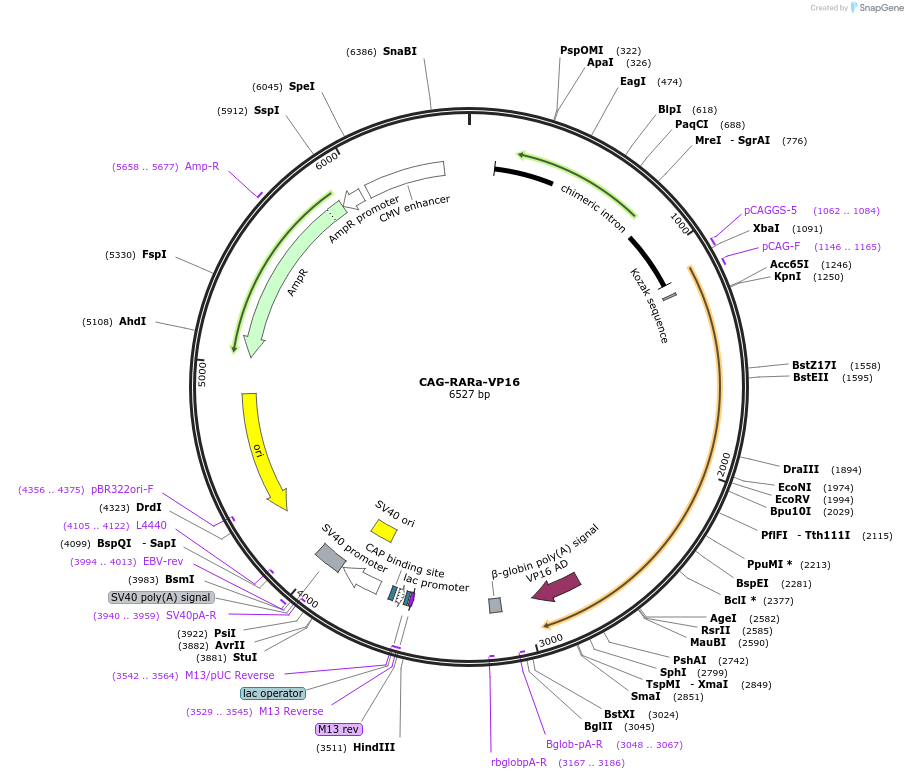
CAG-RARa-VP16
Plasmid#185559PurposeUbiquitous expression of constitutively active Retinoic Acid Receptor alphaDepositorAvailable SinceJuly 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
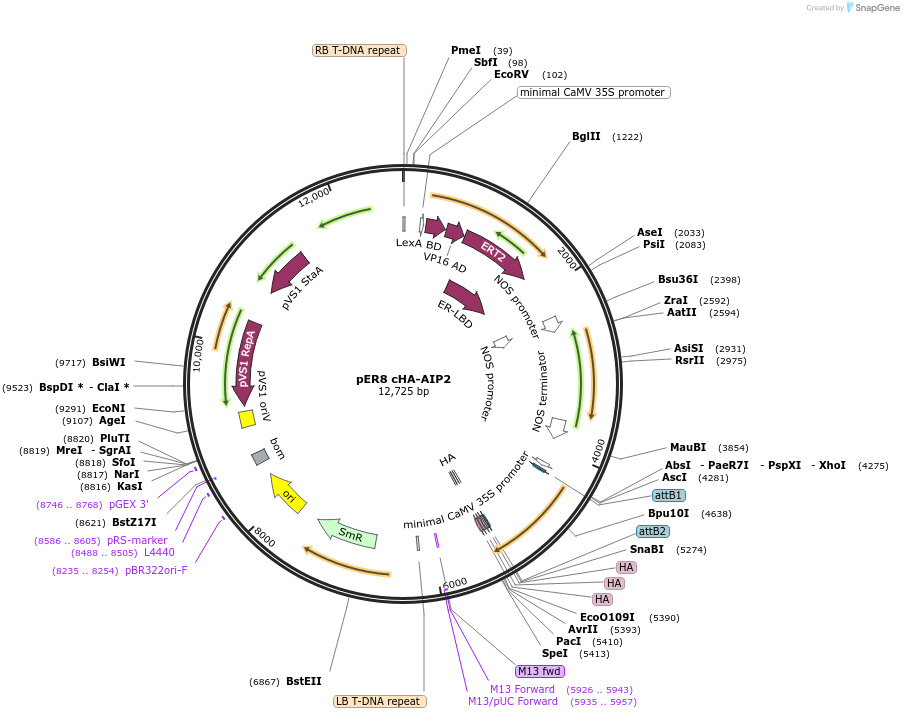
pER8 cHA-AIP2
Plasmid#118687PurposePlant inducible expressionDepositorAvailable SinceApril 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pFastBac-His-Flag-Tipin
Plasmid#22890DepositorAvailable SinceJan. 22, 2010AvailabilityAcademic Institutions and Nonprofits only -
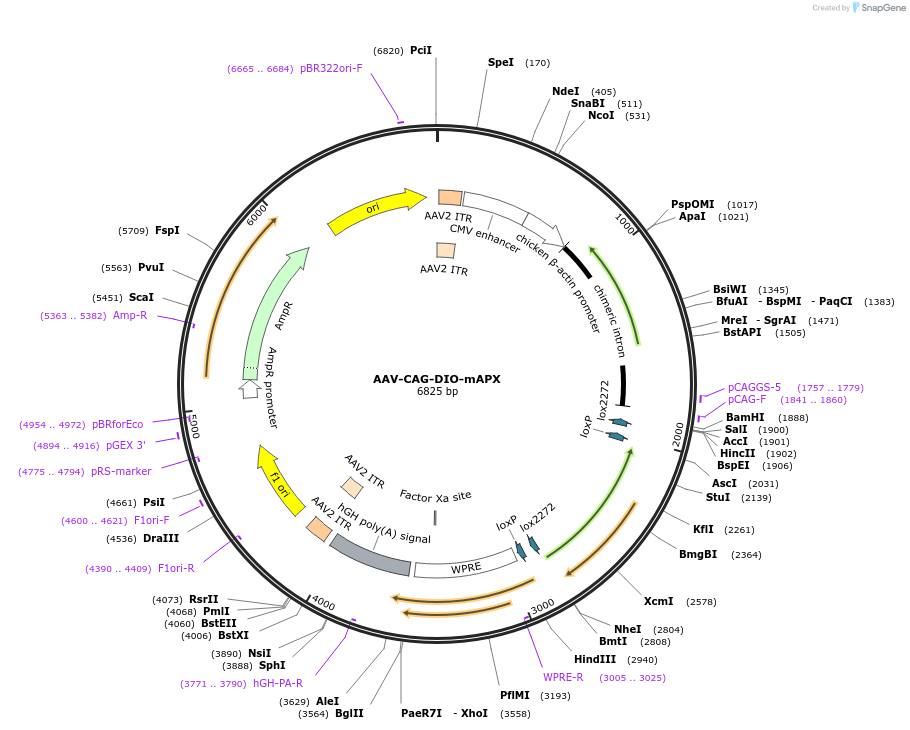
AAV-CAG-DIO-mAPX
Plasmid#79908PurposeAdeno-associated virus plasmid, Cre-dependent FLEX switch, mAPXDepositorInsertmAPX
UseAAV and Cre/LoxExpressionMammalianAvailable SinceJuly 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
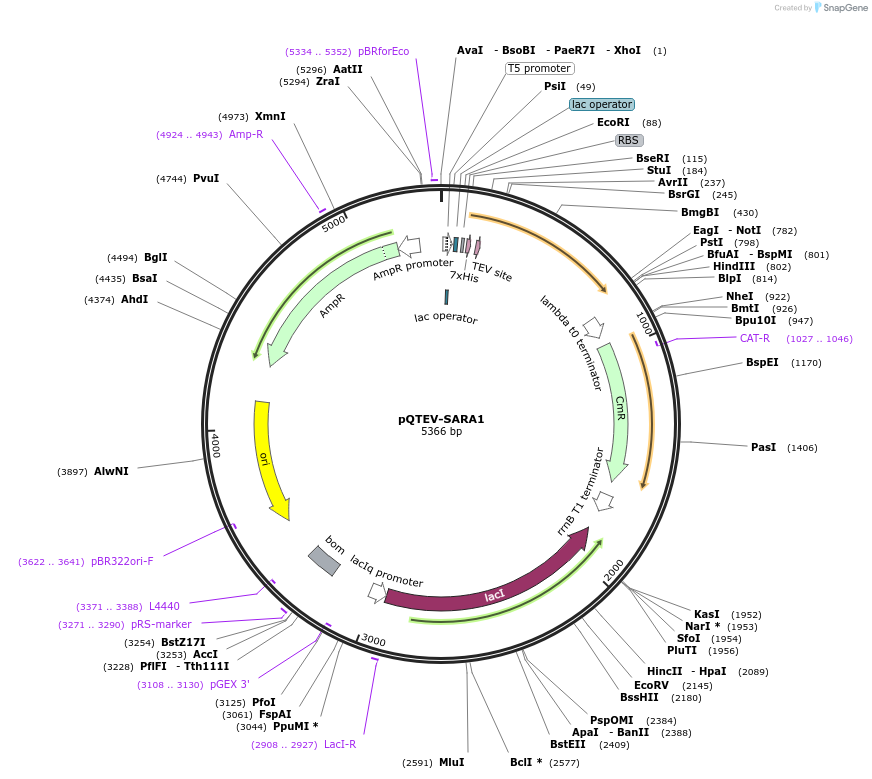
pQTEV-SARA1
Plasmid#34809DepositorAvailable SinceMarch 22, 2013AvailabilityAcademic Institutions and Nonprofits only -
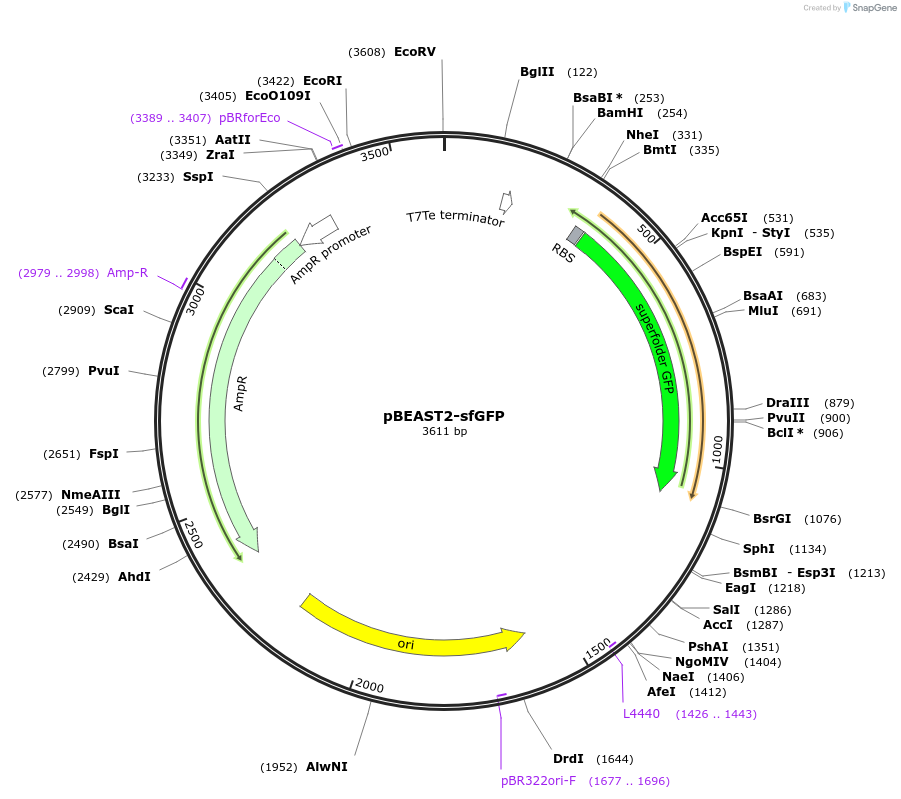
pBEAST2-sfGFP
Plasmid#126575PurposeStrong constitutive expression of super-folder GFP for cell-free expressionDepositorInsertsfGFP
UseSynthetic Biology; Cell-free protein synthesisAvailable SinceJune 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
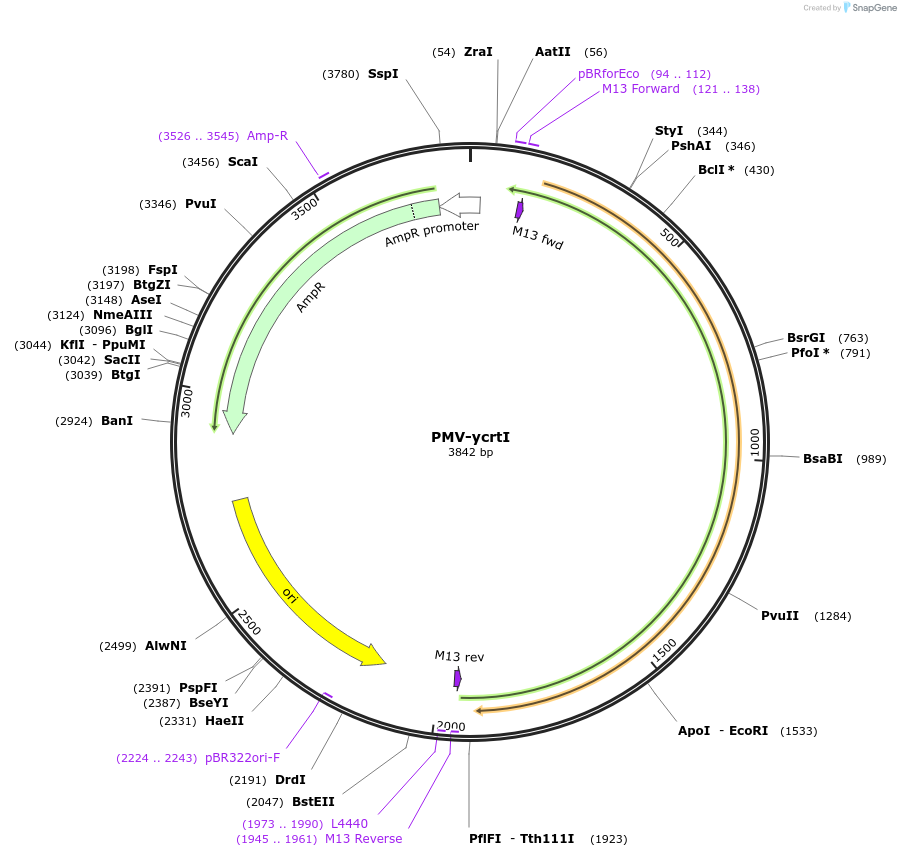
PMV-ycrtI
Plasmid#65342Purposesysthesis of crtI gene in the beta-carotene pathwayDepositorInsertycrtI
UseSynthetic BiologyAvailable SinceFeb. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
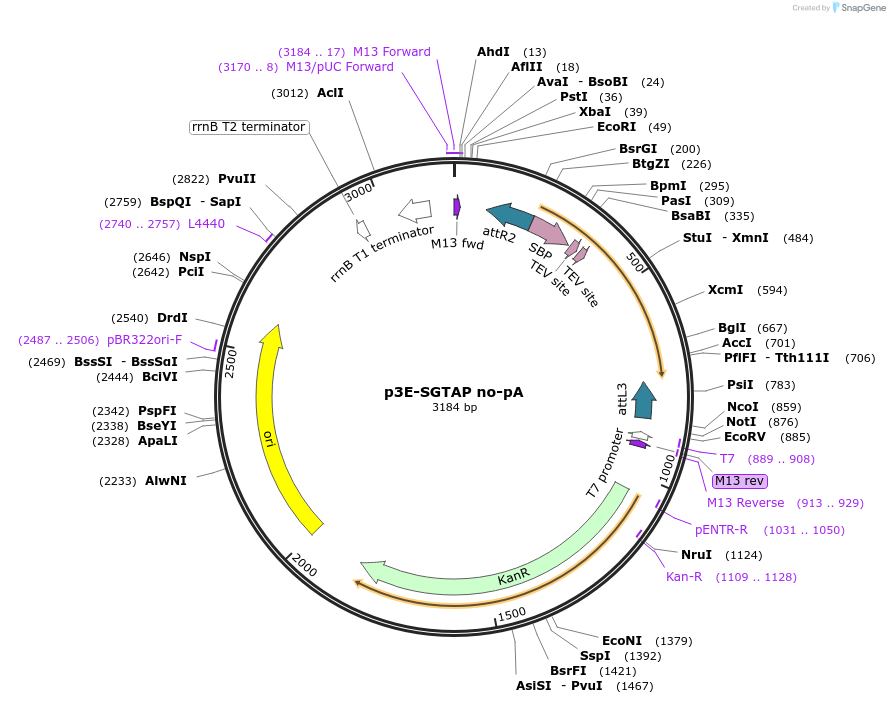
p3E-SGTAP no-pA
Plasmid#82599Purpose3' entry vector containing streptavidin binding protein (SBP) followed by a TEV cleavage site and 2 protein G copies without pA for tandem affinity purificationDepositorInsertSBP-TEV-2x protein G
PromoterNoneAvailable SinceDec. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
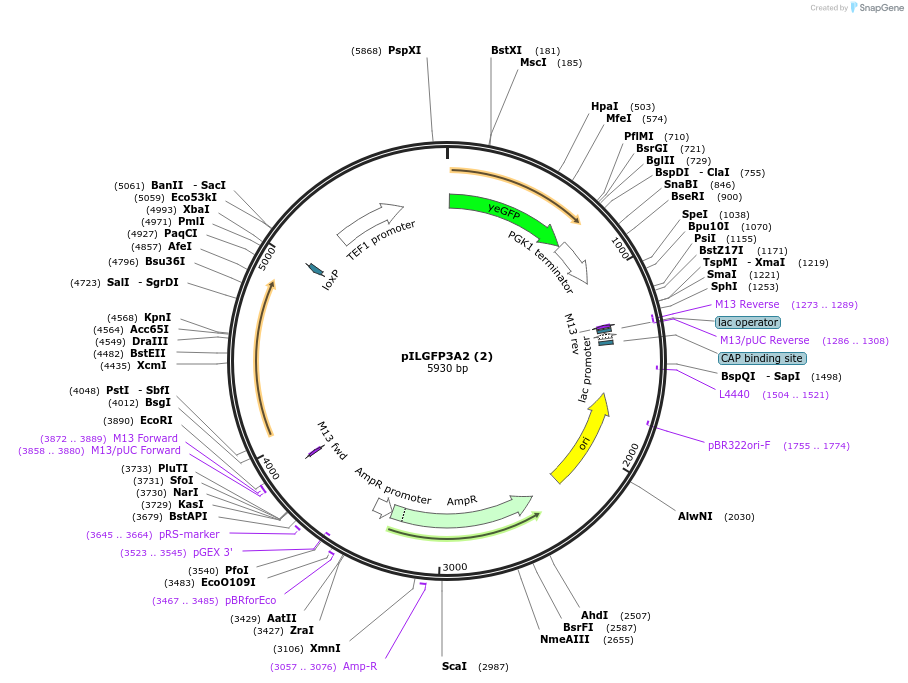
pILGFP3A2 (2)
Plasmid#185852PurposeTesting the TEF1 promoter appended with 3 tetracycline riboswitch using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PTEF1>3*TcRb>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
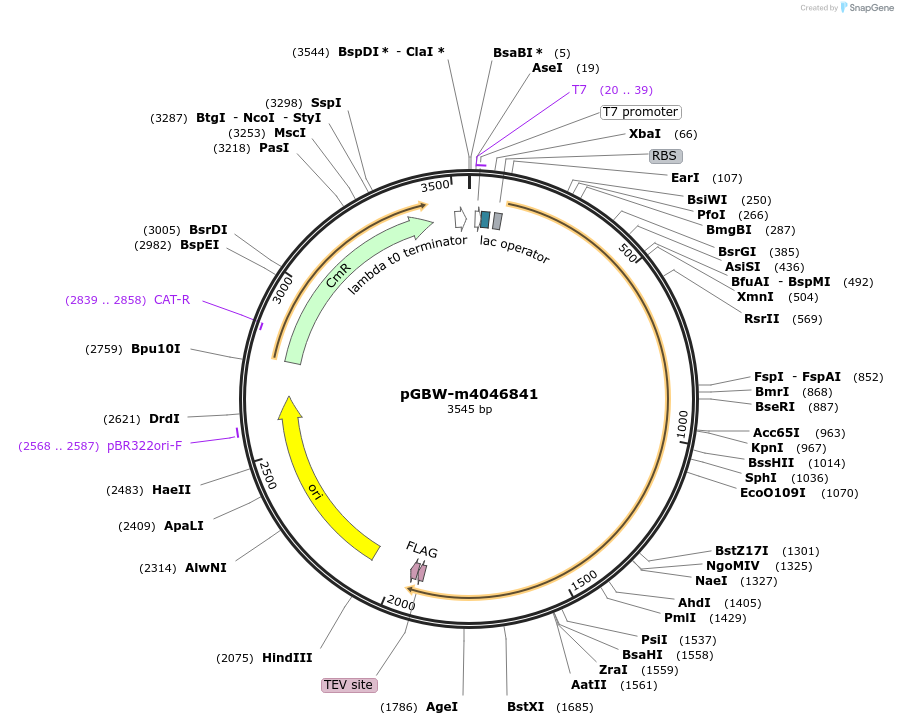
pGBW-m4046841
Plasmid#145578PurposeBacterial Expression plasmid for SARS-CoV-2 helicaseDepositorInsertSARS-CoV-2 helicase (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;FLAG_2ExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
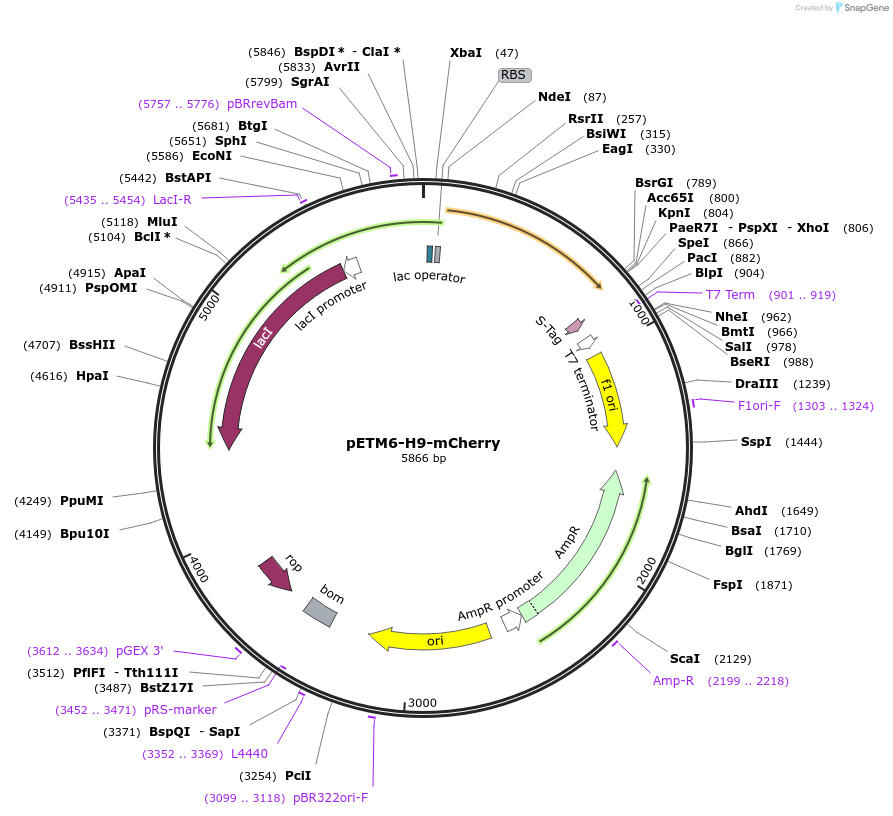
pETM6-H9-mCherry
Plasmid#66532PurposeExpresses mCherry under the control of Mutant T7 Promoter H9DepositorInsertmCherry
UseSynthetic BiologyExpressionBacterialMutationCodon Optimized for E. coliPromoterMutant T7 Promoter - H9Available SinceJuly 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
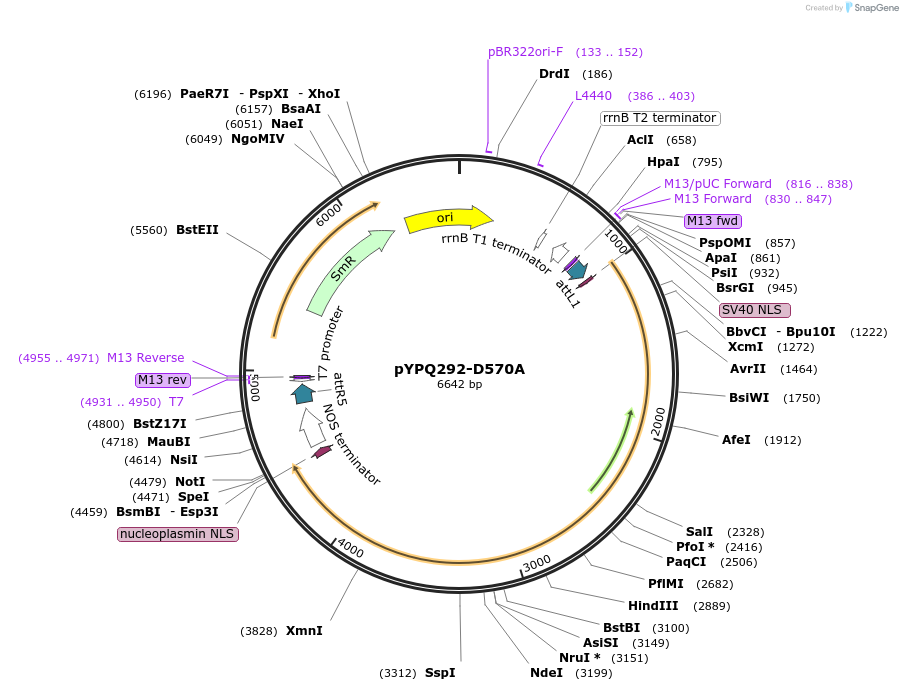
pYPQ292-D570A
Plasmid#129679PurposeGateway entry clone for CRISPR-Cas12b systemsDepositorInsertAaCas12b
UseCRISPR; Gateway compatible aacas12b entry cloneExpressionPlantMutationD570A; AaCas12b is rice codon optimizedAvailable SinceMarch 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
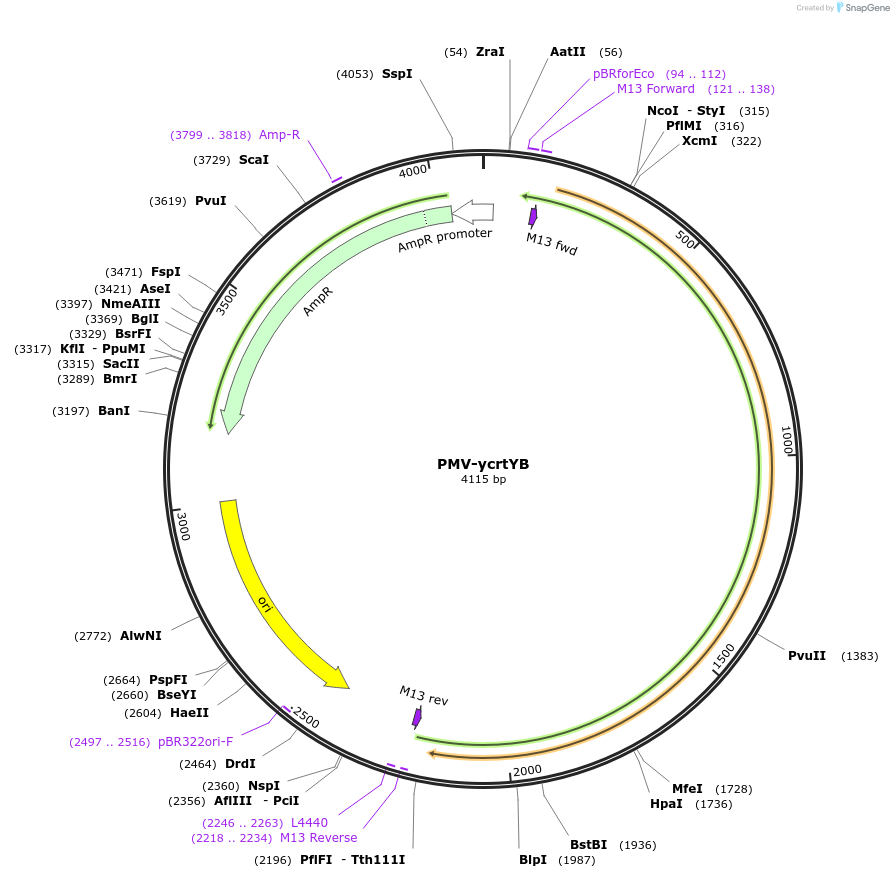
PMV-ycrtYB
Plasmid#65343Purposesysthesis of crtYB gene in the beta-carotene pathwayDepositorInsertycrtYB
UseSynthetic BiologyAvailable SinceFeb. 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
mTFP1-Lysosomes-20
Plasmid#55495PurposeLocalization: Lysosome Membrane, Excitation: 462, Emission: 492DepositorInsertLAMP1
TagsmTFP1ExpressionMammalianPromoterCMVAvailable SinceOct. 2, 2014AvailabilityAcademic Institutions and Nonprofits only -
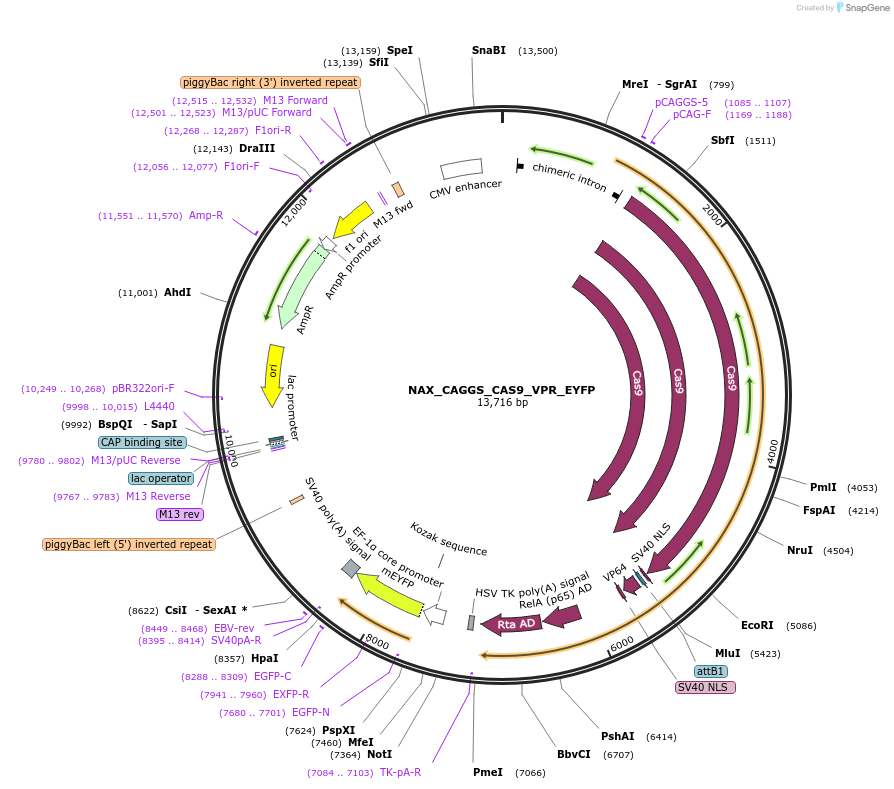
NAX_CAGGS_CAS9_VPR_EYFP
Plasmid#167890PurposePiggyBac compatible plasmid expressing Cas9-VPRDepositorInsertCas9-VPR
UseCRISPRAvailable SinceFeb. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
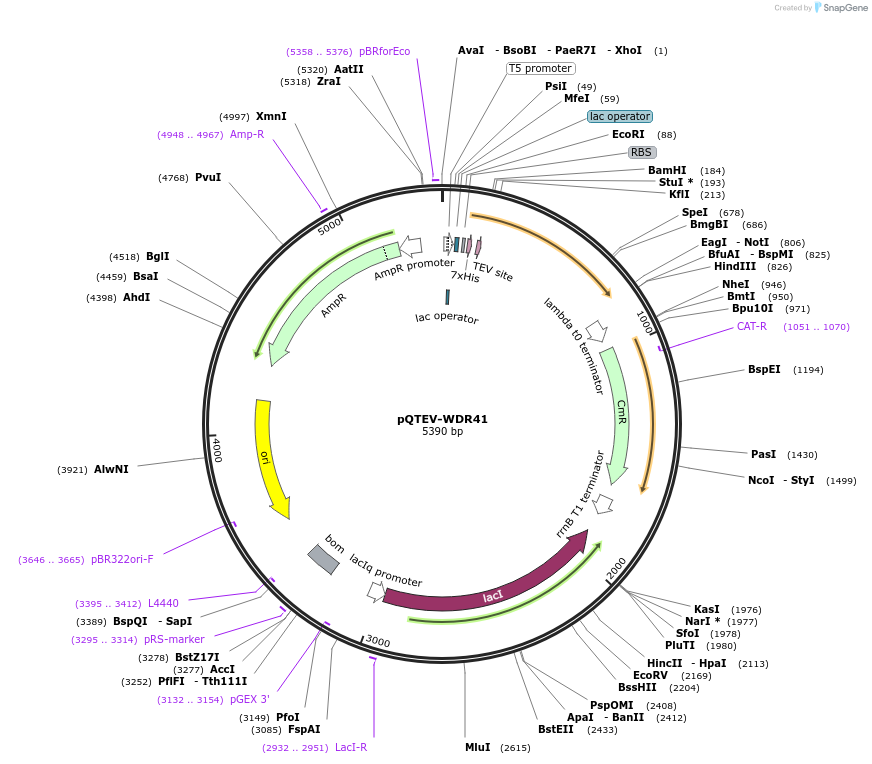
pQTEV-WDR41
Plasmid#34808DepositorAvailable SinceMarch 25, 2013AvailabilityAcademic Institutions and Nonprofits only -
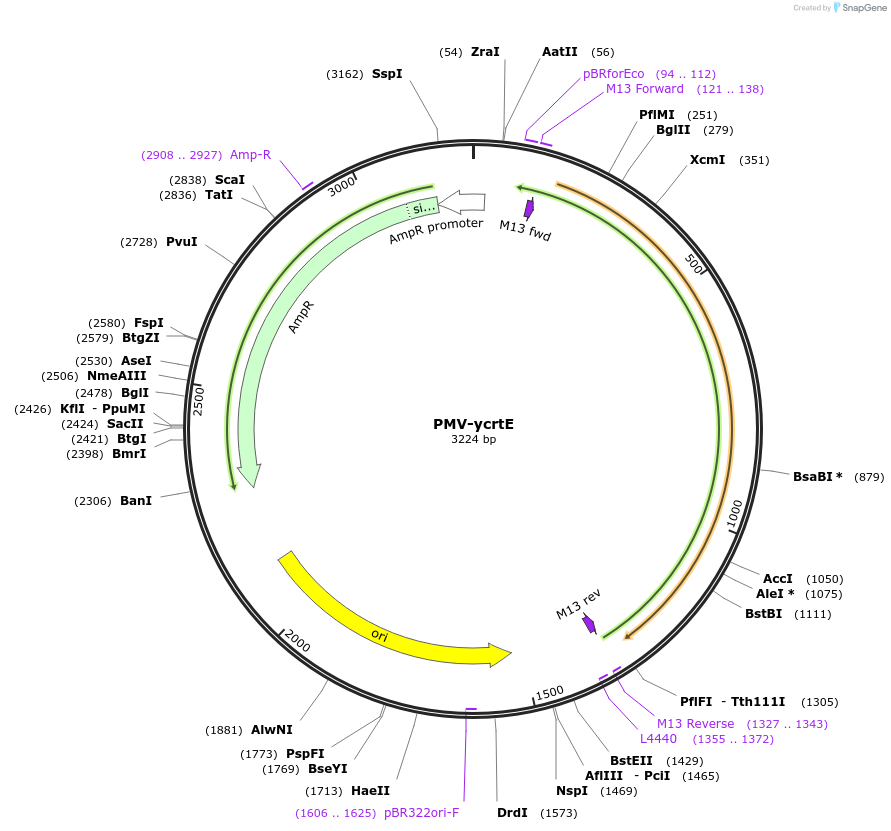
PMV-ycrtE
Plasmid#65341Purposesysthesis of crtE gene in the beta-carotene pathwayDepositorInsertycrtE
UseSynthetic BiologyAvailable SinceFeb. 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
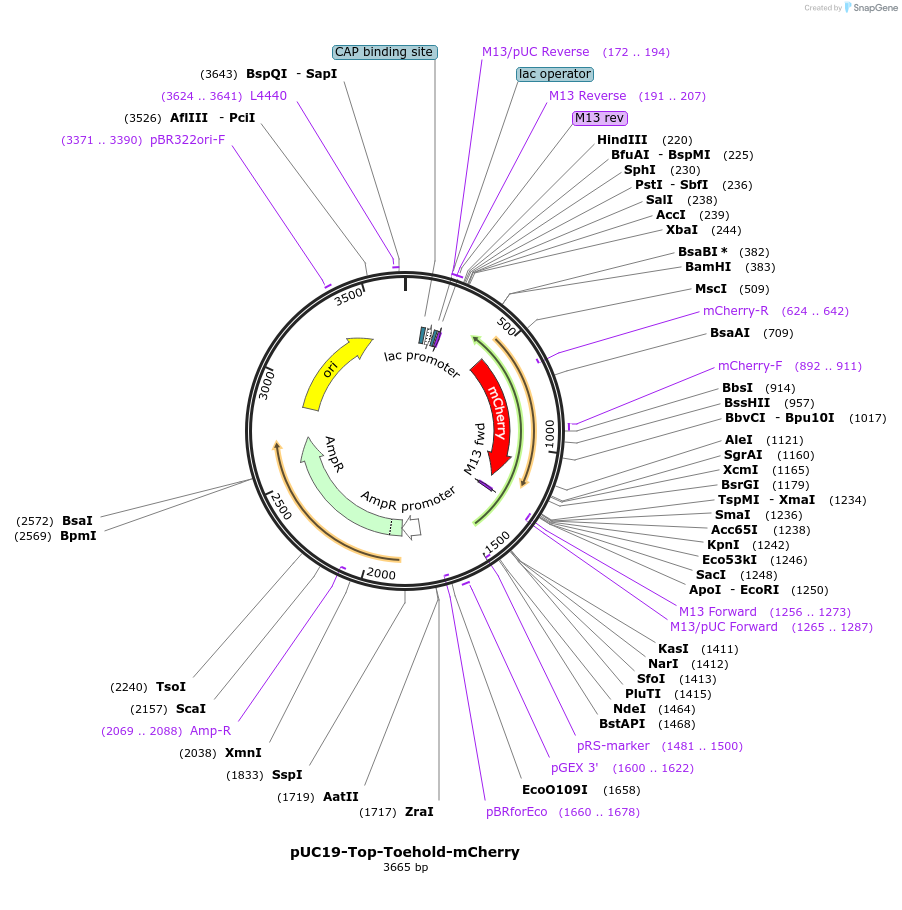
pUC19-Top-Toehold-mCherry
Plasmid#87318PurposeExpresses the "Top" transcript of the Split-Broccoli system upstream of an RNA Toehold switch controlling mCherry.DepositorInsertSplit-Broccoli "Top" transcript upstream of an RNA Toehold (TS2_KS01) controlling mCherry translation.
UseSynthetic BiologyExpressionBacterialPromoterP70aAvailable SinceMay 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
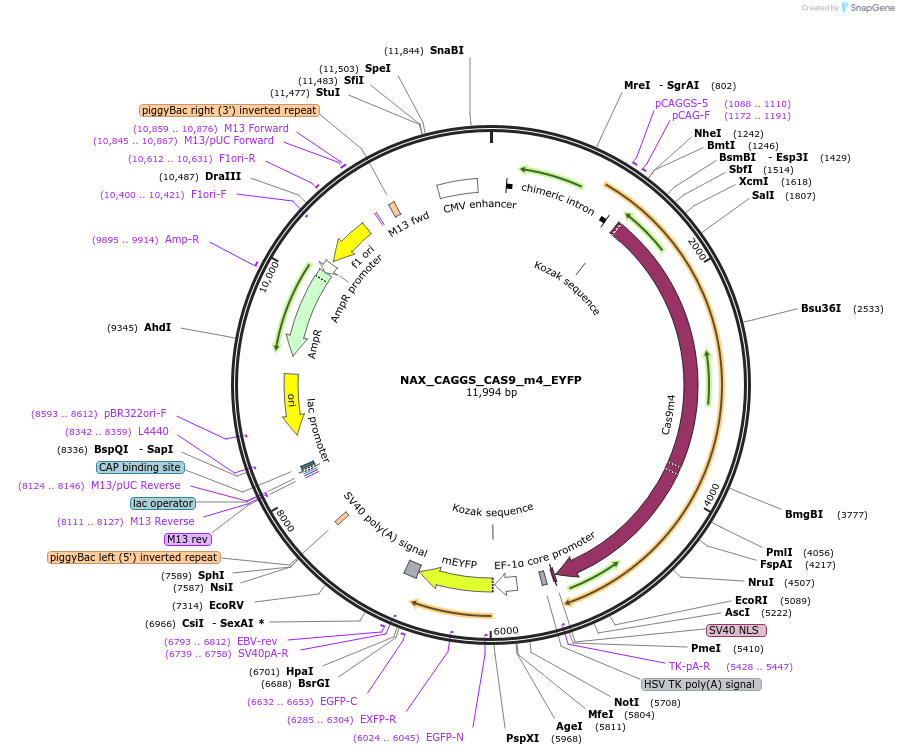
NAX_CAGGS_CAS9_m4_EYFP
Plasmid#167866PurposePiggyBac compatible plasmid expressing dCas9DepositorInsertdCas9
UseCRISPRMutationD10A, D839A, H840A, N863AAvailable SinceFeb. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
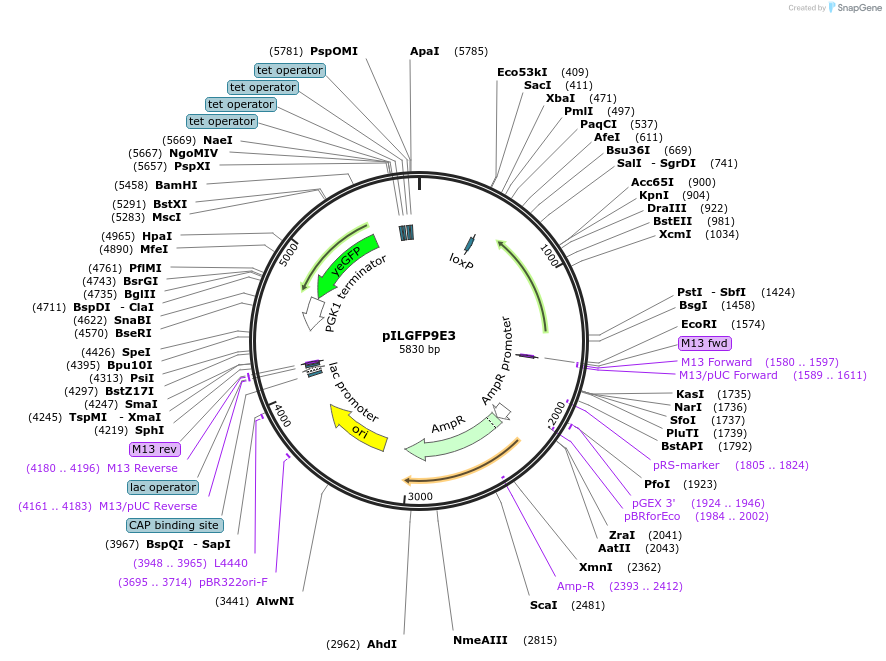
pILGFP9E3
Plasmid#185842PurposeTesting the TEF1 promoter inserted with 4 TetO elements using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PTEF1+[TetO]>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
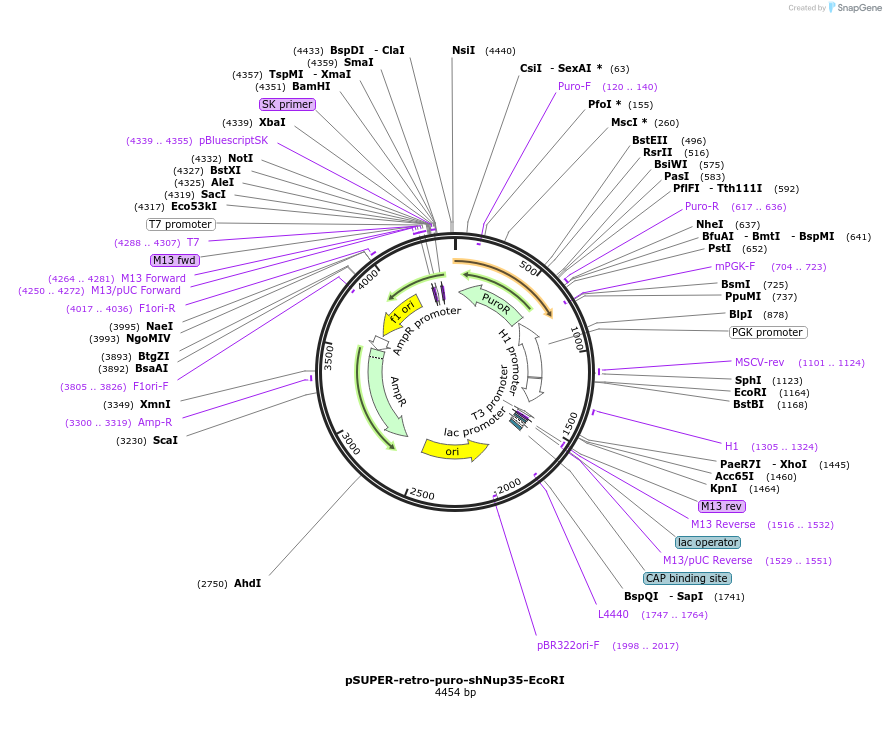
pSUPER-retro-puro-shNup35-EcoRI
Plasmid#87332PurposeTo express shRNA against human Nup35DepositorInsertshRNA against human Nup35
UseRNAiExpressionMammalianPromoterH1Available SinceMay 1, 2017AvailabilityAcademic Institutions and Nonprofits only