We narrowed to 19,978 results for: REN
-
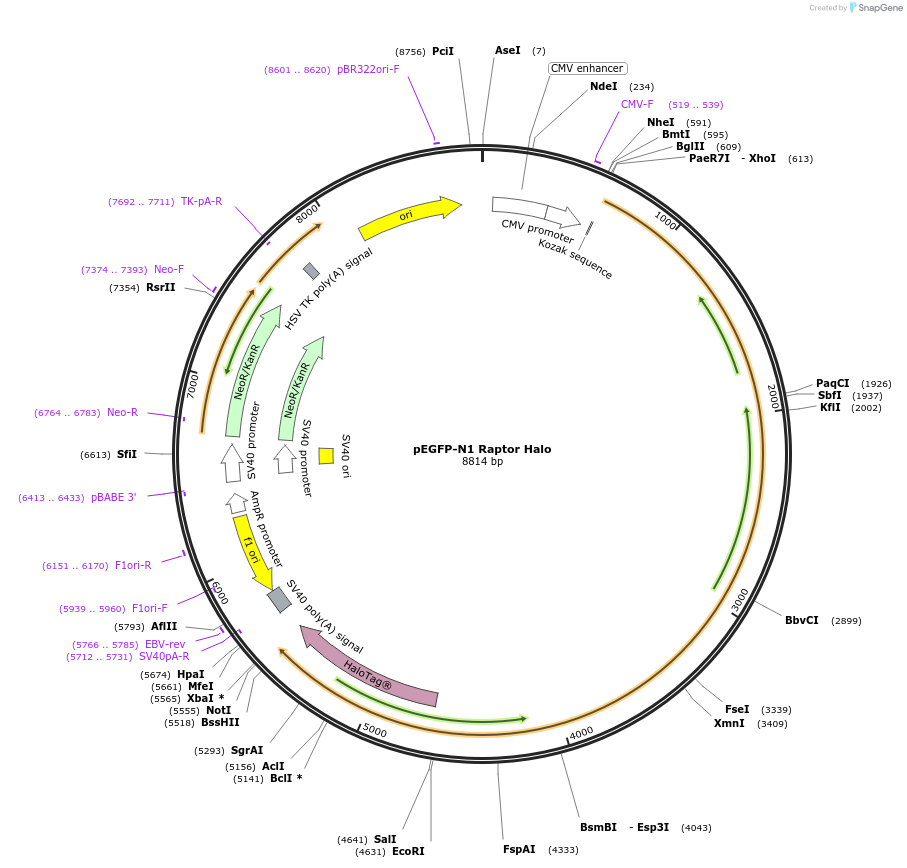
Plasmid#112754Purposeexpress Raptor-HaloDepositorAvailable SinceOct. 31, 2018AvailabilityAcademic Institutions and Nonprofits only
-
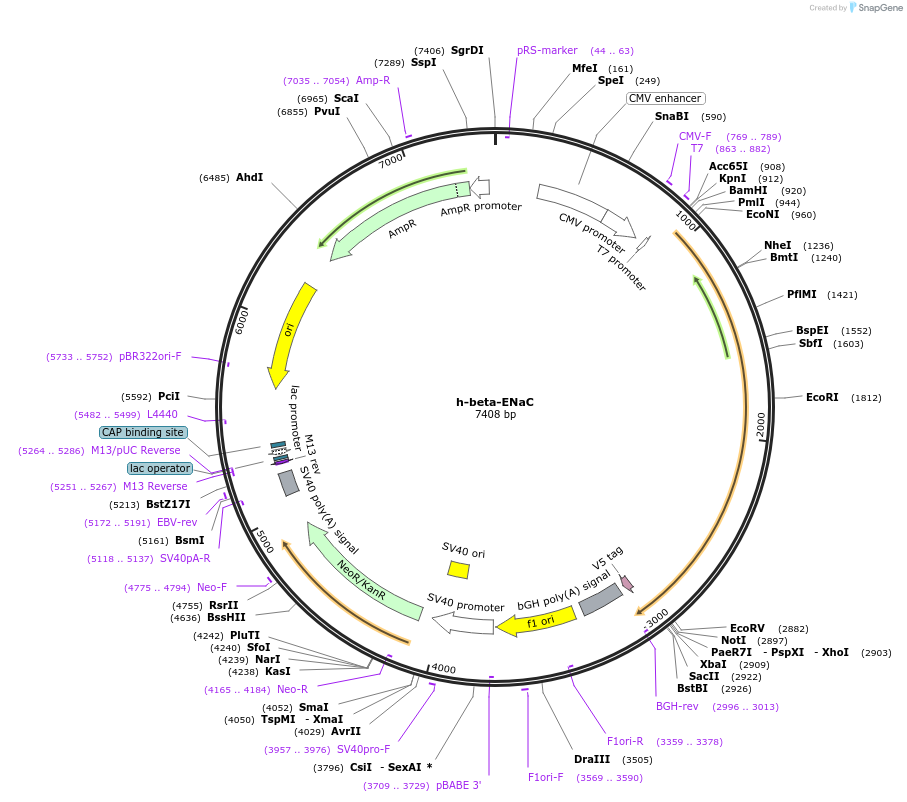
h-beta-ENaC
Plasmid#83429Purposefull length human beta ENaC with V5 tagDepositorAvailable SinceNov. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
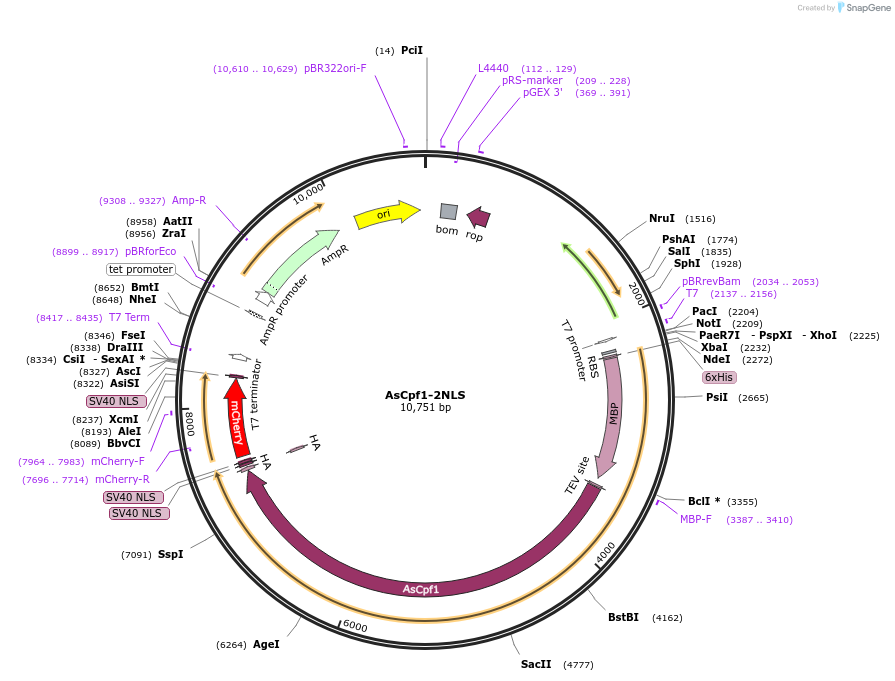
AsCpf1-2NLS
Plasmid#102565Purposebacterial expression of AsCpf1-2NLSDepositorInsertAsCpf1
Tags6xHis-MBP-TEV and HA-2xSV40NLSExpressionBacterialPromoterT7Available SinceDec. 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
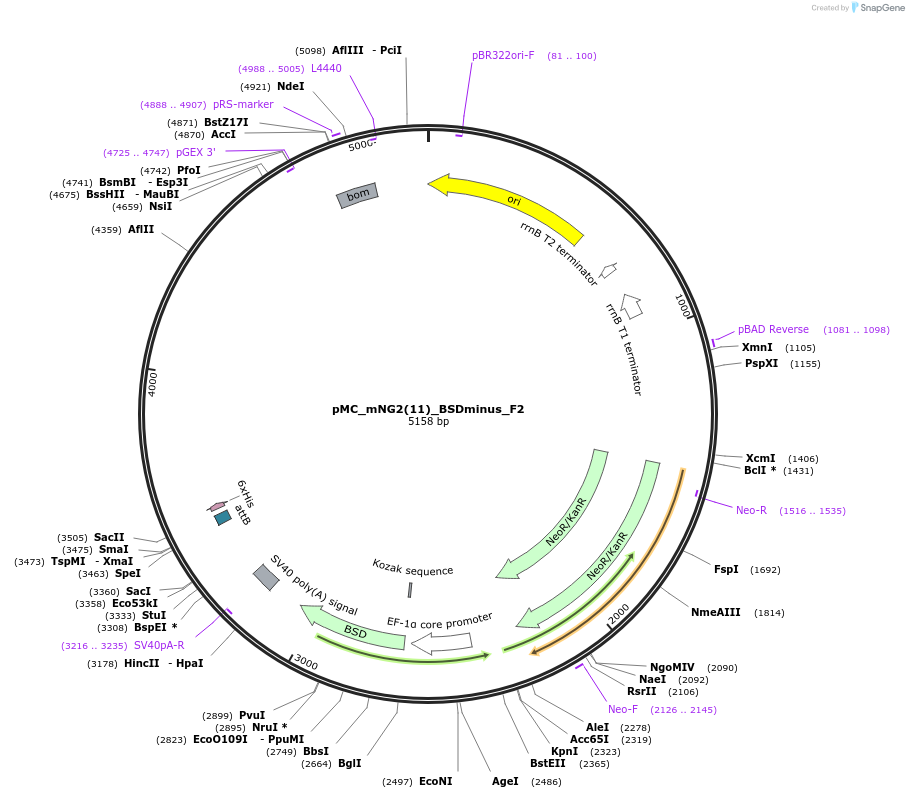
pMC_mNG2(11)_BSDminus_F2
Plasmid#184164PurposeDonor cassette plasmid for high-throughput tagging, encoding an mNG2(11) synthetic exon in frame 2, as well as a blasticidin resistance gene for selection of genomic integrants.DepositorInsertsmNG2(11)
bsd
UseCRISPRAvailable SinceMay 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
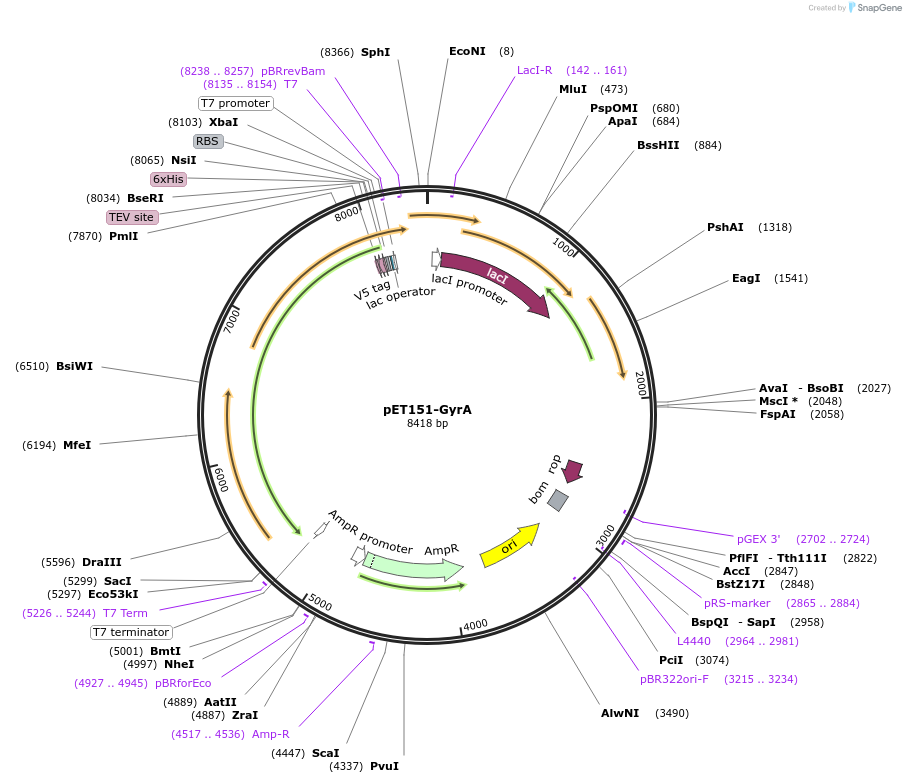
pET151-GyrA
Plasmid#216229PurposeS. aureus USA300 GyrA Codon-optimized for E. coliDepositorInsertDNA gyrase
Tags6xHisExpressionBacterialPromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
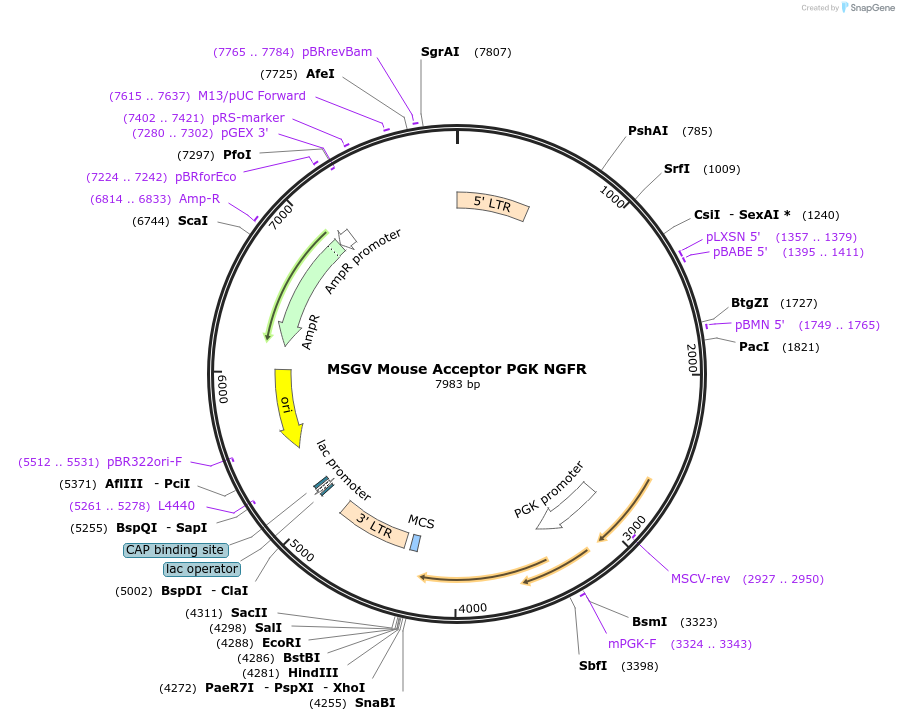
MSGV Mouse Acceptor PGK NGFR
Plasmid#128482PurposeA TCR Variable Region cloning expression vector using Mouse Constant Regions to reduce mis-pairing with endogenous TCR expressing NGFR ref Biotechniques. 2015 Mar 1;58(3):135-9DepositorTypeEmpty backboneUseRetroviralPromoterMSCVAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
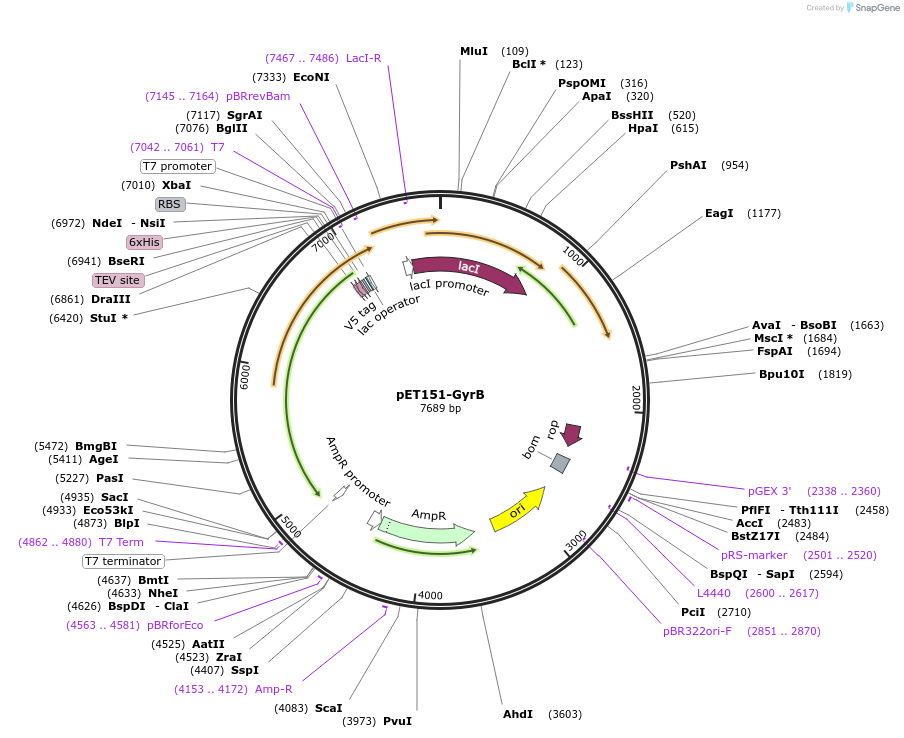
pET151-GyrB
Plasmid#216233PurposeS. aureus USA300 GyrB Codon-optimized for E. coliDepositorInsertDNA gyrase
Tags6xHisExpressionBacterialPromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
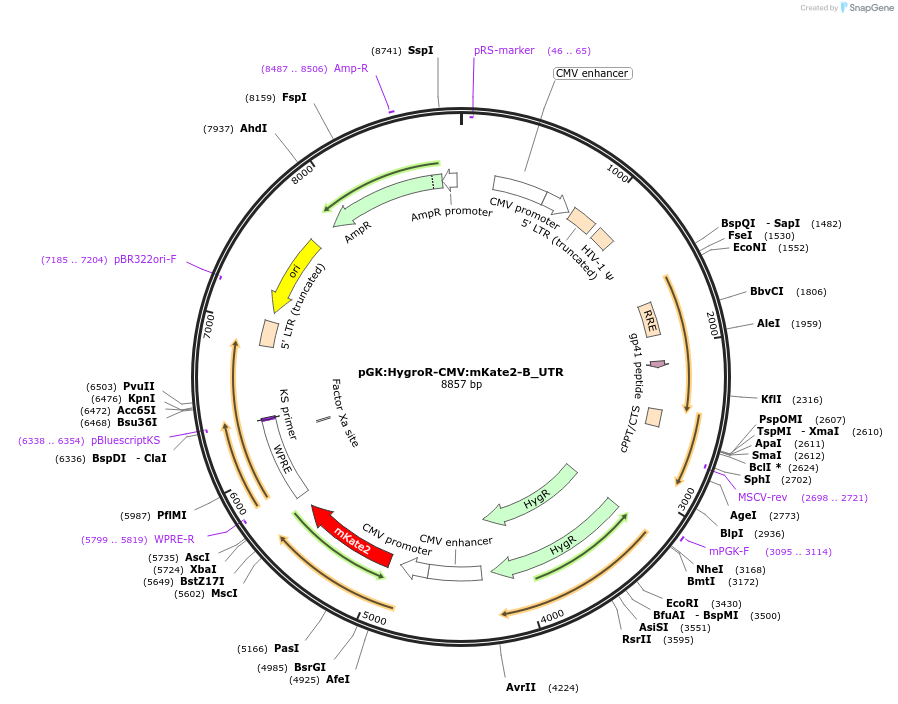
pGK:HygroR-CMV:mKate2-B_UTR
Plasmid#68480PurposeLentivirus expressing mKate2 with UTR sensor cassette and hygromycin resistanceDepositorInsertsHygromycin Resistance
mKate2-B_UTR
UseLentiviralExpressionMammalianMutationshRNA sensor cassette "B" added to 3&#x…PromoterCMV and pGKAvailable SinceFeb. 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
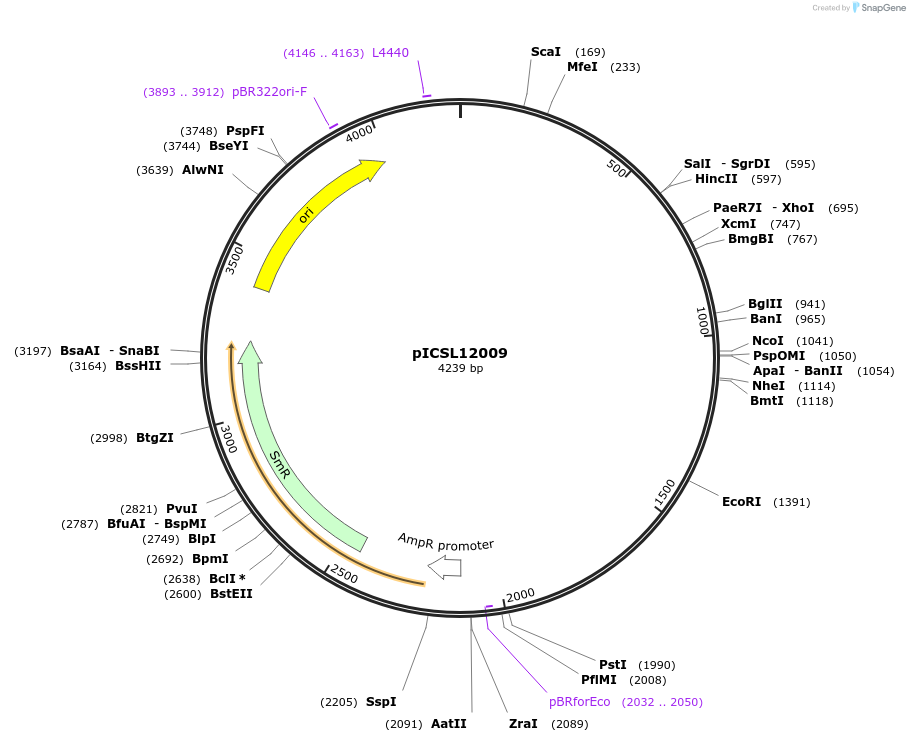
pICSL12009
Plasmid#68257PurposeLevel 0 Golden Gate Part: Promoter and 5UTR Ubiquitin (Zea mays)DepositorInsertPromoter and 5UTR Ubiquitin (Zea mays)
UseSynthetic BiologyAvailable SinceAug. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
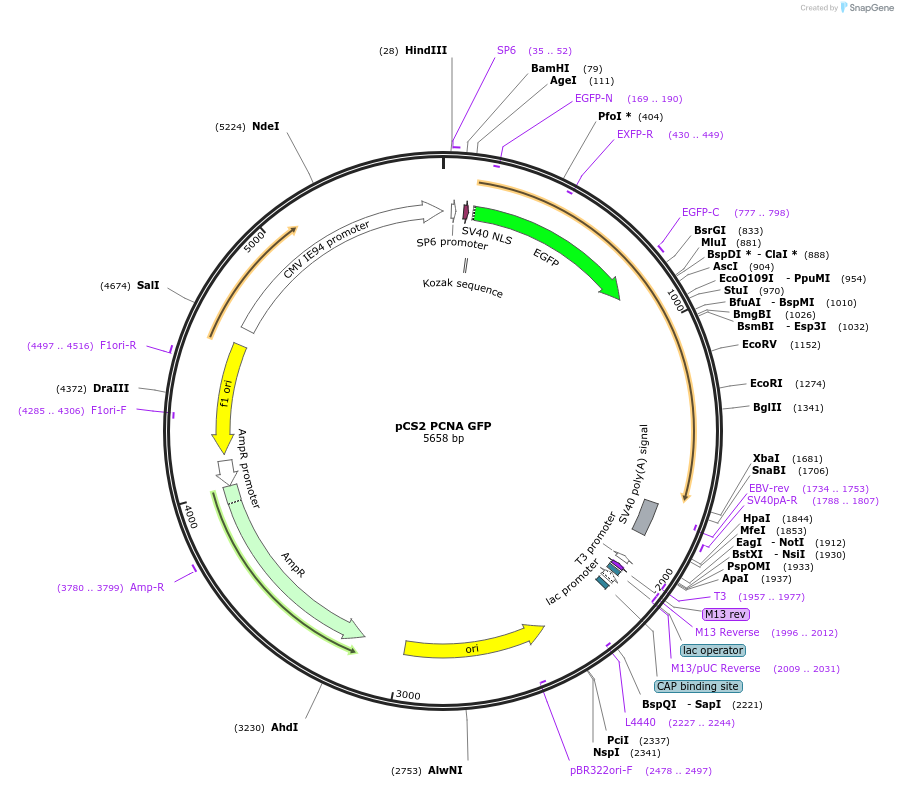
pCS2 PCNA GFP
Plasmid#105937Purposein vitro transcription, cell cycle nuclear markerDepositorAvailable SinceMarch 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
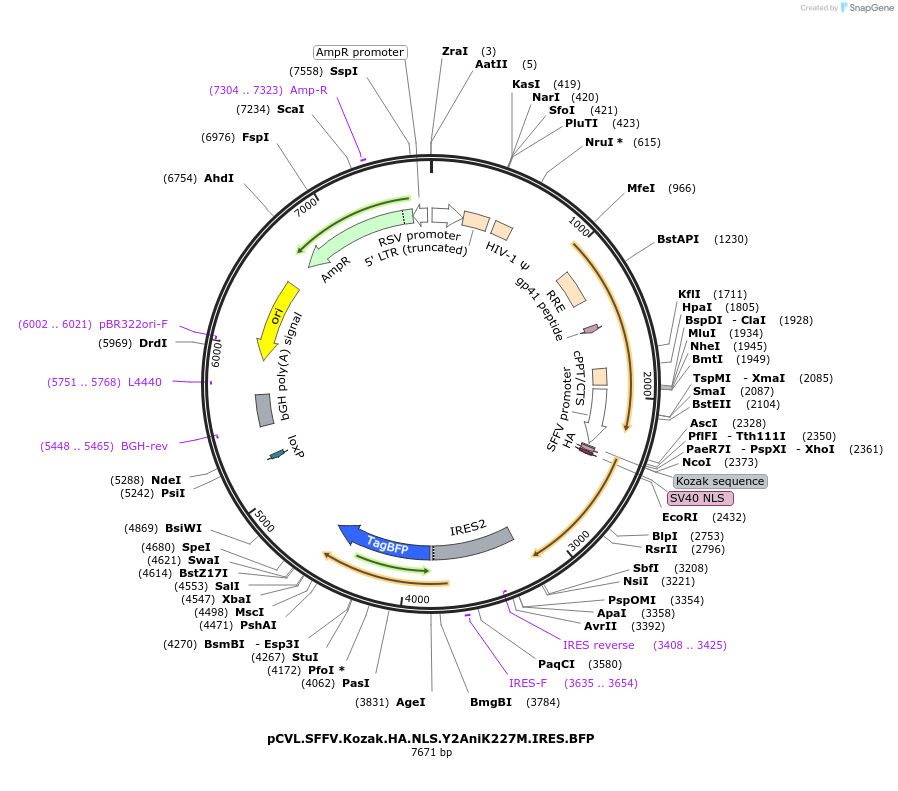
pCVL.SFFV.Kozak.HA.NLS.Y2AniK227M.IRES.BFP
Plasmid#45579PurposeExpresses K227M nickase variant of I-Ani I nuclease in mammalian cells with BFP trackerDepositorInsertcoY2 Ani K227M
UseLentiviralTagsHA, IRES BFP, and NLSExpressionMammalianMutationco indicates codon optimization for mammalian exp…PromoterSFFVAvailable SinceNov. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pEGFP(C3)-Nup153-C (880-1475)
Plasmid#64317PurposeGFP-tagged mammalian expression of C-terminal domain of Nup153DepositorInsertNup153 (NUP153 Human)
TagsGFPExpressionMammalianMutationC-terminal subunit (880-1475)PromoterCMVAvailable SinceApril 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
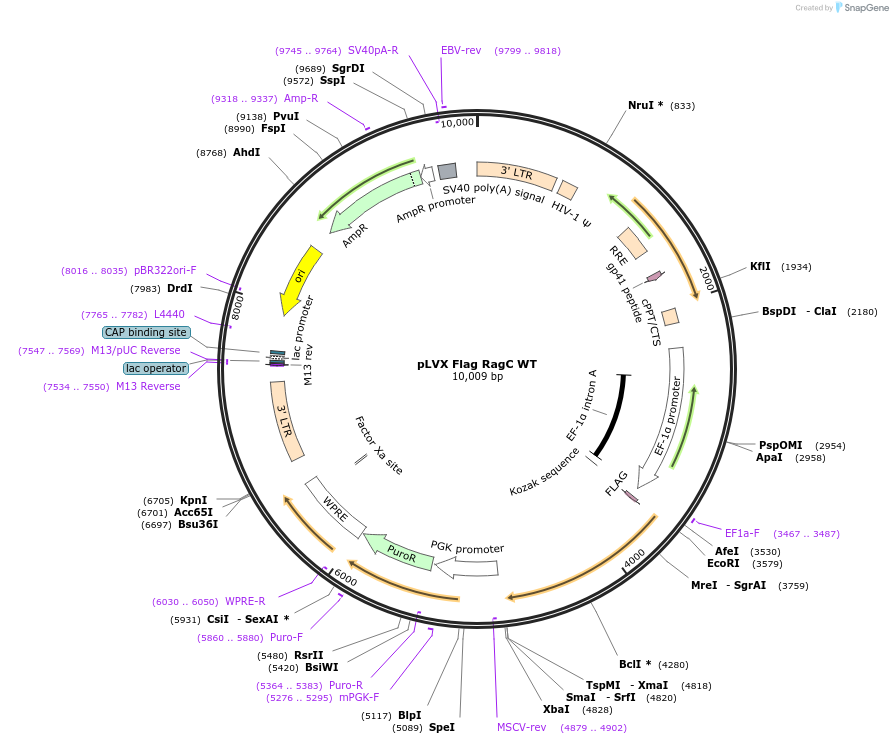
pLVX Flag RagC WT
Plasmid#112756Purposeexpress Flag-RagCDepositorAvailable SinceOct. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
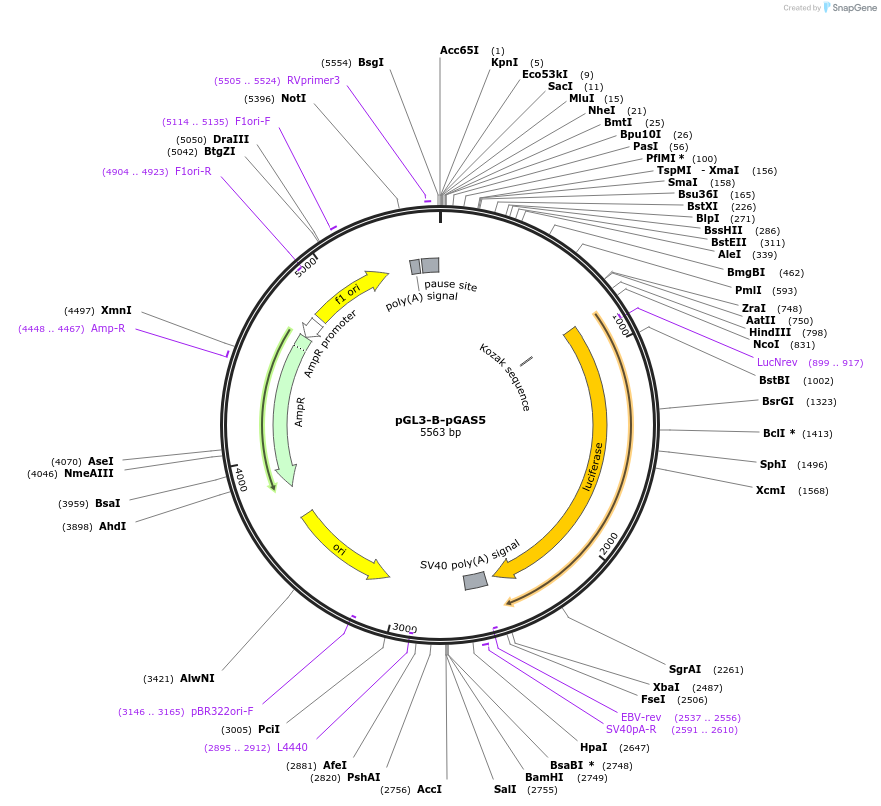
pGL3-B-pGAS5
Plasmid#46371DepositorAvailable SinceMay 19, 2014AvailabilityAcademic Institutions and Nonprofits only -
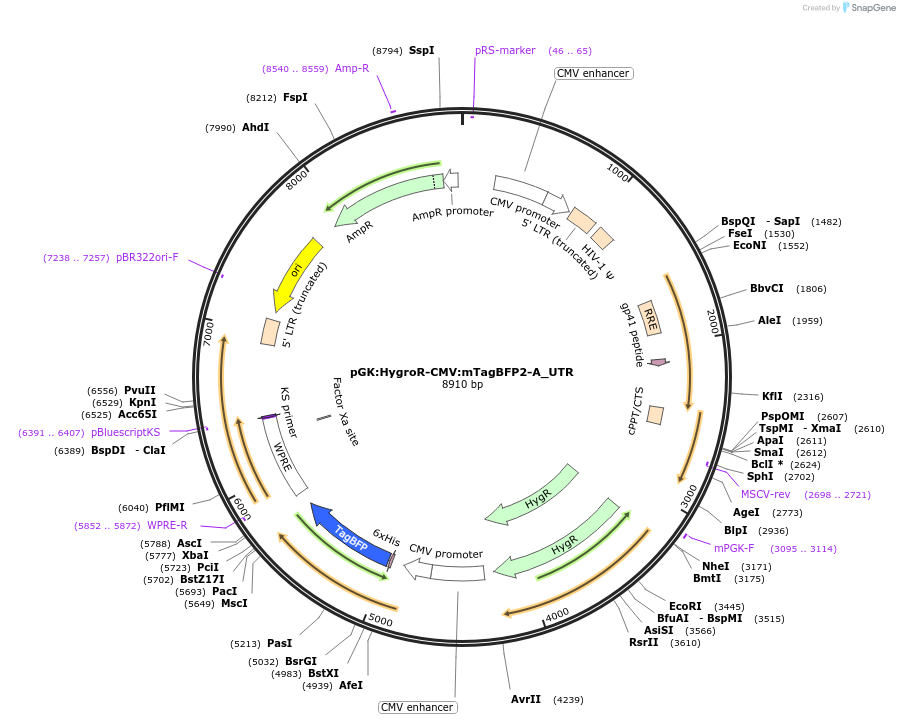
pGK:HygroR-CMV:mTagBFP2-A_UTR
Plasmid#68479PurposeLentivirus expressing mTagBFP2 with UTR sensor cassette and hygromycin resistanceDepositorInsertsHygromycin Resistance
mTagBFP2-A_UTR
UseLentiviralExpressionMammalianMutationshRNA sensor cassette "A" added to 3&#x…PromoterCMV and pGKAvailable SinceNov. 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
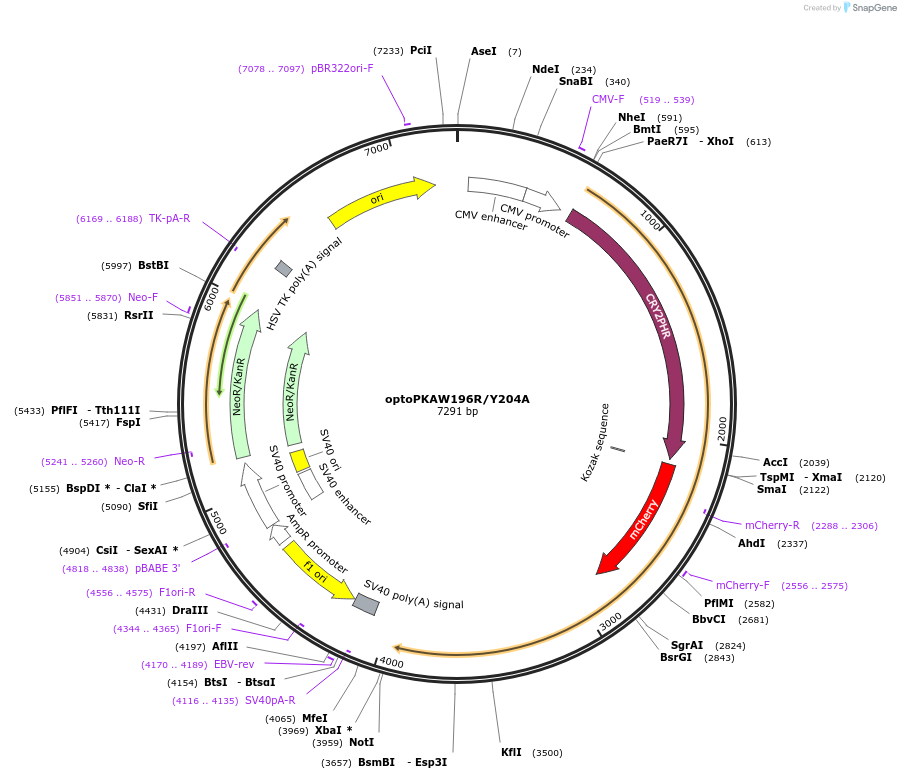
optoPKAW196R/Y204A
Plasmid#104546PurposeExpresses Cry2-mCherry-PKA W196R/Y204A in mammalian cells.DepositorInsertCry2-mCherry-PKA W196R/Y204A
TagsmCherryExpressionMammalianMutationPKA W196R and Y204APromoterCMVAvailable SinceMarch 12, 2018AvailabilityAcademic Institutions and Nonprofits only -
pSUPERIORpuro-shGAS5
Plasmid#46370DepositorAvailable SinceAug. 16, 2013AvailabilityAcademic Institutions and Nonprofits only -
pET14b PHAS-I
Plasmid#15679DepositorInsertPHAS-I (Eif4ebp1 Rat)
Tags6xHisExpressionBacterialMutationNucleotides 1-354 of rat PHAS-I plus 450bp of 3&#…Available SinceSept. 24, 2007AvailabilityAcademic Institutions and Nonprofits only -
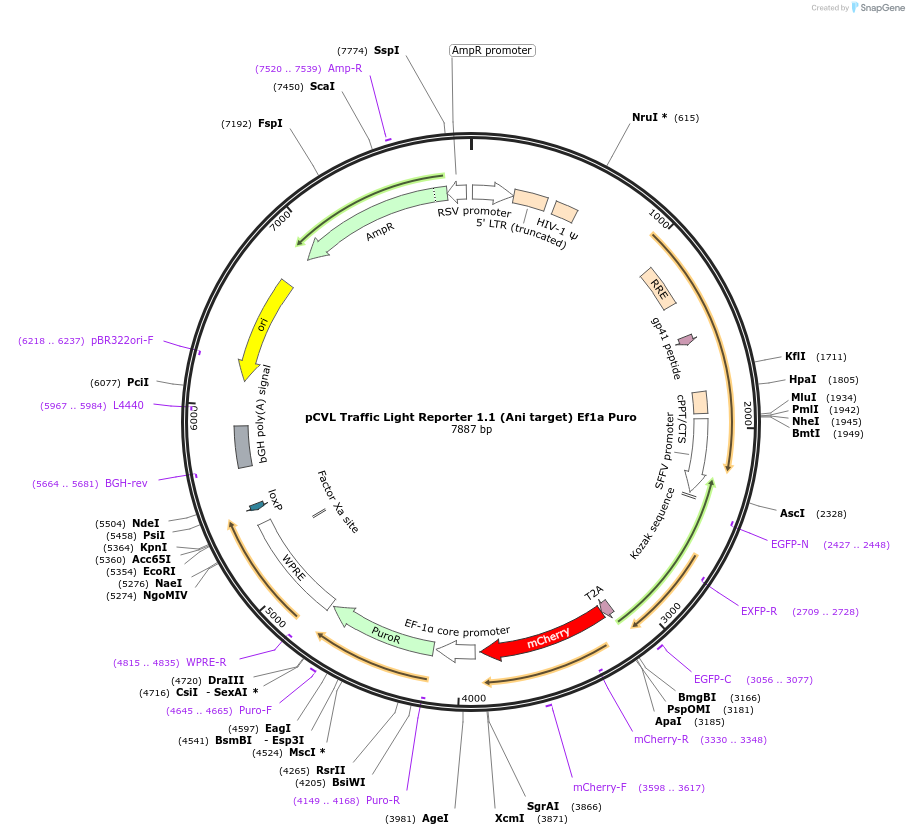
pCVL Traffic Light Reporter 1.1 (Ani target) Ef1a Puro
Plasmid#31480DepositorInsertTraffic Light Reporter 1.1 (ani target)
UseLentiviralMutationmCherry has M9S, M16L to reduce background mCherr…Available SinceAug. 22, 2011AvailabilityAcademic Institutions and Nonprofits only -
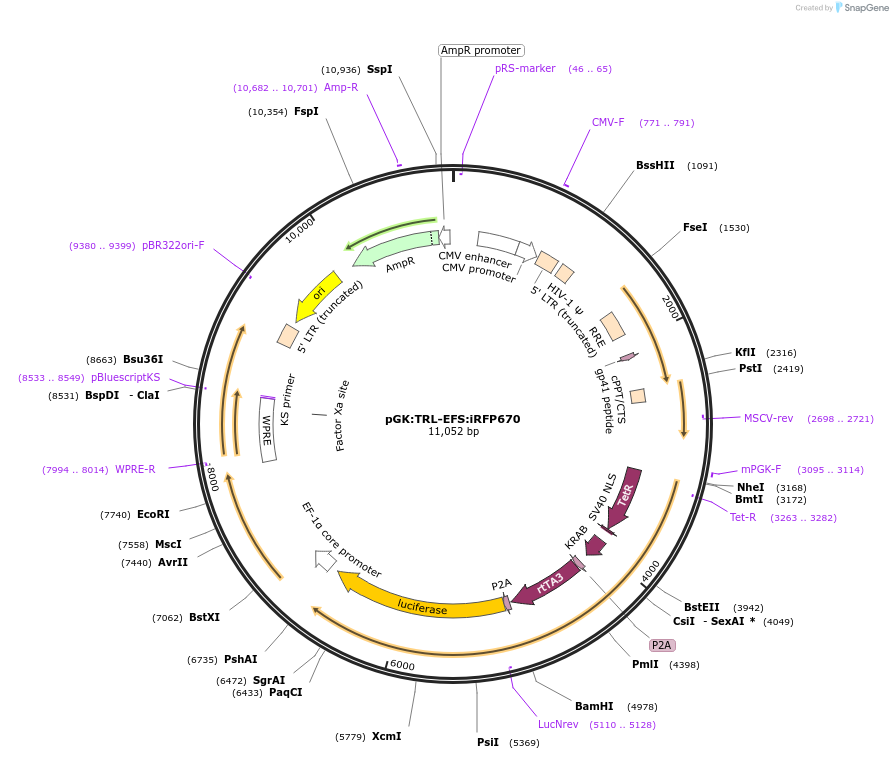
pGK:TRL-EFS:iRFP670
Plasmid#68475PurposeLentiviral plasmid expressing tTR-KRAB-rtTA3-Luc cassette and iRFP670DepositorInsertstTR-KRAB-2A-rtTA3-2A-Luciferase
iRFP670
UseLentiviral and LuciferaseExpressionMammalianPromoterEFS and pGKAvailable SinceFeb. 22, 2016AvailabilityAcademic Institutions and Nonprofits only