We narrowed to 9,280 results for: Pol
-
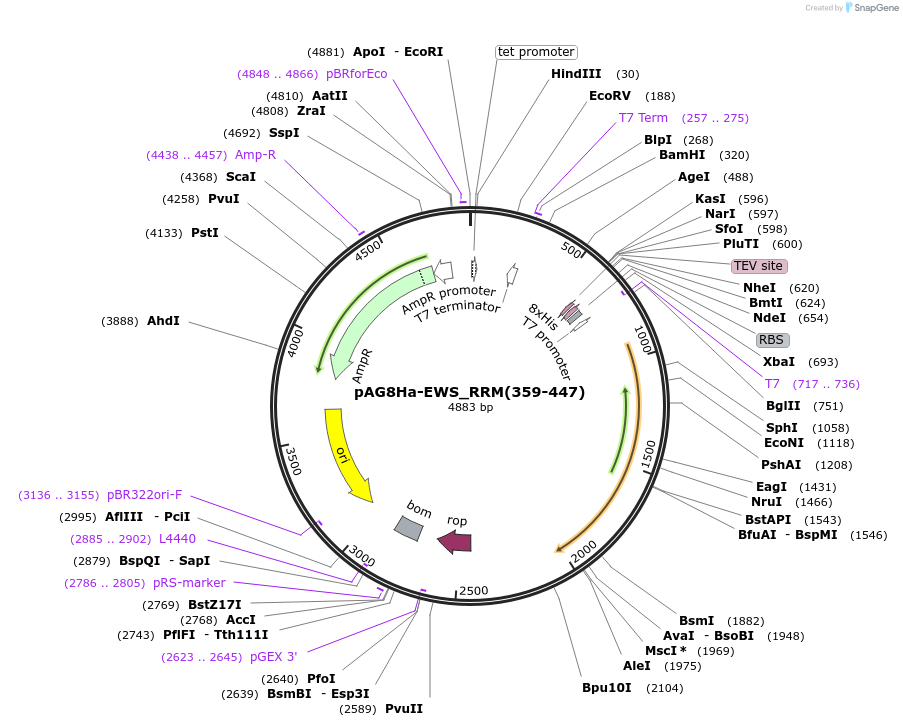
Plasmid#188045PurposeExpresses His8-EWS_RRM(359-447) with a TEV site for removal of the His8 tag.DepositorInsertEWSR1 (EWSR1 Human)
TagsHis8ExpressionBacterialMutationdeleted amino acids 1-358, 446-656PromoterT7Available SinceOct. 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
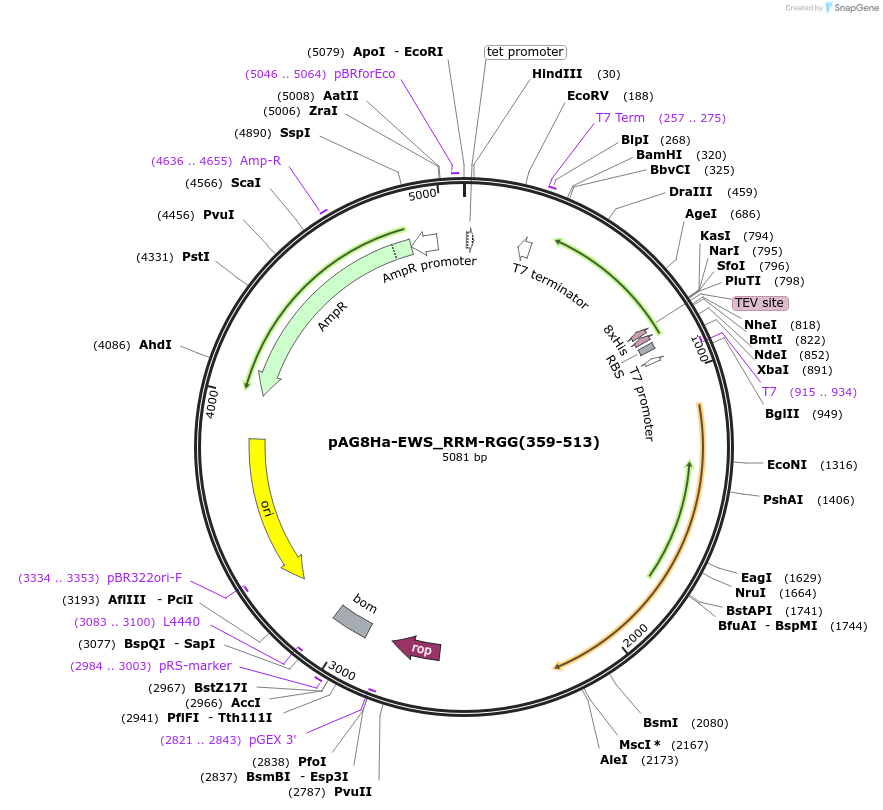
pAG8Ha-EWS_RRM-RGG(359-513)
Plasmid#188046PurposeExpresses His8-EWS_RRM-RGG(359-513) with a TEV site for removal of the His8 tag.DepositorInsertEWSR1 (EWSR1 Human)
TagsHis8ExpressionBacterialMutationdeleted amino acids 1-358, 514-656PromoterT7Available SinceOct. 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
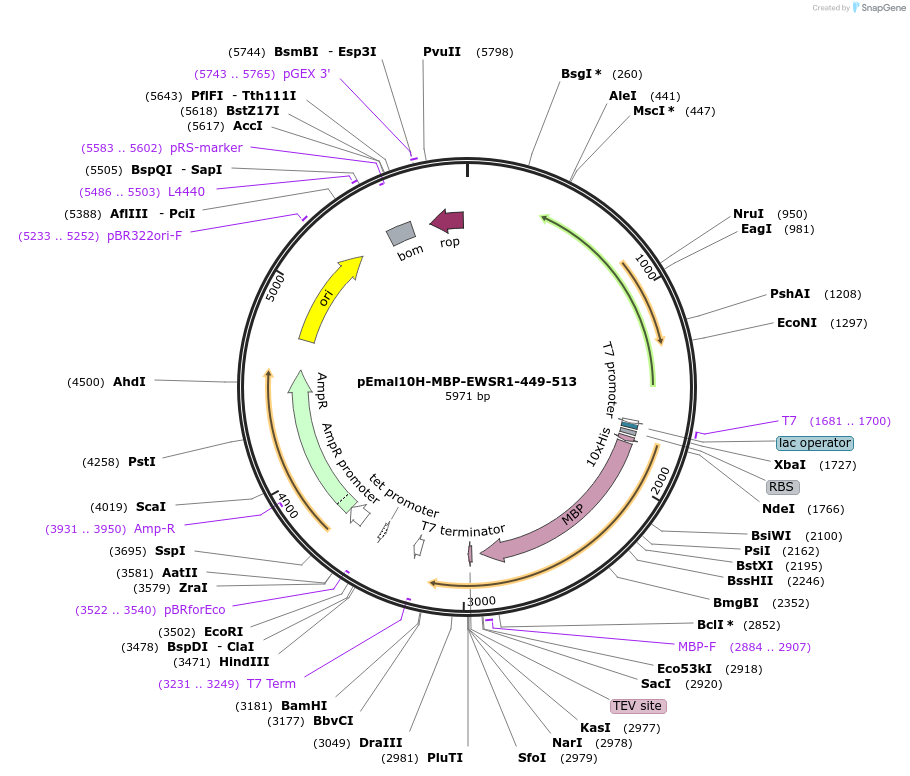
pEmal10H-MBP-EWSR1-449-513
Plasmid#199439PurposeExpresses RG2 (the second RG-rich region, 449-513) of EWSDepositorInsertEWSR1 (EWSR1 Human)
TagsHis10 and MBPExpressionBacterialMutationdeleted amino acids 1-448, 514-656PromoterT7Available SinceOct. 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
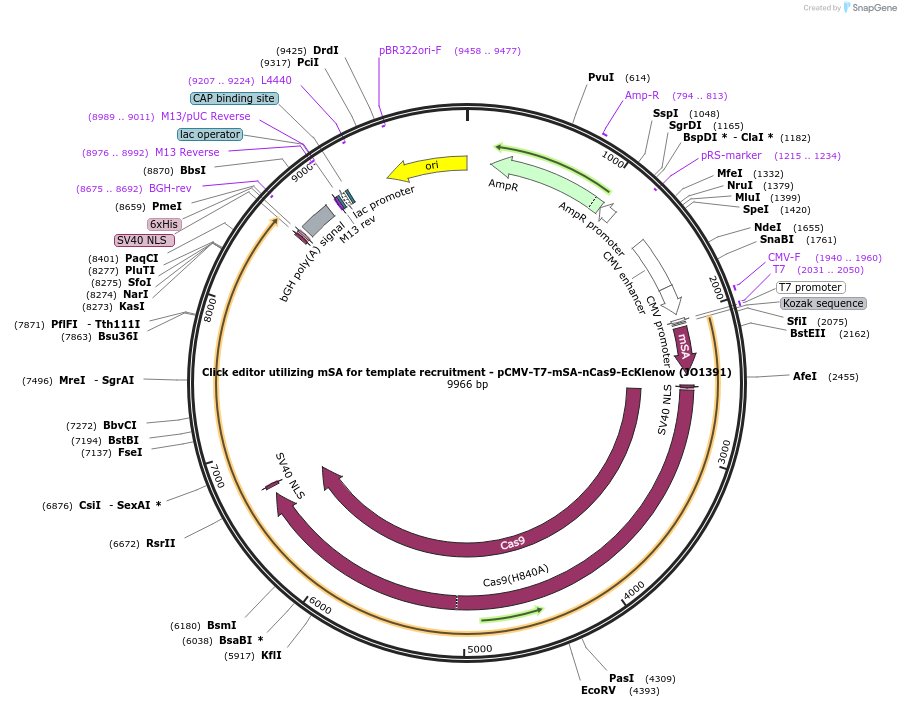
Click editor utilizing mSA for template recruitment - pCMV-T7-mSA-nCas9-EcKlenow (JO1391)
Plasmid#217802PurposeVariant CE1 construct with mSA for template recruitment, expressed from CMV or T7 promoters.DepositorInsertmSA-linker-SV40NLS-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
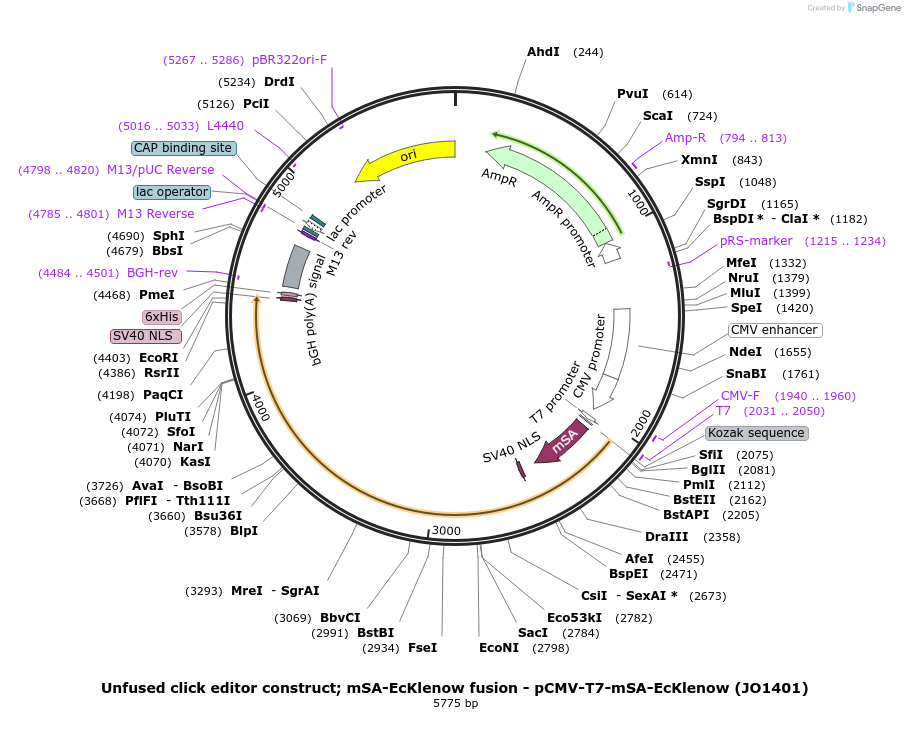
Unfused click editor construct; mSA-EcKlenow fusion - pCMV-T7-mSA-EcKlenow (JO1401)
Plasmid#217806PurposeUnfused click editor construct expressing mSA-EcKlenow, expressed from CMV or T7 promoters.DepositorInsertmSA-linker-SV40NLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationEcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
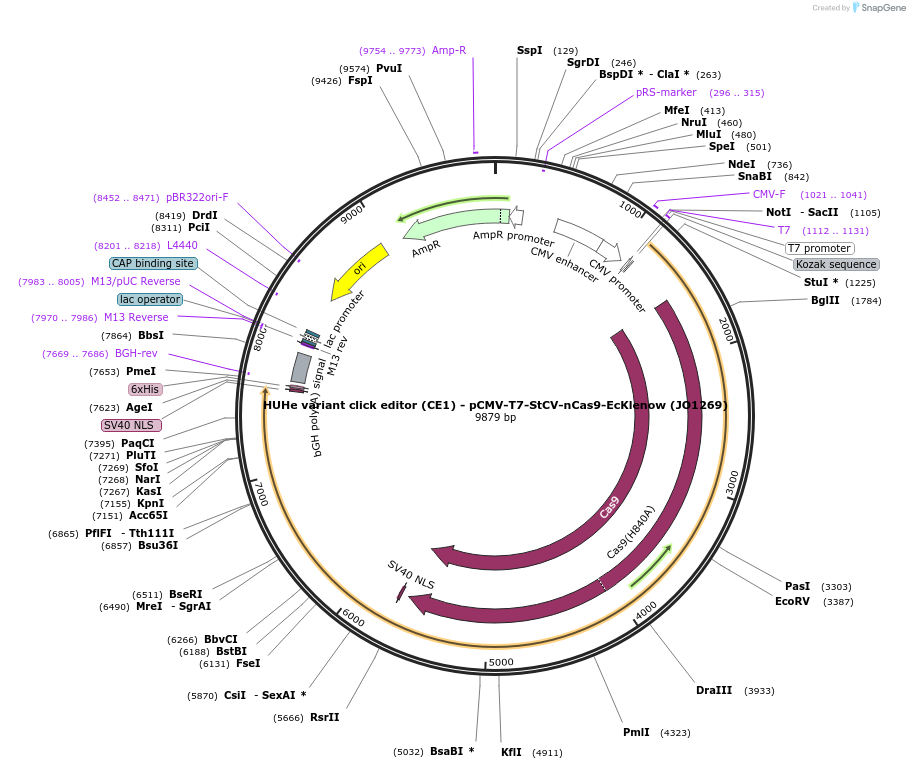
HUHe variant click editor (CE1) - pCMV-T7-StCV-nCas9-EcKlenow (JO1269)
Plasmid#217790PurposeVariant CE1 construct with StCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertStCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
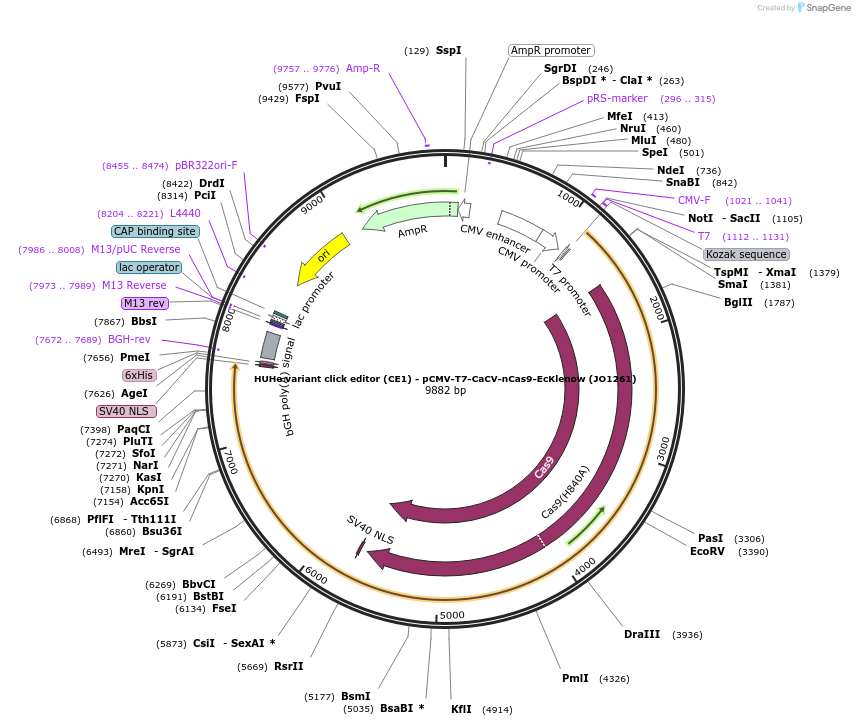
HUHe variant click editor (CE1) - pCMV-T7-CaCV-nCas9-EcKlenow (JO1261)
Plasmid#217791PurposeVariant CE1 construct with CaCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertCaCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
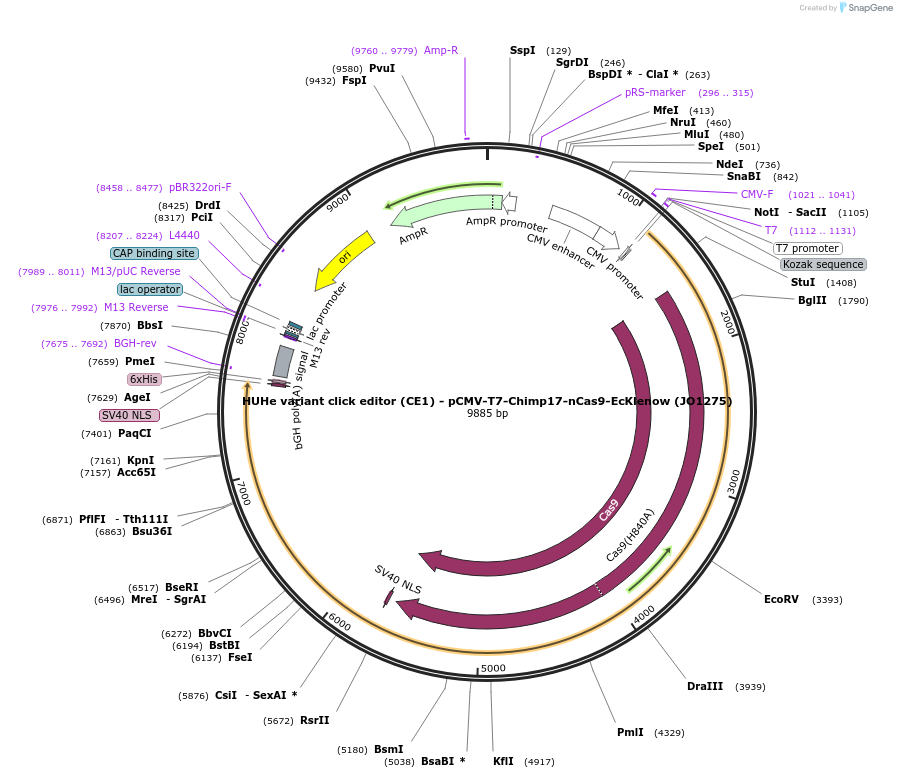
HUHe variant click editor (CE1) - pCMV-T7-Chimp17-nCas9-EcKlenow (JO1275)
Plasmid#217792PurposeVariant CE1 construct with Chim HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertChimp17-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
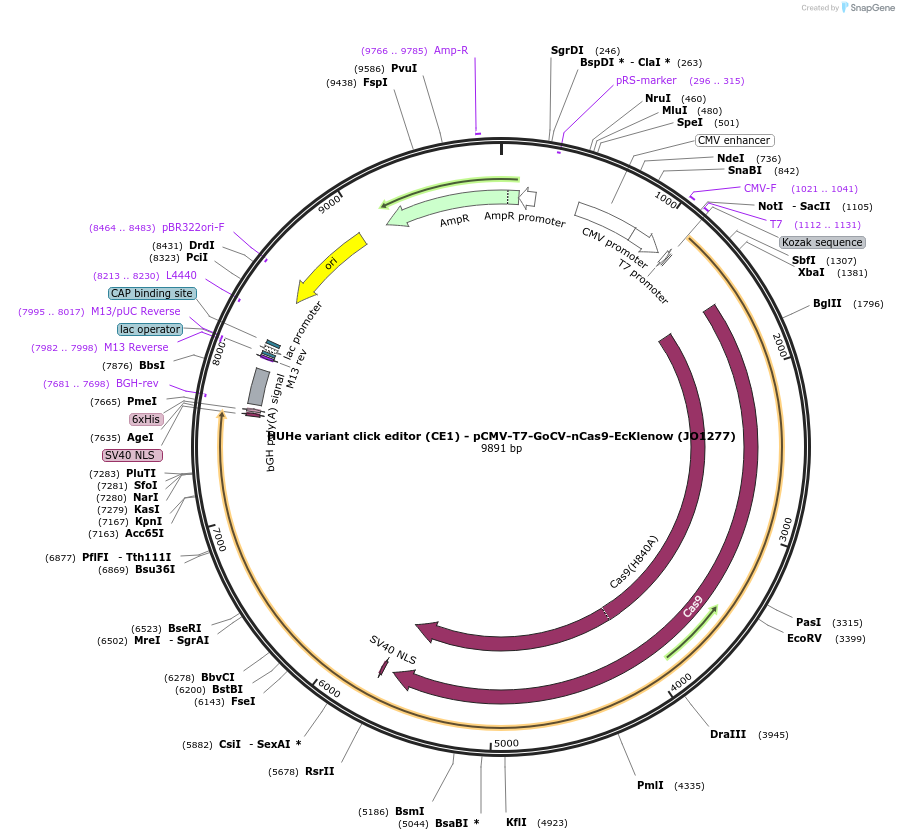
HUHe variant click editor (CE1) - pCMV-T7-GoCV-nCas9-EcKlenow (JO1277)
Plasmid#217793PurposeVariant CE1 construct with GoCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertGoCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
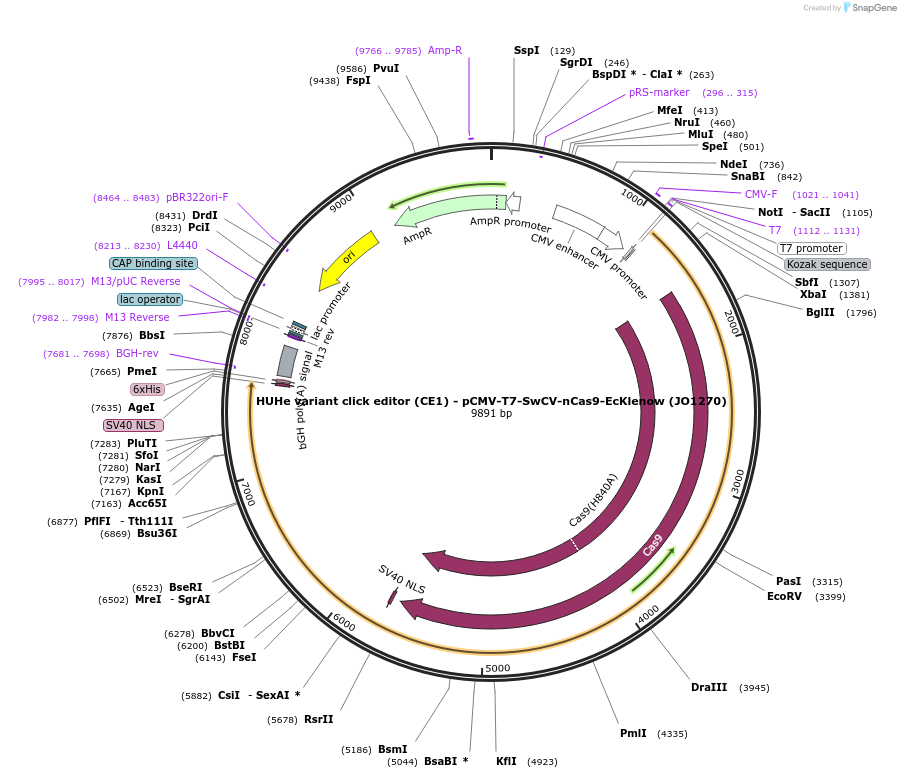
HUHe variant click editor (CE1) - pCMV-T7-SwCV-nCas9-EcKlenow (JO1270)
Plasmid#217794PurposeVariant CE1 construct with SwCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertSwCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
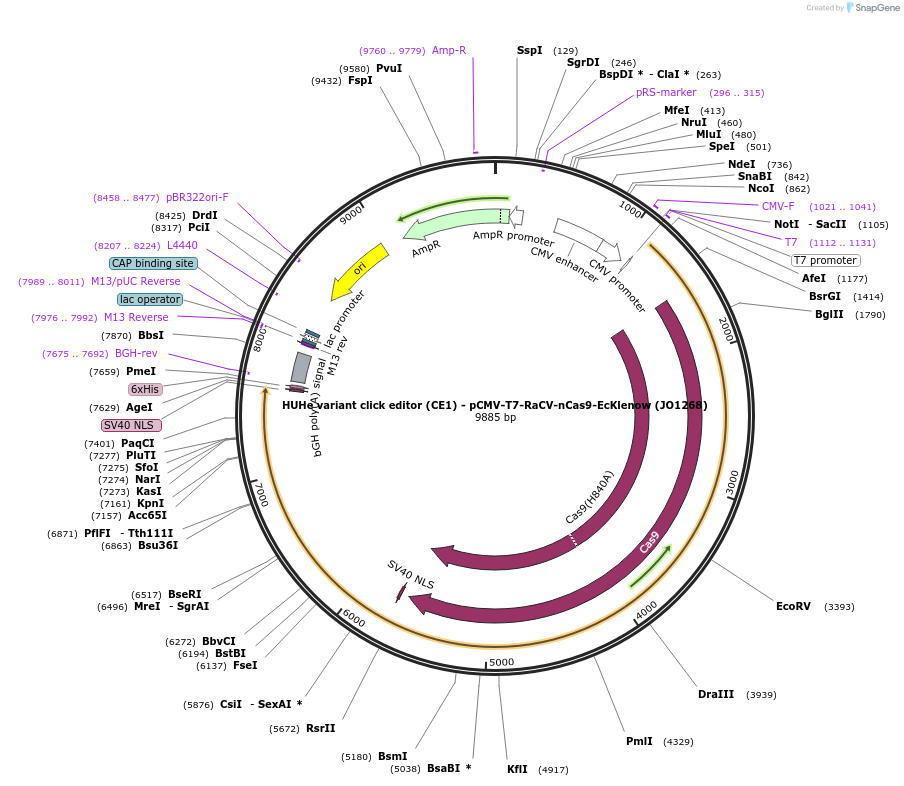
HUHe variant click editor (CE1) - pCMV-T7-RaCV-nCas9-EcKlenow (JO1268)
Plasmid#217795PurposeVariant CE1 construct with RaCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertRaCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
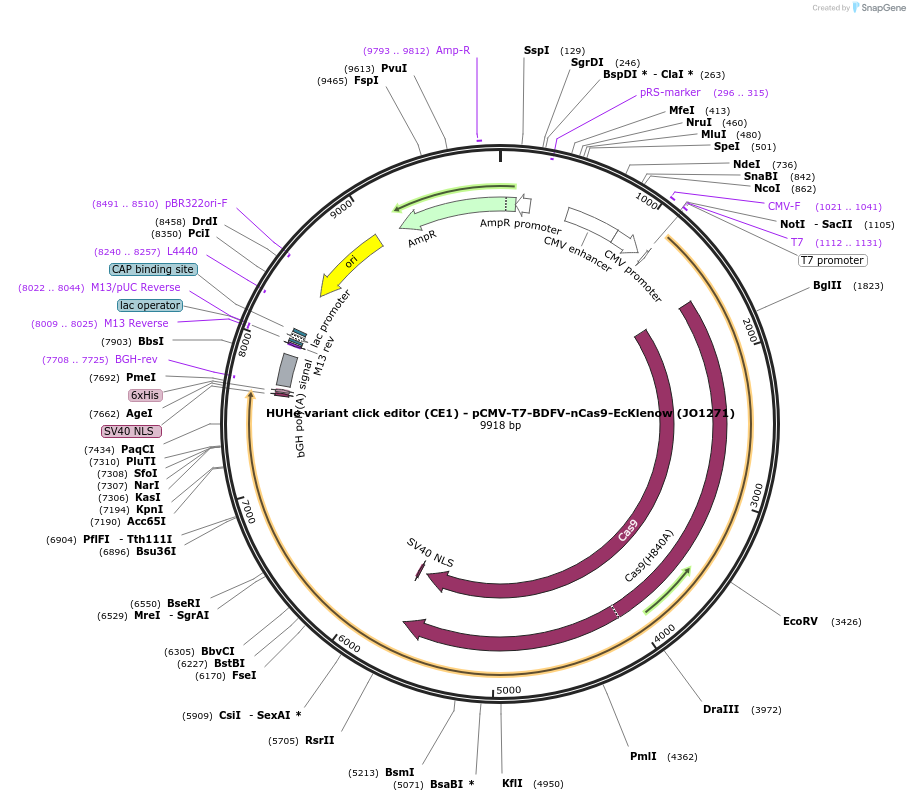
HUHe variant click editor (CE1) - pCMV-T7-BDFV-nCas9-EcKlenow (JO1271)
Plasmid#217785PurposeVariant CE1 construct with BDFV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertBDFV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
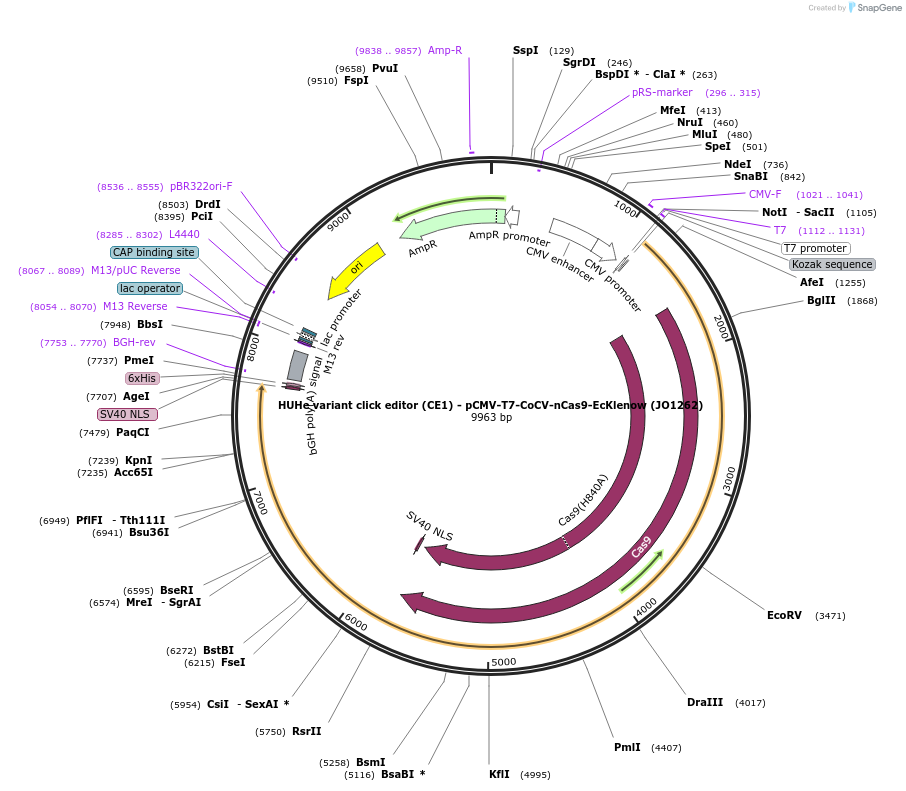
HUHe variant click editor (CE1) - pCMV-T7-CoCV-nCas9-EcKlenow (JO1262)
Plasmid#217786PurposeVariant CE1 construct with CoCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertCoCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
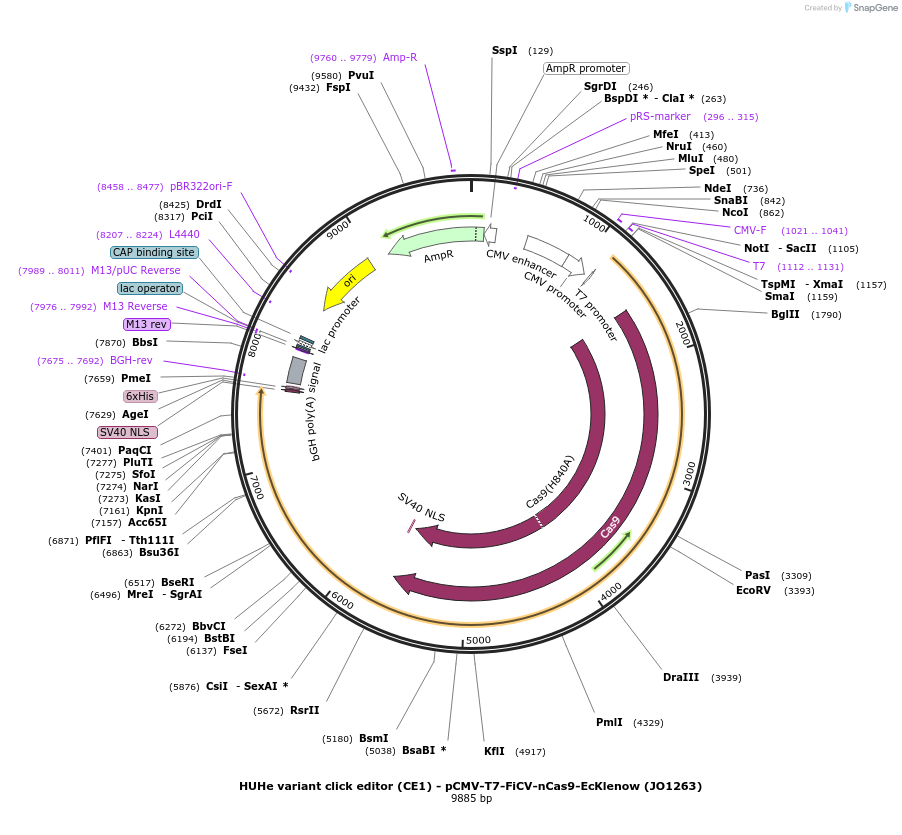
HUHe variant click editor (CE1) - pCMV-T7-FiCV-nCas9-EcKlenow (JO1263)
Plasmid#217787PurposeVariant CE1 construct with FiCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertFiCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
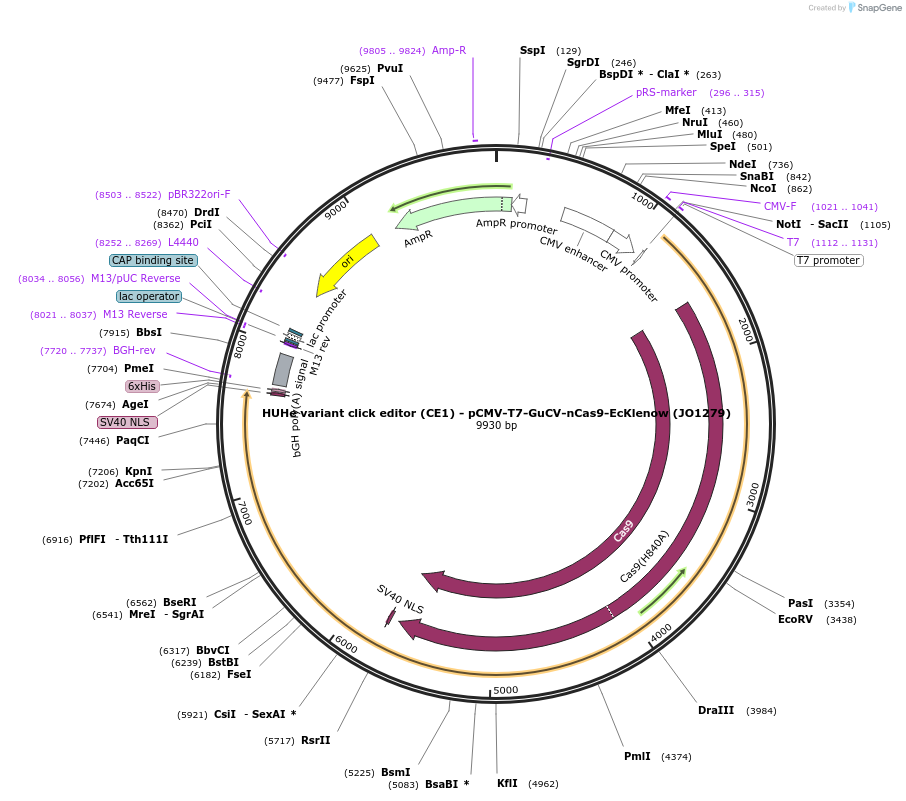
HUHe variant click editor (CE1) - pCMV-T7-GuCV-nCas9-EcKlenow (JO1279)
Plasmid#217788PurposeVariant CE1 construct with GuCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertGuCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
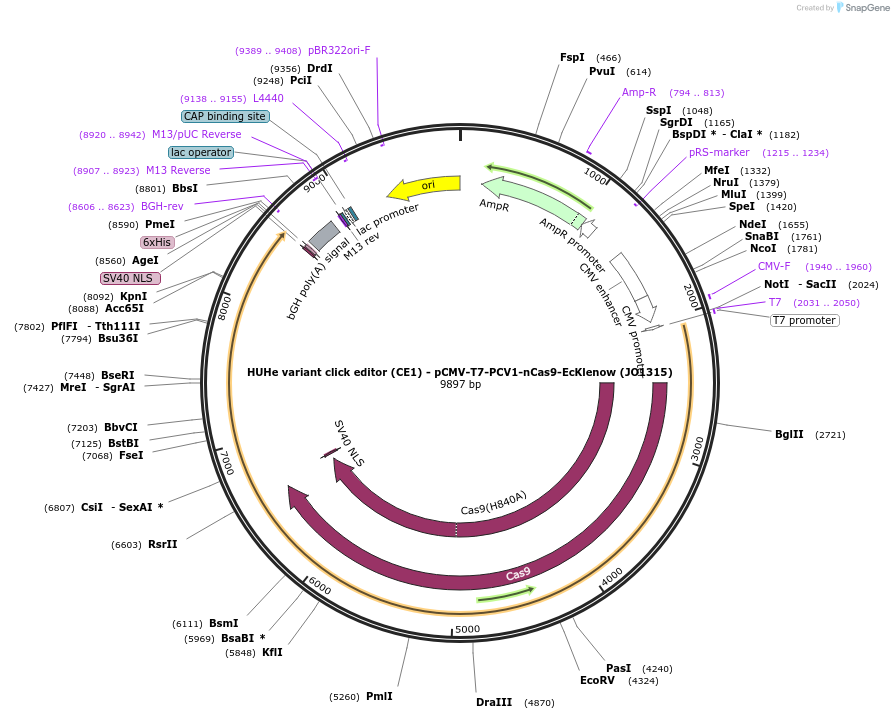
HUHe variant click editor (CE1) - pCMV-T7-PCV1-nCas9-EcKlenow (JO1315)
Plasmid#217789PurposeVariant CE1 construct with PCV1 HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertPCV1-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
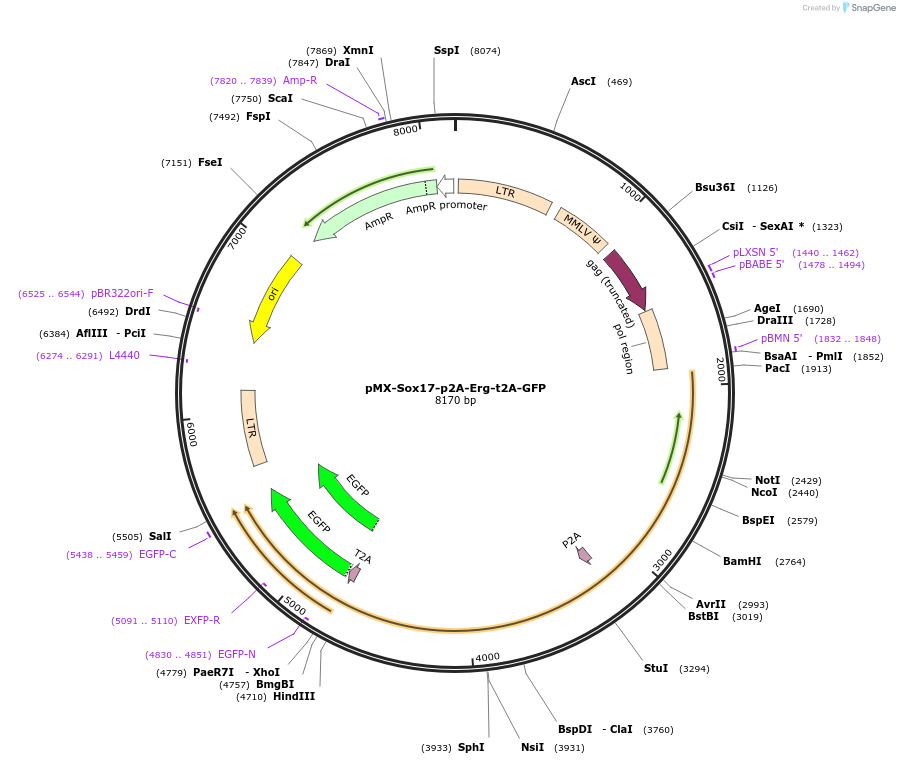
pMX-Sox17-p2A-Erg-t2A-GFP
Plasmid#222496Purposemouse fibroblast-to-endothelial cell direct reprogrammingDepositorAvailable SinceAug. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
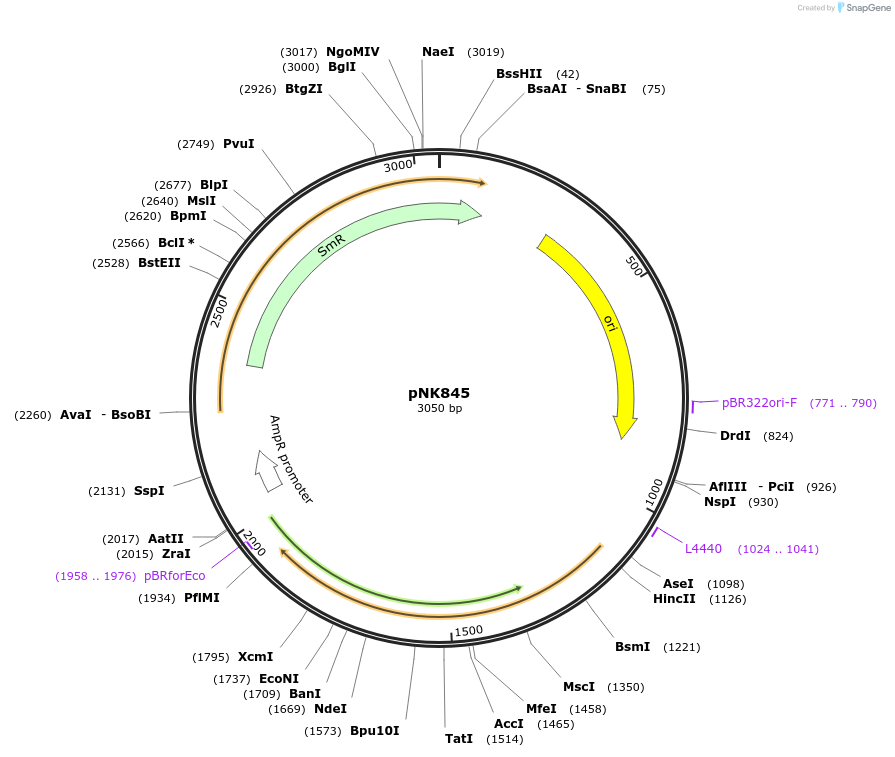
pNK845
Plasmid#219746PurposeMoClo-compatible Level 0 promoterless vector encoding mutant of Neonothopanus nambi luciferase nnLuz_v4 codon-optimised for expression in Pichia pastoris, Homo sapiensDepositorInsertmutant of fungal luciferase
UseLuciferase and Synthetic BiologyMutationI3S, N4T, F11L, I63T, T99P, T192S, A199PAvailable SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
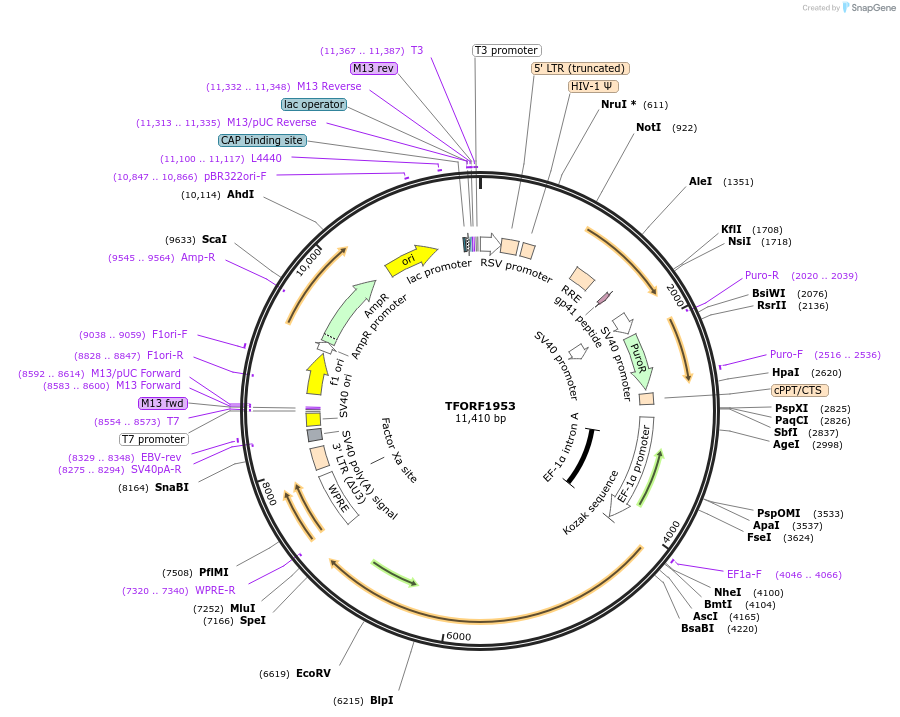
TFORF1953
Plasmid#142897PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceMarch 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
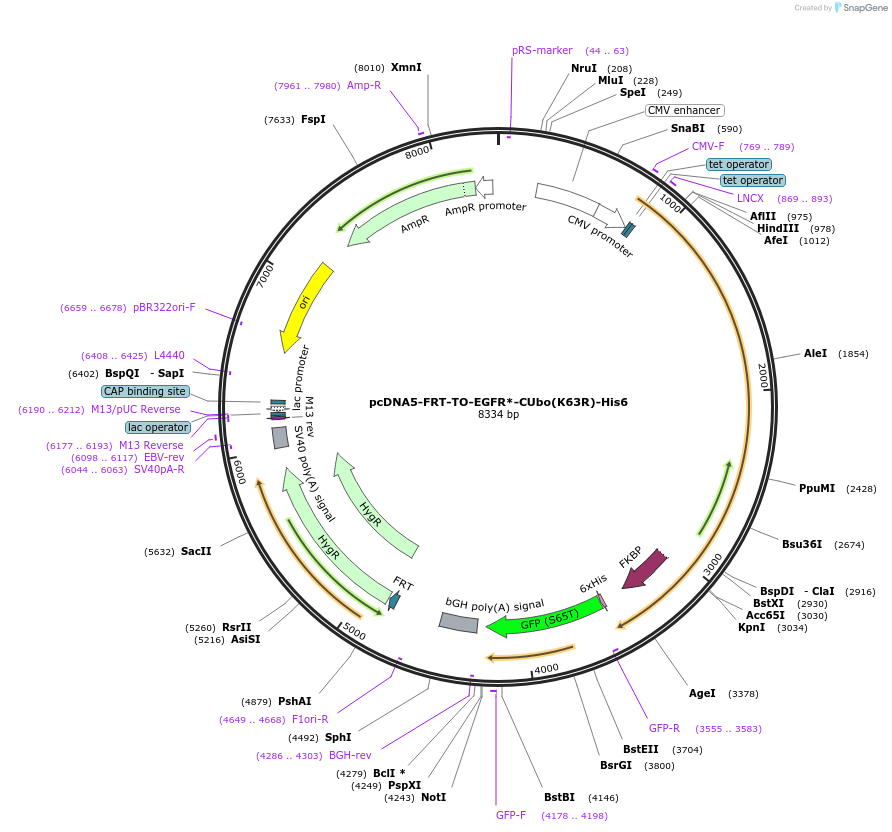
pcDNA5-FRT-TO-EGFR*-CUbo(K63R)-His6
Plasmid#212820PurposeConstitutive or doxycycline-inducible expression of EGFR*-CUbo(K63R)-His6 in mammalian cellsDepositorAvailable SinceFeb. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
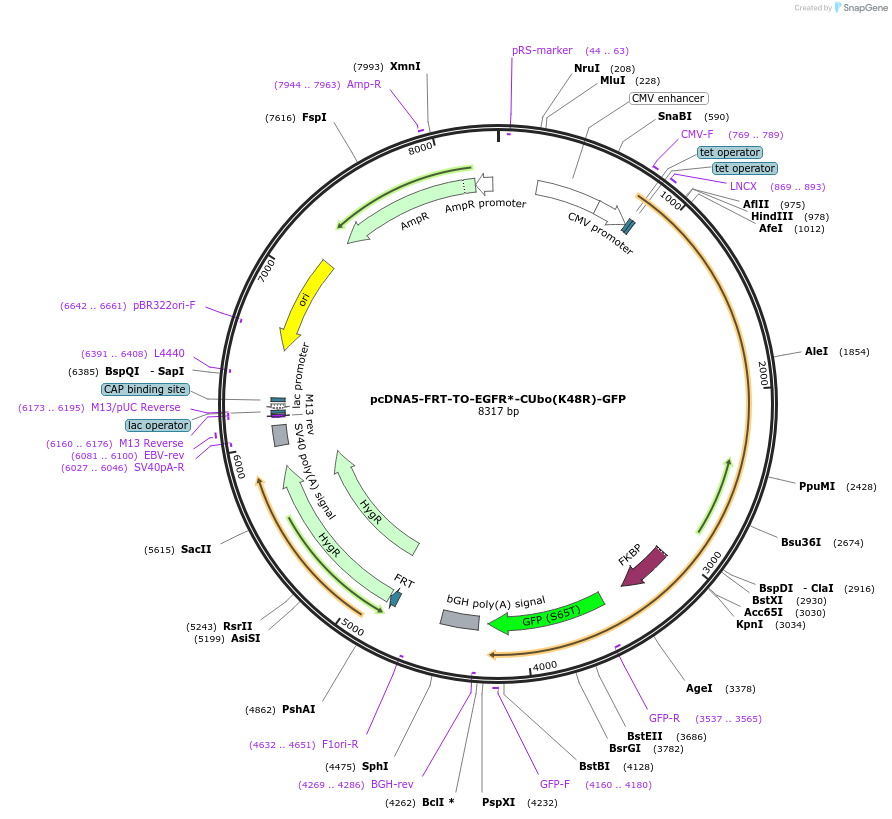
pcDNA5-FRT-TO-EGFR*-CUbo(K48R)-GFP
Plasmid#212816PurposeConstitutive or doxycycline-inducible expression of EGFR*-CUbo(K48R)-GFP in mammalian cellsDepositorAvailable SinceJan. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
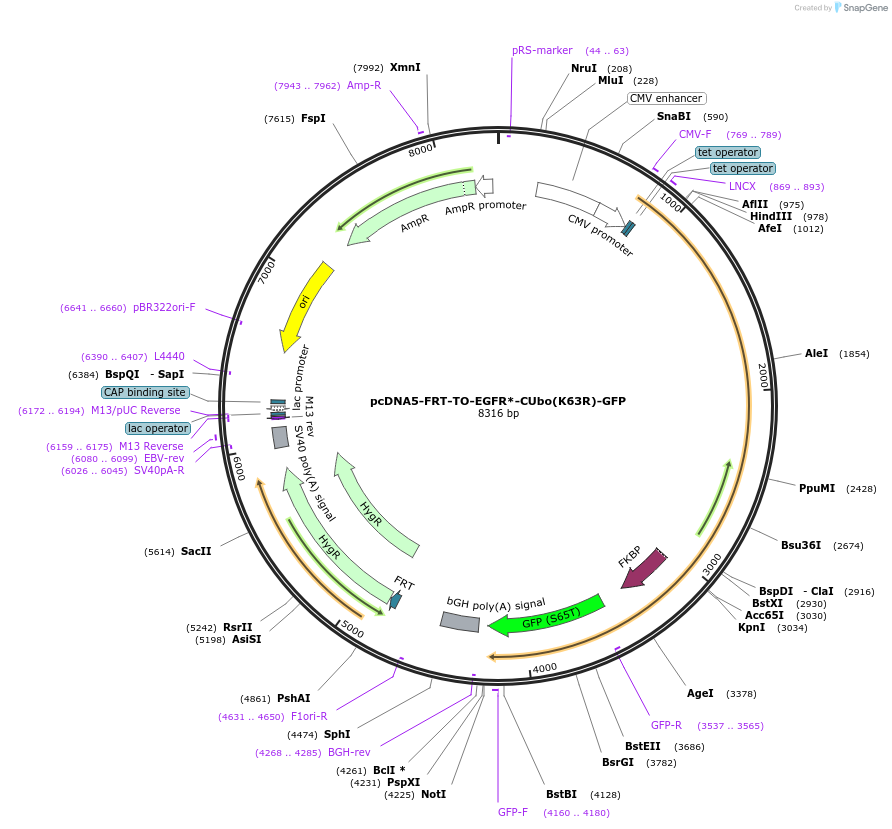
pcDNA5-FRT-TO-EGFR*-CUbo(K63R)-GFP
Plasmid#212817PurposeConstitutive or doxycycline-inducible expression of EGFR*-CUbo(K63R)-GFP in mammalian cellsDepositorAvailable SinceJan. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
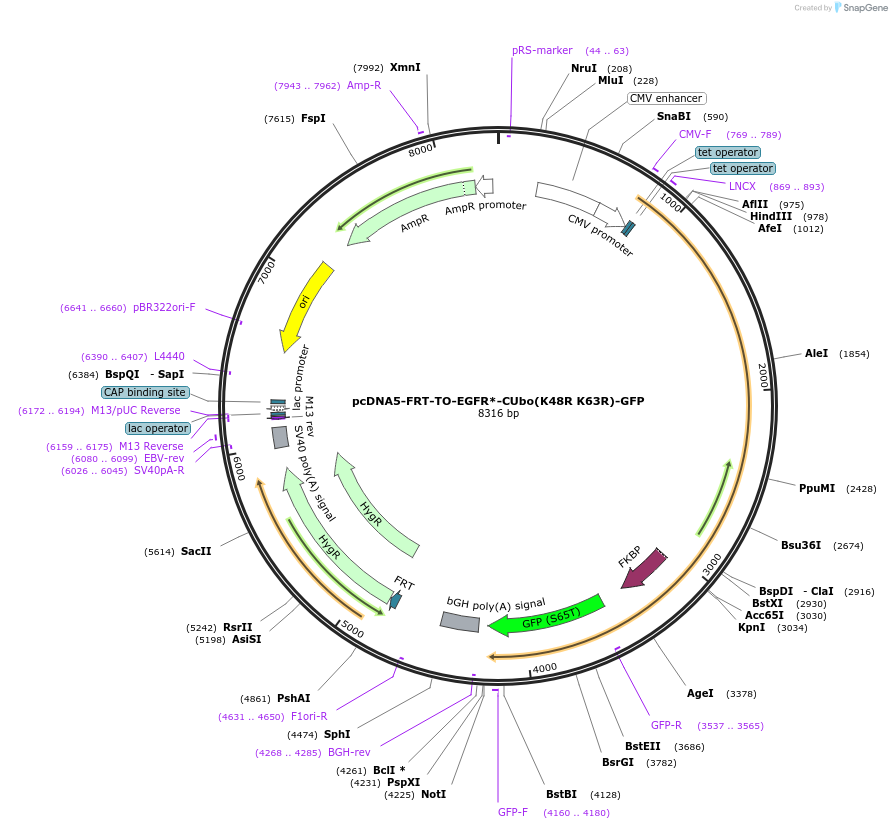
pcDNA5-FRT-TO-EGFR*-CUbo(K48R K63R)-GFP
Plasmid#212818PurposeConstitutive or doxycycline-inducible expression of EGFR*-CUbo(K48R K63R)-GFP in mammalian cellsDepositorAvailable SinceJan. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
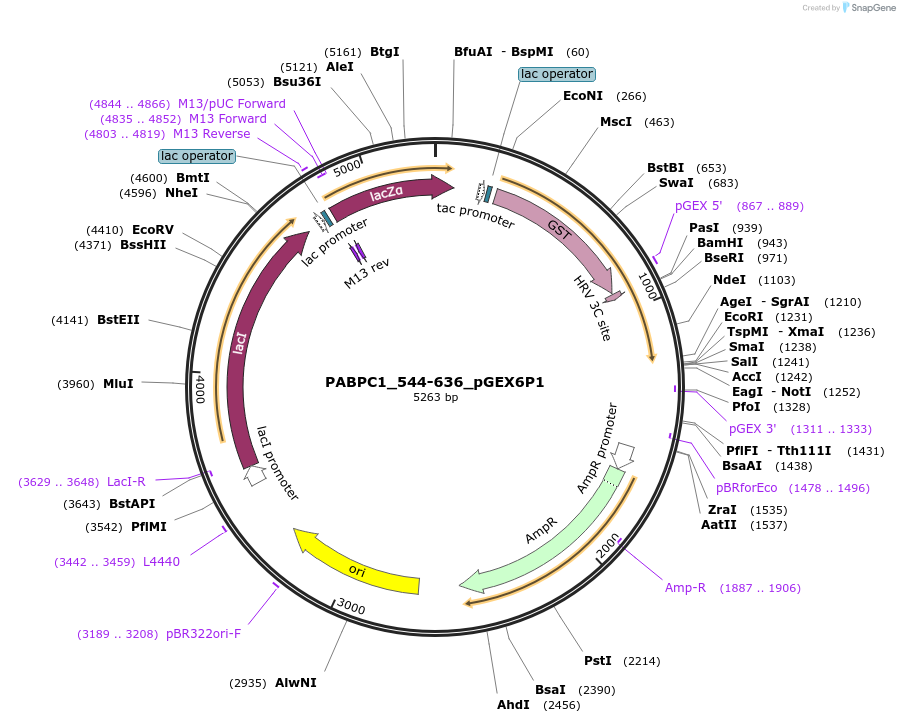
PABPC1_544-636_pGEX6P1
Plasmid#209789PurposeBacterial expression of PABPC1 Mlle domainDepositorInsertMlle domain of PABPC1 (PABPC1 Human)
TagsGSTExpressionBacterialMutationaa 544-636 onlyPromotertacAvailable SinceNov. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
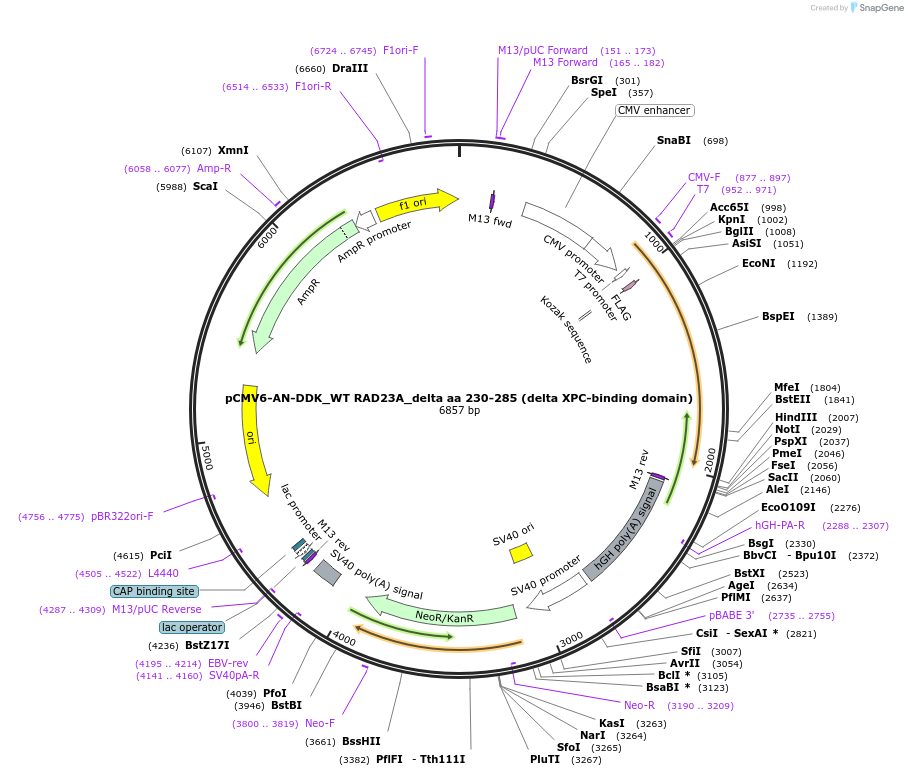
pCMV6-AN-DDK_WT RAD23A_delta aa 230-285 (delta XPC-binding domain)
Plasmid#201464PurposeExpresses a mutant form of human RAD23A that lacks the XPC-binding domain. Variant of plasmid pCMV6-AN-DDK_WT RAD23A.DepositorInsertUV excision repair protein RAD23 homolog A (RAD23A Human)
TagsFLAGExpressionMammalianMutationLacks residues 230-285Available SinceNov. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
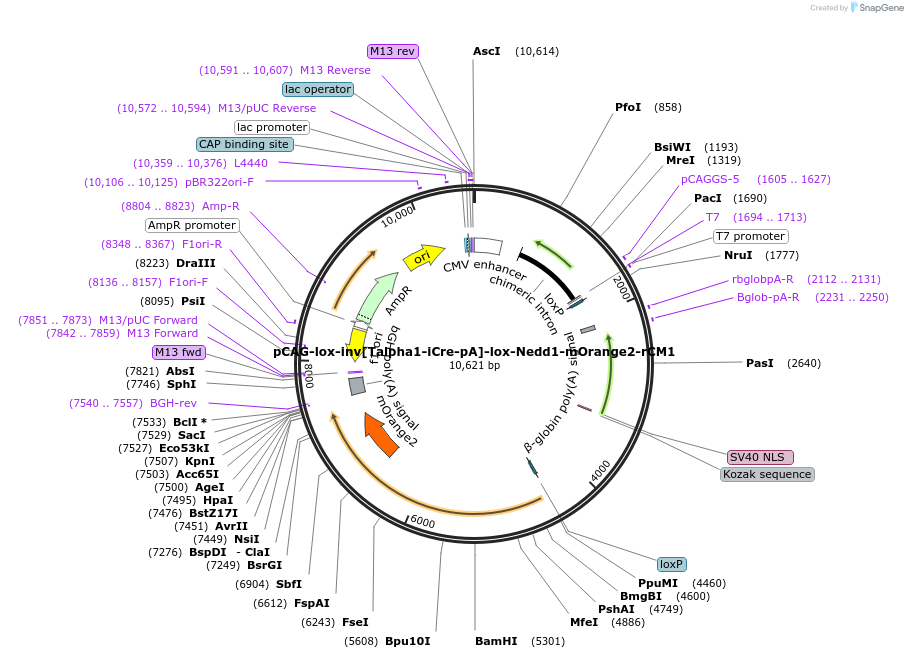
pCAG-lox-inv[Talpha1-iCre-pA]-lox-Nedd1-mOrange2-rCM1
Plasmid#196879PurposeNeuron-specific expression of Need1 fused to mOrange2 and rat (r) Centrosomin motif 1 (CM1). Nedd1 targets CM1 to the centrosome to activate gamma-TuRC.DepositorAvailable SinceSept. 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
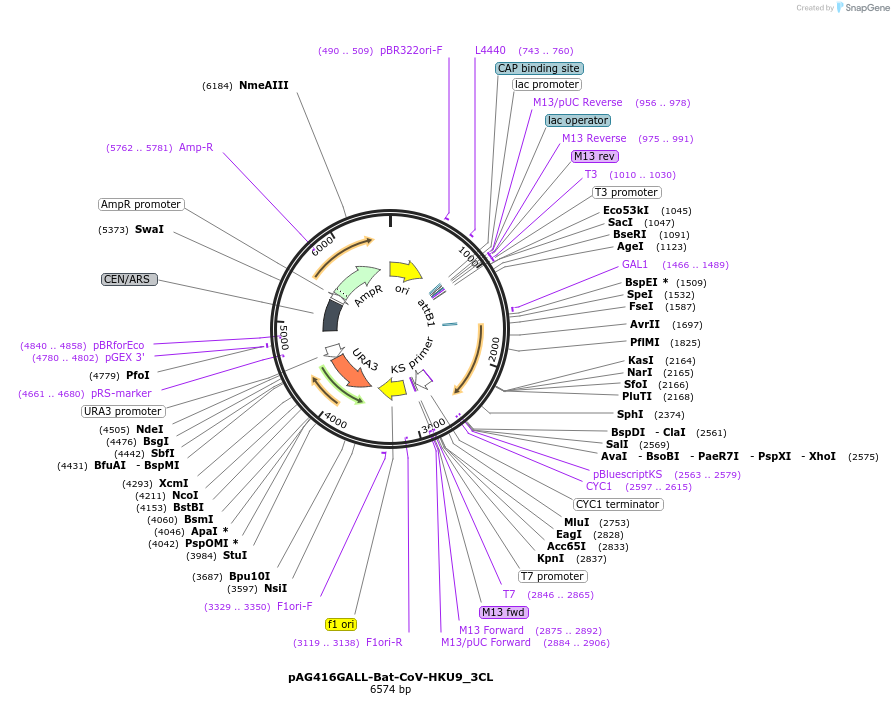
pAG416GALL-Bat-CoV-HKU9_3CL
Plasmid#203478PurposeExpresses Bat-CoV HKU9 3CL protease from a GALL promoter with a URA3 markerDepositorAvailable SinceJuly 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
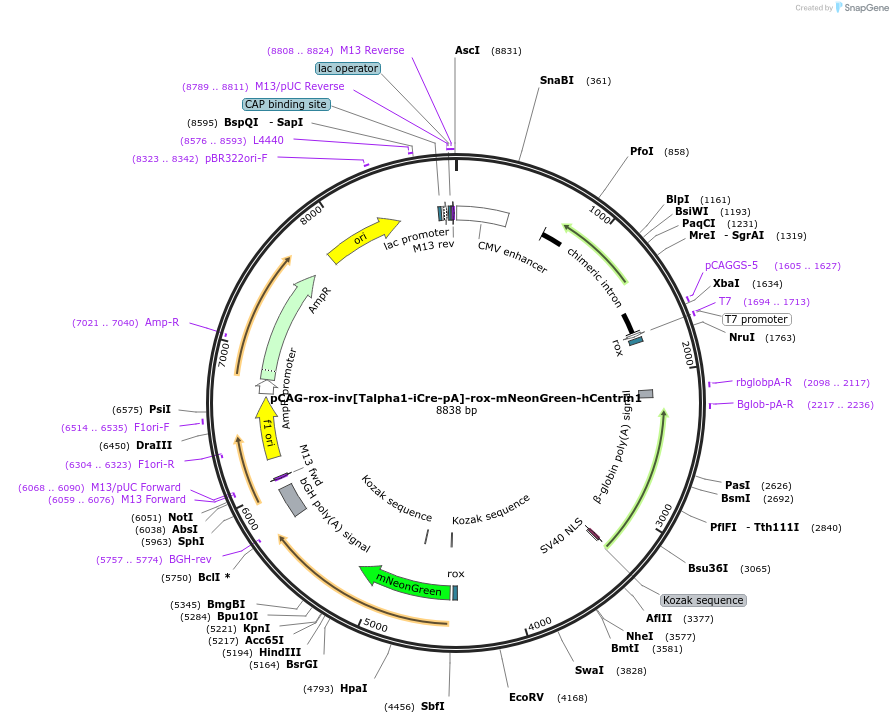
pCAG-rox-inv[Talpha1-iCre-pA]-rox-mNeonGreen-hCentrin1
Plasmid#196892PurposeNeuron-specific expression of human (h)Centrin fused to mNeonGreen. Used in combination with Talpha1-Dre-pA plasmidsDepositorAvailable SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
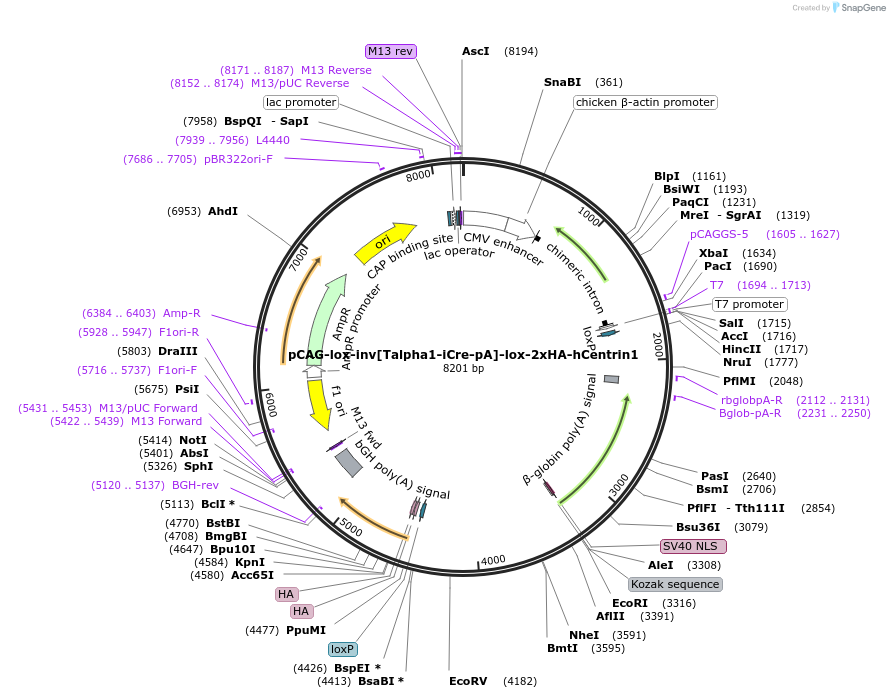
pCAG-lox-inv[Talpha1-iCre-pA]-lox-2xHA-hCentrin1
Plasmid#196891PurposeNeuron-specific expression of human (h)Centrin fused to 2xHA-tagsDepositorAvailable SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
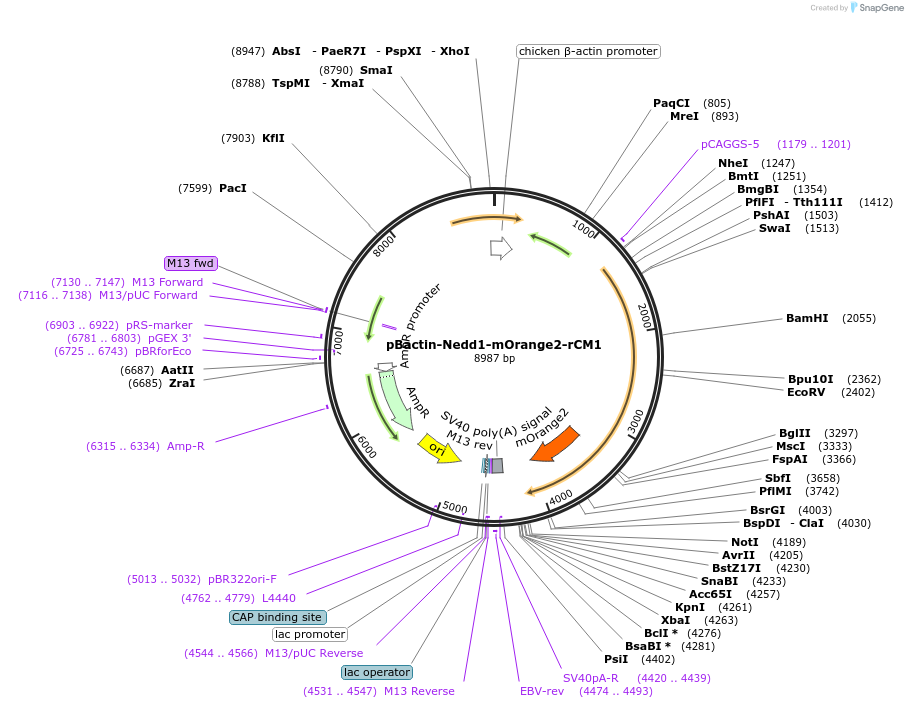
pBactin-Nedd1-mOrange2-rCM1
Plasmid#196861PurposeExpression of neural precursor cell expressed developmentally down-regulated 1 (Nedd1) fused to mOrange2 and rat (r) Centrosomin motif 1 (CM1). Nedd1 targets CM1 to the centrosome to activate gamma-TuRC.DepositorInsertNedd1-mOrange2-rCM1 (Nedd1 Rat)
ExpressionMammalianPromoterChicken bactin (plus Chicken bactin intron)Available SinceMay 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
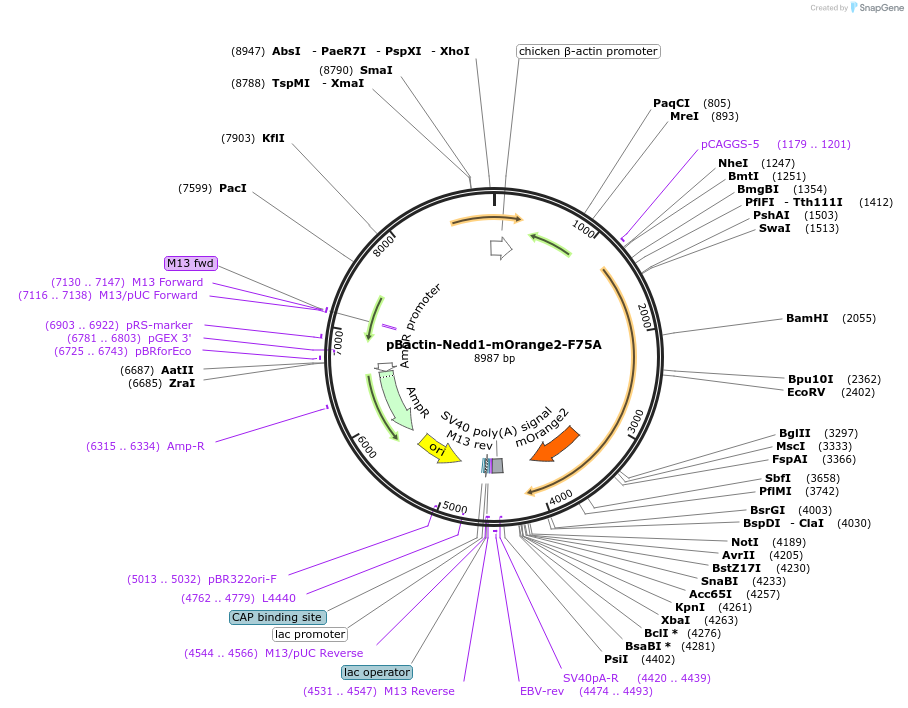
pBactin-Nedd1-mOrange2-F75A
Plasmid#196862PurposeExpression of Need1-rCM1 harboring a (F75A) mutation in the rat (r) Centrosomin motif 1 (CM1) and fused to mOrange2. F75A mutatuon renders CM1 incapable of binding and activating gamma-TuRCDepositorInsertNedd1-mOrange2-F75A (Nedd1 Rat)
ExpressionMammalianMutationF75A in the CM1 domainPromoterChicken bactin (plus Chicken bactin intron)Available SinceMay 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
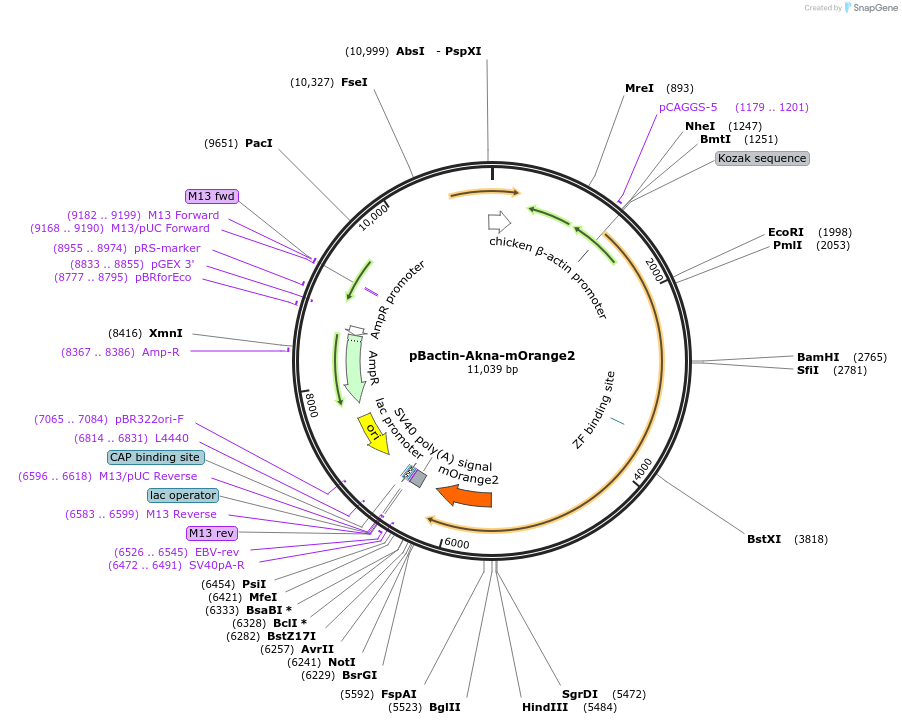
pBactin-Akna-mOrange2
Plasmid#196865PurposeExpression of the centrosomal protein Akna fused to mOrange2DepositorInsertAkna-mOrange2 (Akna Mouse)
ExpressionMammalianPromoterChicken bactin (plus Chicken bactin intron)Available SinceMay 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
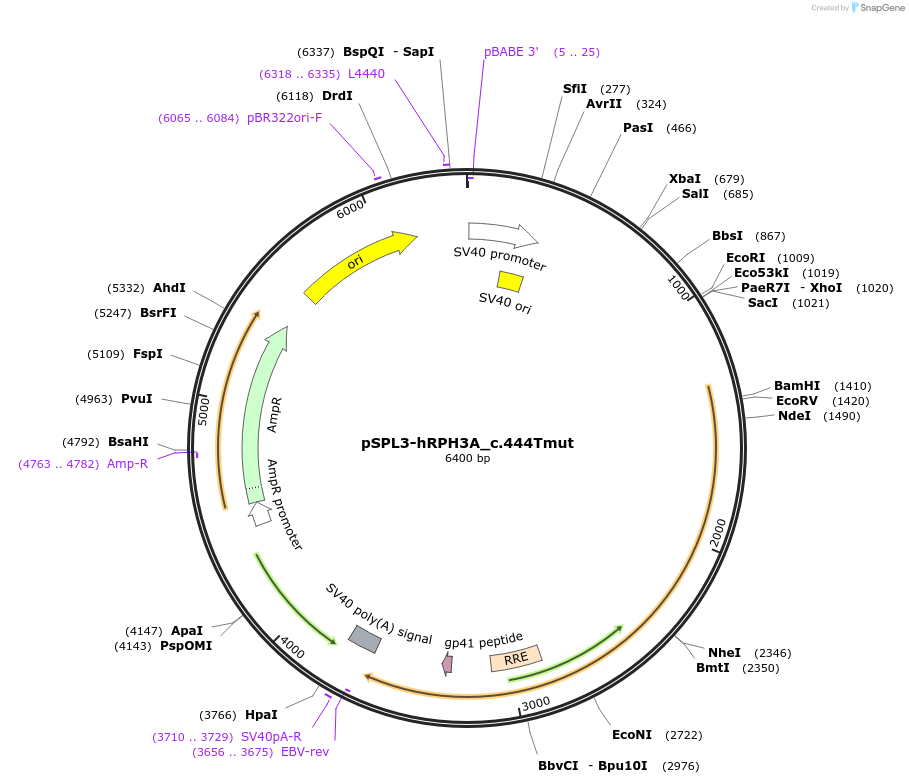
pSPL3-hRPH3A_c.444Tmut
Plasmid#190477Purposeminigene assayDepositorInsertRPH3A (NM_014954.3) c.1853 [exon 20 and part of surrounding introns only] (RPH3A Human)
ExpressionMammalianMutationc.444G>TPromoterSV40Available SinceSept. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
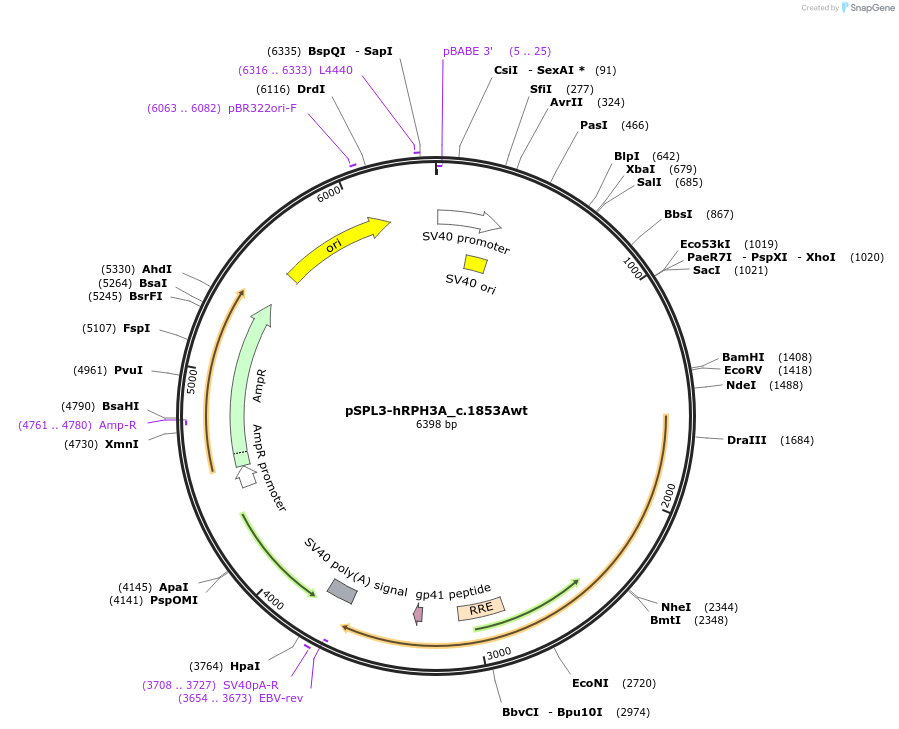
pSPL3-hRPH3A_c.1853Awt
Plasmid#190478Purposeminigene assayDepositorInsertRPH3A (NM_014954.3) c.1853 [exon 20 and part of surrounding introns only] (RPH3A Human)
ExpressionMammalianPromoterSV40Available SinceSept. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
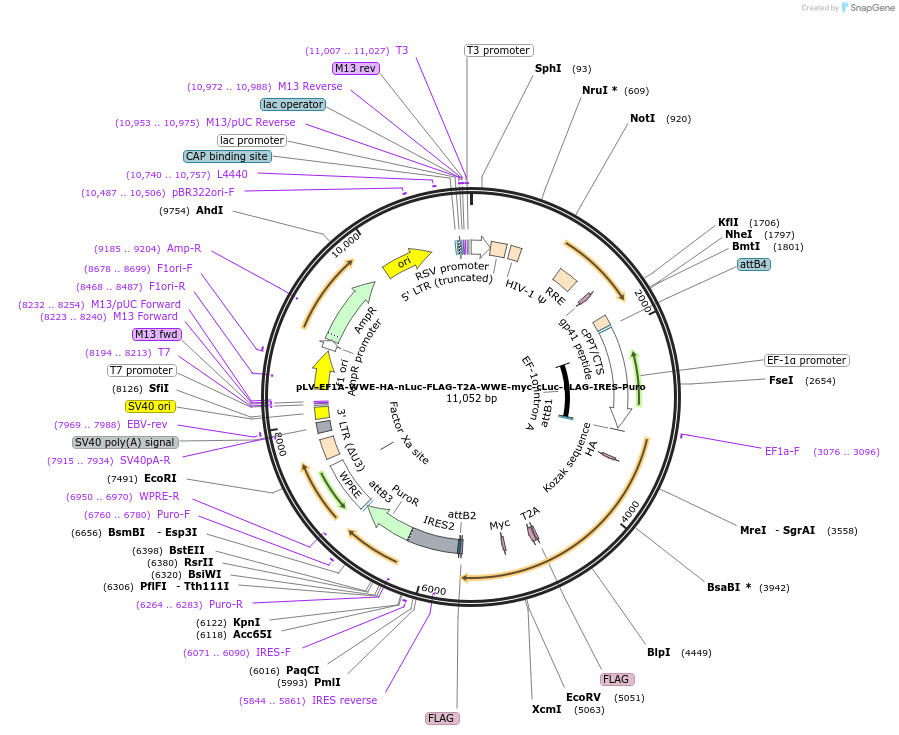
pLV-EF1A-WWE-HA-nLuc-FLAG-T2A-WWE-myc-cLuc-FLAG-IRES-Puro
Plasmid#187611PurposeSplit luciferase construct with nLuc fused to C-terminus of WWE domain, linked by T2A to cLuc fused to C-terminus of WWE domain, linked by IRES to puromycin resistance cassetteDepositorInsertsWWE domain with C-terminal nLuc
WWE domain with C-terminal cLuc
UseLentiviralTagsFLAG, HA, Split luciferase (cLuc), Split lucifera…ExpressionMammalianPromoterEF1AAvailable SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
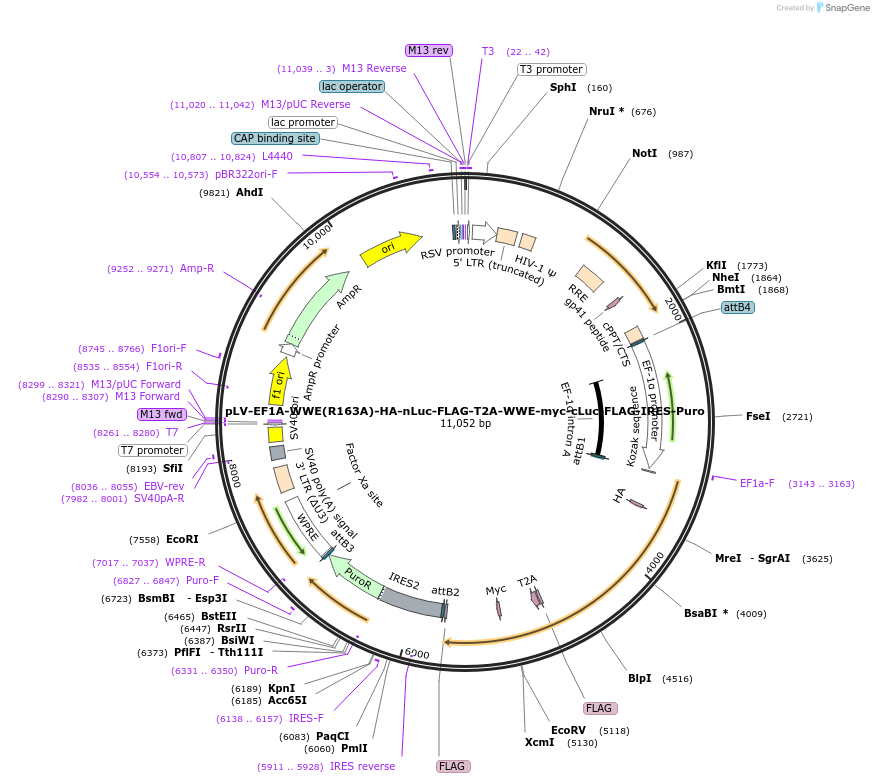
pLV-EF1A-WWE(R163A)-HA-nLuc-FLAG-T2A-WWE-myc-cLuc-FLAG-IRES-Puro
Plasmid#187612PurposeSplit luciferase construct with nLuc fused to C-terminus of WWE domain with Arg163Ala mutation, linked by T2A to cLuc fused to C-terminus of WWE domain, linked by IRES to puromycin resistance cassetteDepositorInsertsWWE domain (R163A) with C-terminal nLuc
WWE domain with C-terminal cLuc
UseLentiviralTagsFLAG, HA, Split luciferase (cLuc), Split lucifera…ExpressionMammalianMutationArg163AlaPromoterEF1AAvailable SinceSept. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
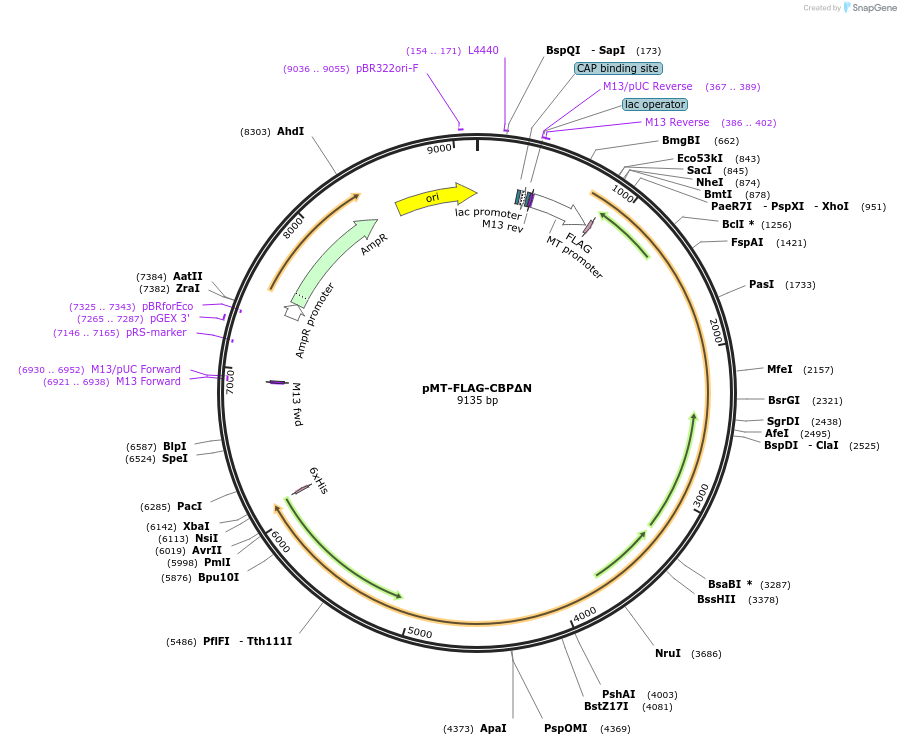
pMT-FLAG-CBPΔN
Plasmid#182840PurposeA 5.2 kb Drosophila CBP cDNA fragment (cut by Nhe I and Nsi I), encoding C-terminal residues 1476-3222, was inserted into the pMT-FLAG vector. Expression in S2 cells by CuSO4 induction.DepositorInsertCBP cDNA fragment (cut by Nhe I and Nsi I) (nej Fly)
TagsFLAG tagExpressionInsectPromoterMTAvailable SinceApril 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
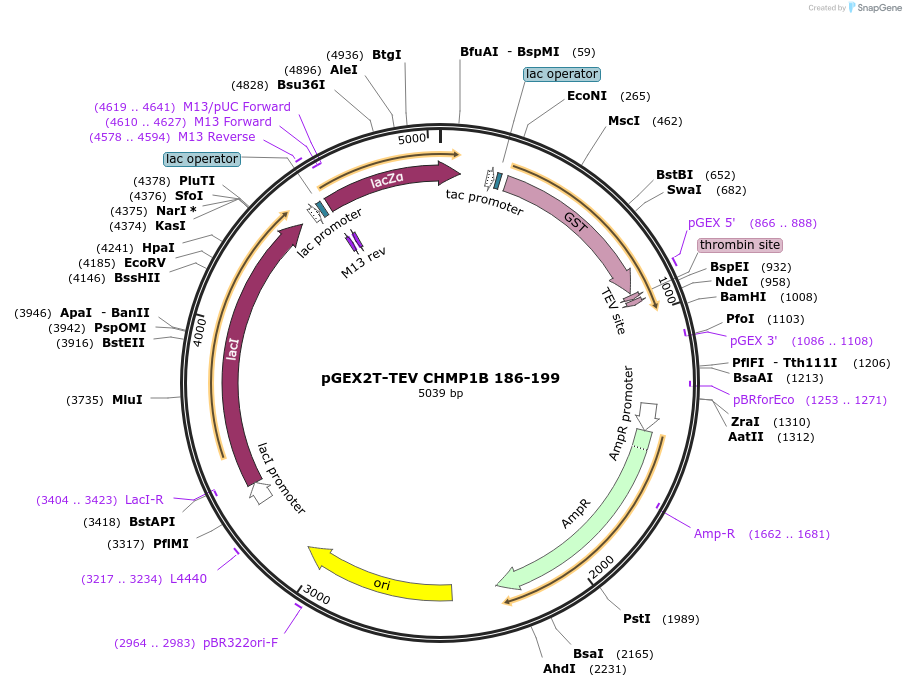
pGEX2T-TEV CHMP1B 186-199
Plasmid#108285PurposeExpresses residues 186-199 of CHMP1B with an N-terminal GST tag and a TEV cleavage site; Internal ID: WISP 09-283DepositorAvailable SinceApril 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
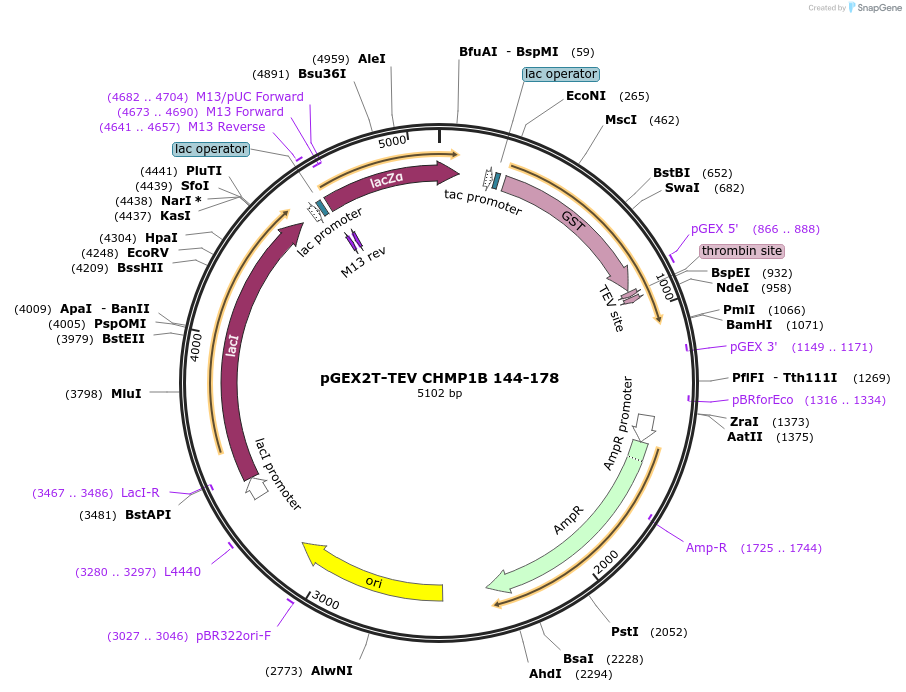
pGEX2T-TEV CHMP1B 144-178
Plasmid#108287PurposeExpresses residues 144-178 of CHMP1B with an N-terminal GST tag and a TEV cleavage site; Internal ID: WISP 09-281DepositorAvailable SinceApril 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
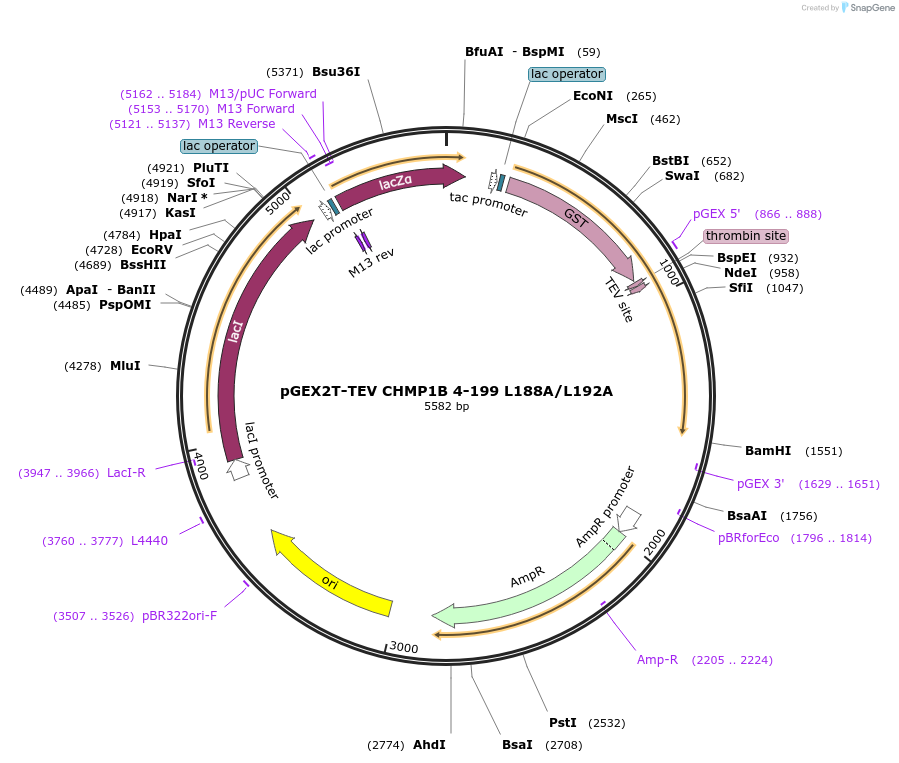
pGEX2T-TEV CHMP1B 4-199 L188A/L192A
Plasmid#108289PurposeExpresses residues 4-199 with L to A mutations at residues 188 and 192 of CHMP1B with an N-terminal GST tag and a TEV cleavage site; Internal ID: WISP 09-284DepositorInsertCHMP1B (CHMP1B Human)
TagsGSTExpressionBacterialMutationchanged L 188 to A, changed L 192 to AAvailable SinceApril 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
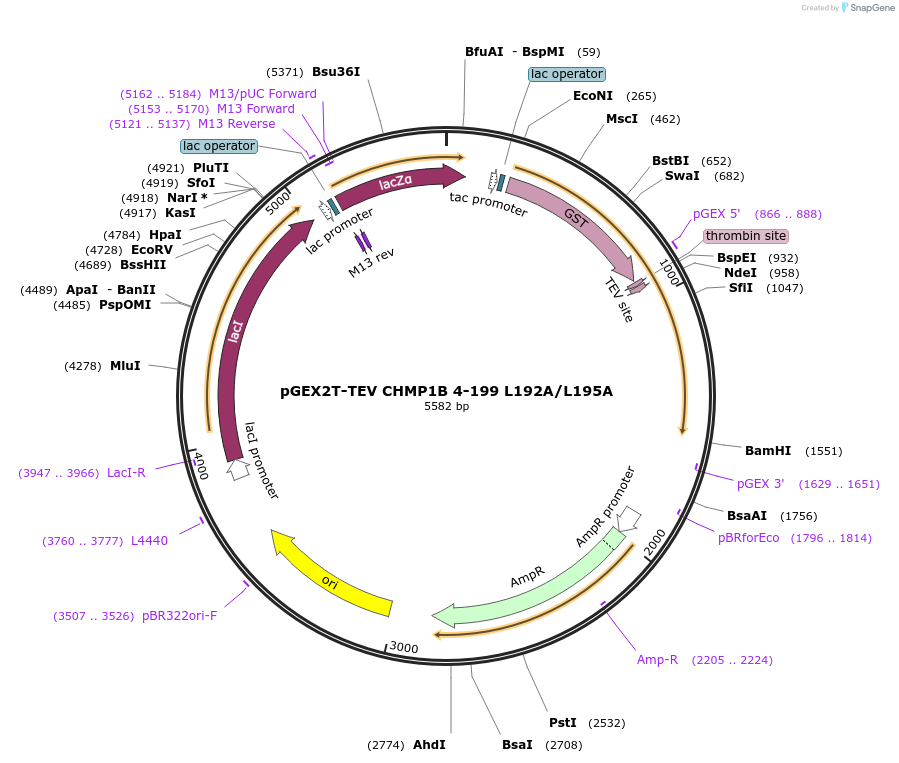
pGEX2T-TEV CHMP1B 4-199 L192A/L195A
Plasmid#108291PurposeExpresses residues 4-199 with L to A mutations at residues 192 and 195 of CHMP1B with an N-terminal GST tag and a TEV cleavage site; Internal ID: WISP 09-285DepositorInsertCHMP1B (CHMP1B Human)
TagsGSTExpressionBacterialMutationchanged L 192 to A, changed L 195 to AAvailable SinceApril 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
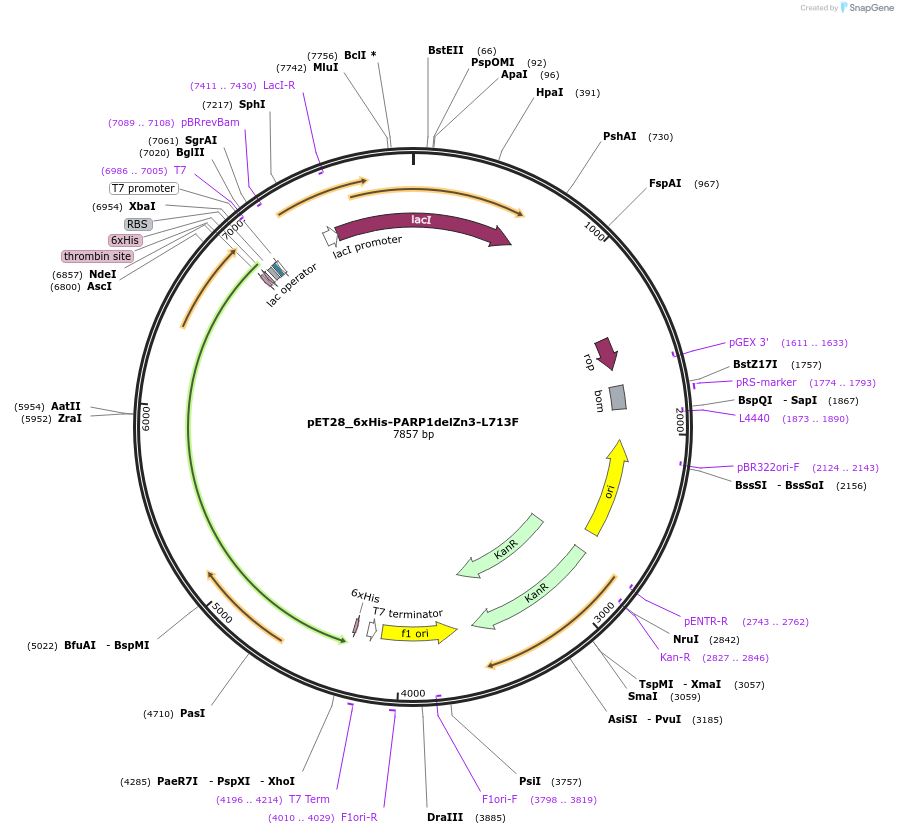
pET28_6xHis-PARP1delZn3-L713F
Plasmid#173934PurposeBacterial expression of PARP1 lacking Zn3 domain but containing L713F gain-of-function mutationDepositorInsertPARP1delZn3-L713F (PARP1 Human)
Tags6xHisExpressionBacterialMutationDeletion of residues 214-373; L713F, V762AAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
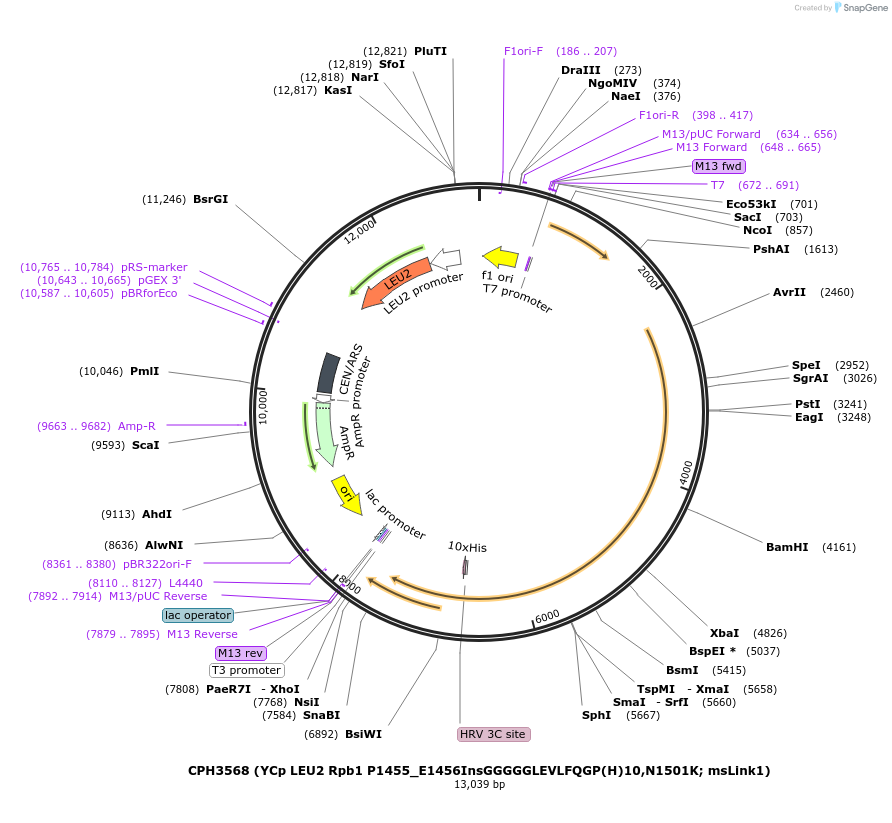
CPH3568 (YCp LEU2 Rpb1 P1455_E1456InsGGGGGLEVLFQGP(H)10,N1501K; msLink1)
Plasmid#91819PurposeYeast expression of S. cerevisiae Rpb1 with an internal PreScission protease site and 10xHis tag for purification of the linker-CTDDepositorInsertRPO21 (RPO21 Budding Yeast)
Tags10xHisExpressionYeastMutation5xGly linker, PreScission Protease site (LEVLFQGP…PromoterEndogenous Rpb1 promoterAvailable SinceMarch 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
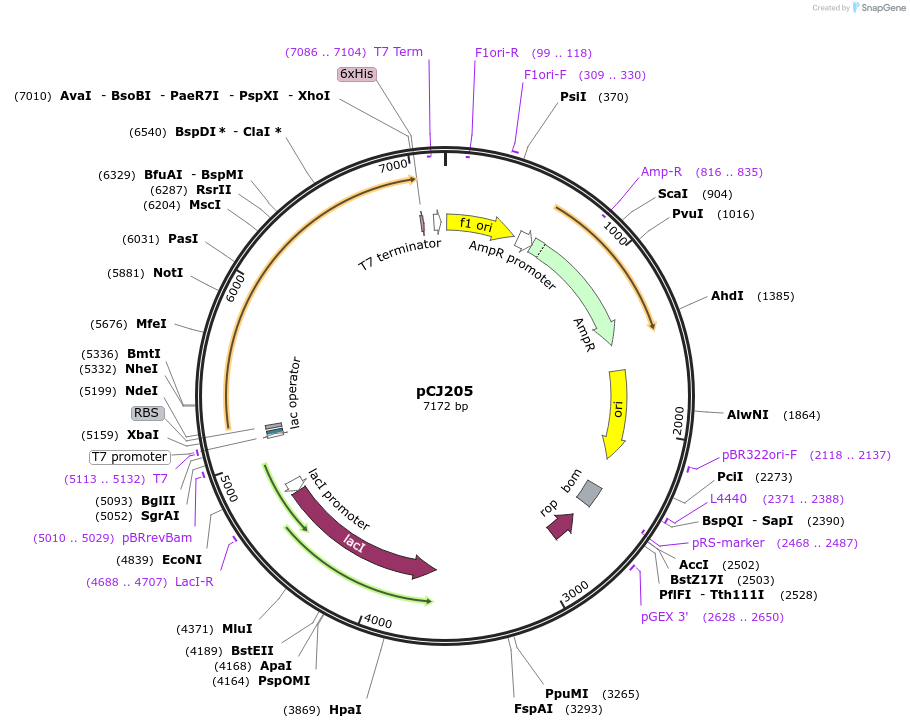
pCJ205
Plasmid#162676PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224A anDepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224A, C529A; codon optimized for expression in E…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCJ198
Plasmid#162671PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating F495I mutation.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationF495I; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
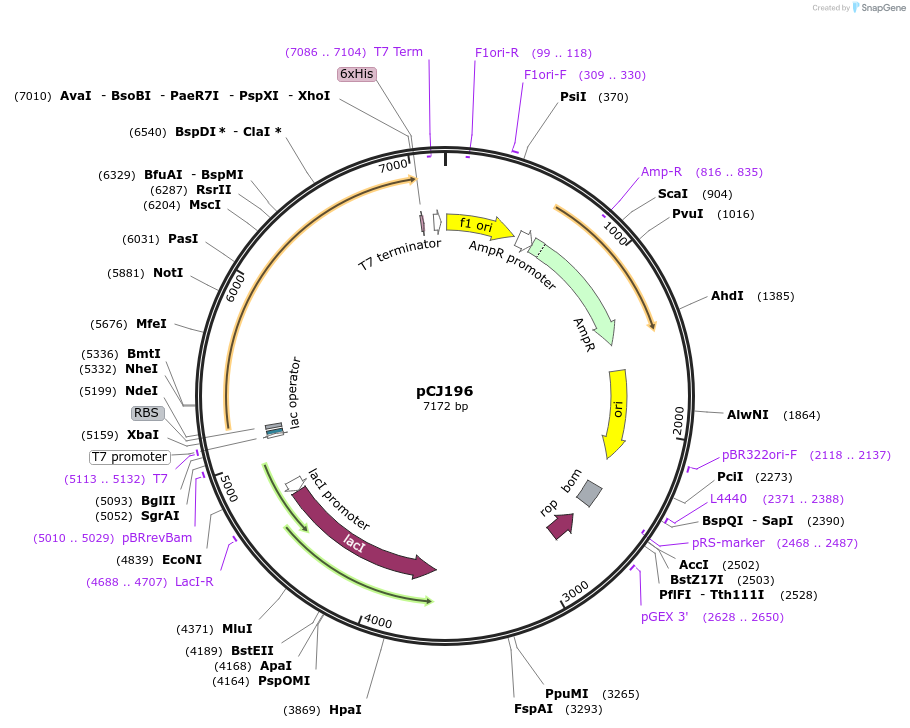
pCJ196
Plasmid#162669PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating catalytic mutation, S225ADepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS225A; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
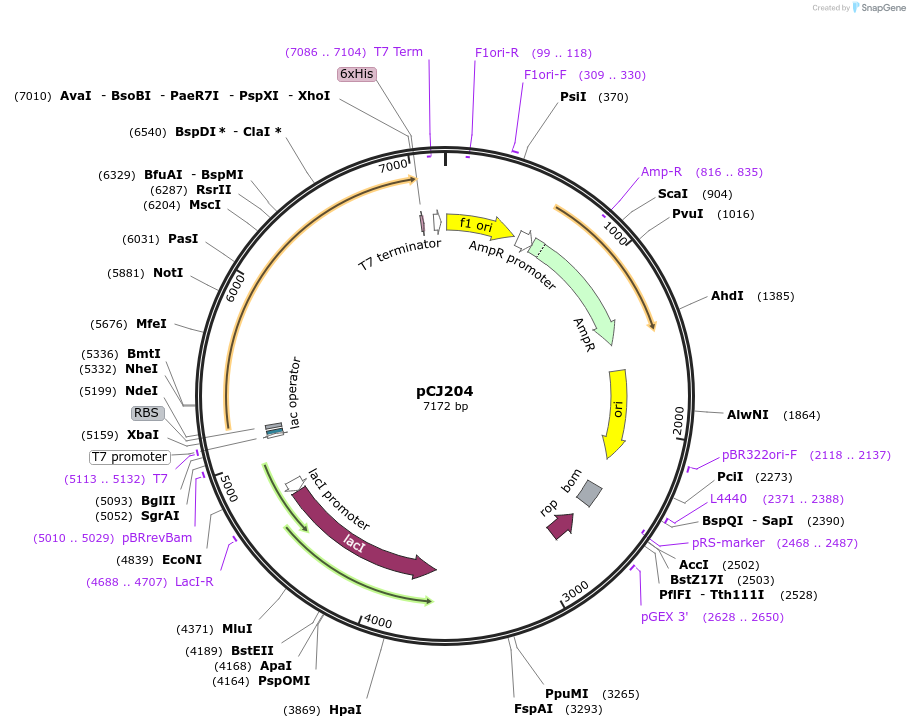
pCJ204
Plasmid#162677PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224H and C529F mutations.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224H, C529F; codon optimized for expression in E…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
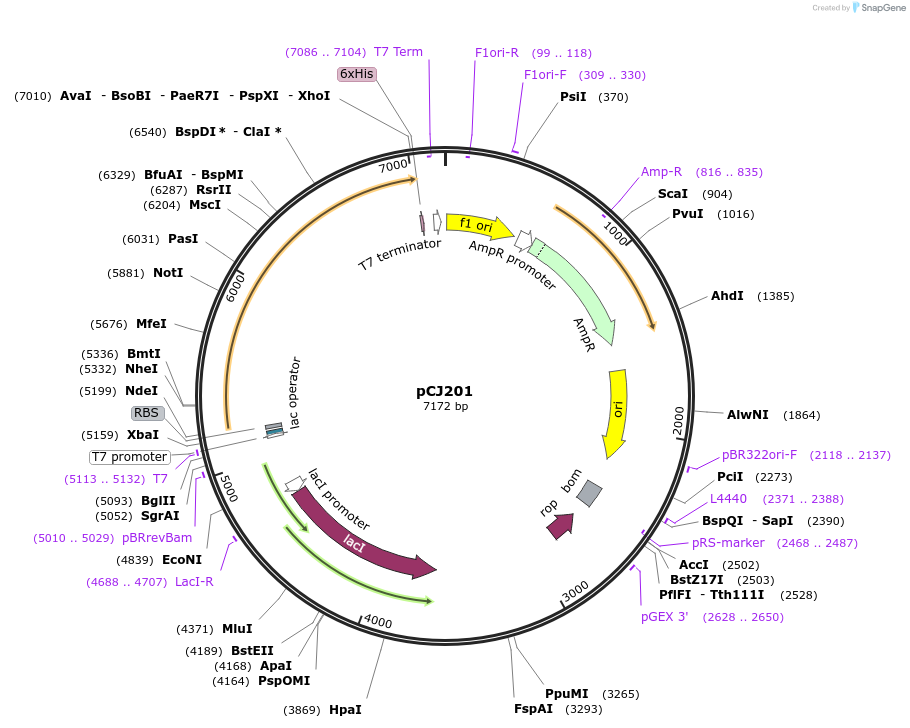
pCJ201
Plasmid#162674PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating C224W and C529SS mutations.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationC224W, C529SS; codon optimized for expression in …PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
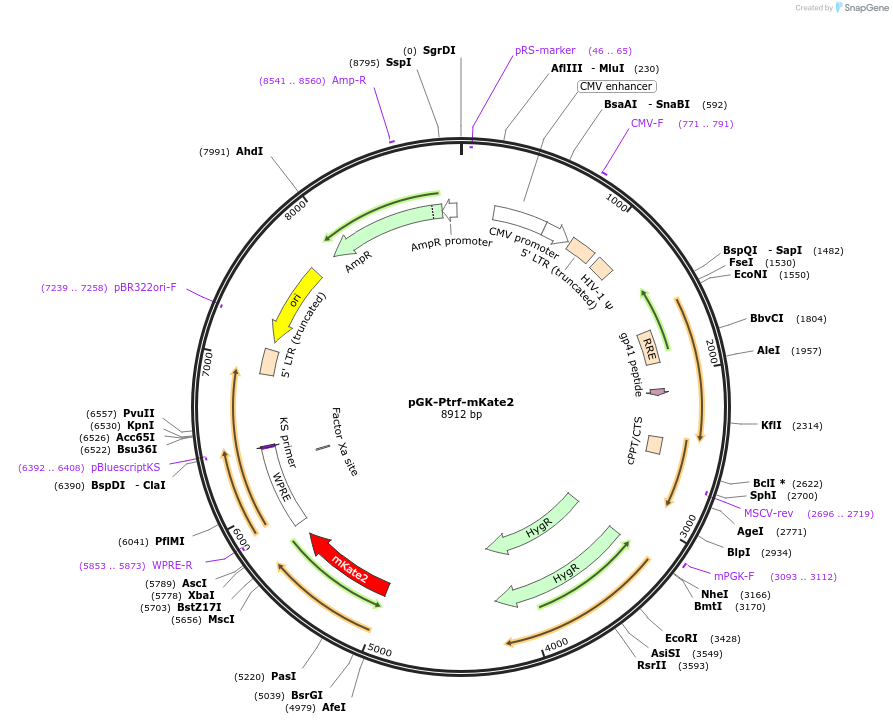
pGK-Ptrf-mKate2
Plasmid#128765PurposeFluorescent mKate reporter for PtrfDepositorAvailable SinceNov. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
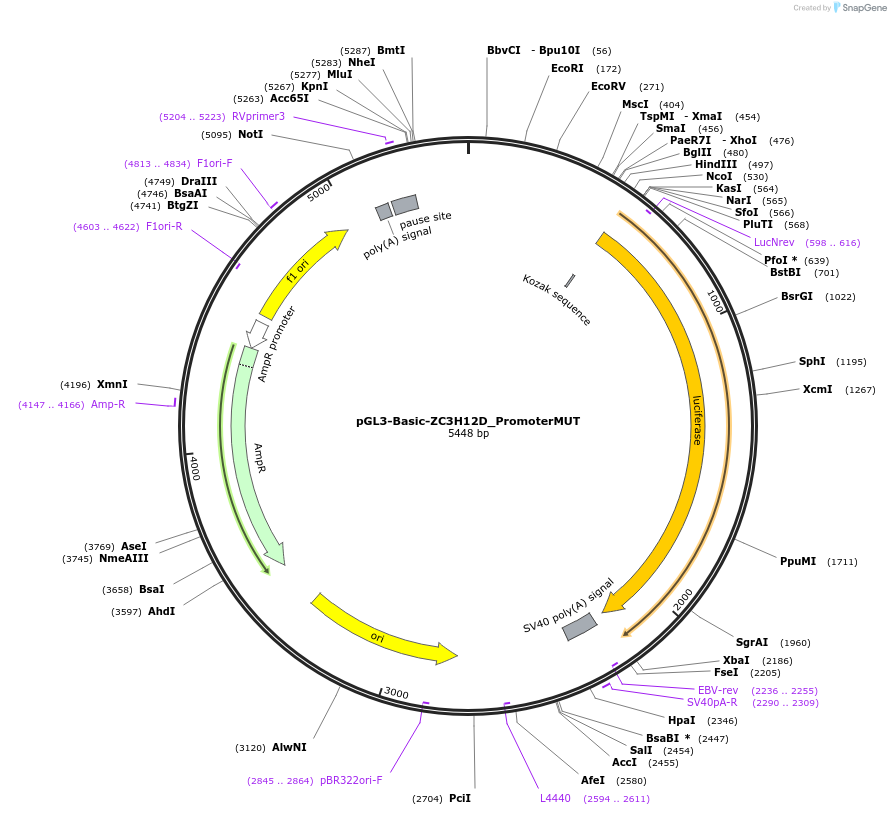
pGL3-Basic-ZC3H12D_PromoterMUT
Plasmid#153067PurposeZC3H12D promoter region carrying mutations in BHLHE40 binding sites, cloned upstream of luciferase reporterDepositorInsertZC3H12D promoter region mutated in BHLHE40 binding sites (ZC3H12D Human)
UseLuciferaseExpressionMammalianMutationMutagenesis to disrupt 3 BHLHE4040 binding sites.…PromoterNoneAvailable SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only