We narrowed to 81,061 results for: Mycs;
-
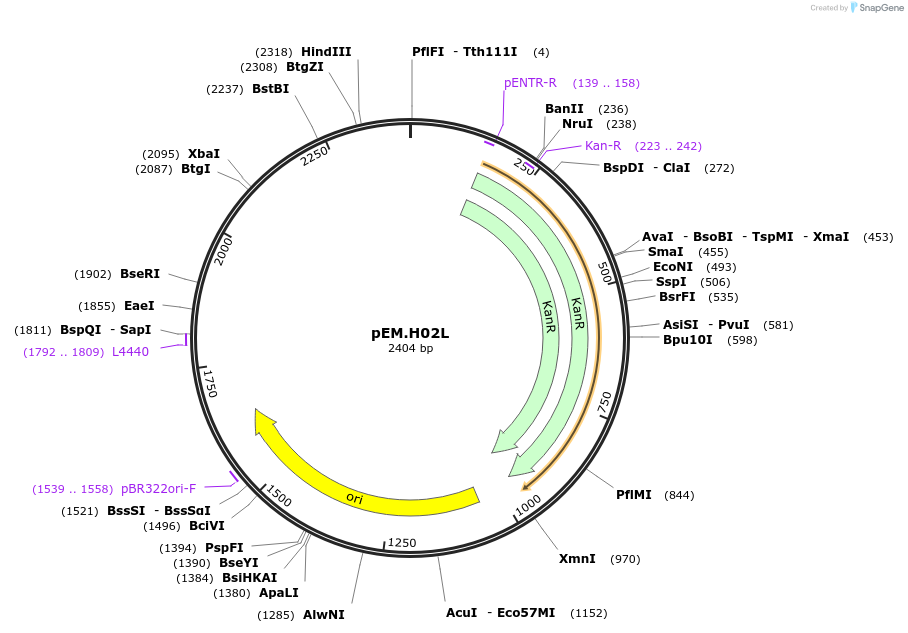
Plasmid#198667PurposeAn Easy Modular Integrative fuSion-ready Expression toolkit (Easy-MISE toolkit) for fast engineering of heterologous productions in Saccharomyces cerevisiaeDepositorInsertXI2_UP
UseSynthetic BiologyExpressionYeastAvailable SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
pEM.H01L
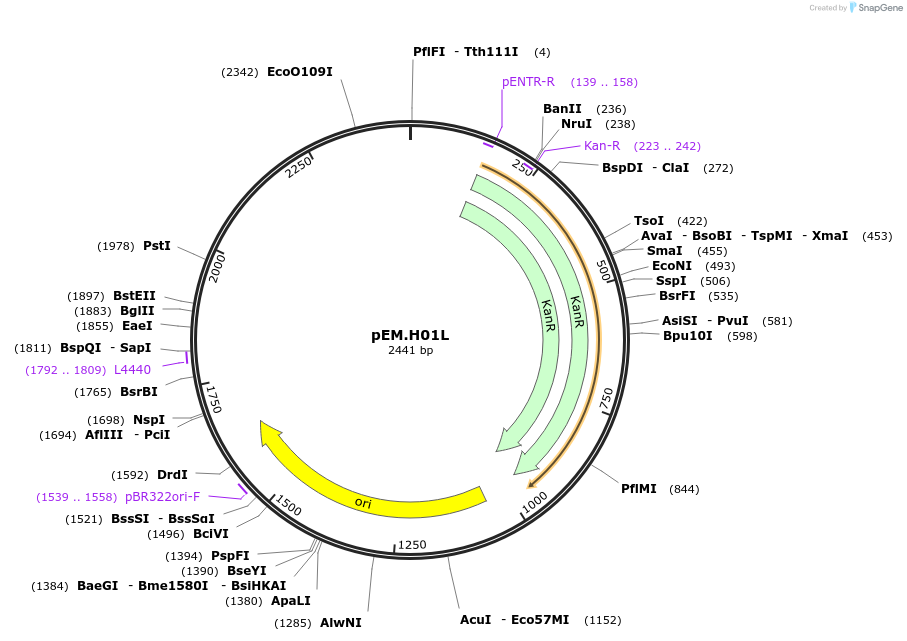
Plasmid#198665PurposeAn Easy Modular Integrative fuSion-ready Expression toolkit (Easy-MISE toolkit) for fast engineering of heterologous productions in Saccharomyces cerevisiaeDepositorInsertX3_UP
UseSynthetic BiologyExpressionYeastAvailable SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
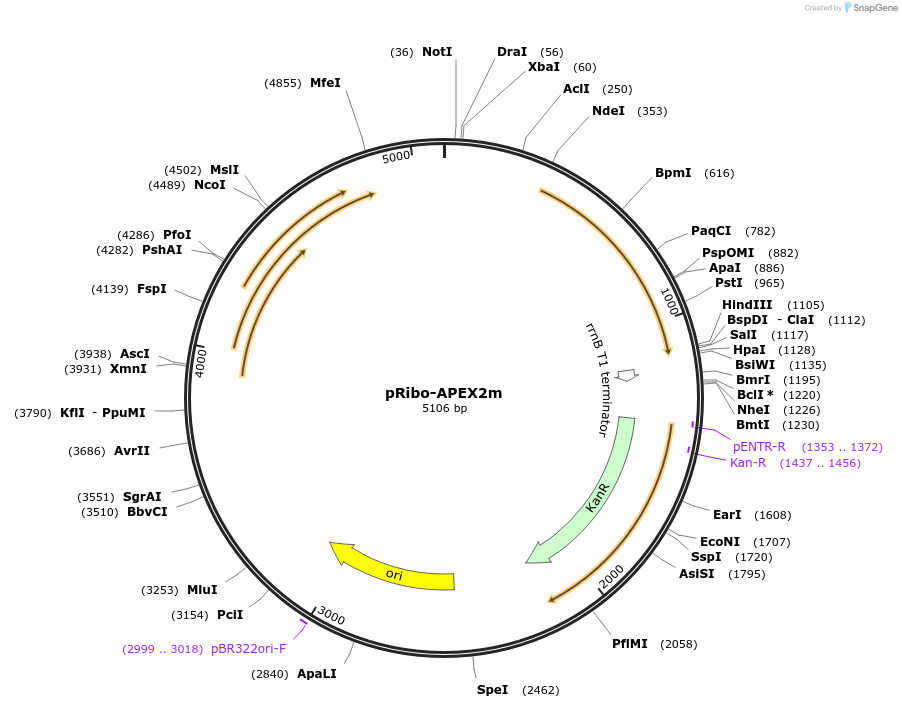
pRibo-APEX2m
Plasmid#176842PurposeExpression of APEX2 from a codon-optimized gene (multi-copy episomal plasmid)DepositorInsertpRibo-APEX2m
ExpressionBacterialPromoterpRibo, theophilline riboswitch inducible hsp60 pr…Available SinceJan. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
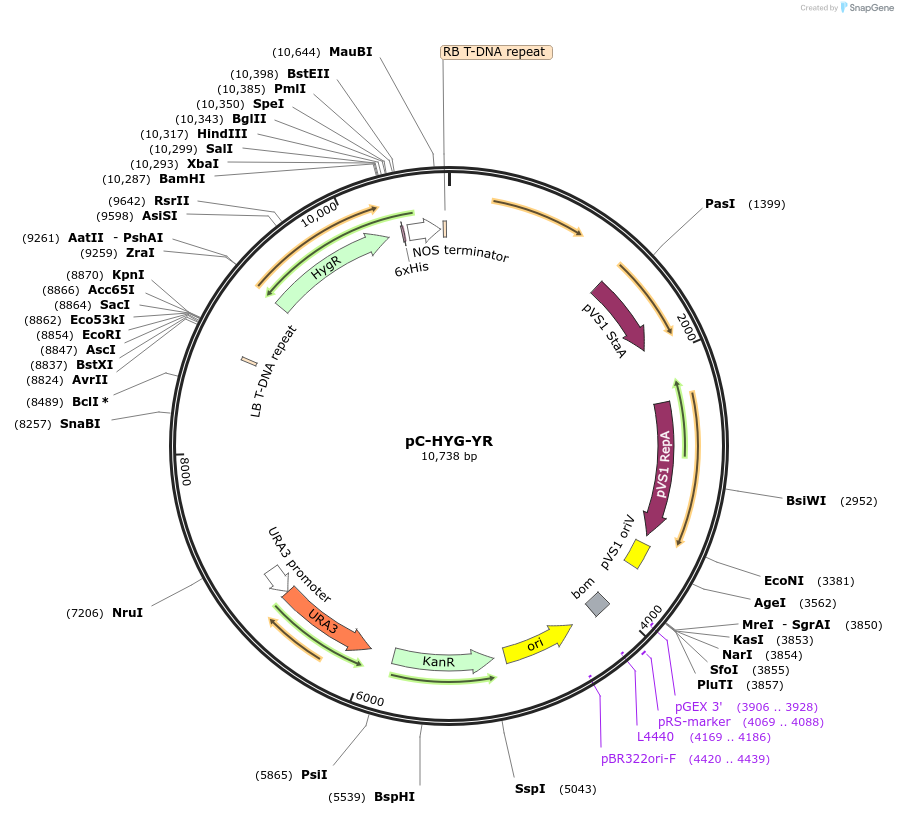
pC-HYG-YR
Plasmid#61765PurposeYeast recombinational cloning compatible Agrobacterium tumefaciens ternary vector containing hygromycin selectable marker on transfer DNA (TDNA).DepositorInsertsHygromycin
2 micron origin or replication and URA3 gene for S. cerevisiae
UseTernary vector for agrobacterium mediated transfo…PromotertrpC promoterAvailable SinceMay 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
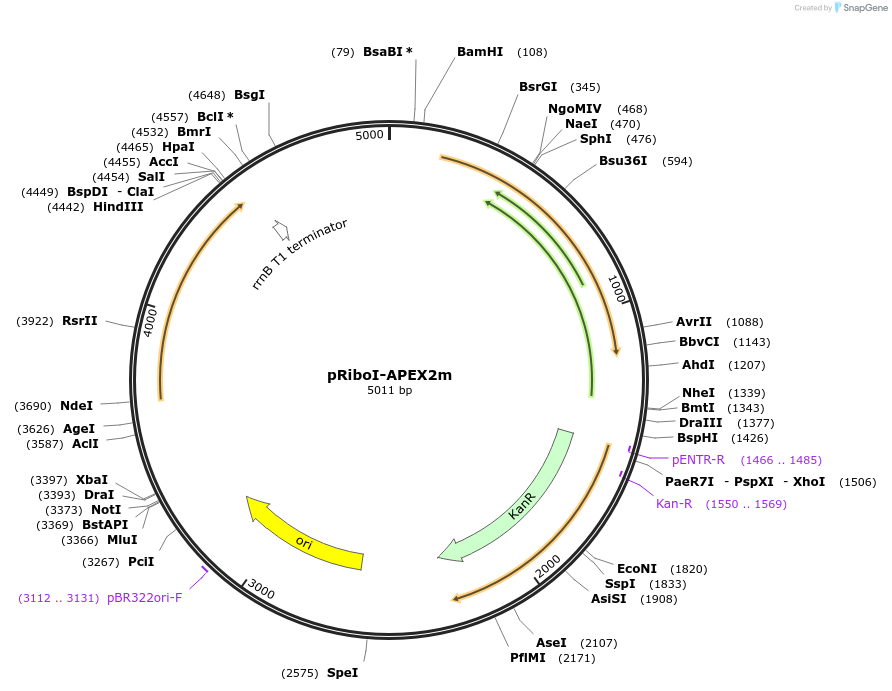
pRiboI-APEX2m
Plasmid#176843PurposeExpression of APEX2 from a codon-optimized gene (single-copy integrating plasmid)DepositorInsertpRibo-APEX2m
UseIntegratingExpressionBacterialPromoterpRibo, theophilline riboswitch inducible hsp60 pr…Available SinceJan. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
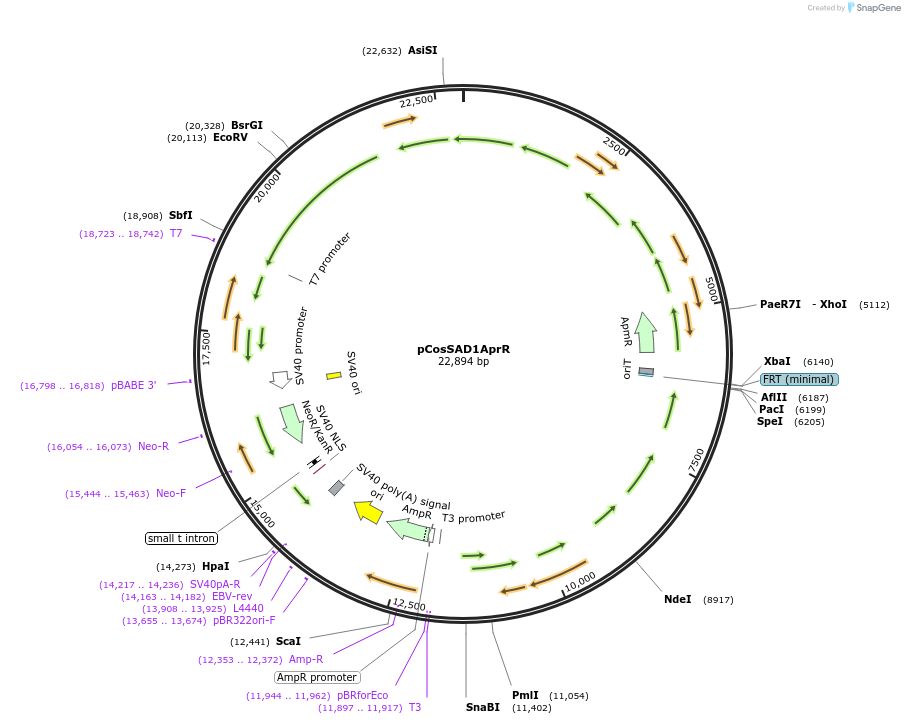
pCosSAD1AprR
Plasmid#190786PurposeAn E. coli cloning vector for the integration of heterologous genes into a linear plasmid pSA3239 in Streptomyces lavendulae subsp. lavendulae CCM 3239 through homologous recombinationDepositorInsertsupstream region of the aur1 cluster for auricin
ampramycin resistance cassette
downstream region of the aur1 cluster for auricin
ExpressionBacterialAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pGEX-2T-FRB-W2027F
Plasmid#73341PurposePoint mutation Trp 2027 → Phe in the FKBP12-rapamycin binding (FRB) domain of mTORDepositorInsertFKBP12-rapamycin binding (FRB) domain of MTOR (MTOR Human)
TagsGST-taggedExpressionBacterialMutationFRB domain only aa2015-2114; W2027FAvailable SinceApril 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pGEX-2T FRB Y2105A
Plasmid#68726PurposePoint mutation Tyr 2105 → Ala in the FKBP12-rapamycin binding (FRB) domain of mTORDepositorInsertFKBP12-rapamycin binding (FRB) domain of MTOR (MTOR Human)
TagsGST-taggedExpressionBacterialMutationFRB domain only aa2015-2114; Y2105AAvailable SinceApril 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
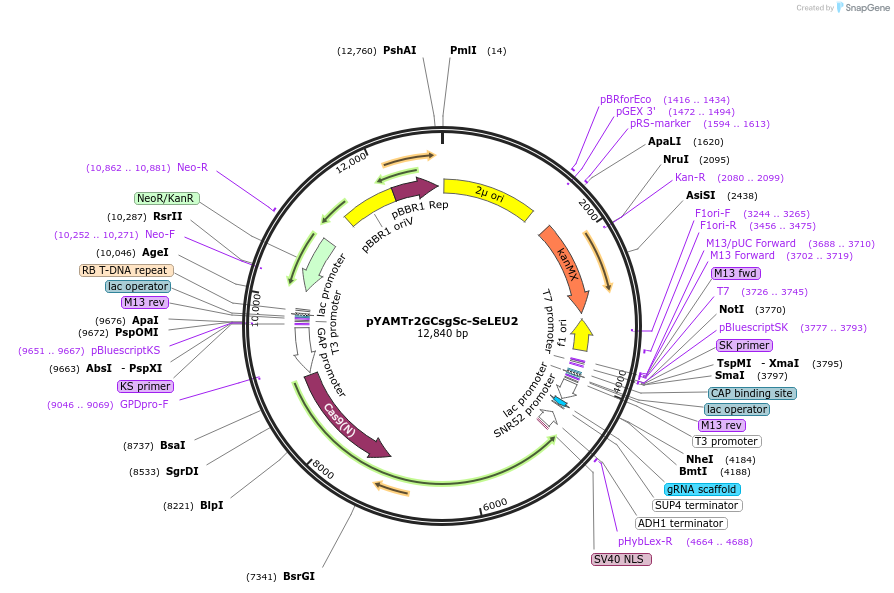
pYAMTr2GCsgSc-SeLEU2
Plasmid#224871PurposeDisruption of LEU2 genes in the Saccharomyces sense strict group speciesDepositorUseCRISPRExpressionYeastAvailable SinceOct. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
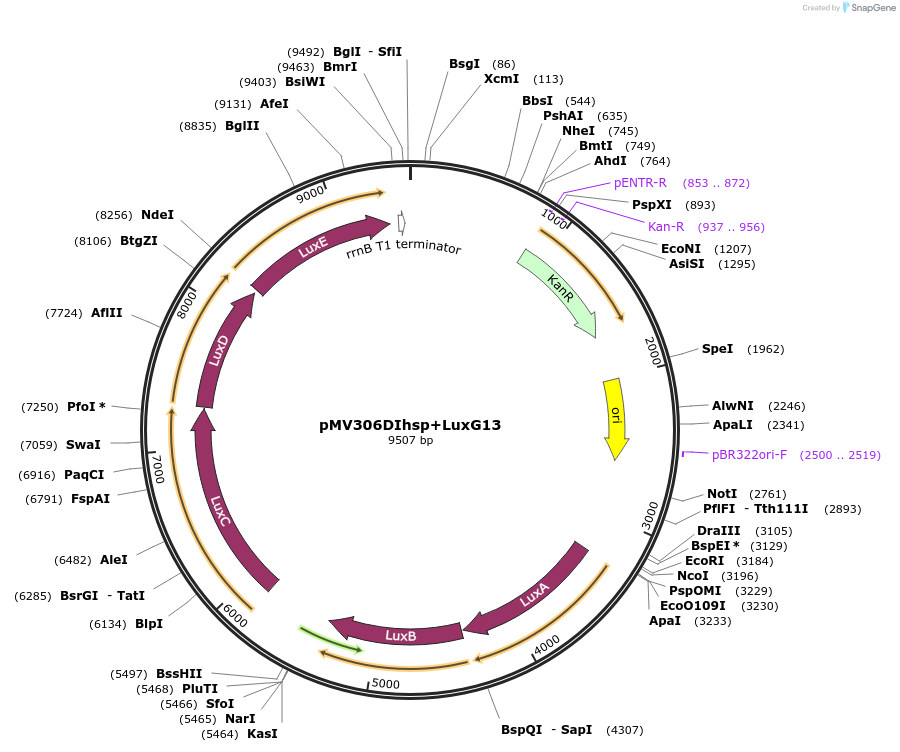
pMV306DIhsp+LuxG13
Plasmid#49999PurposeIntegrase removed to increase stability of the plasmid in mycobacteriaDepositorInsertBacterial luciferase operon + G13 promoter
ExpressionBacterialMutationIt contains a gram-positive enhanced translation …PromoterG13 from M. marinum G13 promoter driving luxCAvailable SinceMay 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
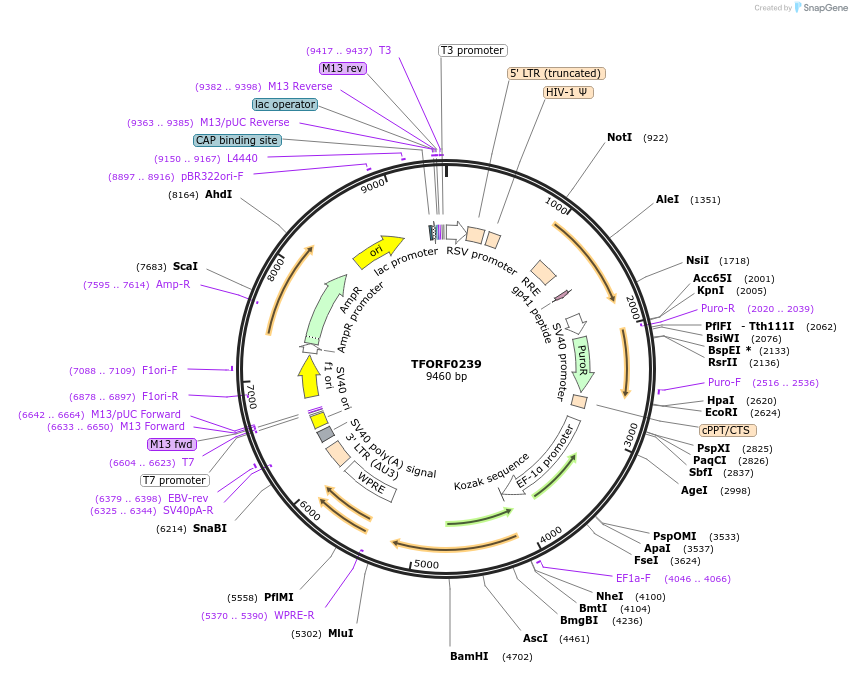
TFORF0239
Plasmid#142385PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceApril 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
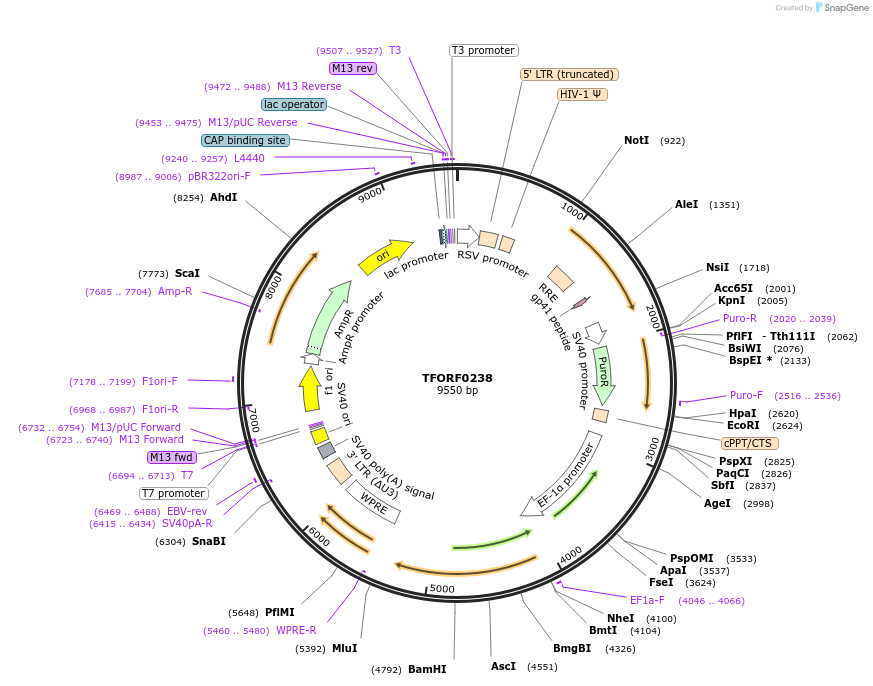
TFORF0238
Plasmid#142490PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
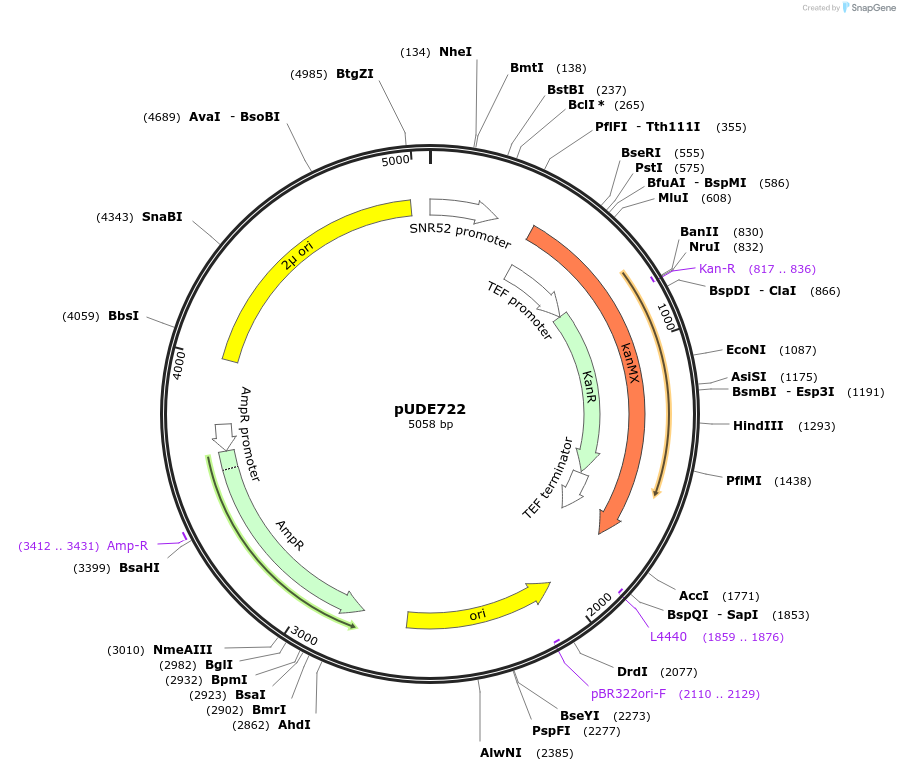
pUDE722
Plasmid#103022Purposeexpression of a Cpf1 programming crRNA targetting CAN1 (crCAN1-4S)DepositorAvailable SinceDec. 1, 2017AvailabilityAcademic Institutions and Nonprofits only -
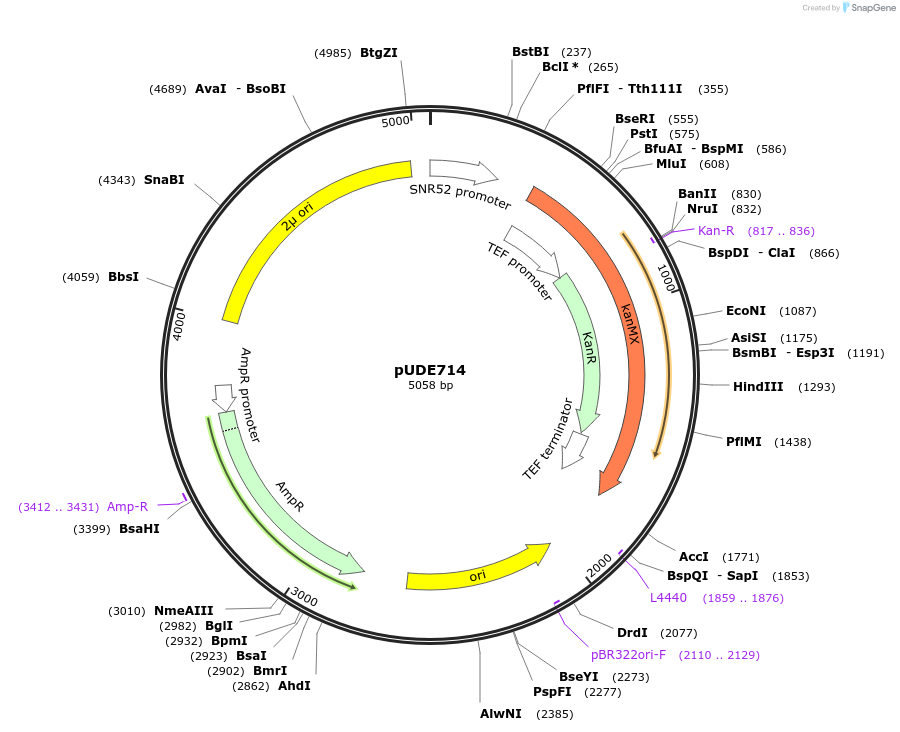
pUDE714
Plasmid#103021Purposeexpression of a Cpf1 programming crRNA targetting HIS4 (crHIS4-4.S)DepositorAvailable SinceDec. 1, 2017AvailabilityAcademic Institutions and Nonprofits only -
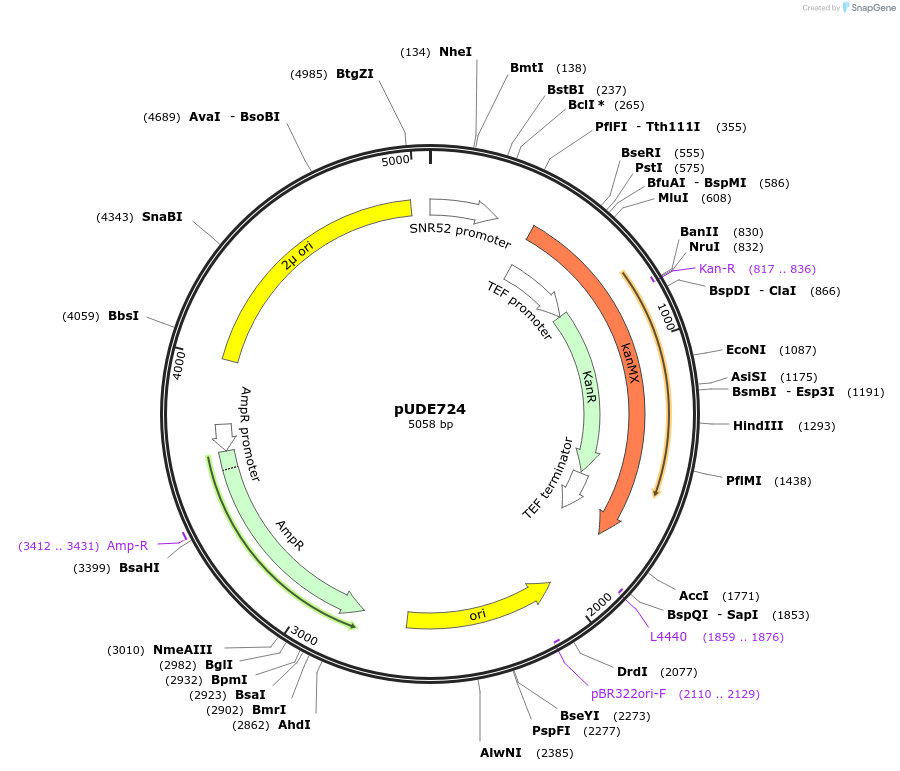
pUDE724
Plasmid#103023Purposeexpression of a Cpf1 programming crRNA targetting PDR12 (crPDR12-3S)DepositorAvailable SinceDec. 1, 2017AvailabilityAcademic Institutions and Nonprofits only -
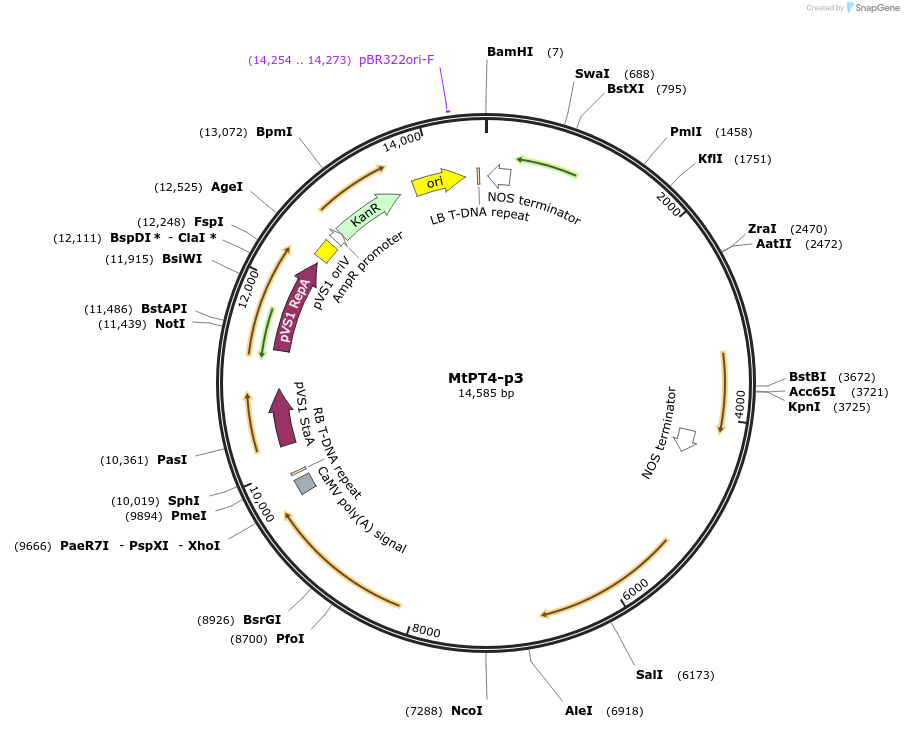
MtPT4-p3
Plasmid#160003PurposeMultigene construct containing the betalain pigment biosynthetic genes under the control of the MtPT4 promoter for visualisation of arbuscular mycorrhizal colonisation in Medicago truncatula roots.DepositorInsertsDsRed (reverse orientation)
DODAα1
CYP76AD1
cDOPA5GT
ExpressionPlantPromoterArabidopsis thaliana Ubiquitin 10 Promoter (AtUb1…Available SinceJuly 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
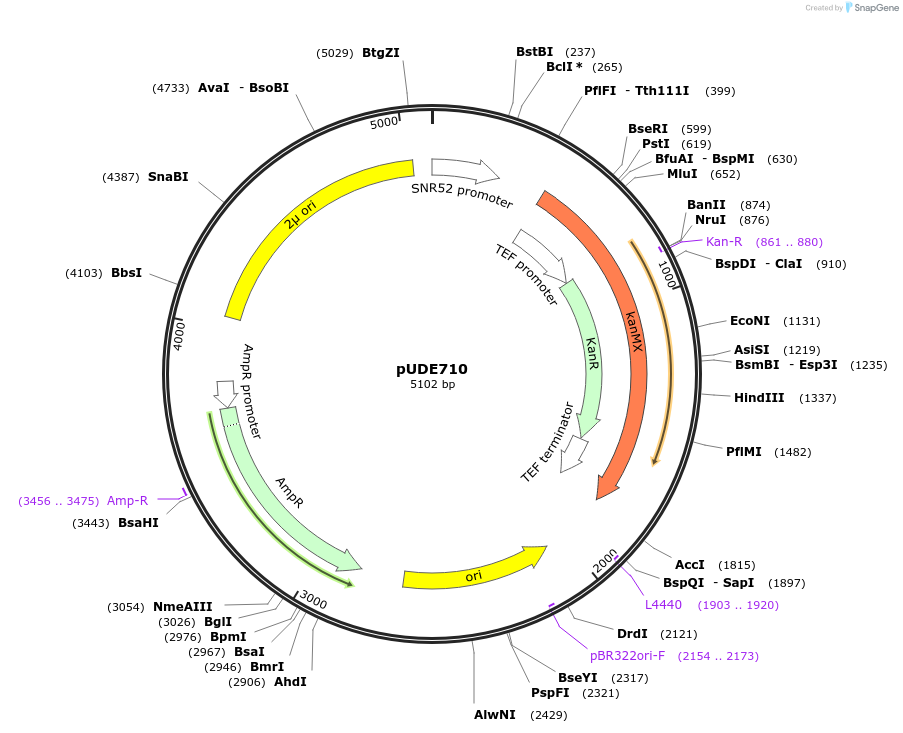
pUDE710
Plasmid#103020Purposeexpression of a Cpf1 programming crRNA targeting ADE2 and HIS4 (crADE2-3 crHIS4-4.S)DepositorAvailable SinceDec. 1, 2017AvailabilityAcademic Institutions and Nonprofits only -
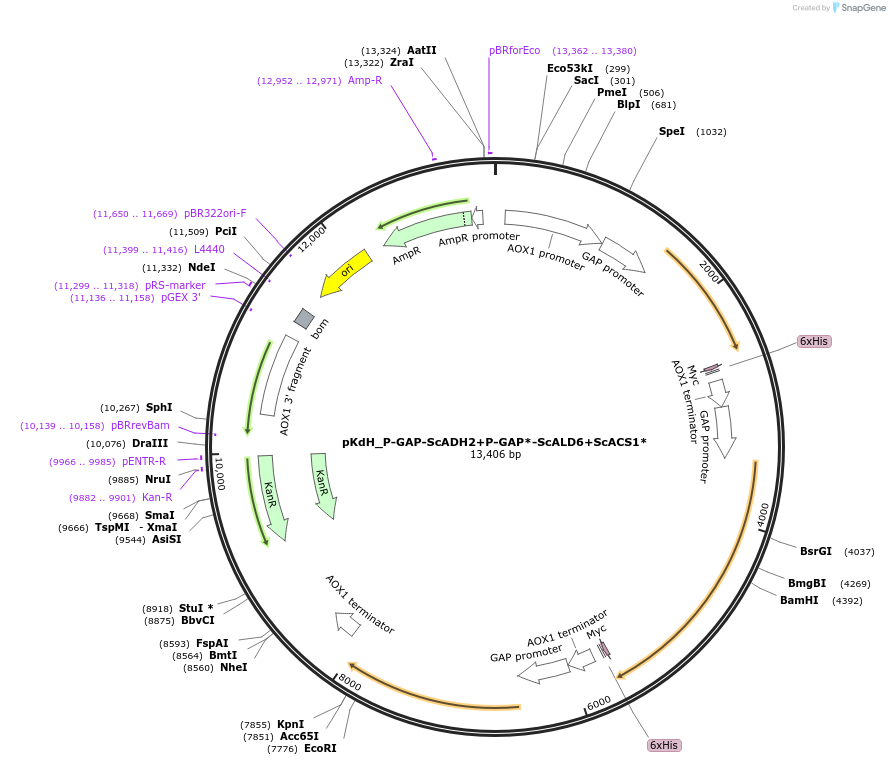
pKdH_P-GAP-ScADH2+P-GAP*-ScALD6+ScACS1*
Plasmid#126739Purposeyeast plasmid overexpressing ADH2,ALD6 and ACS1 from Saccharomyces cerevisiaeDepositorInsertadh2, acs1*, ald6
Tags6xHis,MycExpressionYeastMutationT3542C and A30S, S629N (please see depositor comm…PromoterGAPAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
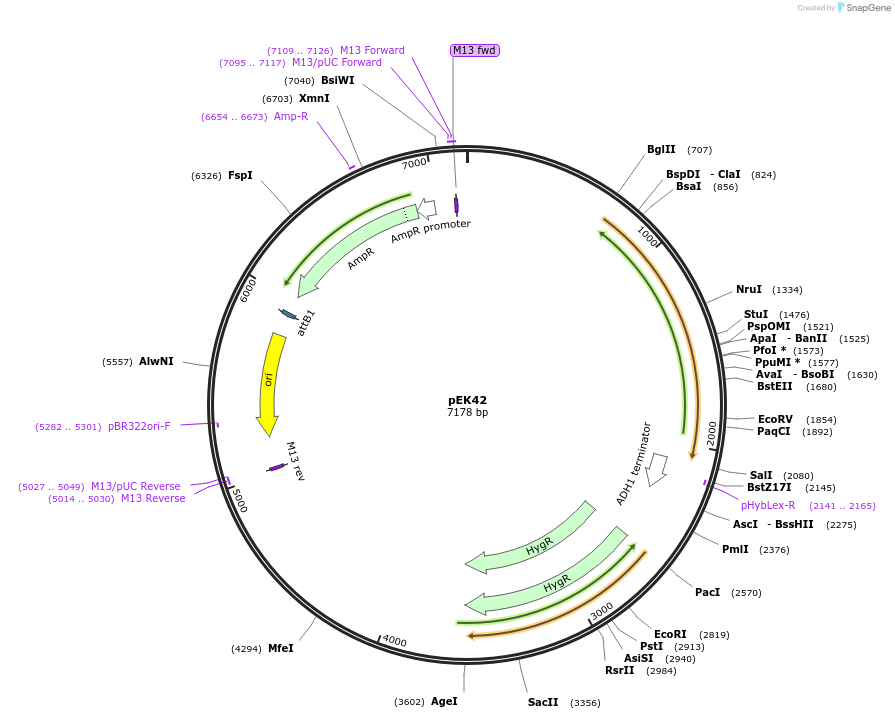
pEK42
Plasmid#214958PurposeEndogenous tagging of B. dendrobatidis GAPDH with a red fluorescent protein, hygromycin B selectionDepositorInsertsBdGAPDH_LHA2-G4S-HsmRuby3-ScADH1ter
SpH2Bpro-Hshph-BdGAPDH_RHA2
UseBd gene targeting vectorExpressionBacterialPromoterSpH2BproAvailable SinceOct. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
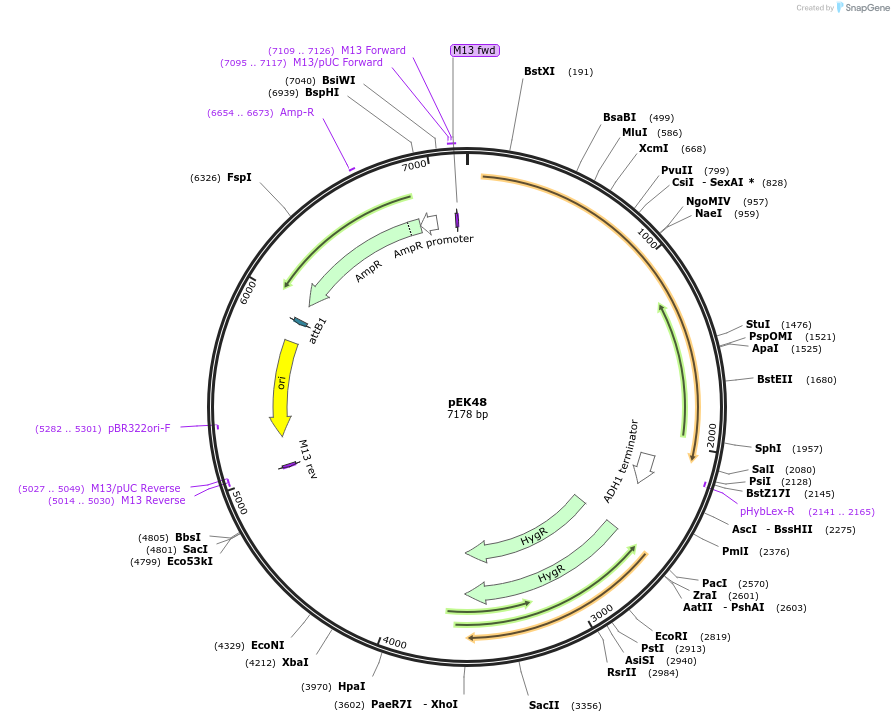
pEK48
Plasmid#214964PurposeEndogenous tagging of B. dendrobatidis Myo17D with a red fluorescent protein, hygromycin B selectionDepositorInsertsBdMyo17D_LHA-G4S-HsmRuby3-ScADH1ter
SpH2Bpro-Hshph-BdMyo17D_RHA
UseBd gene targeting vectorExpressionBacterialPromoterSpH2BproAvailable SinceOct. 20, 2025AvailabilityAcademic Institutions and Nonprofits only