We narrowed to 4,284 results for: abo
-
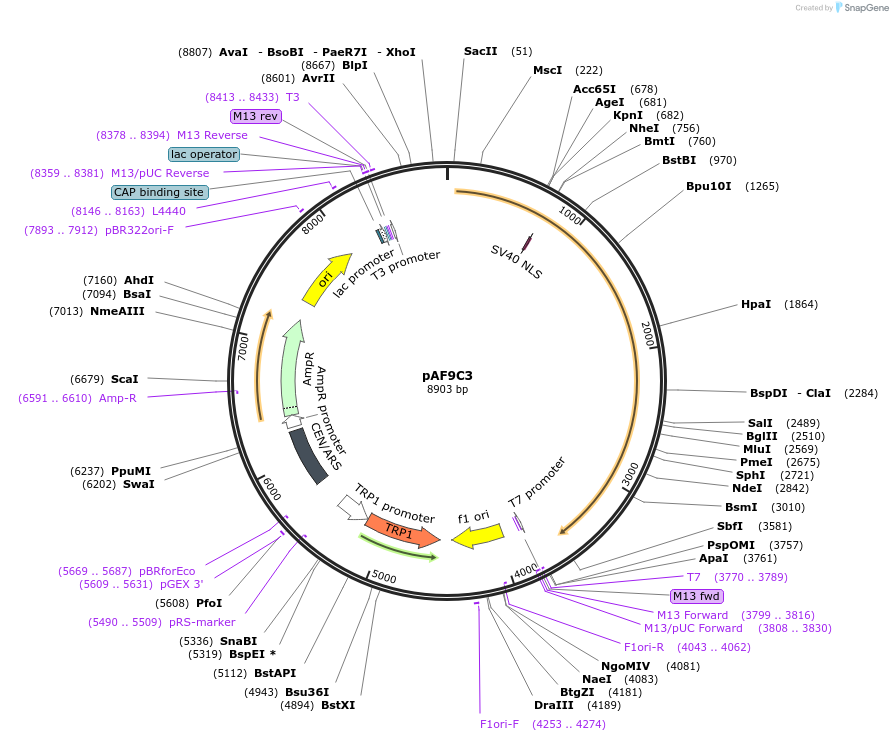
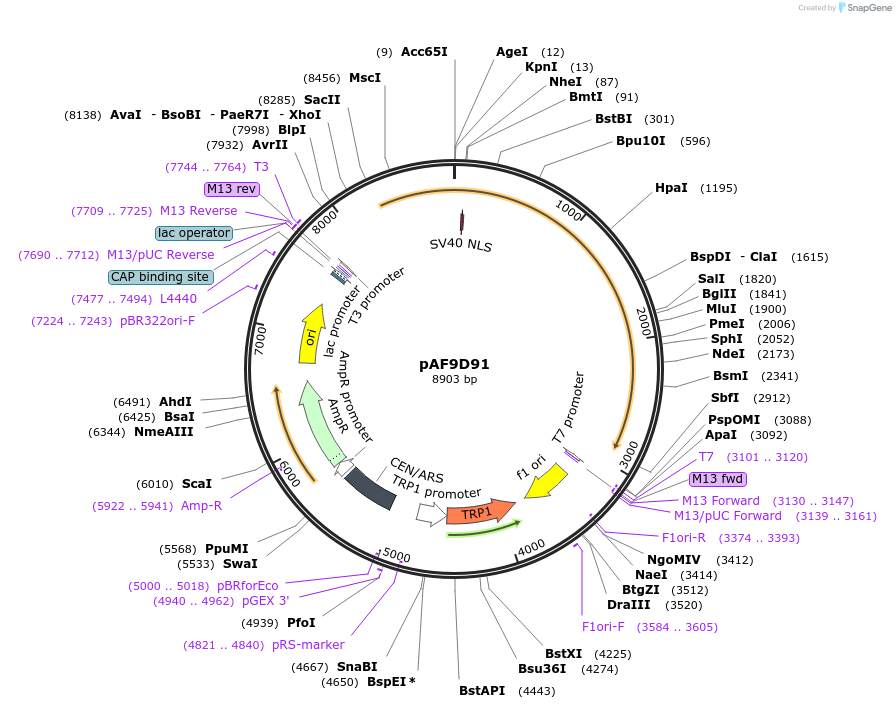
Plasmid#185843PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>RevTetR_r1.7>SIN3C>PABF1
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAF9D91
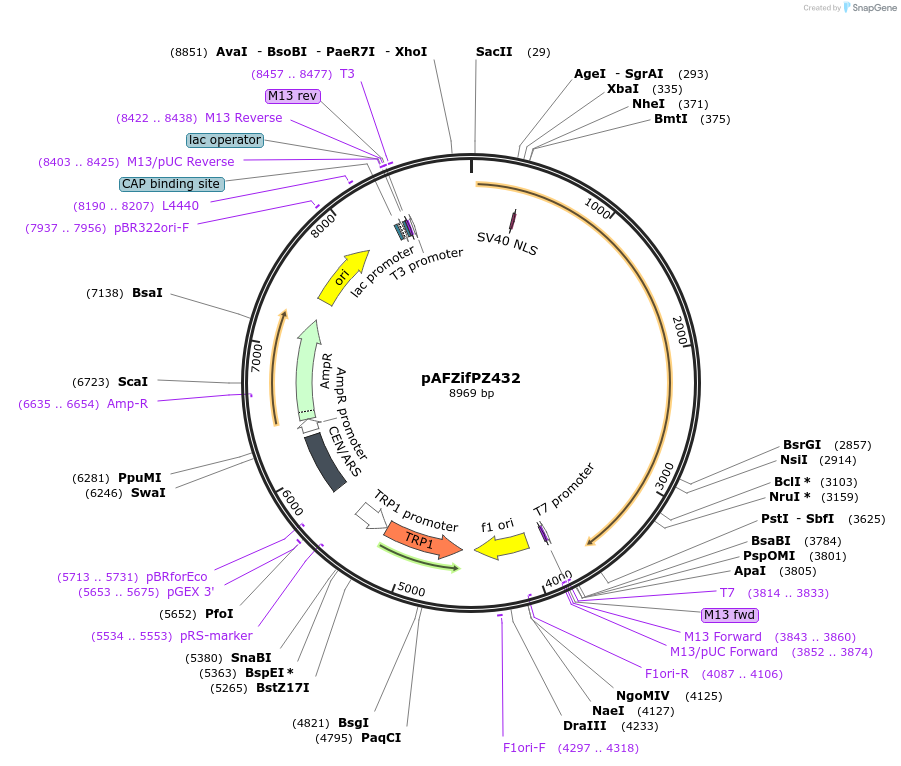
Plasmid#185845PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>TetR>SIN3C>PABF1
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAFZifPZ432
Plasmid#185850PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>ZifPZ43>MED15>PAFB1
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
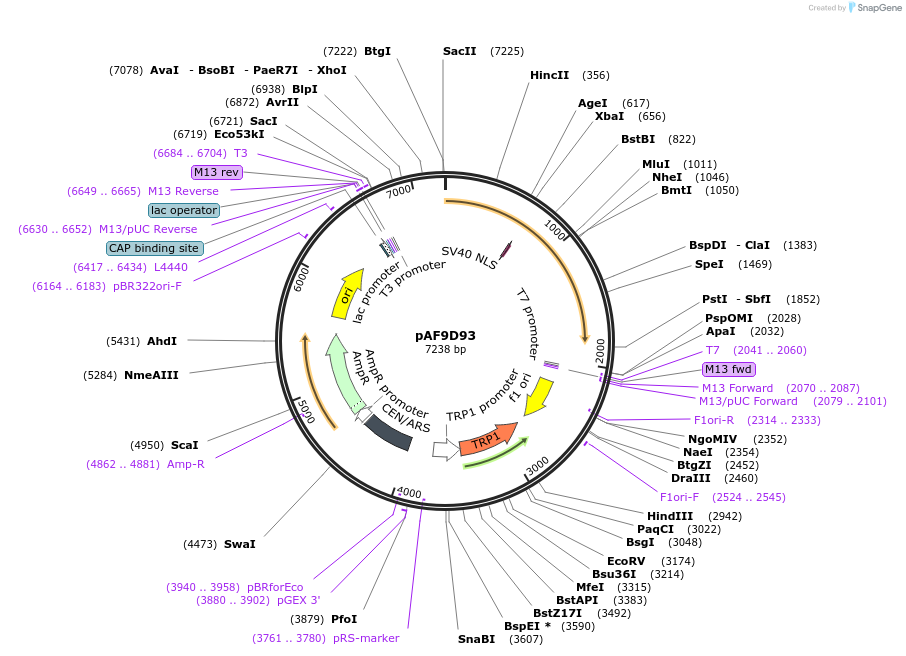
pAF9D93
Plasmid#185847PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>TetR>MIG1C>PABF1
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
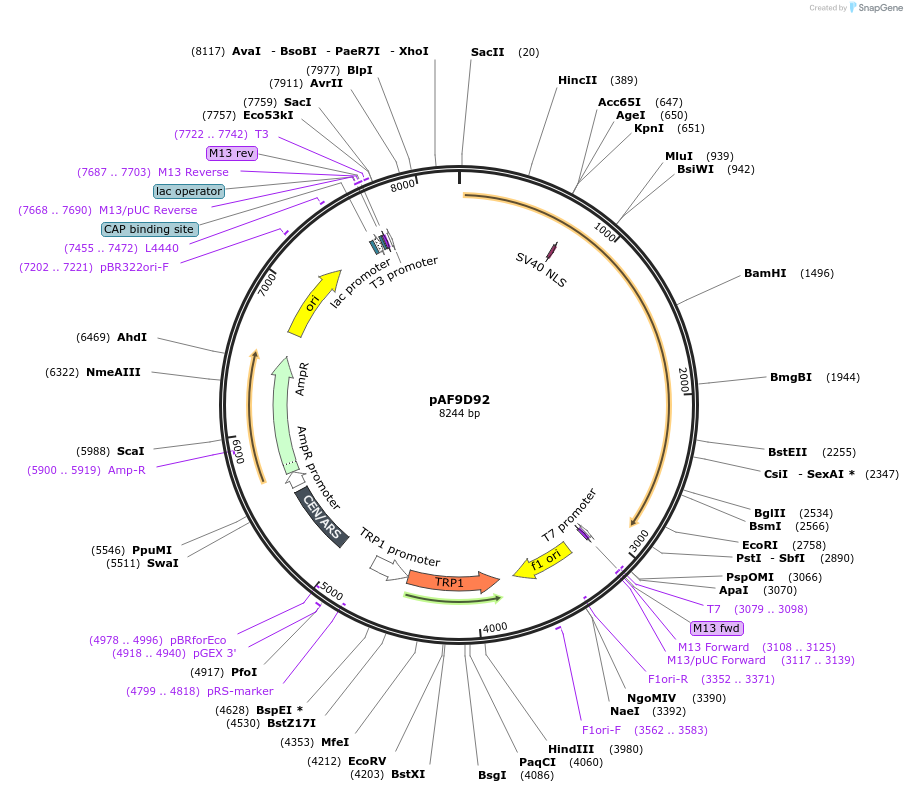
pAF9D92
Plasmid#185846PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>TetR>TUP1>PABF1
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
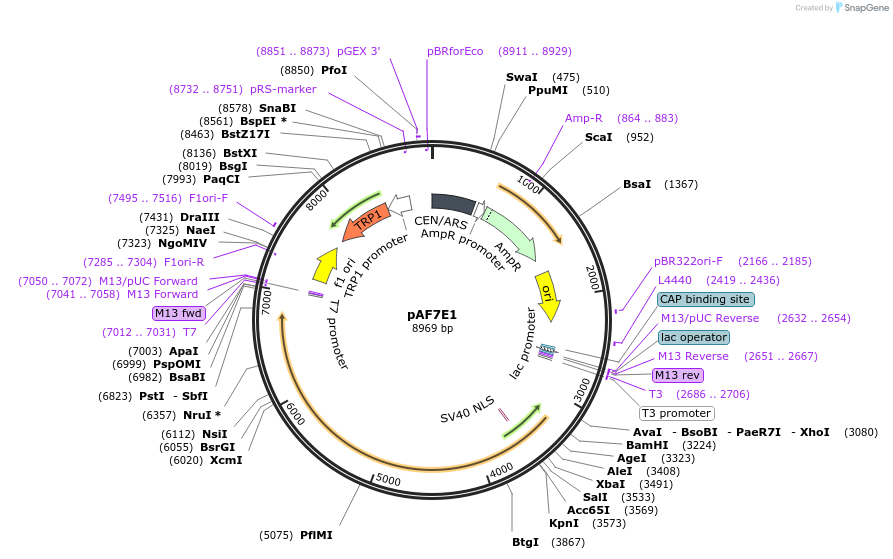
pAF7E1
Plasmid#185839PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>Zif268>MED15>PABF1
ExpressionYeastMutationNoneAvailable SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
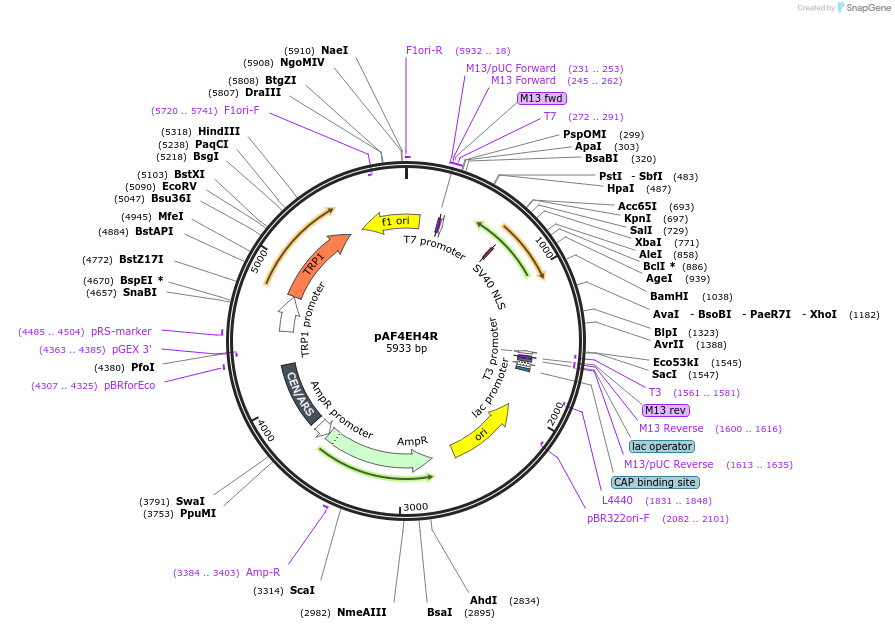
pAF4EH4R
Plasmid#185837PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>Zif268>VP16-PABF1
ExpressionYeastMutationNoneAvailable SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
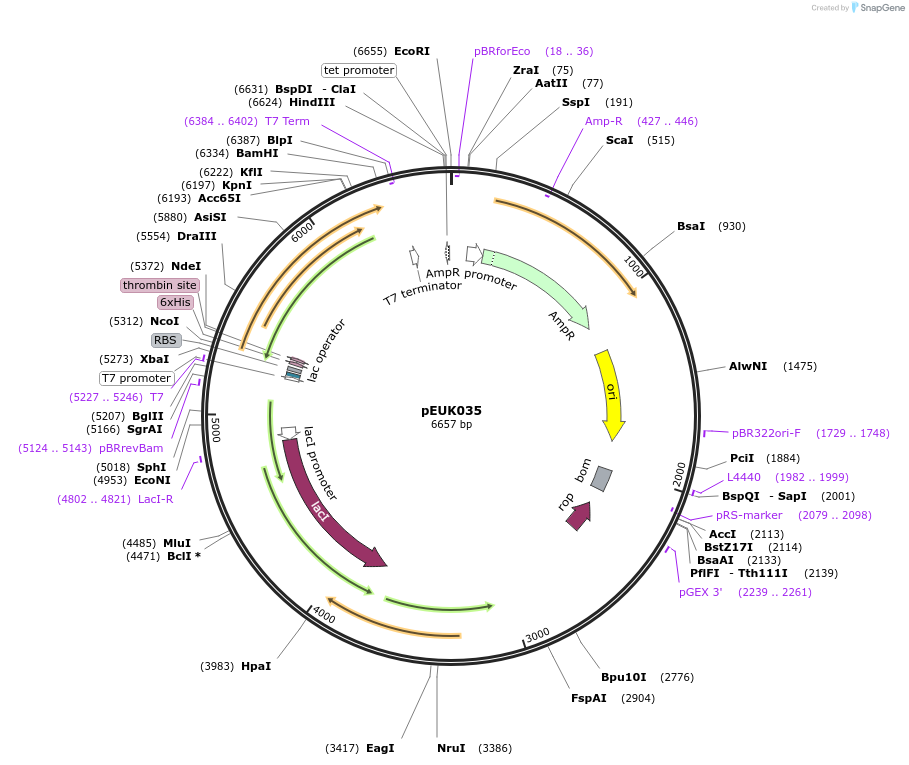
pEUK035
Plasmid#180827PurposeT7-inducible expression construct to produce SwGdmB (HY78_09235) in E. coliDepositorArticleInsertSwGdmB
Tags6X HisExpressionBacterialPromoterT7Available SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
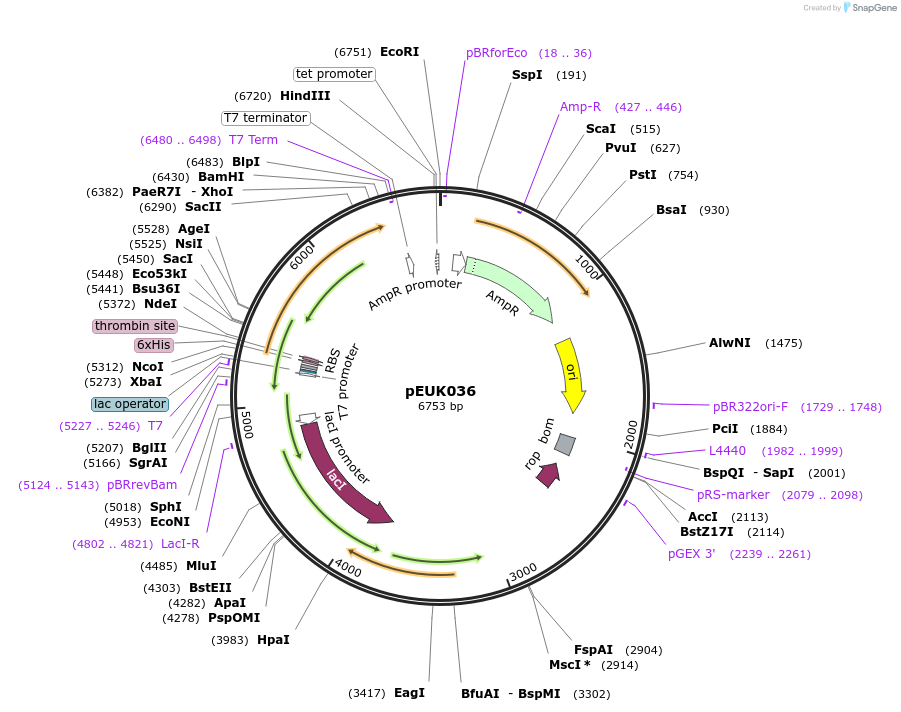
pEUK036
Plasmid#180828PurposeT7-inducible expression construct to produce CnGdmA (CNE_RS35785) in E. coliDepositorArticleInsertCnGdmA
Tags6X HisExpressionBacterialPromoterT7Available SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
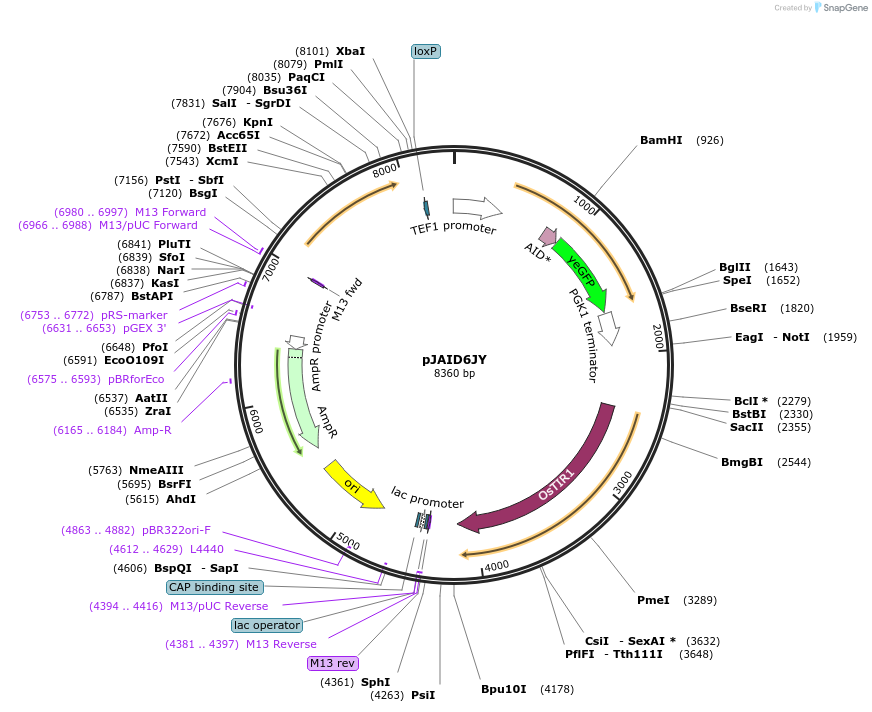
pJAID6JY
Plasmid#165049PurposeIntegrative expression of TRX1-AID* (mini auxin inducible degron)-yEGFP and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>TRX1-AID*>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
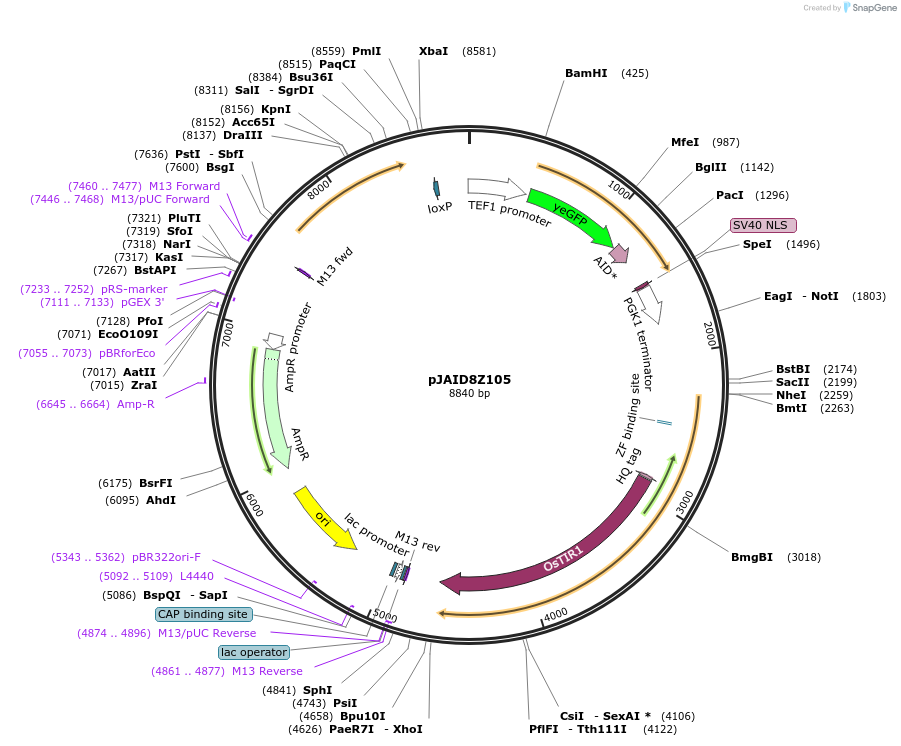
pJAID8Z105
Plasmid#165057PurposeIntegrative expression of nucleus-targeting yEGFP-AID*-CUP1 and SKP1-fusing rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>yEGFP>AID*>CUP1>SV40NSL>T-PGK1-P-ACS2>SKP1>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
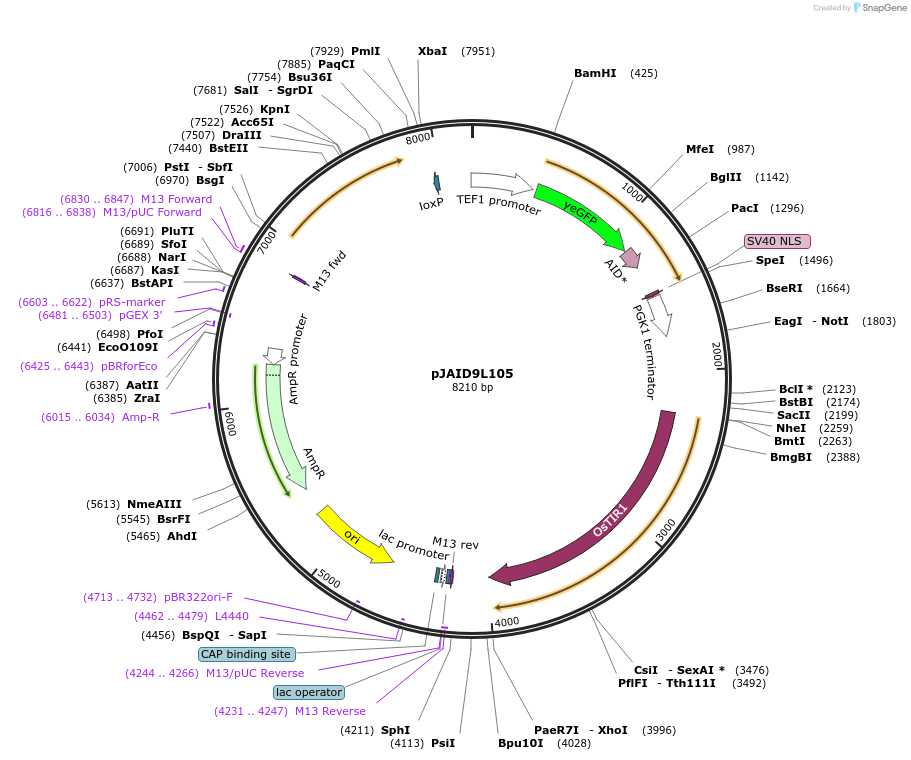
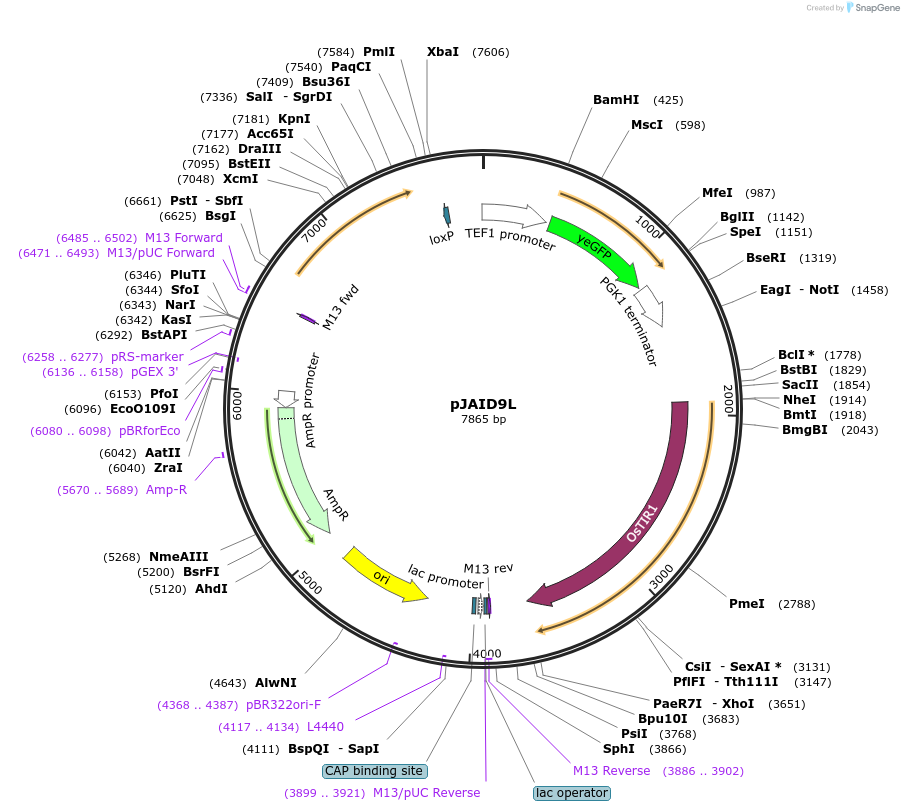
pJAID9L105
Plasmid#165055PurposeIntegrative expression of nucleus-targeting yEGFP-AID*-CUP1 and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>yEGFP>AID*>CUP1>SV40NSL>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJAID9L
Plasmid#165052PurposeIntegrative expression of yEGFP and rice auxin receptor gene OsTIR1. OsTIR1 is codon-optimised for expression in S. cerevisiaeDepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
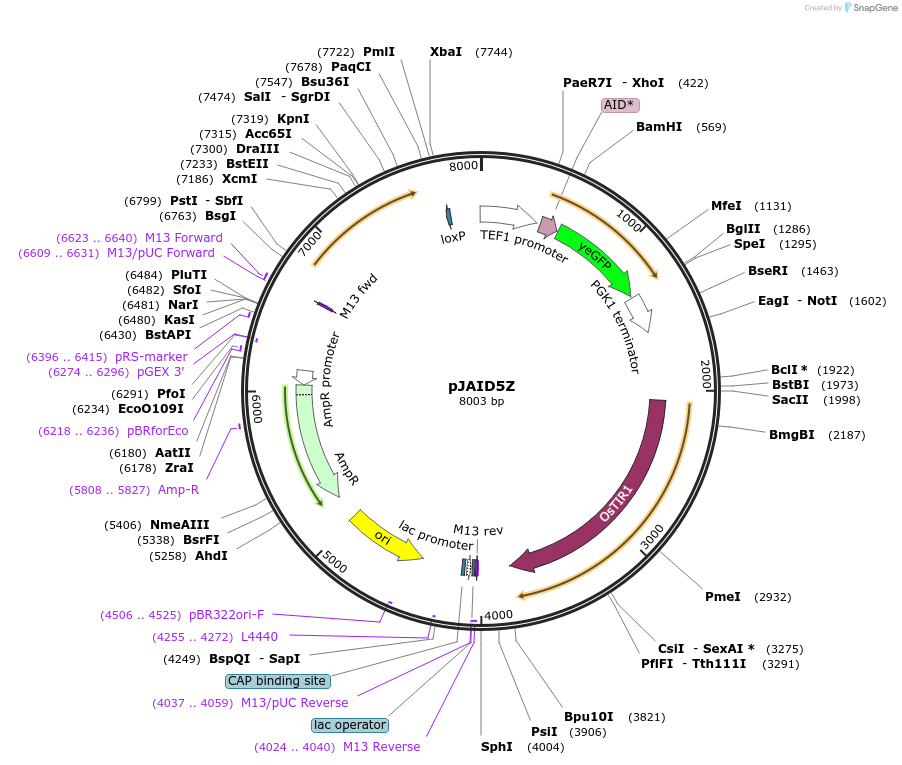
pJAID5Z
Plasmid#165047PurposeIntegrative expression of AID* (mini auxin inducible degron)-yEGFP and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>AID*>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationOsTIR1 is codon-optimised for expression in S. ce…Available SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
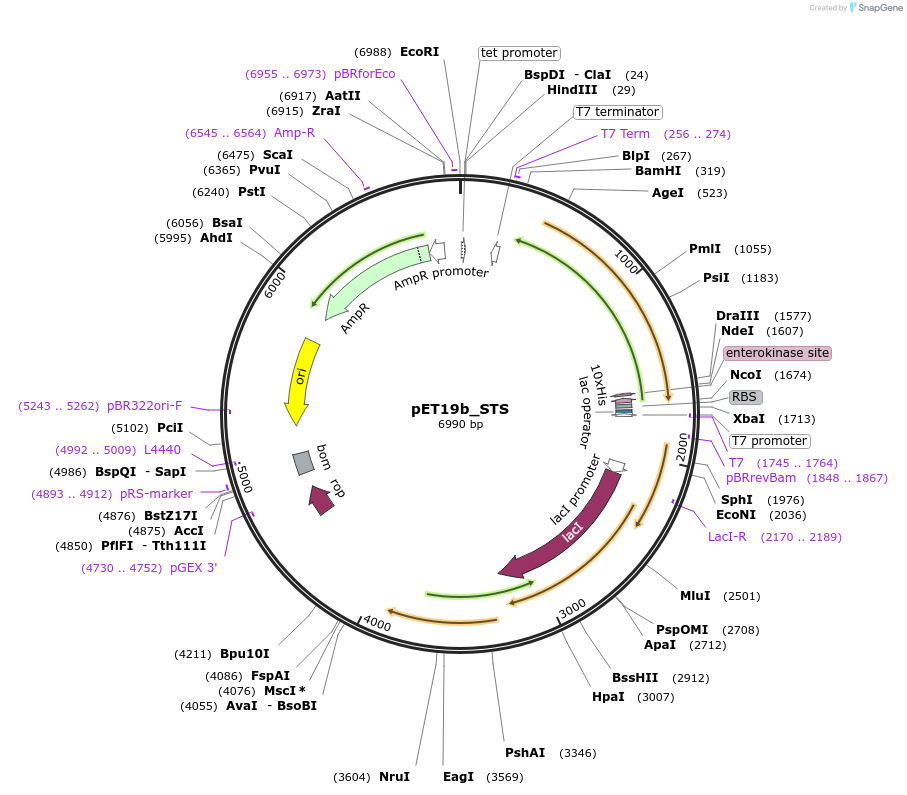
pET19b_STS
Plasmid#139799Purposeexpresses biosynthetic enzyme STSDepositorInsertSTS (STS Mustard Weed, vitis vinifera)
ExpressionBacterialAvailable SinceAug. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
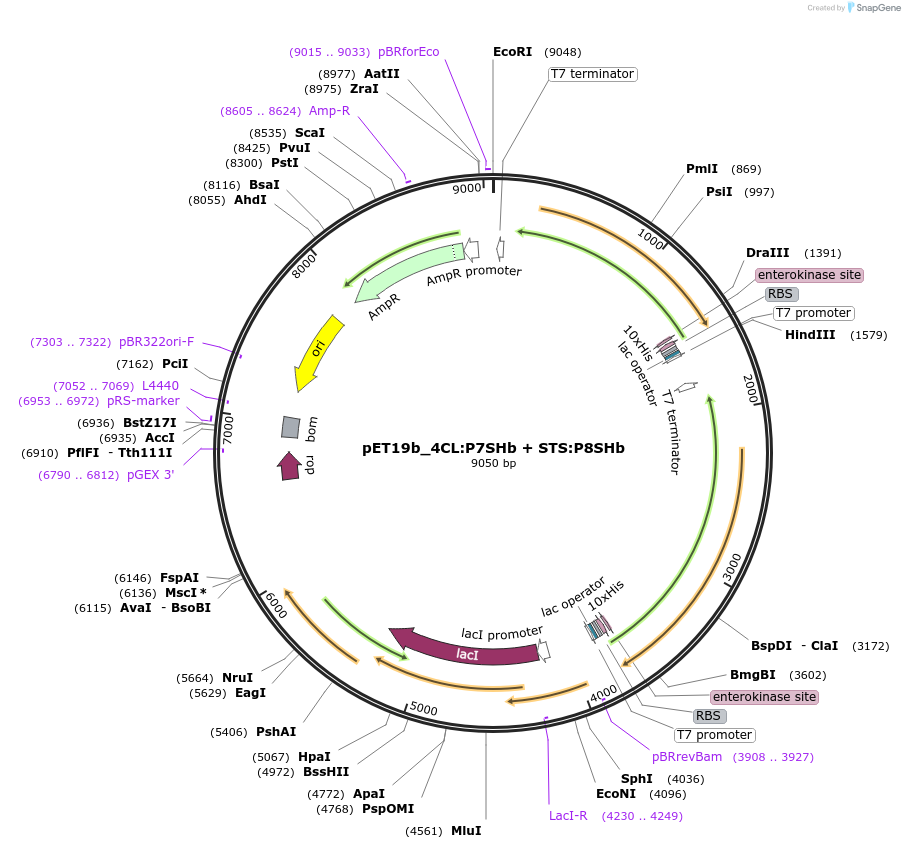
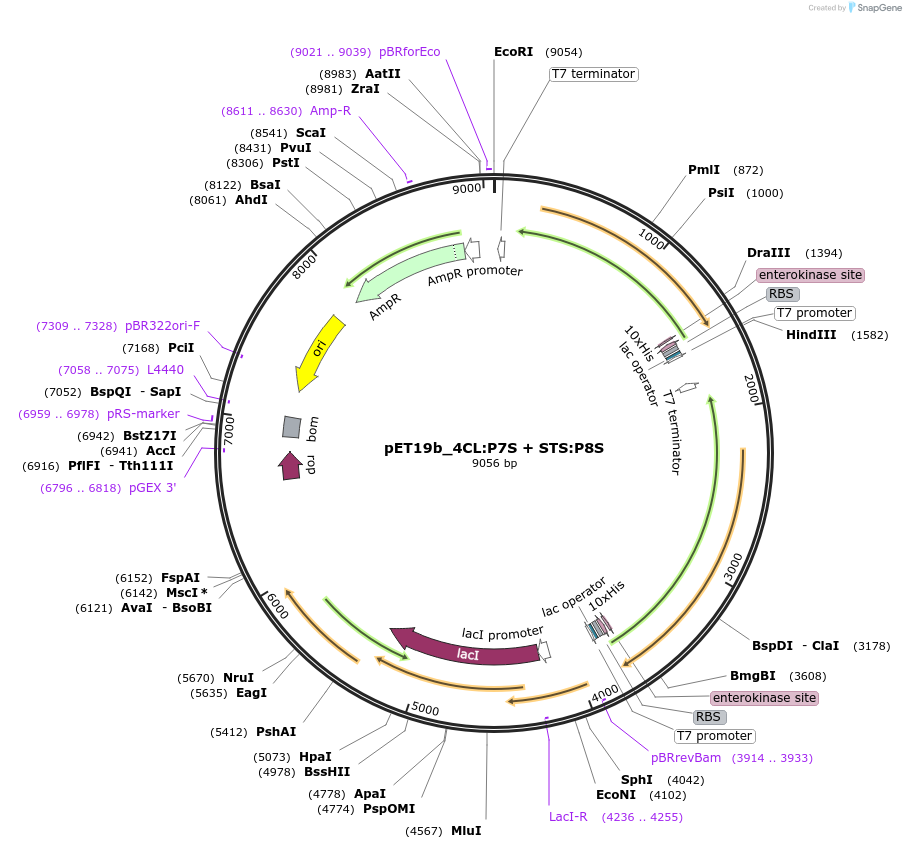
pET19b_4CL:P7SHb + STS:P8SHb
Plasmid#139793Purposeexpression of biosynthetic enzymes 4CL and STS in fusion with CCDepositorInserts4CL:P7SHb
STS:P8SHb
ExpressionBacterialAvailable SinceJune 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
pET19b_4CL:P7S + STS:P8S
Plasmid#139794Purposeexpression of biosynthetic enzymes 4CL and STS in fusion with CCDepositorInserts4CL:P7S
STS:P8S
ExpressionBacterialAvailable SinceJune 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
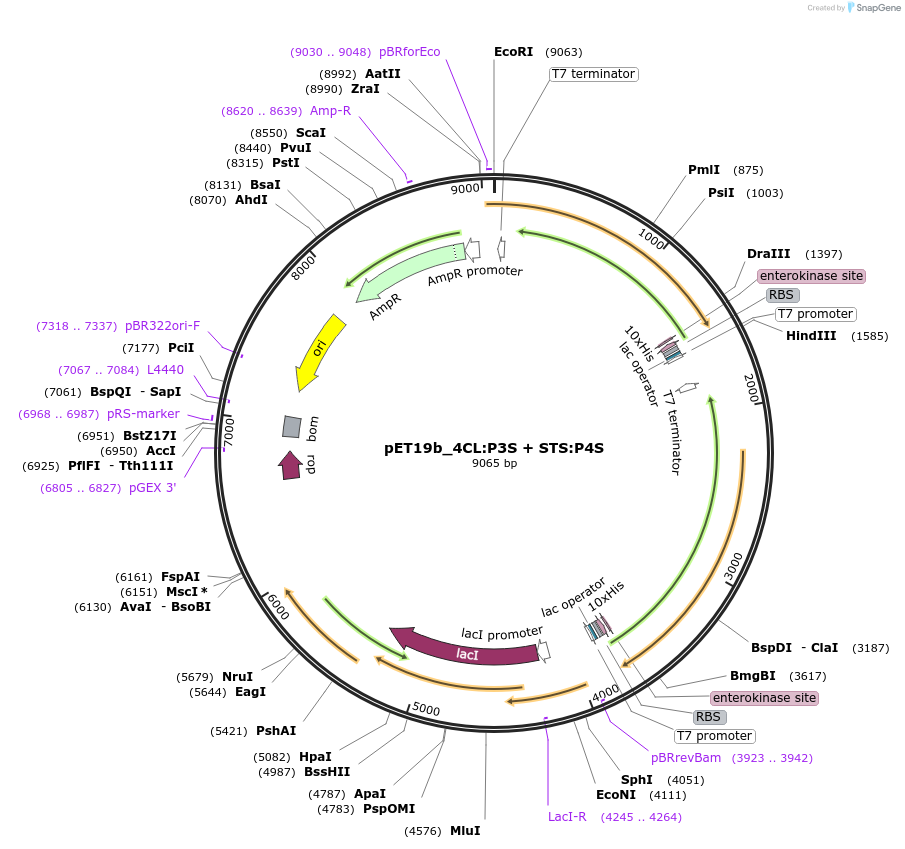
pET19b_4CL:P3S + STS:P4S
Plasmid#139789Purposeexpression of biosynthetic enzymes 4CL and STS in fusion with CCDepositorInsertbiosynthetic enzymes 4CL and STS in fusion with coiled-coil segments
ExpressionBacterialAvailable SinceApril 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
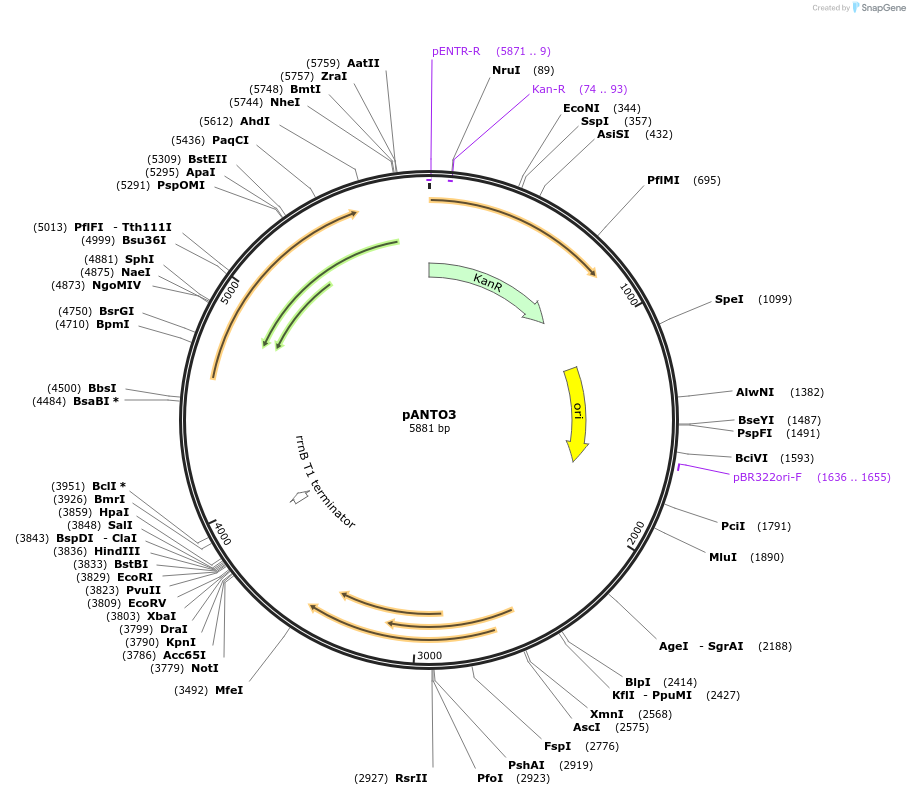
pANTO3
Plasmid#131107PurposeTo generate a replicative plasmid carrying mycobacteriophage L5 integraseDepositorInsertmycobacteriophage L5 integrase
ExpressionBacterialAvailable SinceDec. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
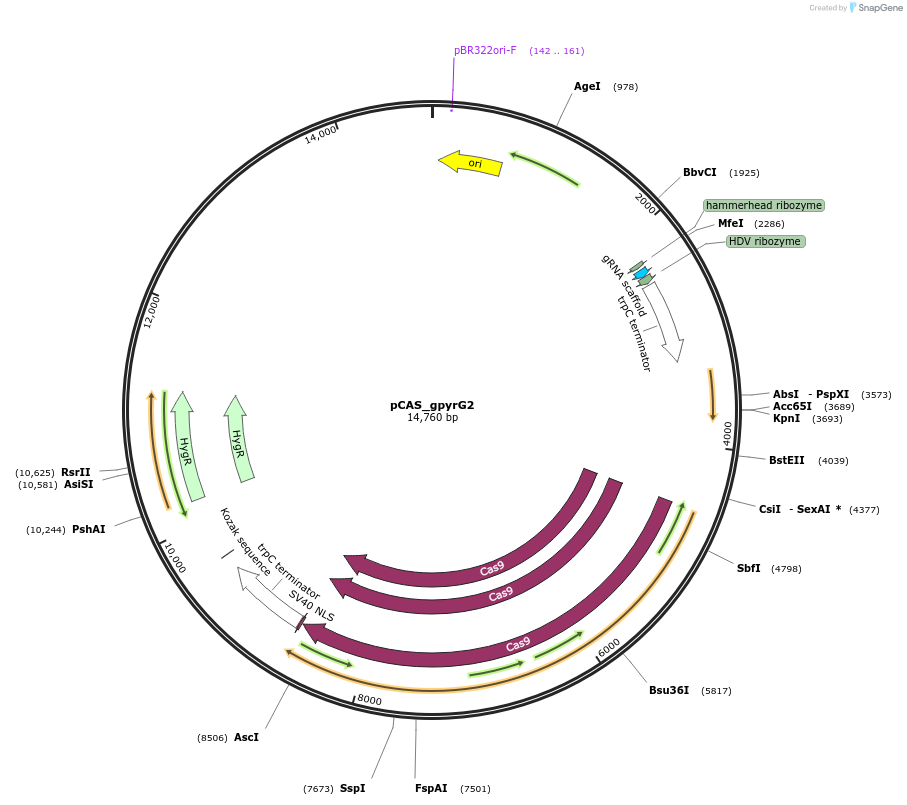
pCAS_gpyrG2
Plasmid#90277Purpose(also pMST620-BB3_gpyrG2_cas9) CRISPR/Cas9 plasmid with gRNA for site pyrG2, Cas9DepositorInsertgRNA (pyrg2)
UseA. nigerExpressionBacterialAvailable SinceDec. 12, 2017AvailabilityAcademic Institutions and Nonprofits only -
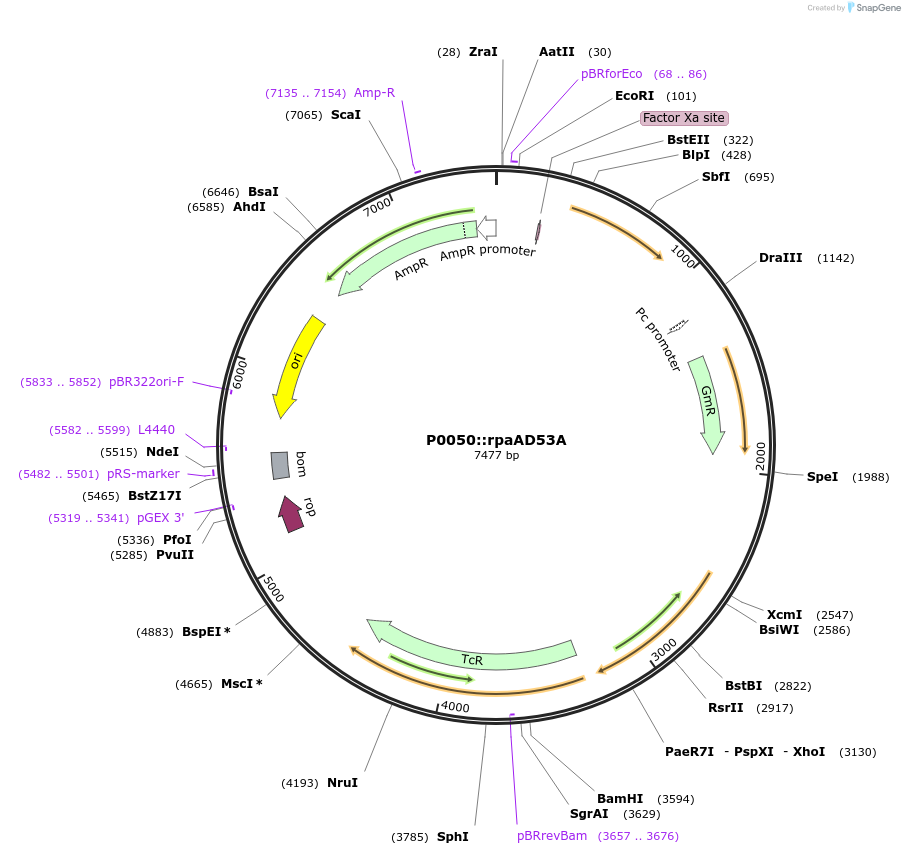
P0050::rpaAD53A
Plasmid#101831Purposedrives expression of rpaA D53A variant from an rpaA-independent promoterDepositorInsertRpaA D53A with p0050 promoter (+ gentamicin resistance casette)
UseCloning vectorMutationD53 mutated to AAvailable SinceOct. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
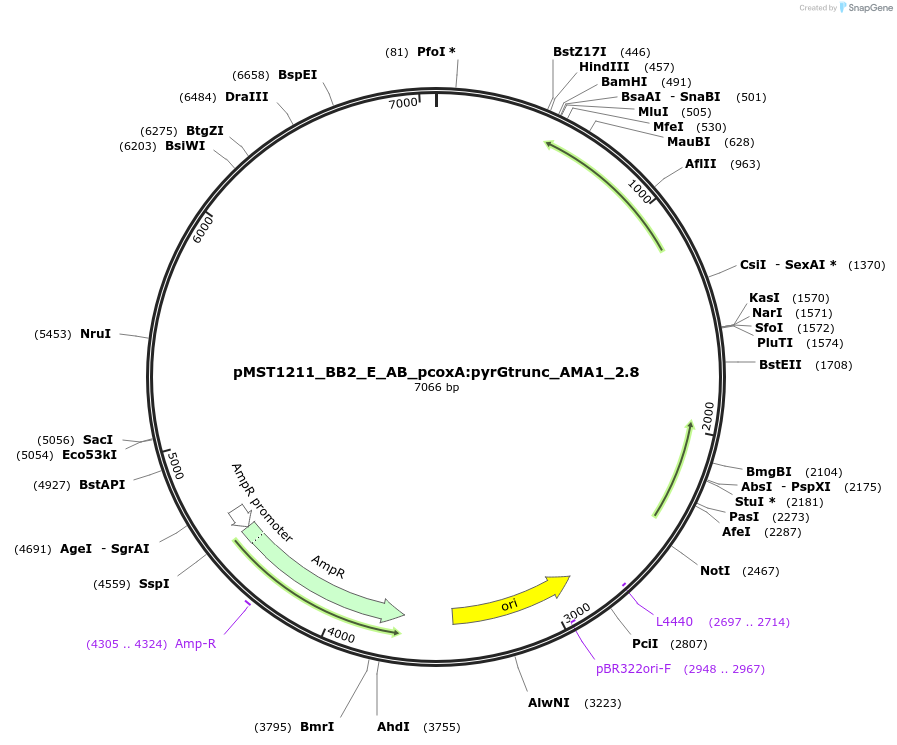
pMST1211_BB2_E_AB_pcoxA:pyrGtrunc_AMA1_2.8
Plasmid#90285PurposeBB2 with split pyrG marker for homologous integration with linker AB (negative control)DepositorInsertpyrGtrunc
ExpressionBacterialMutationtruncated version of pyrG gene (sequence is from …Available SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
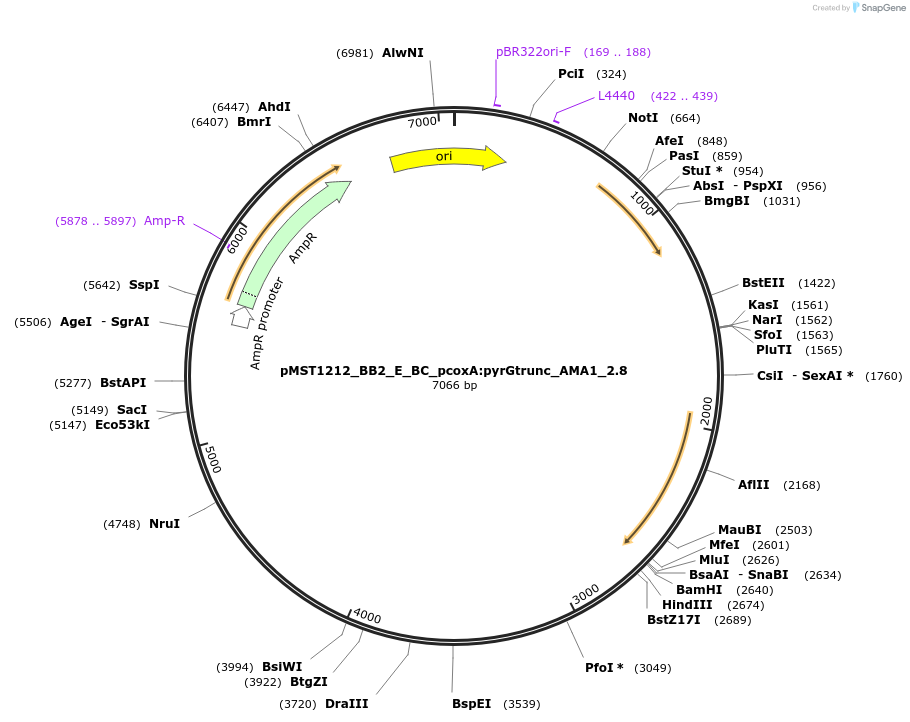
pMST1212_BB2_E_BC_pcoxA:pyrGtrunc_AMA1_2.8
Plasmid#90286PurposeBB2 with split pyrG marker for homologous integration with linker BC (for 1 expression cassette)DepositorInsertpyrGtrunc
ExpressionBacterialMutationtruncated version of pyrG gene (sequence is from …Available SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
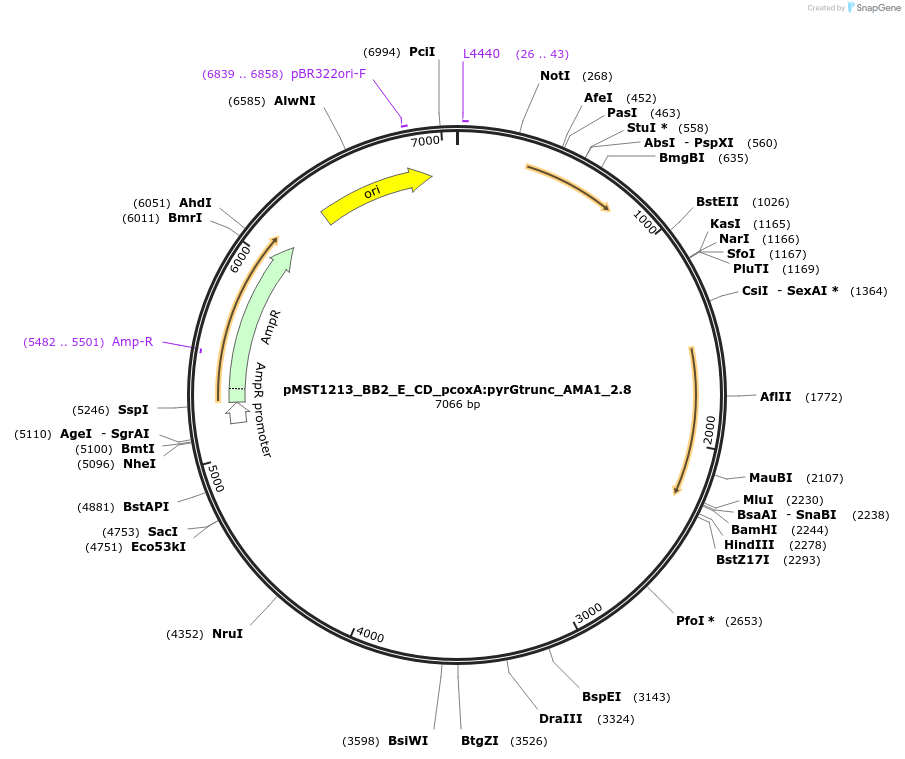
pMST1213_BB2_E_CD_pcoxA:pyrGtrunc_AMA1_2.8
Plasmid#90287PurposeBB2 with split pyrG marker for homologous integration with linker CD (for 2 expression cassettes)DepositorInsertpyrGtrunc
ExpressionBacterialMutationtruncated version of pyrG gene (sequence is from …Available SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
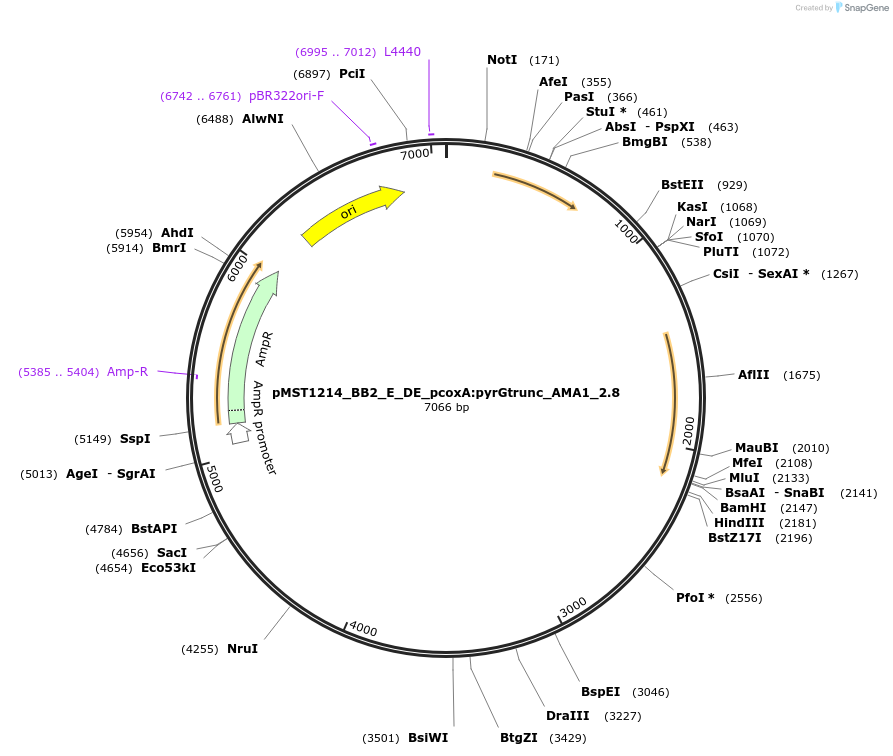
pMST1214_BB2_E_DE_pcoxA:pyrGtrunc_AMA1_2.8
Plasmid#90288PurposeBB2 with split pyrG marker for homologous integration with linker DE (for 3 expression cassettes)DepositorInsertpyrGtrunc
ExpressionBacterialMutationtruncated version of pyrG gene (sequence is from …Available SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
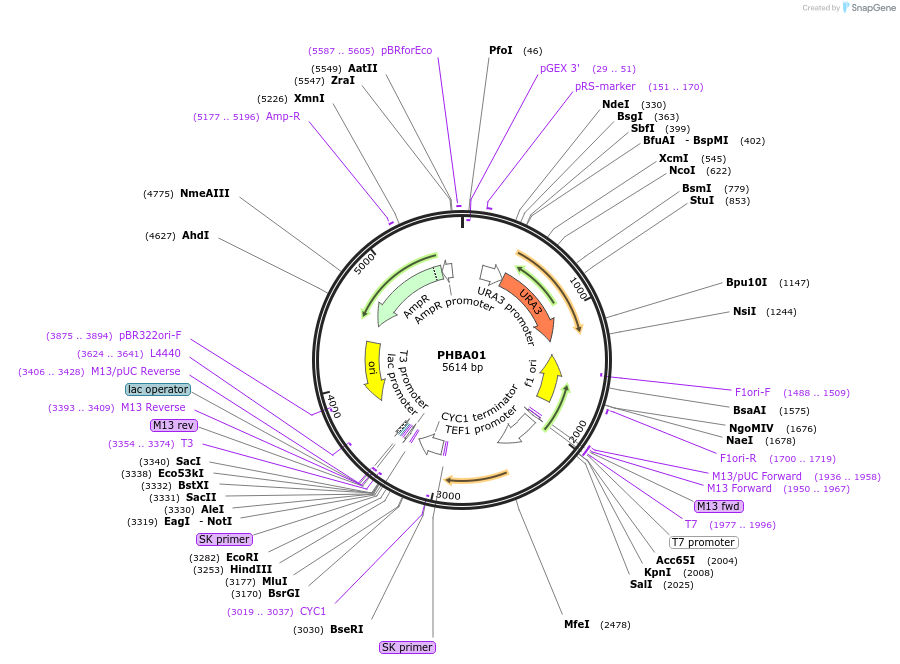
PHBA01
Plasmid#83535PurposeConstitutive TEF1 promoter mediated expression of the UBiC geneDepositorInsertpTEF1-UBiC-CYC1t
ExpressionYeastPromoterpTEF1Available SinceOct. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
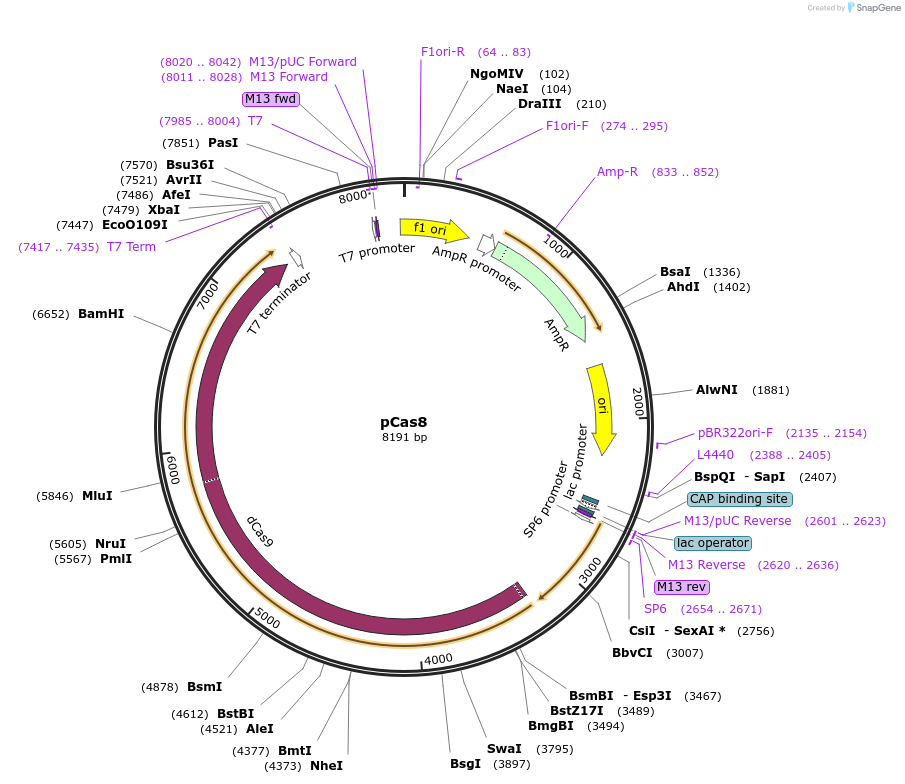
pCas8
Plasmid#82380PurposedCas9 without a promoter in acsADepositorInsertdCas9
PromoternoneAvailable SinceSept. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
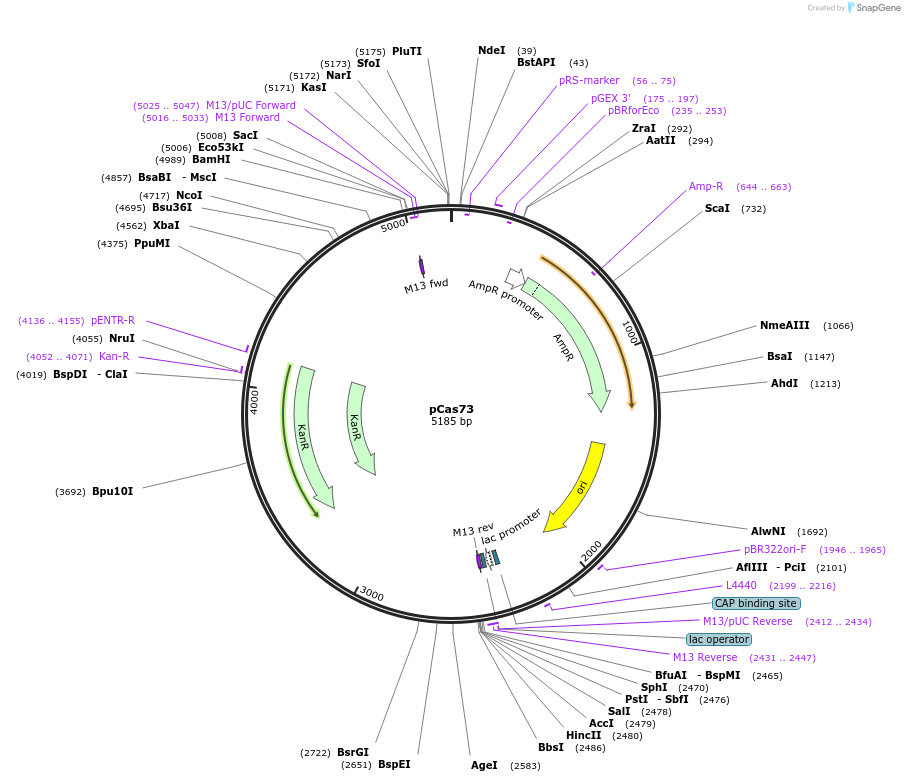
pCas73
Plasmid#82398PurposeΔccm:Km, used to delete ccm operonDepositorInsertccm deletion
PromoternoneAvailable SinceSept. 12, 2016AvailabilityAcademic Institutions and Nonprofits only -
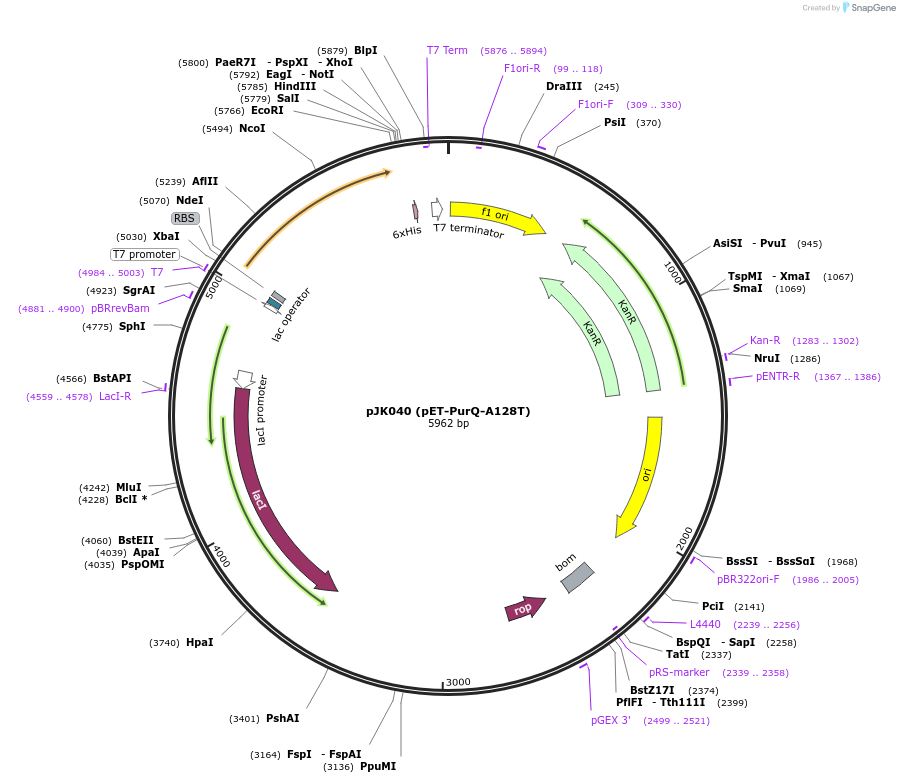
pJK040 (pET-PurQ-A128T)
Plasmid#73840PurposeProduces Bacillus subtilis formylglycinamidine ribonucleotide synthetase glutaminase subunit, A128T mutant (PurQ-A128T)DepositorInsertformylglycinamidine ribonucleotide synthetase, glutaminase subunit
ExpressionBacterialMutationA128T mutantPromoterT7Available SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
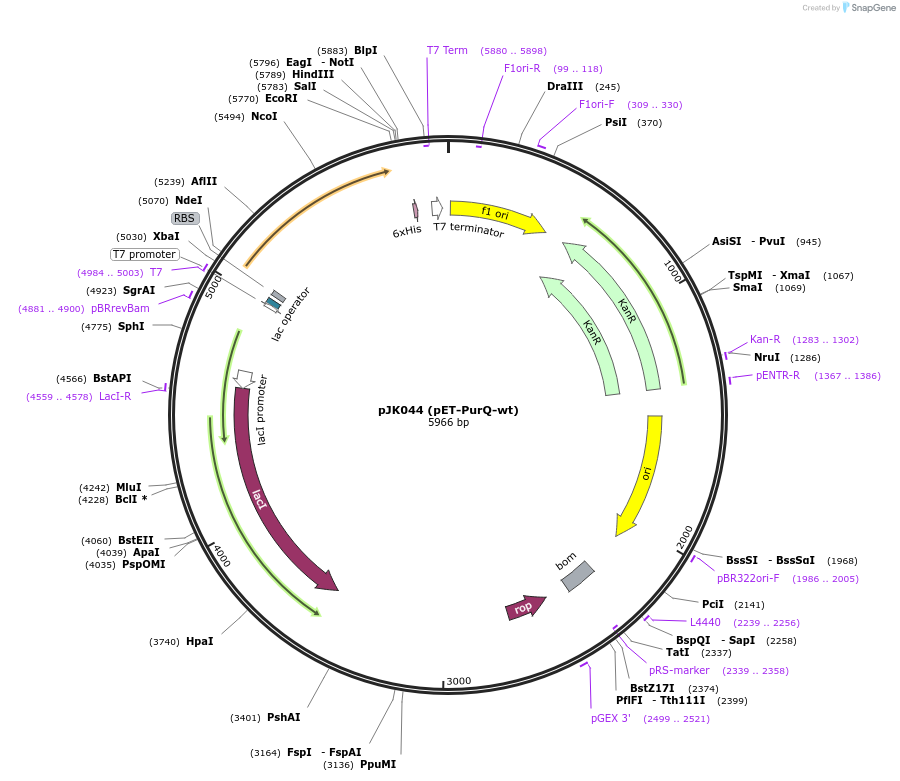
pJK044 (pET-PurQ-wt)
Plasmid#73839PurposeProduces Bacillus subtilis formylglycinamidine ribonucleotide synthetase glutaminase subunit, (PurQ)DepositorInsertformylglycinamidine ribonucleotide synthetase, glutaminase subunit
ExpressionBacterialPromoterT7Available SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
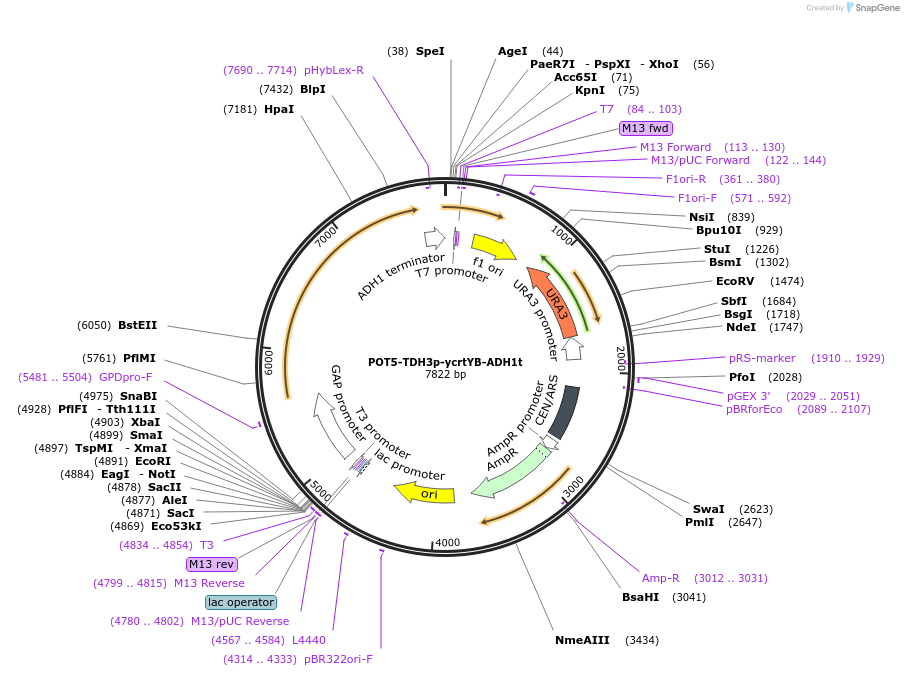
POT5-TDH3p-ycrtYB-ADH1t
Plasmid#65358PurposeTU for combinatorial construction of ?-carotene pathwayDepositorInsertTDH3p-ycrtYB-ADH1t
UseUnspecifiedAvailable SinceFeb. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
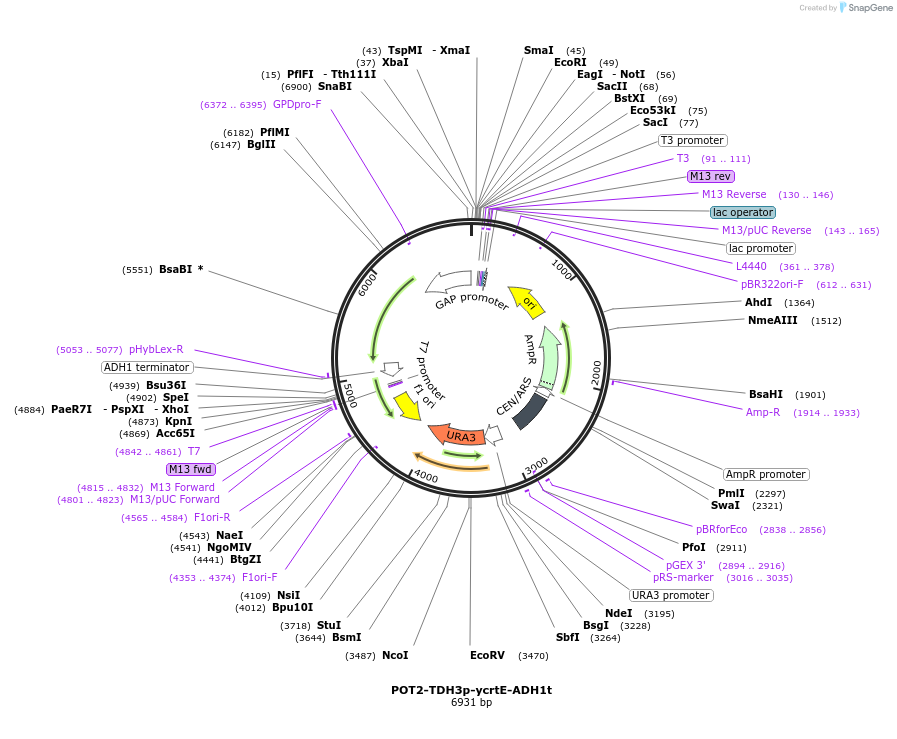
POT2-TDH3p-ycrtE-ADH1t
Plasmid#65352PurposeTU for combinatorial construction of ?-carotene pathwayDepositorInsertTDH3p-ycrtE-ADH1t
UseUnspecifiedAvailable SinceFeb. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
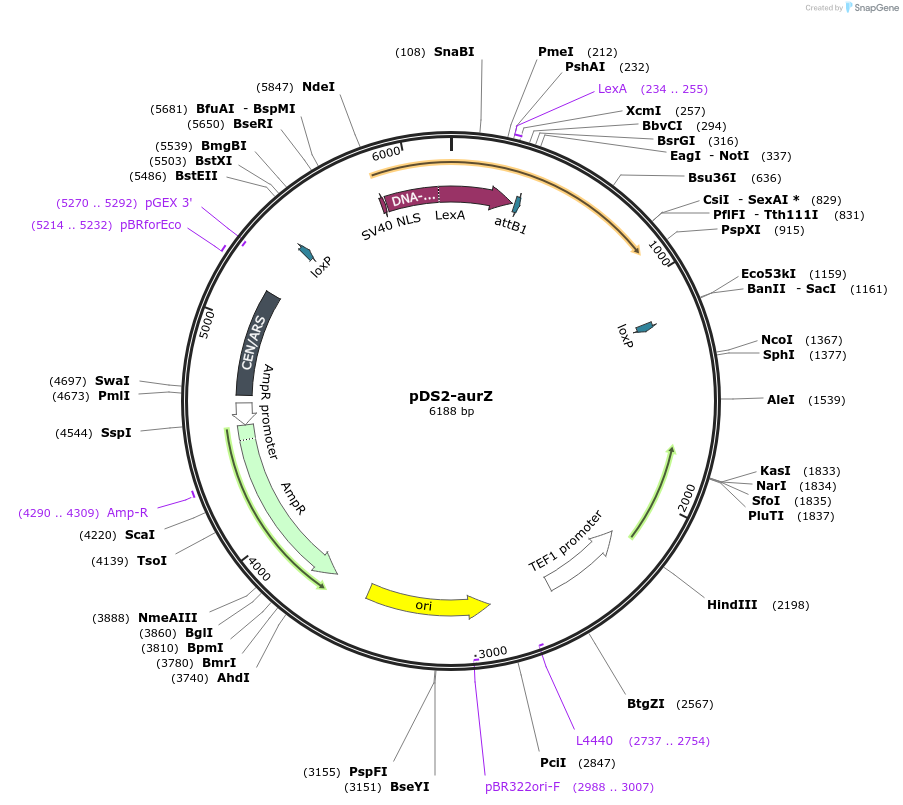
pDS2-aurZ
Plasmid#70151PurposeFusarium graminearum AurZ encoded on CEN/ARS yeast shuttle vector with DS2 selection markerDepositorInsertaurZ
ExpressionYeastPromoterTefAvailable SinceJan. 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
pDS3-aurJ
Plasmid#70152PurposeFusarium graminearum AurJ encoded on CEN/ARS yeast shuttle vector with DS3 selection markerDepositorInsertaurJ
ExpressionYeastPromoterGPD1Available SinceDec. 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
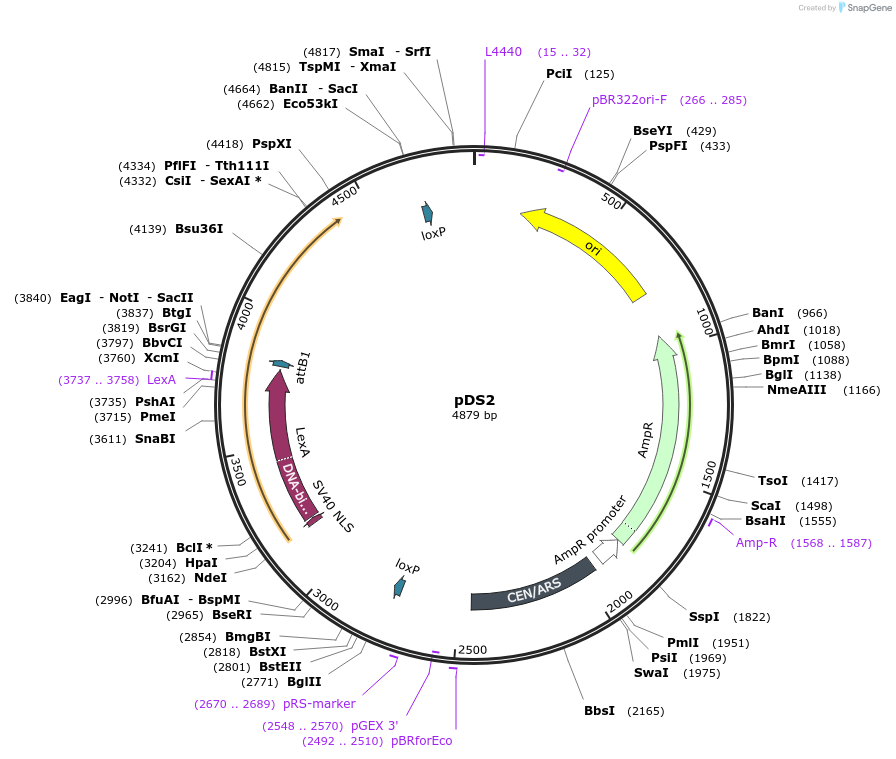
pDS2
Plasmid#70135PurposeCEN/ARS yeast shuttle vector with DS2 selection markerDepositorInsertDS2
UseSynthetic BiologyPromoterAgTEF1Available SinceDec. 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
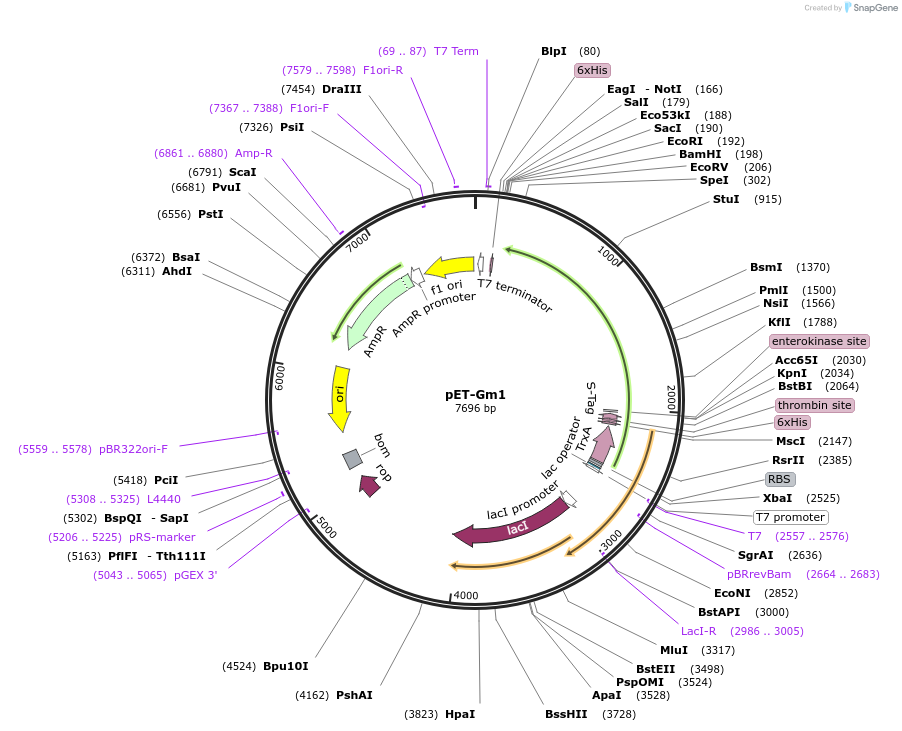
pET-Gm1
Plasmid#53528PurposeExpresses a plant arginyl-tRNA synthetase in E.coliDepositorInsertarginyl-tRNA synthetase
TagsHis Tag and ThioredoxinExpressionBacterialPromoterT7Available SinceJuly 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
-
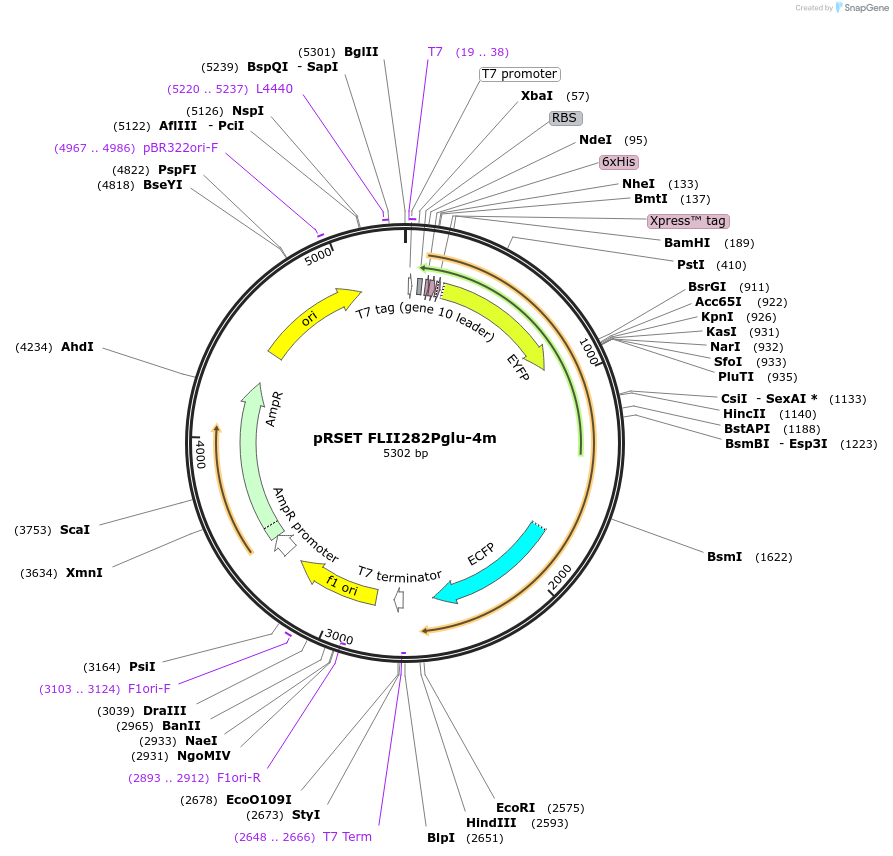
pRSET FLII282Pglu-4m
Plasmid#13564DepositorInsertFLIPglu F16A T282S-ECFP-N283R
TagsCitrineExpressionBacterialMutationF16A, ECFP insert in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
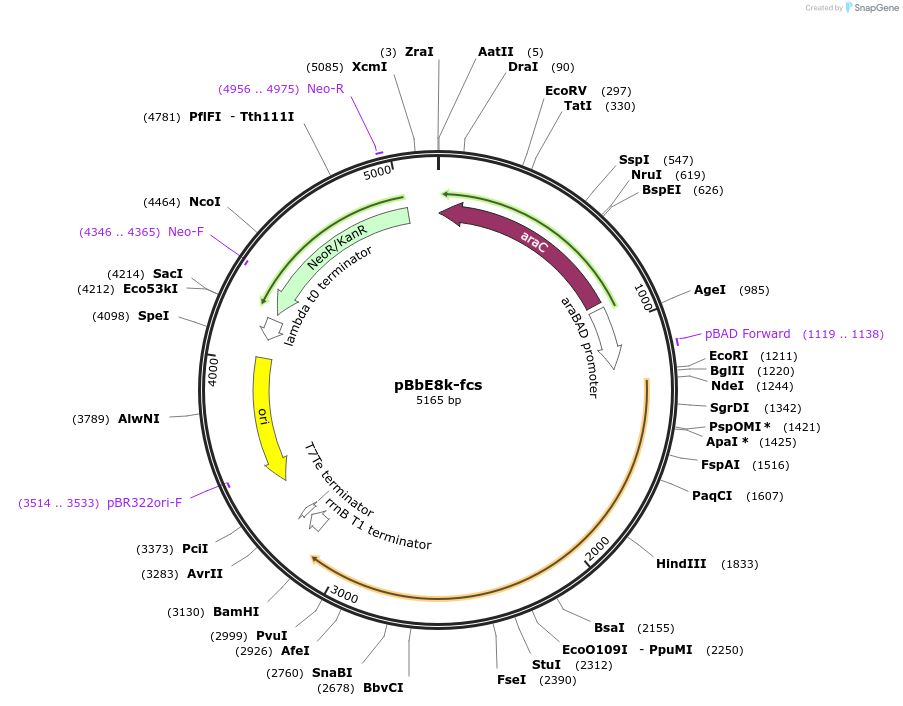
pBbE8k-fcs
Plasmid#83068PurposeExpresses Fcs under pBAD regulationDepositorInsertfcs
UseSynthetic BiologyExpressionBacterialPromoterpBADAvailabilityAcademic Institutions and Nonprofits only -
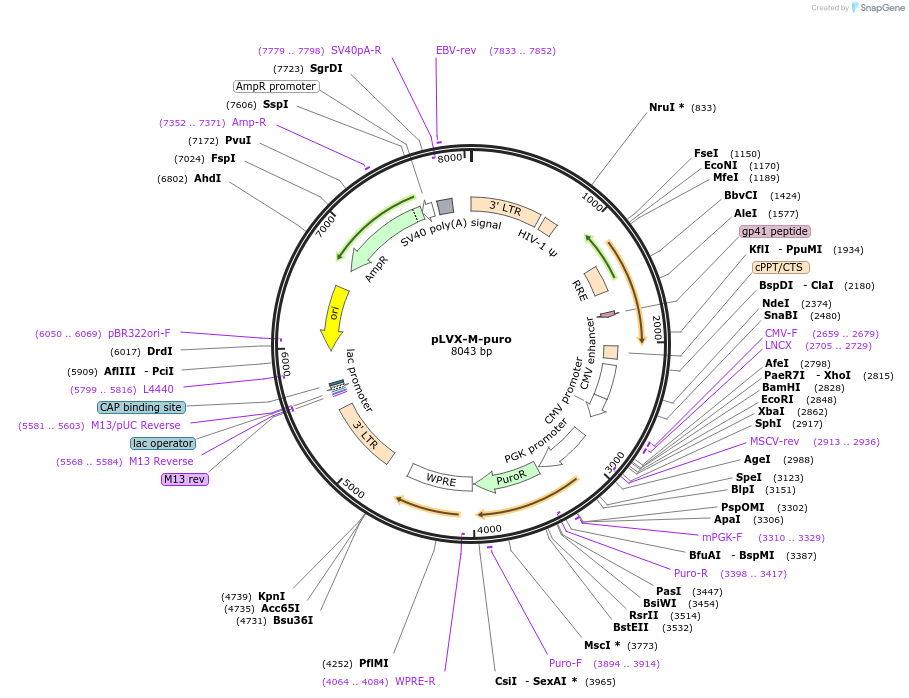
pLVX-M-puro
Plasmid#125839Purposemodifying MCS, replacing EcoRI to BamHI region with BamHI and EcoRIDepositorTypeEmpty backboneUseLentiviralAvailable SinceJune 5, 2019AvailabilityAcademic Institutions and Nonprofits only