We narrowed to 11,014 results for: phen
-
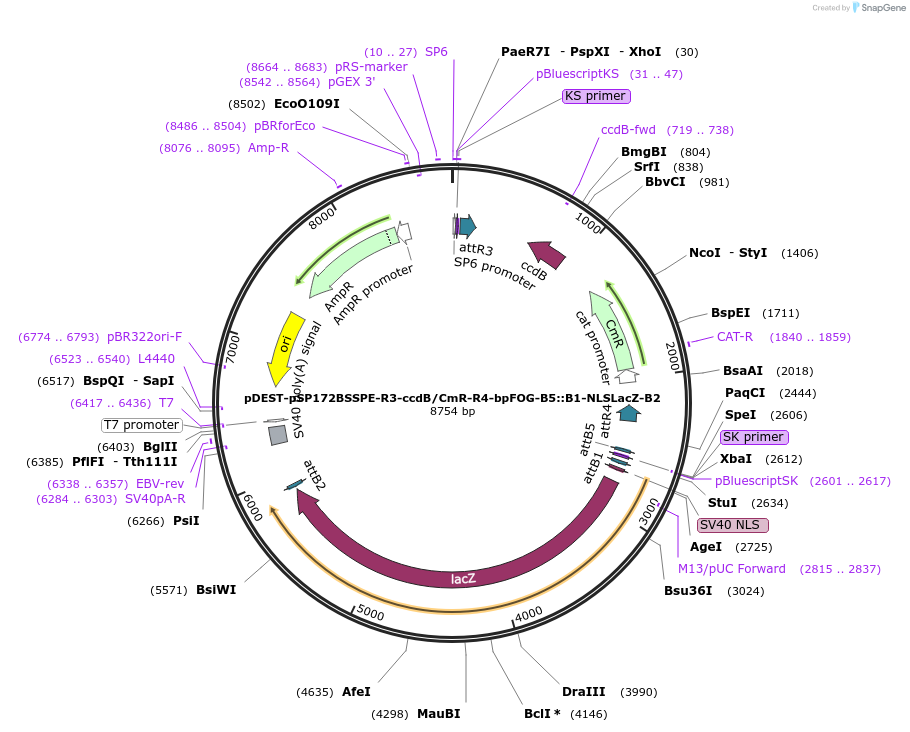
Plasmid#186414PurposeAttR3/R4 destination vector for ascidian electroporation with AttB1/B2 recombined NLSLacZ under control of AttB4/B5-recombined pFOG ectodermal regulatory sequence.DepositorInsertlacZ (lacZ E. coli)
UseGateway destination vectorTagsNuclear localization signalPromoterBasal promoter of the FOG geneAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
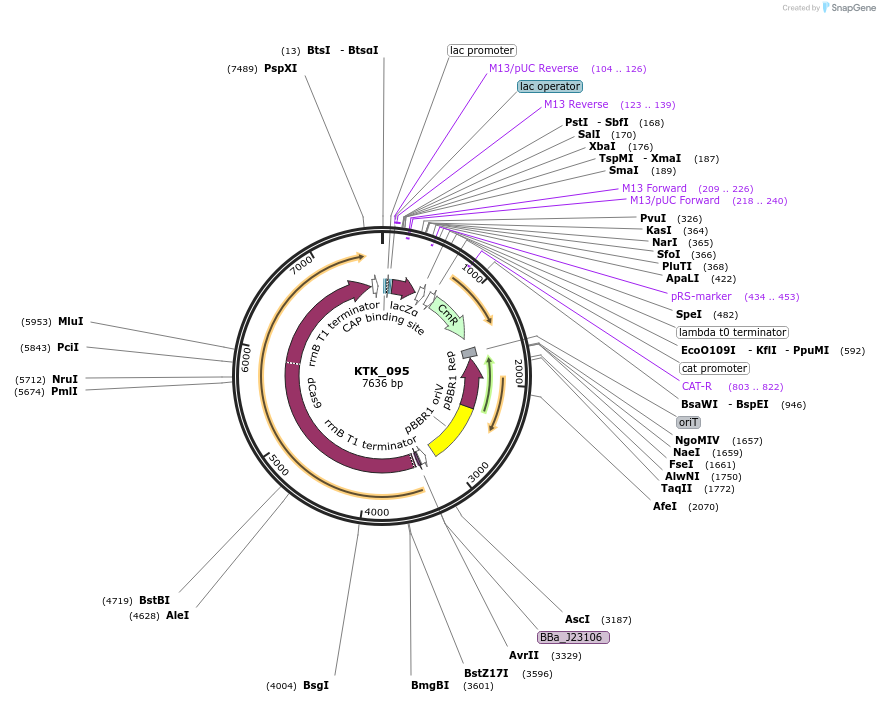
KTK_095
Plasmid#180524PurposedCas9 KTK compatible plasmid. Contains dCas9 and lacZ dropout region with flanking D1.1 overhangs for insertion of gRNA expression assemblyDepositorTypeEmpty backboneExpressionBacterialAvailable SinceApril 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
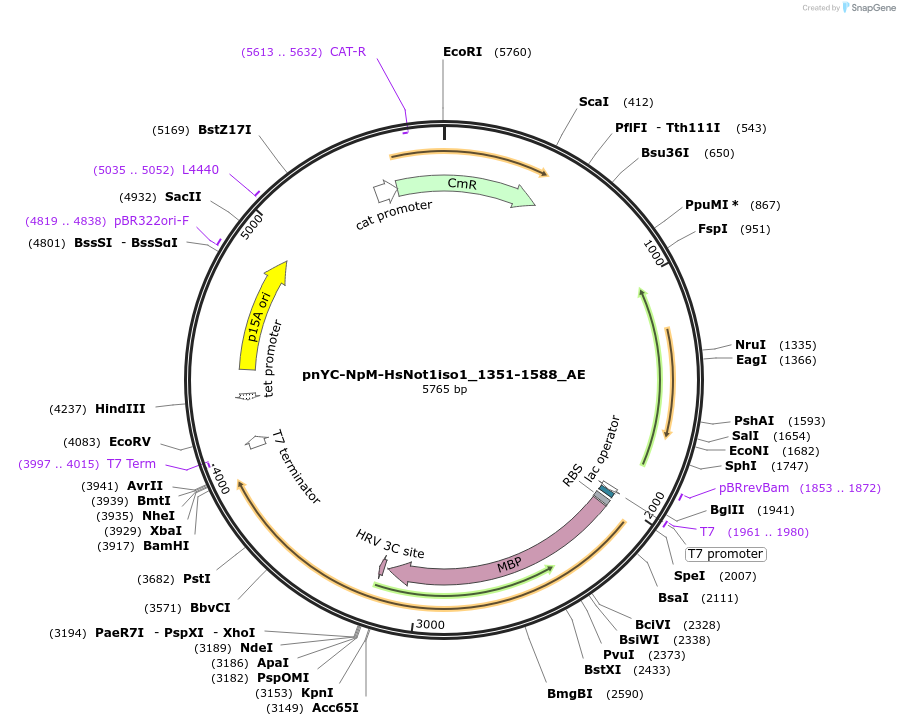
pnYC-NpM-HsNot1iso1_1351-1588_AE
Plasmid#148630PurposeBacterial Expression of HsNot1iso1_1351-1588DepositorInsertHsNot1iso1_1351-1588 (CNOT1 Human)
ExpressionBacterialAvailable SinceMarch 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
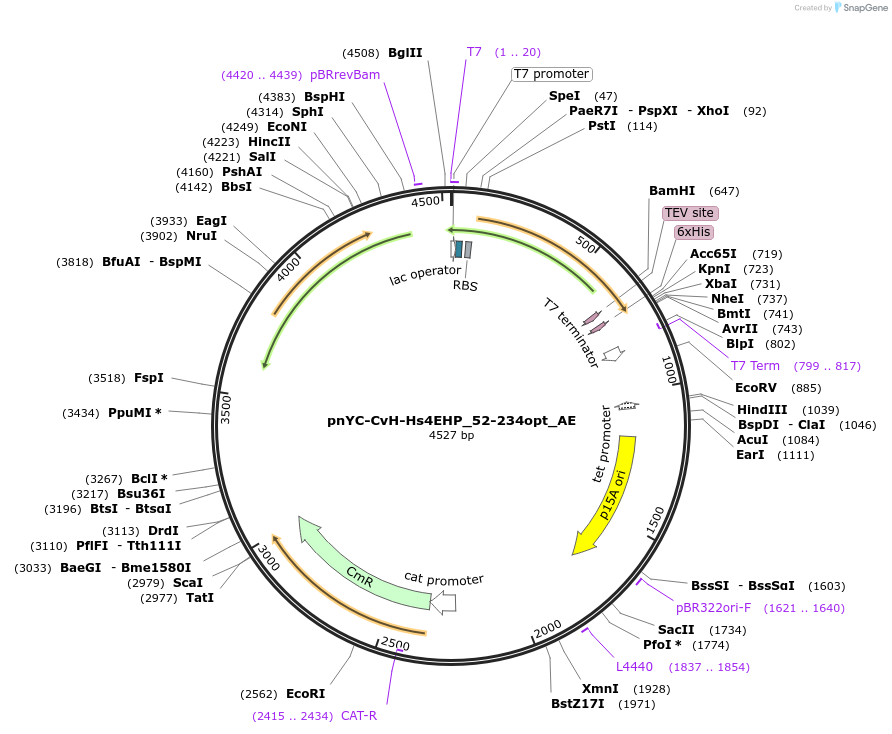
pnYC-CvH-Hs4EHP_52-234opt_AE
Plasmid#148564PurposeBacterial Expression of Hs4EHP_52-234optDepositorInsertHs4EHP_52-234opt (EIF4E2 Human)
ExpressionBacterialAvailable SinceMarch 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
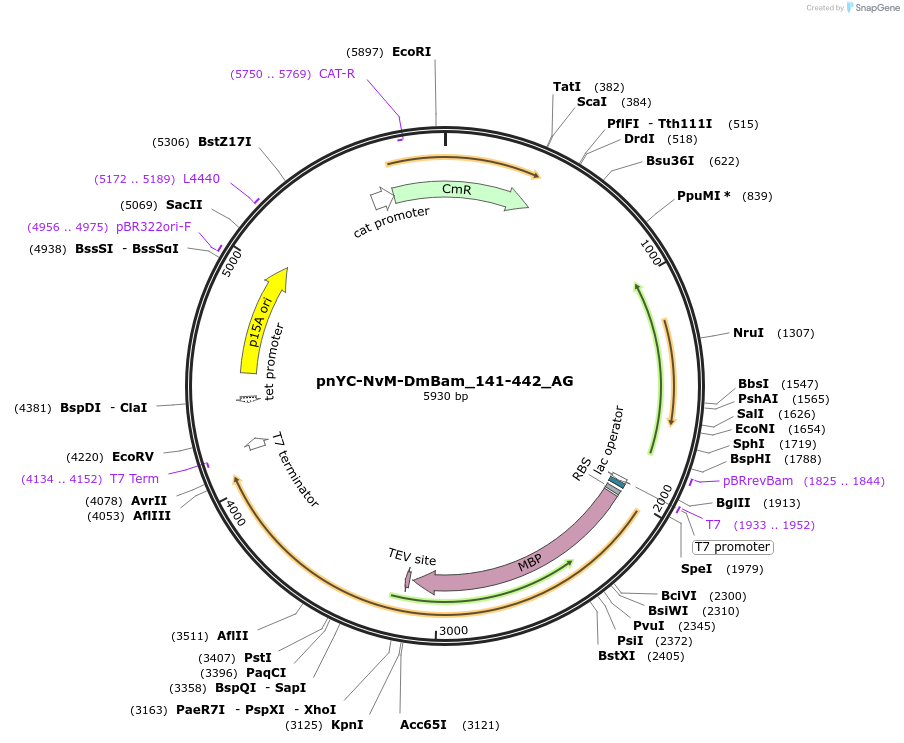
pnYC-NvM-DmBam_141-442_AG
Plasmid#148833PurposeBacterial Expression of DmBam_141-422DepositorInsertDmBam_141-422 (bam Fly)
ExpressionBacterialMutationone non silent mutation A239 to S, described as …Available SinceMarch 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
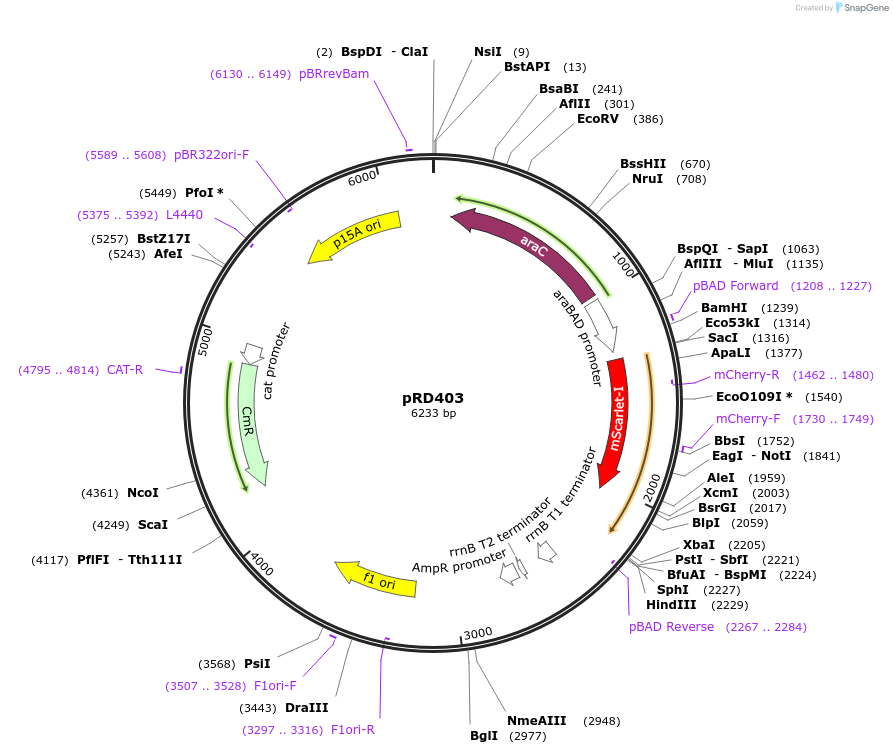
pRD403
Plasmid#163108PurposeExpression of mScarlet-I_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmScarlet-I_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
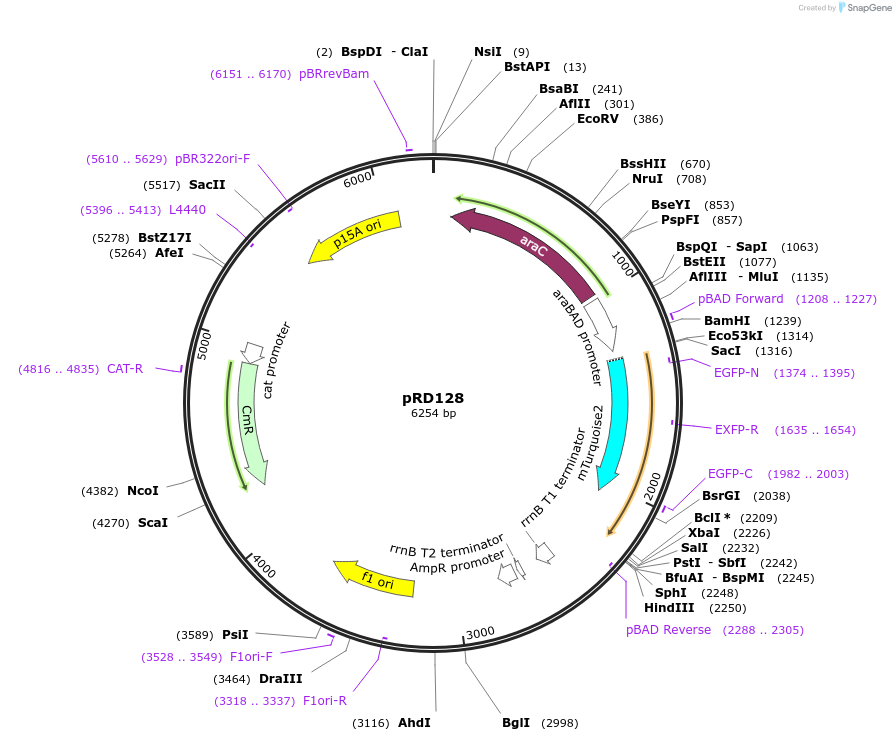
pRD128
Plasmid#163097PurposeExpression of mTurquoise2_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmTurquoise2_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
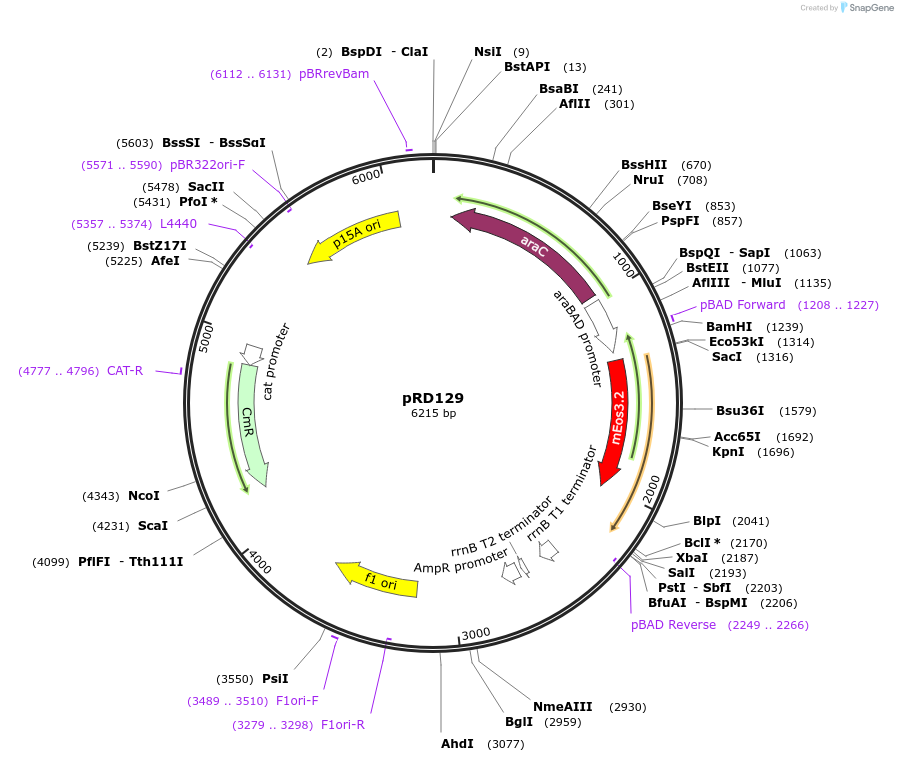
pRD129
Plasmid#163098PurposeExpression of mEos3.2_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmEos3.2_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
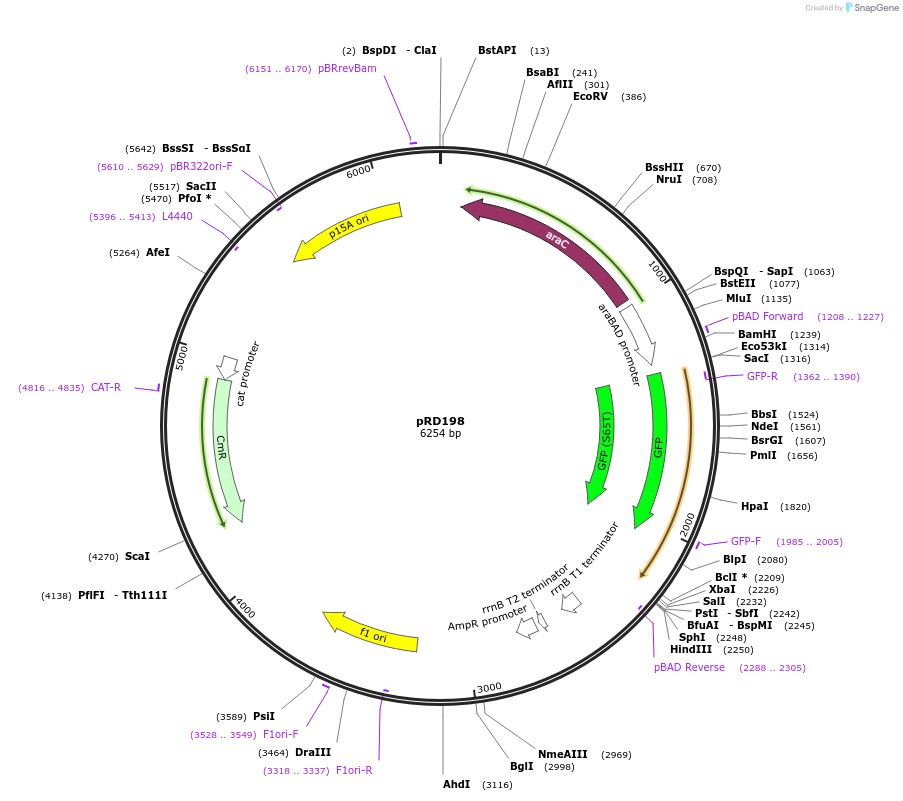
pRD198
Plasmid#163101PurposeExpression of mEGFP_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmEGFP_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
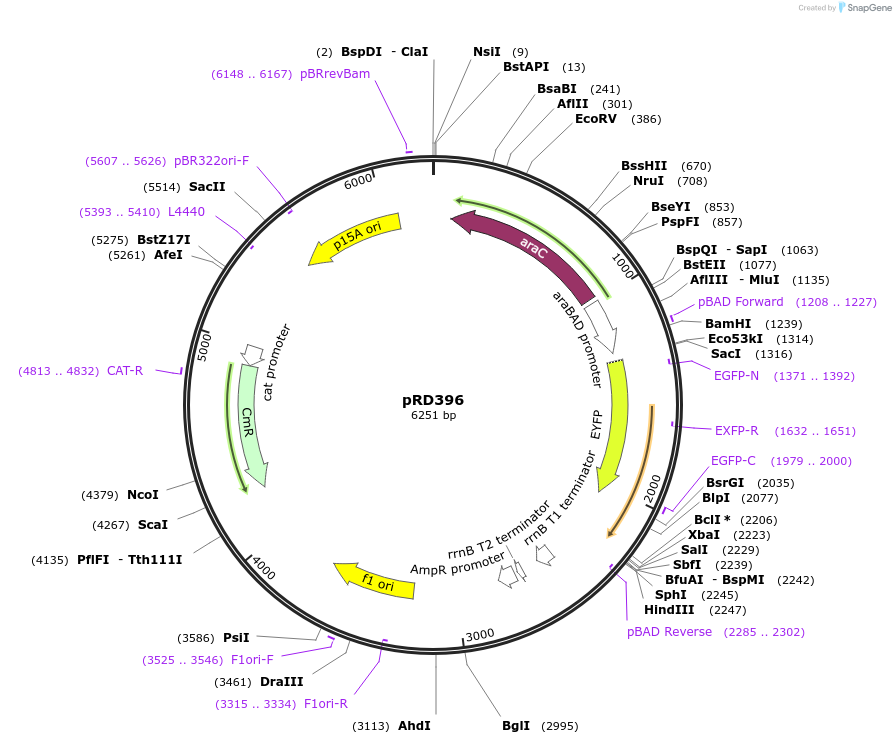
pRD396
Plasmid#163103PurposeExpression of eYFP_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInserteYFP_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
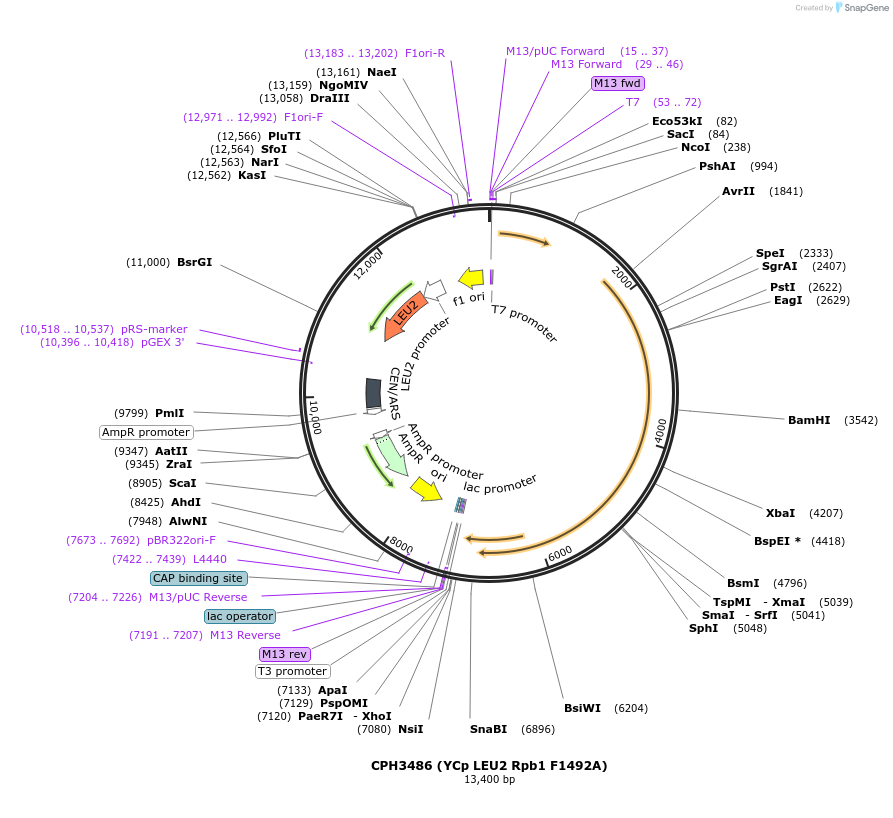
CPH3486 (YCp LEU2 Rpb1 F1492A)
Plasmid#91808PurposeYeast expression of S. cerevisiae Rpb1 with a F1492A mutationDepositorInsertRPO21 (RPO21 Budding Yeast)
ExpressionYeastMutationchanged phenylalanine 1492 to alaninePromoterEndogenous Rpb1 promoterAvailable SinceMarch 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
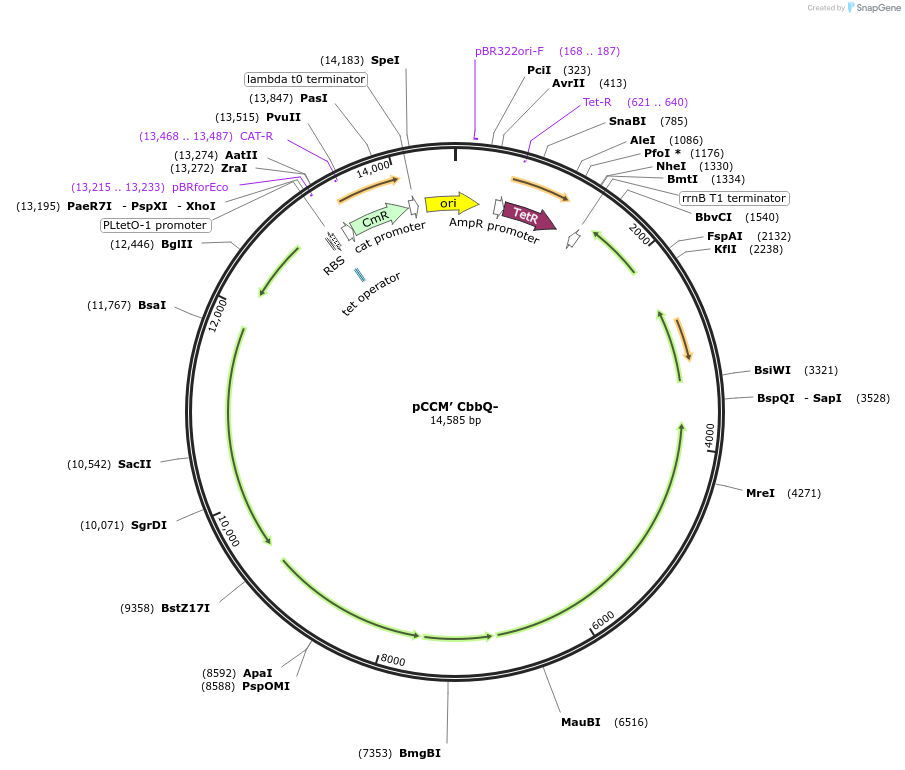
pCCM’ CbbQ-
Plasmid#162711PurposeSecond CCM operon with inactive CbbQ (K46A, E107Q) rubisco activaseDepositorInsertSecond CCM operon cloned from H. neapolitanus
ExpressionBacterialMutationInactivation of CbbQ subunit of CbbOQ rubisco act…PromoterPLtet0-1 promoterAvailable SinceFeb. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
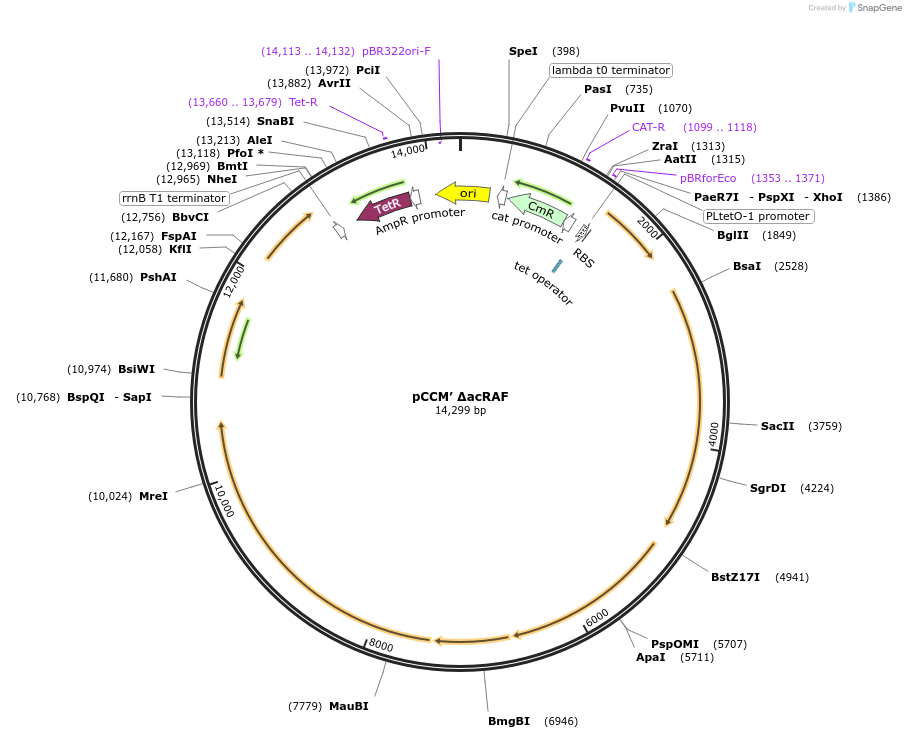
pCCM’ ΔacRAF
Plasmid#162710PurposeSecond CCM operon lacking acRAF; Deletion of putative rubisco chaperone, acRAFDepositorInsertSecond CCM operon cloned from H. neapolitanus
ExpressionBacterialMutationpCCM' with a deletion of putative rubisco ch…PromoterPLtet0-1 promoterAvailable SinceFeb. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
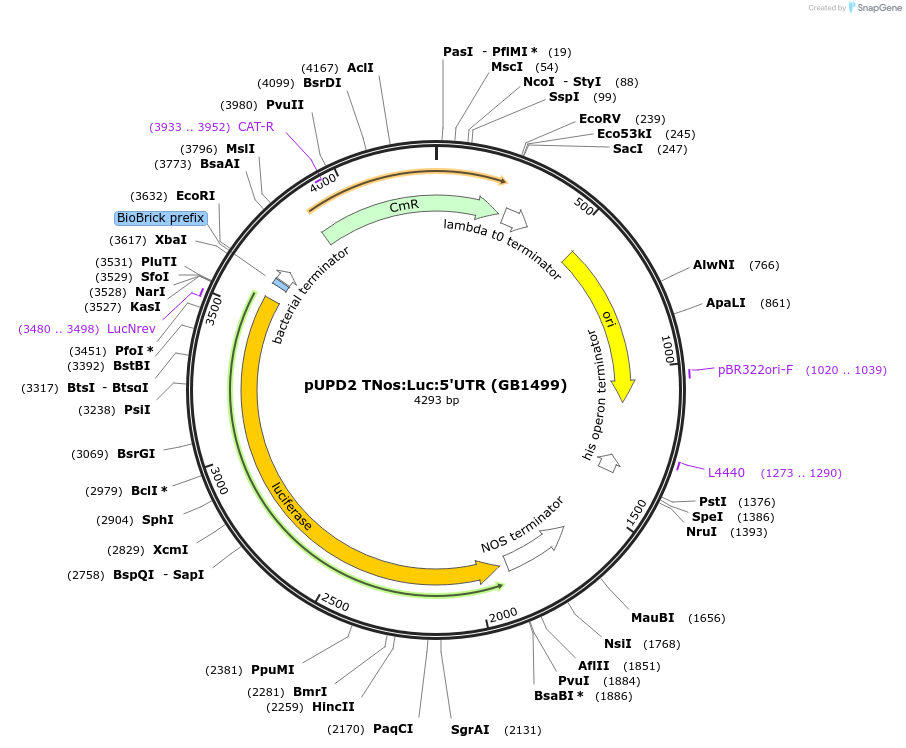
pUPD2 TNos:Luc:5'UTR (GB1499)
Plasmid#160580PurposeReversed sequences of the NOS terminator, Firefly Luciferase CDS and 5' UTR of 35S promoterDepositorInsertLuc
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJan. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
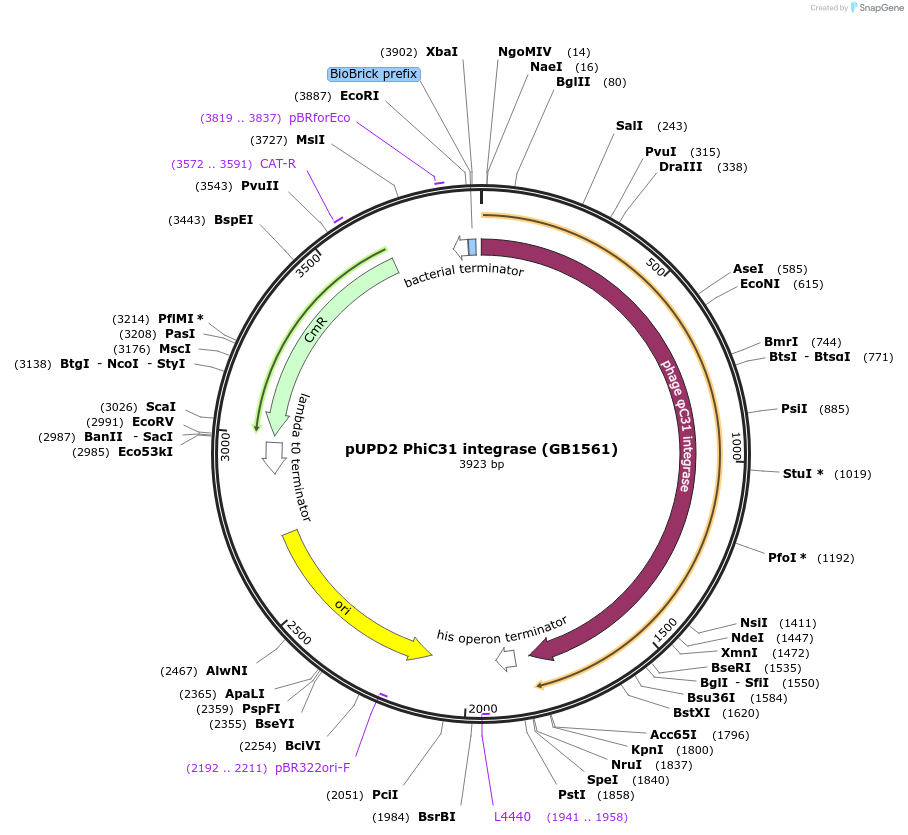
pUPD2 PhiC31 integrase (GB1561)
Plasmid#160586PurposeStreptomyces phage PhiC31 integrase, adapted for C-terminal fusionsDepositorInsertPhiC31
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJan. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
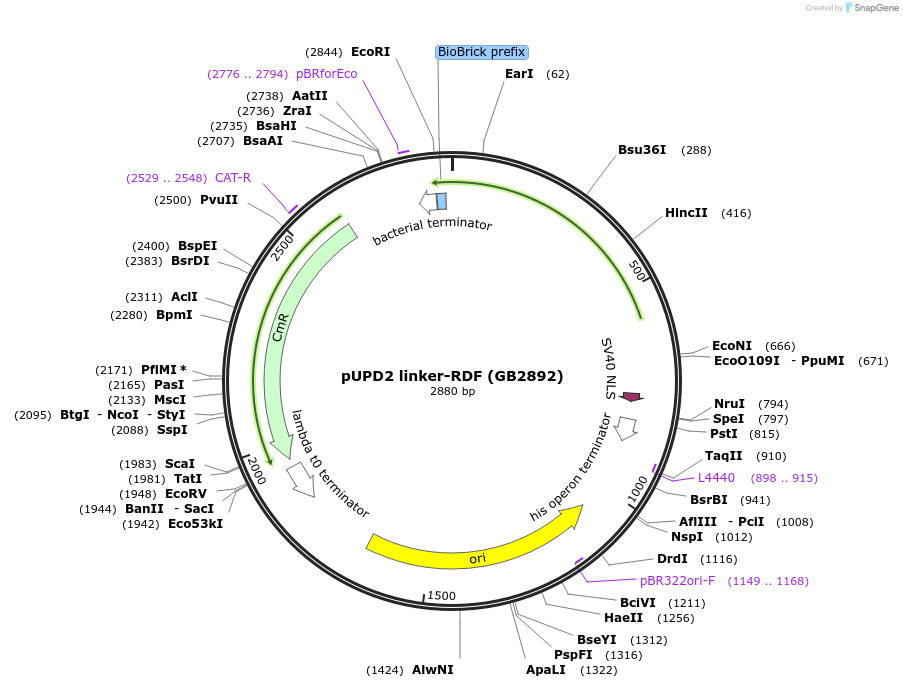
pUPD2 linker-RDF (GB2892)
Plasmid#160604PurposeRDF sequence for C-terminal fusion with PhiC31 recombinase, together with a 18-nt linker sequenceDepositorInsertRDF
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJan. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
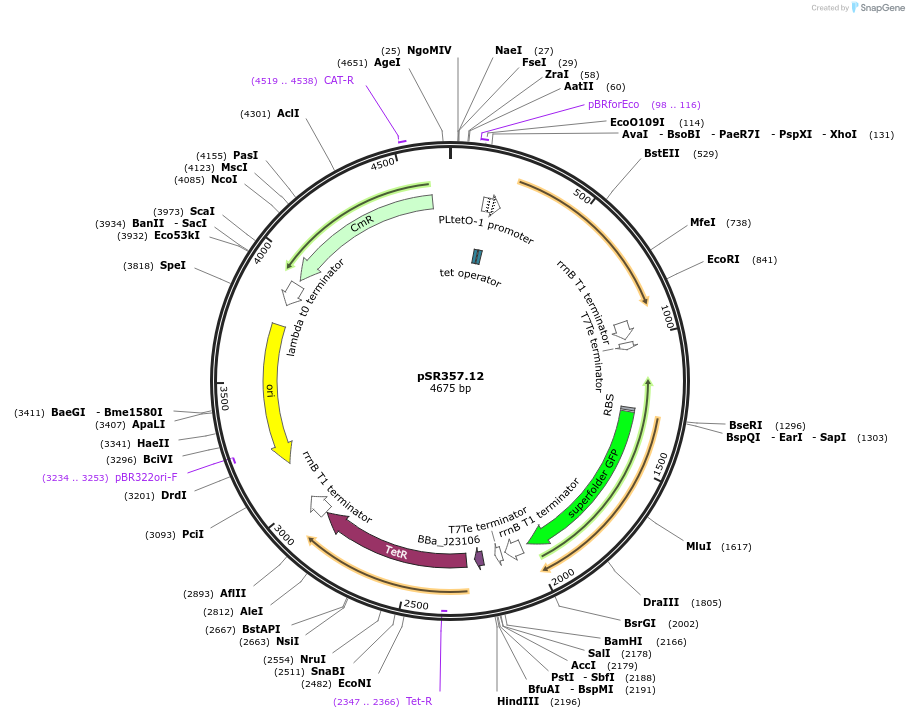
pSR357.12
Plasmid#125107PurposeExpression of narL(1-134aa)-ydfI(129-213aa) chimera under PLtetO-1, output Promoter PydfJ115DepositorInsertsnarL(REC)-ydfI(DBD)134
sfgfp
tetR
UseSynthetic BiologyExpressionBacterialMutationA84T - see depositor comments below and Q183K-see…Available SinceJune 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
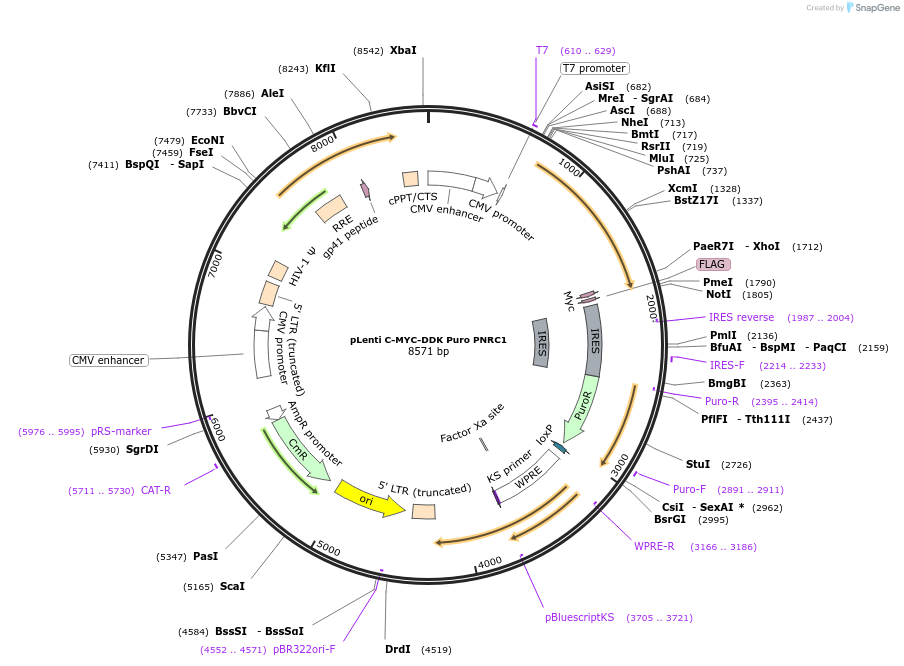
pLenti C-MYC-DDK Puro PNRC1
Plasmid#123300PurposeMammalian expression of PNRC1 C-MYC-FLAGDepositorAvailable SinceMarch 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
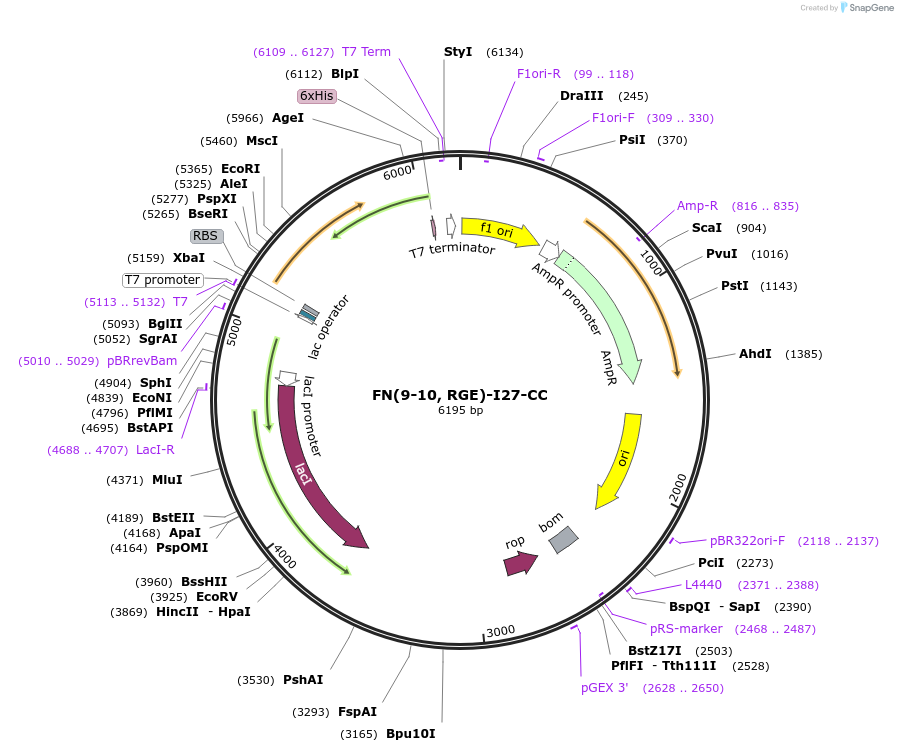
FN(9-10, RGE)-I27-CC
Plasmid#85396PurposeMTFM mechanosensor for integrin. Fibronectin domains 9&10 with RGE mutation fused to titan I27 domain with two cysteines for immobilization, site for p-azidophenylalanine incorporation & Cy3 labelingDepositorInsertFibronectin (FN) domains 9&10 containing RGE mutation fused to titin immunoglobulin domain (I27), containing a TAG (amber) codon
Tags6x HisExpressionBacterialMutationGRGDS motif mutated to GRGESPromoterT7Available SinceMay 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
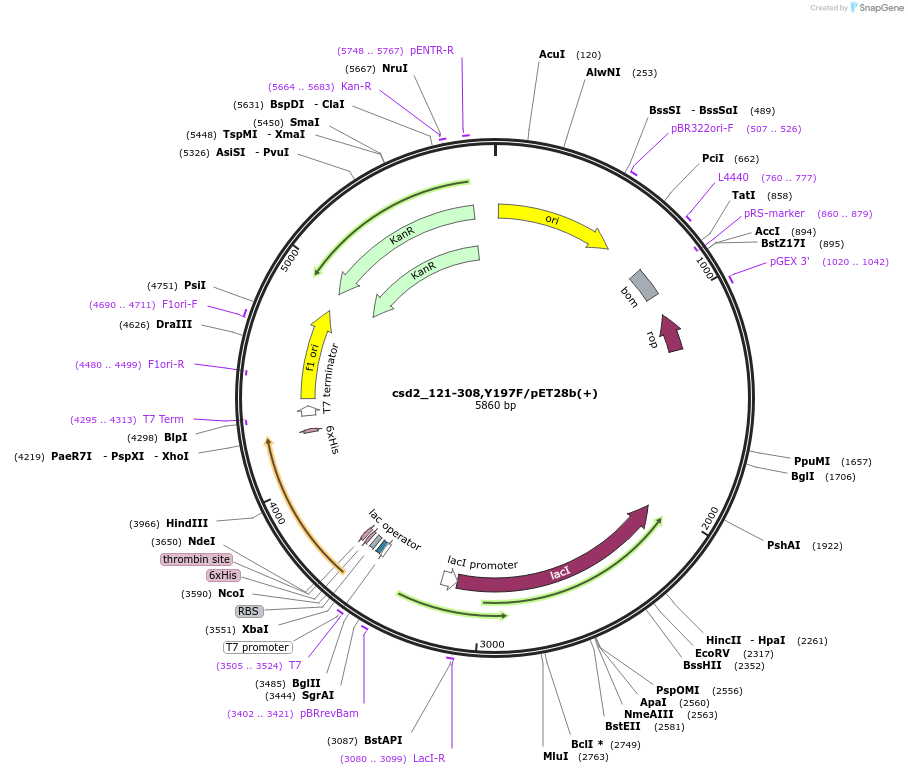
csd2_121-308,Y197F/pET28b(+)
Plasmid#83299Purposecsd2, construct: 121-308, pET28b(+) NC-tag, mutation: Y197FDepositorInserthp1544
TagsHis tagExpressionBacterialMutationdeleted amino acid 1-120, and changed Tyrosine 19…Available SinceOct. 31, 2016AvailabilityAcademic Institutions and Nonprofits only -
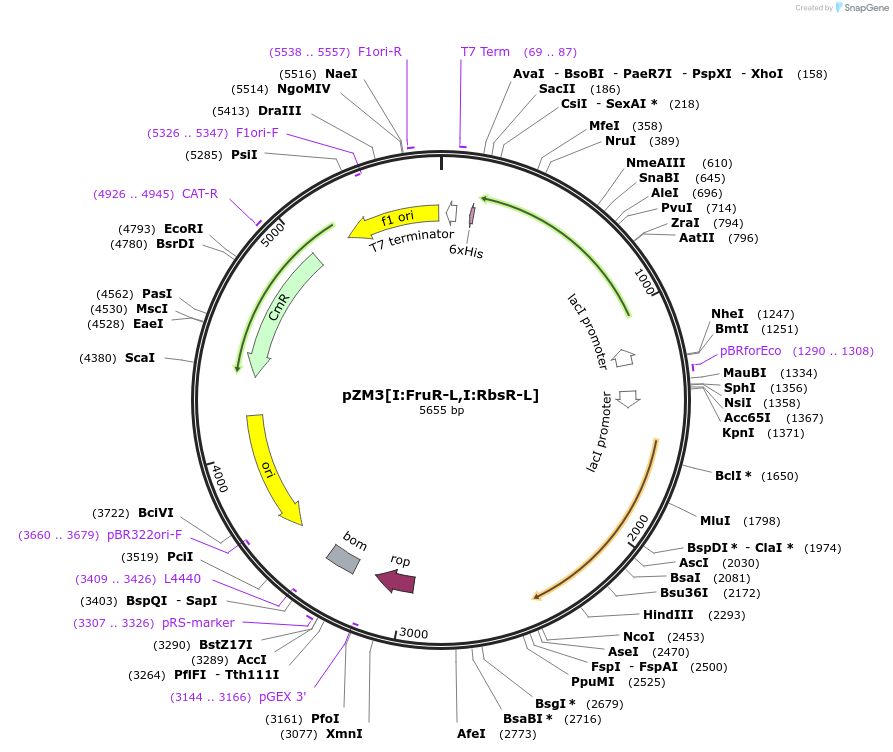
pZM3[I:FruR-L,I:RbsR-L]
Plasmid#60777PurposeContains PI driving expression FruR-L, and PI driving expression of RbsR-L.DepositorInsertsFruR-L
RbsR-L
MutationLacI/GalR repressor chimera. LacI DBD with FruR L…Available SinceJuly 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
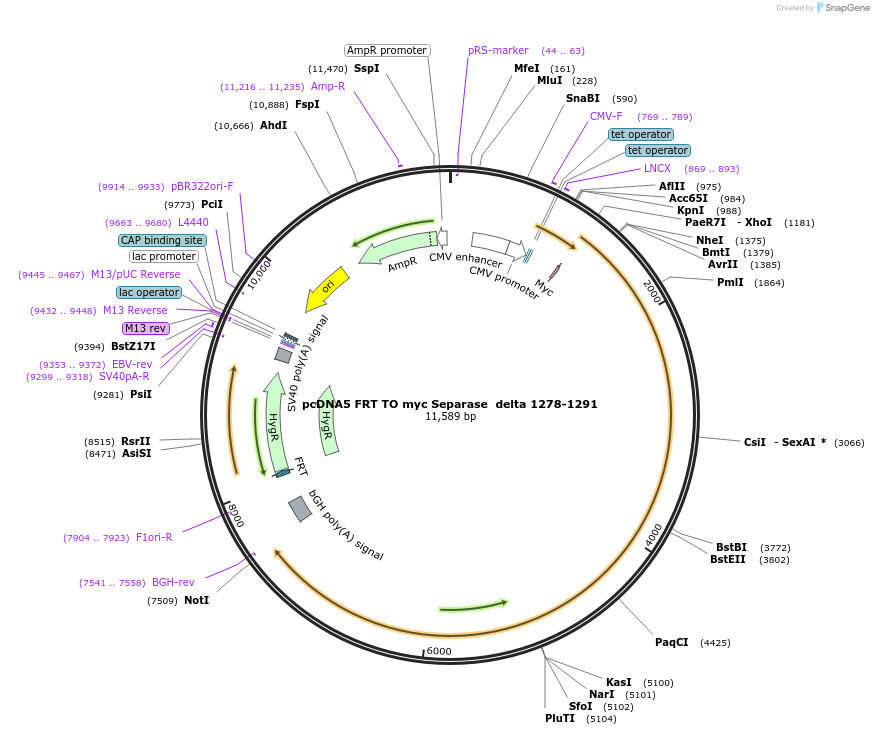
pcDNA5 FRT TO myc Separase delta 1278-1291
Plasmid#59830PurposeAllows the integration of myc Separase delta 1278-1291 in the genome and Tet-inducible expression.DepositorInsertSeparase (ESPL1 Human)
TagsMycExpressionMammalianMutationdelta 1278-1291Promoterhybrid human cytomegalovirus (CMV)/TetO2 promoterAvailable SinceFeb. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
pENTR-MYC-hDIXDC1-L-F630S
Plasmid#61241PurposeEntry clone encoding long isoform of human DIXDC1DepositorInserthuman DIXDC1 (DIXDC1 Human)
UseEntr cloneTagsMYCMutationChanged Phenylalanine 630 to SerineAvailable SinceJan. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
pENTR-FLAG-hDIXDC1-L-F630S
Plasmid#61212PurposeEntry clone encoding long isoform of human DIXDC1DepositorInserthuman DIXDC1 (DIXDC1 Human)
UseEntr cloneTagsFLAGMutationChanged Phenylalanine 630 to SerineAvailable SinceJan. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
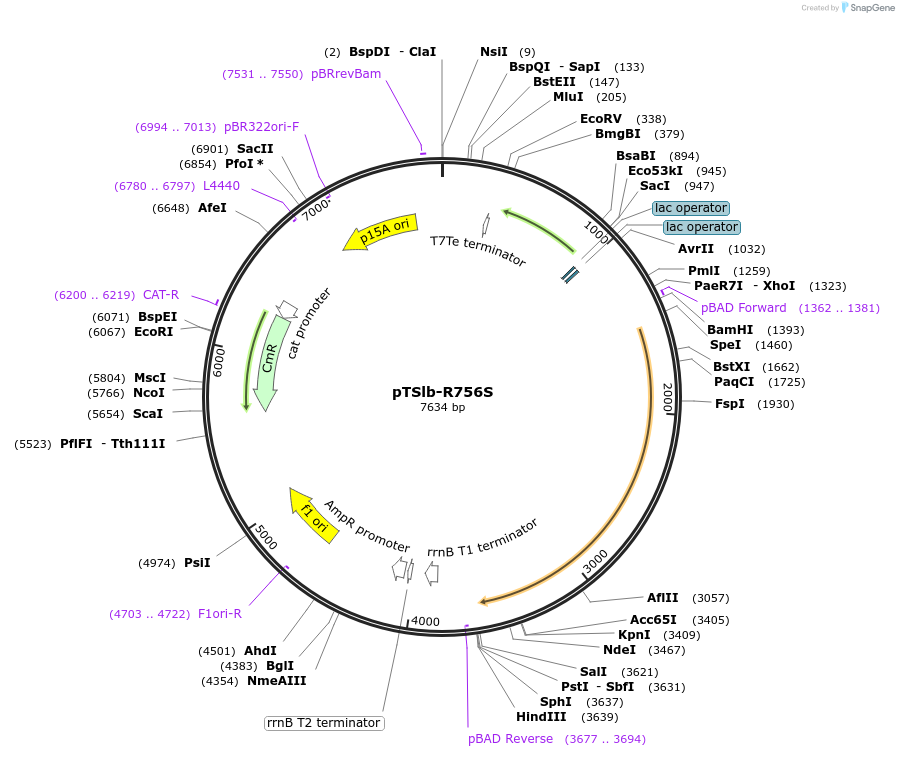
pTSlb-R756S
Plasmid#60730PurposeExpresses both fragments of T7 RNAP split at position 179. The N-terminal fragment is driven by PLac while the C-terminal fragment is driven by PBAD. C-terminal fragment has the point mutations R756SDepositorInsertsResidues 1-179 of split T7 RNAP
Residues 180-880 of split T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationResidues 1-180 of split T7 RNAP and Residues 180-…Available SinceJan. 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
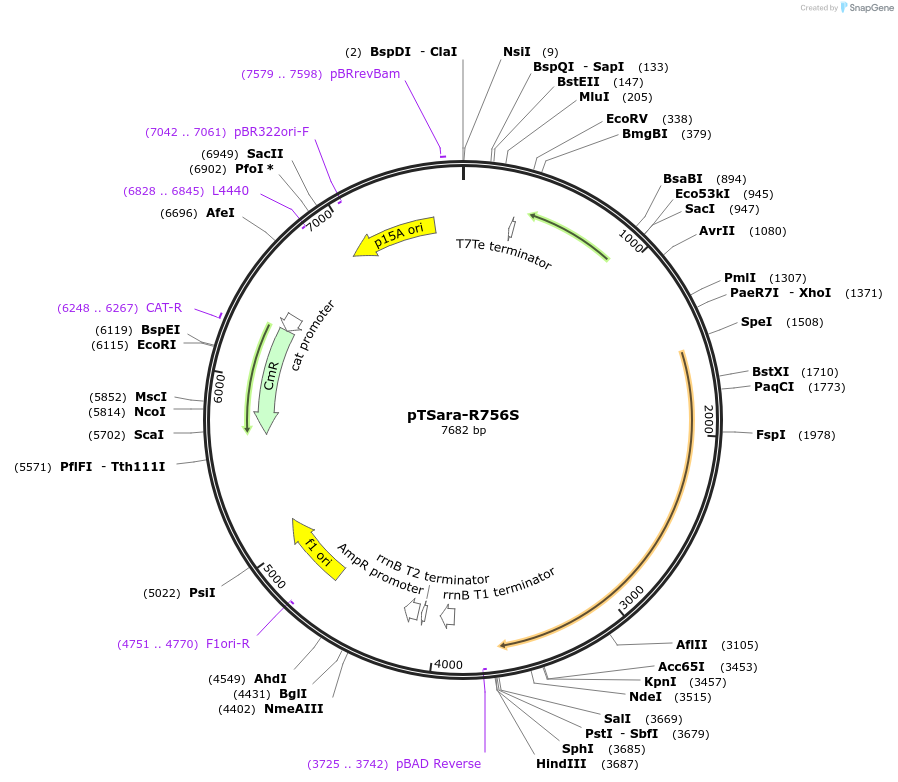
pTSara-R756S
Plasmid#60725PurposeExpresses both fragments of T7 RNAP split at position 179. Both fragments are driven by PBAD. The C-terminal fragment has the point mutation R756S. Contains a constitutive araC ORFDepositorInsertsResidues 1-179 of split T7 RNAP
Residues 180-880 of split T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationResidues 1-180 of split T7 RNAP and Residues 180-…Available SinceDec. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
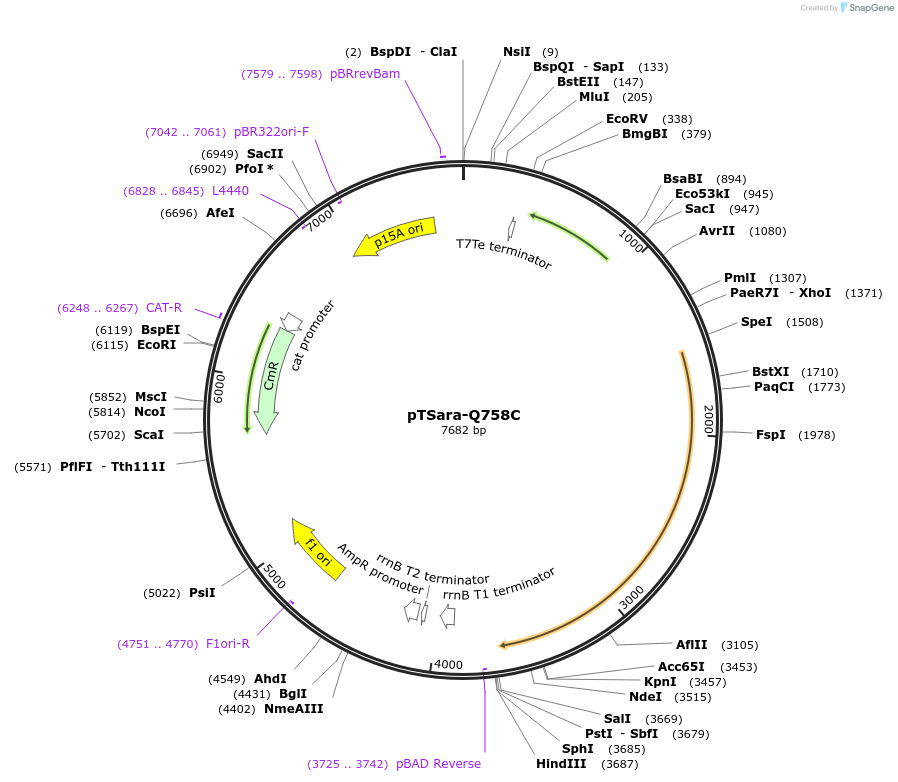
pTSara-Q758C
Plasmid#60724PurposeExpresses both fragments of T7 RNAP split at position 179. Both fragments are driven by PBAD. The C-terminal fragment has the point mutation Q758C. Contains a constitutive araC ORFDepositorInsertsResidues 1-179 of split T7 RNAP
Residues 180-880 of split T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationResidues 1-180 of split T7 RNAP and Residues 180-…Available SinceDec. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
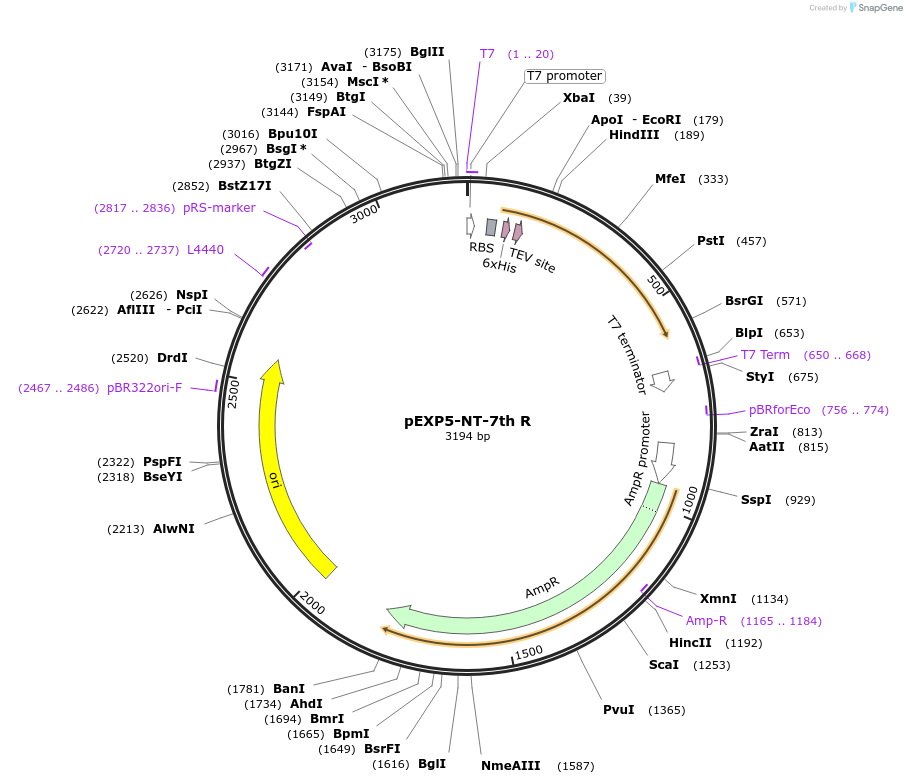
pEXP5-NT-7th R
Plasmid#42573DepositorInsertmutated calmodulin gene
TagsPolyhistidine (6×His) region, TEV recognition sit…ExpressionBacterialMutationchanged Phenylalanine 92 to Glutamine acid, Isole…PromoterT7Available SinceMarch 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
-