We narrowed to 8,018 results for: AMPH
-
Plasmid#68604PurposeGateway binary vector designed for transgenic research with Marchantia polymorpha as well as other plantsDepositorTypeEmpty backboneTagsGlucocorticoid receptor (GR)ExpressionPlantPromoterMpEF1alpha promoterAvailable SinceJune 24, 2016AvailabilityAcademic Institutions and Nonprofits only
-
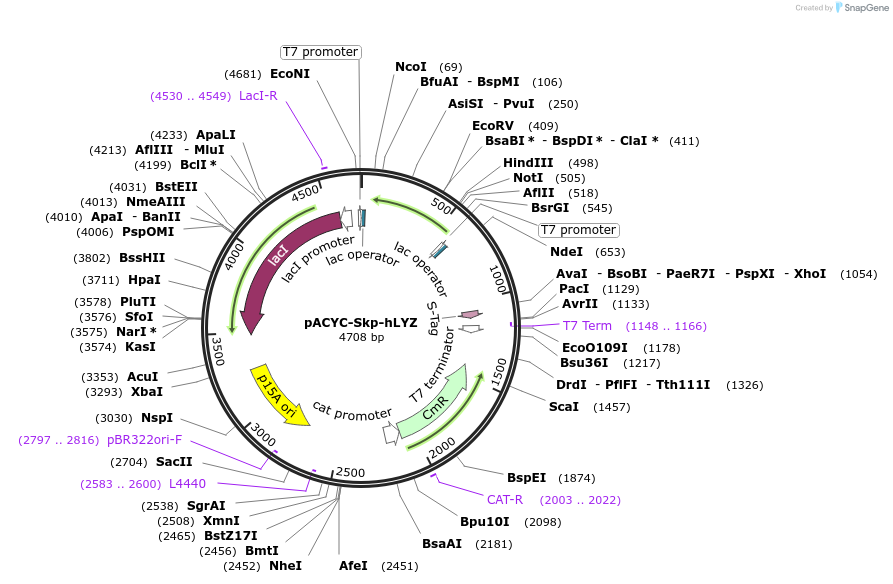
pACYC-Skp-hLYZ
Plasmid#83924PurposeCo-expresses cytoplasmic copies of the Skp protein folding chaperone and wild type human lysozyme..DepositorAvailable SinceNov. 28, 2016AvailabilityAcademic Institutions and Nonprofits only -
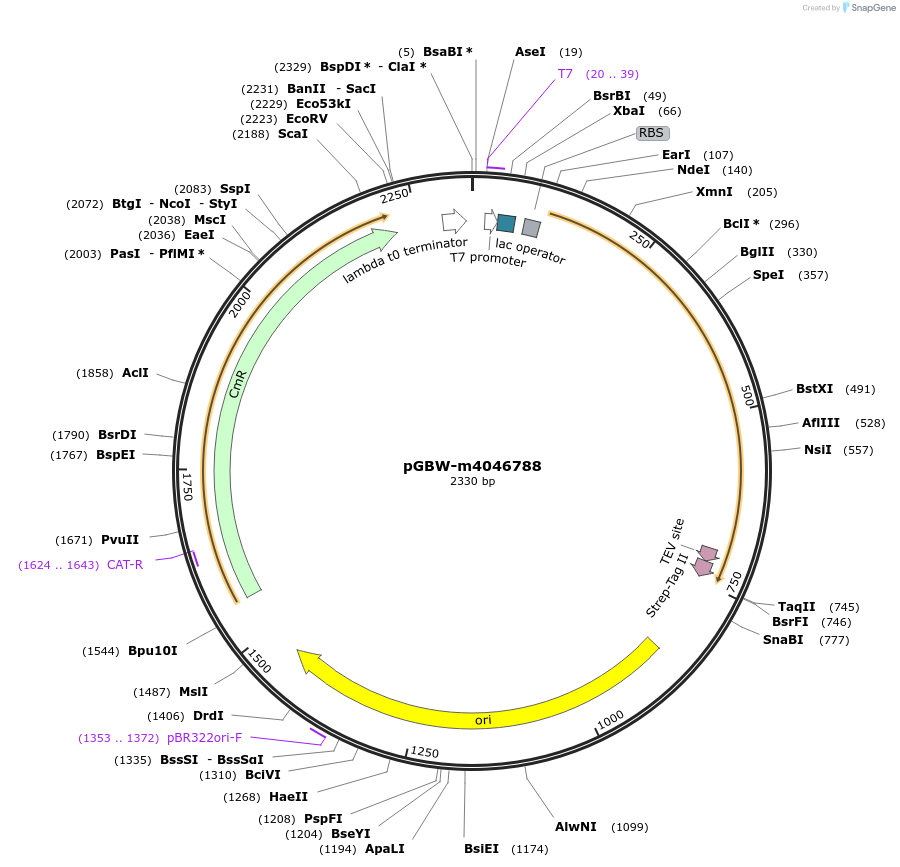
pGBW-m4046788
Plasmid#145675PurposeBacterial Expression plasmid for SARS-CoV-2 nsp8DepositorInsertSARS-CoV-2 nsp8 (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
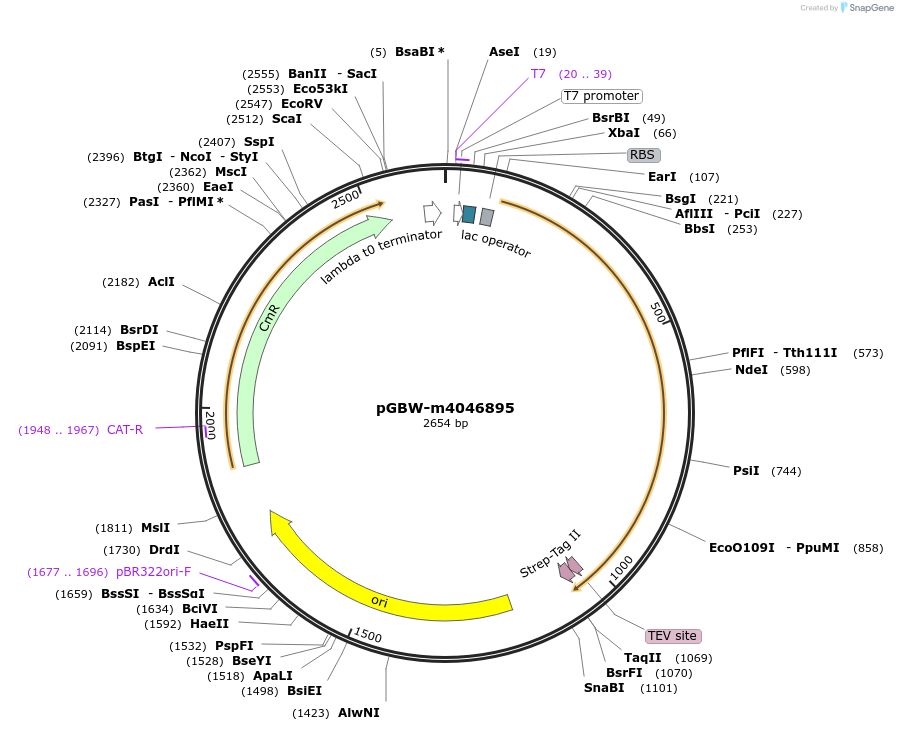
pGBW-m4046895
Plasmid#145596PurposeBacterial Expression plasmid for SARS-CoV-2 3C-like proteinaseDepositorInsertSARS-CoV-2 3C-like proteinase (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
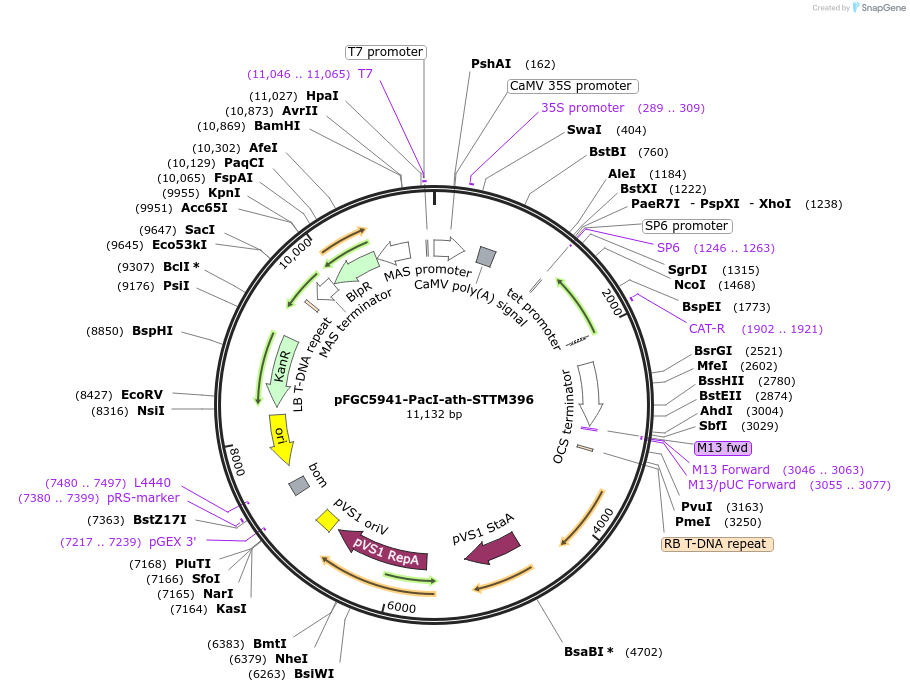
pFGC5941-PacI-ath-STTM396
Plasmid#84122Purposetarget miRNA396 for destruction in Arabidopsis thalianaDepositorInsertSTTM396
ExpressionPlantPromoter2x35SAvailable SinceMarch 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
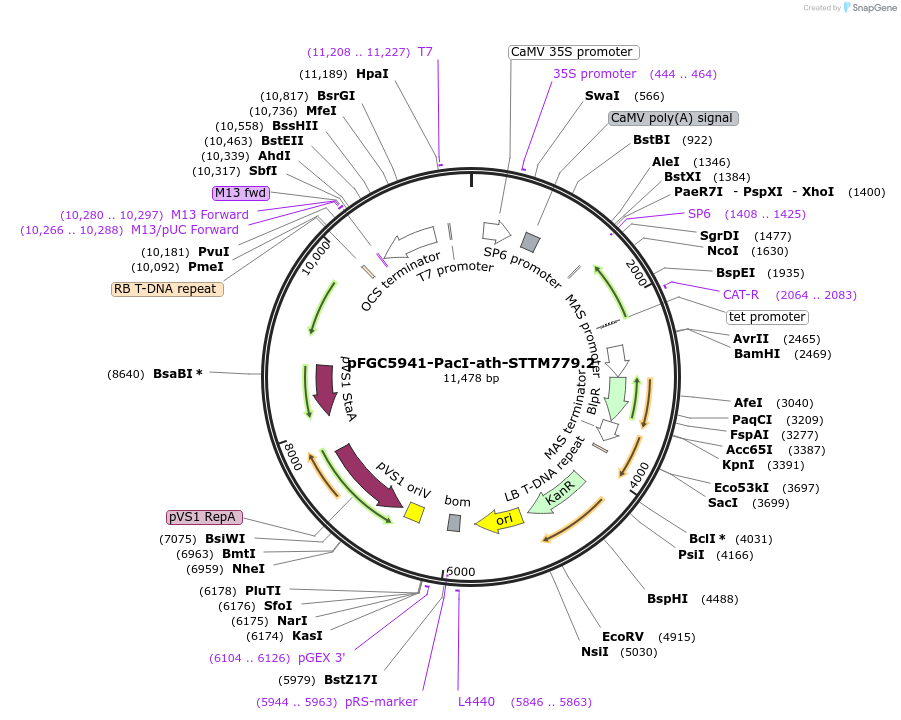
pFGC5941-PacI-ath-STTM779.2
Plasmid#84145Purposetarget miRNA779.2 for destruction in Arabidopsis thalianaDepositorInsertSTTM779.2
ExpressionPlantPromoter2x35SAvailable SinceMarch 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
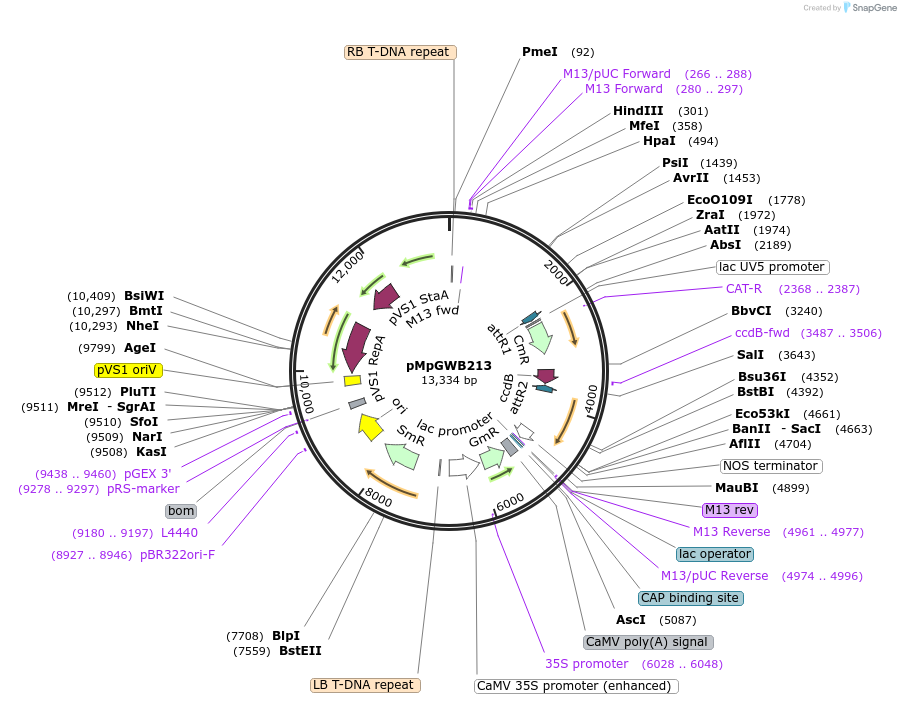
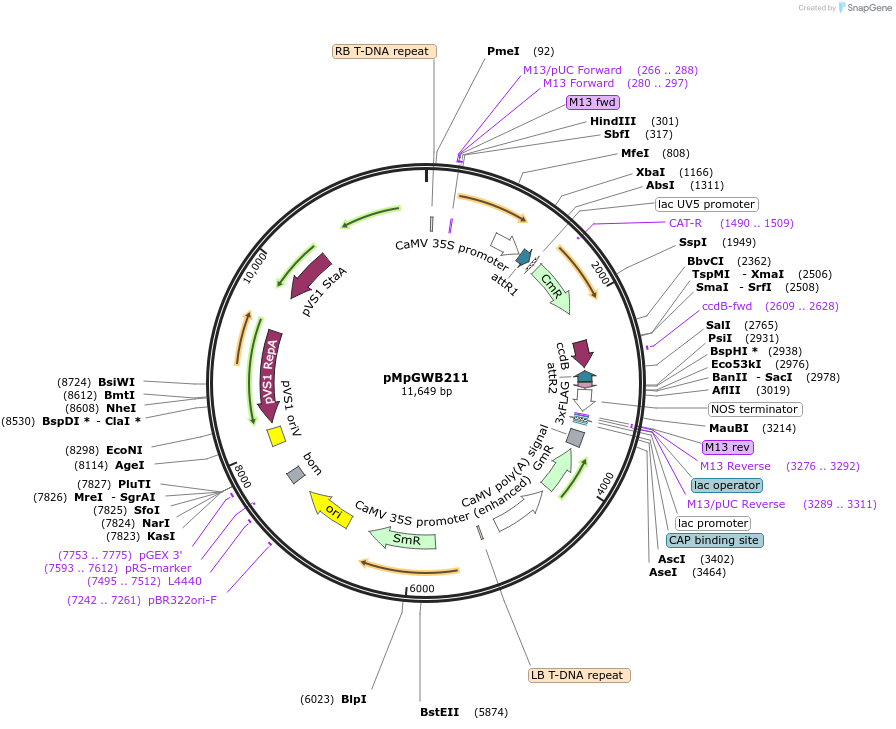
pMpGWB211
Plasmid#68602PurposeGateway binary vector designed for transgenic research with Marchantia polymorpha as well as other plantsDepositorTypeEmpty backboneTags3xFLAGExpressionPlantPromoterCauliflower mosaic virus 35S promoterAvailable SinceJune 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
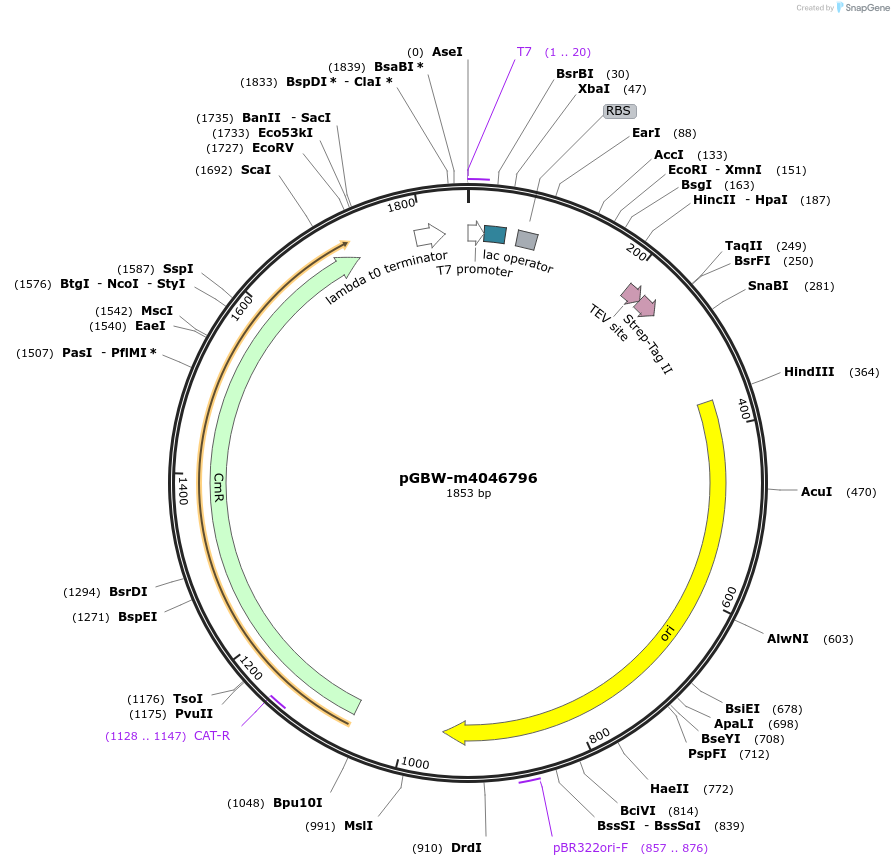
pGBW-m4046796
Plasmid#145666PurposeBacterial Expression plasmid for SARS-CoV-2 ORF10 proteinDepositorInsertSARS-CoV-2 ORF10 protein (ORF10 Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
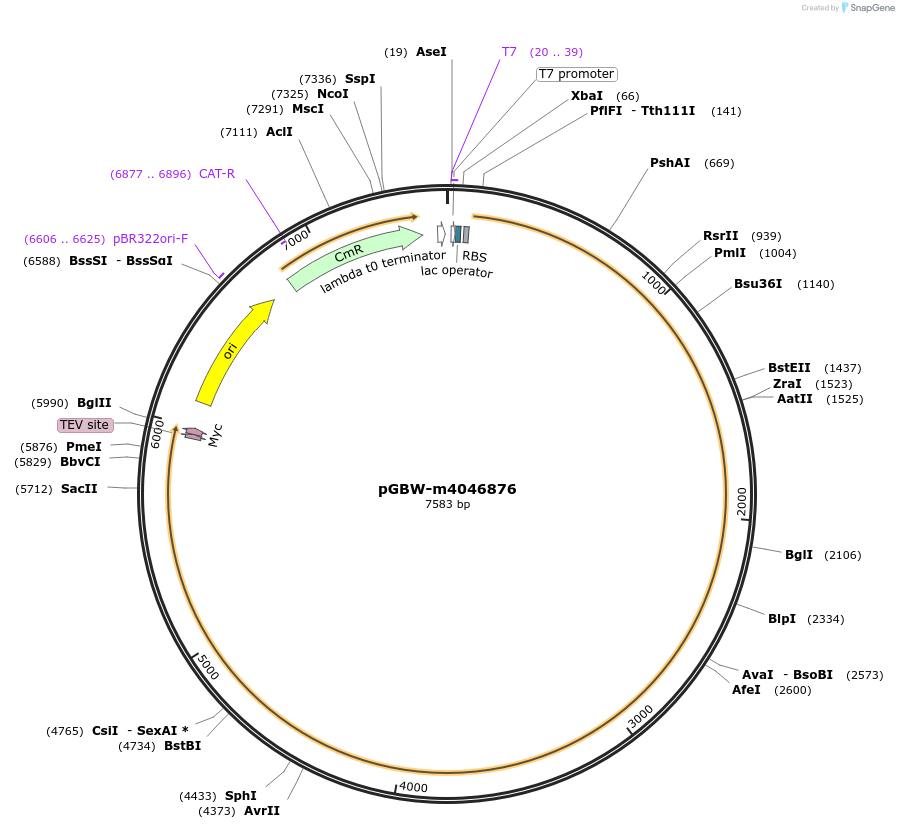
pGBW-m4046876
Plasmid#145669PurposeBacterial Expression plasmid for SARS-CoV-2 nsp3DepositorInsertSARS-CoV-2 nsp3 (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;MycExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
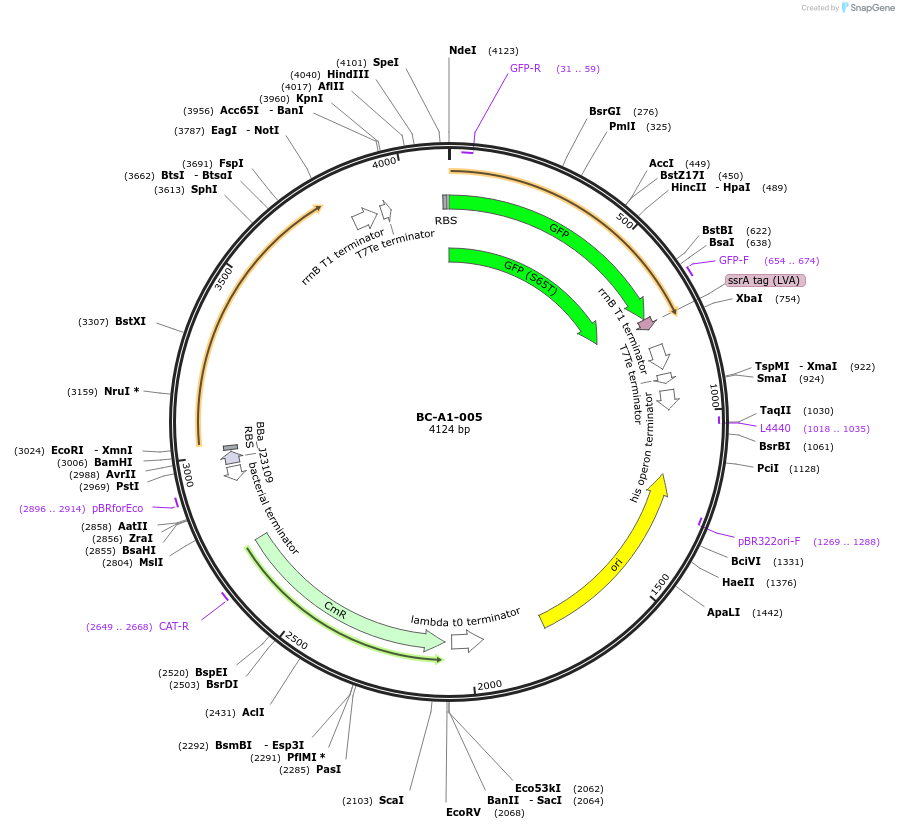
BC-A1-005
Plasmid#78692PurposeA weak promoter drives the expression of wild type luxR, which regulates expression of LVA-tagged GFP under the control of a strong RBS.DepositorInsertBBa_J23109-BBa_B0034-luxR-Terminator-pLux-BBa_B0034-GFP(LVA)-Terminator
UseSynthetic BiologyAvailable SinceJan. 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
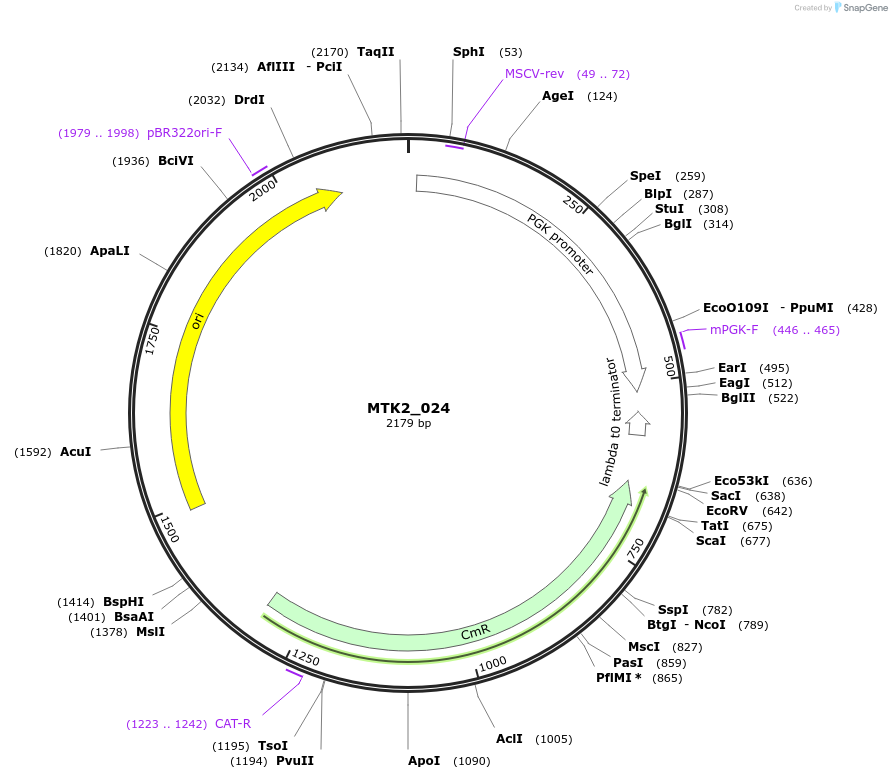
MTK2_024
Plasmid#123716PurposeEncodes crippled mouse pPGK1 as a Type 2 part to be used in the MTK systemDepositorInsertpPGK1c
ExpressionMammalianMutationCrippled Kozac sequenceAvailable SinceDec. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
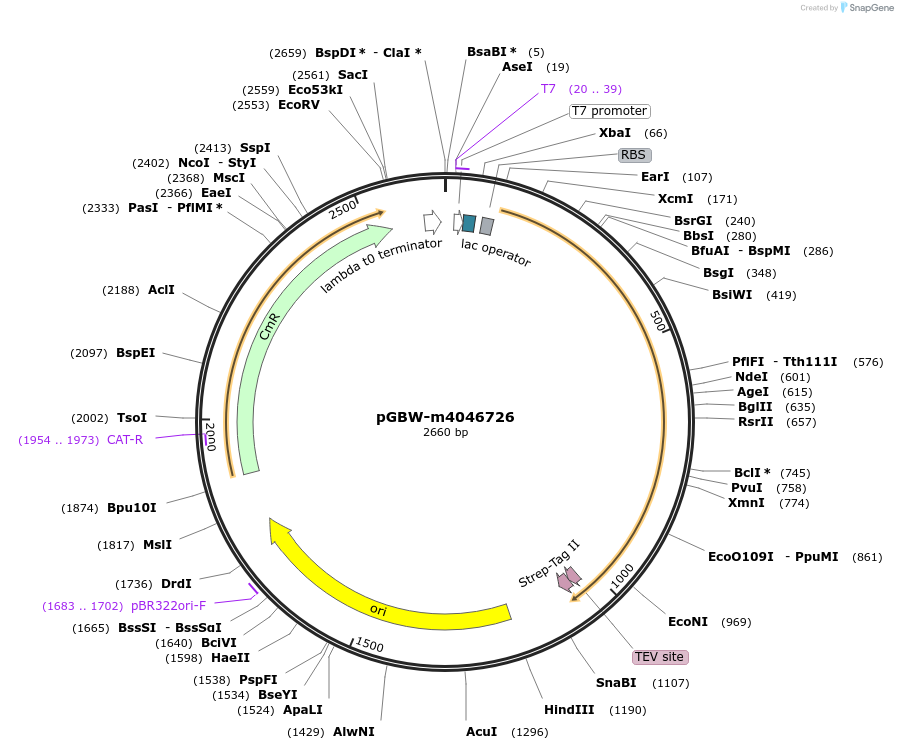
pGBW-m4046726
Plasmid#145756PurposeBacterial Expression plasmid for SARS-CoV-2 3C-like proteinaseDepositorInsertSARS-CoV-2 3C-like proteinase (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
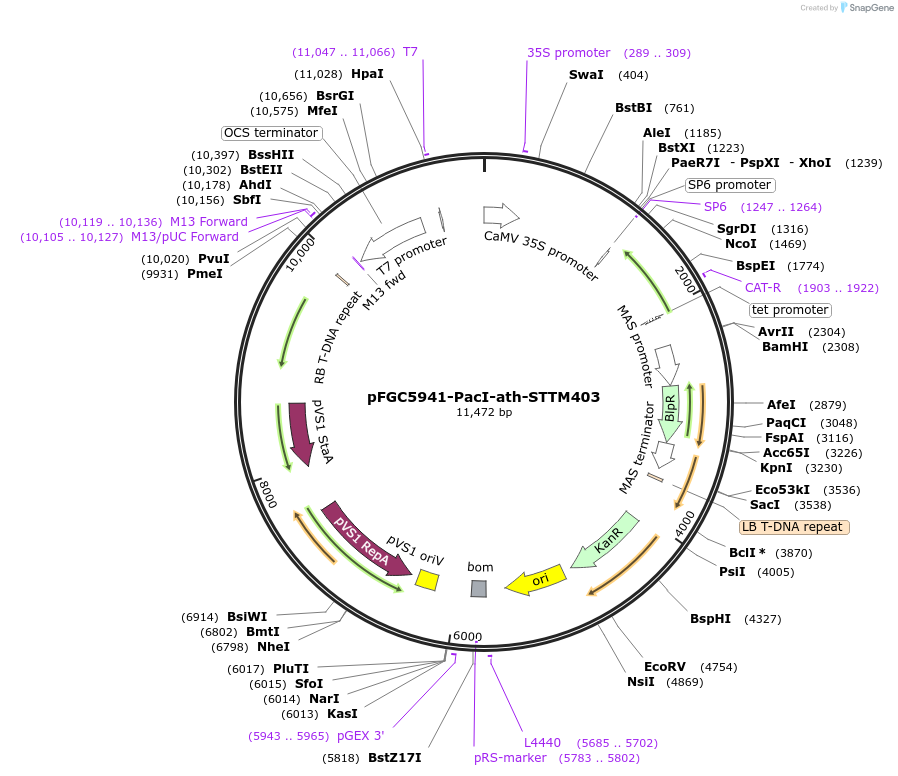
pFGC5941-PacI-ath-STTM403
Plasmid#84128Purposetarget miRNA403 for destruction in Arabidopsis thalianaDepositorInsertSTTM403
ExpressionPlantPromoter2x35SAvailable SinceNov. 30, 2016AvailabilityAcademic Institutions and Nonprofits only -
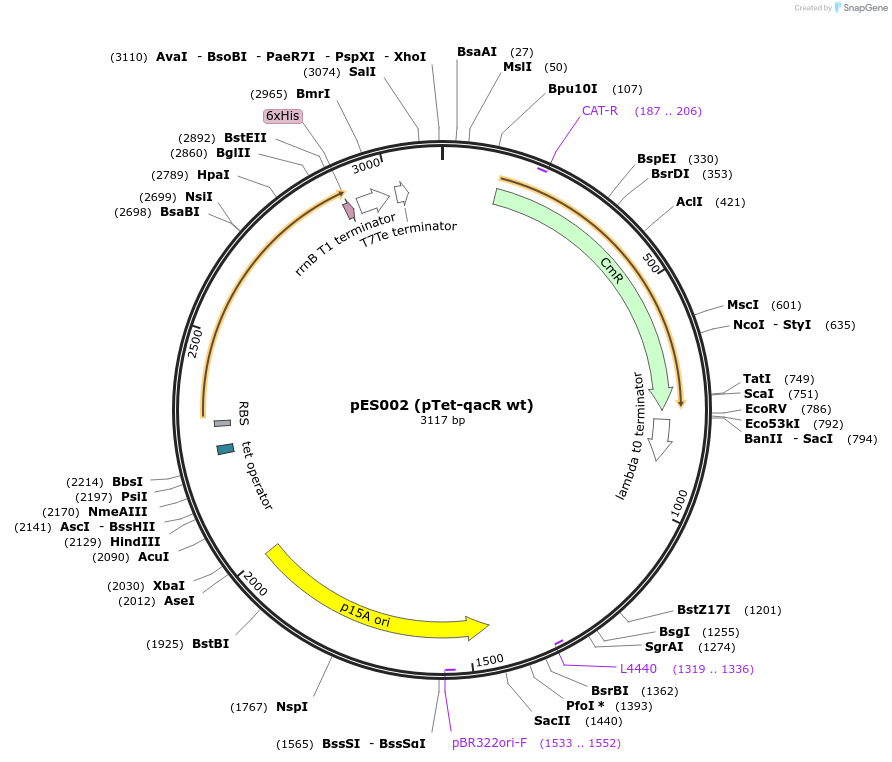
pES002 (pTet-qacR wt)
Plasmid#69031Purposewild-type qacR downstream of a Tet promoterDepositorAvailable SinceOct. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
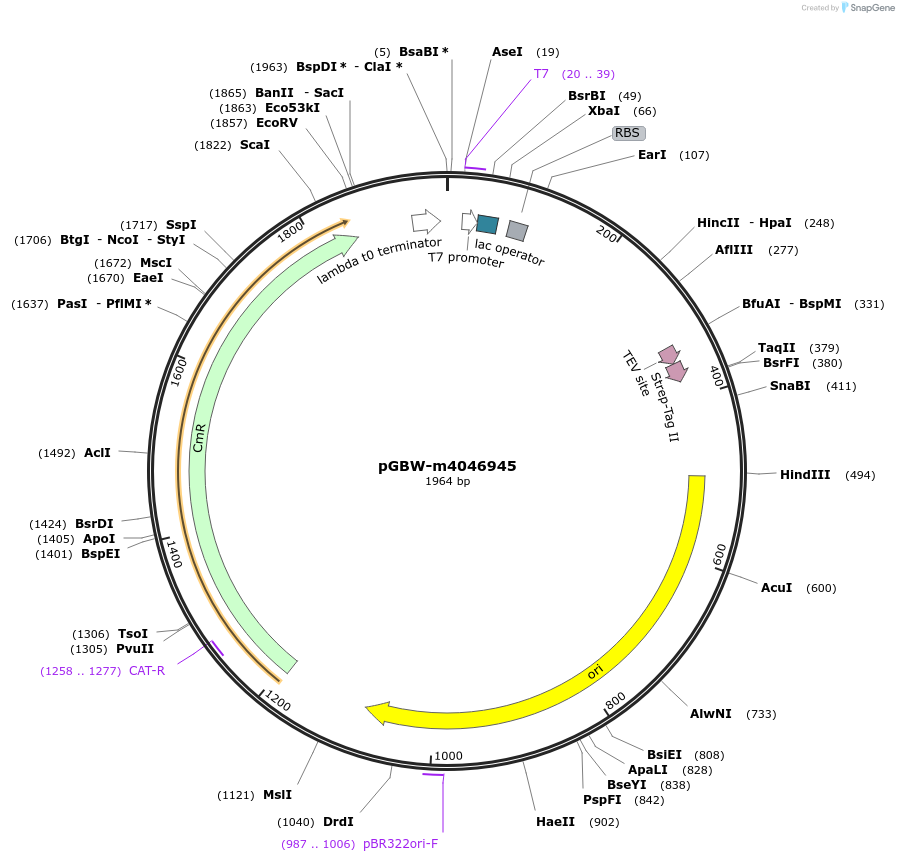
pGBW-m4046945
Plasmid#145668PurposeBacterial Expression plasmid for SARS-CoV-2 E (envelope)DepositorInsertSARS-CoV-2 E (envelope) (E Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
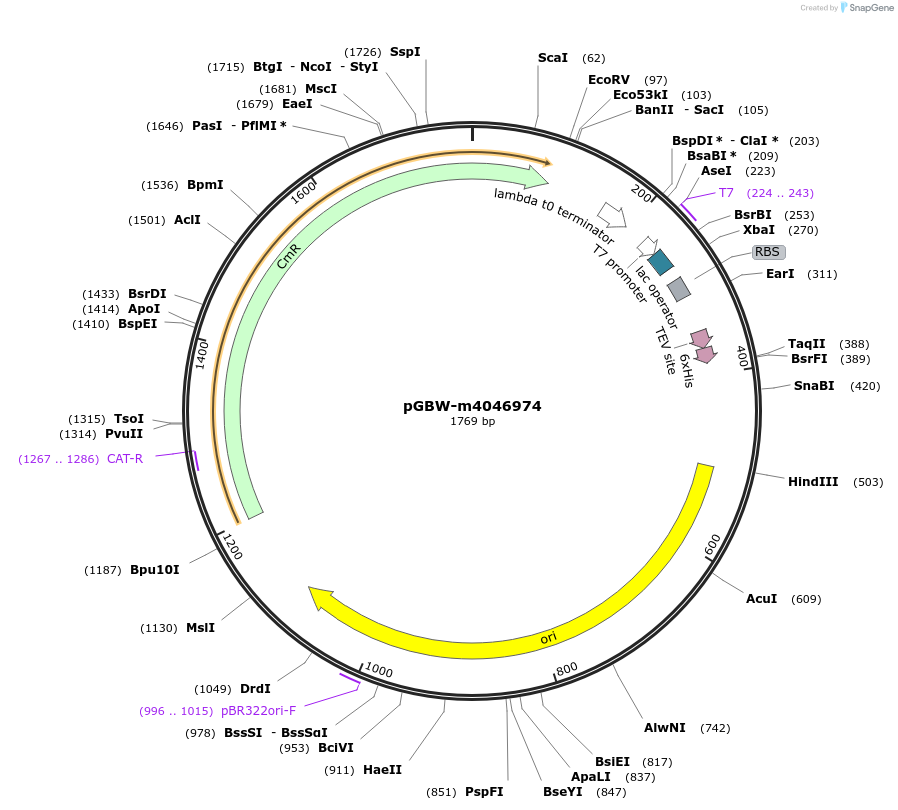
pGBW-m4046974
Plasmid#145593PurposeBacterial Expression plasmid for SARS-CoV-2 nsp11DepositorInsertSARS-CoV-2 nsp11 (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;6xHISExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
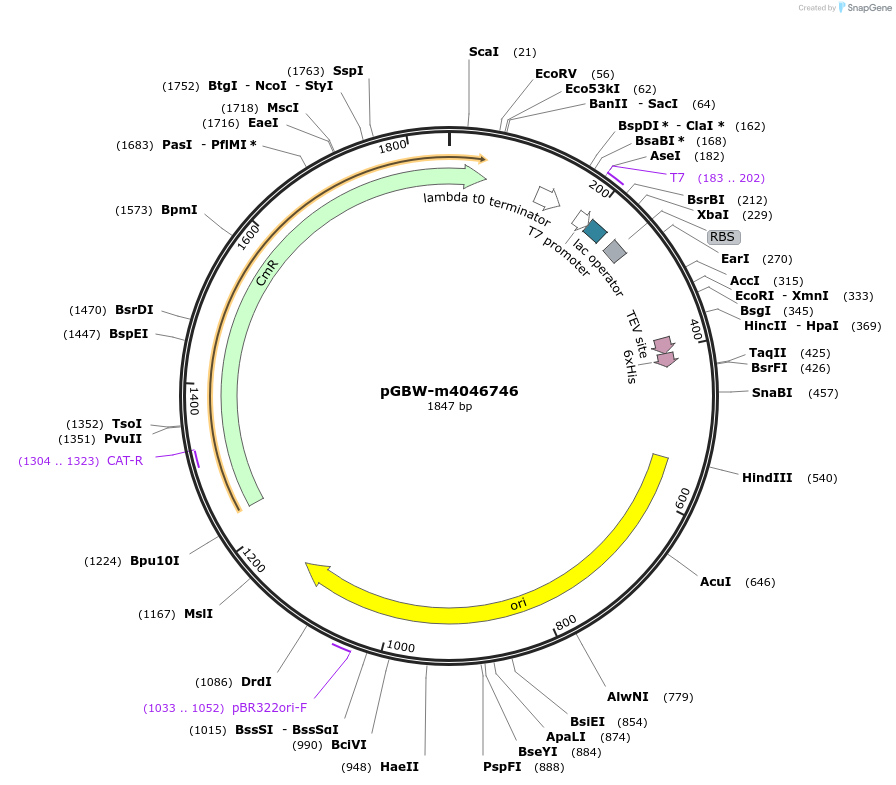
pGBW-m4046746
Plasmid#145641PurposeBacterial Expression plasmid for SARS-CoV-2 ORF10 proteinDepositorInsertSARS-CoV-2 ORF10 protein (ORF10 Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;6xHISExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
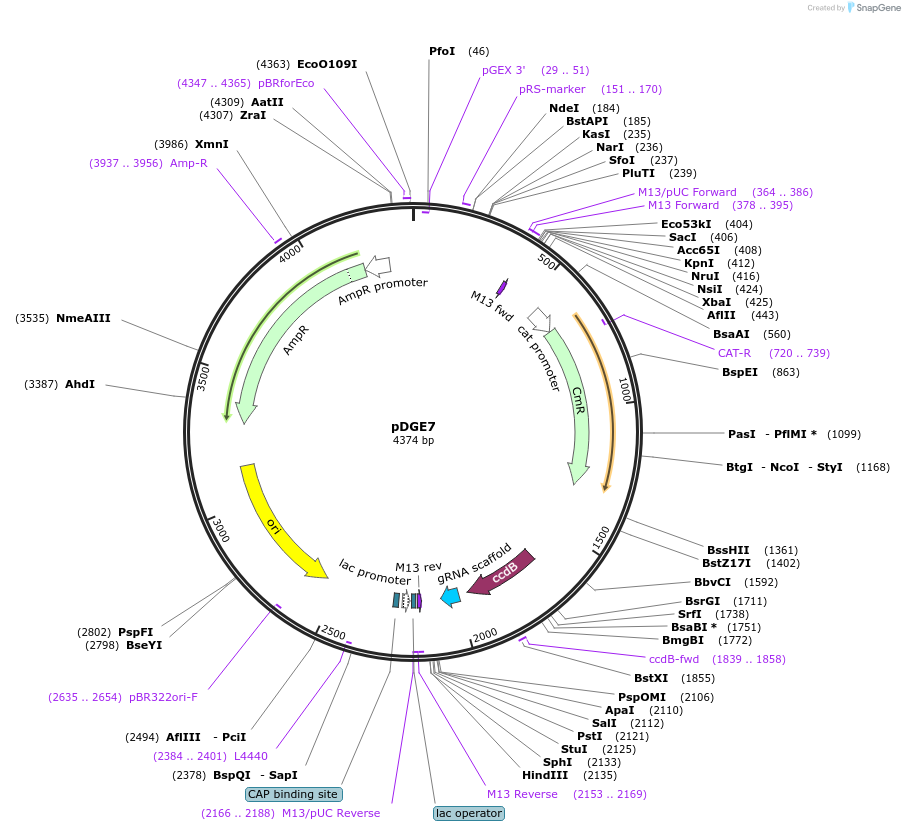
pDGE7
Plasmid#79462PurposesgRNA shuttle vector; module 2DepositorInsertpAtU6-ccdB-sgRNA (ccdB(letD) )
UseCRISPR and Synthetic BiologyMutationBsaI site eliminatedAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
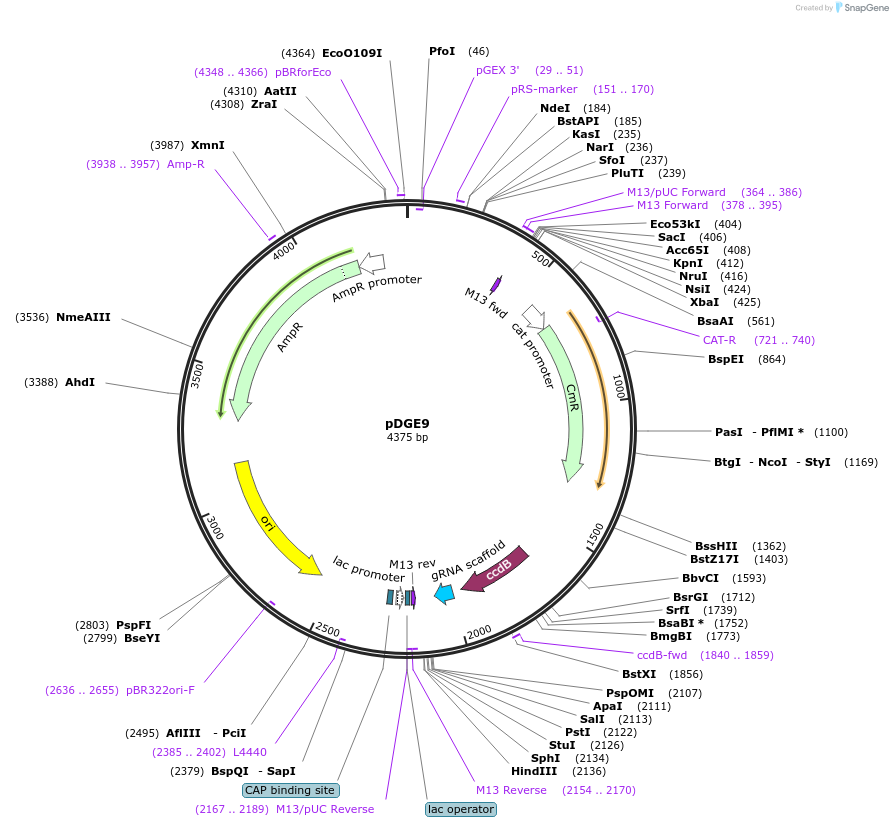
pDGE9
Plasmid#79465PurposesgRNA shuttle vector; module 3DepositorInsertpAtU6-ccdB-sgRNA (ccdB(letD) )
UseCRISPR and Synthetic BiologyMutationBsaI site eliminatedAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
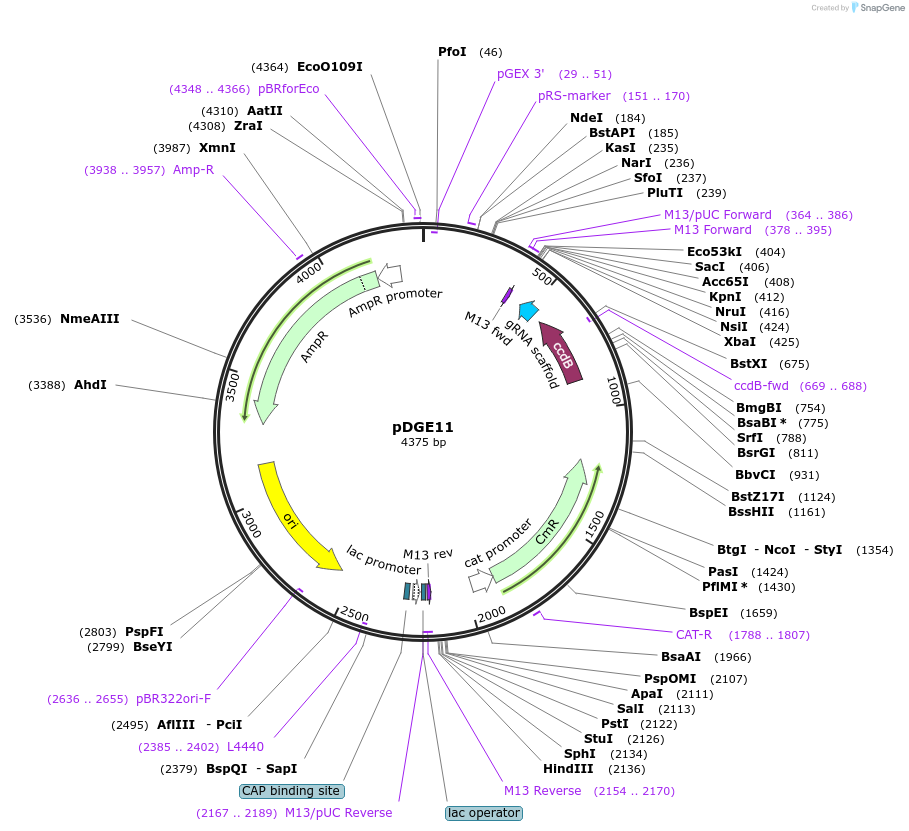
pDGE11
Plasmid#79467PurposesgRNA shuttle vector; module 4EDepositorInsertpAtU6-ccdB-sgRNA (ccdB(letD) )
UseCRISPR and Synthetic BiologyMutationBsaI site eliminatedAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only