We narrowed to 2,494 results for: pcas9
-
Plasmid#101214PurposeBacterial expression plasmid for SpCas9 + K855A, HNH FRET variantDepositorInsertSpCas9 variant C80S/C574S/S355C/S867C/K855A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, S355C, S867C and K855APromoterT7Available SinceOct. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
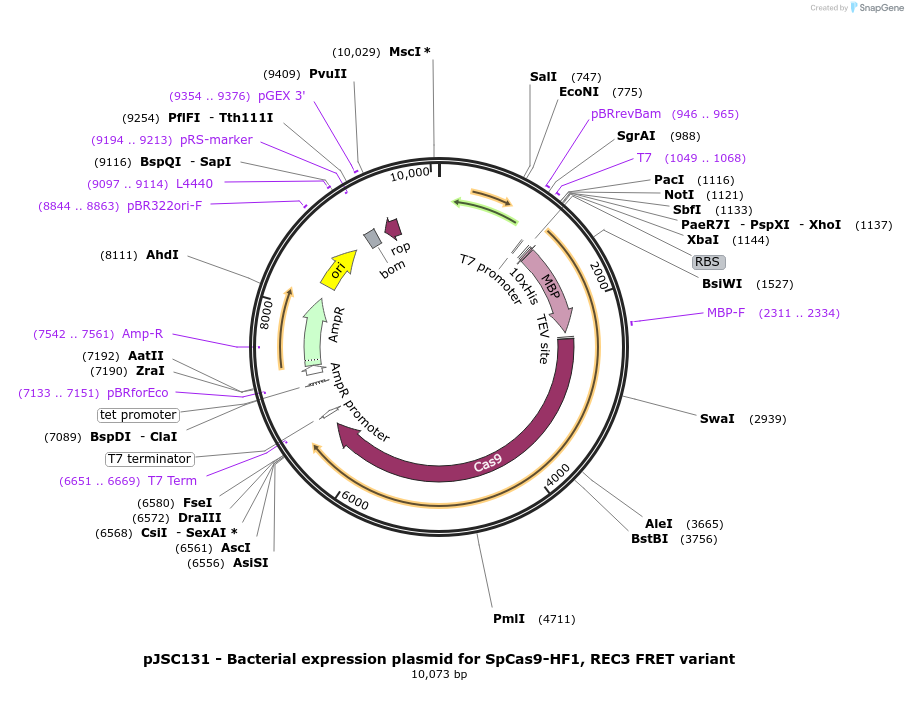
pJSC131 - Bacterial expression plasmid for SpCas9-HF1, REC3 FRET variant
Plasmid#101212PurposeBacterial expression plasmid for SpCas9-HF1, REC3 FRET variantDepositorInsertSpCas9 variant C80S/C574S/S701C/S960C/N497A/R661A/Q695A/Q926A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, S701C, S960C, N497A, R661A, Q695A an…PromoterT7Available SinceOct. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
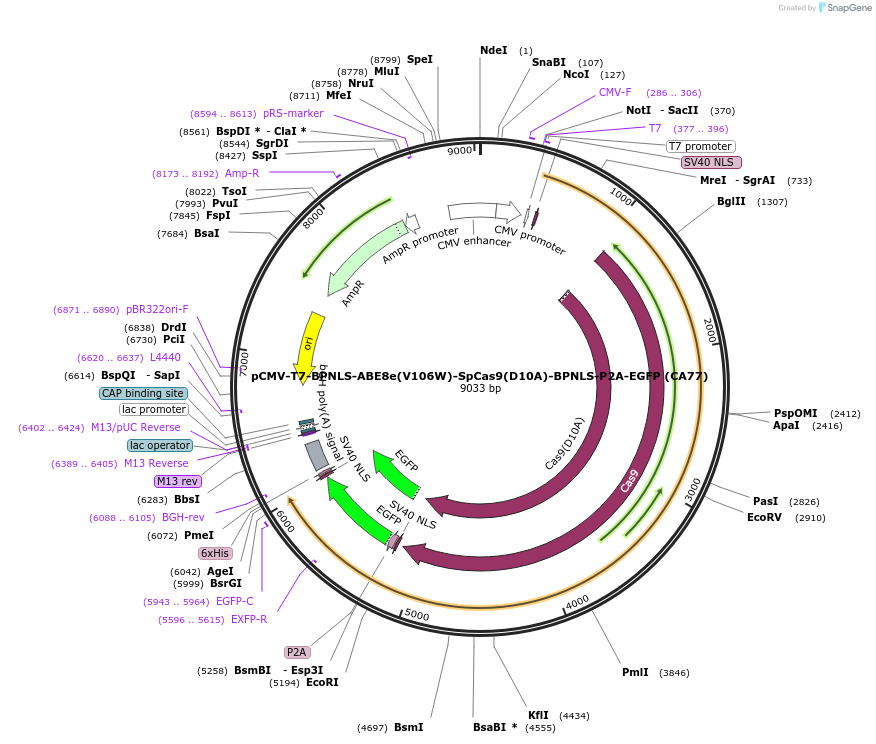
pCMV-T7-BPNLS-ABE8e(V106W)-SpCas9(D10A)-BPNLS-P2A-EGFP (CA77)
Plasmid#208290PurposeCMV and T7 promoter expression plasmid for human codon optimized ABE8e(V106W) A-to-G base editor with SpCas9(D10A) and P2A-EGFPDepositorInsertpCMV-T7-BPNLS-ABE8e(V106W)-SpCas9-BPNLS-P2A-EGFP
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLS and BPNLS-P2A-EGFPExpressionMammalianMutationABE8e mutations and V106W in TadA and nSpCas9(D10…PromoterCMV and T7Available SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
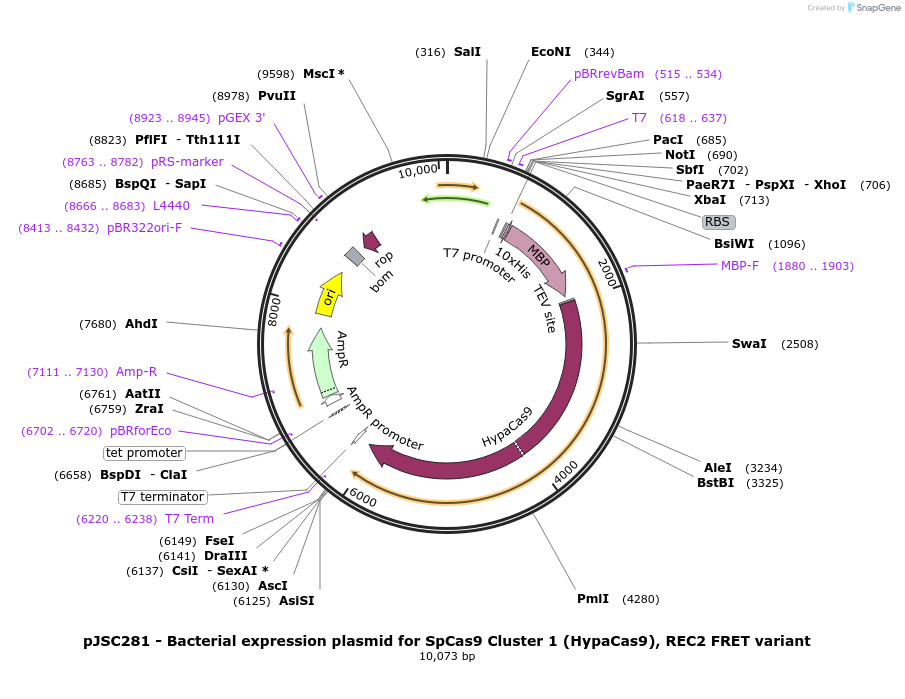
pJSC281 - Bacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC2 FRET variant
Plasmid#101230PurposeBacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC2 FRET variantDepositorInsertSpCas9 variant C80S/C574S/E60C/D273C/N692A/M694A/Q695A/H698A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, E60C, D273C, N692A, M694A, Q695A and…PromoterT7Available SinceNov. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
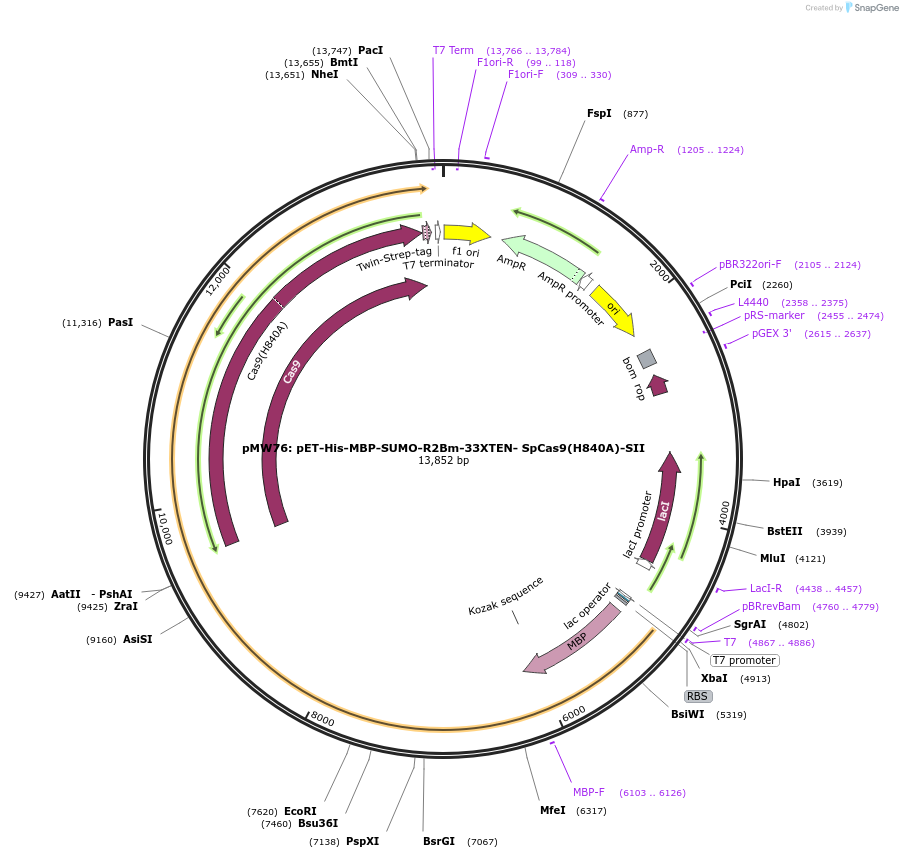
pMW76: pET-His-MBP-SUMO-R2Bm-33XTEN- SpCas9(H840A)-SII
Plasmid#209292PurposeExpresses R2Bm-Cas9(H840A) fusion in E. coliDepositorInsertR2
TagsHis, MBP, SpCas9(H840A), StrepII, and bdSUMOExpressionBacterialPromoterT7Available SinceNov. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
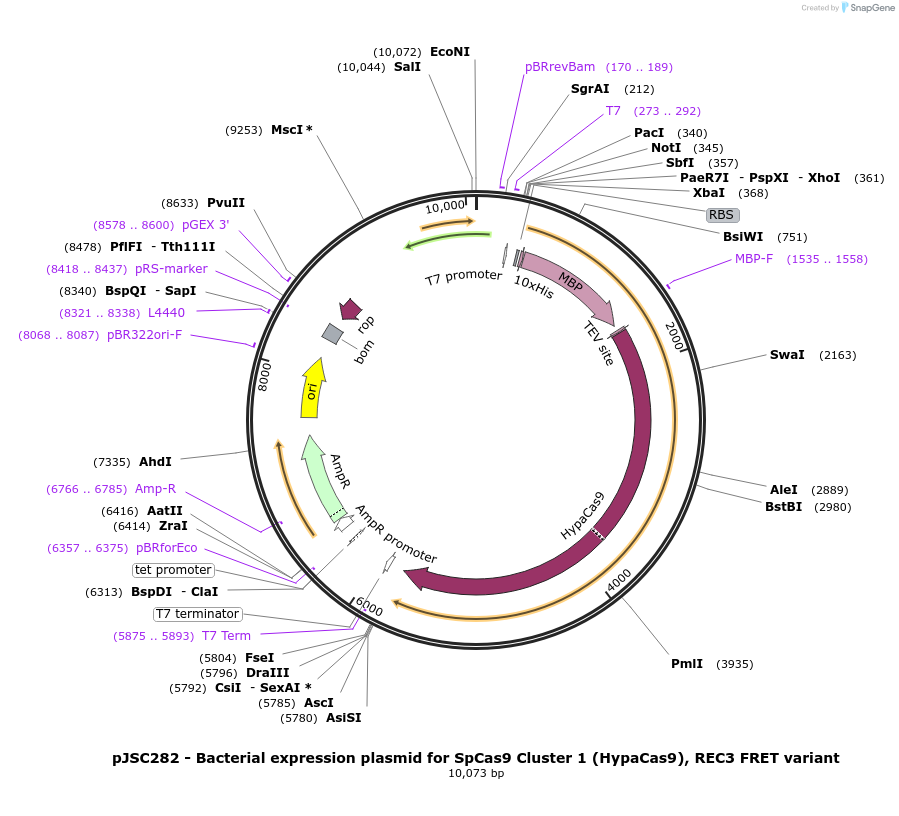
pJSC282 - Bacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC3 FRET variant
Plasmid#101231PurposeBacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC3 FRET variantDepositorInsertSpCas9 variant C80S/C574S/S701C/S960C/N692A/M694A/Q695A/H698A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, S701C, S960C, N692A, M694A, Q695A an…PromoterT7Available SinceNov. 6, 2017AvailabilityAcademic Institutions and Nonprofits only -
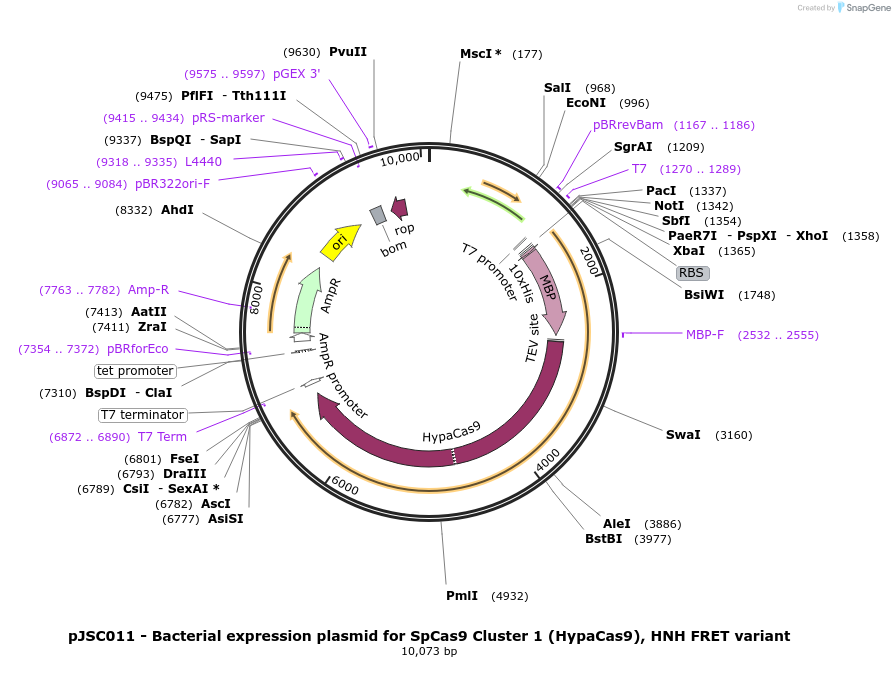
pJSC011 - Bacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), HNH FRET variant
Plasmid#101229PurposeBacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), HNH FRET variantDepositorInsertSpCas9 variant C80S/C574S/S355C/S867C/N692A/M694A/Q695A/H698A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, S355C, S867C, N692A, M694A, Q695A an…PromoterT7Available SinceOct. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
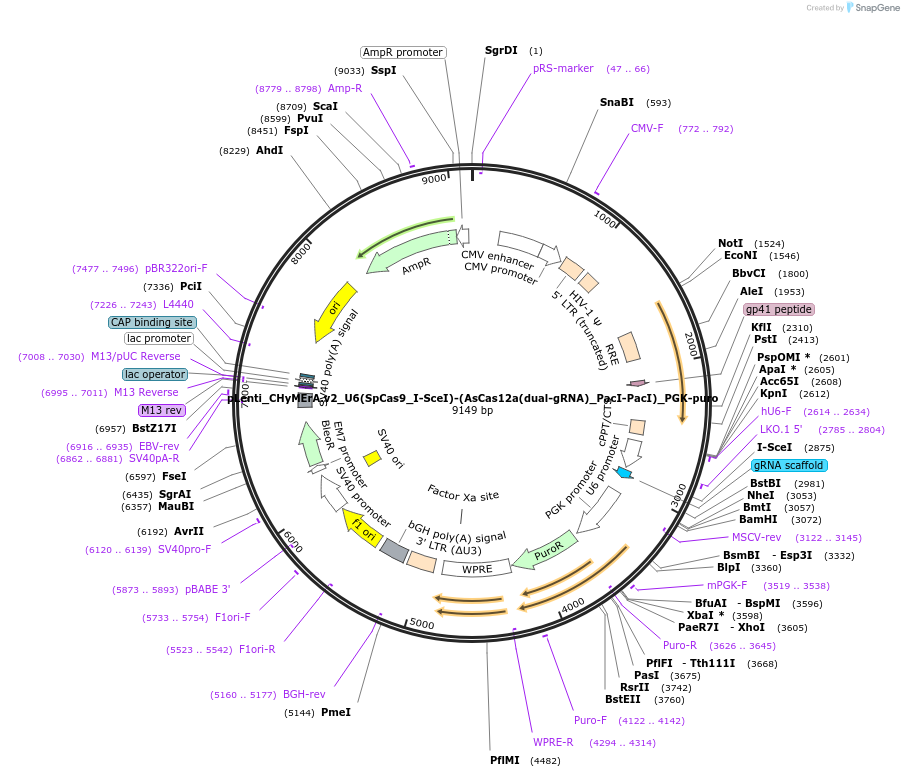
pLenti_CHyMErA.v2_U6(SpCas9_I-SceI)-(AsCas12a(dual-gRNA)_PacI-PacI)_PGK-puro
Plasmid#189636PurposeLentiviral expression of single SpCas9 and dual AsCas12a gRNAs for generating combinatorial CHyMErA.v2 3Cs librariesDepositorInserthU6 Cas9-Cas12a-Cas12a gRNA cassette, PGK puro cassette
UseCRISPR and LentiviralExpressionMammalianAvailable SinceNov. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
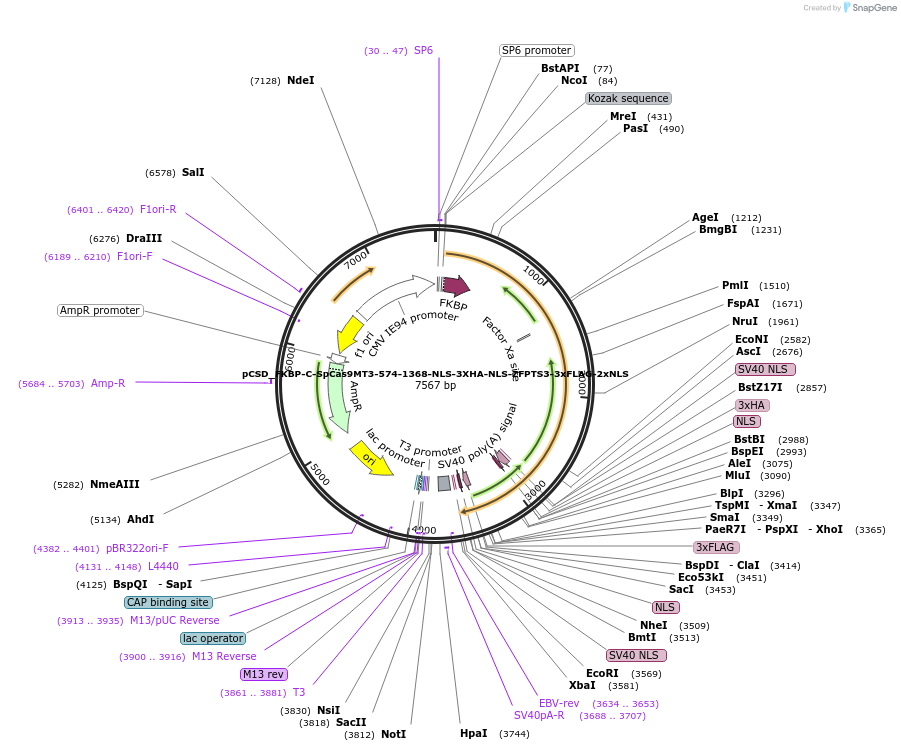
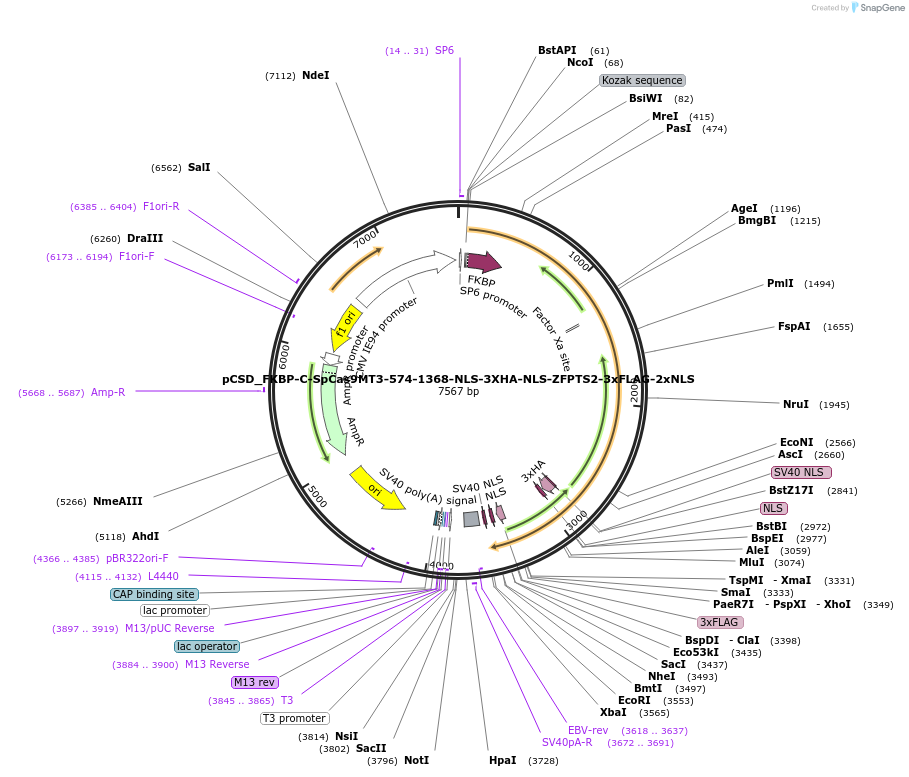
pCSD_FKBP-C-SpCas9MT3-574-1368-NLS-3XHA-NLS-ZFPTS3-3xFLAG-2xNLS
Plasmid#107300PurposeExpresses C terminal (574-1368 aa) R1335K mutant SpCas9 fused to FKBP and ZFP-VEGFA-TS3 in mammalian cellsDepositorInsertTruncated SpCas9 (aa 574-1368) fused to FKBP
UseCRISPRTags3x Flag, 2x NLS, FKBP, SV40NLS, 3xHA, c-Myc NLS, …ExpressionMammalianMutationTruncated SpCas9 (aa 574-1368) with R1335K mutati…PromoterCMV IE94Available SinceNov. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
pCSD_FKBP-C-SpCas9MT3-574-1368-NLS-3XHA-NLS-ZFPTS2-3xFLAG-2xNLS
Plasmid#107299PurposeExpresses C terminal (574-1368 aa) R1335K mutant SpCas9 fused to FKBP and ZFP-VEGFA-TS2 in mammalian cellsDepositorInsertTruncated SpCas9 (aa 574-1368) fused to FKBP
UseCRISPRTags3x Flag, 2x NLS, FKBP, SV40NLS, 3xHA, c-Myc NLS, …ExpressionMammalianMutationTruncated SpCas9 (aa 574-1368) with R1335K mutati…PromoterCMV IE94Available SinceNov. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
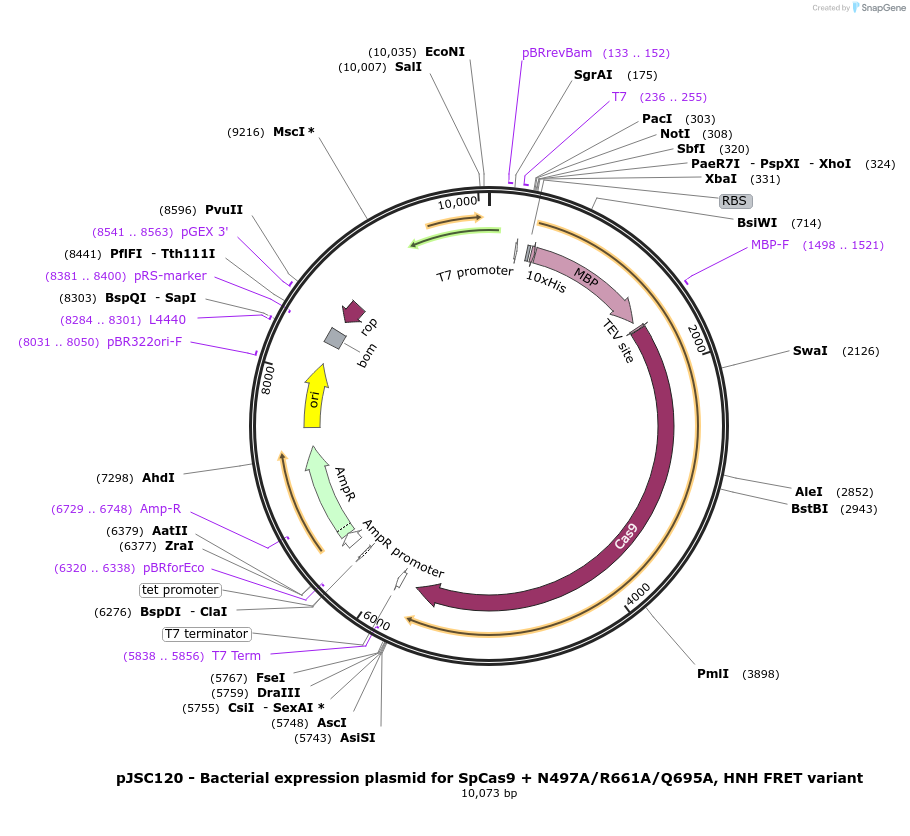
pJSC120 - Bacterial expression plasmid for SpCas9 + N497A/R661A/Q695A, HNH FRET variant
Plasmid#101208PurposeBacterial expression plasmid for SpCas9 + N497A/R661A/Q695A, HNH FRET variantDepositorInsertSpCas9 variant C80S/C574S/S355C/S867C/N497A/R661A/Q695A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, S355C, S867C, N497A, R661A and Q695APromoterT7Available SinceNov. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
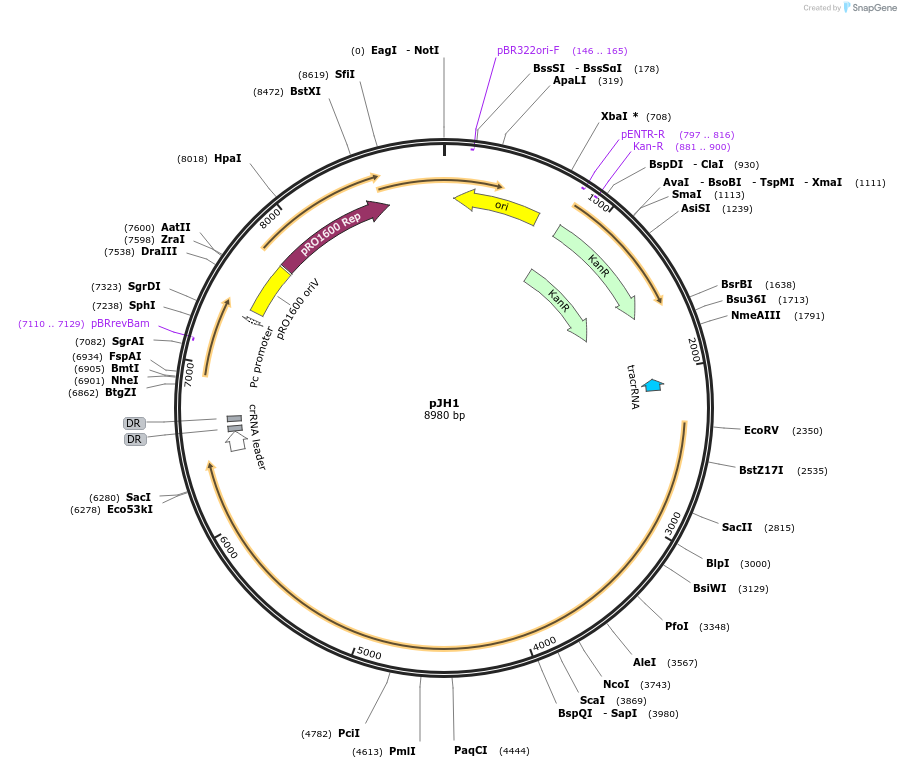
pJH1
Plasmid#154204Purposeencodes CRISPR-Cas9 system derived from plasmid pCas9DepositorTypeEmpty backboneUseCRISPRAvailable SinceJuly 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
The Expanded CRISPEY Kit
Plasmid Kit#1000000257PurposeUse the Expanded CRISPEY Kit for CRISPR-based gene editing of the Saccharomyces cerevisiae genome.DepositorApplicationGenome EditingVector TypeYeast ExpressionEditing TypeCRISPRAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
CRISPaint Gene Tagging Kit
Plasmid Kit#1000000086PurposeCRISPR-assisted insertion tagging (CRISPaint) plasmid collection for modular integration of large DNA fragments into user-defined genomic locations via a ligase-4-dependent mechanism.DepositorApplicationGenome EditingVector TypeMammalian ExpressionEditing TypeCRISPRAvailable SinceSept. 28, 2016AvailabilityAcademic Institutions and Nonprofits only -
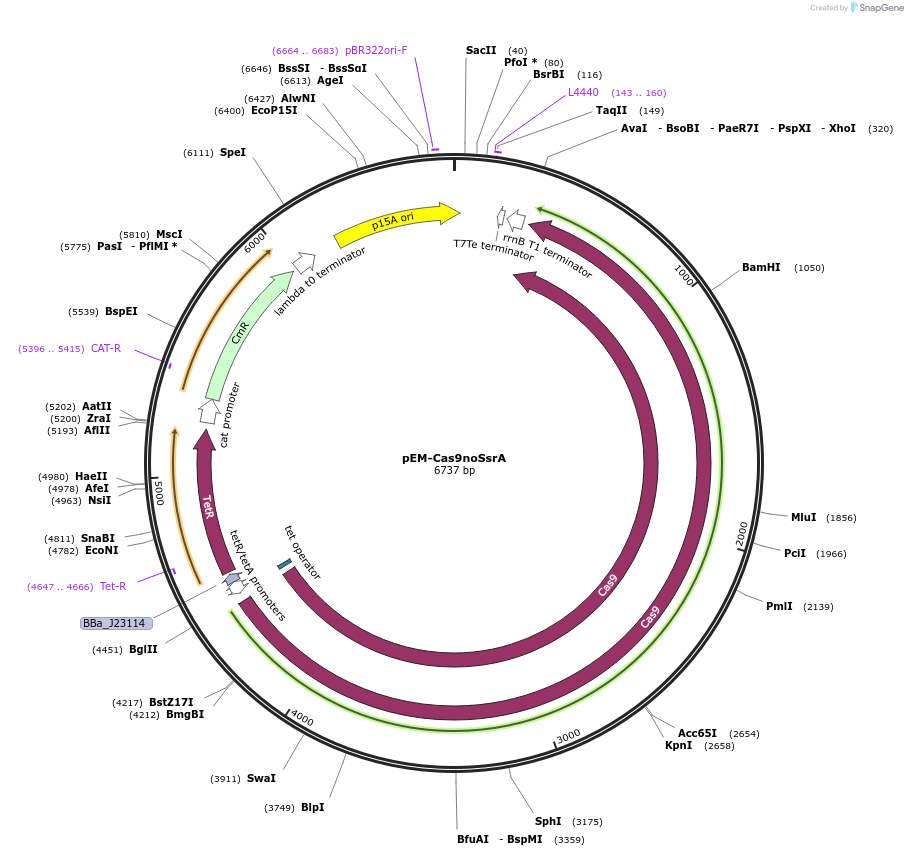
pEM-Cas9noSsrA
Plasmid#102285PurposeModified from pCas9-CR4 (Addgene: 62655) to remove the ssrA tag from Cas9.DepositorInsertCas9
UseCRISPRExpressionBacterialMutationremoved ssrA tagPromoterpTetAvailable SinceApril 12, 2018AvailabilityAcademic Institutions and Nonprofits only -
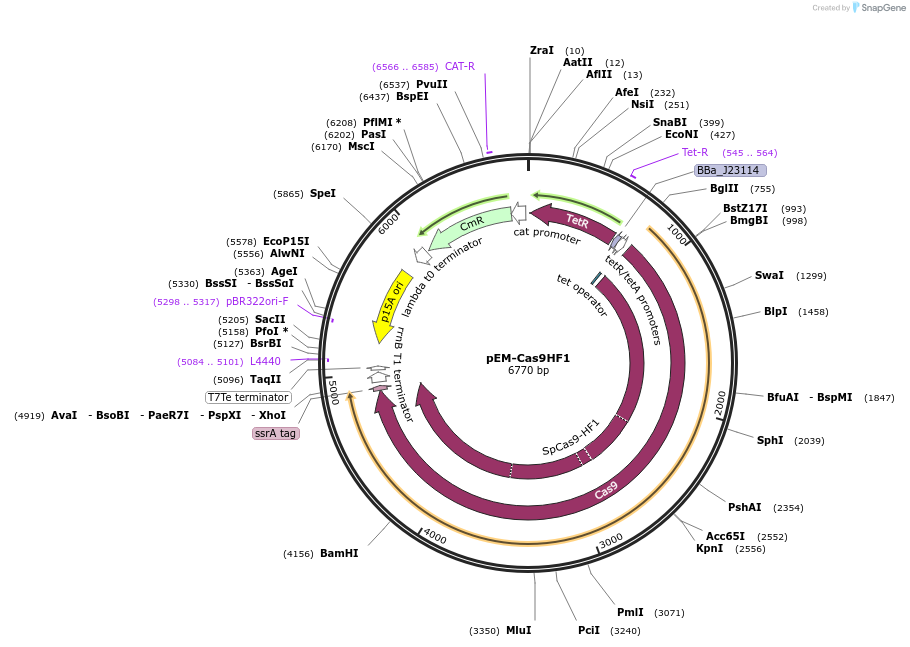
pEM-Cas9HF1
Plasmid#89961PurposeModified from pCas9-CR4 (Addgene: 62655) to use the high-fidelity version of Cas9, SpCas9-HF1 (N497A/R661A/Q695A/Q926A) from Kleinstiver et al 2016.DepositorInsertCas9HF1
ExpressionBacterialMutationN497A/R661A/Q695A/Q926APromoterpTetAvailable SinceJune 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
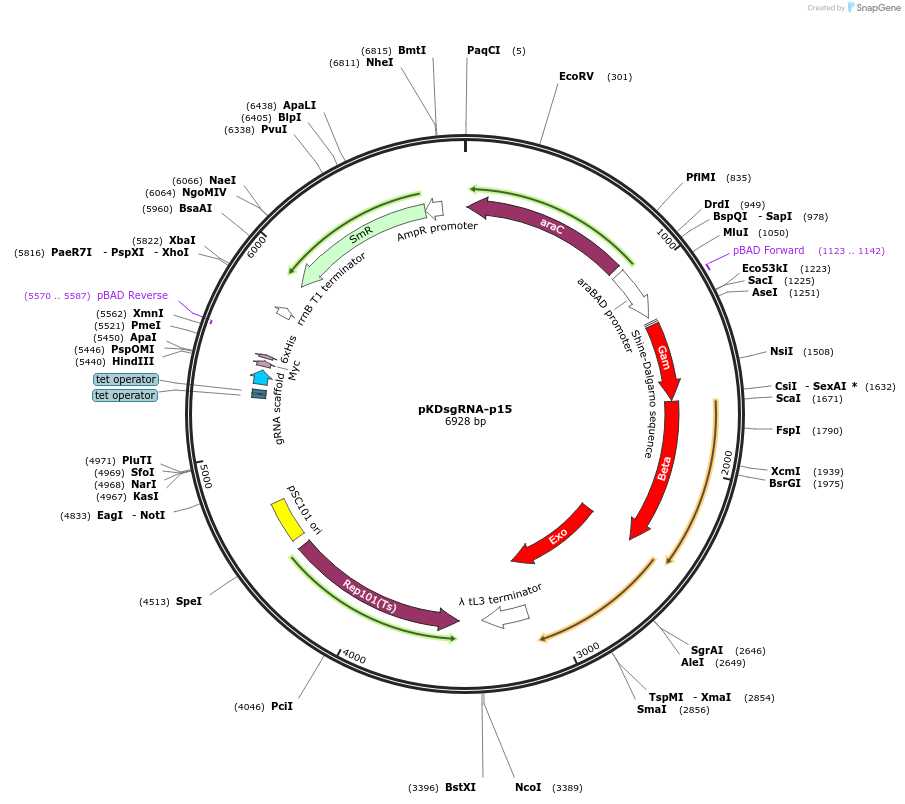
pKDsgRNA-p15
Plasmid#62656PurposeArabinose inducible lambda red and anhydrotetracycline inducible sgRNA expression that targets pCas9 plasmidDepositorInsertexo, beta, gam, sgRNA
ExpressionBacterialAvailable SinceOct. 19, 2015AvailabilityAcademic Institutions and Nonprofits only -
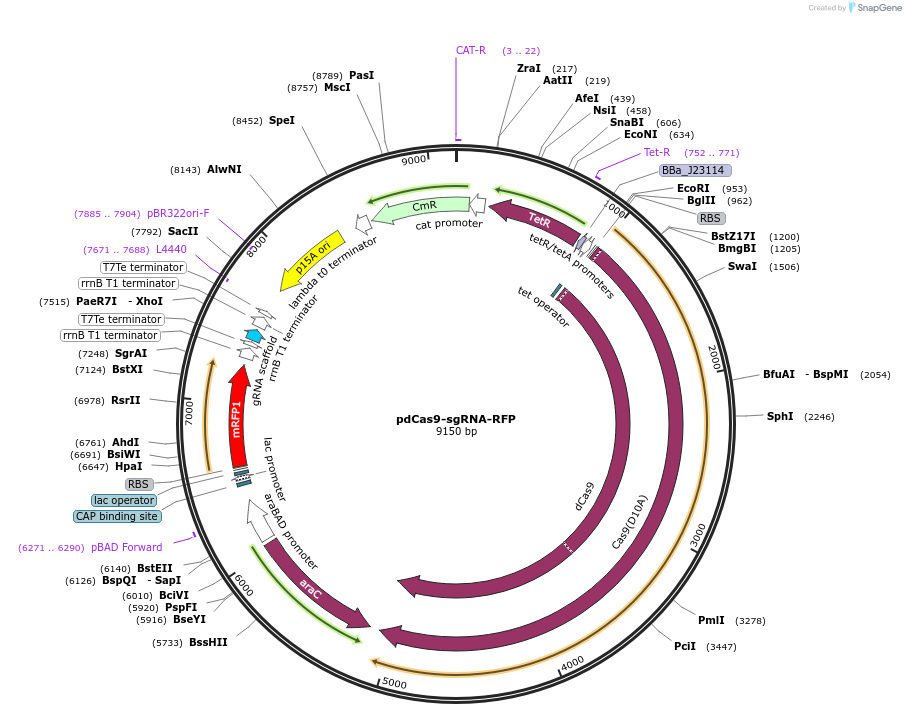
pdCas9-sgRNA-RFP
Plasmid#166005PurposeBacterial expression of dCas9 (aTc control) and sgRNA (arabinose control)DepositorTypeEmpty backboneUseCRISPRExpressionBacterialAvailable SinceApril 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
Toronto KnockOut version 3 library (TKOv3)
Pooled Library#90294PurposeOptimized library offering improved accuracy, efficiency, and scalability for CRISPR screens compared to TKOv1DepositorAvailable SinceMay 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
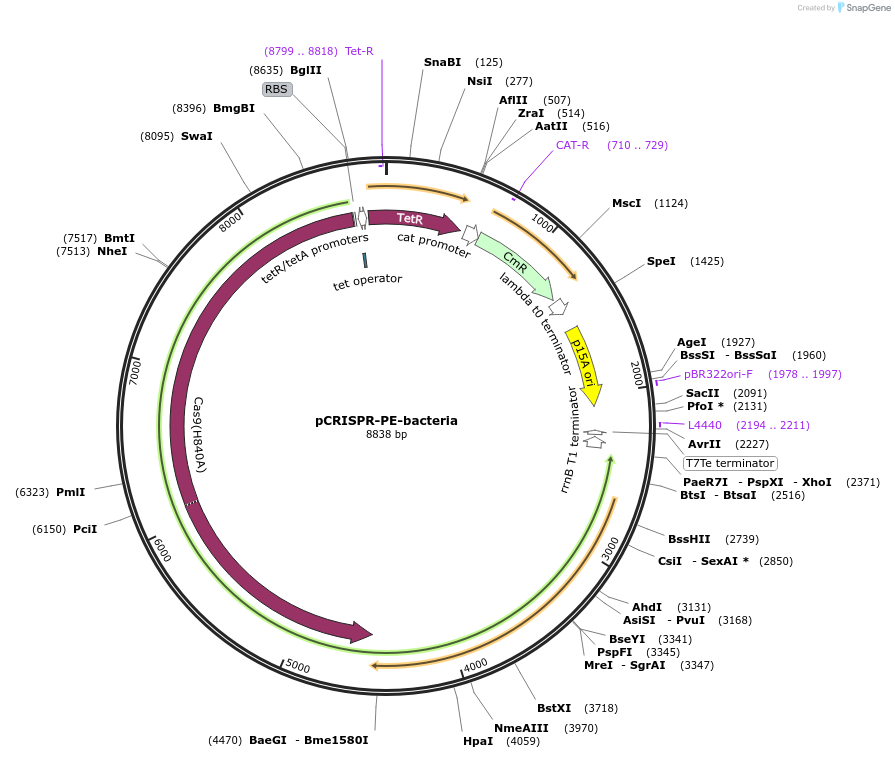
pCRISPR-PE-bacteria
Plasmid#172715PurposeExpression of an E. coli codon optimized fusion protein of Cas9n-linker-M-MLV2 for prime editingDepositorInsertCas9n-linker-M-MLV2
ExpressionBacterialMutationH840A of Cas9Available SinceSept. 10, 2021AvailabilityAcademic Institutions and Nonprofits only