We narrowed to 5,769 results for: chia
-
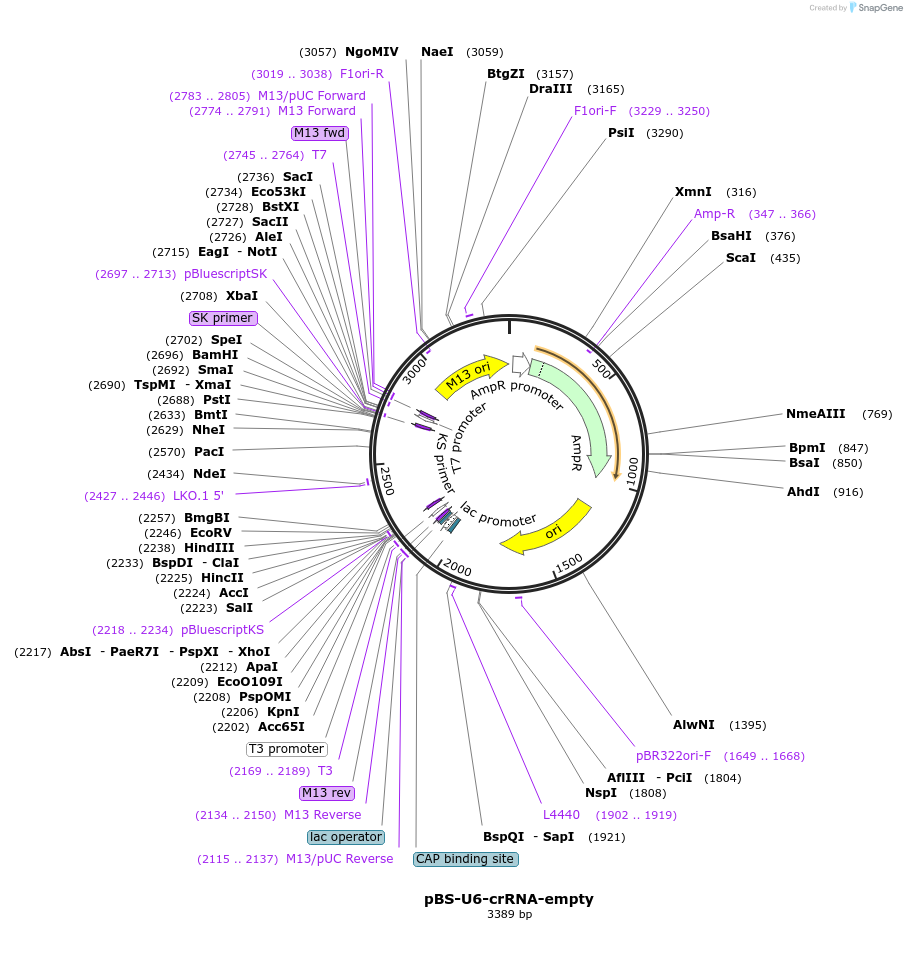
Plasmid#134921PurposeComponents for genome editing in mammalian cells with pPB-CAG-hCas3 and pCAG-All-in-one-hCascade.DepositorInsertcrRNA for CRISPR-Cas3
ExpressionMammalianPromoterU6Available SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
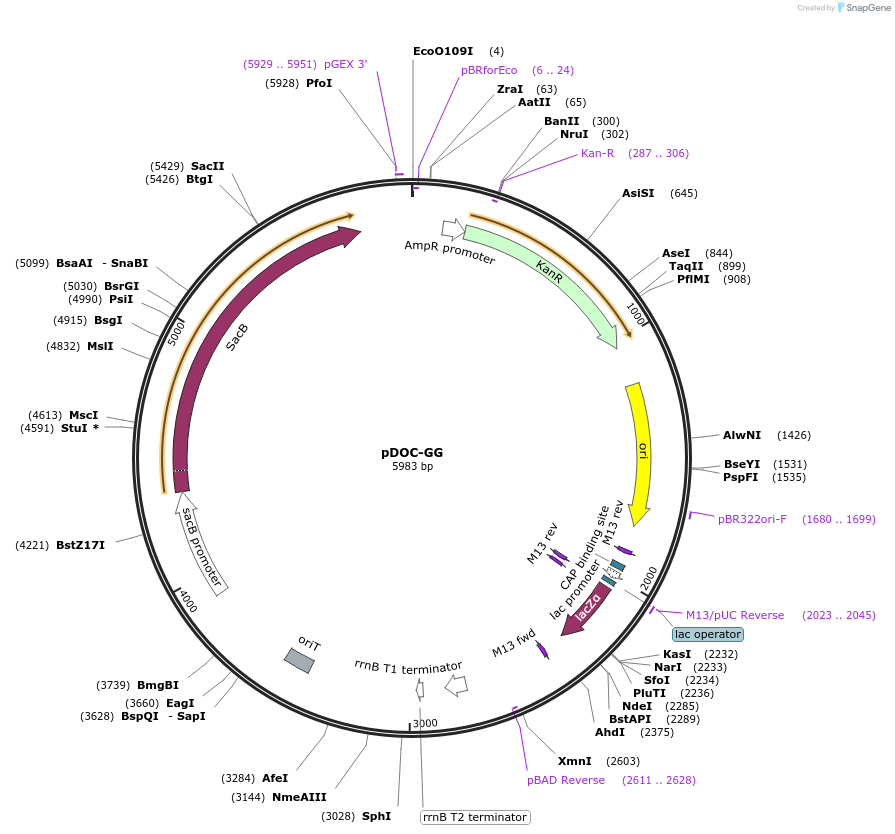
pDOC-GG
Plasmid#149377PurposeChassis plasmid for building mutagenesis cassettes by Golden Gate assembly to form a donor plasmid for recombineering by Gene DoctoringDepositorInsertsFragment containing a kanamycin resistance cassette
Fragment containing pMB1 origin of replication and an I-SceI recognition sequence
Fragment containing a lacZ⍺ expression cassette flanked by BsaI sites
Fragment containing an I-SceI recognition sequence
UseSynthetic BiologyExpressionBacterialAvailable SinceAug. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
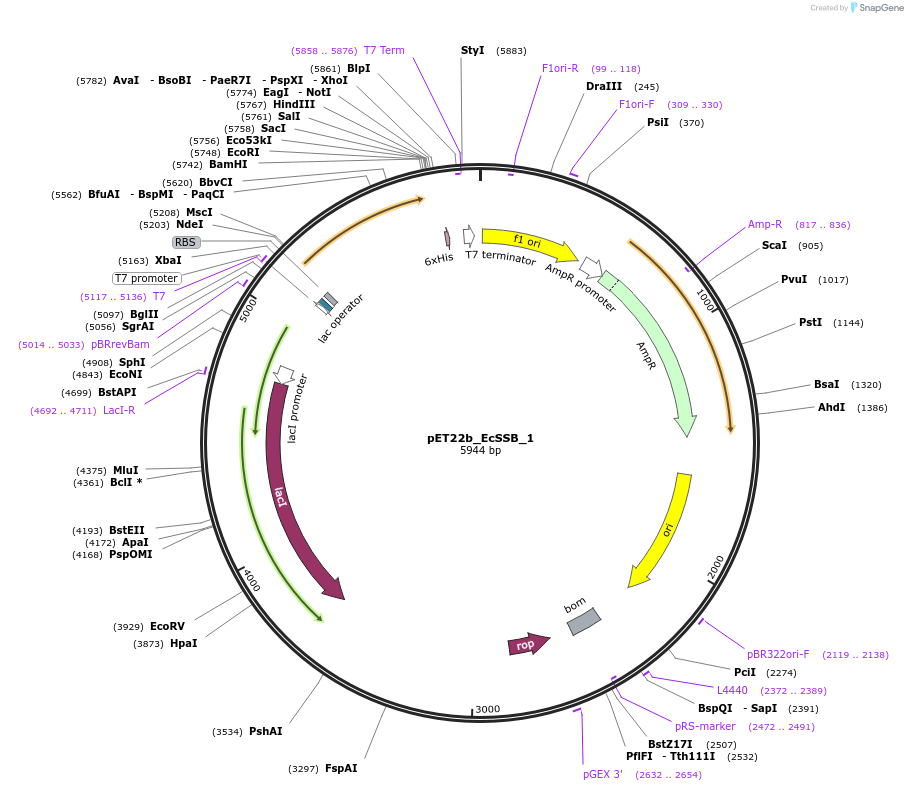
pET22b_EcSSB_1
Plasmid#78203PurposeExpression of single-cysteine variant of single-stranded DNA-binding protein protein, EcSSB, as scaffold for ssDNA biosensorDepositorInsertssb (G26C)
ExpressionBacterialMutationG26C (please see depositor comment below)PromoterT7Available SinceJuly 12, 2016AvailabilityAcademic Institutions and Nonprofits only -
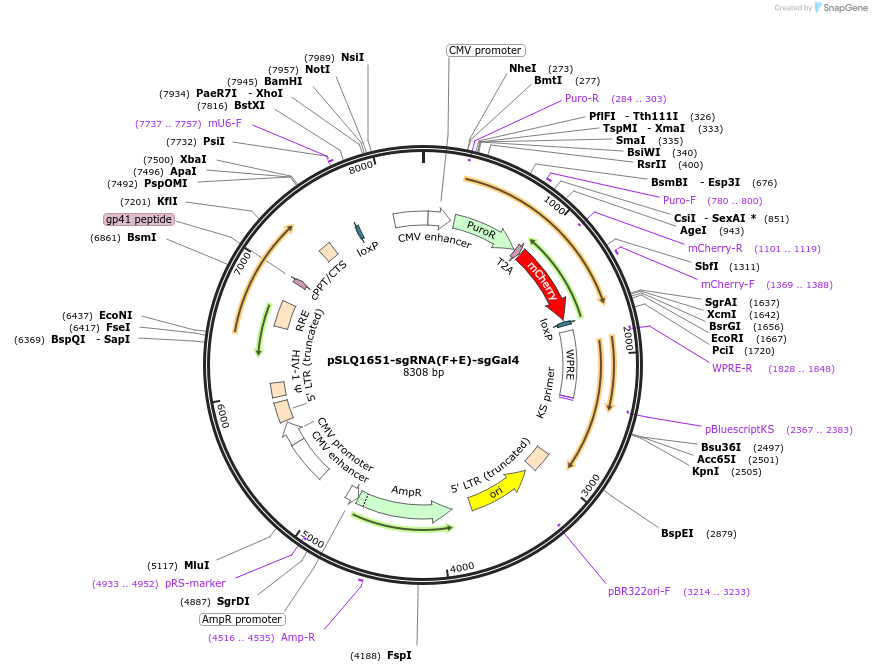
pSLQ1651-sgRNA(F+E)-sgGal4
Plasmid#100549PurposesgGal4-expressing vector modified from Addgene plasmid #51024DepositorInsertsgGal4
UseCRISPRTagssgRNAExpressionMammalianPromoterU6Available SinceSept. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
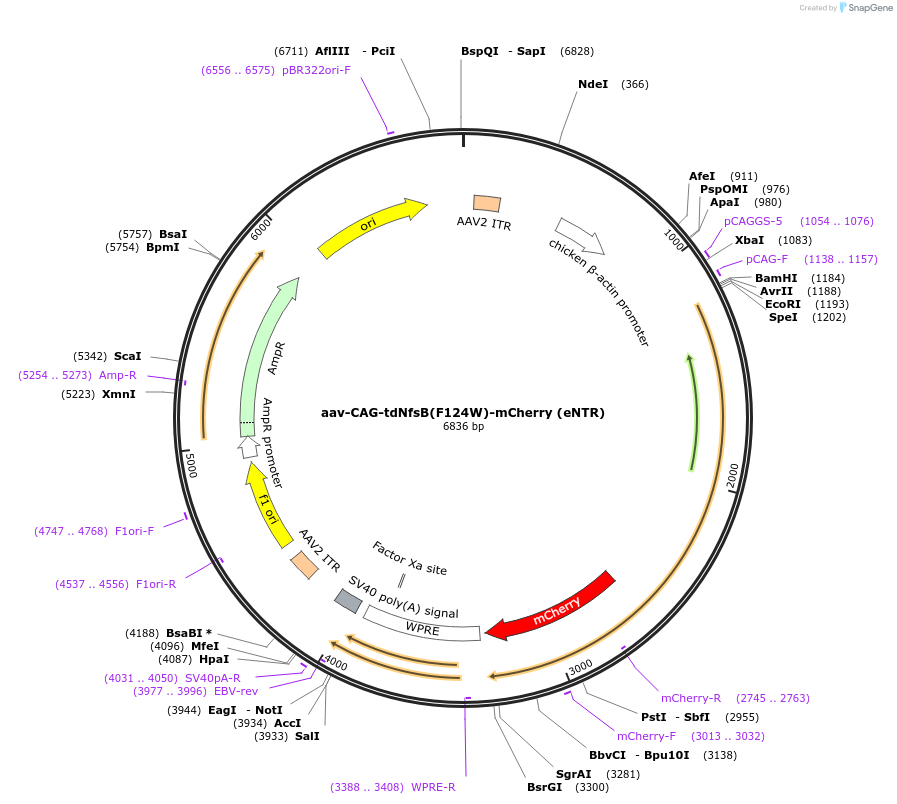
aav-CAG-tdNfsB(F124W)-mCherry (eNTR)
Plasmid#113761PurposeAAV-mediated expression of bacterial tdNfsB-mCherry (eNTR) under the CAG promoterDepositorInserttdNfsB(F124W)-mCherry
UseAAVTagsmCherryExpressionMammalianMutationF124WPromoterCAGAvailable SinceSept. 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
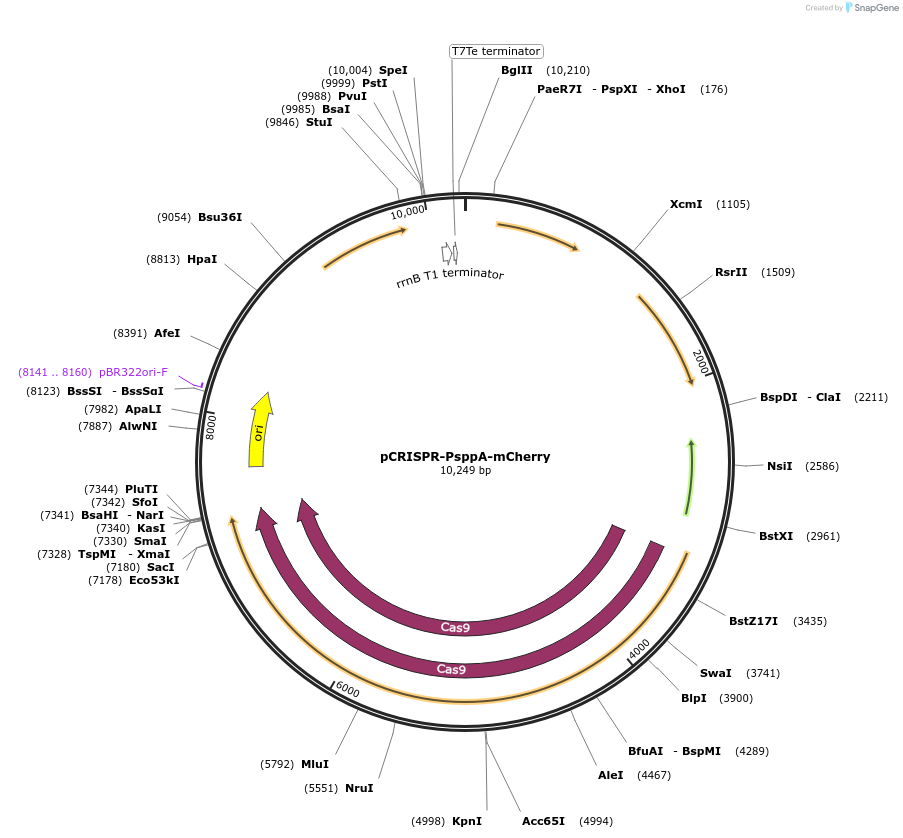
pCRISPR-PsppA-mCherry
Plasmid#225428PurposePlasmid encodes SgRNA, SpCas9 and homologous arms for homologous recombination of mCherryDepositorInsertmCherry, Cas9, SgRNA
UseCRISPRExpressionBacterialPromoterPsppA upstream of mCherry and Cas9, P3 upstream o…Available SinceJan. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
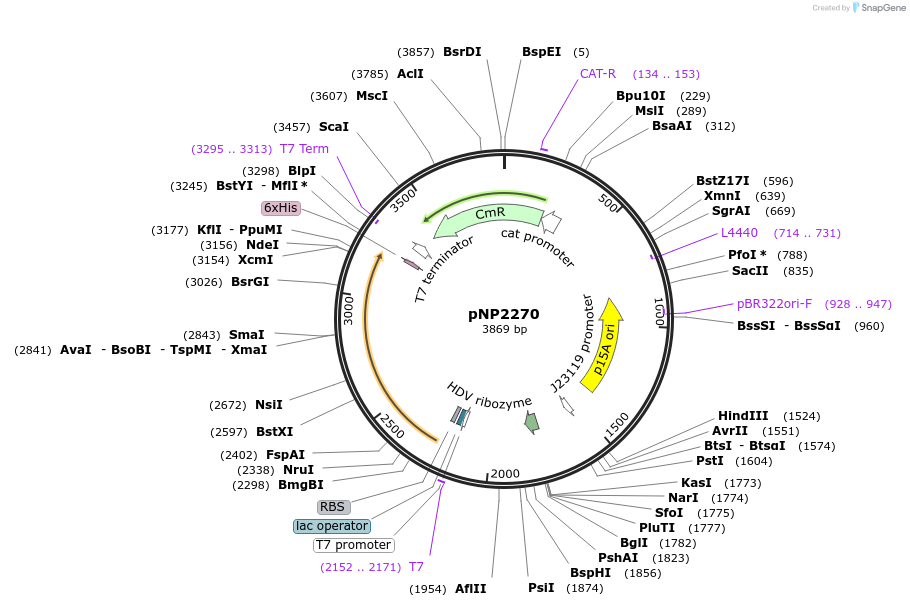
pNP2270
Plasmid#222733PurposepNP1950 - J23119-IS621 bRNA1 TBL-lacZ-1 (Extended RTG), DBL-WT, HDV; T7-IS621 RecombinaseDepositorInsertIS621 Helper (lacZ)
ExpressionBacterialAvailable SinceJuly 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
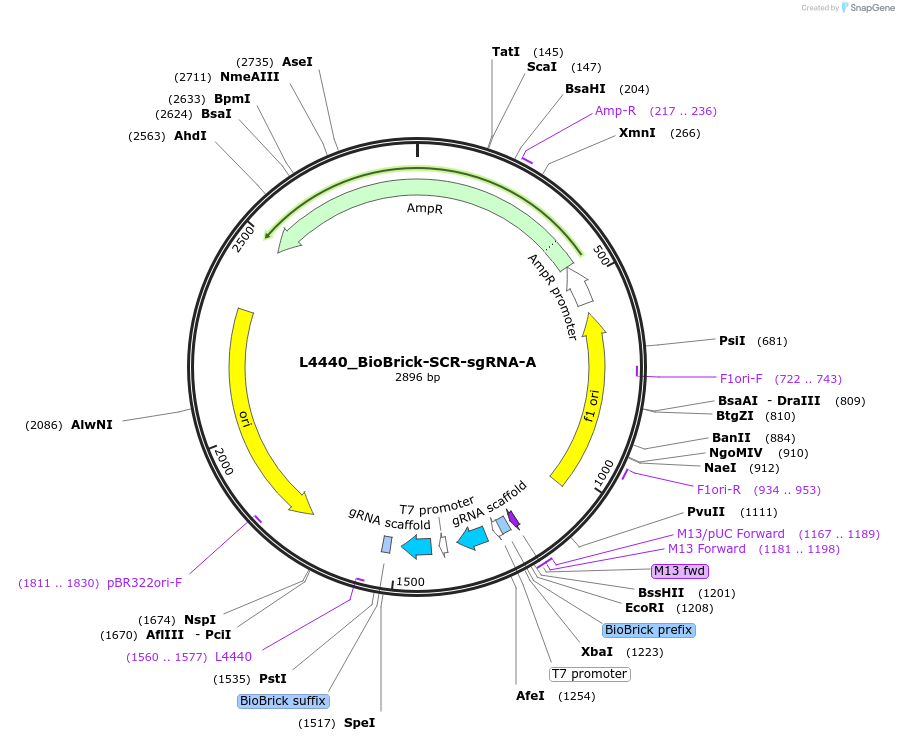
L4440_BioBrick-SCR-sgRNA-A
Plasmid#177811PurposeOverexpression of sgRNAs in E. coli HT115 (scrambled targeting sequences)DepositorInsertScrambled sgRNA targeting sequences
ExpressionBacterialAvailable SinceJune 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
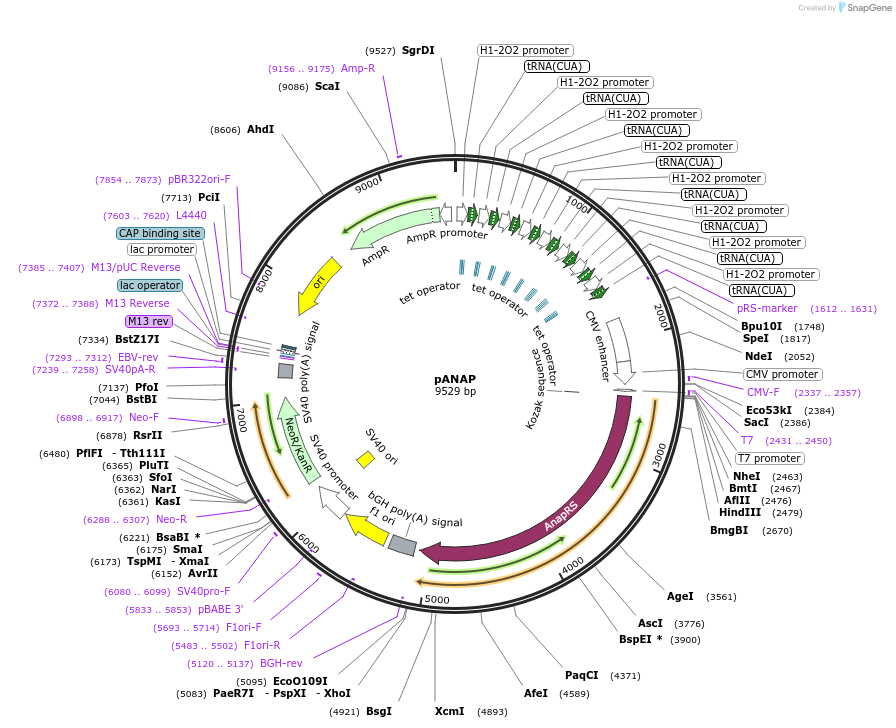
pANAP
Plasmid#48696PurposeAllows co-translational incorporation of the fluorescent ncAA Anap directly into the target protein; expresses amber suppressor tRNA and the synthetase AnapRSDepositorInserts8 copies of the amber suppressor tRNACUA-Leu
AnapRS
ExpressionMammalianMutationmutant E.coli leucyl synthetase specific for Anap…PromoterCMV and human H1 promoterAvailable SinceNov. 25, 2013AvailabilityAcademic Institutions and Nonprofits only -
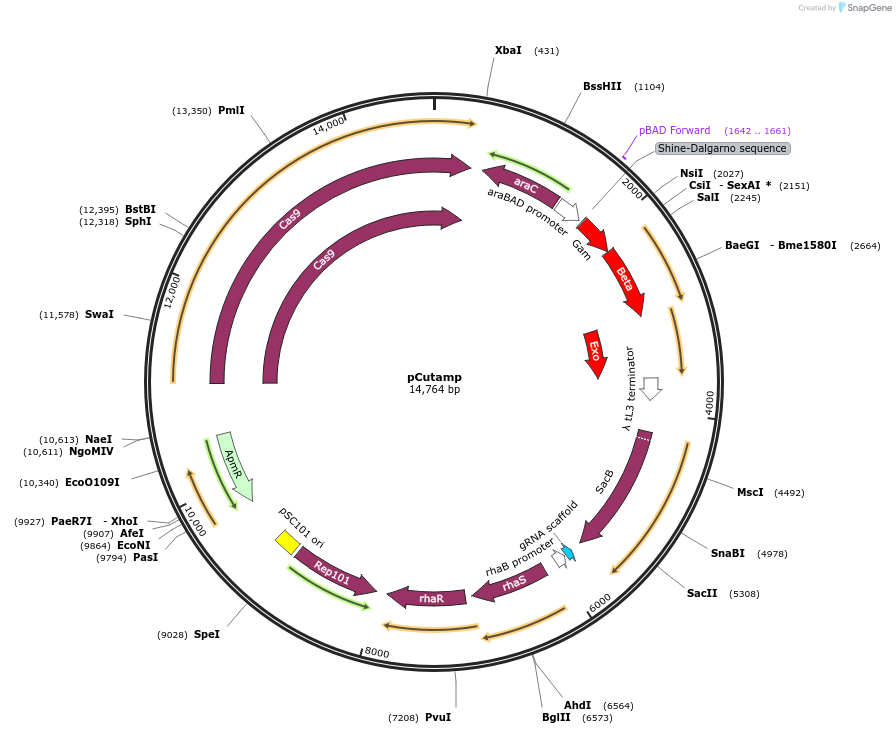
pCutamp
Plasmid#140632PurposePlasmid-curing in Escherichia coli by targeting the AmpR promoterDepositorInsertSpCas9_lambda-RED system, SacB, Rha induction system, sgRNA targeting AmpR promoter
UseCRISPRExpressionBacterialAvailable SinceAug. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
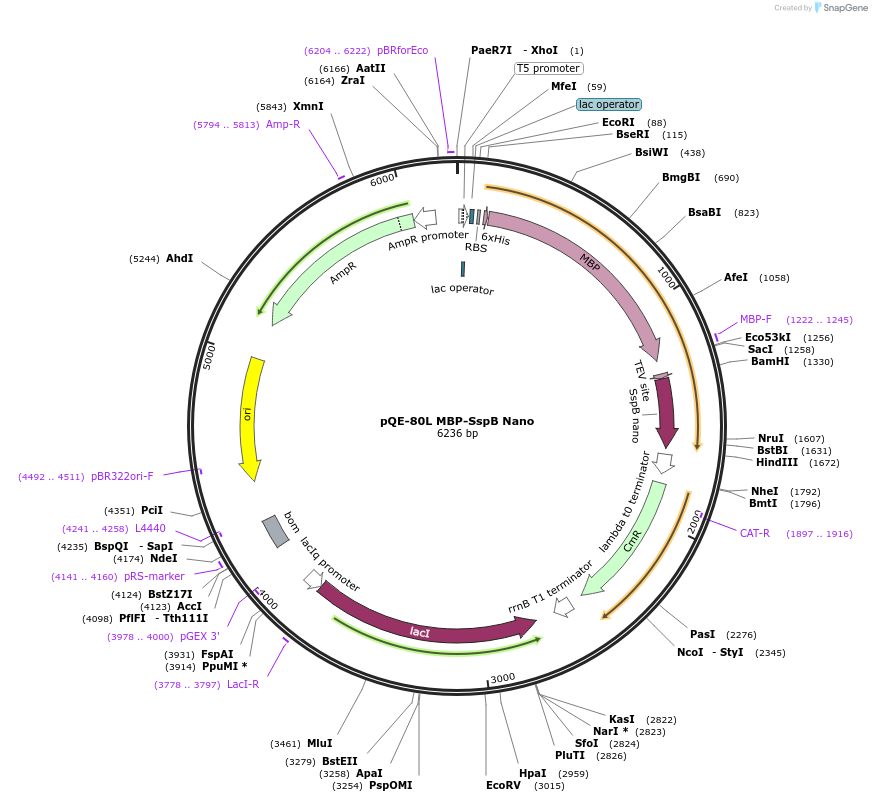
pQE-80L MBP-SspB Nano
Plasmid#60409PurposeE. coli expression of SspB NanoDepositorInsertSspB nano
TagsHis-MBP-TEVExpressionBacterialMutationY11K and A15EAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
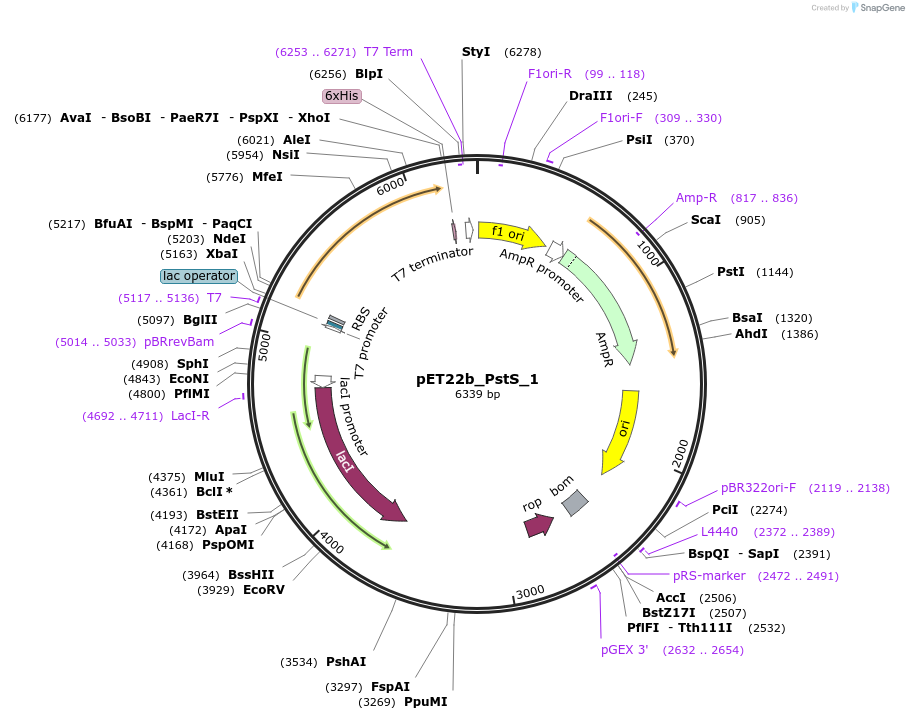
pET22b_PstS_1
Plasmid#78198PurposeExpression of single-cysteine variant of mature phosphate binding protein, PstS, as scaffold for phosphate biosensorDepositorInsertpstS (A197C)
ExpressionBacterialMutationA197C (please see depositors comment below)PromoterT7Available SinceJuly 12, 2016AvailabilityAcademic Institutions and Nonprofits only -
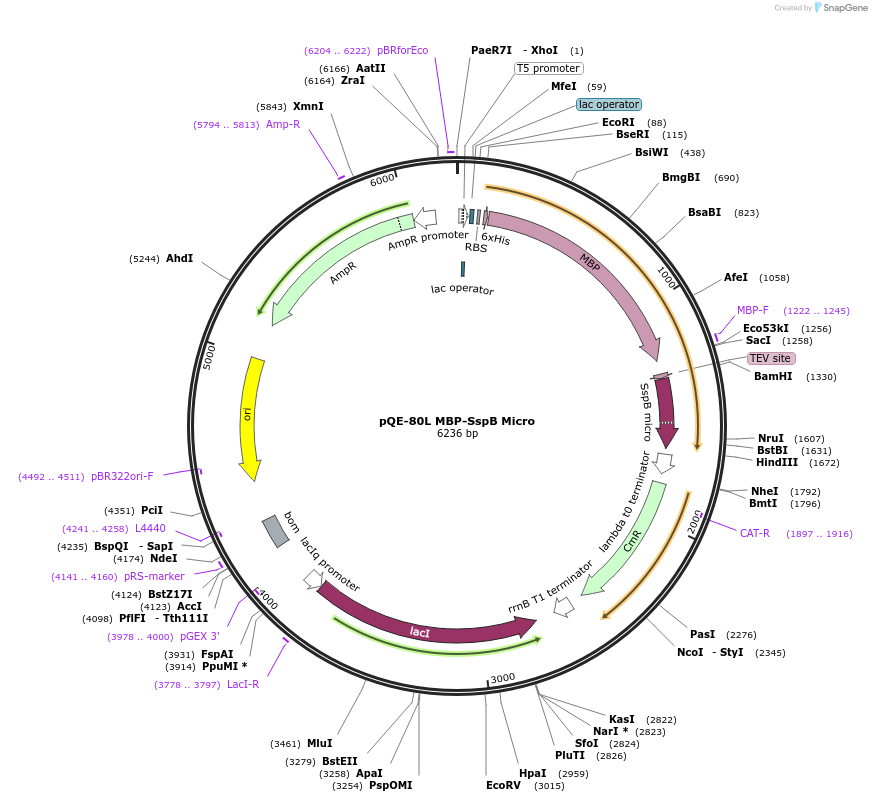
pQE-80L MBP-SspB Micro
Plasmid#60410PurposeE. coli expression of SspB MicroDepositorInsertSspB micro
TagsHis-MBP-TEVExpressionBacterialMutationR73Q, Y11K and A15EAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
pQE-80L iLID (C530M)
Plasmid#60408PurposeE. coli expression of iLID (C530M)DepositorInsertiLID C530M
TagsHisExpressionBacterialMutationC530MAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
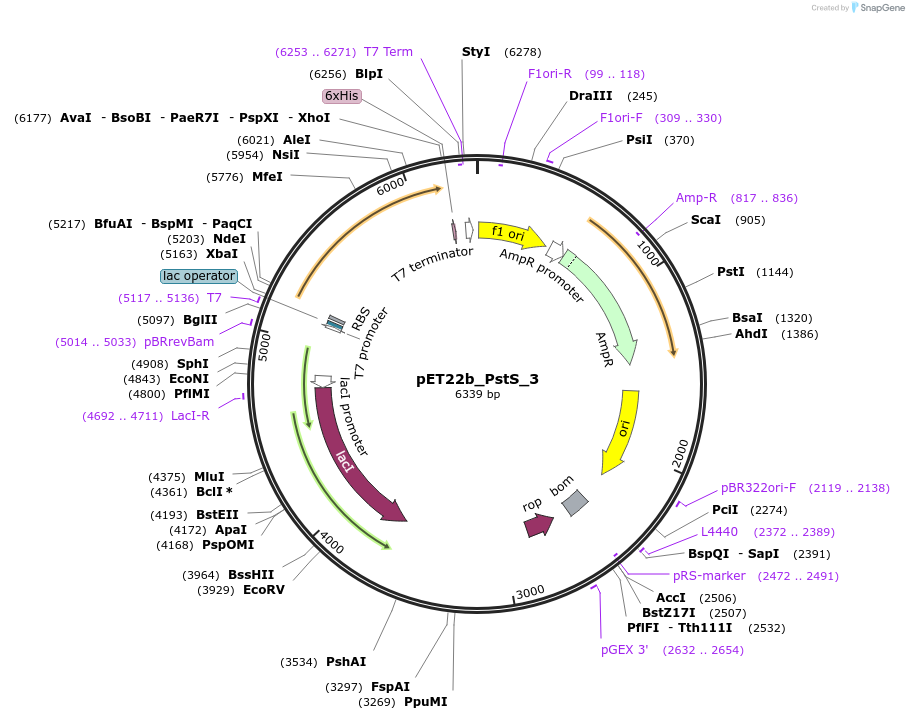
pET22b_PstS_3
Plasmid#78200PurposeExpression of double-cysteine variant of mature phosphate binding protein, PstS, which also has a mutation to weaken phosphate binding, as scaffold for phosphate biosensorDepositorInsertpstS (A17C,I76G,A197C)
ExpressionBacterialMutationA17C,I76G,A197C (please see depositor comment bel…PromoterT7Available SinceJuly 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
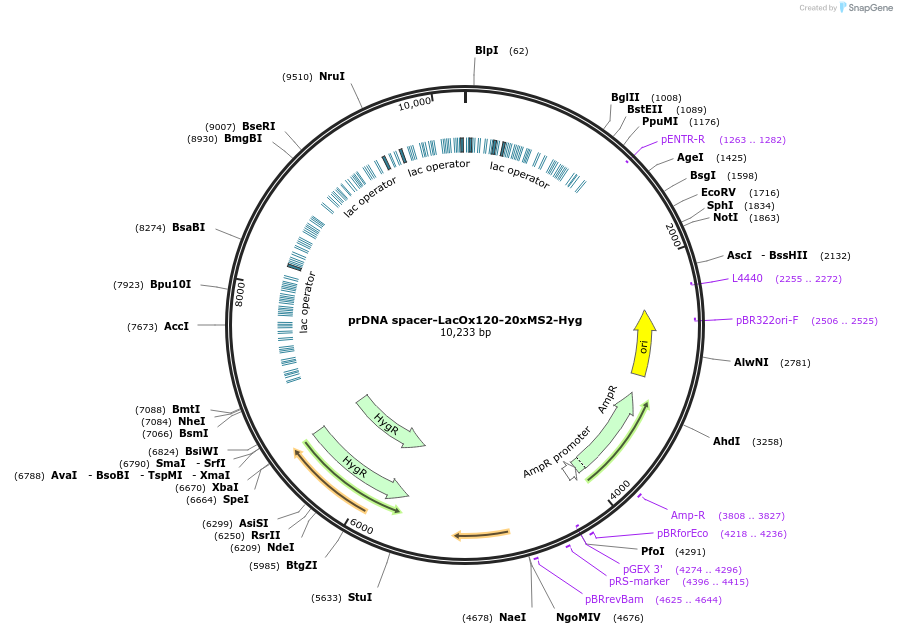
prDNA spacer-LacOx120-20xMS2-Hyg
Plasmid#210220PurposeIntegrates 120xLacO repeats and 20xMS2 repeats with a hygromycin drug selection marker into the rDNA spacer.DepositorInserts120xLacO repeats derived from: PMID: 12864855.
24xMS2 repeats derived from Addgene plasmid # 31865
Available SinceDec. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
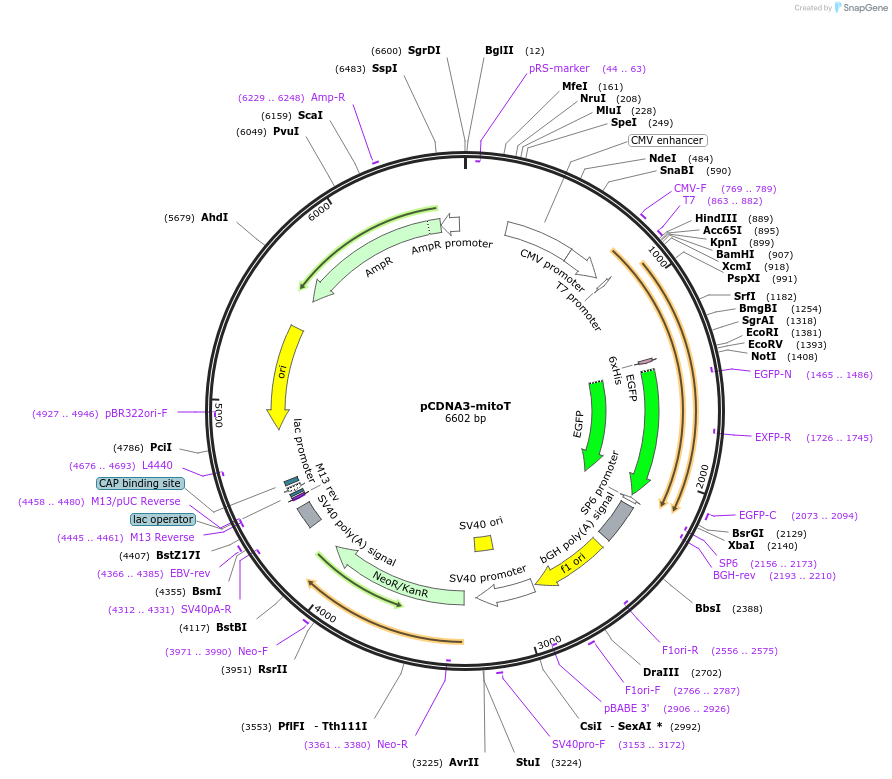
pCDNA3-mitoT
Plasmid#174519PurposeEngineered intermembrane bridge to link the inner and outer mitochondrial membranes in mammalian cellsDepositorInsertMitoT synthetic tether
Tags6xHis and eGFPExpressionMammalianPromoterCMVAvailable SinceSept. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
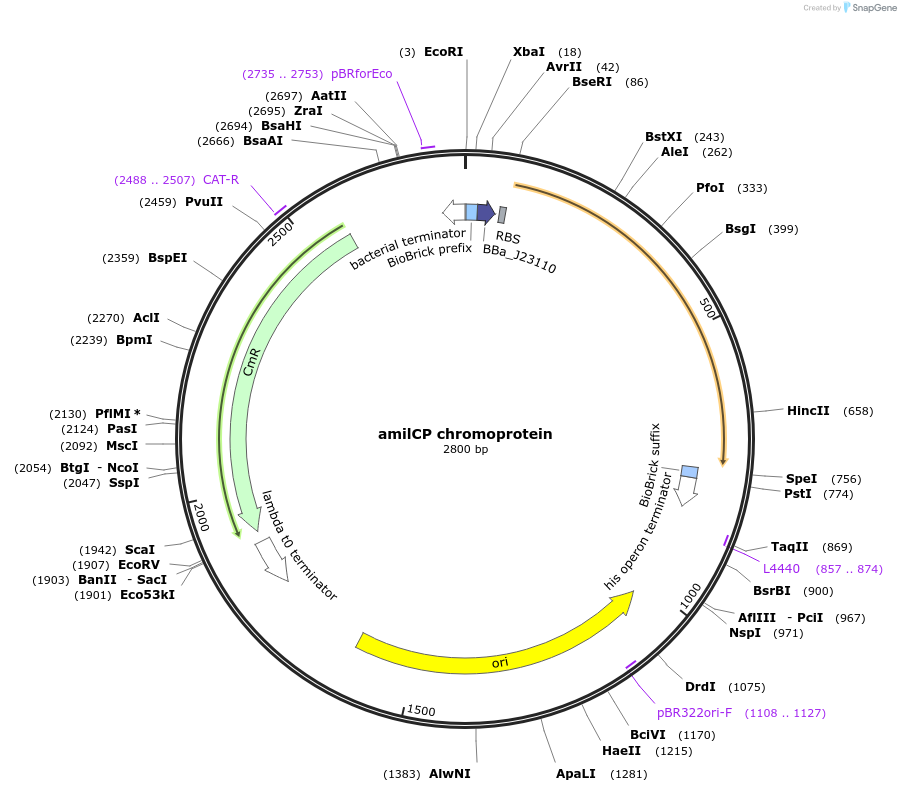
amilCP chromoprotein
Plasmid#117847PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilCP chromoprotein in E. coliDepositorInsertpromoter, RBS, amilCP
UseSynthetic Biology; Escherichia coliAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
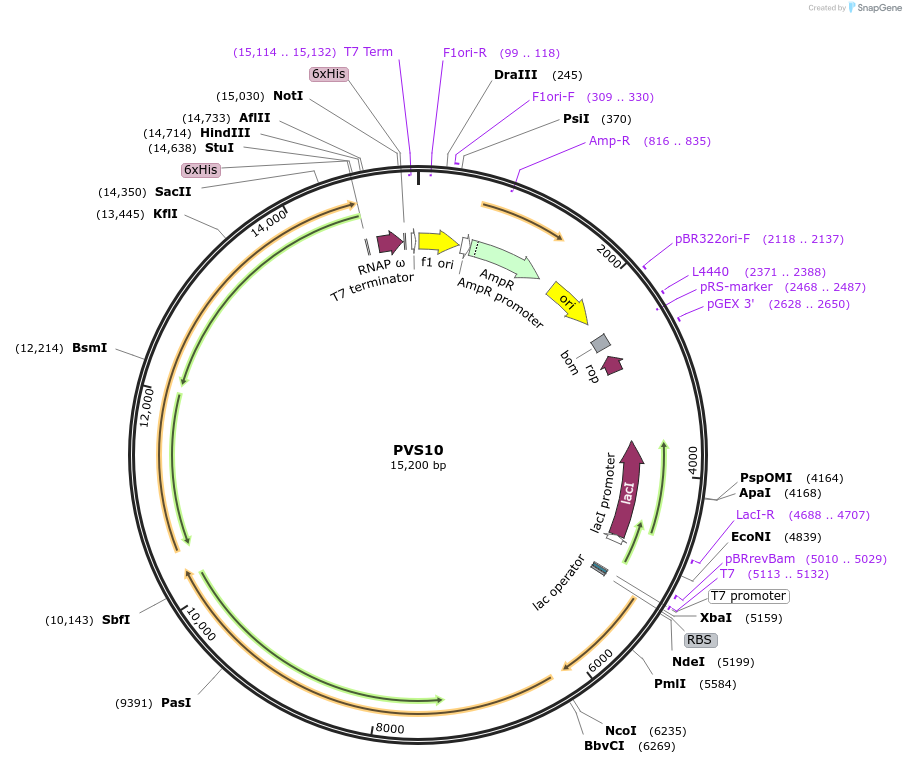
PVS10
Plasmid#104398Purposepolycistronic vector for expression of E. coli core RNAPDepositorTagsHis6ExpressionBacterialPromoterT7Available SinceDec. 13, 2017AvailabilityAcademic Institutions and Nonprofits only -
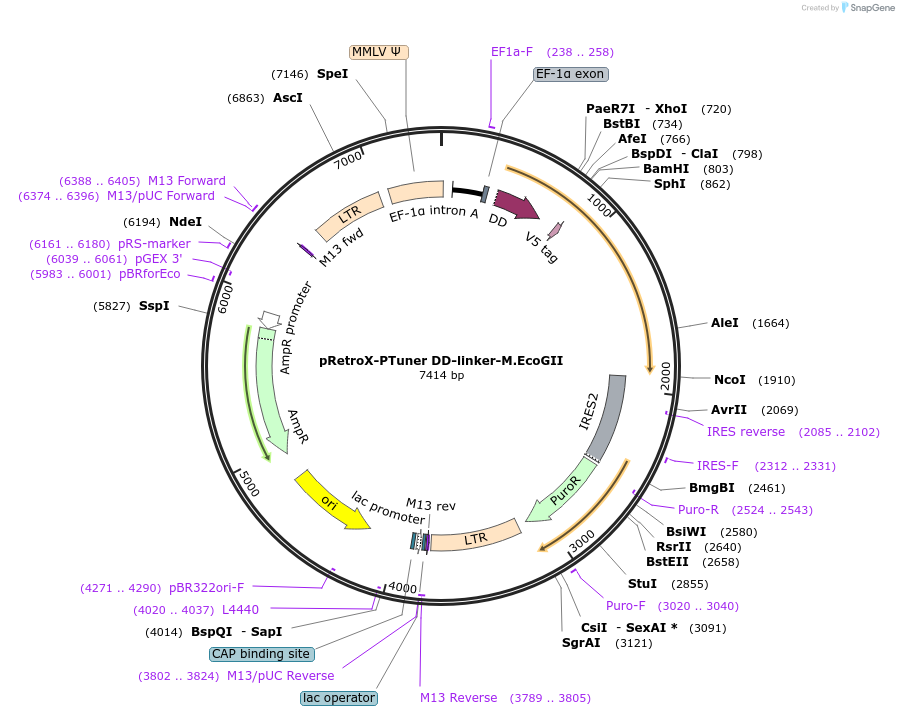
pRetroX-PTuner DD-linker-M.EcoGII
Plasmid#122082PurposeExpress M.EcoGII with detabilization domain - inducibleDepositorInsertDD-linker-M.EcoGII
UseRetroviralTagsV5 tag on Nter of M.EcoGIIExpressionBacterial and MammalianAvailable SinceMarch 13, 2019AvailabilityAcademic Institutions and Nonprofits only