We narrowed to 5,133 results for: puc
-
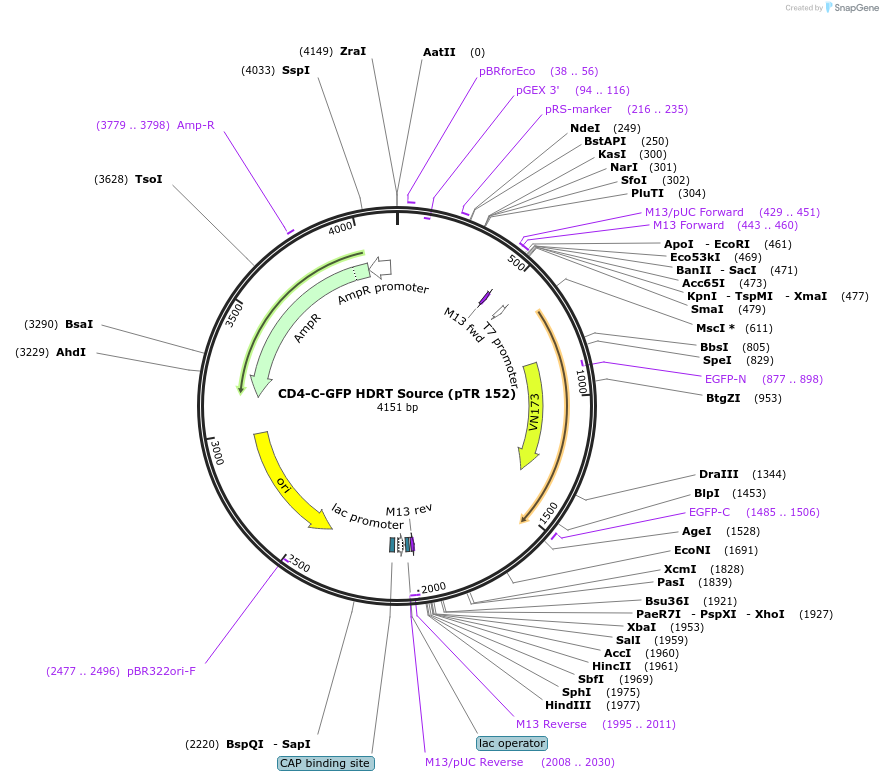
Plasmid#112018PurposeDNA sequence source for amplifying an HDR template to tag endogenous human CD4 gene with GFPDepositorInsertCD4-GFP HDRT (CD4 Human)
UseCRISPR and Synthetic BiologyAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
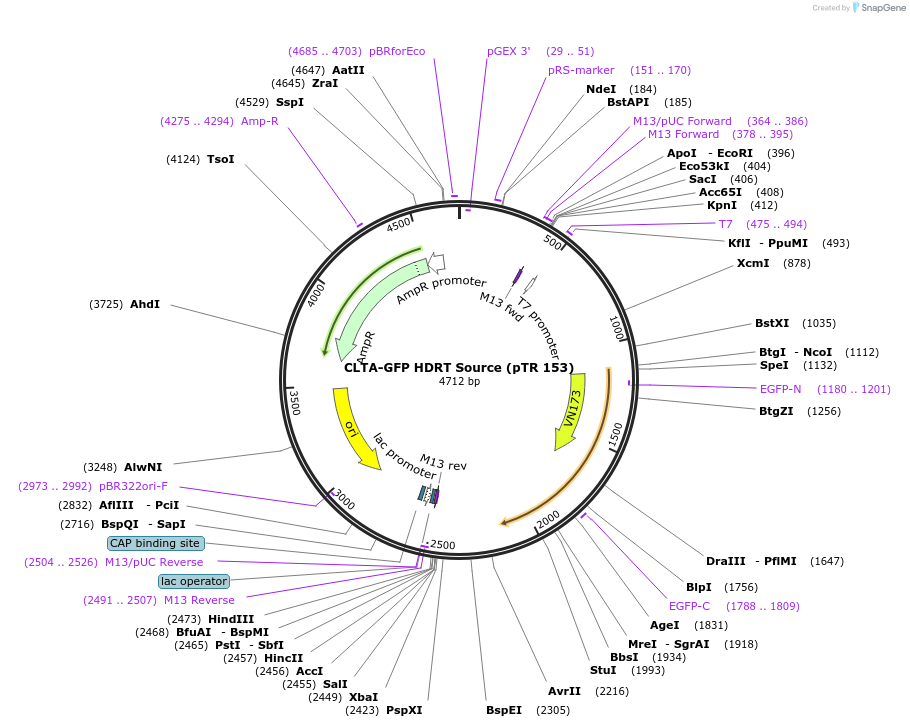
CLTA-GFP HDRT Source (pTR 153)
Plasmid#112016PurposeDNA sequence source for amplifying an HDR template to tag endogenous human CLTA gene with GFPDepositorInsertCLTA-GFP HDRT (CLTA Human)
UseCRISPR and Synthetic BiologyAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
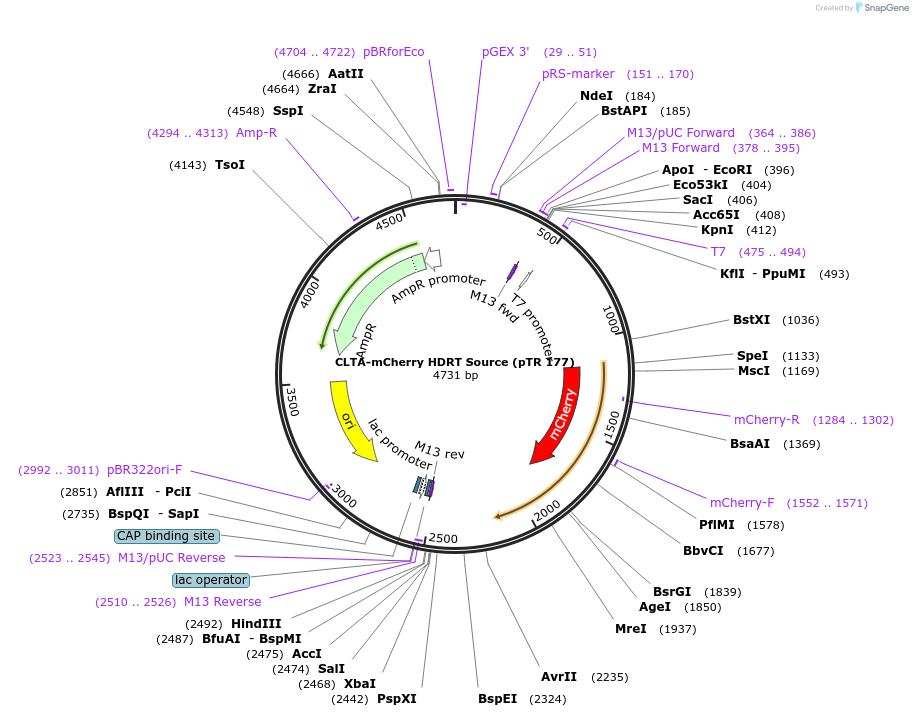
CLTA-mCherry HDRT Source (pTR 177)
Plasmid#112017PurposeDNA sequence source for amplifying an HDR template to tag endogenous human CLTA gene with mCherryDepositorInsertCLTA-mCherry HDRT (CLTA Human)
UseCRISPR and Synthetic BiologyAvailable SinceNov. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
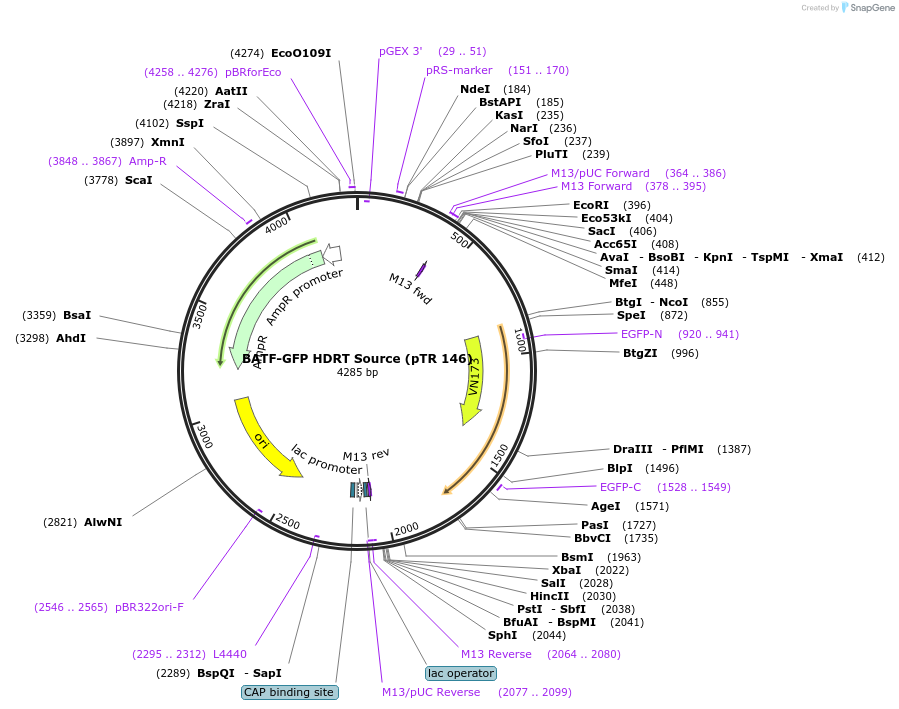
BATF-GFP HDRT Source (pTR 146)
Plasmid#112015PurposeDNA sequence source for amplifying an HDR template to tag endogenous human BATF gene with GFPDepositorInsertBATF-GFP HDRT (BATF Human)
UseCRISPR and Synthetic BiologyAvailable SinceAug. 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
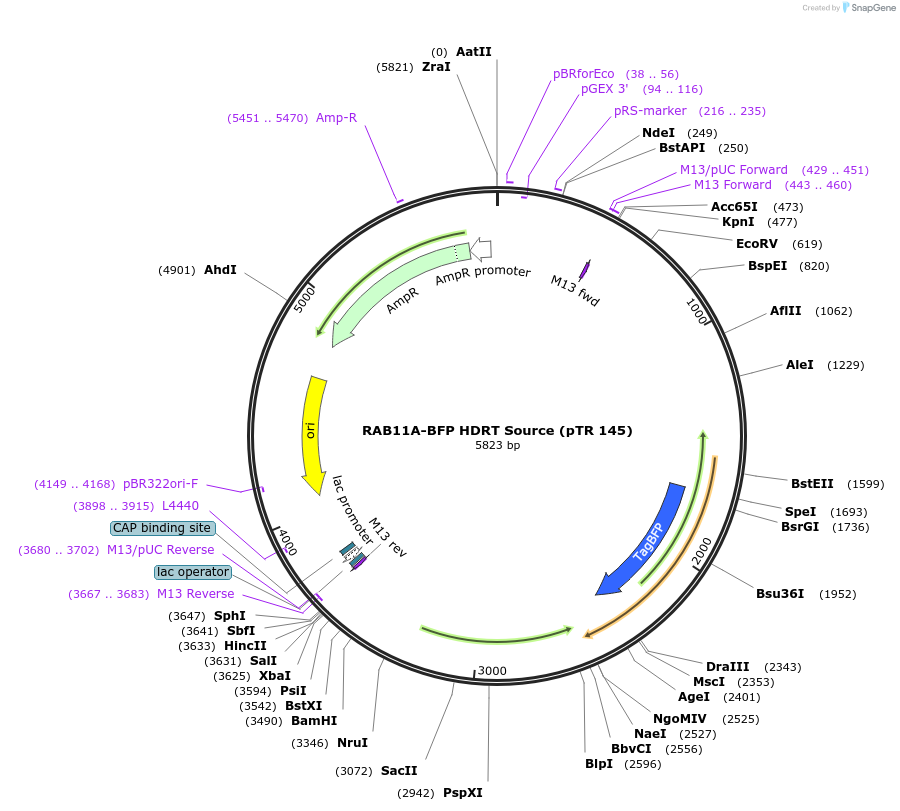
RAB11A-BFP HDRT Source (pTR 145)
Plasmid#112014PurposeDNA sequence source for amplifying an HDR template to tag endogenous human RAB11A gene with BFPDepositorInsertRAB11A-BFP HDRT (RAB11A Human)
UseCRISPR and Synthetic BiologyAvailable SinceMarch 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
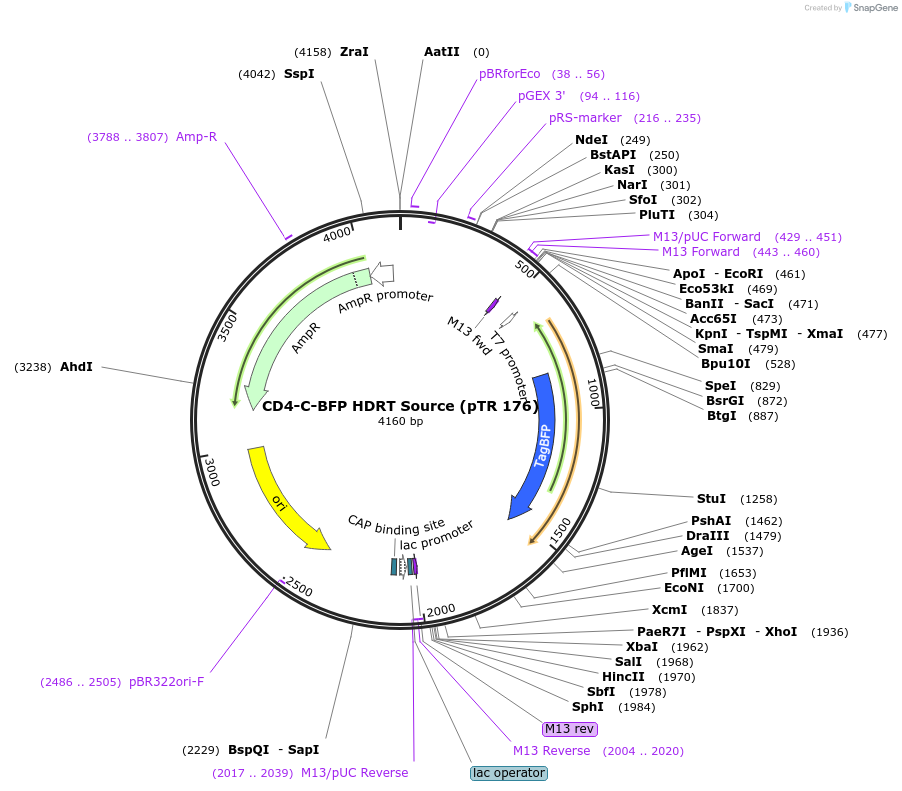
CD4-C-BFP HDRT Source (pTR 176)
Plasmid#112020PurposeDNA sequence source for amplifying an HDR template to tag endogenous human CD4 gene with BFPDepositorInsertCD4-BFP HDRT (CD4 Human)
UseCRISPR and Synthetic BiologyAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
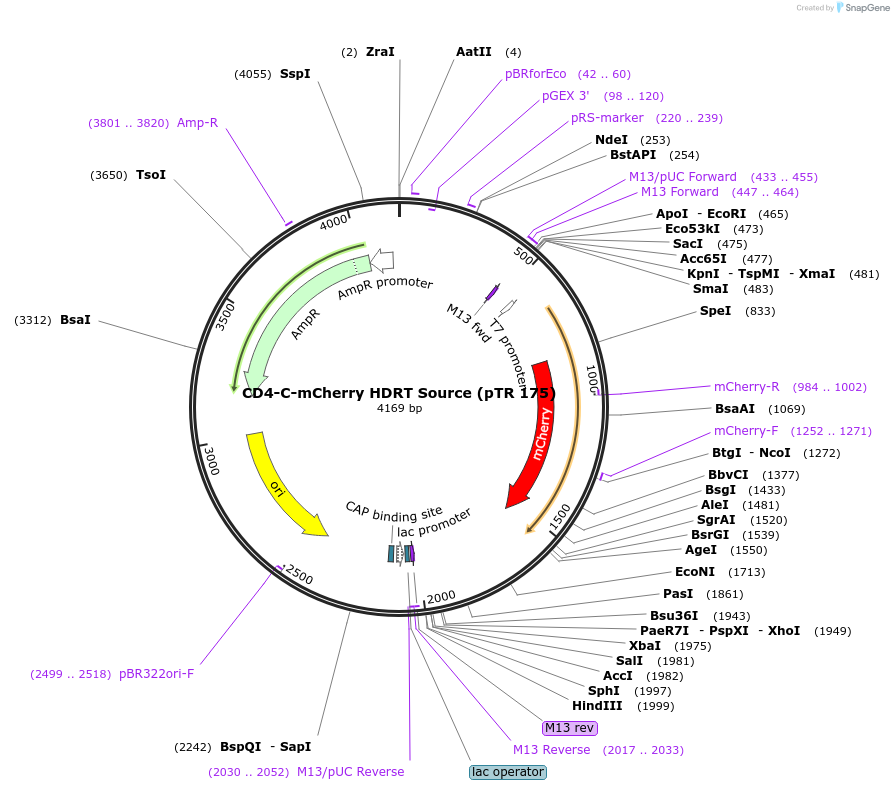
CD4-C-mCherry HDRT Source (pTR 175)
Plasmid#112019PurposeDNA sequence source for amplifying an HDR template to tag endogenous human CD4 gene with mCherryDepositorInsertCD4-mCherry HDRT (CD4 Human)
UseCRISPR and Synthetic BiologyAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
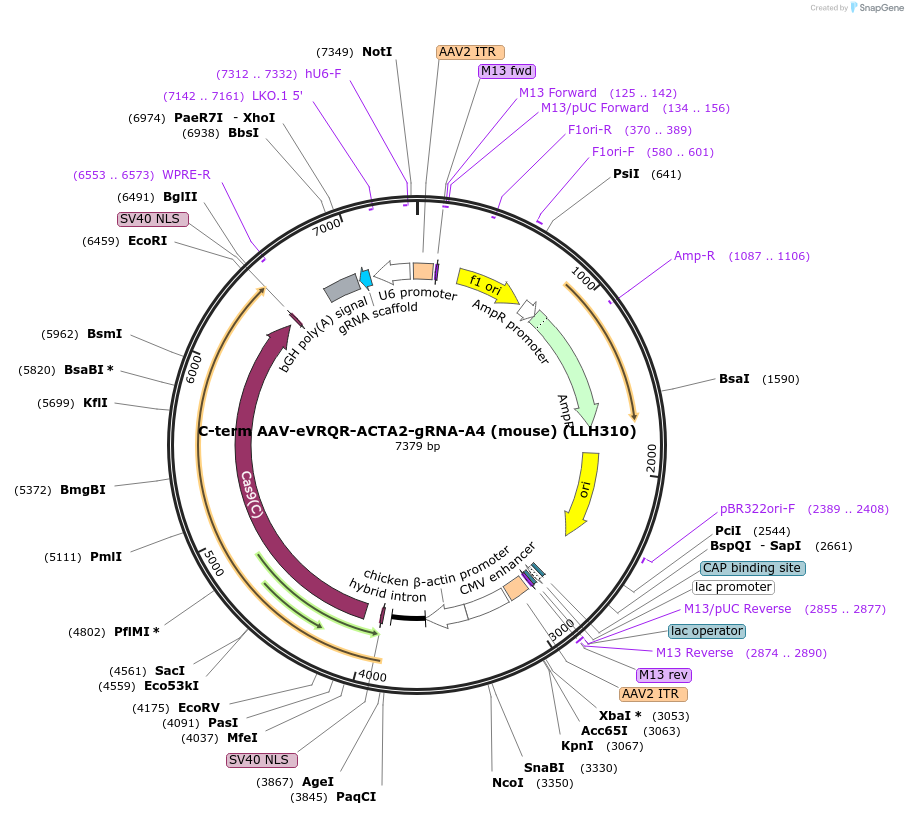
C-term AAV-eVRQR-ACTA2-gRNA-A4 (mouse) (LLH310)
Plasmid#242662PurposeCBh promoter expression plasmid for C-terminal intein-split AAV construct with C-term of SpCas9-VRQR and mouse gRNA-A4DepositorInsertpAAV-pCbh-BPNLS-NpuC-SpCas9-VRQR-BPNLS-WPRE-bGH_PA-ACTA2_mouse_NGA_gRNA-pU6
UseAAV and CRISPRTagsBPNLSMutationVRQR mutations in SpCas9(S55R/D1135V/G1218R/R1335…PromoterCBhAvailable SinceOct. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
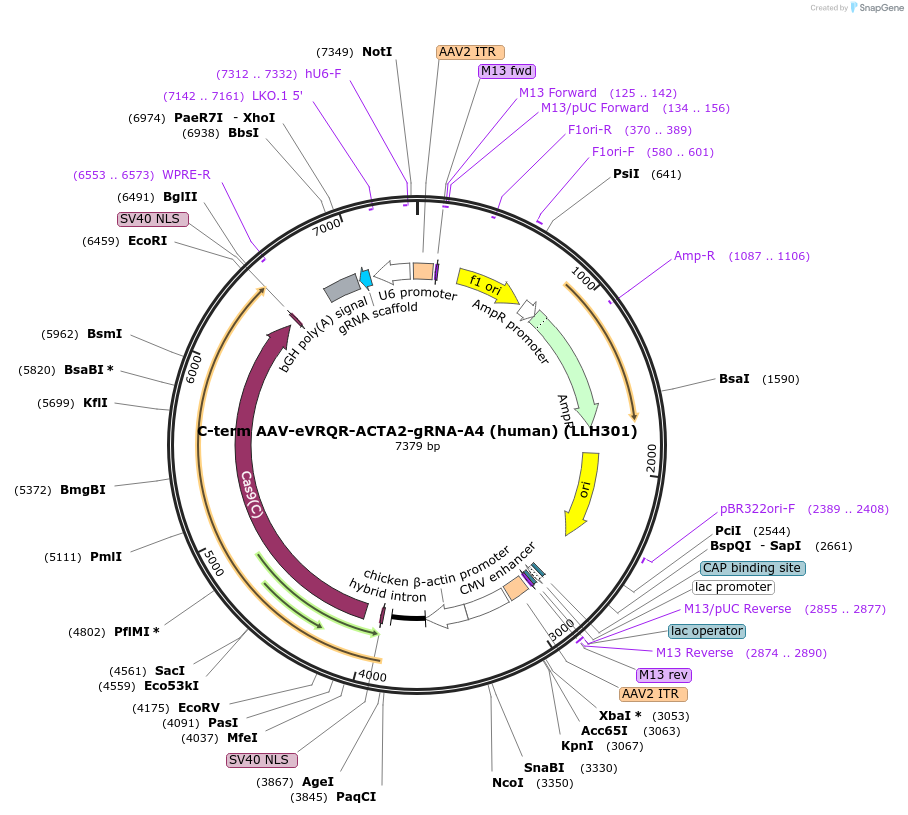
C-term AAV-eVRQR-ACTA2-gRNA-A4 (human) (LLH301)
Plasmid#242661PurposeCBh promoter expression plasmid for C-terminal intein-split AAV construct with C-term of SpCas9-VRQR and human gRNA-A4DepositorInsertpAAV-pCbh-BPNLS-NpuC-SpCas9-VRQR-BPNLS-WPRE-bGH_PA-ACTA2_human_NGA_gRNA-pU6
UseAAV and CRISPRTagsBPNLSMutationVRQR mutations in SpCas9(S55R/D1135V/G1218R/R1335…PromoterCBhAvailable SinceOct. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
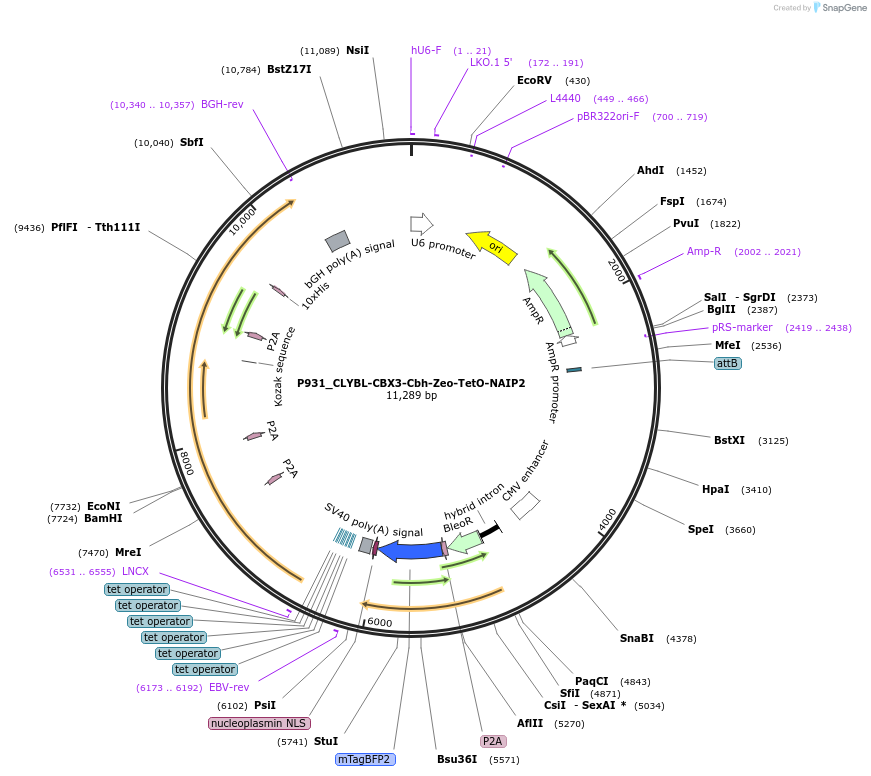
P931_CLYBL-CBX3-Cbh-Zeo-TetO-NAIP2
Plasmid#202761PurposeEncodes four RGC specific transcription factors under the control of a TetOn promoter at the CLYBL safe harborDepositorExpressionMammalianPromoterTetOAvailable SinceFeb. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
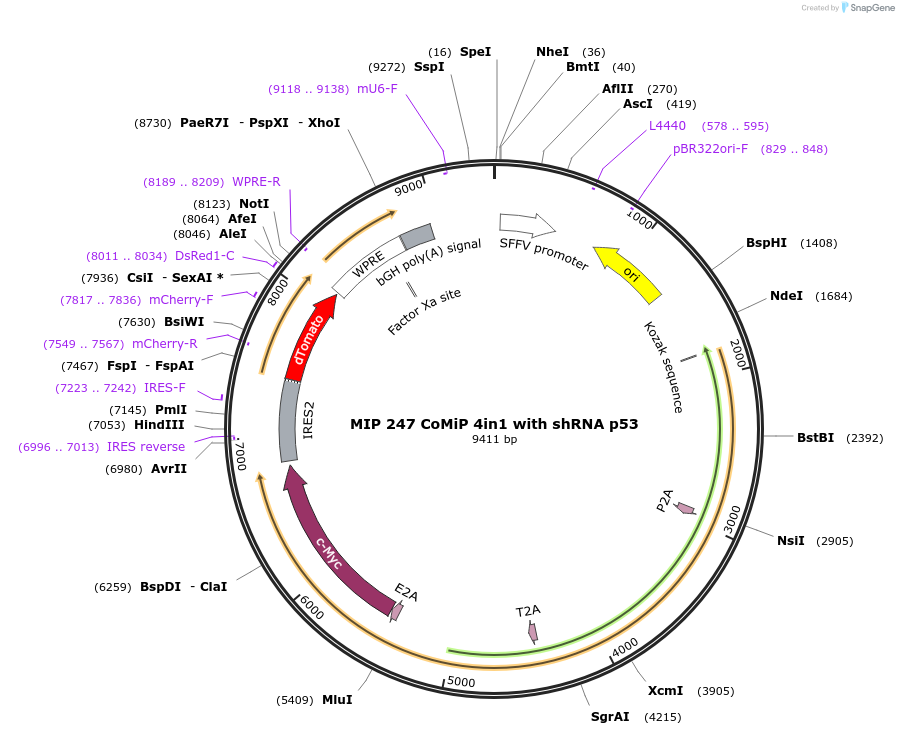
MIP 247 CoMiP 4in1 with shRNA p53
Plasmid#63726PurposeTo express Oct4, Klf4, Sox2, c-Myc and hairpin RNA p53. A single plasmid reprogramming system using a mini-intronic plasmid (MIP).TagsIRES-dtomatoExpressionMammalianMutationcodon-optimisedPromoterSFFV for Pct4, Klf4, Sox2 and c-myc; U6 for p53Available SinceAug. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
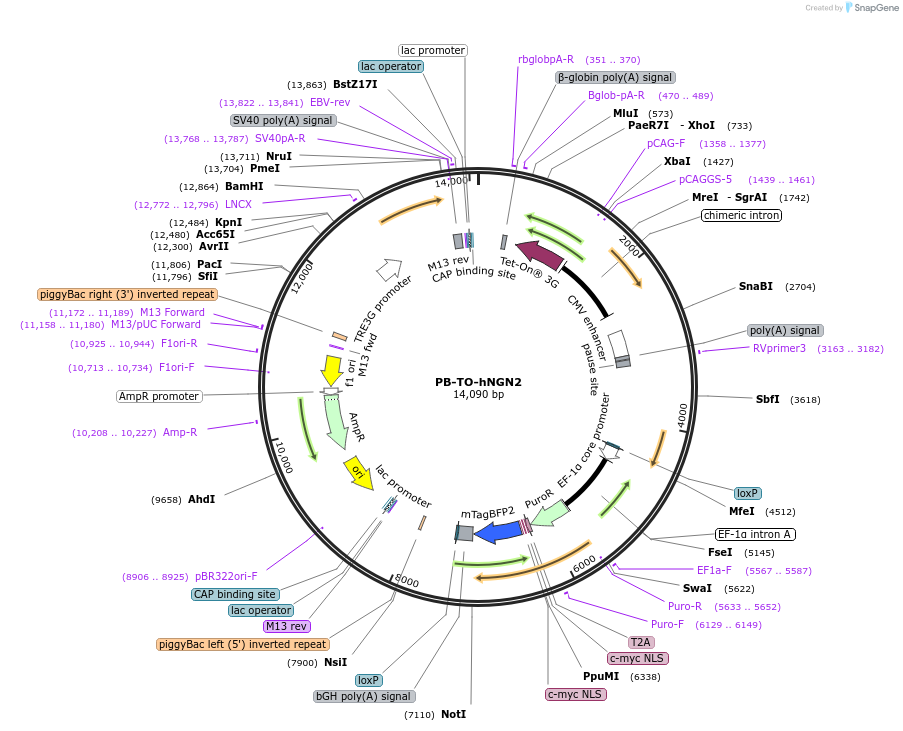
PB-TO-hNGN2
Plasmid#172115PurposePiggybac Tet-ON plasmid for differentiating hiPSCs into glutamatergic neurons via NGN2 expressionInsertsTagsT2A-mycNLS-mTagBFP2ExpressionMammalianPromoterEF1a and TRE3GAvailable SinceApril 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
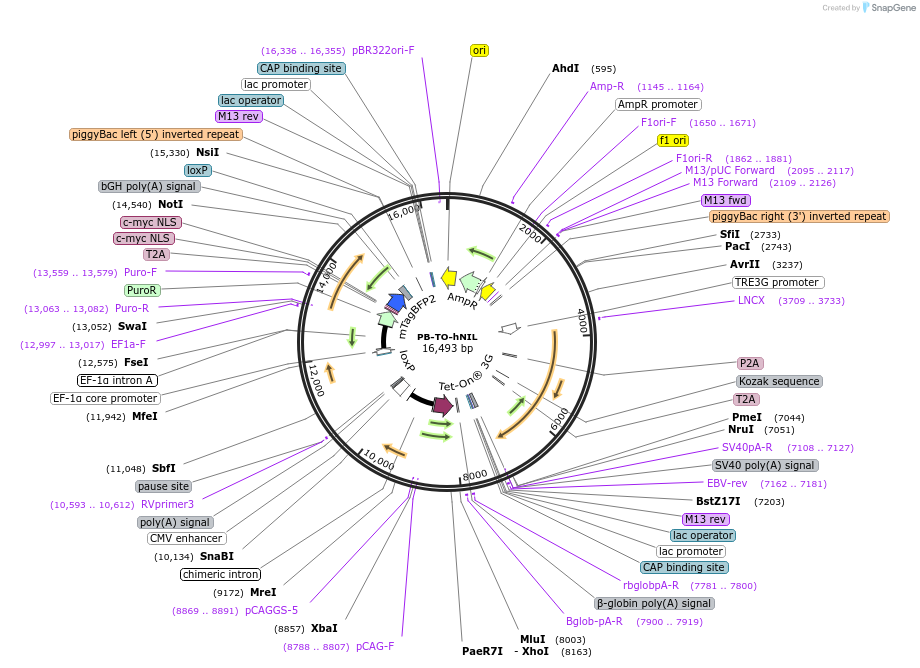
PB-TO-hNIL
Plasmid#172113PurposePiggybac Tet-ON plasmid for differentiating hiPSCs into lower motor neurons via NGN2, ISL1, and LHX3 expressionTagsT2A-mycNLS-mTagBFP2ExpressionMammalianPromoterEF1a and TRE3GAvailable SinceApril 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
Yeast GoldenBraid Cloning System and Toolkit
Plasmid Kit#1000000138PurposeModular cloning system for generation of high expression transcriptional units; Integrates in yeast (S. cerevisiae) genome at two different loci supporting high and stable transgene expressionDepositorAvailable SinceSept. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
XanthoMoClo Xanthobacter Genetic Toolkit
Plasmid Kit#1000000259PurposeUse this modular cloning kit to assemble plasmids for expression in Xanthobacter and Roseixanthobacter.DepositorAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
attB GPCR Library with Unique Molecular Identifiers
Pooled Library#243681PurposeFor stable recombination of nearly all human GPCRs with unique molecular identifiers.DepositorExpressionMammalianSpeciesHomo sapiensAvailable SinceOct. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
attB Barcoded Canonical GPCR Library
Pooled Library#243682PurposeFor stable recombination of nearly all human GPCRs with unique molecular identifiers.DepositorExpressionMammalianSpeciesHomo sapiensAvailable SinceOct. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
pC004 - PFS library in Beta-lactamase gene Target Plasmid
Pooled Library#79153PurposeLibrary of protospacer flanking sites (PFS) inserted at the 5' end of the β-lactamase gene for screening PFS preference for Cas13a/C2c2.DepositorAvailable SinceOct. 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library C
Pooled Library#157973PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library B
Pooled Library#157972PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library A
Pooled Library#157971PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
plIG-123_HA-GD2-28z_CAR_TF_Library
Pooled Library#207478PurposeHA-GD2-28z CAR in combination with 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into thDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-128_HA-GD2-28z_CAR_TFxTF_Library
Pooled Library#207480PurposeHA-GD2-28z CAR in combination with ~10,000 transcription factor combinations (and related proteins) and controls (GFP/RFP combinations). Library can be used to generate HDR templates for modular pooleDepositorUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-106_NY-ESO-1_TCR_SR_Library
Pooled Library#207474PurposeNY-ESO-1 TCR in combination with 129 surface receptors and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into the TRAC locus of human T cellsDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-107_NY-ESO-1_TCR_TF_Library
Pooled Library#207475PurposeNY-ESO-1 TCR in combination with 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into theDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-109_CD19-BBz_CAR_SR_Library
Pooled Library#207476PurposeCD19-BBz CAR in combination with 129 surface receptors and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into the TRAC locus of human T cellsDepositorUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-110_CD19-BBz_CAR_TF_Library
Pooled Library#207477PurposeCD19-BBz CAR in combination with 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into theDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-124_HA-GD2-28z_CAR_SR_Library
Pooled Library#207479PurposeHA-GD2-28z CAR in combination with 129 surface receptors and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into the TRAC locus of human T celDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-132_CD19-28z_CAR_TF_and_SR_Library
Pooled Library#207481PurposeCD19-28z CAR in combination with 129 surface receptors, 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knocDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 1 (positions 333-436)
Pooled Library#174293PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only